Abstract
The YtlI regulator of Bacillus subtilis activates the transcription of the ytmI operon encoding an l-cystine ABC transporter, a riboflavin kinase, and proteins of unknown function. The expression of the ytlI gene and the ytmI operon was high with methionine and reduced with sulfate. Using deletions and site-directed mutagenesis, a cis-acting DNA sequence important for YtlI-dependent regulation was identified upstream from the −35 box of ytmI. Gel mobility shift assays confirmed that YtlI specifically interacted with this sequence. The replacement of the sulfur-regulated ytlI promoter by the xylA promoter led to constitutive expression of a ytmI′-lacZ fusion in a ytlI mutant, suggesting that the repression of ytmI expression by sulfate was mainly at the level of YtlI synthesis. We further showed that the YrzC regulator negatively controlled ytlI expression while this repressor also acted on ytmI expression via YtlI. The cascade of regulation observed in B. subtilis is conserved in Listeria spp. Both a YtlI-like regulator and a ytmI-type operon are present in Listeria spp. Indeed, the Lmo2352 protein from Listeria monocytogenes was able to replace YtlI for the activation of ytmI expression and a lmo2352′-lacZ fusion was repressed in the presence of sulfate via YrzC in B. subtilis. A common motif, AT(A/T)ATTCCTAT, was found in the promoter region of the ytlI and lmo2352 genes. Deletion of part of this motif or the introduction of point mutations in this sequence confirmed its involvement in ytlI regulation.
All living organisms require sulfur for the synthesis of proteins and essential cofactors. Sulfur can be assimilated either from inorganic sources, such as sulfate and thiosulfate, or from organic sources, such as sulfate esters, sulfamates, sulfonates, cysteine, methionine, or their derivatives. In Bacillus subtilis, sulfate is transported into the cell via a sulfate permease, CysP, related to inorganic phosphate transporters (24). Sulfate is subsequently reduced to sulfide probably in four steps involving ATP sulfurylase, adenosine 5′-phosphosulfate (APS) kinase, 3′-phosphoadenosine 5′-phosphosulfate (PAPS) reductase, and sulfite reductase (3, 25, 45). An O-acetyl-l-serine thiol-lyase, the cysK gene product, condenses sulfide and O-acetylserine (OAS) to form cysteine (45). Sulfonates, cysteine, and methionine can be used as sulfur sources by B. subtilis. Several alkanesulfonates are taken up by a sulfonate ABC (ATP-binding cassette) transporter and then converted to sulfite by a FMNH2-dependent monooxygenase (46). The transport of l-cystine, the oxidized form of cysteine, has recently been investigated. Three systems are present in B. subtilis: two ABC transporters, TcyABC and TcyJKLMN, and a symporter, TcyP (5). The TcyJKLMN and TcyP uptake systems are high-affinity transporters with apparent Km values for l-cystine of 2.5 μM and 0.6 μM, respectively. In addition, the TcyJKLMN system is involved in the uptake of cystathionine, S-methylcysteine, djenkolic acid, and other sulfur compounds (5, 39). The tcyJKLMN genes belong to a large operon (operon ytmI), which also encodes a riboflavin kinase, two putative flavin-dependent monooxygenases, a putative acetyltransferase, and a putative amidohydrolase (7, 39, 41, 42). The expression of the ytmI operon and the tcyP gene is regulated in response to sulfur availability, while the level of expression of the tcyABC operon remains low in all the conditions tested (5). In addition, the expression of the ytmI operon is induced by disulfide stress or paraquat (21, 29).
In Escherichia coli and Salmonella enterica serovar Typhimurium, most of the genes involved in sulfate and sulfonate assimilation, cysteine biosynthesis, and l-cystine transport are coordinately regulated in the cysteine regulon (18). The high-level expression of these genes requires CysB, a LysR-type transcriptional activator, the inducer N-acetylserine, and limitation in sulfur availability (18, 19). The interaction of CysB with the inducer allows it to interact with the activation sites present upstream of the −35 promoter region of the positively regulated cys genes (15, 19, 28). Among the genes regulated by CysB, one encodes Cbl, a second LysR-type regulator that controls the expression of the ssu and tau operons involved in aliphatic sulfonate and taurine assimilation. In E. coli, the MetJ repressor and a third LysR-type regulator, MetR, regulate the methionine biosynthetic pathway (12).
In B. subtilis, the S-box transcription antitermination system is involved in the control of expression of genes participating in methionine uptake, biosynthesis, and recycling in response to methionine availability (1, 13, 26, 38). In addition, two LysR-type regulators, CysL and YtlI, play a role in the regulation of sulfur metabolism. CysL positively controls the expression of the cysJI operon encoding the sulfite reductase by binding to its promoter region (14). YtlI is a positive regulator of the ytmI operon encoding the TcyJKLMN l-cystine ABC transporter (5, 7). The expression of both the ytmI and ytlI genes is high in the presence of methionine and low in the presence of sulfate as sulfur source (1, 7). This indicates the existence of a cascade of regulation for the ytmI operon. In this work, we analyzed the complex regulation of ytmI expression by the YtlI activator.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The B. subtilis strains and plasmids are listed in Table 1. B. subtilis was grown in SP medium or in minimal medium (6 mM K2HPO4, 4.4 mM KH2PO4, 0.3 mM trisodium citrate, 5 mM MgCl2, 0.5% glucose, 50 mg liter−1 l-tryptophan, 22 mg liter−1 ferric ammonium citrate, 0.1% l-glutamine) containing 1 mM K2SO4, 1 mM l-methionine, or 0.5 mM l-cysteine as sole sulfur source. OAS was added to the final concentration of 1 mM. Antibiotics were added to the following concentrations: ampicillin, 100 μg ml−1; chloramphenicol, 5 μg ml−1; spectinomycin, 100 μg ml−1; and kanamycin, 5 μg ml−1. Solid media were prepared by addition of 20 g liter−1 Noble agar (Difco). Standard procedures were used to transform E. coli (33) and B. subtilis (20).
TABLE 1.
Bacterial strains used in this studya
| Strain | Genotype | Source |
|---|---|---|
| 168 | trpC2 | Laboratory stock |
| BSIP1214 | trpC2 ytlI::aphA3 | Reference 7 |
| BSIP1215 | trpC2 amyE::pΔFytlI′-lacZ cat | pDIA5575 → 168 |
| BSIP1224 | trpC2 ytlI::aphA3 amyE::pΔFytlI′-lacZ cat | pDIA5575 → BSIP1214 |
| BSIP1256 | trpC2 amyE::pΔAytmI′-lacZ cat | pDIA5599 → 168 |
| BSIP1257 | trpC2 amyE::pΔCytmI′-lacZ cat | pDIA5600 → 168 |
| BSIP1258 | trpC2 amyE::pΔDytmI′-lacZ cat | pDIA5601 → 168 |
| BSIP1259 | trpC2 amyE::pΔGytlI′-lacZ cat | pDIA5602 → 168 |
| BSIP1261 | trpC2 amyE::pΔJytlI′-lacZ cat | pDIA5606 → 168 |
| BSIP1264 | trpC2 amyE::pΔIytlI′-lacZ cat | pDIA5607 → 168 |
| BSIP1519 | trpC2 ytlI::aphA3 amyE::pΔAytmI-lacZ cat | pDIA5599 → BSIP1214 |
| BSIP1521 | trpC2 ytlI::aphA3 amyE::pΔCytmI′-lacZ cat | pDIA5600 → BSIP1214 |
| BSIP1523 | trpC2 ytlI::aphA3 amyE::pΔDytmI′-lacZ cat | pDIA5601 → BSIP1214 |
| BSIP1589 | trpC2 amyE::pΔHytlI′-lacZ cat | pDIA5648 → 168 |
| BSIP1712 | trpC2 amyE::pΔBytmI′-lacZ cat | pDIA5689 → 168 |
| BSIP1722 | trpC2 ytlI::aphA3 amyE::pΔBytmI′-lacZ cat | BSIP1214 → BSIP1712 |
| BSIP1724 | trpC2 cysH::Tn10 amyE::pΔFytlI′-lacZ cat | BSIP1175 → BSIP1215 |
| BSIP1725 | trpC2 ΔcysJI::aphA3 amyE::pΔFytlI′-lacZ cat | BSIP1206 → BSIP1215 |
| BSIP1726 | trpC2 cysK::spc amyE::pΔFytlI′-lacZ cat | BSIP1304 → BSIP1215 |
| BSIP1731 | trpC2 amyE::pΔA(−73 A→G)ytmI′-lacZ cat | pDIA5699 → 168 |
| BSIP1733 | trpC2 amyE::pΔA(−65 T→G)ytmI′-lacZ cat | pDIA5701 → 168 |
| BSIP1735 | trpC2 amyE::pΔA(−61 G→T)ytmI′-lacZ cat | pDIA5703 → 168 |
