Abstract
Mga is a transcriptional regulator in the pathogen Streptococcus pyogenes that positively activates several important virulence genes involved in colonization and immune evasion in the human host. A naturally occurring mutant of Mga that is defective in its ability to activate transcription has been identified in the serotype M50 strain B514-Sm. Sequence alignment of the defective M50 Mga with the fully functional Mga from serotypes M4 and M49 revealed only three amino acid changes that might result in a defective protein. Electrophoretic mobility shift assays using purified M50 and M4 maltose binding protein-Mga found that both exhibited DNA-binding activity towards regulated promoters. Thus, the significance of each residue for the functionality of M50 Mga was explored through introduction of “gain-of-function” mutations based on M4 Mga. Transcriptional studies of the mutant alleles under both constitutive (PrpsL) and autoactivated (Pmga4) promoters illustrated that an arginine-to-methionine change at position 461 of M50 Mga protein fully restored activation of downstream genes. Western blot analyses of steady-state Mga levels suggest that the M461 residue may play a role in overall conformation and protein stability of Mga. However, despite the conservation of the M461 protein among all other Mga proteins, it does not appear to be necessary for activity in a divergent M6 Mga. These studies highlight the potential differences that exist between divergent Mga proteins in this important human pathogen.
The group A streptococcus (Streptococcus pyogenes; GAS) is a gram-positive bacterial pathogen of humans that can colonize various sites throughout the body and elicit a broad range of diseases ranging from mild erythema to life-threatening bacteremia. The capacity to colonize diverse sites stems in part from the bacterium's ability to regulate its genes in response to the different environments encountered during an infection. Transcriptional regulators within a cell are often able to transduce such environmental signals into differential virulence gene expression, facilitating bacterial survival through expression of proteins involved in attachment as well as factors involved in evading the host immune response. In support of this model, GAS possesses numerous regulatory networks that are responsive to environmental stimuli, including 13 two-component signal transduction systems and at least three “stand-alone” transcriptional regulators (for a review see reference 14).
One such transcriptional regulator is the multiple gene regulator of GAS or Mga, which has been defined as a “stand-alone” response regulator because it is able to activate transcription of a number of important virulence genes in response to growth and environmental conditions and yet lacks any identifiable sensory elements (14). Mga-regulated gene products are important for adhesion and immune evasion during early stages of colonization at tissue sites and include M proteins (emm and arp), M-like proteins (mrp), C5a peptidase (scpA), and extracellular matrix binding proteins (sclA [also called scl1] and fba [also called orfX]). Previous studies have shown that Mga binds to specific sites within regulated promoters via two amino-terminal helix-turn-helix (HTH) DNA-binding domains (20), leading to autoactivation of mga expression (22, 23) as well as to the transcriptional activation of other Mga-regulated genes (1, 18). Expression of the Mga regulon is activated in response to different signals such as increased CO2 levels, body temperature, and exponential-phase growth (5, 19, 21, 24), although the mechanisms that Mga utilizes to respond to such cues remain undefined.
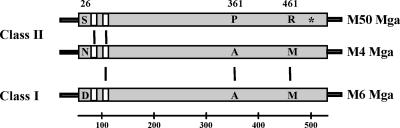
The mga locus on the GAS chromosome includes at least one emm family gene (emm, arp, and emmL) and scpA located immediately downstream, with additional genes encoding M-like proteins (mrp and enn) and a fibronectin-binding protein (fba) being present in certain serotypes (8, 11, 30). The antiphagocytic M protein provides the foundation for serological typing of the group A streptococcus (16) and has been divided into two classes to reflect the ability to react with antibodies against a conserved domain within its surface-exposed region (3). Although not absolute, serotypes containing class I emm genes (e.g., M1, M6, and M24) are generally associated with throat infections whereas those possessing class II emm genes (M2, M4, and M49) are generally associated with infections at both skin and throat sites (4, 13). Using a PCR-based analysis, two major types of mga alleles were found to exist among the different serotypes of GAS that correlate with the pattern of genes found at the mga locus and the class of M protein produced (32). The two divergent types of mga differ in sequence identity by approximately 22%, primarily within the extreme 3′ end (32). Despite the sequence differences, Mga proteins produced from the two types exhibit similar abilities to complement an mga deletion in a class I M6 strain (2).
Initial investigations of Mga from the serotype M50 strain B514-Sm, which was originally isolated from infections in mice (12), revealed a protein that was defective in activation of the downstream genes mrp50, encoding an M-like protein, and emm50, encoding a class II M protein (31, 32). Furthermore, the defective M50 mga allele exhibited 98% nucleotide identity to the mga encoding a fully functional Mga found in other class II serotype M4 and M49 strains (32). In that study, four amino acids were reported to be different between the M50 Mga protein and M4 and M49 Mga. However, our current study finds that only three amino acid differences exist and an amino acid alignment revealed that none of the changes resides within the recently established HTH DNA-binding motifs (20) or at other sites predicted to be important for Mga activity (32). We have used mutagenesis of residues that differ between M4 and M50 Mga to establish those amino acids important for Mga-specific transcription of the downstream gene mrp and autoactivation of mga in these strains.
MATERIALS AND METHODS
Bacterial strains and media.
The S. pyogenes vectors for integration (VIT) strain RTG229 is a derivative of the serotype M6 strain JRS4 (10, 27). B514-Sm is a spontaneous streptomycin-resistant derivative of the serotype M50 GAS strain B514 (17). The clinical isolate AP4 is a serotype M4 GAS strain (28). AL168-mga contains a Tn916 insertion that inactivates the mga22 gene in the M22 GAS isolate AL168 (29). Escherichia coli DH5α (New England Biolabs) was used as a host for all plasmid constructions, while E. coli SA2817 was used for protein purifications (18).
E. coli was grown in Luria-Bertani broth, while GAS strains were grown in Todd-Hewitt medium supplemented with 0.2% yeast extract. Growth of GAS was measured by absorbance on a Klett-Summerson photoelectric colorimeter using the A filter. The indicated concentrations of antibiotics were used: ampicillin at 100 μg/ml for E. coli, spectinomycin at 100 μg/ml for both E. coli and GAS, and erythromycin at 500 μg/ml for E. coli and 1 μg/ml for GAS.
DNA manipulation.
Plasmid DNA was isolated from E. coli using the Wizard Miniprep kit (Promega). Genomic DNA was isolated using the FastDNA Prep kit and a FastPrep cell disruptor (Bio 101). DNA fragments were purified from agarose gels using the QIAquick gel extraction kit (QIAGEN). All PCRs were performed using Pfu Turbo DNA polymerase (Stratagene), and resulting products were purified with the QIAquick PCR purification system (QIAGEN). All site-specific mutations were generated with the QuickChange site-directed mutagenesis kit (Stratagene) using mutagenic oligonucleotides synthesized by Integrated DNA Technologies. DNA sequencing was done by the McDermott Center sequencing core facility at UT Southwestern Medical Center.
Expression and purification of M4 and M50 MBP-Mga from E. coli.
Plasmids expressing amino-terminal fusions of maltose binding protein (MBP) to the wild-type Mga from serotypes M4 and M50 (MBP-Mga) were constructed as follows: a 1.9-kb DNA fragment of only the mga coding sequence was amplified from genomic DNA (gDNA) from both AP4 (M4) and B514-Sm (M50) using the primers DivMga-5′blunt and DivMga-3′HIII (Table 1). The resulting PCR fragments were digested with HindIII and inserted into XmnI/HindIII-digested vector pMal-c2 (New England Biolabs) to generate the malE-mga fusion alleles for purification of M4 (pKSM155) and M50 (pKSM156) MBP-Mga, respectively (Table 2). MBP-Mga proteins were purified from E. coli as previously described (18). Protein purity was assessed by sodium dodecyl sulfate (SDS)-polyacrylamide gels stained with Sypro Ruby (Sigma) and by analysis of Western blots probed with anti-MBP antibodies (New England Biolabs) as described below.
