Abstract
The Staphylococcus aureus cidABC and lrgAB operons have been shown to regulate murein hydrolase activity and affect antibiotic tolerance. The cid operon enhances murein hydrolase activity and antibiotic sensitivity, whereas the lrg operon inhibits these processes. Based on these findings and the structural similarities of the cidA and lrgA gene products to the bacteriophage holin family of proteins, we have proposed that the cid and lrg operons encode holin- and antiholin-like proteins, respectively, that function to control the murein hydrolase activity produced by the bacteria. Analysis of cid operon transcription revealed the presence of two transcripts, one spanning all three cid genes and whose expression is induced by growth in the presence of acetic acid and the other spanning cidB and cidC only that is produced in a sigma B-dependent manner. The cidABC operon lies immediately downstream from the cidR gene, encoding a potential LysR-type transcriptional regulator. In this study, we demonstrate that cidR is involved in the regulation of cidABC expression. Northern blot analyses revealed that the cidR gene product positively regulates cidABC expression by increasing transcription in the presence of acetic acid produced as a result of the metabolism of glucose. As expected for an operon that encodes a positive effector of murein hydrolase activity, the upregulation of cidABC expression resulted in increased murein hydrolase activity produced by these cells. Furthermore, it was demonstrated that antibiotic tolerance and stationary-phase survival of S. aureus are affected by the cidR gene. Taken together, these results demonstrate that the cidR gene product functions as a transcriptional activator of cidABC transcription in response to acetic acid accumulation in the growth medium.
Antibiotic tolerance can be defined as the intrinsic ability of microorganisms to survive the killing effects of antimicrobial agents. The term was originally used to contrast it with antibiotic resistance mechanisms and to describe physiological alterations in bacteria that result in a change in their sensitivities to antibiotics from a bactericidal response to a bacteriostatic one (28). Although the changes that affect antibiotic tolerance were shown to involve the control of murein hydrolase activity, the molecular basis for these changes is uncertain. Indeed, the molecular control of murein hydrolase activity is still undefined and is the subject of considerable research. Moreover, the way in which the bacterial cell wall is assembled and maintained is an important problem in bacteriology that remains to be solved.
Recent studies in our laboratory have demonstrated that the Staphylococcus aureus cidABC and lrgAB operons encode a complex regulatory system that affects murein hydrolase activity and antibiotic tolerance (4, 7, 20-23). It has been proposed that the cid and lrg gene products function in a manner that resembles bacteriophage-encoded holins and antiholins, respectively, involved in the control of cell death and lysis during the lytic cycle of a bacteriophage infection (2, 20, 22). Mutagenesis studies have revealed that a cidA mutation causes decreased extracellular murein hydrolase activity and increased antibiotic tolerance, whereas a lrgAB mutation causes increased murein hydrolase activity and decreased tolerance (7, 21, 22). Although the precise mechanism by which the cid and lrg gene products affect these processes is unknown, it has been hypothesized that they not only function to control the murein hydrolase activity produced by the cell but constitute the molecular components of a cell death mechanism triggered in response to stress or other developmental cues (1, 20).
Expression of the lrgAB operon is positively regulated by a two-component regulatory system encoded by the lytSR operon located immediately upstream of lrgAB (4). By comparison, the cidABC operon lies downstream of an open reading frame (ORF), designated cidR (GenBank accession no. AY581892), whose gene product is homologous to the LysR-type transcriptional regulator (LTTR) family of proteins (26). A recent transcription analysis of the cid operon revealed the presence of two overlapping transcripts: one spanning all three cid genes and the other spanning cidB and cidC only (23). The 2.7-kb cidBC transcript was shown to be expressed maximally during exponential growth and in a sigma B (σB)-dependent manner (23). In contrast, the cidABC transcript was expressed at relatively low levels in the absence of excess glucose and could only be detected using reverse transcriptase PCR (RT-PCR) (23). More recent studies in our laboratory have revealed that cidABC expression is greatly increased by the accumulation of acetic acid in the culture supernatant of S. aureus cultures containing high levels of glucose (22). However, the precise molecular mechanism by which cidABC expression is induced in the presence of acetic acid was not defined.
In this study, the role of the cidR gene product in regulating expression of the cid operon was investigated in detail by creating an isogenic cidR mutant of the previously characterized S. aureus clinical isolate UAMS-1. Northern blot analyses of the cidR mutant indicated that CidR enhances cidABC expression in the presence of acetic acid generated by the metabolism of excess glucose. These data also demonstrate that the cidR gene product affects the control of murein hydrolase activity by enhancing cidABC expression in the presence of acetic acid. Finally, the cidR mutation was also shown to affect the survival of S. aureus in stationary phase. These results demonstrate that the cidR gene product is a major component of this complex regulatory system.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains used in this study are listed in Table 1. All S. aureus strains were grown in either tryptic soy broth (TSB; Difco Laboratories, Detroit, MI) or filter-sterilized NZY broth (3% [wt · vol−1] N-Z Amine A [Sigma Chemical Co., St. Louis, MO], 1% [wt · vol−1] yeast extract [Fisher Scientific, Fair Lawn, NJ], pH 7.5) supplemented as necessary with 1.5% (wt · vol−1) granulated agar (Difco). Escherichia coli DH5α was grown in Luria-Bertani medium (Fisher Scientific). Liquid cultures were grown in Erlenmeyer flasks at 37°C with shaking (250 rpm) in a volume that was no greater than 10% of the flask volume. All antibiotics were purchased from either Sigma Chemical Co. or Fisher Scientific and were used at the following concentrations: ampicillin, 100 μg · ml−1; erythromycin, 3 μg · ml−1; chloramphenicol, 5 μg · ml−1; tetracycline, 5 μg · ml−1.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Descriptiona | Reference or source |
|---|---|---|
| S. aureus | ||
| UAMS-1 | Clinical osteomyelitis isolate; rsbU+ | 6 |
| KB1090 | UAMS-1 cidR::Tc; Tcr | This study |
| KB1050 | UAMS-1 cidA::Em; Emr | 22 |
| E. coli DH5α | Host strain for construction of recombinant plasmids | 9 |
| Plasmids | ||
| pRB474 | Shuttle vector carrying B. subtilis vegII promoter; Cmr | 3 |
| pDG1515 | Source of Tcr cassette; Tcr Ampr | 8 |
| pCR2.1 | E. coli plasmid; Ampr | Invitrogen |
| pSJ10 | pCR2.1 containing 1,465-bp cidR promoter region | This study |
| pSJ11 | cidR ORF cloned into BamHI and EcoRI sites of pRB474 | This study |
Abbreviations: Tcr, tetracycline resistance; Emr, erythromycin resistance; Cmr, chloramphenicol resistance; Ampr, ampicillin resistance.
