Abstract
In members of one of the subfamilies of the bacterial ATP binding cassette (ABC) transporters, the two nucleotide binding domains are fused as a single peptide and the proteins have no membrane-spanning domain partners. Most of the ABC efflux transporters of this subfamily have been characterized in actinomycetes, producing macrolide, lincosamide, and streptogramin antibiotics. Among 40 ABC efflux transporters of Bacillus subtilis, five proteins belong to this subfamily. None of these proteins has been functionally characterized. We examined macrolide, lincosamide, and streptogramin antibiotic resistance in insertional disruptants of the genes that encode these proteins. It was found that only a disruptant of vmlR (formerly named expZ) showed hypersensitivity to virginiamycin M and lincomycin. Expression of the vmlR gene was induced by the addition of these antibiotics in growth medium. Primer extension analysis revealed that transcription of the vmlR gene initiates at an adenosine residue located 225 bp upstream of the initiation codon. From the analysis of the vmlR and lacZ fusion genes, a 52-bp deletion from +159 to +211 resulted in constitutive expression of the vmlR gene. In this region, a typical ρ-independent transcriptional terminator was found. It was suggested that the majority of transcription ends at this termination signal in the absence of antibiotics, whereas under induced conditions, RNA polymerase reads through the terminator, and transcription continues to the downstream vmlR coding region, resulting in an increase in vmlR expression. No stabilization of vmlR mRNA occurred under the induced conditions.
In order to survive in natural environments, an important defense mechanism for Bacillus subtilis, a soil bacteria, may be to acquire resistance to antibiotics produced by other bacteria in the same area. One of the mechanisms of acquiring antibiotic resistance involves the exclusion of antibiotics by a membrane-bound transporter. Recently, it was found that in B. subtilis 168, a two-component regulatory system detects bacitracin, a dodecapeptide antibiotic produced by Bacillus licheniformis. A sensor kinase, BceS, responds to extracellular bacitracin and transmits a signal to a cognate response regulator, BceR, which induces the expression of a bacitracin efflux ATP binding cassette (ABC) transporter, BceAB, resulting in increased resistance to bacitracin (16, 20). These observations prompted us to examine the possibility that the remaining ABC transporters of B. subtilis, where no function has been assigned, are involved in resistance toward antibiotics produced by other bacteria.
ABC transporters constitute one of the most abundant groups of proteins in prokaryotes and eukaryotes. A typical ABC transporter consists of two hydrophobic membrane-spanning domains (MSD) and two hydrophilic domains (nucleotide binding domains [NBD]) that bind and couple ATP hydrolysis to the transport process. The four domains are generally expressed as an independent polypeptide but are occasionally expressed as multidomain polypeptides corresponding to a variety of domain fusions. In one subgroup found in bacteria, the two NBD are fused as a single polypeptide without any MSD partners (25). It is generally assumed that these NBD proteins may work as a drug efflux pump in conjunction with a certain transmembrane protein not yet identified. Most of the ABC efflux transporters of this subfamily have been identified in antibiotic-producing actinomycetes (18). CarA, SrmB, TlrC, OleB, LmrC, and VarM have been identified in Streptomyces producing carbomycin, spiramycin, tylosin, oleandomycin, lincomycin and virginiamycin, respectively (12, 22, 24, 27). MsrA, which has been reported as an inducible erythromycin resistance determinant in Staphylococcus epidermidis (26), and VgA (3) and VgaB (4), which have been reported in virginiamycin M-resistant Staphylococcus aureus, also belong to this subfamily. These drugs are classified as macrolide, lincosamide, and streptogramin antibiotics. Among 40 ABC exporters of B. subtilis, five proteins, ExpZ, YdiF, YfmM, YfmR, and YkpA, belong to this subfamily (25). None of these proteins has been functionally characterized. Therefore, we examined whether five ABC transporters of this subfamily were involved in macrolide-lincosamide-streptogramin antibiotic resistance in B. subtilis 168.
In this study, we report that the deletion of the vmlR gene (formerly named expZ) resulted in hypersensitivity toward virginiamycin M and lincomycin in B. subtilis strain 168. Expression of the vmlR gene was induced in response to these antibiotics in growth medium. Based on the analysis of vmlR and lacZ fusion genes, a cis-acting regulatory sequence was identified within the 225-bp 5′ untranslated region (5′ UTR) of the vmlR gene. In the absence of antibiotics, it was found that the majority of transcription initiated from the vmlR promoter terminated at the transcriptional termination signal located upstream of the vmlR coding region. In contrast, in the presence of antibiotics, some transcription reads through the termination signal, resulting in increased expression of the vmlR gene.
MATERIALS AND METHODS
Construction of mutant strains and plasmids.
B. subtilis 168 (trpC2 glt) was used as the wild-type strain. The pMutin insertion mutants constructed by members of the Japan and European Union consortia of B. subtilis functional genomics were used as disrupted mutants of the following genes: expZ (vmlR), ydiF, yfmM, yfmR ykpA, and ydgF. For assaying the resistance to macrolide-lincosamide-streptogramin antibiotics, the erythromycin resistance gene of the pMutin insertion mutants was disrupted by insertion of a tetracycline resistance gene; resulting strains were named VMLRd::Tcr, YDIFd::Tcr, YFMMd::Tcr, YFMRd::Tcr, YDGFd::Tcr, and YKPAd::Tcr. To obtain a deletion mutant of the vmlR gene, regions 1.0 kb upstream and 1.5 kb downstream of the vmlR gene were amplified by High Fidelity Platinum Taq (Invitrogen) and were cloned into pBR322. A neomycin resistance gene was inserted between these fragments. The resulting plasmids were linearized and transformed into B. subtilis 168, and transformants were selected by neomycin resistance to obtain 168/ΔvmlR::Neor. To construct strains with transcriptional fusion of the gradually deleted 5′ UTR and N terminus of the vmlR coding region and the lacZ reporter gene, five fragments were amplified by High Fidelity Platinum Taq (Invitrogen) using the following primer sets: E−253 and B+43, E−253 and B+105, E−253 and B+159, E−93 and B+198, and E−93 and B+545. The number in the primer name indicates the 5′ end position of the primer relative to the transcriptional start site (+1). Upper and lower primers contain EcoRI (E) and BamHI (B) recognition sites, respectively. The PCR products digested with EcoRI and BamHI were cloned between the corresponding sites of pMutin3 (29). These recombinant plasmids were transformed into strain 168, and the transformants obtained through a single crossover event were selected by erythromycin resistance. Resulting strains were named VMLR43, VMLR105, VMLR159, VMLR198, and VMLR545. To obtain VMLRΔ52 and VMLRΔ135, gene splicing by overlap extension was used (10). Two-step PCRs using the internal overlapping primers del1, del2 or del3, and del4 and the flanking primer pair E−93 and B+545 were carried out. The resulting fragments were cloned into pMutin3, which was then introduced into strain 168. All the pMutin-inserted strains contained an intact vmlR gene downstream of the vmlR-lacZ transcriptional fusion gene. Therefore, resistance to virginiamycin M or lincomycin of these strains is similar to that of strain 168. The same vmlR-lacZ fusion genes of strain VMLR545, VMLRΔ52, and VMLRΔ135 were introduced into the amyE locus on the chromosome of strain 168/ΔvmlR::Neor, using pDL2 as a vector (7). All the constructs used in this study were verified by DNA sequencing with an ABI Prism 310 genetic analyzer (PE Biosystems).