TABLE 1.
PCR primers and their relevant targets
| Target and primer | Sequencea | Reference |
|---|---|---|
| 23S rRNA | ||
| rRNA-23SL | GGAAGGTAAGCCAAAGAGAG | 25 |
| rRNA-23SR | TCCTAGTTGTCTGTGCAACC | 25 |
| mga6 | ||
| M6 M461R-a | GATGTGATCGTGACAGATGTTAgGGTAGGAAAAAGCGATGAG | This study |
| M6 M461R-b | CTCATCGCTTTTTCCTACCcTAACATCTGTCACGATCACATC | This study |
| Mgadel-L2 | CGAGGCCTTAGCTTTTTGATGGCATCATGG | This study |
| Mgadel-R2 | CCCTTGGACTTTCATCGC | This study |
| MgaHis-BglII | gccgagatctGCGAGAAAGG | This study |
| OYR-29 | AAACCAACGCCTATTTGACGCATAC | This study |
| mga4 mga50 | ||
| DivMga-5′blunt | CATGTAAGTAAATTGTTTACTAGCCAACAATGGAG | This study |
| DivMga-3′HIII | gcgtcaaagcttCTGAAATCCTATGATGATGTTGC | This study |
| DivMga-Pet1 | gggcatatgCATGTAAGTAAATTG | This study |
| DivMga-Pet2 | gggctcgagTGATGATGTTGCTTGTTT | This study |
| DivMga-R5 | TGGATCCATCTATTAGATGAG | This study |
| DivMga-R9 | ccccaagcttGATAAGGACATGAAGTTAAT | This study |
| DivMga-Sph | ccgcatgcctatgatgatgttgcttg | This study |
| MgaHis-Sph | cgcatgcCTATGATGATGTTGCTTG | This study |
| Mga50 P361A-a | CACCCTCGCATTTATGAGgCCTTTGTGACAAGTGTCGAGAAGC | This study |
| Mga50 P361A-b | TCTTCTCGACACTTGTCACAAAGGcCTCATAAATGCGAGGGTG | This study |
| Mga50 R461M-a | CAGTACGATGTTATCGTGACAGATGTTAtGGTGGGTAAAAGCGAAG | This study |
| Mga50 R461M-b | CTTCGCTTTTACCCACCaTAACATCTGTCACGATAACATCGTACTG | This study |
| mrp49 | ||
| mrp49-L1 | CAAGCTAAGCTAGATACAGCAACT | This study |
| mrp49-R1 | CGCCTGTTGACGGTAATT | This study |
| Parp | ||
| Parp4-L | CATTGACAGTGATCGCATCT | This study |
| Parp4-R | AAGCGAATACTGTTTATTCG | This study |
| Pmga | ||
| Pmga-B | gcgggatccTAAGTTAACCAGTTCACAAA | 22 |
| Pmga-X | ggctcgagACCTTGTATACCCTTCTTTT | 22 |
| Pmrp | ||
| Pmrp4-L | AGCCAGACAATTCAGTTAAA | This study |
| Pmrp4-R | CTCAGTGAATAGAGTTTGTTTG | This study |
| Pmrp4-EcoRI | gcgggaattcTCAATTTCTAAGAATTGTG | This study |
| Pmrp4-BglII | gcggagatctGGATTTCAGACGTCAT | This study |
| PrpsL | ||
| GASrpsL-EcoRI | gggcgaattcTGTCTAAAATCACATCTTCG | This study |
| GASrpsL-Hind | ggggaagcttGGTTGATATAGCACTTGGTGAC | This study |
| Other | ||
| SpacI | CTACATCCAGAACAACCTCTGC | This study |
| T7-term | CTAGTTATTGCTCAGCGG | Novagen |
Noncomplementary sequences are in lowercase, mutagenic nucleotides are in boldface, and introduced restriction sites are underlined.
TABLE 2.
Plasmids used in this study
| Plasmid | Relevant characteristic(s) | Reference or source |
|---|---|---|
| pBluescriptII KS(−) | Blue/white cloning vector | Stratagene |
| pET21a | Six-His vector | Novagen |
| pMal-c2 | malE MBP fusion vector | New England Biolabs |
| pMga4-4 | WTmga4ain pLZ12-Spec | 2 |
| pMga50-His | WT mga50 gene with C-terminal six-His tag | This study |
| pPmga-blue | Pmga in pBluescript II KS(−) | This study |
| pPmga-gusA | gusA under Pmga6 | This study |
| pPmrp-gusA | gusA under Pmrp | This study |
| pVIT164 | GAS integration vector | 6 |
| pJRS312 | Spectinomycin cassette | 26 |
| pJRS515 | WT mga6 in pBluescript II KS(−) | 18 |
| pJRS547 | Deletion of 2.1 kb of mga and Pmga (mga-16) | This study |
| pJRS9160 | Counterselectable gene replacement vector | 20 |
| pKSM140 | Promoterless gusA in pCIV2 suicide vector | This study |
| pKSM155 | N-terminal MBP fusion to M4 Mga | This study |
| pKSM156 | N-terminal MBP fusion to M50 Mga | This study |
| pKSM162 | WT mga6 under constitutive Pspac | 20 |
| pKSM164 | WT mga6-his under native Pmga6 | 1 |
| pKSM174 | mga-16 deletion allele in pJRS9160 | This study |
| pKSM315 | WT mga50 under constitutive Pspac | This study |
| pKSM318 | WT mga6-his under constitutive Pspac | This study |
| pKSM320 | WT mga4-his under constitutive Pspac | This study |
| pKSM321 | WT mga50-his under constitutive Pspac | This study |
| pKSM317-H | mga50-his (S26N) under constitutive Pspac | This study |
| pP361A-H | mga50-his (P361A) under constitutive Pspac | This study |
| pR461M-H | mga50-his (R461M) under constitutive Pspac | This study |
| pKSM322 | WT mga4-his under native Pmga4 | This study |
| pSKM323 | mga50-his (S26N) under native Pmga4 | This study |
| pKSM324 | WT mga4-his under constitutive PrpsL | This study |
| pKSM325 | WT mga50-his under constitutive PrpsL | This study |
| pKSM326 | mga50-his (S26N) under constitutive PrpsL | This study |
| pKSM327 | mga50-his (P361A) under constitutive PrpsL | This study |
| pKSM328 | mga50-his (R461M) under constitutive PrpsL | This study |
| pKSM329 | WT mga50-his under native Pmga4 | This study |
| pKSM330 | mga50-his (R461M) under native Pmga4 | This study |
| pKSM331 | mga50-his (S26N/R461M) under native Pmga4 | This study |
| pKSM332 | mga50-his (S26N/R461M) under constitutive PrpsL | This study |
| pKSM333 | WT mga6-his under constitutive PrpsL | This study |
| pKSM336 | mga50-his (P361A) under native Pmga4 | This study |
| pKSM337 | mga6-his (M461R) under constitutive Pmga6 | This study |
| pKSM338 | mga6-his (M461R) under constitutive PrpsL | This study |
WT, wild type.
EMSA.
Promoter probes for Parp and Pmrp were generated by PCR amplification from serotype M4 strain AP4 gDNA using the relevant primer pairs Parp4-R and Parp4-L and Pmrp4-R and Pmrp4-L, respectively (Table 1). Electrophoretic mobility shift assay (EMSA) was performed as previously described (18). Briefly, constant amounts of probe end labeled with [γ-32P]ATP were incubated with increasing concentrations of purified MBP-Mga protein for 15 min at 16°C before being separated on a 5% polyacrylamide gel.
Whole-cell GAS protein extracts.
Whole-cell GAS proteins were extracted as previously described (20). Briefly, mid-logarithmic-phase GAS cultures (55 Klett units) were harvested by centrifugation and resuspended in saline containing 1× Complete protease inhibitor cocktail (Roche). Cells were lysed using a FastPrep cell disruptor (Bio 101, Inc.), and soluble lysates were recovered by centrifugation. Total protein concentrations were determined using the Protein Assay kit (Bio-Rad).