DNA manipulations.
Genomic DNA was isolated from S. aureus using the method of Dyer and Iandolo (5). Plasmid DNA purification was performed using Wizard Plus kits from Promega, Inc. (Madison, Wis.). The restriction enzymes and T4 DNA ligase used in this study were purchased from either New England Biolabs (Beverly, Mass.) or Invitrogen Life Technologies (Carlsbad, Calif.). Preparation and transformation of E. coli DH5α were accomplished using the method described by Inoue et al. (12). Electroporation of DNA into S. aureus was carried out using the procedures of Schenk and Laddaga (27).
Allele replacement of the cidR gene in UAMS-1.
A cidR mutation was generated in S. aureus UAMS-1 by allele replacement as follows. A 573-bp DNA fragment spanning a region 5′ to cidR (nucleotides [nt] 2626938 to 2627504 of the S. aureus 8325 genome; http://www.genome.ou.edu/staph.html) was PCR amplified using primers EmhR1 (5′-GGCCGGATCCTCACTTCTCTAGGGAAATTGC-3′) and EmhR2 (5′-GCGCCTGCAGACATGCCCATGTTTATATGTCC-3′) and ligated into the BamHI and PstI sites of plasmid pDG1515 (8) upstream of a Tcr cassette. This plasmid was designated pRB1. Next, a 458-bp DNA fragment spanning a region 3′ of cidR (nt 2626145 to 2626602 of the S. aureus 8325 genome) was PCR amplified using primers cidR-Kpn (5′-CCCGGTACCATCCCTTTCTCGAGATGTCTAAATTG-3′) and cidA-Eco (5′-GGCTTTGTTCCGAATTCTGTAGCGCA-3′) and ligated into the KpnI and EcoRI sites of pRB1 downstream of the Tcr cassette. This plasmid, designated pRB2, was then digested with BamHI and KpnI to liberate a 3.17-kb fragment containing the Tcr cassette along with the flanking cidR sequences, which was subsequently ligated into the BamHI and KpnI sites of the temperature-sensitive shuttle vector pCL10 (25) to generate pRB3. This plasmid was then transformed into strain UAMS-1 by electroporation (27), spread onto tryptic soy agar (TSA) plates containing chloramphenicol, and incubated at 37°C overnight. This was followed by growth at the nonpermissive temperature (43°C) in the presence of tetracycline to select for cells in which the plasmid had integrated into the chromosome via homologous recombination. To promote a second recombination event, a single colony was inoculated into antibiotic-free TSB and grown at 30°C for 5 days with 1:1,000 dilutions into fresh TSB each day. After the fifth day, the culture was diluted and spread on TSA plates containing tetracycline to yield isolated colonies. The colonies were then screened for the Tcr and Cms phenotypes. Verification that the cidR gene had been deleted was carried out by PCR amplification and Southern blot analyses (data not shown). The confirmed mutant strain was designated KB1090 (Table 1).
Complementation of the cidR mutation in KB1090 was achieved by PCR amplifying the cidR ORF (nt 2626254 to 2627413 of the S. aureus 8325 genome) using primers cidR-F-BamHI (5′-CCCGGATCCGTAAAAGCTCAATACCTCACCTCG-3′) and cidR-R-EcoRI (5′-CCCGAATTCGGAAACGCTCTCTAAATTTCAC-3′). The resulting PCR product was ligated into the BamHI and EcoRI sites of the gram-positive expression vector pRB474 (3). This placed the expression of cidR under the control of the vegII promoter, a vegetative promoter from Bacillus subtilis (13). This recombinant plasmid was designated pSJ11.
Isolation of RNA.
For RNA isolation, fresh overnight cultures of S. aureus strains were used to inoculate NZY broth to an optical density at 600 nm (OD600) of 0.1. Cells were harvested during exponential growth (2 h), early stationary phase (6 h), and late stationary phase (12 h). Total RNA was isolated from the cell pellets by using the RNeasy kit (QIAGEN, Valencia, Calif.) and the FASTPREP FP120 instrument (Bio 101, Inc., Vista, Calif.) according to the manufacturer's recommended protocols.
Primer extension analysis.
The transcription start site of cidR was mapped by primer extension analysis as described by Sambrook et al. (24). Specifically, the reverse primer cidR-probe (5′-GCCTCCTTGCTTAACGACTTC-3′), complementary to the 5′ end of the cidR gene (nt 2627185 to 2627205 of the S. aureus 8325 genome), was end labeled with [γ-32P]ATP (6,000 Ci · mmol−1) and used in the primer extension reaction. One hundred micrograms of total bacterial RNA, isolated from an exponential growth phase S. aureus UAMS-1 culture, was used as the template in the primer extension reaction. A DNA sequencing ladder of the cidR promoter region was obtained using the cidR-probe primer and a Sequenase Kit (United States Biochemical Corporation, Cleveland, OH) according to the manufacturer's recommendations for using an end-labeled primer in the sequencing reaction. Plasmid pSJ10, containing 1,465 bp upstream of the putative GTG translational start site of cidR, was used as the template in the sequencing reactions. The sequencing and primer extension products were run simultaneously through an 8% (wt · vol−1) denaturing polyacrylamide gel, and the bands were visualized by autoradiography.