| BSIP1737 | trpC2 amyE::pΔA(−49 T→G)ytmI′-lacZ cat | pDIA5705 → 168 |
| BSIP1738 | trpC2 amyE::pΔA(−45 T→G)ytmI′-lacZ cat | pDIA5706 → 168 |
| BSIP1746 | trpC2 amyE::pΔA(−50 T→G)ytmI′-lacZ cat | pDIA5710 → 168 |
| BSIP1754 | trpC2 amyE::plmo2352′-lacZ cat | pDIA5717 → 168 |
| BSIP1760 | trpC2 amyE::pΔA(−72 T→G)ytmI′-lacZ cat | pDIA5719 → 168 |
| BSIP1762 | trpC2 amyE::pΔEytmI′-lacZ cat | pDIA5721 → 168 |
| BSIP1764 | trpC2 ytlI::aphA3 amyE::pΔEytmI′-lacZ cat | BSIP1214 → BSIP1762 |
| BSIP1767 | trpC2 amyE::pΔA(−71 T→G)ytmI′-lacZ cat | pDIA5727 → 168 |
| BSIP1768 | trpC2 amyE::pΔA(−66 T→G)ytmI′-lacZ cat | pDIA5728 → 168 |
| BSIP1770 | trpC2 amyE::pΔA(−42 T→G)ytmI′-lacZ cat | pDIA5730 → 168 |
| BSIP1773 | trpC2 ytlI::aphA3 amyE::pΔA(−73 A→G)ytmI′-lacZ cat | BSIP1214 → BSIP1731 |
| BSIP1774 | trpC2 ytlI::aphA3 amyE::pΔA(−65 T→G)ytmI′-lacZ cat | BSIP1214 → BSIP1733 |
| BSIP1775 | trpC2 ytlI::aphA3 amyE::pΔA(−61 G→T)ytmI′-lacZ cat | BSIP1214 → BSIP1735 |
| BSIP1776 | trpC2 ytlI::aphA3 amyE::pΔA(−49 T→G)ytmI′-lacZ cat | BSIP1214 → BSIP1737 |
| BSIP1777 | trpC2 ytlI::aphA3 amyE::pΔA(−45 T→G)ytmI′-lacZ cat | BSIP1214 → BSIP1738 |
| BSIP1778 | trpC2 ytlI::aphA3 amyE::pΔA(−50 T→G)ytmI′-lacZ cat | BSIP1214 → BSIP1746 |
| BSIP1779 | trpC2 ytlI::aphA3 amyE::pΔA(−72 T→G)ytmI′-lacZ cat | BSIP1214 → BSIP1760 |
| BSIP1780 | trpC2 ytlI::aphA3 amyE::pΔA(−71 T→G)ytmI′-lacZ cat | BSIP1214 → BSIP1767 |
| BSIP1781 | trpC2 ytlI::aphA3 amyE::pΔA(−66 T→G)ytmI′-lacZ cat | BSIP1214 → BSIP1768 |
| BSIP1782 | trpC2 ytlI::aphA3 amyE::pΔA(−42 T→G)ytmI′-lacZ cat | BSIP1214 → BSIP1770 |
| BSIP1783 | trpC2 ytlI::aphA3 thrC::pxylA-ytlI spc amyE::pΔEytmI′-lacZ cat | pDIA5679 → BSIP1764 |
| BSIP1784 | trpC2 ytlI::aphA3 thrC::pxylA-lmo2352 spc amyE::pΔEytmI′-lacZ cat | pDIA5687 → BSIP1764 |
| BSIP1793 | trpC2 ΔyrzC amyE::pΔFytlI′-lacZ cat | Materials and methods |
| BSIP1797 | trpC2 ΔyrzC amyE::pΔAytmI-lacZ cat | Materials and methods |
| BSIP1798 | trpC2 ΔyrzC amyE::lacZ aphA3 | pAC7 → BSIP1793 |
| BSIP1801 | trpC2 ΔyrzC amyE::plmo2352′-lacZ cat | pDIA5717 → BSIP1798 |
| BSIP1811 | trpC2 ΔyrzC ytlI::aphA3 amyE::pΔAytmI′-lacZ cat | BSIP1214 → BSIP1797 |
Arrows indicate construction by transformation; cat is the pC194 chloramphenicol acetyl transferase gene, aphA3 is the Enterococcus faecalis kanamycin resistance gene, erm is an erythromycin resistance gene, and spc is a spectinomycin resistance gene.
The loss of amylase activity was detected as previously described (43). β-Galactosidase-specific activity was measured as described by Miller (27) with cell extracts obtained by lysozyme treatment. Protein concentrations were determined by the method of Bradford (4). One unit of β-galactosidase is defined as the amount of enzyme that produces 1 nmol min−1 of O-nitrophenol at 28°C. The mean values of at least three independent experiments are presented. Standard deviations are less than 20% of the mean.
DNA manipulations.
Plasmids from E. coli and chromosomal DNA from B. subtilis were prepared according to standard procedures. Restriction enzymes and phage T4 DNA ligase were used as specified by the manufacturers. DNA fragments used for cloning experiments were prepared by PCR using a High Fidelity PCR system (Roche). Amplified fragments were purified with a QIAquick PCR purification kit (QIAGEN).
Plasmid construction.
Plasmid pAC6 (43) was used to construct transcriptional fusions between a series of deletions of the ytmI promoter region and the promoterless lacZ gene. The regions pΔA(−75 to +47), pΔB(−71 to +47), pΔC(−58 to +47), pΔD(−42 to +47), and pΔE(−75 to +17) were amplified by PCR with the creation of 5′-EcoRI and 3′-BamHI sites (Fig. 1). PCR products were inserted into pAC6 to give plasmids pDIA5599 (pΔA), pDIA5689 (pΔB), pDIA5600 (pΔC), pDIA5601 (pΔD), and pDIA5721 (pΔE), respectively. The different fusions were integrated at the amyE locus of strains 168 or BSIP1214 (ytlI::aphA3) (Table 1).
FIG. 1.
The intergenic region between the ytlI gene and the ytmI operon. The transcription start points of the ytlI and the ytmI genes (+1) are indicated by vertical arrows. The −35 and −10 regions for each gene are in uppercase and boxed. The ribosome-binding sites (RBS) are boxed, while the translational start sites of ytlI and ytmI are indicated. These two genes are transcribed divergently. The molecular elements corresponding to the ytmI and ytlI promoter regions were located on the upper and lower strand, respectively. Deletion end points of the different fusions with the lacZ gene are indicated by broken arrows and numbered with respect to the transcriptional start sites of the ytlI and ytmI genes, respectively.
Transcriptional fusions between a series of 5′ and 3′ deletions of the ytlI promoter region and the promotorless lacZ gene were also constructed. The pΔF(−130 to +111), pΔG(−46 to +111), pΔH(−39 to +111), pΔI(−25 to +111), and pΔJ(−130 to −1) regions were amplified by PCR with the creation of EcoRI and BamHI sites (Fig. 1). The PCR products were inserted into pAC6 to give pDIA5575 (pΔF), pDIA5602 (pΔG), pDIA5648 (pΔH), pDIA5607 (pΔI), and pDIA5606 (pΔJ), respectively. The fusions were integrated at the amyE locus of strain 168 (Table 1).
A transcriptional lmo2352′-lacZ fusion was constructed as follows. An EcoRI-BamHI DNA fragment corresponding to the lmo2352 promoter region (nucleotides −266 to +47 relative to the translational start site) was generated by PCR using chromosomal DNA from Listeria monocytogenes strain EGD-e as template. This fragment was cloned into pAC6. The resulting plasmid pDIA5717 was integrated at the amyE locus of B. subtilis 168.
Plasmid pXT (31) was used to express the ytlI and lmo2352 genes under the control of the xylose-inducible promoter (pxylA). The complete coding sequences of ytlI and lmo2352 (nucleotides −45 to +953 or −64 to +958 relative to the translational start site, respectively) were amplified by PCR with the creation of EcoRI and BamHI sites. The amplified fragments were inserted into the BamHI and EcoRI sites of pXT, producing pDIA5632 (ytlI) and pDIA5687 (lmo2352). The ytlI and lmo2352 genes were then integrated by a double-crossover event at the thrC locus (Table 1).
Construction of the yrzC mutant and derivative strains.
To disrupt the yrzC gene, a two-step PCR procedure was used. DNA fragments corresponding to the 5′ end (nucleotides −1199 to +33 relative to the translational start site) or the 3′ end (nucleotides −32 to +1140 relative to the translational stop site) of the yrzC gene were first amplified by PCR with a 30-bp overlapping fragment at one end of each PCR. A second PCR using the yrzC upstream and downstream regions and the two external primers allowed amplification of a single PCR fragment containing an in-frame deletion of yrzC. A truncated YrzC protein containing the first 11 amino acids of YrzC, an insertion of 10 amino acids, and the last 10 amino acids of YrzC was obtained. The wild-type strain was then cotransformed with the ΔyrzC PCR fragment and chromosomal DNA from either the BSIP1215 (amyE::pΔFytlI′-lacZ cat) or the BSIP1256 (amyE::pΔAytmI′-lacZ cat) strain. Cmr transformants carrying these fusions at the amyE locus were obtained on SP plates containing 5 μg ml−1 chloramphenicol and X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside). Blue transformants resulting from the deletion of the yrzC gene were obtained, giving rise to strains BSIP1793 (ytlI) and BSIP1797 (ytmI), respectively (Table 1). The deletion of the central part of the yrzC gene was checked by PCR, while the absence of mutation in the adjacent genes (yrvN and yrvO) was verified by sequencing the corresponding DNA fragments.