Western blot analysis.
Proteins were analyzed by Western blotting as previously described (20). Blots were incubated with a 1:2,000 dilution of either anti-His tag monoclonal antibody (Novagen) or anti-MBP antiserum (New England Biolabs) and then incubated with a 1:25,000 dilution of either anti-mouse (Chemicon) or anti-rabbit (Sigma) horseradish peroxidase-conjugated secondary antibody, respectively, and visualized using the Western Lightning chemiluminescence system (Perkin-Elmer). As a loading control, blots were stripped and reprobed with a 1:50,000 dilution of mouse anti-Hsp60 monoclonal antibody (StressGen Biotechnologies Corp.). All Western blot assays were done at least three times.
Construction of a Δmga allele in the M6 Pemm-gusA reporter strain KSM148.174.
A deletion encompassing the 2.1-kb mga coding sequence and its 493-bp promoter region (Pmga) was generated by inverse PCR from the plasmid pJRS515 (18), which contains the wild-type serotype M6 mga locus, using the diverging primers Mgadel-L2 and Mgadel-R2 (Table 1). The resulting 5.4-kb product was religated to form the mga-16 deletion allele marked with a new StuI restriction site in the plasmid pJRS547. A 2.5-kb BamHI/HindIII fragment containing the mga-16 deletion allele from pJRS547 was subsequently cloned into the counterselectable gene replacement vector pJRS9160 (20) to generate pKSM174 (Table 2). An mga-deleted derivative of the serotype M6 VIT strain KSM148 (25) was constructed through an allelic replacement of the wild-type mga at the native locus with the unmarked mga-16 deletion allele using pKSM174 as previously described (20) to produce KSM148.174.
Construction of an Δmga allele in the M6 Pmrp-gusA reporter strain KSM149.
An mga-deleted derivative of the serotype M6 VIT strain RTG229 (10) was constructed as follows: to allow the use of the erythromycin-resistant pKSM174, the erythromycin cassette found in the VIT locus of RTG229 was replaced with the spectinomycin cassette from pJRS312 (26) to generate VIT230. An mga-deleted VIT230 strain was created by allelic replacement of its wild-type mga at the native locus with the mga-16 deletion allele from pKSM174 as previously described (20) to produce VIT231.
An mga-deleted serotype M6 VIT strain containing a single-copy transcriptional fusion of the serotype M4 Mga-regulated mrp promoter to gusA (Pmrp-gusA) was constructed as follows: a Pmga-gusA fusion was first produced by amplification of a 493-bp Pmga fragment from serotype M6 JRS4 gDNA using the primers Pmga-B and Pmga-X (Table 1). The resulting fragment was digested with BamHI and cloned into BamHI/SmaI-digested pBluescript II KS(−) (Table 2) to produce pPmga-blue. A 1.9-kb EcoRI/HindIII fragment containing the promoterless gusA gene from pKSM140 (25) was inserted into EcoRI/HindIII-digested pPmga-blue to generate pBlue-Gus#2. The resulting 2.4-kb HpaI/SalI Pmga-gusA fragment was excised from pBlue-Gus#2 and cloned into SmaI/SalI-digested pVIT164 (6) to produce pPmga-gusA. Pmrp was amplified from serotype M4 AP4 gDNA using primers Pmrp4-EcoRI and Pmrp4-BglII (Table 1). Pmga was excised from pPmga-gusA following digestion with EcoRI/HpaI and replaced with EcoRI-digested Pmrp to produce pPmrp-gusA. This construct was linearized with XmnI and transformed into the mga-deleted strain VIT231 to generate KSM149.
Construction of the Pspac-mga-his plasmids.
A wild-type mga50 gene containing a 3′ six-His tag under the constitutive promoter Pspac (Pspac-mga50-his) was generated as follows: the mga50 gene was amplified from serotype M50 B514-Sm gDNA using primers DivMga-R9 and DivMga-Sph (Table 1), digested with HindIII/SphI, and cloned downstream of Pspac in HindIII/SphI-digested pKSM162 (21), to create pKSM315. To produce an mga50-his fusion, the mga50 allele was amplified from serotype M50 B514-Sm gDNA using primers DivMga-Pet1 and DivMga-R9 (Table 1), digested with NdeI/XhoI, and cloned into the NdeI/XhoI-digested pET21a (Novagen), creating pMga50-His. The C-terminal 996-bp fragment of the mga50-his gene was amplified from pMga50-His using primers DivMga-R5 and MgaHis-Sph (Table 1), digested with BamHI/SphI, and cloned into the BamHI/SphI-digested pKSM315, producing pKSM321 (Table 2).
The Pspac-mga4-his allele was generated as follows: mga4 was amplified from pMga4-4 (2) using primers DivMga-R9 and DivMga-Pet2 (Table 1), digested with HindIII/XhoI, and cloned into the HindIII/XhoI-digested pKSM321 to produce pKSM320 (Table 2).
The S26N mutation in M50 mga was constructed as follows: the 917-bp HindIII/BamHI fragment from pKSM320 was cloned into HindIII/BamHI-digested pKSM321, producing pKSM317-H. Site-specific mutants of Mga50 P361A and R431M were generated in pKSM321 as described above using the mutagenic primers Mga50 P361A-a, Mga50 P361A-b, Mga50 R461M-a, and Mga50 R461M-b (Table 1), resulting in pP361A-H and pR431M-H, respectively (Table 2).
Construction of the PrpsL-mga-his plasmids.
The wild-type and mutant mga alleles were placed under the constitutive promoter PrpsL as follows: a 385-bp region of the rpsL promoter was amplified from serotype M6 JRS4 gDNA using the primers GASrpsL-EcoRI and GASrpsL-Hind (Table 1). Pspac was excised from pKSM320 following digestion with XmnI/HindIII and replaced with the HindIII-digested PrpsL fragment to produce pKSM324 (Table 2). Plasmids pKSM325, pKSM326, and pKSM327 were all constructed in a similar fashion using pKSM321, pKSM317-H, and pP361A-H, respectively, as template vectors instead of pKSM320. The PrpsL-mga50-his allele containing the R461M mutation was constructed by inserting a 1.9-kb HindIII/SphI fragment containing the R461M mutation from pR431M-H into HindIII/SphI-digested pKSM324 to produce pKSM328 (Table 2). A double mutant of Mga50 (S26N and R461M) was generated in pKSM326, which already contains the S26N mutation, using the mutagenic primers Mga50 R461M-a and Mga50 R461M-b (Table 1), resulting in pKSM332 (Table 2).
Construction of the Pmga4-mga-his plasmids.
An mga4-his fusion under the native mga4 promoter (Pmga4) was generated as follows: the 988-bp BamHI/SphI fragment containing mga4-his from pKSM320 was cloned into BamHI/SphI-digested pMga4-4 (2), which contains Pmga4-mga4, producing pKSM322. An S26N allele was constructed using the 988-bp BamHI/SphI fragment from pKSM321 cloned into the BamHI/SphI-digested pKSM322 to produce pKSM323 (Table 2). Pmga4-mga50-his was generated as follows: the 1.9-kb NsiI/SphI fragment containing the mga50-his fusion from pKSM321 was cloned into NsiI/SphI-digested pMga4-4, producing pKSM329. Plasmid pKSM330 (Pmga4-mga50[R461M]-his) was constructed in a similar fashion by extracting the NsiI/SphI fragment from pR431M-H instead of pKSM321. The Pmga4-mga50[S26N, R461M]-his double mutant was produced by excising a 707-bp BamHI/XhoI fragment from pKSM332 and inserting it into the BamHI/XhoI-digested pKSM322 vector, producing pKSM331 (Table 2). Pmga4-mga50[P361A]-his was generated as described above using the mutagenic primers Mga50 P361A-a and Mga50 P361A-b (Table 1) in pKSM329, resulting in pKSM336.
Construction of M6 mga mutant alleles under Pmga6 and PrpsL.