Northern blot analysis.
Northern blot analyses were performed as described previously (24), with minor modifications (22, 23). Digoxigenin (DIG)-labeled DNA probes were synthesized using a PCR-based DIG Probe Synthesis Kit (Roche) with primers (22) cidA1-F (5′-CCCCATATGCACAAAGTCCAATTA-3′) and cidA1-R (5′-CCCCTCGAGTTCATAAGCGTCTACACC-3′) to synthesize a cidA-specific probe.
Detection of cidABC expression by RT-PCR.
RT-PCR was performed as described previously (21), with the following modifications. Briefly, cidABC cDNA was generated using Moloney murine leukemia virus reverse transcriptase (Fermentas, Hanover, Md.) and the reverse primer cidC1-R (5′-GCCGTTGTCGACAATTGTGATAACCTTTCAATC-3′). The cidABC cDNA products were then detected by PCR using primers cidA1-F (5′-AGACATATTTAGAAAGGGATCCCGCCATGCACAAAGTCC-3′) and cidC1-R. The RT-PCR primers used for the detection of the gyrA transcripts were described previously (21). PCRs for both cidABC and gyrA were carried out in 50-μl aliquots and consisted of 2 μl of cDNA, 0.2 μM each primer, 10× PCR buffer (Invitrogen Life Technologies), 3 mM MgCl2, 0.2 μM deoxynucleoside triphosphate, and 0.5 U of Taq polymerase (Invitrogen Life Technologies). Amplification was performed with initial denaturation at 94°C for 5 min, followed by 25 cycles of denaturation at 94°C for 30 s, annealing at 52°C for 30 s, and extension at 72°C for 3 min, followed by a final extension step of 72°C for 5 min. The cidA1-F and cidC1-R primers amplify a 2.9-kb product spanning cidABC, and the gyrA-specific primers amplify a 100-bp product (GenBank accession no. D10489).
Murein hydrolase assays.
Fresh overnight cultures of S. aureus strains were used to inoculate Erlenmeyer flasks containing 10 ml of NZY broth to an initial OD600 of 0.1, and they were grown for 16 h at 37°C and 250 rpm. The culture supernatants were collected by centrifugation and concentrated approximately sixfold using a Centricon-3 concentrator (Millipore, Bedford, MA). Protein concentrations of the extracellular supernatants were determined using the Bradford assay (Bio-Rad Laboratories, Hercules, CA) according to the manufacturer's recommendation. Quantitative cell wall hydrolysis assays were performed essentially as described by Mani et al. (17).
Determination of stationary-phase survival.
Overnight cultures of S. aureus strains were used to inoculate 40 ml of NZY medium to an OD600 of 0.1. Flasks were capped with bug stoppers (Whatman Inc., Clifton, N.J.), and cultures were grown for up to 2 weeks at 37°C and 250 rpm. Aliquots (0.3 ml) were taken at 24-h intervals, and the CFU per milliliter were determined by plating serial dilutions of each sample on TSA plates. Acetate and glucose concentrations were determined using kits purchased from R-Biopharm, Inc. (Marshall, Mich.) and used according to the manufacturer's directions. The pH of the culture supernatants was determined using a φ300 pH meter (Beckman, Inc., Palo Alto, Calif.).
RESULTS
Identification of the S. aureus cidR gene.
The S. aureus cidABC operon has been previously shown to enhance murein hydrolase activity and cell death (21-23). Its counterpart, the lrgAB operon, encodes a negative effector of these processes and is positively regulated by the LytSR two-component regulatory system. Unlike the lrgAB operon, which lies immediately downstream from the lytS and lytR genes (4), the cidABC operon is located immediately downstream of an ORF, designated cidR, encoding an LTTR (Fig. 1A). The translation start site of the cidR gene is predicted to be a GTG codon (GenBank accession no. AY581892) based on its proximity to a putative ribosome-binding site (Fig. 1A), as well as the high degree of amino acid sequence similarity of the resulting N terminus with other known LTTRs (data not shown). The predicted amino acid sequence of the cidR gene product (CidR) contains 292 residues with a deduced molecular mass of 33.4 kDa and a pI of 5.39 (http://us.expasy.org/tools/pi_tool.html). Analysis of the CidR amino acid sequence revealed that it contains a conserved N-terminal helix-turn-helix motif that is likely to be responsible for DNA binding (26) and a putative C-terminal regulatory domain, to which an inducer may bind (10, 18, 26). Unlike most LTTRs, which are usually divergently transcribed from a promoter that overlaps the promoter of the regulated target gene (26), the cidR gene is transcribed in the same direction as cidABC.
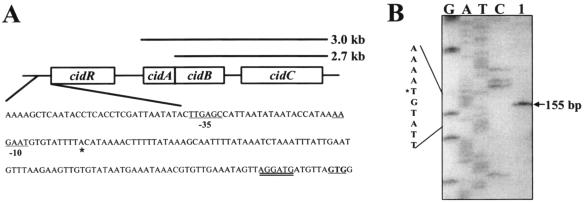
FIG. 1.
Analysis of the cidR promoter region. (A) The cidR transcription start site determined by primer extension analysis (B) is indicated by asterisks, whereas putative −10 and −35 elements are underlined. The putative GTG start codon is in bold, and the predicted ribosome binding site is double underlined. Transcripts spanning cidABC (3.0 kb) and cidBC (2.7 kb) are indicated above the corresponding genes. (B) Primer extension analysis of total cellular RNA (100 μg) (lane 1) from UAMS-1 yielded a 155-bp cDNA product, mapping the cidR transcription start site to an adenine residue located 103 bp upstream of the cidR GTG start codon. The size of the extension product was determined by comparison with the DNA sequencing ladder of the cidR promoter region. Primer extension and sequencing reactions were prepared using the same primer.