Culture conditions.
Bacterial strains were cultured in LB medium (10 g liter−1 bactotrypton, 5 g liter−1 yeast extract, 10 g liter−1 NaCl). Where necessary, the following antibiotics were added to the medium: erythromycin (0.4 μg ml−1), chloramphenicol (5 μg ml−1), tetracycline (12 μg ml−1), and neomycin (15 μg ml−1). Virginiamycin M, oleandomycin, spiramycin, erythromycin, clindamycin, lincomycin, and rifampin were purchased from Sigma Aldrich Co. Purified virginiamycin M and S were prepared by reverse-phase high-performance liquid chromatography (HPLC) as described by Lee et al. (Cosmosr/5018; Nacalai Tesque Co.) (13). With the exception of the cases indicated, the data presented in this study were obtained with commercially available virginiamycin M. The induction of resistance and expression of vmlR were also confirmed with purified virginiamycin M.
Assay of drug resistance phenotype.
To screen antibiotic resistance, a filter disk diffusion method was used. B. subtilis strains were grown in LB medium to mid-log phase. A total of 100 μl of culture was mixed with 2.5 ml of soft LB medium agar and poured onto the LB plate. After cooling, filter paper disks (5-mm diameter) carrying antibiotics were placed on the top of the agar, and the plates were incubated at 37°C overnight. Following incubation, the diameters of the growth inhibition zones produced by the diffusion of antibiotics from the filter paper disks were measured. Determination of relative resistance was carried out using a 96-well titer plate as described previously (21).
Northern hybridization.
Total RNA was extracted from cells grown in LB medium with hot phenol as described previously (1). Five micrograms of total RNA was fractionated by electrophoresis using a 1.5% agarose gel containing 6% formaldehyde. RNA was then blotted onto a nylon membrane (Hybond N+; Amersham Pharmacia. Biotech) in 20× SSC (1× SSC is 0.15 M NaCl, 0.015 M sodium citrate). Alternatively, total RNA (8 μg) was fractionated using a 7 M urea-6% polyacrylamide gel and electroblotted onto a clear blot N-plus membrane (ATTO). Northern hybridization was carried out as described previously (2). PCR-amplified DNA fragments labeled with [α-32P]dCTP were used to probe the blot. Probe A and probe B contained the sequences localized from +213 to +493 bp and +3 to +181 bp, respectively (relative to the transcription initiation site at the +1 position). Digoxigenin (DIG)-labeled RNA probe was synthesized using a DIG RNA Labeling Kit (Sp6/T7; Roche Applied Sciences) according to the procedure recommended by the manufacturers. PCRII-Topo (Invitrogen) containing vmlR sequence from +3 to +181, which was linearized by restriction enzyme digestion, was used as a template. Hybridization was carried out at 68°C using DIG Easy Hyb (Roche Applied Sciences). Determination of the half-life of mRNA was carried out as described previously (2). Samples were removed from early log phase cultures 0, 1, 2, 4, 6, 8, and 12 min after the addition of rifampin (500 μg ml−1).
Primer extension analysis of RNA.
Primer extension analysis was carried out as described previously (21). A rhodamine X-isothiocyanate-labeled oligonucleotide primer complementary to the sequence localized from 43 to 67 bp upstream from the initiation codon of the vmlR gene was used.
In vitro transcription assay.
A 270-bp fragment corresponding to the vmlR sequence between +7 and +277 and a 218-bp fragment of the same region containing a 52-bp deletion of the terminator sequence were amplified by PCR and cloned into PCRII-Topo (Invitrogen). The resulting plasmids, TopovmlR+ter and TopovmlRΔter, were linearized by digestion with EcoRV and used as the template for in vitro transcription by Sp6 RNA polymerase. In vitro transcription assays were carried out using the DIG RNA labeling kit (Sp6/T7; Roche Applied Sciences) according to the procedure recommended by the manufacturer, with some modifications. The DNA template (0.5 μg) was mixed with Sp6 RNA polymerase and incubated for 30 min at 37°C in transcription buffer. After incubation, heparin (final concentration, 200 μg ml−1) and a mixture containing 1 mM concentration of each nucleotide were added to allow a single round of transcription. RNA synthesis was performed for 20 min at 37°C and stopped by adding EDTA to a final concentration of 20 mM. Samples were precipitated with ethanol and fractionated using a 7 M urea-6% polyacrylamide gel. The gel was stained with SYBR Gold (Molecular Probes) and analyzed with FLA3000G Imaging Analyzer (Fuji).
RESULTS
VmlR is involved in virginiamycin M and lincomycin resistance in B. subtilis.
Genome analysis revealed that there are approximately 40 efflux transporters that belong to the ABC family in B. subtilis (25). Among them, five genes, expZ, ykpA, yfmM, yfmR, and ydiF, encode hydrophilic proteins which contain two NBD. Genes encoding putative MSD subunits were not detected in their vicinity. None of these genes has been functionally characterized. ABC transporters of this subfamily have been characterized as determinants of macrolide, lincosamide, or streptogramin resistance (18). Therefore, we examined resistance toward the antibiotics listed in Table 1 in the disruptants of expZ, ykpA, yfmM, yfmR, and ydiF genes (strains VMLRd::Tcr, YKPAd::Tcr, YFMMd::Tcr, YFMRd::Tcr, and YDIFd::Tcr, respectively) by the filter disk diffusion method as described in Materials and Methods. Only the expZ disruptant showed increased sensitivity toward virginiamycin M and lincomycin. Subsequently, the relative resistance to these antibiotics was determined in the expZ disruptant (168/Δvm/R::Neor) (Table 1). The relative resistance for virginiamycin M and lincomycin was 0.025 and 0.058, respectively. We found that growth inhibition by commercially available virginiamycin M varied depending on the lot number used. We also determined the relative resistance value using virginiamycin M purified by HPLC (13), which yielded a constant value. Clindamycin, erythromycin, and oleandomycin resistance decreased to approximately 46 to 66% of those observed in strain 168. Resistance toward virginiamycin S and spiramycin was unchanged, and resistance to rifampin increased 30%. Homology searches revealed that the predicted amino acid sequence of ExpZ showed 31%, 32%, 33%, and 29% identity to VarM (12), VgaB (4), VgA (3), and LmrC (24), respectively. This result indicates that ExpZ is involved in virginiamycin M and lincomycin resistance in B. subtilis. Accordingly, we designated ExpZ as VmlR (virginiamycin M and lincomycin resistance).