A Pmga6-mga6-his plasmid containing the M461R mutation was generated in pKSM164 (1) via site-specific mutagenesis using the mutagenic primers M6 M461R-a and M6 M461R-b (Table 1) to produce pKSM337 (Table 2). The wild-type and mutant mga6 genes were placed under the constitutive expression of PrpsL as follows: a 3′ 1.5-kb region of mga6-his was amplified from pKSM164 (1) using the primers OYR-29 and MgaHis-BglII (Table 1) and digested with SpeI/BglII. The 3′ end of Pspac-mga6 was excised from pKSM162 (20) following digestion with SpeI/BglII and replaced with the digested PCR fragment to produce pKSM318. The mga6-his gene was then amplified from pKSM318 using the primers SpacI and T7-term (Table 1), digested with XhoI/HindIII, and put into XhoI/HindIII-digested pKSM328, producing pKSM333 (Table 2). An M6 Mga M461R mutant was generated using the mutagenic primers M6 M461R-a and M6 M461R-b (Table 1) in pKSM333, resulting in pKSM338 (Table 2).
GusA reporter assays.
Soluble whole-cell lysates were isolated from GAS grown to mid-logarithmic phase (55 Klett units) as described above. GusA activity was determined for each lysate (100 μl) as described by Ribardo and McIver (25). Total protein concentrations were determined as above. GusA units are defined as the optical density at 420 nm/total protein concentration of the lysate.
Northern blot and slot blot analysis.
Total RNA was isolated from GAS grown to mid-logarithmic phase using the FastRNA Pro Blue kit and FastPrep cell disruptor (Bio 101). Northern blot analysis was performed using the NorthernMax system (Ambion) as previously described (25). As a loading control, blots were stripped and reprobed with a 23S rRNA probe (Table 1). Slot blot analysis was performed as previously described (21).
RESULTS
Wild-type M4 and M50 MBP-Mga fusion proteins exhibit DNA-binding activity.
Previous studies have shown that the mga alleles found in serotypes M50 and M4 are almost identical at the amino acid level. Yet, the Mga protein from the M50 strain is defective in activating downstream Mga-regulated genes compared to its fully functional M4 counterpart (31, 32). To date, the specific M50 Mga residues responsible for this reduction in activation have not been characterized. M50 Mga was originally thought to possess four amino acid changes that differed from Mga proteins produced by serotypes M4 and M49 (32). We now report that, after multiple independent rounds of sequencing from the original serotype M50 strain B514-Sm gDNA, only three of these nonconservative amino acid changes were observed (Fig. 1; S26, P361, and R461). Although none of the changes were located within the two recently established helix-turn-helix DNA-binding domains of Mga (20), it was important to rule out the possibility that the three mutations in M50 Mga resulted in a structural alteration affecting its DNA-binding activity, leading to the observed reduction in gene activation.
FIG. 1.
Schematic comparison of class I and class II Mga proteins. Mga homologues are found in all serotypes of GAS and can range in size from 530 amino acids (M6) to 533 amino acids (M4 and M50). Based upon amino acid alignment, both class I and class II Mga proteins contain two N-terminal HTH domains (white boxes). The three nonconservative amino acid differences detected between Mga4 and Mga50 (positions 26, 361, and 461) are indicated with their respective amino acids shown in each homologue. A fourth nonconservative change at position 521 identified in a previous study (32), but not in the present study, is also indicated (*). Solid black lines represent residues or domains relevant to this study that are 100% conserved among the proteins.
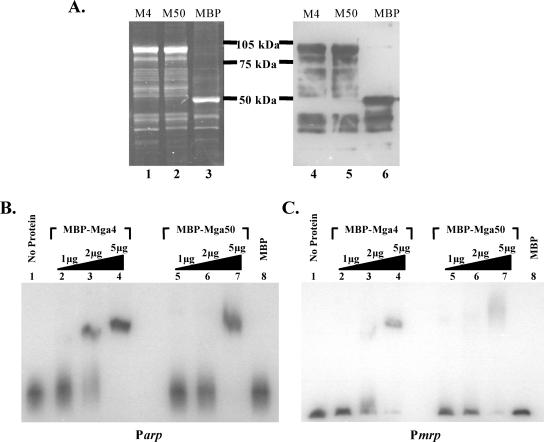
EMSAs were performed to investigate the ability of purified M4 and M50 Mga proteins to bind to DNA targets in vitro. Plasmids containing malE-mga fusion alleles encoding MBP-Mga4 (pKSM155) and MBP-Mga50 (pKSM156) were generated, and Mga was purified from E. coli lysates containing these plasmids using an amylose affinity resin column. Comparable amounts of each 103-kDa protein were assessed for purity by SDS-polyacrylamide gel electrophoresis followed by staining for total protein using Sypro Ruby and specific detection of the fusion protein using Western analysis with anti-MBP antiserum (Fig. 2A). Increasing amounts of the purified MBP-Mga proteins were incubated with a constant amount of radiolabeled promoter probes corresponding to two Mga-regulated promoters from the serotype M4 strain AP4. Because the mga operon of the AP4 strain does not contain an emm gene, the two native promoters Parp (Fig. 2B) and Pmrp (Fig. 2C) were chosen for use as probes. EMSA reaction mixtures containing 5 μg of either M4 MBP-Mga or M50 MBP-Mga showed reduced mobility of each probe, indicative of a protein-DNA interaction compared to 5 μg of the purified MBP control protein alone (Fig. 2B and C). Thus, the activation-defective M50 Mga exhibited the ability to bind to DNA from the promoter regions of the Mga-regulated genes mrp and arp, although the bands resulting from M50 MBP-Mga bound to each promoter probe were more diffuse and required more protein for a definitive shift than observed for M4 MBP-Mga bound to the same targets (Fig. 2B and C).
FIG. 2.
Electrophoretic mobility shift analysis of M4 and M50 MBP-Mga binding to class II Mga-regulated promoters. (A) Purification of M4 and M50 MBP-Mga fusion proteins from E. coli lysates using an amylose affinity resin. Purified protein was assessed with SDS-polyacrylamide gels stained with Sypro Ruby (left) and a Western blot probed with anti-MBP (right). Purified M4 MBP-Mga (lanes 1 and 4), M50 MBP-Mga (lanes 2 and 5), and MBP control (lanes 3 and 6) are shown. (B and C) EMSAs were performed on two M4 Mga-regulated promoters, Parp (B) and Pmrp (C). Identical amounts of each radiolabeled promoter probe were incubated for 15 min at 16°C with an increasing amount (1, 2, and 5 μg) of either the purified M4 MBP-Mga (lanes 2 to 4) or M50 MBP-Mga (lanes 5 to 7) fusion protein before being separated on a 5% polyacrylamide gel. Binding was also assessed in the presence of no protein (lanes 1) or 5 μg MBP alone (lanes 8) for each.
Establishment of PrpsL as a constitutive promoter in GAS.
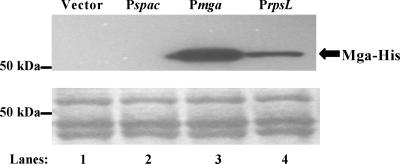
In order to remove any effect that the promoter might have on overall transcript levels as a result of autoregulation by the Mga protein, each allele was placed under the control of a constitutive promoter. Although the spac promoter (Pspac) has been used in other studies for low-level constitutive expression of M6 Mga in GAS (9, 20, 21), expression of M4 and M50 from Pspac did not produce a detectable level of protein (Fig. 3), nor did it show activation of an Mga-regulated GusA reporter to a level significantly above background (data not shown). Therefore, the promoter for the ribosomal protein S12 gene rpsL (PrpsL) from GAS was tested as a constitutive promoter. Expression of mga4 from PrpsL resulted in Mga-regulated GusA expression levels significantly above background (data not shown) and produced a detectable amount of Mga-His, albeit at a lower level than mga4 expressed from its native promoter Pmga (Fig. 3). Thus, different mga alleles were cloned under the control of PrpsL for Mga-independent expression in GAS (Table 2).
