Abstract
The prevalence of insertion sequences IS1548, IS861, IS1381, and ISSa4 and of the group II intron GBSi1 within Streptococcus agalactiae human isolates strongly correlates with the genetic lineages obtained by multilocus sequence typing. Our results yielded an evolutionary scheme for the acquisition of these genetic elements linked to the ecosystems from which the isolates were obtained.
Mobile genetic elements drive bacterial evolution and adaptation via recombination and horizontal transfer events (12), and they can influence the virulence of bacteria by modulating gene expression (9). These mobile genetic elements include bacteriophages, transposons, superintegrons, insertion sequences (ISs), and group II introns. ISs are small genetic elements, usually less than 2.5 kb in size, encoding only their own mobility (15), whereas group II introns have a more complicated mechanism and transpose via an RNA intermediate that acts like a ribozyme (17). Streptococcus agalactiae has emerged, during the last 35 years, as the most common pathogen implicated in neonatal infectious diseases (24). It harbors several mobile genetic elements. Five are well characterized: four ISs, IS1548 (8), IS861 (18), IS1381 (26), and ISSa4 (24), and one group II intron, GBSi1 (7). Some of these elements have been reported to be associated with virulence genes: IS1548 with the hyaluronate lyase gene (8), IS1548 and GBSi1 with the C5a-peptidase gene and the gene encoding the laminin-binding protein Lmb (7), IS861 and IS1548 with the capsule cluster cps gene (21, 22), and ISSa4 with the cylB gene, which encodes the membrane-spanning domain of a putative hemolysin transporter (25). Given the influence of these repeat elements on genome plasticity, the aim of this study was to correlate the presence of IS1548, IS861, IS1381, ISSa4, and GBSi1 with the evolution of the S. agalactiae species analyzed by multilocus sequence typing (MLST).
Our collection was composed of 52 epidemiologically unrelated S. agalactiae isolates, representative of the various human sites where S. agalactiae is found. They were collected in France from 1986 to 2003: 20 were isolated from the cerebrospinal fluid of neonates, 17 from the blood cultures of patients suffering from endocarditis, 10 from the vaginas of asymptomatic women, and 5 from the gastric fluids of neonates. Serotyping was performed with a Pastorex rapid latex agglutination test (Bio-Rad, Hercules, Calif.) and by a molecular serotyping process as described by Kong et al. (13). Thirty isolates were from serotype III (57.7% of isolates), seven from serotype Ib (13.5%), four from serotype II (7.7%), three from serotype Ia (5.8%), two from serotype V (3.8%), and one from serotype IV (1.9%). Five isolates were nontypeable (9.6%).
We evaluated the prevalence of IS1548, IS861, IS1381, ISSa4, and GBSi1 within the 52 isolates by PCR using previously described primers and amplification conditions (1, 4). PCR products corresponding to each IS and GBSi1 were checked by sequencing. Forty of the 52 isolates possessed IS861 (76.9%), 37 possessed IS1381 (71.2%), 19 possessed IS1548 (36.5%), 16 possessed GBSi1 (30.8%), and 4 possessed ISSa4 (7.7%). Based on the presence or absence of each of the five elements, nine genetic variants, designated V1 to V9, were defined among the 52 isolates in our collection (Table 1).
TABLE 1.
Nine genetic variants defined by combination of the presence or absence of each of the four ISs and of the group II intron GBSi1a
| Genetic variant | No. of different ISs and/or group II intron | Result for:
|
No. (%) of isolates | Serotype (no. of isolates)b | ||||
|---|---|---|---|---|---|---|---|---|
| IS1548 | GBSi1 | IS861 | IS1381 | ISSa4 | ||||
| V1 | 0 | − | − | − | − | − | 2 (4) | III (1), NT (1) |
| V2 | 1 | − | − | − | + | − | 6 (11) | NT (2), IV (1), V (1), Ia (1), Ib (1) |
| V3 | 1 | − | − | − | − | + | 3 (6) | III (3) |
| V4 | 2 | − | + | + | − | − | 10 (9) | III (10) |
| V5 | 2 | − | − | + | + | − | 8 (15) | Ib (6), Ia (1), NT (1) |
| V6 | 3 | + | − | + | + | − | 17 (33) | III (15), II (1), NT (1) |
| V7 | 3 | − | + | + | + | − | 3 (6) | Ia (1), II (1), III (1) |
| V8 | 3 | − | + | − | + | + | 1 (2) | II (1) |
| V9 | 4 | + | + | + | + | − | 2 (4) | II (1), V (1) |
Serotypes of the isolates are indicated in the last column.