The replacement of the pΔFytlI′-lacZ by a kanamycine resistance gene at the amyE locus was performed by transformation with linearized pAC7 (carrying the aphA3 gene between two amyE fragments), giving strain BSIP1798 (Table 1). A lmo2352-lacZ fusion was then introduced into this strain by transformation with linearized pDIA5717 and selection for chloramphenicol resistance.
Site-directed mutagenesis of the ytlI and the ytmI promoter regions.
Plasmids pDIA5575 [pΔF(−130 to +111)ytlI′-lacZ] or pDIA5599 [pΔA(−75 to +47)ytmI′-lacZ] were used to perform site-directed mutagenesis with a Quikchange site-directed mutagenesis Kit (Stratagene). Two synthetic oligonucleotides (35 to 40 bp) complementary to opposite strands and containing the mutation were used to amplify plasmid pDIA5575 or pDIA5599. The methylated parental DNA templates were digested by the DpnI endonuclease. DNA of pDIA5575 or pDIA5599 derivative plasmids containing the point mutations was extracted. The presence of the mutation was verified by sequencing the ytlI or ytmI promoter regions. The linearized plasmids were used to transform B. subtilis 168. The ytmI′-lacZ fusions containing point mutations were also introduced into the ytlI mutant, BSIP1214. The strains obtained were listed in Tables 1 and 3.
TABLE 3.
Effect of the sulfur source and of the YrzC regulator on the expression of different ytlI′-lacZ transcriptional fusionsa
| Strain | Relevant genotype | β-Galactosidase activity (nmol ONP min−1 mg of protein−1)
|
|
|---|---|---|---|
| Methionine | Sulfate | ||
| BSIP1215 | pΔF(−130 to +111)ytlI′-lacZ | 215 | 6 |
| BSIP1259 | pΔG(−46 to +111)ytlI′-lacZ | 43 | 4 |
| BSIP1590 | pΔH(−39 to +111)ytlI′-lacZ | 25 | 50 |
| BSIP1264 | pΔI(−25 to +111)ytlI′-lacZ | 2 | 3 |
| BSIP1261 | pΔJ(−130 to −1)ytlI′-lacZ | 165 | 4.5 |
| BSIP1728 | pΔF(−46 A→C)ytlI′-lacZ | 60 | 8 |
| BSIP1714 | pΔF(−45 A→C)ytlI′-lacZ | 78 | 71 |
| BSIP1720 | pΔF(−43 T→C)ytlI′-lacZ | 68 | 7 |
| BSIP1729 | pΔF(−42 C→A)ytlI′-lacZ | 150 | 67 |
| BSIP1721 | pΔF(−41 C→G)ytlI′-lacZ | 128 | 22 |
| BSIP1730 | pΔF(−40 T→A)ytlI′-lacZ | 52 | 3 |
| BSIP1715 | pΔF(−39 A→G)ytlI′-lacZ | 105 | 22 |
| BSIP1716 | pΔF(−38 T→A)ytlI′-lacZ | 260 | 215 |
| BSIP1215 | pΔF(−130 to +111)ytlI′-lacZ | 215 | 6 |
| BSIP1793 | pΔF(−130 to +111)ytlI′-lacZ ΔyrzC | 118 | 98 |
Cells were grown in minimal medium in the presence of sulfate or methionine at 1 mM final concentration. The β-galactosidase activities were obtained from cultures in mid-exponential growth phase. ONP, O-nitrophenol.
RNA isolation and analysis.
Total RNA was isolated from B. subtilis 168 grown in minimal medium containing 1 mM methionine. Exponentially growing cells (optical density at 600 nm, 0.8) were harvested. One gram of 0.1-mm-diameter glass beads (Sigma) was added. The cells were broken by shaking in a Fastprep apparatus (Bio101) two times for 30 s. The Trizol reagent (Gibco-BRL) was used to extract total RNA. For the primer extension experiments, the oligonucleotides IV184 (5′-TCGTATGAAACGTTTTGATTGAGCGTAACTCC-3′) (ytlI) and IV185 (5′-CTCCGATGCATCTTCAACAGTTGCCAGTCG-3′) (ytmI) were labeled with T4 polynucleotide kinase in the presence of [γ-32P]ATP and hybridized with 10 μg of RNA. The primers were extended by use of reverse transcriptase, and the products were analyzed as previously described (33). The same primer was used for the generation of a sequence ladder using the method of Sanger et al. (34).
Overproduction and purification of YtlI.
The ytlI gene (nucleotides (+1 to +923 relative to the translational start site) was amplified by PCR using primers introducing a 5′-NdeI and a 3′-XhoI site and was cloned in the pET20b+ vector (Novagen) digested by NdeI and XhoI. A translational fusion adding six carboxy-terminal histidine residues to YtlI was obtained. The plasmid, pDIA5621 (pET20b+ytlI), was transformed into the E. coli BL21(DE3) strain (Novagen), which contains pDIA17 (30) encoding the lacI repressor. The resulting strain was grown at room temperature in Luria-Bertani medium to an optical density at 600 nm of 2.5. Isopropyl β-d-thiogalactoside (1 mM) was added to induce the expression of ytlI, and incubation was pursued for 3 h. Cells were centrifuged and resuspended in buffer I (50 mM sodium phosphate [pH 8], 300 mM NaCl, 20 mM imidazole). E. coli crude extracts were loaded on a Ni-nitrilotriacetic agarose column. The column was washed and the YtlI-His6 was eluted with an immidazole gradient (30 to 500 mM). The purification of YtlI-His6 was confirmed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (data not shown).
Gel mobility shift assays.
DNA fragments containing various ytmI promoter regions (ΔA, -75 to +47; ΔB, -71 to +47; and ΔA containing a point mutation, A-73G or T-49G) were amplified by PCR using pDIA5599, pDIA5689, pDIA5699, or pDIA5705, respectively, as template. In these plasmids, the ytmI promoter region was flanked by the cat gene in the 5′ end and by the lacZ gene in the 3′ end. PCR products were obtained using [γ-32P]ATP 5′ end-labeled primers located in these two flanking genes. Protein-DNA complexes were formed in 10-μl volumes, by incubating the 32P-labeled DNA fragments with different amounts of YtlI-His6 purified protein in binding buffer (25 mM phosphate buffer [pH 7], 2 mM MgSO4, 150 mM NaCl, 1 mM dithiothreitol, 0.1 mM EDTA, 10% glycerol) in the presence of 1 μg ml−1 of poly(dIdC). The DNA binding reaction was incubated at room temperature for 20 min. Samples were separated on a 6% polyacrylamide native gel in Tris-borate-EDTA buffer at 10 V cm−1 for 1 h. Dried gels were analyzed with a Storm Imager (Molecular Dynamics). Radioactive signal intensities were quantified with the PDQuest software (PDI; Bio-Rad).
RESULTS
Identification of the ytlI and ytmI promoters.
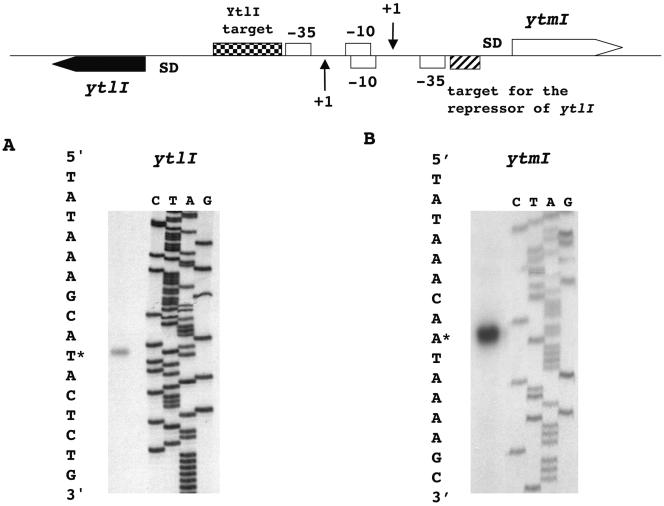
The DNA sequence of the ytlI-ytmI intergenic region is presented in Fig. 1. The translation initiation codons of ytlI and ytmI are a TTG and an ATG preceded by a ribosome-binding site, respectively. The 5′ ends of the ytlI and ytmI transcripts were identified by primer extension analysis using total RNA extracted from a wild-type strain grown in minimal medium in the presence of methionine (Fig. 2). Transcription was initiated at a single A located 57 bp upstream from the translational start point of ytmI. The deduced −35 (TAGTCA) and −10 (TATAGT) boxes of the promoter corresponded to the consensus sequence of σA-dependent promoters (Fig. 1). A T located 78 bp upstream from the TTG was the transcriptional start site of ytlI. The −35 (TTTACT) and −10 (TACTAT) regions of the promoter are quite similar to the consensus of σA-dependent promoters (Fig. 1).
FIG. 2.