TABLE 1.
Increased drug sensitivity of the vmlR deletion mutant, 168/ΔvmlR::Neor
| Drug | Relative resistancea |
|---|---|
| Virginiamycin M | 0.025 |
| Virginiamycin M purified | 0.042 |
| Lincomycin | 0.058 |
| Clindamycin | 0.46 |
| Erythromycin | 0.66 |
| Oleandomycin | 0.66 |
| Spiramycin | 0.95 |
| Virginiamycin S | 1.05 |
| Rifampin | 1.29 |
Relative resistance was determined by dividing the 50% inhibitory concentration for the vmlR deletion mutant by the 50% inhibitory concentration for strain 168. The 50% inhibitory concentrations of the drugs for strain 168 (μg ml−1) were virginiamycin M (Sigma), 12.5; virginiamycin M (purified), 3.71; lincomycin, 5.31; clindamycin, 0.088; erythromycin, 0.119; oleandomycin, 2.78; spiramycin, 5.28; virginiamycin S, 1.47; rifampin, 0.0117.
In bacteria, genes involved in a related function often constitute an operon or are located adjacent to each other on the chromosome. The ydgE gene, located immediately downstream of the vmlR gene, encodes a protein that has no homology to the protein with known function. Northern hybridization analysis indicated that the ydgE gene is divergently transcribed and that the expression was not induced by the addition of virginiamycin M (data not shown). The ydgF gene, located next to the vmlR gene in the upstream region, encodes a predicted membrane protein that has 12 transmembrane segments and shows homology to amino acid permease (23). We examined the possibility that YdgF may function as an MSD partner of VmlR protein. However, a ydgF disruptant (YDGFd::Tcr) showed the wild-type level of resistance to drugs listed in Table 1, indicating that the YdgF is not involved in drug resistance.
Preincubation with a subinhibitory concentration of virginiamycin M or lincomycin induces resistance in B. subtilis 168.
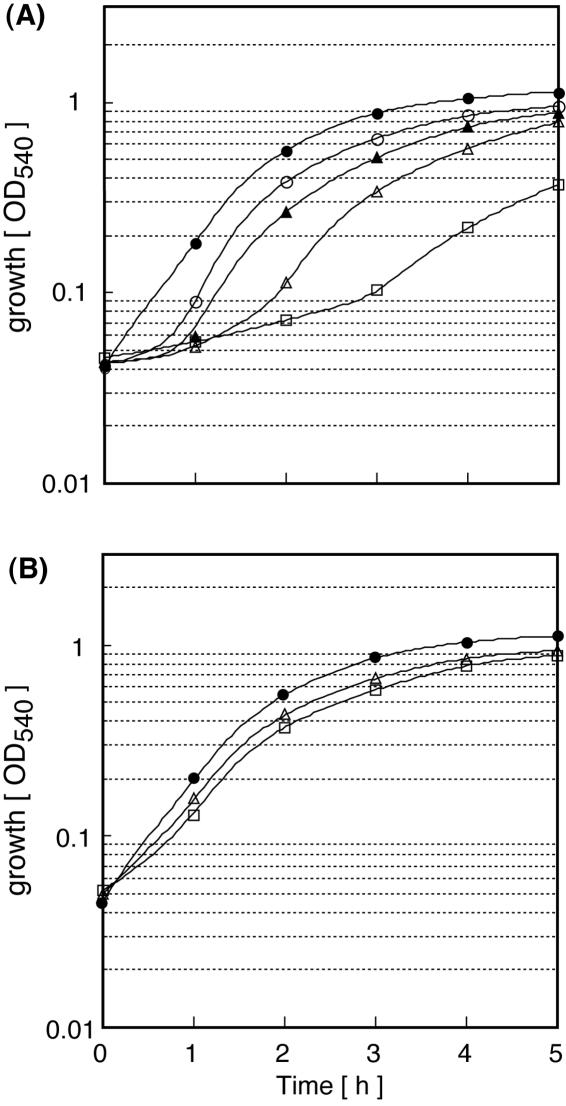
When B. subtilis 168 was grown in LB medium containing various concentrations of virginiamycin M, a lag in growth was observed at virginiamycin M concentrations above 1.9 μg ml−1 (Fig. 1A). The duration of the growth lag varied depending on the concentration of virginiamycin M added to the growth medium. When B. subtilis 168 cells were grown in LB medium containing 1.9 μg ml−1 virginiamycin M for 90 min and then inoculated into LB medium containing various concentrations of virginiamycin M, no lag was observed even at virginiamycin M concentrations where a marked lag was observed without preincubation (1.9 to ∼15 μg ml−1) (Fig. 1B). Resistance to virginiamycin M increased after preincubation compared to experiments performed without preincubation. The 50% inhibitory concentration increased approximately 3.8-fold during a 3-h growth period in the presence of virginiamycin M. Preincubation with a subinhibitory concentration of lincomycin (2 μg ml−1) also resulted in increased resistance to lincomycin.
FIG. 1.
Preincubation with a subinhibitory concentration of virginiamycin M induces virginiamycin M resistance in B. subtilis. An overnight culture of B. subtilis 168 grown in LB medium was inoculated into LB medium (optical density at 530 nm of 0.05) with (B) or without (A) 1.9 μg ml−1 virginiamycin M. When cultures reached early log phase (optical density at 530 nm of 0.5), a 0.1-ml culture was added to a 96-well titer plate that contained 0.1 ml of LB medium with serially diluted concentrations of virginiamycin M, and growth was traced by measuring the optical density at 540 nm over a period of 5 h. (A) No preincubation. Virginiamycin M concentrations (μg ml−1) are indicated as follows: filled circles, 0; open circles, 1.9; filled triangles, 3.8; open triangles, 7.5; open squares, 15. (B) Preincubation with 1.9 μg ml−1 virginiamycin M. Virginiamycin M (μg ml−1) concentrations are indicated as follows: filled circles, 0.95; open triangles, 8.5; open squares, 16.
