Abstract
pSM19035 of the pathogenic bacterium Streptococcus pyogenes is a low-copy-number plasmid carrying erythromycin resistance, stably maintained in a broad range of gram-positive bacteria. We show here that the ω-ɛ-ζ operon of this plasmid constitutes a novel proteic plasmid addiction system in which the ɛ and ζ genes encode an antitoxin and toxin, respectively, while ω plays an autoregulatory function. Expression of toxin Zeta is bactericidal for the gram-positive Bacillus subtilis and bacteriostatic for the gram-negative Escherichia coli. The toxic effects of ζ gene expression in both bacterial species are counteracted by proper expression of ɛ. The ɛ-ζ toxin-antitoxin cassette stabilizes plasmids in E. coli less efficiently than in B. subtilis.
Bacterial plasmids are generally inherited in a very stable manner and independently of the cell chromosome. The specific mechanisms of stable plasmid maintenance have been studied mainly for plasmids replicating in gram-negative bacteria (23, 44). In the majority of high-copy-number plasmids, the copy number and cell division control in combination with the multimer resolution system ensure a very low frequency of plasmid loss. For low-copy-number plasmids, mechanisms exist which enable their maintenance during cell growth in nonselective conditions. While the active partitioning process precisely distributes plasmid copies to each daughter cell at division (33, 64), plasmid addiction systems (also called toxin-antitoxin TA cassettes) kill or reduce the growth of plasmid-free descendant cells (1, 35). The molecular basis of the postsegregational killing (PSK) requires the existence of at least two plasmid genes: one specifying a stable toxic agent and another coding for an unstable factor which prevents the lethal action of the toxin. While the toxin is always a protein, the antidote is either antisense RNA (which inhibits the translation of toxin mRNA) or a protein (35). Many such systems and their chromosomal analogues have been described for different gram-negative bacteria (6, 24, 28, 45). Antisense RNA-regulated stabilization systems constitute a well-conserved group called the hok-sok family (the name reflecting the functions of the host killing and suppression of killing genes from plasmid R1) (22).
Some common features can be indicated for proteic plasmid addiction systems (PPAS): organization in operons, autoregulation, formation of the antidote-toxin complex, and different decay rates of the two proteins involved (23, 29, 67). In contrast to the hok-sok family, there is no significant sequence similarity among PPAS genes. The specific mechanisms leading to the noxious effects of the toxin are known for few systems only. The first identified and best understood is ccd of Escherichia coli F factor (34). The CcdB toxin is a gyrase poison able to bind the free GyrA subunit and to trap cleaved DNA-gyrase complex, leading to the induction of SOS response and subsequent cell death (3). Gyrase is also the target for the ParE toxin of the broad-host-range RK2 plasmid parDE system (37), although the precise mechanism of its action has not been elucidated. The Kid toxin of the R1 plasmid specifically inhibits DnaB-dependent replication (50). Recent investigation showed that PemK toxin, the R100 plasmid homologue of Kid, is a sequence-specific endoribonuclease (65). These toxins have homologues, both sequence and functional, on many prokaryotic chromosomes also involved in RNA turnover (RelE [46], MazF [66]). The long known Doc toxin of the P1 prophage phd/doc system seems to act on translation via an interaction with mazEF (30). Recent reviews concerning the proteic plasmid stabilization systems in connection with chromosomally encoded TA cassettes suggest their possible role in general stress response, bacterial apoptosis, genome shuffling, or cell cycle arrest (12, 31).
Much less is known about the stable plasmid inheritance in gram-positive bacteria. Small replicons do not use any specific mechanism and rely on their high copy number. Multimer resolution systems, contributing to the efficient random plasmid distribution, are frequent. An antisense RNA-regulated stability determinant has been found in Enterococcus faecalis. This cassette, designated par, stabilizes the pAD1 plasmid and has no sequence homology to the hok-sok family (62). The 400-nt-long par region encodes two small, convergently transcribed RNAs (210-nt-long RNA I and 65-nt-long RNA II), with 33 codons for the fst (faecalis plasmid-stabilizing toxin) peptide in the longer RNA I. The smaller, short-lived RNA II shows a high degree of complementarity to RNA I and prevents fst translation. The Fst peptide alters the cell membrane, inhibits macromolecular synthesis, and affects cell division (63).
A proteic stability system in gram-positive bacteria has recently been described for the pRUM plasmid from a clinical isolate of Enterococcus faecium (25). The axe-txe cassette codes for two small proteins able to stabilize plasmids in a broad spectrum of bacteria. Induction of txe expression causes the inhibition of growth and cell division in E. coli which can be alleviated by simultaneous expression of axe. Interestingly, the antitoxin Axe is able to interact with, and counteract to some extent, the noncognate toxin YoeB of the E. coli axe-txe homologue. The Axe antitoxin shares 24% sequence identity with antitoxin YefM.
The most extensively studied group of large plasmids replicating in gram-positive bacteria is the inc18 family (4) consisting of pAMβ1, pIP501, and pSM19035. Despite their low copy number, members of this family are stably kept in their hosts; hence, they should use a specific mechanism(s) assuring their stable maintenance.
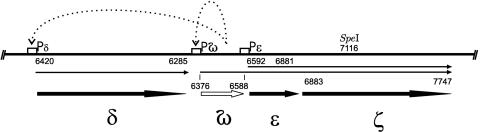
Most of the information on the mechanisms of the stable maintenance of plasmids from the inc18 family comes from studies on pSM19035 and its derivatives. A very intriguing property of this plasmid is the presence of extraordinarily long inverted repeats comprising over 80% of the plasmid genome (2). Two regions (SegA and SegB) involved in the plasmid stable maintenance have been identified (9). These regions are present in the repeated region; hence, the stable maintenance functions are duplicated in pSM19035 (11). The SegA region encodes a site-specific recombinase (gene β) which, by resolving plasmid oligomers into monomers, maximizes random plasmid segregation (10, 48) The SegB region assures better-than-random plasmid segregation. It consists of four genes organized into two transcription units: δ and ω-ɛ-ζ (15) (Fig. 1). Although the protein product of gene δ reveals significant homology to a family of ATPases involved in active plasmid partitioning, most of the stabilization function relies on the remaining part of the SegB region (9).
FIG. 1.
The SegB region of pSM19035 plasmid. Gene products are represented by thick arrows, transcripts by thin arrows, and promoters by the letter P. Dashed lines symbolize the regulatory circuits exerted by Omega protein (15). The nucleotide coordinates, shown as numbers above the arrows, correspond to the pBT233 plasmid sequence (EMBL X64695). The unique SpeI restriction site, used for the construction of truncated ζ, is shown.
Initial analysis (9) of the segregational stability of shortened variants of the pSM19035-derived plasmid pBT233 revealed that region SegB, encompassing genes δ, ɛ, and ζ, is responsible for stable plasmid maintenance. Recently, it was found that the SegB region also contains the gene ω which is part of an operon together with the ɛ and ζ genes. The product of the gene ω acts as a repressor of several transcription units in pSM19035, including the ω-ɛ-ζ operon (15). All the pBT233 derivatives previously tested for segregational stability contained the ω gene.
Previous analysis also reports the use of a plasmid which contains only genes δ and ζ and is segregationally stable (pBT348) (9). Nucleotide sequencing revealed that in pBT348, the intended inactivation of the ɛ gene at the SnaBI site by Bal 31 erosion resulted in the removal of only a single codon from the ɛ open reading frame (ORF), and this most likely did not affect the functionality of the ɛ gene or its product. Further attempts to obtain, either in E. coli or in Bacillus subtilis, plasmids carrying the intact ζ gene without a functional ɛ gene were futile (our unpublished data).
The products of ɛ and ζ genes were shown to form, in vivo and in vitro, a stable ɛ2ζ2 heterotetramer (8), and its crystal structure was determined (43). Camacho et al. (8) showed that inhibition of transcription or translation (by the addition of specific antibiotics to the growth in rich medium of B. subtilis cells harboring a plasmid with the ω-ɛ-ζ operon) reduces the cellular level of epsilon antitoxin, leading to the loss of cell viability.