Mapping of the transcription start site of the ytlI gene and of the ytmI operon by primer extension. Total RNA was extracted from B. subtilis strain 168 grown in minimal medium in the presence of 1 mM methionine as sole sulfur source. Primer extension experiments were performed using (A) oligonucleotide IV184, which hybridized with the ytlI gene, and (B) oligonucleotide IV185, which hybridized with the ytmI gene. For each experiment, the labeled oligonucleotide was loaded as a control. Sequencing reactions (lanes C, T, A, and G) were performed with the IV185 or IV184 oligonucleotides as primers and pDIA5599 (ytmI) or pDIA5575 (ytlI) as templates. Asterisks indicate the 5′ ends of mRNAs. SD, Shine-Dalgarno.
Characterization of a cis-acting target sequence upstream from the ytmI operon.
In order to identify DNA sequences necessary for the sulfur-dependent regulation of the ytmI operon, ytmI promoter regions containing 5′ deletions were fused to the lacZ gene. The fusion end points are indicated in Fig. 1. These fusions were introduced as a single copy at the amyE locus of the wild-type and ytlI::aphA3 strains. β-Galactosidase activity was measured in the different strains grown in minimal medium in the presence of methionine or sulfate as sole sulfur source (Table 2). The expression of the pΔA(−75 to +47)ytmI′-lacZ fusion was increased about 550-fold when methionine was used as the sulfur source instead of sulfate (Table 2, line 1). The level of expression of the pΔB(−71 to +47), pΔC(−58 to +47), and pΔD(−42 to +47)ytmI′-lacZ fusions in the presence of methionine was reduced compared to that of the pΔA(−75 to +47) fusion (Table 2, lines 1 to 4). The introduction of an ytlI gene disruption led to a decrease of expression of the pΔA, pΔB, and pΔCytmI′-lacZ fusions in the presence of methionine to the level observed in the pΔDytmI′-lacZ fusion (Table 2, lines 1 to 4). These results indicated that the DNA fragment located between nucleotides −75 and −71 was necessary to observe a high level of regulation of the ytmI operon by YtlI. Further deletions in the 5′ part of the ytmI promoter region led to a gradual decrease of the YtlI-dependent control in response to the sulfur source. It is worth noting that in an ytlI mutant, all these fusions conserved a low but significant repression by sulfate (Table 2, lines 1 to 4). This suggested the existence of a second regulation in response to sulfur availability independent of YtlI. To determine whether the region downstream from the ytmI promoter was important for this regulation, a pΔE(−75 to +17)ytmI′-lacZ fusion was constructed. The expression of this fusion strongly decreased during growth with sulfate compared with methionine. Moreover, the residual repression by sulfate observed with the pΔA(−75 to +47)ytmI-lacZ fusion in a ytlI background was totally abolished for the pΔE(−75 to +17) fusion (Table 2, lines 1 and 5). This suggested that a second minor site of sulfur-dependent regulation was present downstream from the ytmI promoter (Fig. 2).
TABLE 2.
Effect of the sulfur source and of the YtlI and YrzC regulators on the expression of different ytmI′-lacZ transcriptional fusionsa
| Relevant genotype | β-Galactosidase activity (nmol ONP min−1 mg of protein−1)
|
|||
|---|---|---|---|---|
|
ytlI+
|
ytlI::aphA3
|
|||
| Methionine | Sulfate | Methionine | Sulfate | |
| pΔA(−75 to +47)ytmI′-lacZ | 535 | 0.9 | 6.5 | 0.7 |
| pΔB(−71 to +47)ytmI′-lacZ | 40 | 0.8 | 6.5 | 1 |
| pΔC(−58 to +47)ytmI′-lacZ | 20 | 0.9 | 8 | 0.9 |
| pΔD(−42 to +47)ytmI′-lacZ | 4 | 0.6 | 4 | 0.6 |
| pΔE(−75 to +17)ytmI′-lacZ | 520 | 6.5 | 4.5 | 4.2 |
| pΔE(−75 to +17)ytmI′-lacZ pxylA ytlI | ND | ND | 510 | 530 |
| pΔE(−75 to +17)ytmI′-lacZ pxylA lmo2352 | ND | ND | 1,365 | 1,350 |
| pΔA(−75 to +47)ytmI′-lacZ | 535 | 0.9 | 6.5 | 0.7 |
| pΔA(−75 to +47)ytmI′-lacZ ΔyrzC | 790 | 730 | 6 | 6 |
Cells were grown in minimal medium in the presence of sulfate or methionine at 1 mM final concentration. The β-galactosidase activities were obtained from cultures in mid-exponential growth phase. The mean values of at least three independent experiments are presented. Standard deviations are less than 20% of the mean. For the strains containing the pxylA ytlI at the thrC locus, minimal medium contained fructose instead of glucose, threonine (50 mg liter−1), and xylose 0.2%. ONP, O-nitrophenol. ND, not determined.
Complementation of a ytlI mutant by the lmo2352 gene from L. monocytogenes.
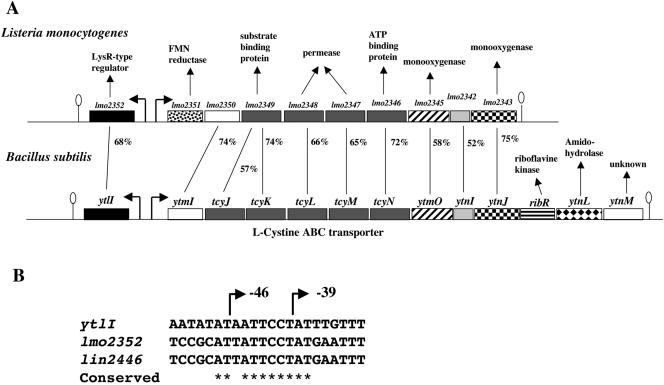
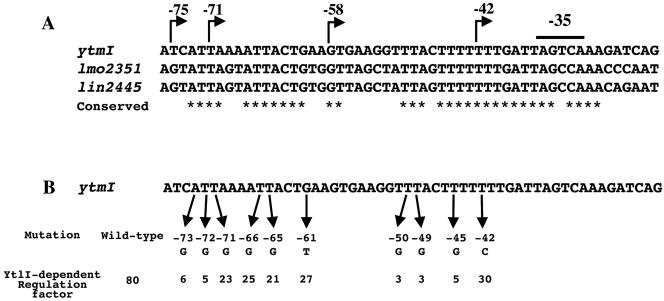
Browsing the genome sequences of A-T-rich Firmicutes spp., we observed that a YtlI-like regulator was present in L. monocytogenes (68% identity) and in Listeria innocua (67% identity) (http://genolist.pasteur.fr). Interestingly, a ytmI-like operon is present adjacent to the gene encoding the regulator in Listeria species (Fig. 3A and data not shown). The similarity between the polypeptides encoded by these operons ranges from 75% identity for YtnJ and Lmo2343 or Lin2437 to 52% identity for YtnI and Lmo2342 or Lin2436. The high level of identity between the YtlI, Lmo2352, and Lin2446 regulators suggested that the regulation was conserved, prompting us to test the ability of the lmo2352 gene to complement a ytlI::aphA3 mutant for the expression of a ytmI′-lacZ fusion. The lmo2352 gene was integrated in the thrC locus of strain BSIP1764 containing a pΔEytmI′-lacZ fusion and a ytlI gene disruption. In the resulting strain BSIP1784, the lmo2352 gene was expressed under the control of the xylA promoter. β-Galactosidase activity was tested after growth of strains BSIP1764 and BSIP1784 in minimal medium containing 1 mM threonine, 0.2% xylose, and methionine as sulfur source. The pΔEytmI′-lacZ fusion was poorly expressed in a ytlI mutant, while the introduction of the lmo2352 gene in trans restored the expression of this fusion (Table 2, lines 5 and 7). This result indicated that the Lmo2352 protein from L. monocytogenes was able to replace YtlI for the activation of transcription of the B. subtilis ytmI operon. A common target for these regulators was therefore searched in the promoter regions of the ytmI, lmo2351, and lin2445 operons. The comparison of the DNA sequence upstream of these operons revealed the presence of a highly conserved motif, ATTANNATTACTGN2GTNAN4TTANTTTTTTTGATTAG (Fig. 4A).
FIG. 3.
The ytlI-ytmI locus from B. subtilis and L. monocytogenes. A: Putative transcription start sites are indicated by broken arrows, and putative terminators are represented by loops. For each gene product of the ytmI operon and its equivalent in the L. monocytogenes lmo2351 operon a percentage of identity is indicated. The genes encoding the ABC transporters are represented by dark gray boxes, the LysR-type regulators by black boxes, and the monooxygenases by crosshatched boxes (ytmO-like) or checkered boxes (ytnJ-like). B: Alignment of the promoter regions of the ytlI, lmo2352, and lin2446 genes. Stars indicate conserved nucleotides in the three promoters. Deletion end points of the different ytlI′-lacZ fusions are indicated by broken arrows.
FIG. 4.
Site-directed mutagenesis in a conserved motif present upstream from the promoter regions of the ytmI, lmo2351, and lin2445 operons. A: Conserved nucleotides in the three promoter regions are indicated by stars. Deletion end points of the different ytmI′-lacZ fusions are indicated by broken arrows. B: Mutations in the cis-acting target of the ytmI promoter region are indicated. For each point mutation, the YtlI-dependent regulation factor was obtained by calculating the ratio between the β-galactosidase activity of the fusion in a wild-type strain and in a ytlI::aphA3 mutant after growth with methionine as sulfur source.