Identification of vmlR transcript and induction of vmlR gene expression at the transcriptional level.
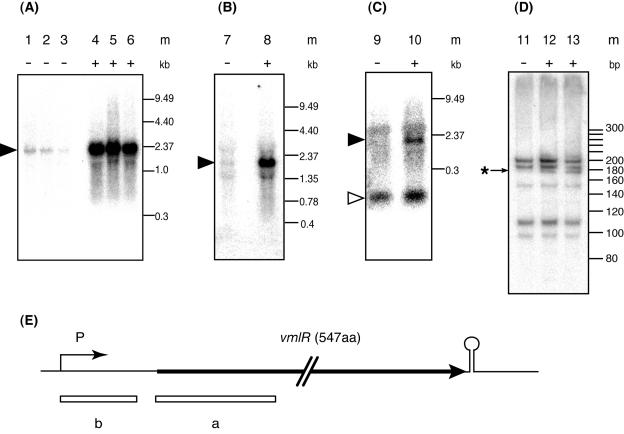
Northern hybridization analysis of total RNA isolated from B. subtilis 168 grown in LB medium with probe A containing the vmlR coding region, identified a 1.9-kb transcript which was low throughout each growth phase. The size of the transcript indicated that the vmlR gene was transcribed as a monocistronic mRNA from a promoter located upstream of the coding region and terminated at the ρ-independent transcriptional terminator found immediately downstream of the vmlR gene (Fig. 2E). The cellular level of vmlR mRNA was higher in early log-phase than in late log-phase cells (Fig. 2A).
FIG. 2.
Induction of vmlR expression by the addition of virginiamycin M or lincomycin in growth medium. An overnight culture of B. subtilis 168 grown in LB medium was inoculated into LB medium (optical density at 530 nm of 0.05) without or with virginiamycin M or lincomycin. Samples were withdrawn during early (optical density at 530 nm of 0.5; lanes 1, 4, 7, 8, 9, 10, 11, 12, and 13), middle (optical density at 530 nm of 1.2; lanes 2 and 5) and late (optical density at 530 nm of 2.0; lanes 3 and 6) log-phase growth. Culture media were the following: LB (lanes 1, 2, 3, 7, 9, and 11), LB containing 1.9 μg ml−1 virginiamycin M (lanes 4, 5, 6, 10, and 12), and LB containing 2 μg ml−1 lincomycin (lanes 8 and 13). m, molecular size standard. Total RNA (5 μg) was loaded per lane. For Northern blotting, a 1.5% agarose gel containing 6% formaldehyde (A, B, and C) and a 6% polyacrylamide gel containing 7 M urea (D) were used. Probe A containing sequences from +213 to 493 was used in panels A and B; probe B containing sequences from +3 to 181 (+1, transcription initiation site) was used in panels C and D. A filled arrow indicates the 1.9-kb vmlR transcript, and an open arrow indicates transcripts less than 300 bp in length. An asterisk indicates an extra band detected under the induced condition. (E) Thick arrow, open reading frame of vmlR; P, promoter; circle on stem, transcriptional terminator; boxes a and b under the open reading frame, probes.
When total RNA was isolated from cells grown in LB medium containing 1.9 μg ml−1 virginiamycin M, the amount of vmlR mRNA was dramatically (more than 10-fold) increased during all growth phases. From this result, we concluded that a low concentration of virginiamycin M in growth medium induced vmlR expression at the transcriptional level, resulting in increased virginiamycin M resistance. Addition of 2 μg ml−1 lincomycin also induced the expression of the vmlR gene (Fig. 2B).
Using probe B containing the 5′ UTR of the vmlR gene, an intense band less than 300 bp in length was detected in the absence of virginiamycin M (Fig. 2C). The intensity of this band slightly increased upon addition of virginiamycin M to the growth medium. When total RNAs were separated by 6% polyacrylamide gel containing 7 M urea and electroblotted to an N-plus (ATTO) membrane, five bands were detected with probe B, both in the presence or absence of antibiotics (Fig. 2D). In addition to these bands, an extra band was detected in RNA samples from cells grown in the presence of virginiamycin M or lincomycin. When a DIG-labeled RNA probe with the antisense sequences covering the same 5′ UTR as probe B was used, the same band patterns were observed as those detected with double-stranded DNA probe B (data not shown). The result showed that these small transcripts originated from the transcription of the antisense strand. Since these bands were not detected with probe A, these transcripts may be produced by premature transcriptional termination or by posttranscriptional processing in the 5′ UTR.
Determination of transcriptional start point by primer extension analysis.
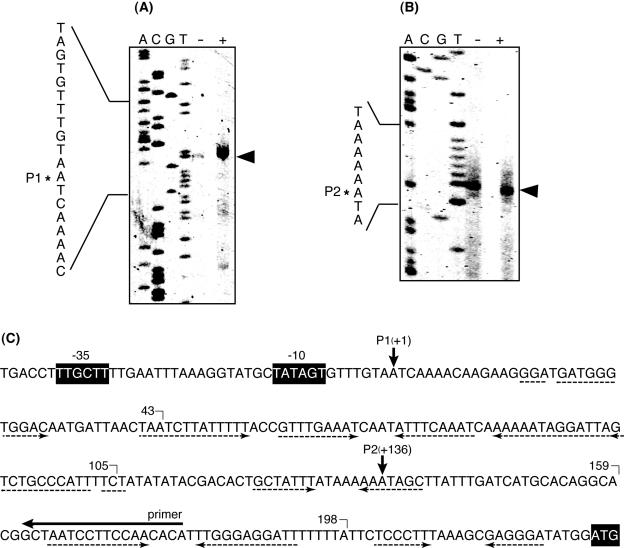
The 5′ end of the vmlR transcript was determined by primer extension. Two bands were detected corresponding to start sites P1 and P2 at an adenosine residue, located 225 bp and 89 bp upstream of the initiation codon for the vmlR gene, respectively (Fig. 3). The intensity of the band at P1 was increased about 10-fold in RNA prepared from cells grown in LB medium containing virginiamycin M, indicating that the 1.9-kb transcript detected in the Northern hybridization analysis has the 5′ end at this position. Typical σA-dependent −10 and −35 consensus sequences were found in the upstream region, as shown in Fig. 3C. The intensity of the band at P2 was similar both in the presence or absence of virginiamycin M, indicating that it might be the 5′ end of the major transcripts detected with probe B. No −10 and −35 consensus sequences were found in the upstream region of P2. Moreover, we confirmed that no promoter activity was found in the region between the +10 and P2 sites when this region was transcriptionally fused to the lacZ reporter gene (data not shown). Therefore, the 5′ end at P2 may be derived from posttranscriptional processing of the transcript initiated from the same promoter located upstream of P1.