In this paper, we characterize the functioning of the ω-ɛ-ζ operon as a stabilizing cassette in B. subtilis and E. coli cells.
MATERIALS AND METHODS
Bacterial strains, media, and culture conditions.
All bacterial strains and plasmids used in this study are listed in Table 1. Bacteria were grown in Luria-Bertani (LB or LBA) medium and minimal M9 or SMM medium (51, 56) supplemented, when necessary, with the appropriate antibiotics at the following concentrations (μg/ml): ampicillin (Amp) at 100 or 30; spectinomycin (Spc) at 20; kanamycin (Km) at 40; erythromycin (Em) at 150 for E. coli and 0.1 to 5 for B. subtilis; chloramphenicol (Cm) at 50 for E. coli and 5 for B. subtilis.
TABLE 1.
Plasmids and strains used in this study
Position numbers in parentheses correspond to the coordinates of the pBT233 plasmid sequence (EMBL X64695). Restriction enzyme recognition sequences are underlined.
Bacterial cultures were grown at a desired temperature (30°C, 37°C, or 41°C) in specially designed glass flasks (equipped with a 0.18-mm side arm) enabling direct measurement of optical density at 600 nm (OD600) using a Kuevetten test spectrophotometer (Dr Lange). This made it possible to work with a constant culture volume. These flasks ensure good aeration thanks to additional invaginations at the bottom.
DNA manipulations and transformation.
Purified plasmid DNA was prepared using the alkaline lysis procedure (51) or was obtained using Nucleobond AX (Macherey Nagel), the QIAGEN plasmid kit, or the Wizard Plus Minipreps DNA purification system (Promega) following the manufacturer's instructions. B. subtilis chromosomal DNA was isolated as previously described (7). Restriction and DNA-modifying enzymes were used according to the supplier's instructions.
Transformation of E. coli was performed according to the standard calcium chloride method or by electrotransformation with a Gene-Pulser (Bio-Rad) (51).
Electrotransformation of B. subtilis with plasmid DNA was performed as described previously (5), and transformation of competent B. subtilis cells with chromosomal or ligated DNA was done according to the method described by Rottlander and Trautner (49).
Chromosomal integration of a DNA fragment carrying the ɛ or ζ gene fused to the xylose-inducible promoter was achieved by transforming competent B. subtilis cells with the plasmid pX-ɛ or pX-ζ. Integration occurred via double crossover between two parts of amyE locus sequences, flanking the expression cassette present in pX and the homologous sequences in the host chromosome. The unique BamHI site downstream of an efficient synthetic ribosome binding site (RBS) sequence was used for ɛ or ζ gene cloning. Transformants were selected on LBA plates containing Cm (5 μg/ml).
Transfer of the recA4 mutation from B. subtilis YB1015 to YB886 strain.
The YB886 strain carrying the ɛ gene integrated into its chromosome (YBE01) was made rec negative by the “congression” technique (19). This technique was necessary because the recA4 trait cannot be selected for directly. A mixture of chromosomal DNA from the recA4 strain YB1015 and plasmid pIL253 (ratio, 10:1, wt/wt) was used for transformation of competent cells, and plasmid transformants were selected on LBA plates supplemented with 5 μg/ml Em. Transformants were transferred onto LBA plates supplemented with methyl methanesulfonate (MMS) (260 μg/ml) and parallel on master LBA plates. Colonies not growing on MMS-LBA plates were taken from the master plates and restreaked onto an MMS-LBA plate to confirm the MMS susceptibility. The MMS-sensitive bacteria were then cultured for several generations in the absence of any selection pressure in order to “cure” the pIL253 plasmid. The transfer of the recA allele was confirmed by comparison of the ability to resume the growth after the UV treatment of these strains to the parental one (with control of growth of the correspondent RecA YB1015 and RecA+ YB886 strains).
Plasmid stability assay.
The apparent plasmid stability was determined as follows. Cells (E. coli or B. subtilis) containing plasmid to be tested were grown overnight in suitable conditions (30°C or 37°C) in liquid media supplemented with an appropriate antibiotic. Five microliters of overnight culture was transferred into 5 ml of fresh, antibiotic-free medium. After 12 h of incubation, the culture was diluted in the same way and left to grow again. Each 12-h cycle (about 10 generations) was repeated until 80 or 100 generations of growth under nonselective conditions were reached. Every 10 generations, dilutions of cultures were plated on nonselective LB agar. After incubation, colonies were examined for resistance to the given antibiotic by replica plating on selective LBA. The percentage of plasmid-free cells was estimated from the ratio of antibiotic-sensitive colonies to all growing colonies.
The loss frequency (LF) values were calculated according to the method of Gerdes et al. (21).
Preparation of cell extracts.
Cell extracts were prepared according to the method of Silhavy et al. (52). E. coli TG1(pCUE3, pCUZ2) was grown in LB medium supplemented with appropriate antibiotics until the OD600 reached 0.2 and then divided into two portions. To one of them, isopropyl-β-d-thiogalactopyranoside (IPTG; 1 mM) was added. Both cultures were grown at 30°C. Samples were taken every 1 h, and the harvested cells were mixed with 2× loading buffer and boiled for 5 min.
B. subtilis YBZ01 was grown in SMM medium supplemented with 0.8% fructose and Cm (5 μg/ml) to the mid-log phase. Then, the culture was divided in two portions and to one, 0.5% d-xylose was added. Suitable aliquots were taken after 1, 2, and 3 h. Samples were incubated at 37°C with 2 mg/ml of lysozyme for 30 min before boiling.
Western blot analysis.
Samples of 10 μl were run on sodium dodecyl sulfate (SDS)-12% polyacrylamide gels and electroblotted as described previously (51). Anti-Zeta antibodies (1:5,000 dilution), kindly provided by J. C. Alonso, were used as the primary antibody. Anti-rabbit class II alkaline phosphatase-conjugated antibody (1:7,500 dilution) detection was performed using nitroblue tetrazolium (NBT) and 5-bromo-4-chloro-3-indolylphosphate (BCIP). The chromatic reaction was stopped by the addition of EDTA when the intensity of the band corresponding to purified Zeta reached its maximum. Band intensity on Western blots was estimated using ImageQuant (Molecular Dynamics) or Phoretix1D Plus NonLinearDynamics (Photometrix).
Microscopic observations.
DAPI (4′,6′-diamidino-2-phenylindole) staining of fixed bacterial cells was carried out essentially as described by Hiraga et al. (32). After methanol fixation, washing with water, and drying (the treatment with poly-l-lysine was omitted), 10 μl of DAPI solution (5 μg/ml of saline) in combination with a small amount of ethidium bromide was dropped on the samples, which were then covered with a glass coverslip and examined immediately.
Vital staining of bacterial cells was performed using a 5-μg/ml DAPI solution. The preparation method for fluorescence microscopy with immobilization and spreading of cells in one focal plane was adopted from the method of Van Helvoort and Woldringh (60). Four microliters of cell suspension was spotted on a “microslab” made in two-layer 2% agarose using a siliconized coverslip (22 by 22 mm).
Preparations were studied under a Nikon Microphot-SA fluorescence microscope using a Plan Apo 60×/1.40 or 100×/1.40 lens in combination with Nomarski contrast. Images were captured using 100 ASA Tmax Kodak film or a cooled charge-coupled device (CCD) CH350A camera and processed with Lucia Laboratory Imaging Ltd. software.
Cotransormation assay.
An approximately equimolar mixture of plasmids was used for bacterial transformation. After transformation, equal volumes of bacterial suspension were plated separately on media supplemented with the appropriate antibiotic selecting for one of the two plasmids only. Transformants from both types of plates were then checked for the presence of the other plasmid by replica plating on the medium supplemented with the antibiotic selecting for the other plasmid.
RESULTS
Determination of the minimal gene set from the SegB region of pSM19035 sufficient for stable plasmid maintenance.