FIG. 3.
Steady-state protein levels of Mga produced from both the native and various constitutive promoters. Western analysis (top) was performed on whole-cell lysates using an anti-His antibody for the detection of Mga-His protein in samples either not producing Mga (lane 1) or producing M4 Mga from the constitutive promoters Pspac (lane 2) and PrpsL (lane 4) or the native promoter Pmga4 (lane 3). Amido black stain (bottom) of total protein on membranes was used as a loading control. All blots shown are representative of data from three independent experiments.
Functional importance of residue 461 for M4 and M50 Mga expressed from a constitutive promoter (PrpsL).
To determine the effect of the three amino acid differences found in the M50 Mga (Fig. 1; S26, P361, and R461) on its reduced transcriptional activation in vivo, each residue was mutated to the corresponding residue found in M4 Mga to test for a “gain-of-function” phenotype. Plasmids were constructed which contained an mga4 or mga50 gene or an mga50 gene in which either one or two of the targeted amino acids in M50 Mga were changed to resemble M4 Mga (Table 2). Since antibodies recognizing M4 and M50 Mga are not available, each construct also contains a carboxy-terminal fusion to the six-His tag to allow for the detection of protein by Western blot assays using anti-His monoclonal antibodies (Novagen). Transformation of the plasmids into a class I mga-deleted GAS reporter strain (KSM149) containing gusA fused to the promoter of a native Mga-regulated gene mrp from the M4 strain AP4 (Pmrp-gusA) in single copy (Table 3) allowed for direct quantitation of Mga-regulated activity by measurement of the level of GusA activity in cell lysates (see Materials and Methods).
TABLE 3.
Reporters/strains and constructs
| Name | Relevant characteristic(s) | Figure(s) used |
|---|---|---|
| Reporters/strains | ||
| KSM149 | Δmga M6 strain containing a single-copy Pmrp-gusA reporter in the chromosome | 4A, 5A, 6B |
| KSM148.174 | Δmga M6 strain containing a single-copy Pemm-gusA reporter in the chromosome | 6A |
| AL168-mga | Heterologous Δmga M22 strain | 4B, 4C, 5B |
| Constructs | ||
| PrpsL-mga | Constitutive rpsL promoter upstream of mga constructs on a plasmid | 4A, 4B, 4C |
| Pmga4-mga | Native mga4 promoter upstream of mga constructs on a plasmid | 5A, 5B |
| Pmga6-mga | Native mga6 promoter upstream of mga constructs on a plasmid | 6A, 6B |
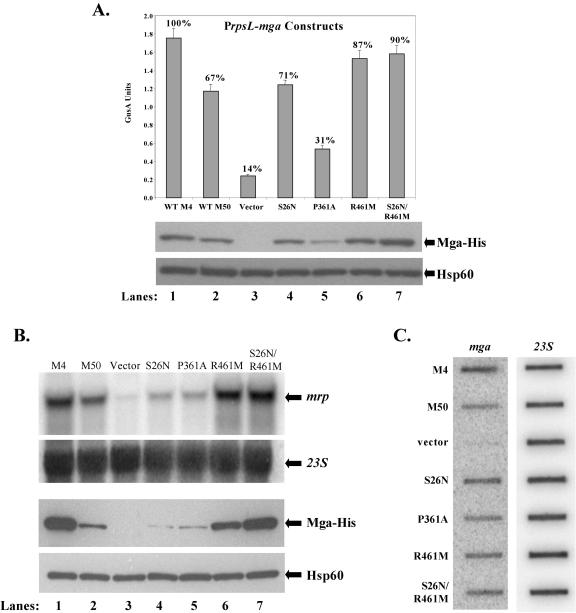
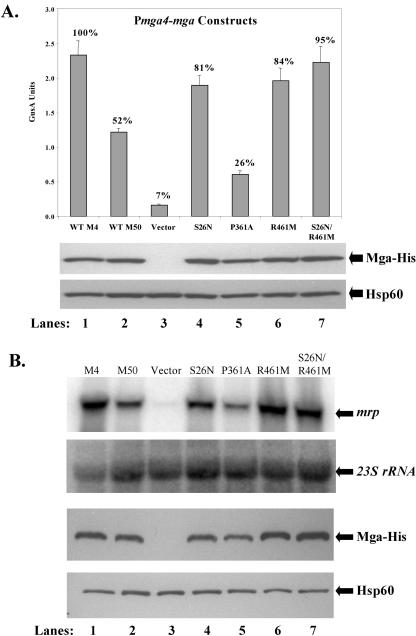
Mga-regulated GusA activity and steady-state levels of Mga-His were assessed in the Pmrp-gusA reporter strain KSM149 expressing the different PrpsL-mga-his plasmids (Fig. 4A). Expression of M50 Mga from PrpsL in the reporter strain produced reduced activity (ca. 67%) compared to the M4 Mga but well above the background seen for vector alone (Fig. 4A). This agrees with previous studies comparing M50 Mga activity to that for M4 Mga (32). Two of the mutated mga50 alleles, producing amino acid changes S26N (pKSM326) and P361A (pKSM327), did not substantially increase the level of GusA activity above that seen for the native M50 Mga (pKSM325), indicating that these mutations did not improve the ability of M50 Mga to activate Pmrp-gusA. In fact, the P361A mutation actually resulted in a more defective protein (Fig. 4A). In contrast, GusA levels similar to those observed for the M4 Mga were seen for the strain containing both R461M and the S26N/R461M double mutation (Fig. 4A). The levels of Mga-His protein detected in each strain were not equivalent compared to the loading control Hsp60 (Fig. 4A). These data suggest that the amino acid change at residue 461 is important for the reduced activity of M50 Mga compared to M4 Mga and may affect its steady-state levels in the cell when it is expressed from the constitutive promoter PrpsL.
FIG. 4.
In vivo transcriptional activity of mutant M50 mga alleles expressed from a constitutive PrpsL promoter. (A) GusA activity of whole-cell lysates (top). Production of β-glucuronidase activity was determined for lysates from an mga-deleted Pmrp-gusA reporter strain KSM149 containing plasmids expressing mga alleles from the PrpsL promoter (M4 Mga [lane 1], M50 Mga [lane 2], vector only [lane 3], and M50 Mga mutants [S26N [lane 4], P361A [lane 5], R461M [lane 6], and S26N/R461M [lane 7]). GusA units represents a measure of absorbance (A420)/protein concentration (μg/ml), and values are the averages of at least three independent experiments. The percent activity compared to M4 Mga is indicated above each bar. Western analysis was performed on whole-cell lysates using both an anti-His antibody for Mga-His protein levels (middle) and anti-Hsp60 antibodies as a control for loading (bottom). WT, wild type. (B) Northern analysis of Mga-specific transcriptional activation. Transcript levels for the Mga-regulated gene mrp were determined using total RNA (5 μg) isolated from an mga-inactivated M22 strain, AL168-mga, containing plasmids expressing Mga alleles from the PrpsL promoter (M4 Mga [lane 1], M50 Mga [lane 2], vector only [lane 3], and M50 Mga mutants S26N [lane 4], P361A [lane 5], R461M [lane 6], and S26N/R461M [lane 7]). Blots were stripped and reprobed with 23S rRNA to serve as a loading control (directly below). Western analysis was performed on whole-cell lysates using both an anti-His antibody for Mga-His protein levels (third panel from the top) and with antibodies to Hsp60 as a control for loading (bottom). All blots shown are representative of data from three independent experiments. (C) Slot blot analysis of mga transcripts produced from the constitutive PrpsL promoter. Total RNA (1 μg) was isolated from the mga-inactivated M22 strains above and probed for M50 mga (left) and 23S rRNA as a loading control (right).