NT, nontypeable.
The genomic positions of the copies of each IS and GBSi1 were determined by Southern blot analysis for each of the 52 isolates. Briefly, DNA was digested with EcoRI, for which there were no cleavage sites in the four ISs or GBSi1. Hybridizations were carried out as previously described (6), with probes specific for each IS and GBSi1. Vector NTI v 9.0 software was employed to estimate the number and the size of EcoRI fragments by using the whole genome sequence of S. agalactiae NEM316 (GenBank accession number AL732656). The Smart Ladder molecular-size standard (Eurogentec), which was run in parallel, allowed us to locate the copies of the ISs and of GBSi1 on the restriction fragments generated in silico.
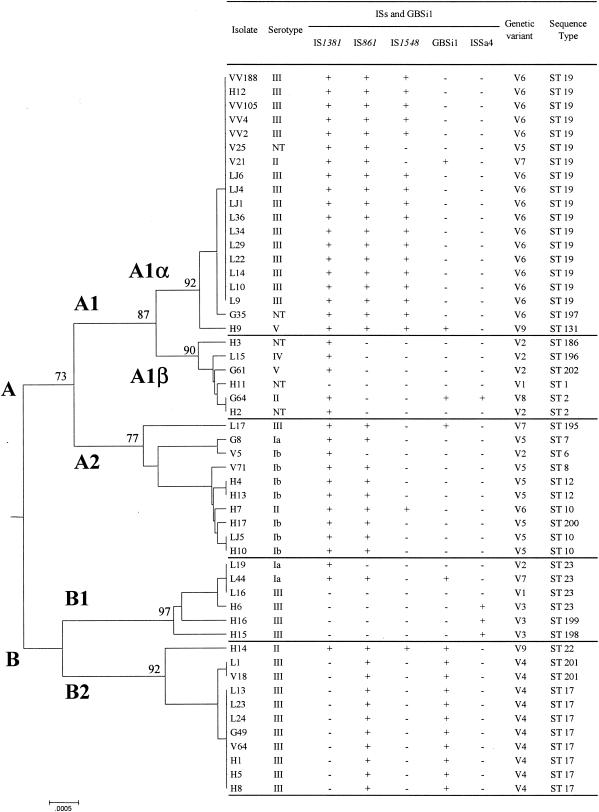
Phylogenetic analysis was performed by the MLST technique with seven housekeeping genes (adhP, pheS, atr, glnA, sdhA, glcK, and tkt) as described by Jones et al. (11). Nucleotide sequences were compared with those available on the MLST database (http://pubmlst.org/sagalactiae/). After assigning an allele number to each locus, the sequence type (ST), which takes into account the allele combinations of the seven loci, was determined for each isolate. Between three (genes pheS and glnA) and eight (gene adhP) alleles were present for each locus (Table 2). Based on the combinations of the alleles for the seven loci, 21 STs were identified, of which 14 corresponded to a single isolate; three STs, ST 19 (n = 17), ST 17 (n = 8), and ST 23 (n = 4), accounted for 55.8% of the whole collection. These STs corresponded to three of the four major STs previously defined by Jones et al., confirming the worldwide spread of ST 17, ST 19, and ST 23. These results also confirm the close correlation of ST 19 and ST 17 with serotype III (11). Moreover, nucleotide sequences of the seven genes were concatenated into a single “supergene” (3,456 bp), which allows the construction of a phylogenetic tree by use of the MEGA software version 2.1 (http://www.megasoftware.net) with the UPGMA algorithm and the Kimura two-parameter mutation model of genetic distance. The level of statistical support for the nodes on the tree was evaluated by examining their percentage of recovery in 1,000 resampled trees by use of the bootstrap test (23). The MEGA software grouped the isolates into two main genomic divisions: division A, which could be subdivided into A1α, A1β, and A2, and division B, which could be subdivided into B1 and B2 (Fig. 1).