As shown in Fig. 1A and B, the transcription start site (+1) for the cidR gene was determined by primer extension analysis to be an adenine residue located 103 bp upstream of the predicted cidR start codon. No canonical −10 and −35 elements (11) (5′-TATAAT-3′ and 5′-TTGACA-3′, respectively) were identified in the sequences preceding the cidR transcription start site. However, the sequences 5′-AAGAAT-3′ (9 bp upstream of the cidR +1 site) and 5′-TTGAGG-3′ (19 bp upstream of the putative −10 element) both containing four out of six matches to the predicted σA-dependent −10 and −35 consensus sequences, respectively, were identified upstream of the +1 site. The suboptimal spacing between the putative −10 and −35 sequences may partially explain the low-level expression of the cidR transcript both in the presence and in the absence of glucose (see Fig. 3).
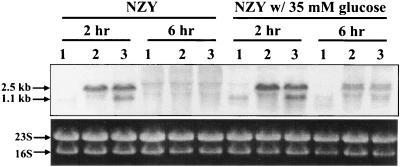
FIG. 3.
Northern blot analysis of cidR transcription. Total cellular RNAs from UAMS-1 (lane 1), KB1090 (lane 2), and KB1090(pSJ11) (lane 3) cells grown in either NZY broth or NZY broth with 35 mM glucose were isolated at 2 and 6 h postinoculation. Five micrograms of each RNA sample was separated in a 1% (wt/vol) agarose-formaldehyde gel, transferred to a nylon membrane, and hybridized to a cidR-specific, DIG-labeled probe.
Positive regulation of cidABC transcription by CidR.
Previous studies of transcription of the S. aureus cid operon revealed the presence of two major overlapping transcripts: a cidABC transcript that is expressed at low levels during the early exponential growth phase and a cidBC transcript that is produced at relatively high levels during exponential growth and whose expression is dependent on rsbU-mediated activation of S. aureus sigma factor B (σB) (21, 23). Furthermore, it has been recently demonstrated that cidABC transcription is strongly upregulated by the acetic acid that accumulates in the culture medium during growth in the presence of excess glucose (22). Since the cidR gene is predicted to encode an LTTR, it is possible that expression of one or both of the cid operon transcripts is regulated by the cidR gene product. To address this, a cidR mutant derivative of UAMS-1 (designated KB1090) was generated by replacing 335 bp from the internal region of this gene with a Tcr cassette. To determine the effect of the cidR mutation on expression of the cidR and cidABC genes, Northern blot analyses were performed on RNA samples isolated from cultures of either UAMS-1, KB1090 (cidR mutant), or KB1090(pSJ11) (the cidR mutant containing the cidR complementation plasmid) grown in NZY broth in either the presence or the absence of 35 mM glucose. As shown in Fig. 2A, cidABC transcription was not detected in either UAMS-1 or the isogenic cidR mutant KB1090 when the cultures were grown in NZY broth in the absence of glucose. In agreement with previous findings (22), cidABC expression was dramatically induced during the transition into stationary phase (6 h growth) when UAMS-1 was grown in the presence of 35 mM glucose (Fig. 2B). However, under these same growth conditions the cidR mutant displayed a complete absence of cidABC transcripts at 6 h of growth, suggesting that the dramatic increase in cidABC expression observed in the presence of 35 mM glucose is dependent on the cidR gene product. The expression of cidABC in strain KB1090(pSJ11) was detectable at low levels at 6 h in the presence of 35 mM glucose, suggesting that the cidR mutation could be partially complemented by expression of cidR from a plasmid. This low-level complementation is likely due to the fact that cidR expression is under the control of the vegII promoter that is maximally active during exponential growth (13). In contrast to cidABC expression, the expression of cidBC was not affected by the cidR mutation both in the presence and in the absence of glucose (data not shown). Also, the CidR-dependent transcription of cidABC was not affected by σB (data not shown).
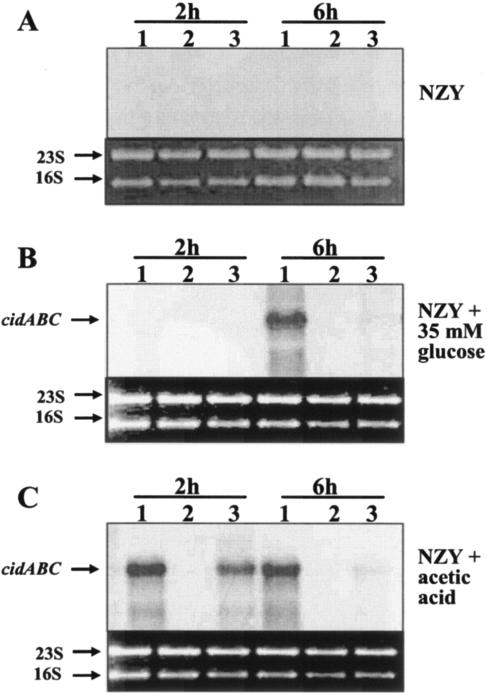
FIG. 2.
Northern blot analyses of cidABC transcription. Total cellular RNAs from UAMS-1 (lane 1), KB1090 (lane 2), and KB1090(pSJ11) (lane 3) cells grown in either NZY broth (A), NZY broth with 35 mM glucose (B), or NZY broth with 26 mM acetic acid (C) were isolated at 2 and 6 h postinoculation. Five micrograms of each RNA sample was separated in a 1% (wt/vol) agarose-formaldehyde gel, transferred to a nylon membrane, and hybridized to a cidA-specific, DIG-labeled probe. The sizes of the transcripts were determined by comparison to an RNA ladder (Invitrogen) run on the same gel.