To investigate the effect of the different M50 Mga mutants on transcriptional activation in a class II background, the PrpsL-mga-his plasmids (Table 2) were introduced into the mga-inactivated M22 strain AL168-mga (Table 3) (29). Northern blot analysis was performed on total RNA isolated from each strain using a radiolabeled probe to the Mga-regulated gene mrp22. As observed above, the strain expressing M50 Mga was reduced in activation of mrp22 compared to wild-type M4 Mga. Furthermore, mrp22 transcript levels in only those strains containing the R461M or the S26N/R461M mutations were comparable to the levels found with M4 Mga (Fig. 4B). In this system, both the S26N and P361A mutations appeared to further reduce activation of mrp22 compared to wild-type M50 Mga (Fig. 4B). Again, the steady-state levels of Mga-His detected in each strain directly correlated with the observed transcript levels for mrp22, although this result was more apparent in the M22 strain AL168-mga (Fig. 4B). This variation in levels of Mga-His protein was not reflected at the level of transcription, since slot blot analysis of mga-specific transcripts in the samples exhibited equivalent expression in contrast to the variable protein levels detected (Fig. 4C). Taken together, changing only amino acid 461 from arginine to methionine in the defective M50 Mga resulted in a restoration of activity comparable to M4 Mga. Furthermore, the varying level of Mga-His protein produced among the mutant alleles, all of which were expressed from the constitutive PrpsL promoter at similar levels, may reflect a destabilized conformation when certain residues are present.
Both residues 26 and 461 are important for transcriptional activation by M4 and M50 Mga expressed from the native promoter (Pmga4).
To determine whether the varying protein levels observed for the mutants expressed from PrpsL reflected in vivo expression levels, each allele was placed under the control of the native Pmga4 found upstream of the fully functional M4 mga. The resulting plasmids (Table 2) were transformed into the mga-deleted Pmrp-gusA reporter strain KSM149, and the level of Mga-regulated GusA activity was measured. In contrast to the PrpsL results (Fig. 4A), levels of Mga-His protein resulting from Pmga4 were comparable across strains, excluding the vector-alone negative control (Fig. 5A and B). Similar to previous results, Pmrp-gusA expression was reduced in the strain expressing M50 Mga to approximately 52% of the levels seen for the M4 Mga-expressing strain (Fig. 5A). However, only the amino acid change P361A (pKSM336) failed to restore Mga-regulated GusA levels to those seen in M4 Mga (pKSM322). As before, the P361A mutation appeared to further reduce M50 Mga activity. Under Pmga4, mutations in both R461M (pKSM330) and S26N (pKSM323) resulted in GusA levels comparable to those for M4 Mga (Fig. 5A), indicating that both positions are involved in transcriptional activation of Mga-regulated genes when expressed from Pmga4. In addition, the S26N/R461M double mutant had a slightly increased activation of Pmrp-gusA over either the S26N or R461M mutant alone.
FIG. 5.
In vivo transcriptional activity of mutant M50 mga alleles expressed from a native Pmga4 promoter. (A) GusA activity of whole-cell lysates (top). Production of β-glucuronidase activity was determined for lysates from an mga-deleted Pmrp-gusA reporter strain KSM149 containing plasmids expressing Mga alleles from the Pmga4 promoter (M4 Mga [lane 1], M50 Mga [lane 2], vector only [lane 3], and M50 Mga mutants S26N [lane 4], P361A [lane 5], R461M [lane 6], and S26N/R461M [lane 7]). GusA units represents a measure of absorbance (A420)/protein concentration (μg/ml), and values are an average of at least three independent experiments. The percent activity compared to M4 Mga is indicated above each bar. Western analysis was performed on whole-cell lysates using both an anti-His antibody for Mga-His protein levels (middle) and antibodies to Hsp60 as a control for loading (bottom). WT, wild type. (B) Northern analysis of Mga-specific transcriptional activation. Transcript levels for the Mga-regulated mrp were determined using total RNA (5 μg) isolated from an mga-inactivated M22 strain, AL168-mga, containing plasmids expressing mga alleles from the Pmga4 promoter (M4 Mga [lane 1], M50 Mga [lane 2], vector only [lane 3], and M50 Mga mutants S26N [lane 4], P361A [lane 5], R461M [lane 6], and S26N/R461M [lane 7]). Blots were stripped and reprobed with 23S rRNA to serve as a loading control (directly below). Western analysis was performed on whole-cell lysates using both an anti-His antibody for Mga-His protein levels (third panel from the top) and antibodies to Hsp60 as a control for loading (bottom). All blots shown are representative of data from three independent experiments.
The results of the Pmga4 studies in the GusA reporter strain were confirmed via Northern analysis of transcript levels of mrp22 in the mga-inactivated M22 strain AL168-mga. Transcript levels of mrp22 in each strain exhibited the same profile seen in the GusA reporter (Fig. 5B), with the R461M mutant and the S26N/R461M double mutant producing mrp transcript levels similar to those seen for strains expressing M4 Mga (Fig. 5B). The S26N mutant did show a positive effect on M50 Mga activity but to a somewhat lesser degree than R461M. Once again, levels of protein produced from Pmga4 were consistent from strain to strain. Overall, these data indicate that both residues 461 and 26 are responsible for the reduced transcriptional activation of M50 Mga when expressed from the native Pmga4. Furthermore, they suggest that expression levels achieved from the autoactivated Pmga4 may mask the potential instability seen for several M50 mutants when expressed from the constitutive Mga-independent rpsL promoter.
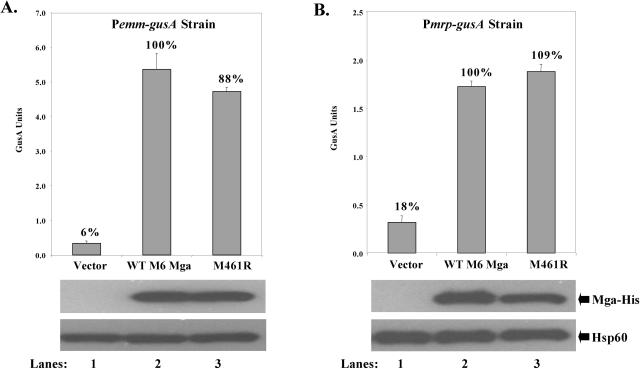
Residue 461 is not important for activity in a divergent serotype M6 Mga.
Upon alignment of primary amino acid sequences for Mga from multiple GAS strains, it was noted that the methionine residue at position 461, but not the serine at residue 26, was conserved among all Mga proteins, with the exception of M50 Mga (Fig. 1). To test whether the conserved M461 was important for full activity and stability in a divergent Mga protein, a site-specific mutation leading to an M461R change was made in the mga from the serotype M6 strain JRS4 under its native promoter Pmga6 (pKSM337). Since mrp is not a native target in the M6 strain, the resulting M461R mutant plasmid (Table 2), a wild-type Pmga6-mga6 (pKSM164), and a vector control (pLZ12-Spc) were transformed into the mga-deleted Pemm6-gusA reporter strain KSM148.174 (Table 3) to assess activation of a single-copy Mga-regulated promoter (Pemm6) in its native M6 background. No significant differences in Mga-regulated GusA levels were seen between the wild-type M6 Mga protein and the M461R Mga (Fig. 6A). To ensure that the activation defect observed in M50 Mga was not restricted to certain Mga-regulated genes, the plasmids were also transformed into the Pmrp-gusA reporter strain KSM149 and analyzed for GusA activity (Fig. 6B). Alteration of M461R in M6 Mga again did not significantly change its ability to activate expression of Mga-regulated Pmrp (Fig. 6B). Western analysis of all samples showed that steady-state levels of Mga were comparable for each strain, indicating that the M461R mutation does not appear to play a significant role in the stability of M6 Mga. Thus, the conserved M461 residue does not appear to be important in a divergent M6 Mga.
FIG. 6.