Site-directed mutagenesis of a conserved motif present in the promoter regions of ytmI, lmo2351, and lin2445.
This motif, that covered the DNA fragment located between nucleotides −75 and −71 previously identified as crucial for YtlI-dependent regulation, was likely to be involved in the binding of this activator to the ytmI promoter region. To confirm this hypothesis, point mutations were introduced in the conserved sequence using site-directed mutagenesis (see Materials and Methods). The pΔAytmI′-lacZ fusions containing various mutations were inserted at the amyE locus of a wild-type strain or a ytlI mutant. The level of β-galactosidase activity in the corresponding strains was determined after growth with methionine or sulfate. The introduction of point mutations did not significantly modify the level of expression of the fusions in the presence of sulfate with β-galactosidase activities ranging from 0.5 to 1 U (mg of protein)−1. In contrast, the expression of the modified pΔAytmI′-lacZ fusions with methionine was decreased compared to that of the wild-type fusion in a ytlI+ strain (data not shown). This was expected for modifications in the binding site of an activator. The contribution of the different nucleotides in the YtlI-dependent regulation of ytmI expression was then estimated. To this purpose, we calculated the ratio of the β-galactosidase activity measured in a wild-type strain and in a ytlI mutant after growth with methionine (Fig. 4B). Mutations in the ytmI promoter region at position −73 (A→G), −72 (T→G), −50 (T→G), −49 (T→G), or −45 (T→G) had a drastic effect on the YtlI-dependent activation with a weak residual control of ytmI expression by YtlI compared to the wild-type promoter. An intermediate level of regulation by YtlI was observed with modifications at position −71 (T→G), −66 (T→G), −65 (T→G), −61 (G→T), or −42 (T→C).
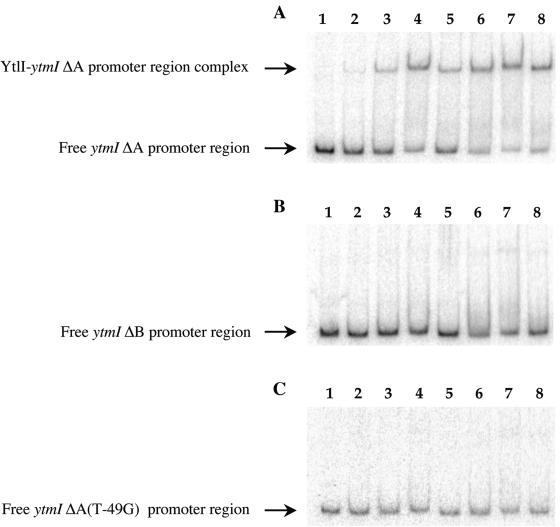
Binding of the YtlI regulator to the ytmI promoter region.
To determine whether YtlI controlled ytmI by direct binding to the promoter region of this operon, the YtlI protein fused with six histidyl residues was overproduced in E. coli and purified. The YtlI-His6 protein was then used in gel shift DNA binding assays. A radiolabeled DNA fragment containing the wild-type ΔA ytmI promoter region (positions −75 to +47) was incubated with increasing amounts of YtlI. The YtlI regulator bound to this DNA fragment, forming a single complex (Fig. 5A). Gel mobility shift assays were also performed to determine more precisely the importance of some nucleotides for this binding. To this purpose, a fragment including the ΔB(−71 to +47) promoter region and two ΔA(−75 to +47) regions carrying a point mutation (A-73→G or T-49→G) were used. The binding of YtlI to the ΔB promoter region or to the ΔA(T-49→G) DNA sequence was almost undetectable (Fig. 5B and C). In gel shift experiments, the YtlI binding was more than 70-fold lower for these two promoter regions than for the wild-type ΔA DNA fragment. Furthermore, YtlI binding to the ΔA(A-73→G) DNA fragment was decreased approximatively 15 times compared to the wild-type ΔA promoter region (data not shown). This indicated that YtlI specifically interacted with the DNA region located upstream from the ytmI promoter. The deletion of the DNA fragment located between positions −75 and −71 or the modification of T in position −49 virtually abolished YtlI binding, while the mutation of A in position −73 strongly reduced it. These results substantiated and extended the data obtained in vivo using lacZ fusions (Table 2, Fig. 4B).
FIG. 5.
Binding of the YtlI regulator to a ytmI promoter region fragment in a mobility shift assay. Gel mobility shift experiments were performed by incubating YtlI with 5′-radiolabeled DNA fragments containing different ytmI promoter regions: ΔA(−75 to +47) (A), ΔB(−71 to +47) (B), or ΔA(T-49→G) (C) DNA fragment. Lane 1, free probe; lanes 2 to 8, increasing amounts of YtlI-His6 (12.5, 25, 50, 100, 200, 400, and 800 ng, respectively).
YtlI-dependent regulation in response to sulfur availability.
As previously shown, the expression of the ytlI gene was higher after growth with methionine than with sulfate (1). To determine whether the activity of YtlI as transcriptional activator was also directly modulated in response to sulfur availability, the sulfur-regulated ytlI promoter was replaced by the xylA promoter. The expression of a pΔE(−75 to +17)ytmI′-lacZ fusion was then tested in a wild-type strain (BSIP1762) and in a ytlI mutant with or without the pxylA-ytlI gene at the thrC locus (BSIP1783 and BSIP1764, respectively). The strains were grown in minimal medium containing 1 mM threonine and 0.2% xylose in the presence of methionine or sulfate. While a ytlI gene disruption abolished the transcriptional activation of the pΔEytmI′-lacZ fusion, the insertion of pxylA-ytlI in the ytlI mutant restored the expression of this fusion after growth in the presence of methionine (Table 2, lines 1 and 6). However, the ytmI expression, which was repressed with sulfate in BSIP1762 (pΔEytmI-lacZ), was constitutive in strain BSIP1783 (pΔEytmI′-lacZ ytlI::aphA3 thrC::pxylA-ytlI). The introduction of pxylA-lmo2352 instead of pxylA-ytlI into BSIP1764 also led to constitutive ytmI expression (Table 2, line 7). In the conditions tested, the transcriptional activation of ytmI by YtlI or Lmo2352 in B. subtilis did not involve modulation of their activity due to the presence of methionine or sulfate.
Identification of a sequence necessary for sulfur-dependent regulation of the ytlI gene.
The ytlI expression was high in the presence of methionine and reduced with sulfate (1). In a ytlI mutant, the sulfur-dependent regulation of ytlI was conserved, indicating a YtlI-independent mechanism of control of ytlI expression (data not shown). To identify the DNA region necessary for the sulfur-mediated control, ytlI promoter regions containing various 5′ or 3′ deletions were fused to the lacZ gene (Fig. 1). These fusions were introduced as a single copy at the amyE locus of B. subtilis 168. β-Galactosidase activity was measured in the different strains grown in minimal medium with methionine or sulfate as sole sulfur source. The pΔI(−25 to +111) fusion gave very low β-galactosidase activity in the presence of either methionine or sulfate (Table 3, line 4). Deletion of the mapped promoter abolished ytlI transcription, confirming the existence of a single promoter. The pΔF(−130 to +111), pΔJ(−130 to −1), and pΔG(−46 to +111)ytlI′-lacZ fusions were 10- to 35-fold more expressed with methionine than with sulfate (Table 3, lines 1 to 2 and 5). In contrast, the pΔH(−39 to +111)ytlI′-lacZ fusion was constitutively expressed (Table 3, line 3). These results strongly suggested that the DNA region located between positions −46 and −39 upstream from the transcriptional start site was necessary to observe a repression by sulfate of ytlI transcription.
To confirm the role of the DNA sequence located upstream from the −35 box in the regulation of the ytlI gene, point mutations were introduced in this region using site-directed mutagenesis. The pΔFytlI′-lacZ fusions containing various mutations were inserted at the amyE locus of the wild-type strain. The level of β-galactosidase activity in the corresponding mutants was determined after growth with methionine or sulfate (Table 3, lines 6 to 13). The modification of the A nucleotide at position −45 or the T at position −38 in the ytlI promoter region led to constitutive expression of the corresponding fusion, while a pΔFytlI′-lacZ fusion containing a replacement of a C by an A in position −42 was still twofold repressed in the presence of sulfate. For the mutations C-41→G and A-39→G, the expression of the ytlI′-lacZ fusion increased in the presence of sulfate and decreased in the presence of methionine alone compared to the wild-type fusion. The level of expression with sulfate for fusions containing modifications at position −46 and −43 was similar to the wild-type strain, while the level of expression with methionine decreased. These results confirmed that the region located between positions -45 and −38 was important for the regulation of expression of the ytlI gene in response to sulfur availability. However, the intermediate level of expression observed with several mutations in the presence of methionine suggested that the sulfur-dependent regulation of the ytlI gene was probably complex.
YrzC is a repressor controlling the ytlI expression.