It was also previously found that the effect of high levels of glucose on cidABC transcription is attributable to the accumulation of acetic acid in the culture medium as a result of glucose catabolism (22). Therefore, a Northern blot analysis was also performed on RNA samples from UAMS-1, KB1090, and KB1090(pSJ11) grown in NZY supplemented with 26 mM acetic acid to determine whether acetic acid induction of high-level cidABC expression is mediated via CidR. Consistent with previously observed results (21), growth of UAMS-1 in NZY broth supplemented with acetic acid resulted in dramatically increased expression of cidABC at 2 h and 6 h after inoculation (Fig. 2C). In contrast, growth of KB1090 in acetic acid-supplemented NZY broth showed no detectable cidABC transcripts at either time point. Furthermore, cidABC expression was restored in KB1090(pSJ11) grown in medium supplemented with acetic acid, especially at 2 h of growth, when the vegII promoter is thought to be most active (Fig. 2C). Collectively, these results indicate that the induction of cidABC expression by acetic acid previously observed (22) is dependent on an intact cidR gene.
As shown in Fig. 3, a Northern blot analysis revealed that the cidR gene is transcribed as a 1.1-kb transcript whose expression is induced only during early exponential growth (2 h growth) in wild-type S. aureus UAMS-1 both in the presence and in the absence of glucose. In the cidR mutant (KB1090), a 2.5-kb readthrough transcript spanning the 5′ end of the cidR ORF and the Tcr cassette was also detected only during early exponential growth, indicating the readthrough transcript is under the control of the cidR promoter. Expression of the readthrough transcript in KB1090 containing the complementation plasmid pSJ11 was not affected by the complementation of cidR in trans. These data suggest that cidR expression is not negatively autoregulated.
Effect of the cidR mutation on murein hydrolase activity.
Recently, it was shown that the cid operon enhances the murein hydrolase activity produced by the cells (21-23), whereas the lrgAB operon has a negative regulatory effect on this activity (7). Therefore, we speculated that the cidR mutant KB1090, as a positive regulator of cidABC expression, would exhibit reduced extracellular murein hydrolase activity. In agreement with previous studies (22), the extracellular murein hydrolase activity of UAMS-1 was dramatically increased when cells were grown in the presence of glucose, whereas the activity of KB1050 was almost undetectable and unaffected by the presence of glucose in the growth medium (Fig. 4). In contrast, the KB1090 strain produced normal levels of extracellular murein hydrolase activity compared to UAMS-1 when both strains were grown in the absence of glucose. However, when grown in the presence of glucose, the KB1090 strain displayed a moderate decrease in murein hydrolase activity compared to the activity produced by UAMS-1. KB1090 and UAMS-1 display similar growth rates in the presence and in the absence of glucose (unpublished data), ruling out the possibility that the differences in extracellular murein hydrolase activity observed are due to growth rate effects. These observations correlated well with the Northern blot analyses in Fig. 2A and B, in that cidABC expression is induced only in UAMS-1 in the presence of glucose, whereas KB1090 showed no induction of cidABC transcription in the presence of glucose. The decreased extracellular murein hydrolase activity of KB1090 in the presence of glucose was partially complemented in a strain, KB1090(pSJ11), that expresses cidR from the plasmid (unpublished data), consistent with the partial complementation of glucose-inducible cidABC transcription observed in this strain (Fig. 2B and C).
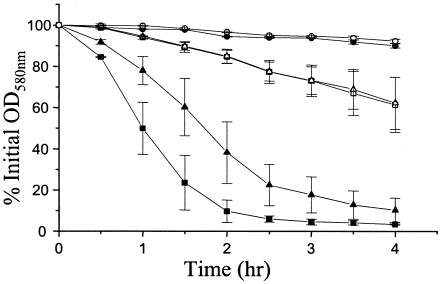
FIG. 4.
Quantitative murein hydrolase assays. One hundred micrograms of extracellular proteins isolated from 16-h cultures of UAMS-1 (wild-type; squares), KB1050 (cidA mutant; circles), and KB1090 (cidR mutant; triangles) grown either in the presence of 35 mM glucose (closed symbols) or in the absence of glucose (open symbols) was added to a 1.0-mg/ml suspension of Micrococcus luteus cells, and the murein hydrolase activity of each sample was measured as a decrease in turbidity over a 4-h time course. These data are the average of three independent experiments. The error bars on the graph correspond to the standard errors of the means.
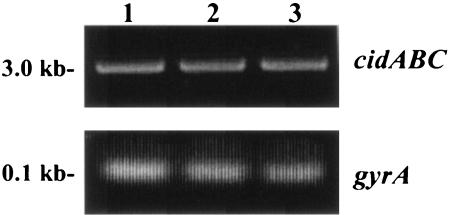
It is important to note that despite its inability to induce cidABC expression, the KB1090 strain still displayed an increase in murein hydrolase activity when grown in the presence of glucose (Fig. 4). One possible explanation to account for this is that the cidR gene product is required for high-level expression of cidABC but that glucose-inducible expression is mediated by some other regulatory protein. To test whether the low-level expression of cidABC previously observed in the absence of glucose (21) is affected by the cidR mutation, an RT-PCR analysis was performed on RNA samples isolated from UAMS-1, KB1090, and KB1090(pSJ11) grown in the absence of glucose. As shown in Fig. 5, cidABC expression was identical in all three strains tested, similar to the presumably constitutively expressed gyrA gene, indicating that low-level transcription of cidABC still occurs in the cidR mutant. Control reactions, lacking RT enzyme, failed to generate RT-PCR products, indicating the absence of contaminating genomic DNA (data not shown). Thus, we conclude that the cidR gene product does not affect the basal level cidABC transcription.
FIG. 5.