TABLE 2.
STs assigned on the basis of allelic profilesa
| ST | Allelic profile | No. (%) of:
|
Total no. (%) of isolates (n = 52) | Serotype (no. of isolates)d | ||
|---|---|---|---|---|---|---|
| CSFb isolates (n = 20) | Endocarditis isolates (n = 17) | Colonizing isolatesc (n = 15) | ||||
| 19 | 1, 1, 3, 2, 2, 2, 2 | 10 (50) | 1 (5.9) | 6 (40) | 17 (32.7) | III (15), NT (1), II (1) |
| 17 | 2, 1, 1, 2, 1, 1, 1 | 3 (15) | 3 (17.7) | 2 (13.3) | 8 (15.4) | III (8) |
| 23 | 5, 4, 6, 3, 2, 1, 3 | 3 (15) | 1 (5.9) | 0 (0) | 4 (7.7) | Ia (2), III (2) |
| 10 | 9, 1, 4, 1, 3, 3, 2 | 1 (5) | 2 (11.8) | 0 (0) | 3 (5.81) | Ib (2), II (1) |
| 12 | 10, 1, 4, 1, 3, 3, 2 | 0 (0) | 2 (11.8) | 0 (0) | 2 (3.85) | Ib (2) |
| 2 | 1, 1, 3, 1, 1, 2, 2 | 0 (0) | 1 (5.9) | 1 (6.6) | 2 (3.85) | II (1), NT (1) |
| 201 | 2, 1, 25, 2, 1, 1, 1 | 1 (5) | 0 (0) | 1 (6.6) | 2 (3.85) | III (2) |
| 1 | 1, 1, 2, 1, 1, 2, 2 | 0 (0) | 1 (5.9) | 0 (0) | 1 (1.92) | NT (1) |
| 6 | 9, 1, 2, 1, 3, 2, 2 | 0 (0) | 0 (0) | 1 (6.6) | 1 (1.92) | Ib (1) |
| 7 | 10, 1, 2, 1, 3, 2, 2 | 0 (0) | 0 (0) | 1 (6.6) | 1 (1.92) | Ia (1) |
| 8 | 4, 1, 4, 1, 3, 3, 2 | 0 (0) | 0 (0) | 1 (6.6) | 1 (1.92) | Ib (1) |
| 22 | 13, 3, 1, 3, 1, 1, 1 | 0 (0) | 1 (5.9) | 0 (0) | 1 (1.92) | II (1) |
| 131 | 1, 1, 3, 2, 2, 3, 2 | 0 (0) | 1 (5.9) | 0 (0) | 1 (1.92) | V (1) |
| 186 | 1, 1, 2, 1, 1, 2, 5 | 0 (0) | 1 (5.9) | 0 (0) | 1 (1.92) | NT (1) |
| 195 | 9, 1, 1, 1, 3, 2, 1 | 1 (5) | 0 (0) | 0 (0) | 1 (1.92) | III (1) |
| 196 | 1, 1, 3, 1, 1, 12, 2 | 1 (5) | 0 (0) | 0 (0) | 1 (1.92) | IV (1) |
| 197 | 1, 1, 3, 1, 2, 2, 2 | 0 (0) | 0 (0) | 1 (6.6) | 1 (1.92) | NT (1) |
| 198 | 13, 4, 6, 3, 2, 1, 3 | 0 (0) | 1 (5.9) | 0 (0) | 1 (1.92) | III (1) |
| 199 | 5, 4, 6, 3, 2, 1, 2 | 0 (0) | 1 (5.9) | 0 (0) | 1 (1.92) | III (1) |
| 200 | 9, 1, 4, 1, 3, 3, 5 | 0 (0) | 1 (5.9) | 0 (0) | 1 (1.92) | Ib (1) |
| 202 | 40, 1, 3, 1, 1, 2, 2 | 0 (0) | 0 (0) | 1 (6.6) | 1 (1.92) | V (1) |
The allelic profiles, available on the MLST database website (http://pubmlst.org/sagalactiae/), were obtained by combination of each of the seven alleles, in the following order: adhP, pheS, atr, glnA, sdhA, glcK, and tkt. The ecological origins and serotypes of the S. agalactiae isolates are also indicated for each ST.