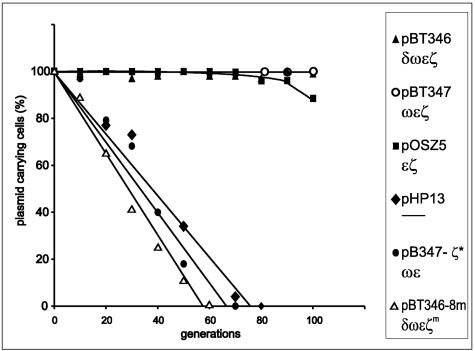
To determine the minimal set of genes from the SegB region of pSM19035 sufficient for the stable maintenance of heterologous plasmid, we tested several derivatives containing various combinations of genes from this region for segregation stability in recA mutant B. subtilis cells (Fig. 2). All studied plasmids were constructed using the same pHP13 replicon known to be unstable in this strain (5). Removal of the gene δ (pBT347) (see reference 9) does not affect plasmid stability. Also, inactivation of the gene ω (pOSZ5) (I. Sitkiewicz, unpublished) has very little effect on stable plasmid maintenance. This plasmid is stably kept for about 90 generations. However, upon further growth in the absence of selection pressure, plasmid-free cells appear. On the other hand, plasmids pBT346-8m and pBT347-ζ* (carrying a mutated or truncated ζ gene, respectively) (Table 1), encoding an inactive product, are not segregationally stable. Both plasmids mentioned above were able to be transformed into E. coli cells without an active ɛ gene, proving the nontoxicity of ζ genes present on them. These data and the previous observations on the inability to inactivate the ɛ gene when the intact ζ gene is present indicate that the minimal set of genes required for stable plasmid maintenance can be narrowed down to the gene pair ɛ and ζ.
FIG. 2.
Maintenance of plasmid pHP13 derivatives carrying various combinations of genes from pSM19035 SegB region. B. subtilis strain YB1015 carrying one of the following plasmids: pHP13 (vector), pBT346 (δωɛζ), pBT346-8m (δωɛ, mutated ζ), pBT347 (ωɛζ), pBT347- ζ* (ωɛ, truncated ζ), or pOSZ5 (ɛζ) was continuously grown in LB at 37°C without antibiotic selection for 100 generations. Plasmid stability was determined as the percentage of erythromycin-resistant colonies, as described in Materials and Methods. The data shown represent mean values of three experiments.
The Zeta protein is a toxin.
To study the effects of Zeta protein on B. subtilis cells, we used the bifunctional, integrative vector pX (38) carrying the xyl promoter-repressor gene cassette for construction of the strain overproducing Zeta protein. The cassette originates from the Bacillus megaterium operon for xylose utilization, and in the pX vector, it is sandwiched between the 5′ and 3′ ends of the amyE locus. The replication origin and the ampicillin resistance gene from pBR322 present on pX allow genetic manipulations in E. coli. We constructed the B. subtilis strain overproducing Zeta protein using this system by previous preparation of the expression cassette containing the cloned intact ζ gene sequence in E. coli (pX-ζ) (Table 1). The ligation mixture of the pX vector and correspondent DNA fragment with intact ζ gene was introduced into E. coli DH5α competent cells already containing the pACE1 plasmid (Table 1) as the yielding source of ɛ. Since the ζ gene DNA inserted into the pX vector was generated by PCR, the correctness of the ζ sequence was checked by sequencing. The functionality of the ζ gene (in terms of toxicity of the Zeta protein product toward E. coli cells) was verified in a biological cotransformation assay. The plasmid DNA mixture isolated from E. coli(pACE1/pX-ζ) was used to transform plasmidless E. coli competent cells selecting for pX-ζ plasmid. The presence of the ɛ carrying plasmid in these transformant cells was confirmed (data not shown).
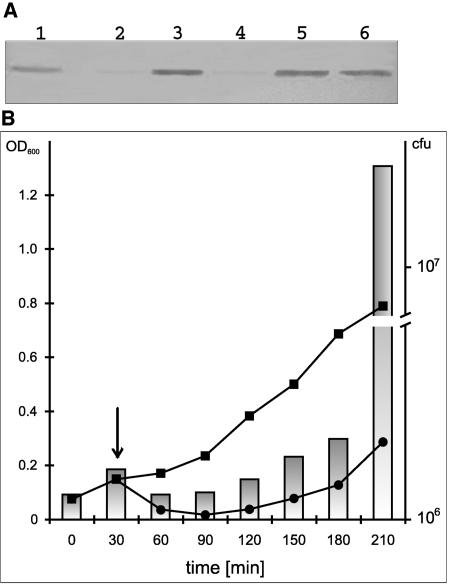
Prior to manipulations of the ζ gene in B. subtilis, a recipient strain was constructed to carry the ɛ gene on a plasmid. The respective plasmid, pATE1 (Table 1), based on the bifunctional pAT18 vector, is replicating and maintained at about five copies in gram-positive bacteria. We used the YB886(pATE1) strain for the integration of the expression cassette from the pX-ζ plasmid to give YBZ01 (Table 1). In this strain, the ability to induce expression of the ζ gene fused to the xylA promoter was analyzed by Western immunoblotting. Clearly, the Zeta protein is produced after induction of the xylA promoter by xylose (Fig. 3). One hour of incubation after the addition of the inducer was sufficient for full induction (Fig. 3, lanes 6 and 8). Similarly as in the control (Fig. 3, lanes 1 and 2), there was no detectable protein Zeta band in samples prepared from uninduced cells (Fig. 3, lanes 5 and 7).
FIG. 3.
Immunodetection of Zeta protein overexpressed in B. subtilis YBZ01 by Western blot analysis. The lysates were prepared and analyzed as described in Materials and Methods. Samples of cultures of YBU01(WT) and YBZ01(xylA-ζ) grown in SMM at 37°C were removed before and after xylose induction. Protein extracts were run on an SDS-12% polyacrylamide gel, and Western blotting was performed using anti-Zeta antibodies. Lanes: 1, extract of uninduced cells of YB886::xylA; 2, extract of xylose-induced cells of YB886::xylA, after 1 h; 3, Rainbow molecular mass standard; 4, 10 ng of purified Zeta protein; 5 and 7, extract of uninduced cells of YB886::xylA-ζ after 1 h and 2 h, respectively; 6 and 8, extract of xylose-induced cells of YB886::xylA-ζ after 1 h and 2 h, respectively.
The effects of Zeta protein overproduction on growing B. subtilis cells.
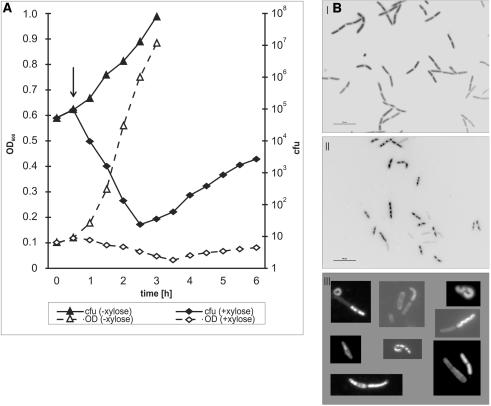
We used the B. subtilis YBZ01 strain expressing the ζ gene to show the influence of the Zeta protein on cell growth and viability. During the classical growth experiment, optical densities of the culture subjected to the induction of Zeta protein production were successively measured and the number of CFU at the same time was also assayed (Fig. 4A).
FIG. 4.
Effects of Zeta overexpression on B. subtilis YBZ01 cells. (A) Inhibition of culture growth after the induction of Zeta protein overproduction. Bacterial culture was divided into two, and after 0.5 h, 0.5% xylose was added (arrow) to one of the subcultures (open diamonds); the other was cultivated in the same conditions but in the absence of xylose (triangles). Continuous lines represent viable counts and dashed lines optical densities. The graphs represent a typical experiment. (B) DAPI-stained cells of B. subtilis YBZ01. Samples were taken from the culture induced for Zeta overproduction at an OD600 of 0.3. Morphology of cells was observed directly by fluorescence using vital DAPI staining and differential interference contrast. Panels: I, without induction of the Zeta protein overproduction; II, 2 h after induction of the Zeta protein overproduction; III, examples of abnormal cells found in xylose-induced YBZ01 culture. The same volume (4 μl) of cultures was spotted. Magnification, ×600; bar, 10 μm. Supplementary material is available at http://www.ibb.waw.pl/zielenkiewicz-jbac187
Very soon after the inducer addition, the culture ceased to grow; instead, a gradual drop in OD600 was observed for 3.5 h and then a slight increase. In contrast, control (uninduced) culture grew rapidly, showing a typical log-rate growth up to saturation. As determined by the CFU assay, after the induction of Zeta protein production, the number of viable cells dropped by several orders of magnitude. This indicates massive cell killing by the Zeta protein.