In vivo transcriptional activity of a mutant M6 mga allele at different promoters. GusA activity of whole-cell lysates is shown (top). Production of β-glucuronidase activity was determined for lysates from either mga-deleted Pemm-gusA reporter strain KSM148.174 (A) or mga-deleted Pmrp-gusA reporter strain KSM19 (B). Both strains contained plasmids expressing wild-type M6 Mga (lanes 1), mutant M461R M6 Mga (lanes 2), and vector alone (lanes 3) from the native Pmga6 promoter. GusA units represents a measure of absorbance (A420)/protein concentration (μg/ml), and values are the average of at least three independent experiments. The percent activity compared to M6 Mga is indicated above each bar. Western analysis was performed on whole-cell lysates using both an anti-His antibody for Mga-His protein levels (middle) and antibodies to Hsp60 as a control for loading (bottom). All blots shown are representative of data from three independent experiments. WT, wild type.
DISCUSSION
Historically, the M50 serotype of GAS has caused several outbreaks of fatal cervical lymphadenitis in mouse colonies at various institutions dating back to the 1930s (12). However, serotype M50 B514 has been isolated only from the throats of asymptomatic individuals working directly with the organism, demonstrating that it retains the capacity to colonize humans although these are not its natural reservoir (15). Characterization of M50 B514-Sm revealed an Mga regulatory protein defective in activation of its Mga-regulated virulence genes (mrp and arp) even though it was 98% identical to other fully functional Mga proteins expressed in other similar class II serotypes such as M4 and M49 (32). Yung et al. previously proposed that the activation defect of M50 Mga might result from an inability to bind properly to target promoter DNA based on the observation that one of the three amino acid differences in M50 Mga (S26N) was located within a putative HTH DNA-binding motif (32). However, the DNA-binding domains have since been shown to be located elsewhere (20) and do not overlap any of the amino acids in question. Furthermore, our results indicate that M50 Mga does retain its ability to bind to DNA (Fig. 2). Thus, M50 Mga is a naturally occurring mutant that could provide insight into the relationship between the primary amino acid sequence of Mga and novel aspects of its ability to function as a transcriptional activator of virulence genes critical for pathogenesis in humans.
Amino acids important for full activity in Mga proteins.
To determine which of the three amino acid changes alone or in combination affected the ability of Mga to activate target gene transcription, “gain-of-function” mutants were introduced in M50 Mga by changing amino acids to their corresponding residues in the functional M4 Mga. All in vivo analyses of the proteins, from the transcriptional reporter fusion in the serotype M6 to the Northern analysis in the M22 serotype, were performed in heterologous systems to rule out the possibility that a defect other than the M50 Mga protein alone was the cause of the loss of transcriptional activity seen in the B514-Sm M50 strain.
Transcription levels resulting from a change in Mga at position 26 from the serine to an asparagine varied based upon the promoter used to express the mutant mga. When the constitutive rpsL promoter was used, transcription remained at a level equal to that of M50 Mga and protein levels were variable. In contrast, when the same mga allele was transcribed from its native promoter, transcriptional activation was elevated and protein levels did not vary. This fluctuation of steady-state protein levels under constitutive expression could be a result of a less stable protein conformation, leading to a more rapid degradation of the protein in the cell. Since a change at position 26 restored full activation under the native mga4 promoter, where protein levels are higher, one can assume that in the native bacterium residue 26 plays a more significant role in producing a functional Mga. This also stresses the importance of Pmga for proper Mga production in a virulent GAS cell.
It was suggested by Yung et al. that the second amino acid difference, which changed residue 361 from a flexible alanine to a constrained proline, could alter the protein conformation of M50 Mga and possibly disrupt its activity (32). However, altering residue 361 to an alanine never resulted in a gain-of-function phenotype. Instead, the outcome of a change at residue 361 was independent of the specific amino acid at this position. Interestingly, the most detrimental effect was seen with an alanine, not a proline, at position 361 in the context of the M50 Mga (i.e., P361A M50 mutant). This mutation resulted in a decrease of transcriptional activation below that of the already deficient native M50 Mga to only slightly above the level seen for background vector alone. These data suggest that this area may be acting in concert with the other parts of the protein because the level of activation can fall depending upon the context in which it is placed and not solely on the amino acid residue at this exact location.
This is not the case for the third mutation at position 461. Changing residue 461 from an arginine to a methionine in M50 Mga always restored the activity to wild-type levels despite the promoter or context in which the mutant was placed. Therefore, this position must play an essential role in the activation of mrp and possibly other Mga-regulated genes.
R461 plays an important role in the conformation of Mga.
Several lines of evidence point to a methionine at position 461 as an amino acid important for correct conformation and thus overall stability of the Mga protein. The first piece of data comes from a comparison of the R461M mutation under the constitutive PrpsL promoter. When the level of transcript is constitutive and M50 Mga is mutated to contain a methionine at position 461, an increase in overall Mga-His protein levels is observed compared to all other constructs containing an arginine at this position, even though the level of mga transcription did not increase among them (Fig. 4). This would imply that a methionine at position 461 is essential for correct conformation and that the substitution of an arginine leads to a protein more sensitive to degradation. However, the mga promoter is able to mask the effects of degradation (Fig. 5), possibly because Pmga undergoes a self-amplification process and thus produces more protein than PrpsL (Fig. 3). This could possibly conceal the low-level degradation apparent only with constitutive PrpsL.
The second line of evidence stems from the EMSAs performed in this study comparing wild-type M50 and M4 MBP-Mga. These data clearly demonstrate that both proteins bind to DNA from the promoter regions of relevant Mga-regulated genes (Fig. 2). Thus, it was inferred that the reduction in M50 Mga activity in vivo is not due simply to a loss of overall DNA binding ability. Although both M50 and M4 MBP-Mga each bound to the promoter probes when 5 μg of protein was used, differences in the overall binding pattern were observed. For instance, at lower levels of M50 MBP-Mga protein (2 μg), binding to Parp was not detected compared to M4 MBP-Mga (Fig. 2B). The bound complex formed by M50 MBP-Mga and promoter DNA also appeared more diffuse with both targets than those observed for M4 MBP-Mga, suggesting that differences may exist in how the two Mga proteins interact at regulated promoters. One possibility for this difference could be a slight structural change that does not disrupt the ability to bind but does alter the overall shape of the bound complex and therefore its binding pattern.
Because no higher-order structural data exist for the Mga protein, the exact location of amino acid 461 in relation to the rest of the protein cannot be determined. The polarity of the position is altered when the nonpolar methionine is changed to a polar arginine; however, the possible consequences of this change do not appear to be overly dramatic in a region predicted to have less than 25% but greater than 5% solvent accessibility according to JNET (7). In addition, the consensus secondary structure prediction generated from the Jpred server (http://www.compbio.dundee.ac.uk/∼www-jpred) found amino acid 461 to be located within a beta sheet, in which case the presence of a methionine over an arginine would not appear to have a significant effect.
Differences between divergent Mga proteins.
Because secondary structural predictions did not reveal a compelling need for a methionine at position 461, a primary sequence alignment of all sequenced Mga proteins was performed to determine the conservation of methionine at this position. Surprisingly, even though the Mga proteins diverge the most from one another in the extreme carboxy terminus, the primary amino acid alignment of all Mga proteins revealed that M461 was 100% conserved with the exception of M50 Mga. Since this was the only amino acid of the three investigated in this study that demonstrated 100% conservation and was shown to be essential for activation in an M50 Mga protein, this residue was mutated to an arginine in a divergent M6 strain to determine if a loss of activation would be seen. However, this mutation did not show a transcriptional defect at either the native M6 Mga-regulated gene emm or the M4/M50 Mga-regulated gene mrp. This illustrates that the importance of M461 is not universal, contrary to what may be predicted based on its conservation and the fact that the Mga proteins have been shown to be functionally equivalent in vivo (2). These data also reinforce the divergence between the two classes of Mga, implying that fundamental differences may also exist between the functional residues used by each class to activate transcription of Mga-regulated virulence genes.