CSF, cerebrospinal fluid.
Isolated from vaginas of asymptomatic women and from the gastric fluids of neonates.
NT, nontypeable.
FIG. 1.
UPGMA dendrogram showing the genetic relationships between the 52 S. agalactiae isolates. The dendrogram was constructed with the MEGA 2.1 software by using the nucleotide sequences of the seven housekeeping genes (pheS, atr, tkt, glcK, sdhA, glnA, and adhP) concatenated into a single “supergene.” Bootstrap values are shown at the major nodes. Isolates can be divided into two divisions (A and B) and five subdivisions (A1α, A1β, A2, B1, and B2). For each isolate, the serotype, ST, presence or absence of the four ISs and of the group II intron GBSi1, and genetic variants (deduced from the combination of the four ISs and GBSi1 and named V1 to V9) are indicated. L and LJ refer to meningitis isolates, H to endocarditis isolates, G to gastric fluid isolates, and V and VV to vaginal isolates.
A strong correlation was observed between the distribution of the ISs and GBSi1 and the divisions obtained by MLST (Fig. 1). IS1381 was significantly associated with division A (34 of the 37 isolates [92%] harboring IS1381 were distributed in division A), whereas this IS was almost completely absent from division B (P < 0.0001, chi-square test) (Fig. 1). IS861 was significantly associated with subdivisions A1α, A2, and B2 (39 of the 40 isolates [98%] harboring IS861 were distributed in subdivisions A1α, A2, and B2) (P < 0.0001) (Fig. 1). IS1548 was significantly associated with subdivision A1α (17 of 19 [89%] isolates harboring IS1548 were distributed in subdivision A1α) (P < 0.0001) (Fig. 1). Similarly, GBSi1 was significantly associated with subdivision B2 (11 of 16 [69%] isolates harboring GBSi1 were distributed in subdivision B2) (P < 0.0001); conversely, 100% of the isolates from subdivision B2 were GBSi1 positive (Fig. 1). Finally, ISSa4 was significantly associated with subdivision B1 (3 of 4 isolates harboring ISSa4 were distributed in subdivision B1) (P < 0.001), although the small number of isolates in B1 and the small number of ISSa4-positive isolates must be taken into account (Fig. 1). The rare exceptions we noticed in the correlation of IS distribution within MLST lineages were probably due to horizontal gene transfers.
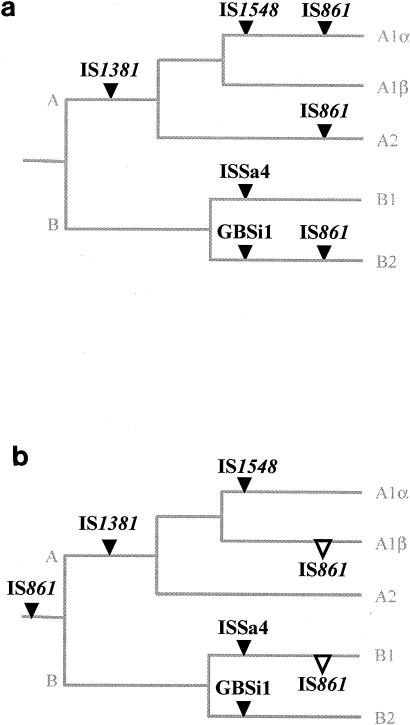
Therefore, based on the prevalence of each IS and GBSi1 in each MLST group, an evolutionary scheme corresponding to the acquisition of these mobile genetic elements by S. agalactiae during its evolution can be proposed. Indeed, IS1381 may have been acquired during the differentiation of the common ancestor (Fig. 2). IS1548, GBSi1, and ISSa4 seem to have been acquired more recently by subdivisions A1α, B2, and B1, respectively. The hypothesis that ISSa4 was acquired recently is consistent with the empirical observation of Spellerberg et al., in that all ISSa4-positive isolates in their study were isolated after 1996 (25). IS861, which is present in both divisions A and B but not in every subdivision, may have been acquired recently by subdivisions A1α, A2, and B2 (Fig. 2a). Alternatively, the earliest common ancestor may have harbored IS861, which was subsequently lost by subdivisions A1β and B1 (Fig. 2b).