Figure 4B shows that after xylose addition (4BII and III), the number and morphology of the cells change(s) compared to uninduced cells (4BI). Those from the culture induced for Zeta protein production become statistically significantly smaller: shorter and finer (3.89 ± 1.13 μm and 0.98 ± 0.15 μm; mean value of 350 measurements) in comparison to B. subtilis fast-growing cells (6.11 ± 0.15 μm and 1.21 ± 0.11 μm, respectively). Some cells have a particular (diverse) shape and often are not stained with DAPI. The number of visible cells is decreasing with time of the Zeta overproduction (about 15% after 2 h and 80% after 4 h; the latter not shown), in accordance with the observed drop in CFU values (Fig. 4A). In the case of the control strain YB886::pX, the cells were uniformly stained, with atypical forms practically absent.
Preliminary characterization of B. subtilis cells which survived Zeta overproduction.
As can be seen in Fig. 4A, prolonged incubation of the culture subjected to induction of Zeta protein production leads to growth recovery of the very few surviving cells. In the course of several growth experiments, about 100 colonies of YBZ01 selected at the time of the lowest OD600 due to the killing by overproduced Zeta were collected on xylose-supplemented LBA. The ability of these cells to grow in the presence of xylose can be explained by any of the following phenomena: 1—the inability of those cells to overproduce (or produce at all) Zeta, due to mutations in the xylA promoter or in genes responsible for xylose uptake; 2—mutations within the ζ gene that render the protein product inactive; 3—mutations in the gene(s) responsible (directly or indirectly) for the production of the target for Zeta action.
An analysis of the ζ gene from six “survivors” showed that in all cases, the ζ gene sequence had been altered. In three cases, a deletion of one A in the AAAAAAA tract (which begins at the 78-nt position) resulting in a frameshift and the stop codon formation after 51 codons were found. Two of them showed additional changes: insertion of G after A32 and A111C substitution in one case and A248G substitution in the second one. The A248G mutation was also found as a single one (but with no immunologically detectable level of the Zeta protein). The deletion of A492, also leading to the formation of the stop codon (after next three triplets), was found in two clones. With the exception of the A492 deletion, all analyzed mutations localize in the N-terminal part of the ζ gene and similarly to the mutants described by Meinhart et al. (43), their correspondent sequences could be cloned into E. coli vectors without ɛ gene expression.
Εpsilon overproduction abolishes stable maintenance of plasmids carrying the ω-ɛ-ζ operon.
In order to examine the biological effects of an excess of Epsilon on the maintenance of plasmids carrying the ω-ɛ-ζ operon, we constructed a B. subtilis strain overexpressing the ɛ gene. The expression cassette xylR-xylA with the ɛ gene was inserted into the B. subtilis chromosome via the shuttle integration pX-derivative plasmid pX-ɛ (Table 1). The resulting new strain, YB886::xylA-ɛ, was designated YBE01 (Table 1) The presence of the ɛ gene in this strain was checked by generating a PCR product of the correct size and restriction pattern, using chromosomal DNA as a template and PCR primers specific for the ɛ gene. Immunodetection of overproduced Epsilon protein in the xylA-ɛ integrants was not possible since no specific anti-Epsilon antibodies were available.
In order to prevent recombination between the homologous (ɛ and cat) genes located, respectively, on the chromosome of strain YB886::xylA-ɛ and the derivatives of the pHP13 vector used in the subsequent stable maintenance assay, prior to transformation with those derivatives, the strain YBE01 was converted to the RecA mutant (designated YUE01) (Table 1).
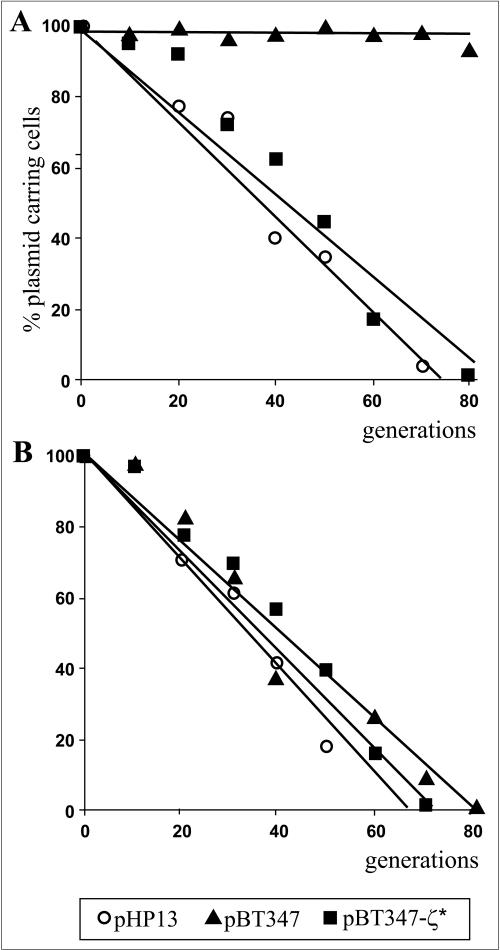
The following plasmids were introduced into the strain YUE01: vector pHP13, pBT347, and pBT347-ζ* (Table 1). Only pBT347 is stably maintained in B. subtilis recA cells (9). The stability assay for the above plasmids was performed in parallel for two types of cultures permanently grown under conditions of xylA promoter induction or repression, for 80 generations in LB medium without antibiotic selection. As shown in Fig. 5, the pHP13 vector was equally unstable in the YB886::xylA-ɛ strain independently of the induction of xylA-ɛ. In contrast, the pBT347 plasmid was stable when xylA-ɛ was repressed but completely lost after 80 generations of growth under the conditions of continuous excess of the Epsilon protein. A similar plasmid encoding a nonfunctional ζ gene (pBT347-ζ*) (Table 1) was lost with the same frequency from both cultures. This shows that under conditions of constant excess of Epsilon, the ω-ɛ-ζ operon fails to act as a plasmid stabilization system. According to the general principle of PSK functioning, the toxicity of poison will be manifested only by free (unbound by an antidote) protein. Constant production of antidote Epsilon probably titrates the Zeta toxin preventing its action even under condition of plasmid loss.
FIG. 5.
The influence on an excess of the Epsilon protein on plasmid stability in B. subtilis. Cultures of B. subtilis YUE01 (YB886::xylA-ɛ, recA4) containing plasmid pHP13 (circles), pBT347 (triangles), or pBT347-ζ* (squares) were grown in LB medium at 37°C for 80 generations without (A) or under (B) continuous xylose (0.5%) induction. Plasmid stability was determined as the percentage of erythromycin resistant colonies, as described in Materials and Methods. The results depicted show averages of at least three assays.
The use of Escherichia coli to study the functioning of the ɛ and ζ genes.
To study the functioning of the ɛ and ζ genes in gram-negative bacteria, we cloned the ɛ and ζ genes separately in Escherichia coli-compatible, moderate copy vectors with different selective markers: a pSC101 derivative pLDR8 (ɛ) and pACYC184 (ζ). Basing on the assumption that the functional ζ gene is toxic to bacterial cells, the ligation mixture, consisting of pACYC184 and a DNA fragment carrying the ζ gene (leading to the construction of pCUZ1; Cmr) (Table 1), was introduced into competent cells of E. coli TG1 already containing the ɛ gene carrying plasmid pCUE1 (Kmr, Table 1). Transformants were selected on LBA medium with chloramphenicol and later tested for kanamycin resistance. The presence of the two plasmids in transformant cells was confirmed by restriction analysis (data not shown). This experiment shows that the ɛ and ζ genes can be separated and can act in trans.