FIG. 3.
Mapping of the 5′ end of vmlR transcript by primer extension analysis. Total RNA was isolated from early log-phase cultures (optical density at 530 nm of 0.5) of B. subtilis 168 grown in LB medium with (+) or without (−) 3.8 μg ml−1 virginiamycin M. Five micrograms of total RNA was used for primer extension analysis. The potential transcription start site is marked with arrows. The A, C, G, and T dideoxy sequencing ladder was obtained with the same primer used for primer extension. The two 5′ ends detected are marked by P1 (A) and P2 (B). The results shown in panels A and B are the upper and lower parts of the same polyacrylamide gel. (C) Nucleotide sequence in the 5′ UTR of the vmlR gene. Putative promoter −10 and −35 consensus sequences and initiation codon are shown in black boxes. Rhodamine X-isothiocyanate-labeled primer used for primer extension is indicated by a horizontal arrow above the sequences. Inverted repeats are indicated by dotted arrows below the sequences. The 3′ ends of the vmlR-lacZ fusion genes (Fig. 4) are marked.
cis-Acting region necessary for induction by virginiamycin M.
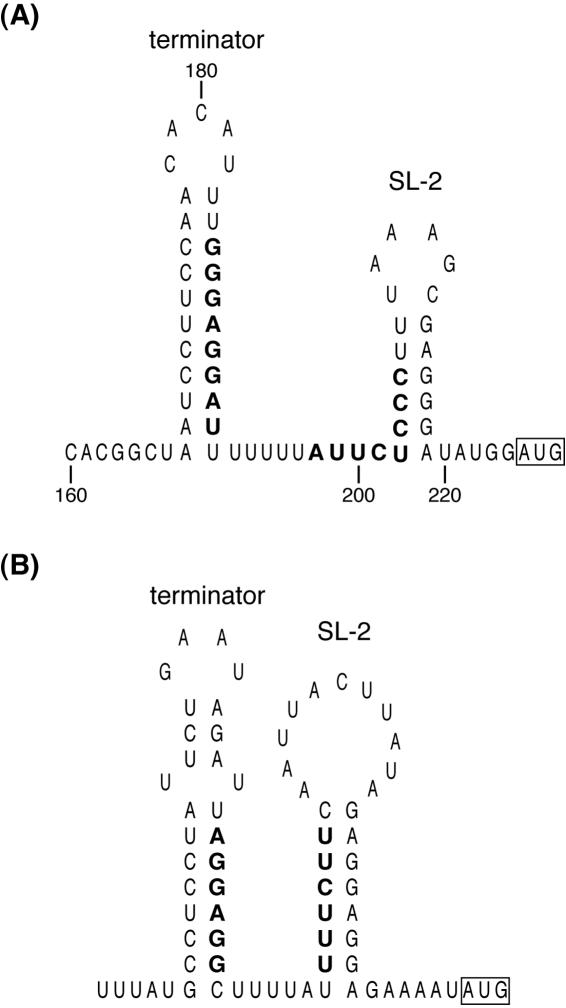
Primer extension analysis revealed that the 5′ UTR of the vmlR is 225 nucleotides in length. Several inverted repeat sequences were detected in the 5′ UTR (Fig. 3C), and one of them located from +165 to +192 was predicted to fold into a typical ρ-independent terminator (see Fig. 6). From the results obtained in the Northern hybridization analysis, we assumed that in the absence of virginiamycin M, transcription from the vmlR promoter prematurely ended at the terminator upstream of the vmlR coding region. Premature termination followed by posttranscriptional processing resulted in several transcripts less than 300 bp in length detected by probe B. The largest band, which was 200 bp in length, corresponds to a transcript from the P1 promoter that terminated at the predicted ρ-independent terminator. In the presence of virginiamycin M, at least a partial transcription read-through of the terminator continued through the vmlR coding region and terminated at the ρ-independent terminator downstream of the vmlR coding region, resulting in an increase of the 1.9-kb transcript detected by probe A as well as an increase in virginiamycin M resistance. The efficiency of the read-through of the terminator was not high, since the level of small transcripts in the presence of virginiamycin M was similar to that under noninduced conditions.
FIG. 6.
Secondary structures in the 5′ UTR of vmlR from B. subtilis (A) and vgA from S. aureus (B) were predicted by the mfold program. Complementary sequences, which potentially fold an alternative stem-loop structure, are shown in bold letters.
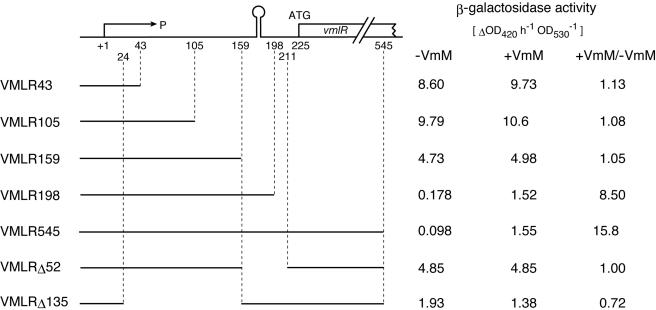
In order to verify this assumption, we constructed a series of transcriptional fusions with progressive deletions of the vmlR 5′ UTR and lacZ gene (Fig. 4). The end points of the vmlR sequence fused to the lacZ gene are indicated in Fig. 3C. The fusion genes cloned into pMutin3 were inserted into the chromosome of strain 168 by a single crossover event. Cells were grown to late log phase in LB medium in the presence or absence of virginiamycin M. In strains VMLR43 and VMLR105, β-galactosidase activities were constitutively high either in the presence or absence of virginiamycin M. In strain VMLR159, which contained an additional 54 bp of the downstream region, the level of β-galactosidase decreased to approximately 50% of the levels in VMLR43 and VMLR105, and expression was still constitutive. The observations indicate that the vmlR promoter itself is constitutively active. In strain VMLR198, induction by virginiamycin M was clearly observed. In strain VMLR545, which contained an additional 347 bp of the downstream region including the N terminus of vmlR coding sequences, the expression pattern of the fusion gene was similar to that in strain VMLR198, indicating that a cis-acting sequence that is indispensable for induction by virginiamycin M resides on the 5′ UTR which is common to both strains. β-Galactosidase activities in VMLR198 and VMLR545 grown in the presence of virginiamycin M were approximately 15% of the activity in VMLR43 or VMLR105. In strain VMLRΔ52, which has a 52-bp deletion of the entire terminator region from the 5′ UTR, β-galactosidase activity increased to a similar level to that observed in VMLR159, and no induction was observed, indicating that the response to the virginiamycin M addition was dependent on the presence of the terminator. In strain VMLRΔ135, which has a 135-bp deletion from +25 to +160, β-galactosidase activity was constitutive. The level of β-galactosidase activity was approximately 20% of that in VML43 and was similar to that of VMLR198 or VMLR545 grown in the presence of virginiamycin M, indicating that the 135-bp region upstream of the terminator may contribute to the increased efficiency of termination in the absence of virginiamycin M. The same vmlR-lacZ fusion genes of strains VMLR545, VMLRΔ52, and VMLRΔ135 were also introduced into the amyE locus on the chromosome of the strain 168/ΔvmlR::Neor. The expression patterns of β-galactosidase in the presence or absence of 0.2 μg ml−1 virginiamycin M (these strains have no functional vmlR gene; therefore a low concentration of virginiamycin M was used) were similar to those presented in Fig. 4 (data not shown). These results indicate that the integration locus of the fusion gene as well as the copy number of the vmlR 5′ UTR region (two in the pMutin-derived strains and one in the derivatives of 168/ΔvmlR::Neor) have no effect on the control of fusion gene expression.