Solovieva et al. (42) have recently characterized mutations leading to an increase of transcription of the ribR gene, which belongs to the ytmI operon and encodes a riboflavine kinase (41). Mutations are located either in the ribosome-binding site or in the coding sequence (substitution of a Ser for an Asn at position 58) of the yrzC gene (42). YrzC acts as a negative regulator of the ytmI operon. To analyze the role of YtlI and YrzC in the control of ytmI expression, we have obtained a strain BSIP1797 containing an in-frame deletion of the yrzC gene and a pΔAytmI′-lacZ fusion by double transformation of the wild-type B. subtilis strain (see Materials and Methods). Strain BSIP1811 (ΔyrzC ytlI::aphA3 pΔAytmI′-lacZ) was then constructed by transforming strain BSIP1797 with the chromosomal DNA of BSIP1214 (ytlI::aphA3) (Table 1). The β-galactosidase activity was assayed after growth of strains BSIP1256, BSIP1797, and BSIP1811 in minimal medium with methionine or sulfate as sole sulfur source (Table 2, lines 8 to 9). The introduction of a deletion in the yrzC gene led to constitutive expression of the pΔAytmI′-lacZ fusion. This result was in agreement with the up-expression of the ytmI operon observed in a ΔyrzC mutant by Solovieva et al. (42). In a ΔyrzC ytlI::aphA3 double mutant, the level of expression of the pΔAytmI′-lacZ fusion was strongly reduced compared to that observed in a ΔyrzC mutant (Table 2, line 9). An epistatic effect of the ytlI mutation over the yrzC deletion was observed. This strongly suggested that the role of YrzC on ytmI expression was indirect and mediated by YtlI. In addition, the expression of a pΔAytmI′-lacZ fusion was no more repressed in the presence of sulfate in a ΔyrzC ytlI::aphA3 double mutant while a residual repression was observed in a ytlI mutant (Table 2, lines 8 to 9). The YrzC regulator could therefore be involved in ytmI regulation via the second minor site of sulfur-dependent regulation proposed before.
The possible involvement of YrzC in the control of ytlI expression was also explored. To this purpose, a strain containing both a yrzC deletion and a pΔFytlI′-lacZ fusion was constructed (see Materials and Methods). The expression of this fusion in a yrzC+ and in a ΔyrzC background was measured after growth of the corresponding strains in the presence of methionine or sulfate (Table 3, lines 14 to 15). The pΔFytlI′-lacZ fusion, which was repressed by sulfate in a wild-type strain, was constitutively expressed in a ΔyrzC mutant. This indicated that YrzC participated in the repression of ytlI expression.
The expression of lmo2352 is regulated by YrzC in B. subtilis.
The activity of Lmo2352 as transcriptional activator does not seem to be modulated in response to sulfur availability in B. subtilis. This asked the question whether the synthesis of Lmo2352 was increased in the presence of methionine as observed for YtlI. A lmo2352′-lacZ fusion was then constructed and inserted at the amyE locus of B. subtilis strain 168. β-Galactosidase activity was 85 U (mg of protein)−1 after growth with methionine and 4 U (mg of protein)−1 after growth with sulfate. In B. subtilis, the transcription of lmo2352 was subject to a sulfur-dependent regulation. Since YrzC acts as a negative regulator of ytlI transcription, we tested the effect of a yrzC deletion on the expression of a lmo2352′-lacZ fusion. β-Galactosidase activity was 98 and 128 U (mg of protein)−1 after growth with methionine or sulfate, respectively. A YrzC gene disruption also led to constitutive expression of lmo2352.
Expression of the ytlI gene in cysteine biosynthesis mutants.
In order to get more insight into the nature of the effector(s) involved in the regulation of the ytlI gene, the expression of the pΔFytlI′-lacZ fusion was tested in mutants deficient in cysteine biosynthesis. The pΔFytlI′-lacZ fusion was introduced at the amyE locus of a cysH::Tn10 mutant or a ΔcysJI::aphA3 mutant lacking APS-PAPS reductase and sulfite reductase, respectively (3, 14, 25). The expression of the pΔFytlI′-lacZ fusion was then tested after growth of the corresponding strains in the presence of 1 mM methionine, 1 mM methionine plus 1 mM sulfate, or 0.5 mM cysteine (Table 4). For strain BSIP1215, β-galactosidase activity was high after growth with methionine and reduced after growth with methionine plus sulfate or cysteine (Table 4, line 1). In a cysH or a ΔcysJI background, the expression of this fusion was similar in the presence of methionine or methionine plus sulfate. However, a repression was still observed in the presence of cysteine (Table 4, lines 2 and 3). This suggested that sulfite or one of the intermediates in the sulfate reduction pathway from sulfate to sulfite did not play a major role in the regulation of the ytlI gene. The pΔFytlI′-lacZ fusion was also introduced at the amyE locus of strain BSIP1304, in which the cysK gene, encoding OAS thiol-lyase is inactivated (2, 45). In the resulting strain, β-galactosidase activity was high in the presence of methionine, methionine plus sulfate, and cysteine (Table 4 line 4). In a ΔcysK mutant, neither sulfate nor cysteine repressed the expression of ytlI. The lack of OAS thiol-tyase probably led to higher OAS and sulfide concentrations and to a lower level of cysteine into the cell. We therefore tested the possible involvement of OAS in the regulation of the ytlI gene. The strain BSIP1215 containing the pΔFytlI′-lacZ transcriptional fusion was grown in minimal medium in the presence of 0.5 mM cysteine until an optical density of 0.6 was reached. The culture was then separated into two samples: one containing 1 mM OAS, the second without OAS. β-Galactosidase was measured 1 h after the addition of OAS. The presence of OAS in the growth medium resulted in an increase in pΔFytlI′-lacZ expression (86 U [mg of protein]−1) compared to the level observed without OAS (24 U [mg of protein]−1).
TABLE 4.
Expression of a pΔF ytlI′-lacZ fusion in cysteine biosynthesis mutantsa
| Relevant genotype | β-Galactosidase activity (nmol ONP min−1 mg−1)
|
||
|---|---|---|---|
| Methionine | Methionine and sulfate | Cysteine | |
| pΔF(−130 to +111)ytlI′-lacZ | 215 | 4 | 4 |
| pΔF(−130 to +111)ytlI′-lacZ cysH::Tn10 | 225 | 205 | 20 |
| pΔF(−130 to +111)ytlI′-lacZ ΔcysJI::aphA3 | 229 | 205 | 7 |
| pΔF(−130 to +111)ytlI′-lacZ cysK::spc | 200 | 205 | 170 |
Cells were grown in minimal medium in the presence of 1 mM methionine, 1 mM methionine and 1 mM sulfate, or 0.5 mM cysteine. The β-galactosidase activities were obtained from cultures in mid-exponential growth. The mean values of at least three independent experiments are presented. Standard deviations are less than 20% of the mean. ONP, O-nitrophenol.
DISCUSSION
Several LysR-type regulators are involved in the control of sulfur metabolism in both gram-negative and gram-positive bacteria (9, 14, 16-18, 40, 49, 50). In this work, we characterized in more detail the role of the LysR-type regulator, YtlI, the transcriptional activator of the B. subtilis complex operon ytmI (5, 7, 41). The ytlI gene and the ytmI operon are transcribed divergently. The ytlI and ytmI promoters overlapped with the −10 boxes located at the same position (Fig. 2), an organization found for several LysR-like regulators (35). A cis-acting sequence important for the sulfur-dependent regulation of ytmI expression was identified using both deletion and site-directed mutagenesis. As observed for other members of this family of regulators (35), two regions seemed to be critical for the transcriptional control by YtlI: the AT at position −73/−72 and a second region around −50. The YtlI protein was able to bind to the ytmI promoter region (−75 to +47) in gel shift experiments (Fig. 5). In contrast, the affinity of YtlI for the promoter region with the −75 to −71 DNA sequence deleted or modified in positions −49 or −73 strongly decreased. In silico analysis suggested that the YtlI-binding site was not found upstream from other B. subtilis sulfur-regulated genes in agreement with the absence of a ytlI-dependent control for most of these genes (1, 7) (S. Auger, unpublished results). This indicated that YtlI was probably not a global regulator of sulfate assimilation, cysteine biosynthesis, and cystine transport in B. subtilis, in contrast to the CysB regulator from E. coli. In addition, neither the key residues important for CysB activity nor the CysB-binding sites are conserved for YtlI (18, 22). This activator shares only weak similarities (less than 30%) with the other LysR-type regulators involved in the control of sulfur metabolism including CysB but also CmbR and MtaR from Lactococcus lactis and group B streptococci (9, 11, 40). This indicates the existence of differences in sulfur-dependent regulation in these microorganisms.