The different two-plasmid system described by Camacho et al. (8) was used to show the ability of the ɛ gene after the induction of its expression to protect E. coli cells from the toxicity of the ζ gene.
The results of the experiment described above and our earlier observations show that the ζ gene can be introduced only into bacterial cells when the ɛ gene is provided either in cis (e.g., plasmids pSM19035 and pBT346) or in trans (plasmids pACE1, pATE1, and pX-ζ). If so, after simultaneous transformation with two plasmids (one carrying the ɛ gene and the other carrying the ζ gene), selection for the plasmid carrying the ζ gene should allow the growth of only those cells which had also been transformed with the plasmid containing the ɛ gene.
To test this, we performed the cotransormation assay using a mixture of plasmids isolated from E. coli(pCUE1, pCUZ1). All Cm-resistant transformants selected for the presence of plasmid pCUZ1 were also resistant to Km (Table 2), the antibiotic marker of plasmid pCUE1. In contrast, only a small fraction (about 11%) of transformants selected for plasmid pCUE1 were also resistant to Cm, the antibiotic marker of plasmid pCUZ1. The percentage of double transformants in this case was approximately the same as in the control transformation with the vector pACYC184 used instead of pCUZ1. This means that a cell carrying a plasmid with the ζ gene is viable only when it also carries the ɛ gene. This observation can be further explored in the development of a specific biological assay for the functionality of the ζ gene.
TABLE 2.
Cotransformation frequency of plasmids carrying separated ɛ and ζ genesa
| Plasmid pair used | % Double transformant selection for:
|
||
|---|---|---|---|
| pCUE1 | pCUZ1 | pACYC184 | |
| pCUE1 and pCUZ1 | 11 | 100 | NA |
| pCUE1 and pACYC184 | 10 | NA | 16 |
About 100 ng of plasmid DNA mixture per 100 μl of competent E. coli TG1 was used. Equal volumes of the same sample were plated on selective media (containing Cm or Km). One hundred transformant colonies from each kind of selective plate were checked for the presence of the other plasmid by replica plating. The results represent averages of at least three independent experiments. NA, does not apply.
The use of a thermosensitive replicon.
The plasmid vector pLDR8 is thermosensitive in replication (17). Thus, cultivation of bacteria containing plasmids pCUE1 (pLDR8 derived) and pCUZ1 at elevated temperature would lead to a situation when only pCUZ1 would replicate, whereas the replication of pCUE1 should be arrested. We analyzed the consequences of the disturbance in the ɛ/ζ gene ratio obtained in this way for E. coli cells.
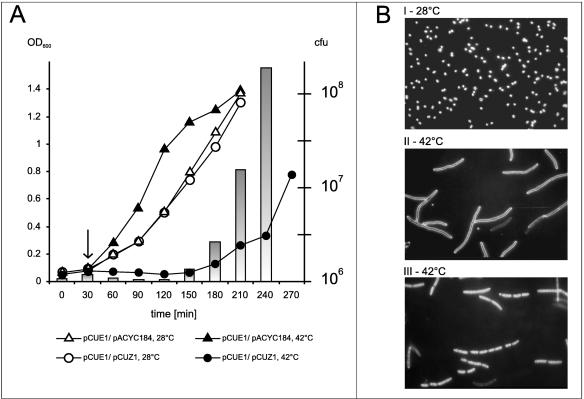
As shown in Fig. 6A, the growth curves of cells carrying the pCUE1 and pCUZ1 plasmid pair or pCUE1 and pACYC184, measured by OD readings, show no differences when the cells were grown at 28°C. The same was true for the CFU numbers determined for these cultures (data not shown). In contrast, at 42°C, while the control strain [E. coli(pCUE1, pACYC184)] grew very well with an exponential rate, cultures of cells containing the pCUE1 and pCUZ1 plasmids showed a lag in OD increase. After approximately 2 h, the cells began to grow, reaching the exponential rate about 1.5 h later. However, the numbers of CFU, determined for E. coli(pCUE1, pCUZ1) at the same time points as the OD measurement, were about two to four times lower during the period of the growth inhibition than the initial value. At the time of growth restoration, the number of CFU was increasing dramatically at a rate about twofold higher than the OD values measured for the same culture.
FIG. 6.
The effects of impaired stoichiometry between the ɛ and ζ genes in E. coli. (A) Growth of E. coli carrying pCUZ1 or pACYC184 after replication arrest of pCUE1. Overnight liquid cultures of E. coli TG1(pCUE1, pCUZ1) (circles) and TG1(pCUE1, pACYC184) (triangles) were cultured in M9 medium at 28°C under antibiotic (Km and Cm) selection, diluted 1:100 in fresh prewarmed medium supplemented with Cm, and allowed to grow at 28°C with aeration. After 30 min of incubation, each culture was divided into two portions: one was left to grow at 28°C (open symbols) and the other was incubated at 42°C (filled symbols). Cell growth was estimated every 30 min by measurement of optical density and the ability to form colonies on LBA. The arrow indicates the moment of temperature shift. Bars representing CFU are shown only for the culture of TG1(pCUE1, pCUZ1) grown at 42°C. (B) Morphology of cells after pCUE1 replication arrest. For microscopy, samples were taken 60 min after the temperature shift. Morphology of cells was observed directly by dark-field phase contrast (I and II) or by fluorescence using vital DAPI staining (III). Magnification, ×600.
During the growth experiment, systematic microscopic observations showed (Fig. 6B) that after the replication arrest of the pCUE1 plasmid, normal-sized cells became filamentous. The percentage of filaments increased rapidly in parallel with the decrease of CFU. When the culture restarted to grow, these filaments disappeared successively. The filaments were practically absent from the culture grown at 28°C, as well as from the control strain [E. coli(pCUE1, pACYC184)] growing at both the permissive and restrictive temperatures (data not shown). It seems reasonable to connect the changes in the OD versus CFU ratios for the E. coli(pCUE1, pCUZ1) strain grown at 42°C with the filamentation and subsequent disintegration of cell filaments.
Expression of epsilon counteracts the toxicity of Zeta.
The decrease in the ɛ gene dosage caused by the thermosensitive plasmid replication arrest results in a transient growth retardation and filament formation in cells carrying the ζ gene. This effect is most likely due to the concomitant decrease in the amount of the protein product specified by the ɛ gene. If so, a similar effect should be observed when the stoichiometry of the ɛ and ζ gene products is disturbed by the overproduction of Zeta.
To test this, we placed the ɛ and ζ genes separately in E. coli vectors in which their expression could be regulated. The first plasmid vector, pGB2 λ (SpcR) (Table 1), is based on a pSC101 derivative, containing the tandem pR and pL promoter artificial cassette together with the cI857 repressor gene from λ phage. The second plasmid vector was a low-copy-number pLEX5AA (Ampr) expression vector bearing the replication origin from the p15A plasmid and an artificial, LacI-regulated PA1-04/03 promoter (40) closely linked to an efficient ribosome binding site.
The resultant constructs (one, designated pCUE3, carrying the ɛ gene and the other, pCUZ2, carrying the ζ gene) (Table 1) were simultaneously introduced into competent E. coli TG1 cells. In the cotransformation assay, we proved that the presence of a functional ζ gene in a bacterial cell requires the presence of the ɛ gene as well. The Ampr and Spcr transformants were tested for growth upon temperature repression/induction of the λ promoters (30°C/41°C) and repression/induction of the PA1-04/03 promoter (no IPTG/1 mM IPTG). Under conditions of induction of ζ gene expression and simultaneous ɛ gene repression, no bacterial growth on agar plates was observed. In the reciprocal situation (i.e., ζ gene repression and ɛ gene induction), good growth of bacteria was visible.
Effects of overproduction of the Zeta protein.
To show that under conditions of ζ gene induction, there is an increase in the production of the Zeta protein, Western blotting analysis was performed for induced and uninduced E. coli(pCUE2, pCUZ3) cultures (Fig. 7A). As measured with the ImageQuant software, induction of the ζ gene results in an at least 10-fold increase in the Zeta protein amount after 1 h. Longer induction times do not lead to a further increase in Zeta protein production.