FIG. 2.
Evolutionary scheme showing the chronology of acquisition of the four ISs and of the group II intron GBSi1 during S. agalactiae evolution. This scheme was deduced from the prevalence of each mobile genetic element within the divisions and subdivisions obtained by MLST as indicated in Fig. 1. IS1381 may have been acquired (acquisition of ISs or GBSi1, ▾) by division A, IS1548 by subdivision A1α, ISSa4 by subdivision B2, and GBSi1 by subdivision B2. IS861 may have been acquired by subdivisions A1α, A2, and B2 (Fig. 2a), or it may have been acquired by the common ancestor and then lost (loss of ISs or GBSi1, ▿) by subdivisions A1β and B1 (Fig. 2b). Branch values are not representative of genetic distances.
One striking finding, already pointed out by Granlund et al. (8) and Bohnsack et al. (3), is the rarity of IS1548+ GBSi1+ isolates (only two isolates in our collection harbored both). To date, three insertion sites on the S. agalactiae chromosome are known for IS1548, named site X, site Y, and site Z (7, 14, 22). Site X corresponds to the intergenic fragment located between the scpB and lmb genes, encoding the C5a-peptidase and the laminin-binding protein, respectively. As IS1548 and GBSi1 may share this insertion site, the simultaneous insertion of IS1548 and GBSi1 at the same insertion site is unlikely. Indeed, in our two IS1548+ GBSi1+ isolates, IS1548 was not found at site X or within hylB. The IS1548-free site X could therefore be explained by the presence of GBSi1 at site X, as confirmed by Southern blot analysis.
Interestingly, the evolutionary scheme presented here suggests another explanation for the mutually exclusive distribution of IS1548 and GBSi1. In fact, subdivisions A1α (carrying IS1548) and B2 (carrying GBSi1) separated early during the evolutionary process (when the ancestors A and B separated) (Fig. 1 and Fig. 2). Furthermore, GBSi1 was associated with IS1381 only once within subdivision B2 isolates (Fig. 1); similarly, ISSa4 was associated with other ISs only once (Fig. 1), in agreement with Dmitriev et al. (4). Conversely, GBSi1 and IS1548 were always associated with IS861, except for one isolate (Fig. 1). Such a preferential association has already been observed in the Escherichia coli reference (ECOR) collection (19). The mutually exclusive distribution of ISs, as well as the preferential association of ISs, is consistent with the delivery of ISs by a vector carrying multiple ISs. Although transmission of ISs by phages has not yet been reported for S. agalactiae, it has been observed for E. coli, Pseudomonas aeruginosa, and Bacillus thuringiensis (16). Despite the low prevalence of plasmids within the species S. agalactiae (10), many of the genomic islands flanked by ISs in S. agalactiae are associated with genes related to phages, integrative plasmids, and conjugative transposons (5, 27). Therefore, the peculiar distribution of ISs (mutually exclusive mode and preferential association mode) could be explained by the link between the isolates and the ecosystems from which they were isolated. It is tempting to consider that ISs and GBSi1 reflect the original ecosystem of the isolates and that many of the IS-mediated mutations contribute to the adaptation of bacterial populations to their environment (20). This hypothesis is in accordance with a recent study reporting that the ST 17 complex, that is, our subdivision B2 characterized by GBSi (Fig. 1), was closer to bovine allelic profiles, whereas the ST 19 complex, i.e., our subdivision A1α characterized by IS1548 (Fig. 1), was closer to allelic profiles initially described for S. agalactiae human isolates (2).