RT-PCR analysis of cidABC expression. RNA samples were isolated from early exponential growth phase cultures of UAMS-1 (lane 1), KB1090 (lane 2), and KB1090(pSJ11) (lane 3) grown in NZY broth and subjected to RT-PCR (see Materials and Methods) to detect transcription of cidABC and gyrA. The corresponding gels are labeled cidABC and gyrA, respectively.
Effect of the cidR mutation on antibiotic sensitivity.
It was previously shown that the cidA mutation resulted in increased tolerance to antibiotics, including penicillin, vancomycin, and rifampin (21, 22). Thus, we also compared antibiotic-induced killing of exponentially growing UAMS-1 and KB1090 cells grown either in the presence or in the absence of glucose. In the presence of glucose, both of these strains displayed decreased tolerance to rifampin and vancomycin compared to cells grown in the absence of glucose as previously observed (22). Surprisingly, the KB1090 strain did not display a difference in tolerance to rifampin challenge compared to UAMS-1 either in the presence or in the absence of glucose (unpublished data). However, growth in the absence of glucose revealed a slight increase in the tolerance of KB1090 to vancomycin relative to UAMS-1 grown under the same conditions. Specifically, incubation in the presence of vancomycin reduced the viable cell counts of the UAMS-1 culture by 90% in 6 h compared to a reduction of 75% in the KB1090 culture during this same time. This increased tolerance to vancomycin was not observed when the cidR mutant was grown in the presence of glucose. Collectively, these results indicate that tolerance to the antibiotics vancomycin and rifampin is only moderately affected by disruption of the cidR gene.
Effect of cidR on stationary-phase survival.
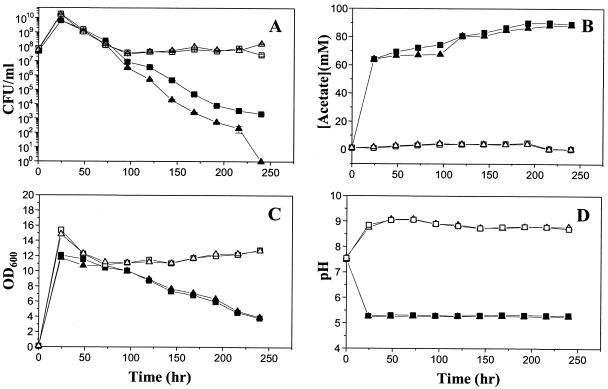
Previous studies also demonstrated that mutations in either the cidA or cidC gene had a dramatic impact on the stationary-phase survival, lysis, and acetate metabolism of cells grown in the presence of excess glucose (19). To determine the effects of cidR on stationary-phase survival, the UAMS-1 and KB1090 strains were incubated for 240 h either in the presence or in the absence of 35 mM glucose, and aliquots of these cultures were taken at 24-h intervals to determine the pH, acetate concentration, OD600, and CFU per milliliter. As shown in Fig. 6A, when the cultures were grown in the absence of glucose, both UAMS-1 and KB1090 retained similar levels of cell viabilities throughout the experiment. However, in the presence of 35 mM glucose, KB1090 consistently displayed approximately 1-log-lower viable cell counts relative to UAMS-1 at time points measured after 125 h of growth. In this experiment, the number of viable cells present in the KB1090 culture had dropped to undetectable levels by 240 h, whereas the viability of the UAMS-1 culture remained at approximately 103 CFU/ml. It should be noted that we chose to present representative data in Fig. 6 since the rates of killing in stationary phase varied slightly from experiment to experiment. Despite this variation, the differences in cell viability between the KB1090 and UAMS-1 cultures were consistently observed in four independent experiments. UAMS-1 and KB1090 grown in the absence of glucose displayed similar rates of lysis during the time course of this assay, as measured by OD600 (Fig. 6C). The OD600 was also similar for both strains grown in the presence of glucose, but these cultures displayed increased rates of lysis compared to the no-glucose cultures, as previously observed (19). These results indicate that the cidR mutation has a moderate effect on stationary-phase viability but not lysis.
FIG. 6.
Effect of cidR on survival in stationary phase. At 24-h intervals, aliquots of UAMS-1 (wild-type; squares) or KB1090 (cidR mutant; triangles) cultures grown either in the presence (closed symbols) or in the absence (open symbols) of 35 mM glucose were removed and the CFU per milliliter (A), acetate concentration (B), OD600 (C), and pH of the culture supernatants (D) were measured over a 240-h time course. The number of CFU per milliliter of each culture (A) was determined by dilution plating on TSA in triplicate, and the error bars in panel A represent standard errors of the means from a single experiment. All these data are from a single representative experiment that was repeated four times.
As observed in previous studies (22), growth in the presence of 35 mM glucose resulted in the accumulation of high levels of acetate (Fig. 6B) and acidification of the culture medium to pH 5.3 (Fig. 6D). The cidC gene, which encodes a pyruvate oxidase that converts pyruvate to acetate, was shown to affect acetate metabolism and, ultimately, the viability of the cells (19). As shown in Fig. 6C and D, the cidR mutation had no effect on the ability of the cells to secrete acetate into the culture medium and thus also did not affect the pH. As the cidR mutation has been shown to be involved in the expression of cidABC (see above), the lack of effect on acetate accumulation and pH is likely due to the constitutive, high-level, sigma B-dependent expression of the cidBC transcripts.
DISCUSSION
LTTRs, first described by Henikoff et al., make up one of the largest families of prokaryotic regulatory proteins (10, 16, 18). Despite this, no S. aureus LTTRs have been characterized to date. In the present study, we have demonstrated that the previously reported effect of acetic acid (produced as a consequence of aerobic glucose metabolism) on cidABC transcription (22) is dependent on the product of the cidR gene, a homologue of the LTTRs (Fig. 2B and C). The products of the cidABC operon have been proposed to be involved in the regulation of murein hydrolase activity and, ultimately, bacterial programmed cell death (20). Therefore, it is concluded that the cidR gene product also comprises part of the murein hydrolase regulatory system by regulating cidABC expression at the transcriptional level in the presence of its as yet unidentified coinducer molecule. In contrast to the cidABC transcript, expression of the 2.7-kb cidBC transcript, whose expression is dependent on the stress sigma factor σB (23), was not affected by the cidR mutation either in the presence or in the absence of glucose (unpublished data). Therefore, it appears that the two overlapping transcripts of the cid locus each respond to different regulatory signals.