FIG. 7.
Effects of the overproduction of the Zeta protein on E. coli cells. (A) Immunodetection of Zeta protein in E. coli TG1 carrying the pCUE3 and pCUZ2 plasmid pair. Samples of TG1(pCUE3, pCUZ2) cultures grown in LB medium at 30°C without or in presence of IPTG (1 mM) were removed after 1, 2, and 3 h. Protein extracts were run on SDS-12% polyacrylamide gel and Western blotting was performed using anti-Zeta antibodies. Lanes: 1, 100 ng of purified Zeta protein; 2 and 4, extracts of uninduced cells after 1 h and 2 h, respectively; 3, 5, and 6, extracts of IPTG induced cell after 1 h, 2 h, and 3 h, respectively. (B) The effect of ζ gene overexpression on the growth of E. coli. TG1(pCUE3, pCUZ2) was cultured in M9 medium at 30°C until an OD600 of 0.1, divided into two portions, and IPTG (1 mM) was added to one (circles). Cell growth was estimated by measuring the optical density (line graphs) and the ability to form colonies on LBA without IPTG. The arrow indicates the moment of IPTG addition. Bars representing CFU are shown only for the IPTG-treated culture (logarithmic scale).
When cells containing plasmids pCUE3 and pCUZ2 are grown in liquid media, the induction of ζ gene expression with simultaneous ɛ gene repression leads to temporary growth inhibition (Fig. 7B). This inhibition lasts for about 2.5 h, and later, the growth is restored in spite of the presence of increased amounts of the Zeta protein in the cells (Fig. 7A, lane 6). The restoration of bacterial growth observed about 3 h after ζ gene induction (shown as 210 min in Fig. 7B) was much more profound when the CFU numbers were measured than in the case of OD values (3.5-fold versus 2-fold, respectively). Microscopic examination of the cultures revealed massive filamentation of bacterial cells during the 2.5-h period after ζ gene induction. After restoration of growth, the number of filaments decreased gradually (data not shown). Hence, also in this experiment, the difference between OD values and CFU numbers could be attributed to the formation and disintegration of filaments. From these experiments, we conclude that Zeta protein is responsible for the inhibition of growth and for filamentation of E. coli cells.
Cell filamentation is one of the phenomena related to the SOS response, a diverse set of physiological effects in E. coli cells exposed to DNA-damaging agents. If Zeta causes induction of the SOS system, one should expect an increased level of mutagenesis in a RecA+ strain. Comparison of the measurements of mutagenesis in strains proficient in SOS induction (recA+) and deficient in SOS induction (recA430) did not show any statistically significant differences (data not shown). The ability to invoke cell filamentation by the induction of ζ gene expression, both in the recA and the recA+ strain, was also confirmed. This suggests that cell filamentation upon expression of the ζ gene is not connected with the SOS response.
Stabilization of heterologous replicons by the ω-ɛ-ζ operon.
The ω-ɛ-ζ operon efficiently stabilizes segregationally unstable plasmids in B. subtilis (Fig. 2). As shown above using E. coli cells, the ζ gene could not be introduced into recipient cells unless the ɛ gene was also provided in cis or in trans. Moreover, overexpression of the ζ gene was toxic for the cells and this toxic effect was counteracted by overexpression of the ɛ gene. Taking this into account, it seems reasonable to expect that ɛ and ζ might also stabilize plasmids in E. coli.
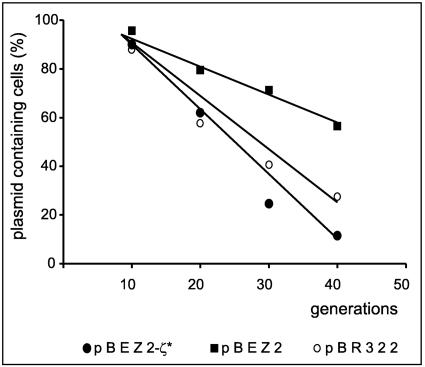
To test this possibility, the segregational stability of plasmid pBR322 carrying the entire ω-ɛ-ζ operon (plasmid pBEZ2) (Table 1) was determined. As a negative control, a derivative of pBR322 (pBEZ2-ζ*) (Table 1) with a nonfunctional ζ gene was used. The experiment was performed using a pcnB mutant of E. coli, strain BR5826 (Table 1). The pcnB mutation leads to a considerable reduction in the copy number of ColE1 plasmid-derived replicons (42), which in consequence lowers the segregational stability of these plasmids. The presence of the ω-ɛ-ζ operon only slightly improved the segregational stability of pBR322 (Fig. 8). This is evident both from the time course experiment and from the calculation of plasmid loss frequencies after 40 generations of growth without selection (4.8 × 10−2 versus 1.4 × 10−2). Similarly, only about twofold stabilization was observed for the mini-R1 derivative plasmid pOU82 (36) carrying the ω-ɛ-ζ operon (data not shown).
FIG. 8.
Stabilization of pBR322 plasmid by ω-ɛ-ζ operon in E. coli. E. coli BR5826 containing pBEZ2 (wild-type ω-ɛ-ζ operon; squares), pBEZ2-ζ* (operon with a nonfunctional ζ gene; circles) or pBR322 (vector; empty circles) was grown at 37°C in LB medium without antibiotic selection. Plasmid stability was determined as the percentage of ampicillin-resistant colonies, as described in Materials and Methods. The data shown represent mean values of three experiments.
DISCUSSION
In this study, we present evidence that the ω-ɛ-ζ operon of the streptococcal low-copy-number plasmid pSM19035 constitutes a novel PPAS, with ɛ and ζ encoding an antitoxin and a toxin, respectively. The genetic organization of PPASs is paradigmatic: the antitoxin and toxin are encoded by adjacent genes organized in an operon (15). The antitoxin usually exerts negative transcriptional autoregulation on the operon and, in many cases, the toxins act as corepressors of transcription (35). In this respect, the organization of the ω-ɛ-ζ operon is unique among the PPASs: neither the antidote-encoding gene ɛ nor the toxin-encoding gene ζ is involved in the transcriptional regulation of the ω-ɛ-ζ operon. Transcription of this operon is controlled by the third element of the system, the gene ω (15).
Autoregulation is believed to play an important role in proper PPAS functioning. For instance, the replacement of the promoter from the pTF-FC2 pas operon by the IPTG-inducible tac promoter strongly decreased the segregational stability of plasmids carrying the tac-pas cassette, irrespective of the presence of IPTG in the medium (55). Also, in the case of the ω-ɛ-ζ operon, inactivation of the autoregulatory gene ω affects to a detectable extent the plasmid segregational stability. When the maintenance of plasmids carrying either the wild-type operon or the one with mutated gene ω is compared (Fig. 2, pBT347 versus pOSZ5), both plasmids are stably kept in B. subtilis cells for 80 to 90 generations. After 100 generations, however, while 100% of cells still carry the plasmid pBT347, the loss of pOSZ5 becomes detectable, about 10% of cells become plasmid free.
The importance of the whole ω-ɛ-ζ operon in efficient stabilization of segregationally unstable plasmids in B. subtilis can be explained in two ways. (i) Gene ω is not directly involved in the ɛ-ζ-mediated killing-antikilling process, and its role in the proper PPAS functioning is restricted to autogeneous regulation of the ω-ɛ-ζ operon. As shown previously (16), it binds to the promoter of the entire operon as a dimer. It has been shown that in the absence of repression by Omega, the intensity of transcription from the ω promoter is increased about 40-fold (15). Hence, in the case of plasmid pOSZ5, mutation in the gene ω may result in the synthesis of increased amounts of the Epsilon and Zeta proteins. The presence of additional, nonphysiological amounts of Epsilon may titrate out to some extent the cellular protease that renders Epsilon unstable, and some molecules of this antidote may escape proteolytic degradation. As a consequence, this may lead to prolonged neutralization of the toxin and survival of some cells that had lost the plasmid.