The expression of the ytlI gene itself was regulated in response to sulfur availability independently of YtlI, indicating that another regulator was involved. A cis-acting target important for the sulfur-dependent regulation of ytlI expression was identified just upstream from the −35 box of this gene (Fig. 3B and Table 3). The deletion or some point modifications of this sequence led to constitutive expression of ytlI, suggesting that a repressor was probably required in this control. The YrzC regulator, which was recently identified as a negative regulator of the ytmI operon (42), was a good candidate to play this role. In this work, we showed that an in-frame deletion of yrzC led to a high level of expression of a ytmI-lacZ or a ytlI-lacZ fusion after growth with either sulfate or methionine. In addition, the presence of the YtlI activator was necessary to observe constitutive expression of ytmI expression in a yrzC mutant (Table 2, line 9). Our genetic analyses strongly suggested that the YrzC repressor mediated sulfur-dependent regulation of this operon by controlling the synthesis of the YtlI activator. The direct or indirect effect of YrzC on ytlI transcription remains to be determined.
A large operon encoding an l-cystine ABC transporter (5), two putative monooxygenases, a putative acetyltransferase (YtmI), and a protein sharing weak similarities with glutaredoxine (YtnI) is present in B. subtilis and in Listeria spp. The high level of conservation between the ytmI, lmo2351, and lin2445 operons suggested a common function, the uptake of l-cystine and related products by the ABC transporter and the degradation of one or several sulfur compounds (5, 39). B. subtilis and Listeria spp. are usually present in soils or in contact with plants (32). These conserved operons could be involved in the assimilation of sulfur compounds present in a common biotope. The lmo2352 and lin2446 genes, adjacent to the lmo2351 and lin2445 operons, encode regulators highly similar to the YtlI activator. Several data strongly suggested a conserved cascade of regulation for the ytmI-type operons in B. subtilis and Listeria: (i) the Lmo2352 regulator was able to replace the YtlI protein to activate ytmI transcription; (ii) a conserved motif was found upstream from the −35 box of the ytmI, lmo2351, and lin2445 operons (Fig. 4A) in the region important for the YtlI-dependent regulation of ytmI; (iii) a sulfur-dependent regulation of the lmo2352 gene was observed in B. subtilis; (iv) YrzC was a negative regulator of lmo2352 expression in this bacterium. In addition, YrzC shares 69% identity with Lmo1515 and Lin1550, which probably play the same role as this repressor in Listeria. A conserved sequence, AT(A/T)ATTCCTAT, was found in the promoter regions of ytlI, lmo2352, and lin2446 (Fig. 3B). The YrzC repressor or another regulator could bind to this motif.
The uptake of l-cystine by TcyJKLMN in B. subtilis and the assimilation of taurine and sulfonate in E. coli are controlled by a cascade of regulation. In the latter, the CysB activator controls the expression of a second regulatory gene cbl by binding to its promoter region (16). In addition, both CysB and Cbl directly bind to the ssu and tau promoter regions to modulate their expression in response to cysteine and sulfate availability (6, 47, 48). In the same way, the organization of the ytmI promoter region is complex, with the existence of two cis-acting targets: the YtlI-binding site present upstream from the −35 box and a second minor site located downstream from the promoter, which could correspond to a direct or indirect target for YrzC (Fig. 2). A major difference was observed for the YtlI-dependent regulation compared to Cbl- or CysB-dependent regulation. Like most of the LysR-type regulators, an effector (APS or N-acetylserine) modulates the transcriptional activation by Cbl or CysB (6, 22). In contrast, our results suggested that the repression by sulfate controlled only the synthesis of YtlI and not its activity. Similarly, the nitrogen assimilation control regulator, NAC, of Klebsiella aerogenes activates the trancription of the hut operon or represses gdh expression independent of the nitrogen supply (36). The expression of the nac gene itself is under the control of the NTR system involving transcriptional activation of the nac gene by the σ54-dependent regulator NtrC (8).
In addition to the regulation of ytlI transcription by the YrzC repressor, we also unraveled some features of the control of ytlI expression in response to sulfur availability. As observed for the cysH and ssu operons involved in sulfate or sulfonate assimilation (23, 44), the addition of OAS to the culture medium led to a partial derepression of ytlI transcription. In addition, constitutive expression of this gene was obtained in a cysK mutant (inactivated for OAS thiol-lyase), which could accumulate OAS. These results suggested a role for OAS in the signaling pathway controlling ytlI, ssu, and cysH expression. Mansilla et al. (23) have proposed that the cysH operon is regulated by the binding of an unknown repressor, CysR, to a sequence located upstream of the −35 box. Despite the absence of a common motif in the cysH and ytlI promoter regions, the possibility that YrzC participated in cysH regulation remains to be seen.
OAS is synthesized by the serine transacetylase, the cysE gene product. In B. subtilis, cysE expression is regulated by transcription antitermination at a cysteine-specific T-box (10). The level of OAS is therefore correlated to the level of uncharged cysteinyl-tRNA, which signals the cysteine status in the cell. YrzC shares 31% identity with IscR, the repressor of the iscRSUA operon of E. coli involved in Fe-S cluster biogenesis (37). [2Fe-2S]-IscR is the active form of the repressor. Interestingly, the [2Fe-2S] cluster may be coordinated by three cysteine residues of IscR (37), which are absent in YrzC. The binding of a [2Fe-2S] cluster to YrzC seems therefore unlikely while its interaction with an effector like OAS could be involved.
Acknowledgments
We are grateful to M. Mazet and M. Mercadié for their help during this work and to E. Rocha, P. Glaser, D. Rodionov, and M. Gelfand for helpful discussions. We thank S. Even and G. Rapoport for the critical reading of the manuscript.
This research was supported by grants from the “Ministère de l'Education Nationale de la Recherche et de la Technologie,” the “Centre National de la Recherche Scientifique” (URA 2171), the “Institut Pasteur,” the “Université Paris 7,” the “Fondation pour la recherche médicale,” and the European Biotech Program (contract QLG2 CT9901455).
REFERENCES
- 1.Auger, S., A. Danchin, and I. Martin-Verstraete. 2002. Global expression profile of Bacillus subtilis grown in the presence of sulfate or methionine. J. Bacteriol. 184:5179-5186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Auger, S., M. P. Gomez, A. Danchin, and I. Martin-Verstraete. 2005. The PatB protein of Bacillus subtilis is a C-S-lyase. Biochimie 87:231-238. [DOI] [PubMed] [Google Scholar]
- 3.Berndt, C., C. H. Lillig, M. Wollenberg, E. Bill, M. C. Mansilla, D. de Mendoza, A. Seidler, and J. D. Schwenn. 2004. Characterization and reconstitution of a 4Fe-4S adenylyl sulfate/phosphoadenylyl sulfate reductase from Bacillus subtilis. J. Biol. Chem. 279:7850-7855. [DOI] [PubMed] [Google Scholar]
- 4.Bradford, M. M. 1976. Rapid and sensitive method for quantitation of microgram quantities of protein utilizing principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 5.Burguiere, P., S. Auger, M. F. Hullo, A. Danchin, and I. Martin-Verstraete. 2004. Three different systems participate in l-cystine uptake in Bacillus subtilis. J. Bacteriol. 186:4875-4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bykowski, T., J. R. van der Ploeg, R. Iwanicka-Nowicka, and M. M. Hryniewicz. 2002. The switch from inorganic to organic sulphur assimilation in Escherichia coli: adenosine 5′-phosphosulphate (APS) as a signalling molecule for sulphate excess. Mol. Microbiol. 43:1347-1358. [DOI] [PubMed] [Google Scholar]
- 7.Coppee, J. Y., S. Auger, E. Turlin, A. Sekowska, J. P. Le Caer, V. Labas, V. Vagner, A. Danchin, and I. Martin-Verstraete. 2001. Sulfur-limitation-regulated proteins in Bacillus subtilis: a two-dimensional gel electrophoresis study. Microbiology 147:1631-1640. [DOI] [PubMed] [Google Scholar]
- 8.Feng, J. L., T. J. Goss, R. A. Bender, and A. J. Ninfa. 1995. Activation of transcription initiation from the nac promoter of Klebsiella aerogenes. J. Bacteriol. 177:5523-5534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fernandez, M., M. Kleerebezem, O. P. Kuipers, R. J. Siezen, and R. van Kranenburg. 2002. Regulation of the metC-cysK operon, involved in sulfur metabolism in Lactococcus lactis. J. Bacteriol. 184:82-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gagnon, Y., R. Breton, H. Putzer, M. Pelchat, M. Grunberg-Manago, and J. Lapointe. 1994. Clustering and co-transcription of the Bacillus subtilis genes encoding the aminoacyl-tRNA synthetases specific for glutamate and for cysteine and the first enzyme for cysteine biosynthesis. J. Biol. Chem. 269:7473-7482. [PubMed] [Google Scholar]