Acknowledgments
We thank Sandra Bourdon, Anne Bouvet, Christophe Burucoa, Julien Loubinoux, Christian Martin, Didier Tandé, Anne Tristan, and François Vandenesch for providing some of the isolates used in this study. We thank Nicola Jones for providing us with new allele numbers and new sequence type numbers. We are indebted to Alain Goudeau for critical reading of the manuscript. We also thank Alain Moreau and Pascal Vaudin for technical advice.
REFERENCES
- 1.Bidet, P., N. Brahimi, C. Chalas, Y. Aujard, and E. Bingen. 2003. Molecular characterization of serotype III group B-streptococcus isolates causing neonatal meningitis. J. Infect. Dis. 188:1132-1137. [DOI] [PubMed] [Google Scholar]
- 2.Bisharat, N., D. W. Crook, J. Leigh, R. M. Harding, P. N. Ward, T. J. Coffey, M. C. Maiden, T. Peto, and N. Jones. 2004. Hyperinvasive neonatal group B streptococcus has arisen from a bovine ancestor. J. Clin. Microbiol. 42:2161-2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bohnsack, J. F., A. A. Whiting, G. Martinez, N. Jones, E. E. Adderson, S. Detrick, A. J. Blaschke-Bonkowsky, N. Bisharat, and M. Gottschalk. 2004. Serotype III Streptococcus agalactiae from bovine milk and human neonatal infections. Emerg. Infect. Dis. 10:1412-1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dmitriev, A., A. Shen, X. Shen, and Y. Yang. 2004. ISSa4-based differentiation of Streptococcus agalactiae strains and identification of multiple target sites for ISSa4 insertions. J. Bacteriol. 186:1106-1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Glaser, P., C. Rusniok, C. Buchrieser, F. Chevalier, L. Frangeul, T. Msadek, M. Zouine, E. Couve, L. Lalioui, C. Poyart, P. Trieu-Cuot, and F. Kunst. 2002. Genome sequence of Streptococcus agalactiae, a pathogen causing invasive neonatal disease. Mol. Microbiol. 45:1499-1513. [DOI] [PubMed] [Google Scholar]
- 6.Gousset, N., A. Rosenau, P.-Y. Sizaret, and R. Quentin. 1999. Nucleotide sequences of genes coding for fimbrial proteins in a cryptic genospecies of Haemophilus spp. isolated from neonatal and genital tract infections. Infect. Immun. 67:8-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Granlund, M., F. Michel, and M. Norgren. 2001. Mutually exclusive distribution of IS1548 and GBSi1, an active group II intron identified in human isolates of group B streptococci. J. Bacteriol. 183:2560-2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Granlund, M., L. Oberg, M. Sellin, and M. Norgren. 1998. Identification of a novel insertion element, IS1548, in group B streptococci, predominantly in strains causing endocarditis. J. Infect. Dis. 177:967-976. [DOI] [PubMed] [Google Scholar]
- 9.Hacker, J., and J. B. Kaper. 2000. Pathogenicity islands and the evolution of microbes. Annu. Rev. Microbiol. 54:641-679. [DOI] [PubMed] [Google Scholar]
- 10.Horodniceanu, T., D. H. Bouanchaud, G. Bieth, and Y. A. Chabbert. 1976. R plasmids in Streptococcus agalactiae (group B). Antimicrob. Agents Chemother. 10:795-801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jones, N., J. F. Bohnsack, S. Takahashi, K. A. Oliver, M. S. Chan, F. Kunst, P. Glaser, C. Rusniok, D. W. Crook, R. M. Harding, N. Bisharat, and B. G. Spratt. 2003. Multilocus sequence typing system for group B streptococcus. J. Clin. Microbiol. 41:2530-2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kazazian, H. H., Jr. 2004. Mobile elements: drivers of genome evolution. Science 303:1626-1632. [DOI] [PubMed] [Google Scholar]