(ii) Gene ω is directly involved in the killing-antikilling process, and the proper functioning of pSM19035-encoded PPAS requires complex formation by all three proteins, Epsilon, Zeta, and Omega. Since Epsilon and Zeta (without Omega) stabilize plasmids quite efficiently (Fig. 2, plasmids pOSZ5 versus pBT347-ζ* or the vector pHP13), the role of Omega would be to serve as an accessory protein that somehow enhances the action of Epsilon and Zeta. A three-component system of this type has been described for plasmid pTF-FC2 (54). In pTF-FC2, however, in agreement with the common features of PPAS, the autoregulatory functions are played by the antidote PasA and are enhanced in the presence of the toxin PasB. The accessory protein PasC plays no role in the autoregulation (55).
Although the second possibility cannot be ruled out, in view of the necessity for autoregulation in all PPASs on the one hand and the well-documented involvement of the ω gene in the autoregulation of the ω-ɛ-ζ operon on the other, the first possibility seems to be more likely.
The importance of the proper stoichiometry between the toxin Zeta and the antitoxin Epsilon has been clearly seen when replication of the vector used for ɛ cloning was arrested (Fig. 6). Also, note that the regulatory circuit of the ω gene in this two-plasmid system is broken.
The ω-ɛ-ζ cassette very efficiently stabilizes plasmids in B. subtilis and rather weakly in E. coli. There are several possibilities explaining the inability of this cassette to function properly in E. coli. Among the most probable ones, one may entertain the synthesis of a large excess of Epsilon or its increased stability. In either case, the biological effect would be manifested in a decreased ability of the ω-ɛ-ζ cassette to function as a PPAS.
The results of experiments presented in this work clearly indicate that the product of the ζ gene is a toxin. Zeta is composed of 287 amino acids; this means that it is much larger than other PPAS-encoded toxins, which usually are composed of about 100 amino acids (23). A homology search of the Zeta amino acid sequence against all protein sequences deposited in databases has not shown any significant identity to proteins of known function. The only meaningful amino acid motif present in Zeta is the Walker ATP/GTP binding motif. Since this motif is present in many families of proteins with diverse functions, the possible mode of Zeta action cannot be predicted from the homology searches. The structure of the inactive complex between the Zeta and Epsilon proteins has been solved by X-ray crystallography (43). No functional assignment of the Zeta protein can be made directly from its structure, except for fold similarity to phosphotransferases, which may suggest similar function. Neither Epsilon nor Zeta structure is similar to any of the known structures of other PPAS proteins.
The Zeta protein exerts a toxic effect on the gram-positive and gram-negative bacteria B. subtilis and E. coli, respectively, and also on the yeast Saccharomyces cerevisiae (data not shown). It should be mentioned, however, that the most extensively studied toxins are encoded by narrow-host-range plasmids such as F, P1, and R1. An interesting and as yet unanswered question may concern the spectrum of sensitive hosts for the toxin ParE from the very-broad-host-range plasmid RK2. An intriguing case in this respect is the chromosomally encoded PPAS relBE from E. coli and the spirochaetal chpK. Their toxins have been shown to exert a toxic phenotype in yeast cells (39, 47). The Kid toxin of the R1 plasmid parD system was demonstrated to affect yeast and even human cells (14). The recently described axe-txe segregational stability system of pRUM was shown to be functional not only in its Gram-positive host but also in Bacillus thuringiensis and the gram-negative E. coli (25). It needs to be added that homologues of these genes are widely distributed among prokaryotic genomes.
A comparison of the toxic effects exerted by Zeta in various microorganisms reveals several differences. In B. subtilis, the action of Zeta is strongly bactericidal. Microscopic examination of cells subjected to Zeta activity suggests some changes in cell morphology and massive cell lysis. A preliminary analysis of cells that survived Zeta overproduction indicates as a reason of their immunity mutations in the ζ gene rather than accumulation of host mutants. In E. coli, the activity of Zeta is bacteriostatic rather than bactericidal. A decrease in CFU numbers in cells subjected to the action of Zeta is mainly correlated with their tendency to form filaments. Since the ζ gene seems not to induce the SOS response in E. coli, the presence of filaments may suggest that in this bacterium, Zeta may act at the stage of cell division. E. coli cells exposed to Zeta for a prolonged time become insensitive to its activity. Contrary to what was observed for B. subtilis, when such insensitive cells are again subjected in their early log phase of growth to ζ gene overexpression, the toxic effects of Zeta are again clearly visible (data not shown). This means that only young, growing E. coli cells are susceptible to Zeta. In the yeast S. cerevisiae, the physiological effects of Zeta overproduction have not been studied yet.
The ɛ gene encodes the 90-amino-acid protein Epsilon that prevents the toxic action of Zeta. Also in the case of Epsilon, homology searches against the protein amino acid sequences deposited in databases have not brought any helpful results. There is a growing number of nucleotide sequences with homology to the epsilon gene, but in no case are the resultant proteins functionally described.
According to the general principle of PPAS action, the antidote should be less stable than the toxin. Camacho et al. (8) have estimated the in vitro and in in vivo Epsilon protein stability as <18 min in contrast to >60 min for protein Zeta. In agreement with these data, we observed that under conditions of excess of Epsilon produced from the B. subtilis chromosome, the ω-ɛ-ζ operon fails to act as a stabilization cassette (Fig. 5).
It is known that the antidote proteins are unstable because they are rapidly degraded by intracellular proteases. In E. coli, the antidote proteins CcdA (61), PemI/Kis (58), PasA (55), and RelB (26) are degraded by the protease Lon and the Phd antitoxin is cleaved by the ClpXP protease (41). Preliminary experiments with the use of B. subtilis protease-deficient mutants indicate an involvement of ClpP protease with the chaperone ClpX in degradation of the Epsilon antidote. Deciphering of the mechanism of Epsilon degradation in E. coli may help explain the observed differences in the efficiency of the ω-ɛ-ζ operon functioning.
Given the medical importance of the inc18 plasmid family, identification of the target of Zeta and unraveling its molecular mechanism of action are of great interest. Also, whether Zeta exhibits an identical or similar mode of action in all susceptible microorganisms remains to be elucidated. Regardless of our progress in solving these problems, with such a broad spectrum of sensitive microorganisms, the ζ gene might already be used in several applications, e.g., as a target gene in direct selection vectors, in containment control for diverse microbial hosts, or in the design of new antimicrobial agents.
Acknowledgments
This work was supported by KBN grant no. 6P04B00820.
REFERENCES
- 1.Aizenman, E., H. Engelberg-Kulka, and G. Glaser. 1996. An Escherichia coli chromosomal “addiction module” regulated by guanosine 3′,5′-bispyrophosphate: a model for programmed bacterial cell death. Proc. Natl. Acad. Sci. USA 93:6059-6063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Behnke, D., H. Malke, M. Hartmann, and F. Walter. 1979. Post-transformational rearrangement of an in vitro reconstructed group-A streptococcal erythromycin resistance plasmid. Plasmid 2:605-616. [DOI] [PubMed] [Google Scholar]
- 3.Bernard, P., and M. Couturier. 1992. Cell killing by the F plasmid CcdB protein involves poisoning of DNA-topoisomerase II complexes. J. Mol. Biol. 226:735-745. [DOI] [PubMed] [Google Scholar]
- 4.Brantl, S., D. Behnke, and J. C. Alonso. 1990. Molecular analysis of the replication region of the conjugative Streptococcus agalactiae plasmid pIP501 in Bacillus subtilis. Comparison with plasmids pAM beta 1 and pSM19035. Nucleic Acids Res. 18:4783-4790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bron, S. 1990. Molecular biological methods for bacillus. John Wiley & Sons Ltd., Chichester, England.