- 11.Golic, N., M. Schliekelmann, M. Fernandez, M. Kleerebezem, and R. van Kranenburg. 2005. Molecular characterization of the CmbR activator-binding site in the metC-cysK promoter region in Lactococcus lactis. Microbiology 151:439-446. [DOI] [PubMed] [Google Scholar]
- 12.Greene, R. C. 1996. Biosynthesis of methionine, p. 542-560. In F. C. Neidhardt (ed.), Escherichia coli and Salmonella, cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 13.Grundy, F. J., and T. M. Henkin. 1998. The S box regulon: a new global transcription terminaison control system for methionine and cysteine biosynthesis genes in gram-positive bacteria. Mol. Microbiol. 30:737-749. [DOI] [PubMed] [Google Scholar]
- 14.Guillouard, I., S. Auger, M. F. Hullo, F. Chetouani, A. Danchin, and I. Martin-Verstraete. 2002. Identification of Bacillus subtilis CysL, a regulator of the cysJI operon, which encodes sulfite reductase. J. Bacteriol. 184:4681-4689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hryniewicz, M. M., and N. M. Kredich. 1994. Stoichiometry of binding of CysB to the cysJIH, cysK, and cysP promoter regions of Salmonella typhimurium. J. Bacteriol. 176:3673-3682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iwanicka-Nowicka, R., and M. M. Hryniewicz. 1995. A new gene, cbl, encoding a member of the LysR family of transcriptional regulators belongs to Escherichia coli cys regulon. Gene 166:11-17. [DOI] [PubMed] [Google Scholar]
- 17.Kahnert, A., P. Mirleau, R. Wait, and M. A. Kertesz. 2002. The LysR-type regulator SftR is involved in soil survival and sulphate ester metabolism in Pseudomonas putida. Environ. Microbiol. 4:225-237. [DOI] [PubMed] [Google Scholar]
- 18.Kredich, N. M. 1996. Biosynthesis of cysteine, p. 514-527. In F. C. Neidhardt (ed.), Escherichia coli and Salmonella, cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 19.Kredich, N. M. 1992. The molecular basis for positive regulation of cys promoters in Salmonella typhimurium and Escherichia coli. Mol. Microbiol. 6:2747-2753. [DOI] [PubMed] [Google Scholar]
- 20.Kunst, F., and G. Rapoport. 1995. Salt stress is an environmental signal affecting degradative enzyme synthesis in Bacillus subtilis. J. Bacteriol. 177:2403-2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leichert, L. I. O., C. Scharf, and M. Hecker. 2003. Global characterization of disulfide stress in Bacillus subtilis. J. Bacteriol. 185:1967-1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lochowska, A., R. Iwanicka-Nowicka, D. Plochocka, and M. M. Hryniewicz. 2001. Functional dissection of the LysR-type CysB transcriptional regulator: regions important for DNA binding, inducer response, oligomerization and positive control. J. Biol. Chem. 276:2098-2107. [DOI] [PubMed] [Google Scholar]
- 23.Mansilla, M. C., D. Albanesi, and D. de Mendoza. 2000. Transcriptional control of the sulfur-regulated cysH operon, containing genes involved in l-cysteine biosynthesis in Bacillus subtilis. J. Bacteriol. 182:5885-5892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mansilla, M. C., and D. de Mendoza. 2000. The Bacillus subtilis cysP gene encodes a novel sulphate permease related to the inorganic phosphate transporter (Pit) family. Microbiology 146:815-821. [DOI] [PubMed] [Google Scholar]
- 25.Mansilla, M. C., and D. de Mendoza. 1997. l-Cysteine biosynthesis in Bacillus subtilis: identification, sequencing, and functional characterization of the gene coding for phosphoadenylylsulfate sulfotransferase. J. Bacteriol. 179:976-981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McDaniel, B. A. M., F. J. Grundy, I. Artsimovitch, and T. M. Henkin. 2003. Transcription termination control of the S box system: direct measurement of S-adenosylmethionine by the leader RNA. Proc. Natl. Acad. Sci. USA 100:3083-3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 28.Monroe, R. S., J. Ostrowski, M. M. Hryniewicz, and N. M. Kredich. 1990. In vitro interactions of CysB protein with the cysK and cysJIH promoter regions of Salmonella typhimurium. J. Bacteriol. 172:6919-6929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mostertz, J., C. Scharf, M. Hecker, and G. Homuth. 2004. Transcriptome and proteome analysis of Bacillus subtilis gene expression in response to superoxide and peroxide stress. Microbiology 150:497-512. [DOI] [PubMed] [Google Scholar]
- 30.Munier, H., A. M. Gilles, P. Glaser, E. Krin, A. Danchin, R. Sarfati, and O. Barzu. 1991. Isolation and characterization of catalytic and calmodulin-binding domains of Bordetella pertussis adenylate cyclase. Eur. J. Biochem. 196:469-474. [DOI] [PubMed] [Google Scholar]
- 31.Nair, S., I. Derre, T. Msadek, O. Gaillot, and P. Berche. 2000. CtsR controls class III heat shock gene expression in the human pathogen Listeria monocytogenes. Mol. Microbiol. 35:800-811. [DOI] [PubMed] [Google Scholar]
- 32.Ryser, E. T., and E. H. Marth. 1991. Occurrence and survival of Listeria monocytogenes in natural environments, p. 22-33. In E. T. Ryser (ed.), Listeria, listeriosis and food safety. Marcel Dekker, Inc., New York, N.Y.
- 33.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 34.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schell, M. A. 1993. Molecular biology of the LysR family of transcriptional regulators. Annu. Rev. Microbiol. 47:597-626. [DOI] [PubMed] [Google Scholar]
- 36.Schwacha, A., and R. A. Bender. 1993. The product of the Klebsiella aerogenes-nac (nitrogen assimilation control) gene is sufficient for activation of the hut operons and repression of the gdh operon. J. Bacteriol. 175:2116-2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schwartz, C. J., J. L. Giel, T. Patschkowski, C. Luther, F. J. Ruzicka, H. Beinert, and P. J. Kiley. 2001. IscR, an Fe-S cluster-containing transcription factor, represses expression of Escherichia coli genes encoding Fe-S cluster assembly proteins. Proc. Natl. Acad. Sci. USA 98:14895-14900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sekowska, A., and A. Danchin. 2002. The methionine salvage pathway in Bacillus subtilis. BMC Microbiol. 2:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sekowska, A., S. Robin, J. J. Daudin, A. Henaut, and A. Danchin. 2001. Extracting biological information from DNA arrays: an unexpected link between arginine and methionine metabolism in Bacillus subtilis. Genome Biol. 2:1-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shelver, D., L. Rajagopal, T. O. Harris, and C. E. Rubens. 2003. MtaR, a regulator of methionine transport, is critical for survival of group B streptococcus in vivo. J. Bacteriol. 185:6592-6599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Solovieva, I. M., R. A. Kreneva, D. J. Leak, and D. A. Perumov. 1999. The ribR gene encodes a monofunctional riboflavin kinase which is involved in regulation of the Bacillus subtilis riboflavin operon. Microbiology 145:67-73. [DOI] [PubMed] [Google Scholar]
- 42.Solovieva, I. M., R. A. Kreneva, L. E. Lopes, and D. A. Perumov. 2005. The riboflavin kinase encoding gene ribR of Bacillus subtilis is a part of a 10 kb operon, which is negatively regulated by the yrzC gene product. FEMS Microbiol. Lett. 243:51-58. [DOI] [PubMed] [Google Scholar]
- 43.Stülke, J., I. Martin-Verstraete, M. Zagorec, M. Rose, A. Klier, and G. Rapoport. 1997. Induction of the Bacillus subtilis ptsGHI operon by glucose is controlled by a novel antiterminator, GlcT. Mol. Microbiol. 25:65-78. [DOI] [PubMed] [Google Scholar]
- 44.van der Ploeg, J. R., M. Barone, and T. Leisinger. 2001. Expression of the Bacillus subtilis sulphonate-sulphur utilization genes is regulated at the levels of transcription initiation and termination. Mol. Microbiol. 39:1356-1365. [PubMed] [Google Scholar]
- 45.van der Ploeg, J. R., M. Barone, and T. Leisinger. 2001. Functional analysis of the Bacillus subtilis cysK and cysJI genes. FEMS Microbiol. Lett. 201:29-35. [DOI] [PubMed] [Google Scholar]
- 46.van der Ploeg, J. R., N. J. Cummings, T. Leisinger, and I. F. Connerton. 1998. Bacillus subtilis genes for the utilization of sulfur from aliphatic sulfonates. Microbiology 144:2555-2561. [DOI] [PubMed] [Google Scholar]
- 47.van der Ploeg, J. R., R. Iwanicka-Nowicka, T. Bykowski, M. M. Hryniewicz, and T. Leisinger. 1999. The Escherichia coli ssuEADCB gene cluster is required for the utilization of sulfur from aliphatic sulfonates and is regulated by the transcriptional activator Cbl. J. Biol. Chem. 274:29358-29365. [DOI] [PubMed] [Google Scholar]
- 48.van der Ploeg, J. R., R. Iwanicka-Nowicka, M. A. Kertesz, T. Leisinger, and M. M. Hryniewicz. 1997. Involvement of CysB and Cbl regulatory proteins in expression of the tauABCD operon and other sulfate starvation-inducible genes in Escherichia coli. J. Bacteriol. 179:7671-7678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vermeij, P., C. Wietek, A. Kahnert, T. Wuest, and M. A. Kertesz. 1999. Genetic organization of sulphur-controlled aryl desulphonation in Pseudomonas putida S-313. Mol. Microbiol. 32:913-926. [DOI] [PubMed] [Google Scholar]
- 50.Weissbach, H., and N. Brot. 1991. Regulation of methionine synthesis in Escherichia coli. Mol. Microbiol. 5:1593-1597. [DOI] [PubMed] [Google Scholar]