Acknowledgments
We thank June Scott, Susan Hollingshead, Gunnar Lindahl, and Lars Heden for bacterial strains used in this study. Critical reading of the manuscript by Deborah Ribardo, Audry Almengor, Temekka Leday, and Traci Kinkel is also greatly appreciated.
This work was supported by NIH/NIAID Public Health Service award R01-AI47928 to K.S.M.
REFERENCES
- 1.Almengor, A. C., and K. S. McIver. 2004. Transcriptional activation of sclA by Mga requires a distal binding site in Streptococcus pyogenes. J. Bacteriol. 186:7847-7857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andersson, G., K. McIver, L. O. Heden, and J. R. Scott. 1996. Complementation of divergent mga genes in group A streptococcus. Gene 175:77-81. [DOI] [PubMed] [Google Scholar]
- 3.Bessen, D., K. F. Jones, and V. A. Fischetti. 1989. Evidence for two distinct classes of streptococcal M protein and their relationship to rheumatic fever. J. Exp. Med. 169:269-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bessen, D. E., C. M. Sotir, T. L. Readdy, and S. K. Hollingshead. 1996. Genetic correlates of throat and skin isolates of group A streptococci. J. Infect. Dis. 173:896-900. [DOI] [PubMed] [Google Scholar]
- 5.Caparon, M. G., R. T. Geist, J. Perez-Casal, and J. R. Scott. 1992. Environmental regulation of virulence in group A streptococci: transcription of the gene encoding M protein is stimulated by carbon dioxide. J. Bacteriol. 174:5693-5701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caparon, M. G., and J. R. Scott. 1991. Genetic manipulation of pathogenic streptococci. Methods Enzymol. 204:556-586. [DOI] [PubMed] [Google Scholar]
- 7.Cuff, J. A., and G. J. Barton. 2000. Application of multiple sequence alignment profiles to improve protein secondary structure prediction. Proteins 40:502-511. [DOI] [PubMed] [Google Scholar]
- 8.Cunningham, M. W. 2000. Pathogenesis of group A streptococcal infections. Clin. Microbiol. Rev. 13:470-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eichenbaum, Z., M. J. Federle, D. Marra, W. M. de Vos, O. P. Kuipers, M. Kleerebezem, and J. R. Scott. 1998. Use of the lactococcal nisA promoter to regulate gene expression in gram-positive bacteria: comparison of induction level and promoter strength. Appl. Environ. Microbiol. 64:2763-2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Geist, R. T., N. Okada, and M. G. Caparon. 1993. Analysis of Streptococcus pyogenes promoters by using novel Tn916-based shuttle vectors for the construction of transcriptional fusions to chloramphenicol acetyltransferase. J. Bacteriol. 175:7561-7570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hollingshead, S. K., J. Arnold, T. L. Readdy, and D. E. Bessen. 1994. Molecular evolution of a multigene family in group A streptococci. Mol. Biol. Evol. 11:208-219. [DOI] [PubMed] [Google Scholar]
- 12.Hook, E. W., R. R. Wagner, and R. C. Lancefield. 1960. An epizootic in Swiss mice caused by a group A Streptococcus, newly designated type 50. Am. J. Hyg. 72:111-119. [DOI] [PubMed] [Google Scholar]
- 13.Kalia, A., B. G. Spratt, M. C. Enright, and D. E. Bessen. 2002. Influence of recombination and niche separation on the population genetic structure of the pathogen Streptococcus pyogenes. Infect. Immun. 70:1971-1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kreikemeyer, B., K. S. McIver, and A. Podbielski. 2003. Virulence factor regulation and regulatory networks in Streptococcus pyogenes and their impact on pathogen-host interactions. Trends Microbiol. 11:224-232. [DOI] [PubMed] [Google Scholar]
- 15.Kurl, D. N. 1981. Laboratory-acquired human infection with group A type 50 streptococci. Lancet ii:752. [DOI] [PubMed] [Google Scholar]
- 16.Lancefield, R. C. 1962. Current knowledge of type-specific M antigens of group A streptococci. J. Immunol. 89:307-313. [PubMed] [Google Scholar]
- 17.Limbago, B., K. S. McIver, V. Penumalli, B. Weinrick, and J. R. Scott. 2001. Restoration of Mga function to a Streptococcus pyogenes strain (M type 50) that is virulent in mice. Infect. Immun. 69:1215-1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McIver, K. S., A. S. Heath, B. D. Green, and J. R. Scott. 1995. Specific binding of the activator Mga to promoter sequences of the emm and scpA genes in the group A streptococcus. J. Bacteriol. 177:6619-6624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McIver, K. S., A. S. Heath, and J. R. Scott. 1995. Regulation of virulence by environmental signals in group A streptococci: influence of osmolarity, temperature, gas exchange, and iron limitation on emm transcription. Infect. Immun. 63:4540-4542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McIver, K. S., and R. L. Myles. 2002. Two DNA-binding domains of Mga are required for virulence gene activation in the group A streptococcus. Mol. Microbiol. 43:1591-1602. [DOI] [PubMed] [Google Scholar]
- 21.McIver, K. S., and J. R. Scott. 1997. Role of mga in growth phase regulation of virulence genes of the group A streptococcus. J. Bacteriol. 179:5178-5187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McIver, K. S., A. S. Thurman, and J. R. Scott. 1999. Regulation of mga transcription in the group A streptococcus: specific binding of Mga within its own promoter and evidence for a negative regulator. J. Bacteriol. 181:5373-5383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Okada, N., R. T. Geist, and M. G. Caparon. 1993. Positive transcriptional control of mry regulates virulence in the group A streptococcus. Mol. Microbiol. 7:893-903. [DOI] [PubMed] [Google Scholar]
- 24.Podbielski, A., J. A. Peterson, and P. Cleary. 1992. Surface protein-CAT reporter fusions demonstrate differential gene expression in the vir regulon of Streptococcus pyogenes. Mol. Microbiol. 6:2253-2265. [DOI] [PubMed] [Google Scholar]
- 25.Ribardo, D. A., and K. S. McIver. 2003. amrA encodes a putative membrane protein necessary for maximal exponential phase expression of the Mga virulence regulon in Streptococcus pyogenes. Mol. Microbiol. 50:673-685. [DOI] [PubMed] [Google Scholar]
- 26.Saile, E., and T. M. Koehler. 2002. Control of anthrax toxin gene expression by the transition state regulator abrB. J. Bacteriol. 184:370-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Scott, J. R., P. C. Guenthner, L. M. Malone, and V. A. Fischetti. 1986. Conversion of an M− group A streptococcus to M+ by transfer of a plasmid containing an M6 gene. J. Exp. Med. 164:1641-1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stenberg, L., P. O'Toole, and G. Lindahl. 1992. Many group A streptococcal strains express two different immunoglobulin-binding proteins, encoded by closely linked genes: characterization of the proteins expressed by four strains of different M-type. Mol. Microbiol. 6:1185-1194. [DOI] [PubMed] [Google Scholar]
- 29.Thern, A., M. Wastfelt, and G. Lindahl. 1998. Expression of two different antiphagocytic M proteins by Streptococcus pyogenes of the OF+ lineage. J. Immunol. 160:860-869. [PubMed] [Google Scholar]
- 30.Whatmore, A. M., V. Kapur, D. J. Sullivan, J. M. Musser, and M. A. Kehoe. 1994. Non-congruent relationships between variation in emm gene sequences and the population genetic structure of group A streptococci. Mol. Microbiol. 14:619-631. [DOI] [PubMed] [Google Scholar]
- 31.Yung, D. L., and S. K. Hollingshead. 1996. DNA sequencing and gene expression of the emm gene cluster in an M50 group A streptococcus strain virulent for mice. Infect. Immun. 64:2193-2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yung, D. L., K. S. McIver, J. R. Scott, and S. K. Hollingshead. 1999. Attenuated expression of the mga virulence regulon in an M serotype 50 mouse-virulent group A streptococcal strain. Infect. Immun. 67:6691-6694. [DOI] [PMC free article] [PubMed] [Google Scholar]