FIG. 4.
Mapping of the cis-acting sequences required for the inducible expression of the vmlR gene by virginiamycin M. The top diagram represents the 5′ UTR and N terminus of the vmlR coding regions. P, promoter; open circle on stem, termination signal. A set of transcriptional lacZ fusion genes containing fragments of various lengths of the vmlR sequences were constructed. Bars below represent sequences that are present in the strains shown in the left columns. Numbers in the strain name represent the location of the 3′ end of the vmlR sequences relative to the transcription initiation site at the +1 position (Fig. 3C). β-Galactosidase activity in cells grown in LB medium with (+) or without (−) 6 μg ml−1 of virginiamycin M are listed in the right column. VmM, virginiamycin M.
Predicted terminator functions in vitro.
In order to determine whether the predicted terminator structure in the 5′ UTR acts as a transcriptional terminator, in vitro transcription analysis with Sp6 RNA polymerase was performed. When TopovmlR+ter was used as a template, two bands, approximately 370 bp and 270 bp in length, were detected (Fig. 5). These bands correspond to a full-length runoff transcript and a transcript terminated at the predicted terminator, respectively. The amount of the 270-bp band was 70% of that of the 370-bp band. When TopovmlRΔter, which has a deletion of the terminator region, was used as a template, a single 314-bp band was detected. The size of the transcript corresponded to the full-length runoff transcript. These results demonstrate that heterologous Sp6 RNA polymerase recognizes the predicted terminator and stops transcription.
FIG. 5.
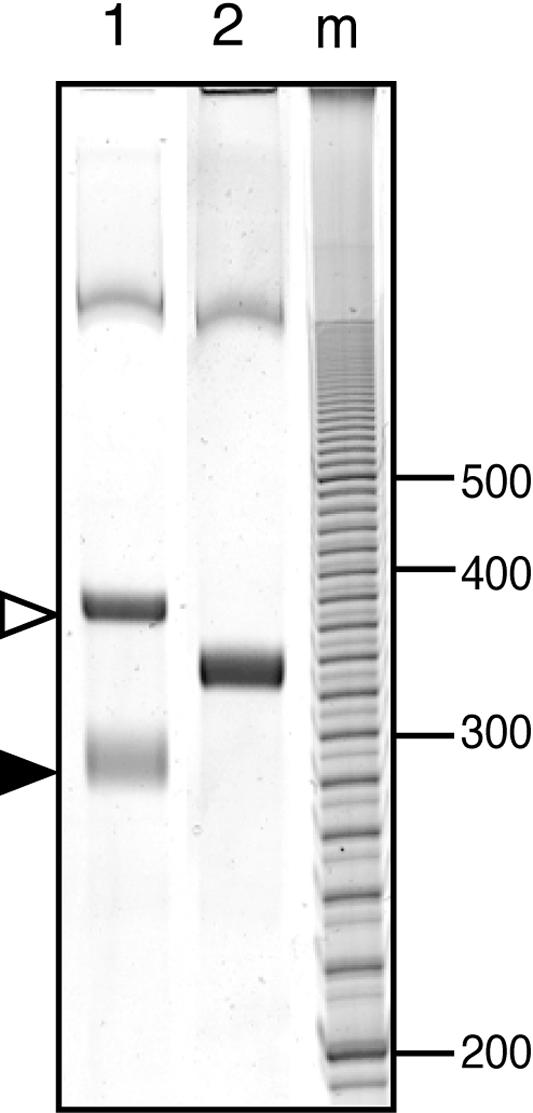
In vitro transcription assay of the vmlR gene controlled by the terminator in the 5′ UTR. The DNA templates used were TopovmlR+ter (lane 1) and TopovmlRΔter (lane 2). The positions of the runoff and terminated transcripts are indicated by open and filled arrows, respectively. m, molecular standard (bp).
No stabilization of vmlR mRNA occurred by the addition of virginiamycin M.
The steady level of mRNA in a cell is a function of its rate of synthesis and degradation. In order to determine whether increased vmlR mRNA levels in the presence of virginiamycin M in growth medium were due to the stabilization of vmlR mRNA, cells were grown to early log phase in LB medium in the presence or absence of virginiamycin M. Rifampin (500 μg ml−1) was added, and samples were removed at set time intervals. Total RNA was isolated, electrophoresed, and hybridized with probe A. The half-life was 2.1 and 1.9 min in the presence or absence of virginiamycin M, respectively (Table 2). The results showed that the increased levels of vmlR mRNA were not due to the stabilization of vmlR mRNA in the presence of virginiamycin M. Transcript hybridized to probe B was not detected 1 min after the addition of rifampin, indicating that it was a very unstable transcript. The results are consistent with the assumption that these small transcripts were derived from premature termination and posttranscriptional processing.
TABLE 2.