Based on previous reports from our laboratory, we proposed a model in which the cidA and lrgA gene products regulate murein hydrolase activity in a manner similar to those of holins and antiholins, respectively (2, 20, 22). In agreement with previously published findings (22), the cidA mutant (KB1050) displayed a nearly complete loss of extracellular murein hydrolase activity in the presence or in the absence of glucose (Fig. 4). In contrast, KB1090 (the cidR mutant) displayed murein hydrolase activity that was at an intermediate level compared to UAMS-1 and KB1050 when grown in the presence of glucose. This “intermediate” phenotype observed in KB1090 may be explained by the fact that low-level cidABC expression occurs in the cidR mutant (Fig. 5). Interestingly, despite the lack of glucose-inducible cidABC expression in the absence of cidR, murein hydrolase activity was still induced by growth in the presence of glucose, albeit not to the same extent as the wild-type strain (Fig. 4). This increase could be a result of either the activities of other glucose-induced and/or CidR-regulated genes or an increase in the activity of the low levels of CidA protein that are present in the cidR mutant strain. The observation that the murein hydrolase activity produced by the cidA mutant (KB1050) was not induced by glucose seems to support the latter model. Furthermore, the low-level expression of cidABC observed in KB1090 may account for the fact that, unlike the cidA mutant KB1050, which displays a clumping phenotype (22), the cidR mutant appears to divide and separate normally (unpublished data). This indicates that the murein hydrolase activity produced by the KB1090 strain is biologically relevant, being sufficient to mediate daughter cell separation. Collectively, these results reinforce an important role for CidA in regulating murein hydrolase activity and suggest that, in addition to upregulating transcription of cidABC, growth of S. aureus in the presence of glucose may also influence the activity of the CidA protein.
Previous studies in our laboratory have also suggested that the cidA gene product contributes to the sensitivity of the cells to several different antibiotics (21, 22). Surprisingly, the antibiotic tolerance of the cidR mutant was virtually identical to that of the wild-type S. aureus UAMS-1 strain, with the exception that a slight increase in tolerance to vancomycin was observed when KB1090 was grown in the absence of glucose (unpublished data). It is possible that the low-level expression of cidABC in KB1090 is physiologically sufficient to induce cell death in response to antibiotics to levels comparable to those observed in UAMS-1. Another possibility stems from the fact that some of LTTRs form a regulon by controlling a group of unlinked genes (14, 15, 26). In addition to the cidABC upregulation, the cidR gene product may regulate the expression of other genes that affect cell death in opposition to the cidA gene product.
Recently, our laboratory has shown that the cidC gene encodes a pyruvate oxidase that affects acetate metabolism under excess glucose conditions and, consequently, cell viability in stationary phase (19). This study also demonstrated that acetic acid-induced cell death in stationary phase is mediated, at least in part, by the product of the cidA gene. In the study presented here, the effect of the cidR gene on stationary-phase survival of S. aureus was also investigated. As shown in Fig. 6, growth of the bacteria in the presence of 35 mM glucose dramatically increased cell death in stationary phase. The cidR mutant reproducibly exhibited decreased viability compared to the wild-type strain despite the lack of differences in acetate accumulation and the pH of the culture medium. In contrast, both the strains maintained similar high levels of viability (>107 CFU/ml) throughout the survival assay in the absence of glucose. The observation that the cidR mutant culture lacking high-level expression of cidABC displayed a dramatic decrease in cell viability and increased cell lysis in the presence of glucose relative to growth in the absence of glucose (Fig. 6A and C) indicates that the cidA gene is not the only factor that affects cell death and lysis in stationary phase.
In the present study, the cidR gene product was shown to positively regulate cid operon expression by increasing transcription of cidABC in the presence of acetic acid produced by glucose catabolism. Since most LTTRs activate transcription of target promoters only in the presence of a small signal molecule (coinducer), identification of the coinducer molecule required for acetic acid-dependent, CidR-mediated cidABC expression is currently in progress. It is also envisioned that the effect of the cidR gene product on murein hydrolase activity in the presence of glucose occurs via a cidABC-mediated mechanism. As the cidR gene was shown to have only moderate effects on the antibiotic tolerance and stationary-phase survival of S. aureus, the simultaneous regulation of multiple pathways that control tolerance to antibiotics and stationary-phase cell death may also occur. Indeed, recent studies in our laboratory have revealed the presence of additional CidR-regulated genes that alter murein hydrolase activity by affecting the expression of both the cid and lrg operons. A more detailed understanding of molecular mechanisms employed by a cidR regulon should provide important insight into the regulation of murein hydrolase activity, as well as the programmed cell death pathway of S. aureus.
Acknowledgments
We thank the Staphylococcus aureus Genome Sequencing Project (University of Oklahoma Health Sciences Center) for providing the sequence of cidR.
This work was funded by NIH grant R01AI038901, NIH-NRRI grant P20RR15587, and DOD grant DAAD 19-03-1-0191.