- 13.Kong, F., S. Gowan, D. Martin, G. James, and G. L. Gilbert. 2002. Serotype identification of group B streptococci by PCR and sequencing. J. Clin. Microbiol. 40:216-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luan, S. L., M. Granlund, and M. Norgren. 2003. An inserted DNA fragment with plasmid features is uniquely associated with the presence of the GBSi1 group II intron in Streptococcus agalactiae. Gene 312:305-312. [DOI] [PubMed] [Google Scholar]
- 15.Mahillon, J., and M. Chandler. 1998. Insertion sequences. Microbiol. Mol. Biol. Rev. 62:725-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mahillon, J., C. Leonard, and M. Chandler. 1999. IS elements as constituents of bacterial genomes. Res. Microbiol. 150:675-687. [DOI] [PubMed] [Google Scholar]
- 17.Michel, F., and J. L. Ferat. 1995. Structure and activities of group II introns. Annu. Rev. Biochem. 64:435-461. [DOI] [PubMed] [Google Scholar]
- 18.Rubens, C. E., L. M. Heggen, and J. M. Kuypers. 1989. IS861, a group B streptococcal insertion sequence related to IS150 and IS3 of Escherichia coli. J. Bacteriol. 171:5531-5535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sawyer, S. A., D. E. Dykhuizen, R. F. DuBose, L. Green, T. Mutangadura-Mhlanga, D. F. Wolczyk, and D. L. Hartl. 1987. Distribution and abundance of insertion sequences among natural isolates of Escherichia coli. Genetics 115:51-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schneider, D., and R. E. Lenski. 2004. Dynamics of insertion sequence elements during experimental evolution of bacteria. Res. Microbiol. 155:319-327. [DOI] [PubMed] [Google Scholar]
- 21.Sellin, M., S. Hakansson, and M. Norgren. 1995. Phase-shift of polysaccharide capsule expression in group B streptococci, type III. Microb. Pathog. 18:401-415. [DOI] [PubMed] [Google Scholar]
- 22.Sellin, M., C. Olofsson, S. Hakansson, and M. Norgren. 2000. Genotyping of the capsule gene cluster (cps) in nontypeable group B streptococci reveals two major cps allelic variants of serotypes III and VII. J. Clin. Microbiol. 38:3420-3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sitnikova, T., A. Rzhetsky, and M. Nei. 1995. Interior-branch and bootstrap tests of phylogenetic trees. Mol. Biol. Evol. 12:319-333. [DOI] [PubMed] [Google Scholar]
- 24.Spellerberg, B. 2000. Pathogenesis of neonatal Streptococcus agalactiae infections. Microbes Infect. 2:1733-1742. [DOI] [PubMed] [Google Scholar]
- 25.Spellerberg, B., S. Martin, C. Franken, R. Berner, and R. Lutticken. 2000. Identification of a novel insertion sequence element in Streptococcus agalactiae. Gene 241:51-56. [DOI] [PubMed] [Google Scholar]
- 26.Tamura, G. S., M. Herndon, J. Przekwas, C. E. Rubens, P. Ferrieri, and S. L. Hillier. 2000. Analysis of restriction fragment length polymorphisms of the insertion sequence IS1381 in group B streptococci. J. Infect. Dis. 181:364-368. [DOI] [PubMed] [Google Scholar]
- 27.Tettelin, H., V. Masignani, M. J. Cieslewicz, J. A. Eisen, S. Peterson, M. R. Wessels, I. T. Paulsen, K. E. Nelson, I. Margarit, T. D. Read, L. C. Madoff, A. M. Wolf, M. J. Beanan, L. M. Brinkac, S. C. Daugherty, R. T. DeBoy, A. S. Durkin, J. F. Kolonay, R. Madupu, M. R. Lewis, D. Radune, N. B. Fedorova, D. Scanlan, H. Khouri, S. Mulligan, H. A. Carty, R. T. Cline, S. E. Van Aken, J. Gill, M. Scarselli, M. Mora, E. T. Iacobini, C. Brettoni, G. Galli, M. Mariani, F. Vegni, D. Maione, D. Rinaudo, R. Rappuoli, J. L. Telford, D. L. Kasper, G. Grandi, and C. M. Fraser. 2002. Complete genome sequence and comparative genomic analysis of an emerging human pathogen, serotype V Streptococcus agalactiae. Proc. Natl. Acad. Sci. USA 99:12391-12396. [DOI] [PMC free article] [PubMed] [Google Scholar]