Half-life of vmlR transcript
| Virginiamycin M (μg ml−1) | Half-life (min) |
|---|---|
| 0 | 1.9 |
| 1.9 | 2.1 |
DISCUSSION
We have shown that the vmlR gene encodes an ABC transporter involved in inducible virginiamycin M and lincomycin resistance in B. subtilis. To date, no ABC transporter that is specific to virginiamycin M and lincomycin has been reported. In addition to the hypersensitivity toward virginiamycin M and lincomycin, the relative resistance toward macrolide antibiotics, erythromycin, clindamycin, and oleandomycin decreased to approximately 50% in the vmlR deletion mutant. Therefore, it is more appropriate to characterize VmlR as a multidrug-resistant efflux transporter with a narrow drug specificity rather than as a specific efflux transporter for virginiamycin M and lincomycin. Both streptogramin A and lincosamide antibiotics induce the expression of the vmlR gene. Expression of MsrA, which is reported to involve macrolide and streptogramin B resistance in S. epidermidis, is induced by erythromycin, but not by streptogramin B (26). The msrA gene is immediately preceded by a 320-bp 5′ UTR containing four inverted repeat sequences and an open reading frame predicted to encode a short peptide. These 5′ UTR structures resemble those found in the macrolide-lincosamide-streptogramin resistance gene, the expression of which is regulated by translational attenuation (17). However, experimental evidence for such regulation of the msrA gene has not been reported.
The results presented in this study clearly show that in the absence of inducer, the majority of the transcription initiated from the vmlR promoter ends at the terminator in the 5′ UTR. In the presence of inducer, partial transcription reads through the terminator and continues to the vmlR coding region. Northern hybridization and vmlR-lacZ fusion gene expression analysis showed that the efficiency of read-through was not high.
A variety of mechanisms for controlling gene expression by premature termination of transcription have been reported in bacteria. Transcription termination is commonly controlled by selective formation of either of two alternate folding patterns, which are called terminator and antiterminator. RNA folding can be regulated through interaction with some regulatory factors, such as ribosomes translating leader peptides, as seen in the E. coli trp operon (32), or an RNA binding protein, as seen in the B. subtilis trp, hut, and pyr operons (5, 6, 14, 19, 28) or the E. coli bgl operon (11). In the regulation of the T-box gene, tRNA directly modulates the folding of leader RNA (8). Recently, modulation of RNA structures by a small effector molecule (without a regulatory protein) that is known as a riboswitch has been demonstrated for the riboflavin, thiamine, and methionine biosynthetic operons (9, 15, 30, 31).
In the 5′ UTR of vmlR mRNA, no small open reading frame that is preceded by the stringent ribosome-binding site is detected, indicating that the vmlR gene is not regulated by ribosome-mediated transcription attenuation like the E. coli trp operon. Several inverted repeat sequences that may be able to fold in alternate stem-loop structures were found in the 5′ UTR of vmlR mRNA (Fig. 3C). However, we were unable to find a typical antiterminator structure that was located upstream of the terminator and also partially overlapped with the terminator. Instead, another stem-loop structure (SL-2) was detected in the downstream region of the terminator (Fig. 6). The 5′ end sequence of the stem in SL-2 is complementary to the 3′ end sequence of the terminator stem, indicating that an alternative stem-loop structure can be folded. The result that no difference in the expression patterns of the vmlR-lacZ fusion gene could be detected between VMLR189 and VMLR545 suggests that SL-2 is not involved in the control of transcriptional termination of vmlR expression. As shown in Fig. 6, both the terminator and SL-2 in the 5′ UTR are conserved in VgA of S. aureus. The ribosome-binding site of the vmlR gene or the vgA gene is included in the stem region of SL-2, which raises the possibility that SL-2 may be involved in the translational control of these genes.
It remains to future work to elucidate a precise mechanism of induction by virginiamycin M or lincomycin. Further experiments are under way to determine whether a trans-acting protein factor such as an RNA-binding protein is involved in the control of transcriptional termination of vmlR gene expression.
Acknowledgments
We thank T. Nihira for the generous gifts of virginiamycin M and S purified by HPLC. We also greatly appreciate all of the members of the Japan and EU consortia of B. subtilis functional genomics who provided the pMutin-based mutants used in this study.
This work was supported by a grant in aid from the Scientific Research Promotion Fund of the Japan Private School Promotion Foundation.
REFERENCES
- 1.Aiba, H., S. Adhya, and B. de Crombrugghe. 1981. Evidence for two functional gal promoters in intact Escherichia coli cells. J. Biol. Chem. 25:11905-11910. [PubMed] [Google Scholar]
- 2.Aiso, T., and R. Ohki. 2003. Instability of sensory histidine kinase mRNAs in Escherichia coli. Genes Cells 8:179-187. [DOI] [PubMed] [Google Scholar]
- 3.Allignet, J., V. Loncle, and N. E. Solh. 1992. Sequence of a staphylococcal plasmid gene, vga, encoding a putative ATP-binding protein involved in resistance to virginiamycin A-like antibiotics. Gene 117:45-51. [DOI] [PubMed] [Google Scholar]
- 4.Allignet, J., and N. E. Solh. 1997. Characterization of a new staphylococcal gene, vgaB, encoding a putative ABC transporter conferring resistance to streptogramin A and related compounds. Gene 202:133-138. [DOI] [PubMed] [Google Scholar]
- 5.Babitzke, P., J. T. Stults, S. J. Shire, and C. Yanofsky. 1994. TRAP, the trp RNA-binding attenuation protein of Bacillus subtilis, is a multisubunit complex that appears to recognize G/UAG repeats in the trpEDCFBA and trpG transcripts. J. Biol. Chem. 269:16597-16604. [PubMed] [Google Scholar]
- 6.Bonner, E. R., J. N. D'Elia, B. K. Billips, and R. L. Switzer. 2001. Molecular recognition of pyr mRNA by the Bacillus subtilis attenuation regulatory protein PyrR. Nucleic Acids Res. 29:4851-4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fukuchi, K., Y. Kasahara, K. Asai, K. Kobayashi, S. Moriya, and N. Ogasawara. 2000. The essential two-component regulatory system encoded by yycF and yycG modulates expression of the ftsAZ operon in Bacillus subtilis. Microbiology 146:1573-1583. [DOI] [PubMed] [Google Scholar]