REFERENCES
- 1.Bayles, K. W. 2003. Are the molecular strategies that control apoptosis conserved in bacteria? Trends Microbiol. 11:306-311. [DOI] [PubMed] [Google Scholar]
- 2.Bayles, K. W. 2000. The bactericidal action of penicillin: new clues to an unsolved mystery. Trends Microbiol. 8:274-278. [DOI] [PubMed] [Google Scholar]
- 3.Bruckner, R. 1992. A series of shuttle vectors for Bacillus subtilis and Escherichia coli. Gene 122:187-192. [DOI] [PubMed] [Google Scholar]
- 4.Brunskill, E. W., and K. W. Bayles. 1996. Identification of LytSR-regulated genes from Staphylococcus aureus. J. Bacteriol. 178:5810-5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dyer, D. W., and J. J. Iandolo. 1983. Rapid isolation of DNA from Staphylococcus aureus. Appl. Environ. Microbiol. 46:283-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gillaspy, A. F., S. G. Hickmon, R. A. Skinner, J. R. Thomas, C. L. Nelson, and M. S. Smeltzer. 1995. Role of the accessory gene regulator (agr) in pathogenesis of staphylococcal osteomyelitis. Infect. Immun. 63:3373-3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Groicher, K. H., B. A. Firek, D. F. Fujimoto, and K. W. Bayles. 2000. The Staphylococcus aureus lrgAB operon modulates murein hydrolase activity and penicillin tolerance. J. Bacteriol. 182:1794-1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guerout-Fleury, A.-M., K. Shazand, N. Frandsen, and P. Stragier. 1995. Antibiotic-resistance cassettes for Bacillus subtilis. Gene 167:335-336. [DOI] [PubMed] [Google Scholar]
- 9.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 10.Henikoff, S., G. W. Haughn, J. M. Calvo, and J. C. Wallace. 1988. A large family of bacterial activator proteins. Proc. Natl. Acad. Sci. USA 85:6602-6606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huerta, A. M., and J. Collado-Vides. 2003. Sigma70 promoters in Escherichia coli: specific transcription in dense regions of overlapping promoter-like signals. J. Mol. Biol. 333:261-278. [DOI] [PubMed] [Google Scholar]
- 12.Inoue, H., H. Nojima, and H. Okayama. 1990. High efficiency transformation of Escherichia coli with plasmids. Gene 96:23-28. [DOI] [PubMed] [Google Scholar]
- 13.Jankovic, I., O. Egeter, and R. Bruckner. 2001. Analysis of catabolite control protein A-dependent repression in Staphylococcus xylosus by a genomic reporter gene system. J. Bacteriol. 183:580-586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ko, M., and C. Park. 2000. H-NS-dependent regulation of flagellar synthesis is mediated by a LysR family protein. J. Bacteriol. 182:4670-4672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kovacikova, G., and K. Skorupski. 1999. A Vibrio cholerae LysR homolog, AphB, cooperates with AphA at the tcpPH promoter to activate expression of the ToxR virulence cascade. J. Bacteriol. 181:4250-4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lochowska, A., R. Iwanicka-Nowicka, D. Plochocka, and M. M. Hryniewicz. 2001. Functional dissection of the LysR-type CysB transcriptional regulator: regions important for DNA binding, inducer response, oligomerization, and positive control. J. Biol. Chem. 276:2098-2107. [DOI] [PubMed] [Google Scholar]
- 17.Mani, N., P. Tobin, and R. K. Jayaswal. 1993. Isolation and characterization of autolysis-defective mutants of Staphylococcus aureus created by Tn917-lacZ mutagenesis. J. Bacteriol. 175:1493-1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Muraoka, S., R. Okumura, N. Ogawa, T. Nonaka, K. Miyashita, and T. Senda. 2003. Crystal structure of a full-length LysR-type transcriptional regulator, CbnR: unusual combination of two subunit forms and molecular bases for causing and changing DNA bend. J. Mol. Biol. 328:555-566. [DOI] [PubMed] [Google Scholar]
- 19.Patton, T. G., K. C. Rice, M. K. Foster, and K. W. Bayles. 2005. The Staphylococcus aureus cidC gene encodes a pyruvate oxidase that affects acetate metabolism and cell death in stationary phase. Mol. Microbiol. 56:1664-1674. [DOI] [PubMed] [Google Scholar]
- 20.Rice, K. C., and K. W. Bayles. 2003. Death's toolbox: examining the molecular components of bacterial programmed cell death. Mol. Microbiol. 50:729-738. [DOI] [PubMed] [Google Scholar]
- 21.Rice, K. C., B. A. Firek, J. B. Nelson, S.-J. Yang, T. G. Patton, and K. W. Bayles. 2003. The Staphylococcus aureus cidAB operon: evaluation of its role in regulation of murein hydrolase activity and penicillin tolerance. J. Bacteriol. 185:2635-2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rice, K. C., J. B. Nelson, T. G. Patton, S.-J. Yang, and K. W. Bayles. 2005. Acetic acid induces expression of the Staphylococcus aureus cidABC and lrgAB murein hydrolase regulator operons. J. Bacteriol. 187:813-821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rice, K. C., T. Patton, S.-J. Yang, A. Dumoulin, M. Bischoff, and K. W. Bayles. 2004. Transcription of the Staphylococcus aureus cid and lrg murein hydrolase regulators is affected by sigma factor B. J. Bacteriol. 186:3029-3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 25.Sau, S., J. Sun, and C. Y. Lee. 1997. Molecular characterization and transcriptional analysis of type 8 capsule genes in Staphylococcus aureus. J. Bacteriol. 179:1614-1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schell, M. A. 1993. Molecular biology of the LysR family of transcriptional regulators. Annu. Rev. Microbiol. 47:579-626. [DOI] [PubMed] [Google Scholar]
- 27.Schenk, S., and R. A. Laddaga. 1992. Improved method for electroporation of Staphylococcus aureus. FEMS Microbiol. Lett. 94:133-138. [DOI] [PubMed] [Google Scholar]
- 28.Tomasz, A., A. Albino, and E. Zanati. 1970. Multiple antibiotic resistance in a bacterium with suppressed autolytic system. Nature 227:138-140. [DOI] [PubMed] [Google Scholar]