- 8.Grundy, F. J., and T. M. Henkin. 1993. tRNA as a positive regulator of transcription antitermination in B. subtilis. Cell 74:475-482. [DOI] [PubMed] [Google Scholar]
- 9.Grundy, F. J., and T. M. Henkin. 1998. The S box regulon: a new global transcription termination control system for methionine and cysteine biosynthesis genes in gram-positive bacteria. Mol. Microbiol. 30:737-749. [DOI] [PubMed] [Google Scholar]
- 10.Horton, R. M. 1993. In vitro recombination and mutagenesis of DNA, p. 251-261. In B. A. White (ed.), Methods in molecular biology, vol. 15. PCR protocols: current methods and applications. Humana Press Inc., Totowa, N.J. [DOI] [PubMed] [Google Scholar]
- 11.Houman, F., M. R. Diaz-Torres, and A. Wright. 1990. Transcriptional antitermination in the bgl operon of E. coli is modulated by a specific RNA binding protein. Cell 62:1153-1163. [DOI] [PubMed] [Google Scholar]
- 12.Kawachi, R., T. Akashi, Y. Kamitani, A. Sy, U. Wangchaisoonthorn, T. Nihira, and Y. Yamada. 2000. Identification of an AfsA homologue (BarX) from Streptomyces virginiae as a pleiotropic regulator controlling autoregulator biosynthesis, virginiamycin biosynthesis and virginiamycin M1 resistance. Mol. Microbiol. 36:302-313. [DOI] [PubMed] [Google Scholar]
- 13.Lee, C.-K., M. Minami, S. Sakuda, T. Nihira, and Y. Yamada. 1996. Stereospecific reduction of virginiamycin M1 as the virginiamycin resistance pathway in Streptomyces virginiae. Antimicrob. Agents Chemother. 40:595-601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu, Y., R. J. Turner, and R. L. Switzer. 1996. Function of RNA secondary structures in transcriptional attenuation of the Bacillus subtilis pyr operon. Proc. Natl. Acad. Sci. USA 93:14462-14467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mandal, M., B. Boese, J. E. Barrick, W. C. Winkler, and R. R. Breaker. 2003. Riboswitches control fundamental biochemical pathways in Bacillus subtilis and other bacteria. Cell 113:577-586. [DOI] [PubMed] [Google Scholar]
- 16.Mascher, T., N. G. Margulis, T. Wang, R. W. Ye, and J. D. Helmann. 2003. Cell wall stress responses in Bacillus subtilis: the regulatory network of the bacitracin stimulon. Mol. Microbiol. 50:1591-1604. [DOI] [PubMed] [Google Scholar]
- 17.Mayford, M., and B. Weisblum. 1989. ermC leader peptide: amino acid sequence critical for induction by translational attenuation. J. Mol. Biol. 206:69-79. [DOI] [PubMed] [Google Scholar]
- 18.Méndez, C., and J. A. Salas. 2001. The role of ABC transporters in antibiotic-producing organisms: drug secretion and resistance mechanisms. Res. Microbiol. 152:341-350. [DOI] [PubMed] [Google Scholar]
- 19.Oda, M., N. Kobayashi, M. Fujita, Y. Miyazaki, Y. Sadaie, Y. Kurusu, and S. Nishikawa. 2004. Analysis of HutP-dependent transcription antitermination in the Bacillus subtilis hut operon: identification of HutP binding sites on hut antiterminator RNA and the involvement of the N terminus of HutP in binding of HutP to the antiterminator RNA. Mol. Microbiol. 51:1155-1168. [DOI] [PubMed] [Google Scholar]
- 20.Ohki, R., Giyanto, K. Tateno, W. Masuyama, S. Moriya, K. Kobayashi, and N. Ogasawara. 2003. The BceRS two-component regulatory system induces expression of the bacitracin transporter, BceAB, in Bacillus subtilis. Mol. Microbiol. 49:1135-1144. [DOI] [PubMed] [Google Scholar]
- 21.Ohki, R., K. Tateno, Y. Okada, H. Okajima, K. Asai, Y. Sadaie, M. Murata, and T. Aiso. 2003. A bacitracin-resistant Bacillus subtilis gene encodes a homologue of the membrane-spanning subunit of the Bacillus licheniformis ABC transporter. J. Bacteriol. 185:51-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Olano, C., A. M. Rodríguez, C. Méndez, and J. A. Salas. 1995. A second ABC transporter is involved in oleandomycin resistance and its secretion by Streptomyces antibioticus. Mol. Microbiol. 16:333-343. [DOI] [PubMed] [Google Scholar]
- 23.Parro, V., M. S. Román, I. Galindo, B. Purnelle, A. Bolotin, A. Sorokin, and R. P. Mellado. 1997. A 23911 bp region of the Bacillus subtilis genome comprising genes located upstream and downstream of the lev operon. Microbiology 143:1321-1326. [DOI] [PubMed] [Google Scholar]
- 24.Peschke, U., H. Schmidt, H.-Z. Zhang, and W. Piepersberg. 1995. Molecular characterization of the lincomycin-production gene cluster of Streptomyces lincolnensis 78-11. Mol. Microbiol. 16:1137-1156. [DOI] [PubMed] [Google Scholar]
- 25.Quentin, Y., G. Fichant, and F. Denizot. 1999. Inventory, assembly and analysis of Bacillus subtilis ABC transport systems. J. Mol. Biol. 287:467-484. [DOI] [PubMed] [Google Scholar]
- 26.Ross, J. I., E. A. Eady, J. H. Cove, W. J. Cunliffe, S. Baumberg, and J. C. Wootton. 1990. Inducible erythromycin resistance in staphlyococci is encoded by a member of the ATP-binding transport super-gene family. Mol. Microbiol. 4:1207-1214. [DOI] [PubMed] [Google Scholar]
- 27.Schoner, B., M. Geistlich, P. Rosteck Jr., R. N. Rao, E. Seno, P. Reynolds, K. Cox, S. Burgett, and C. Hershberger. 1992. Sequence similarity between macrolide-resistance determinants and ATP-binding transport proteins. Gene 115:93-96. [DOI] [PubMed] [Google Scholar]
- 28.Shimotsu, H., and D. J. Henner. 1984. Characterization of the Bacillus subtilis tryptophan promoter region. Proc. Natl. Acad. Sci. USA 81:6315-6319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vagner, V., E. Dervyn, and S. D. Ehrlich. 1998. A vector for synthetic gene inactivation in Bacillus subtilis. Microbiology 144:3097-3104. [DOI] [PubMed] [Google Scholar]
- 30.Winkler, W., A. Nahvi, and R. R. Breaker. 2002. Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature 419:952-956. [DOI] [PubMed] [Google Scholar]
- 31.Winkler, W. C., S. Cohen-Chalamish, and R. R. Breaker. 2002. An mRNA structure that controls gene expression by binding FMN. Proc. Natl. Acad. Sci. USA 99:15908-15913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yanofsky, C. 2004. The different roles of tryptophan transfer RNA in regulating trp operon expression in E. coli versus B. subtilis. Trends Genet. 20:367-374. [DOI] [PubMed] [Google Scholar]