- 6.Brown, J. M., and K. J. Shaw. 2003. A novel family of Escherichia coli toxin-antitoxin gene pairs. J. Bacteriol. 185:6600-6608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burdett, V., J. Inamine, and S. Rajagopalan. 1982. Heterogeneity of tetracycline resistance determinants in Streptococcus. J. Bacteriol. 149:995-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Camacho, A. G., R. Misselwitz, J. Behlke, S. Ayora, K. Welfle, A. Meinhart, B. Lara, W. Saenger, H. Welfle, and J. C. Alonso. 2002. In vitro and in vivo stability of the epsilon2zeta2 protein complex of the broad host-range Streptococcus pyogenes pSM19035 addiction system. Biol. Chem. 383:1701-1713. [DOI] [PubMed] [Google Scholar]
- 9.Ceglowski, P., A. Boitsov, N. Karamyan, S. Chai, and J. C. Alonso. 1993. Characterization of the effectors required for stable inheritance of Streptococcus pyogenes pSM19035-derived plasmids in Bacillus subtilis. Mol. Gen. Genet. 241:579-585. [DOI] [PubMed] [Google Scholar]
- 10.Ceglowski, P., A. Boitsov, S. Chai, and J. C. Alonso. 1993. Analysis of the stabilization system of pSM19035-derived plasmid pBT233 in Bacillus subtilis. Gene 136:1-12. [DOI] [PubMed] [Google Scholar]
- 11.Ceglowski, P., and J. C. Alonso. 1994. Gene organization of the Streptococcus pyogenes plasmid pDB101: sequence analysis of the orf eta-copS region. Gene 145:33-39. [DOI] [PubMed] [Google Scholar]
- 12.Christensen, S. K., K. Pedersen, F. G. Hansen, and K. Gerdes. 2003. Toxin-antitoxin loci as stress-response-elements: ChpAK/MazF and ChpBK cleave translated RNAs and are counteracted by tmRNA. J. Mol. Biol. 332:809-819. [DOI] [PubMed] [Google Scholar]
- 13.Churchward, G., D. Belin, and Y. Nagamine. 1984. A pSC101-derived plasmid which shows no sequence homology to other commonly used cloning vectors. Gene 31:165-171. [DOI] [PubMed] [Google Scholar]
- 14.de la Cueva-Méndez, G., A. D. Mills, L. Clay-Farrace, R. Díaz-Orejas, and R. A. Laskey. 2003. Regulatable killing of eukaryotic cells by the prokaryotic proteins Kid and Kis. EMBO J. 22:246-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de la Hoz, A. B., S. Ayora, I. Sitkiewicz, S. Fernandez, R. Pankiewicz, J. C. Alonso, and P. Cegłowski. 2000. Plasmid copy-number control and better-than-random segregation genes of pSM19035 share a common regulator. Proc. Natl. Acad. Sci. USA 97:728-733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de la Hoz, A. B., F. Pratto, R. Misselwitz, C. Speck, W. Weihofen, K. Welfle, W. Saenger, H. Welfle, and J. C. Alonso. 2004. Recognition of DNA by ω protein from the broad-host range Streptococcus pyogenes plasmid pSM19035: analysis of binding to operator DNA with one to four heptad repeats. Nucleic Acids Res. 32:3136-3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Diederich, L., L. J. Rasmussen, and W. Messer. 1992. New cloning vectors for integration in the lambda attachment site attB of the Escherichia coli chromosome. Plasmid 28:14-24. [DOI] [PubMed] [Google Scholar]
- 18.Diederich, L., A. Roth, and W. Messer. 1994. A versatile plasmid vector system for the regulated expression of genes in Escherichia coli. BioTechniques 16:916-923. [PubMed] [Google Scholar]
- 19.Erickson, R. J., and J. C. Copeland. 1973. Congression of unlinked markers and genetic mapping in the transformation of Bacillus subtilis 168. Genetics 73:13-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Friedman, B. M., and R. E. Yasbin. 1983. The genetics and specificity of the constitutive excision repair system of Bacillus subtilis. Mol. Gen. Genet. 190:481-486. [DOI] [PubMed] [Google Scholar]
- 21.Gerdes, K., J. E. Larsen, and S. Molin. 1985. Stable inheritance of plasmid R1 requires two different loci. J. Bacteriol. 161:292-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gerdes, K., A. P. Gultyaev, T. Franch, K. Pedersen, and N. D. Mikkelsen. 1997. Antisense RNA-regulated programmed cell death. Annu. Rev. Genet. 31:1-31. [DOI] [PubMed] [Google Scholar]
- 23.Gerdes, K. 2000. Toxin-antitoxin modules may regulate synthesis of macromolecules during nutritional stress. J. Bacteriol. 182:561-572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gotfredsen, M., and K. Gerdes. 1998. The Escherichia coli relBE genes belong to a new toxin-antitoxin gene family. Mol. Microbiol. 29:1065-1076. [DOI] [PubMed] [Google Scholar]
- 25.Grady, R., and F. Hayes. 2003. Axe-Txe, a broad-spectrum proteic toxin-antitoxin system specified by a multidrug-resistant, clinical isolate of Enterococcus faecium. Mol. Microbiol. 47:1419-1432. [DOI] [PubMed] [Google Scholar]
- 26.Gronlund, H., and K. Gerdes. 1999. Toxin-antitoxin systems homologous with relBE of Escherichia coli plasmid P307 are ubiquitous in prokaryotes. J. Mol. Biol. 285:1401-1415. [DOI] [PubMed] [Google Scholar]
- 27.Haima, P., S. Bron, and G. Venema. 1987. The effect of restriction on shotgun cloning and plasmid stability in Bacillus subtilis Marburg. Mol. Gen. Genet. 209:335-342. [DOI] [PubMed] [Google Scholar]
- 28.Hayes, F. 1998. A family of stability determinants in pathogenic bacteria. J. Bacteriol. 180:6415-6418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hayes, F. 2003. Toxins-antitoxins: plasmid maintenance, programmed cell death, and cell cycle arrest. Science 301:1496-1499. [DOI] [PubMed] [Google Scholar]
- 30.Hazan, R., B. Sat, M. Reches, and H. Engelberg-Kulka. 2004. Postsegregational killing mediated by the P1 phage “addiction module” phd-doc requires the Escherichia coli programmed cell death system mazEF. J. Bacteriol. 183:2046-2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hazan, R., B. Sat, and H. Engelberg-Kulka. 2004. Escherichia coli mazEF-mediated cell death is triggered by various stressful conditions. J. Bacteriol. 186:3663-3669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hiraga, S., H. Niki, T. Ogura, C. Ichinose, H. Mori, B. Ezaki, and A. Jaffe. 1989. Chromosome partitioning in Escherichia coli: novel mutants producing anucleate cells. J. Bacteriol. 171:1496-1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hiraga, S. 1992. Chromosome and plasmid partition in Escherichia coli. Annu. Rev. Biochem. 61:283-306. [DOI] [PubMed] [Google Scholar]
- 34.Jaffé, A., T. Ogura, and S. Hiraga. 1985. Effects of the ccd function of the F plasmid on bacterial growth. J. Bacteriol. 163:841-849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jensen, R. B., and K. Gerdes. 1995. Programmed cell death in bacteria: proteic plasmid stabilization systems. Mol. Microbiol. 17:205-210. [DOI] [PubMed] [Google Scholar]
- 36.Jensen, R. B., E. Grohmann, H. Schwab, R. Diaz-Orejas, and K. Gerdes. 1995. Comparison of ccd of F, parDE of RP4, and parD of R1 using a novel conditional replication control system of plasmid R1. Mol. Microbiol. 17:211-220. [DOI] [PubMed] [Google Scholar]
- 37.Jiang, Y., J. Pogliano, D. R. Helinski, and I. Konieczny. 2002. ParE toxin encoded by the broad-host-range plasmid RK2 is an inhibitor of Escherichia coli gyrase. Mol. Microbiol. 44:971-979. [DOI] [PubMed] [Google Scholar]
- 38.Kim, L., A. Mogk, and W. Schumann. 1996. A xylose-inducible Bacillus subtilis integration vector and its application. Gene 181:71-76. [DOI] [PubMed] [Google Scholar]
- 39.Kristoffersen, P., G. B. Jensen, K. Gerdes, and J. Piskur. 2000. Bacterial toxin-antitoxin gene system as containment control in yeast cells. Appl. Environ. Microbiol. 66:5524-5526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lanzer, M., and H. Bujard. 1988. Promoters largely determine the efficiency of repressor action. Proc. Natl. Acad. Sci. USA 85:8973-8977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lehnherr, H., and M. B. Yarmolinsky. 1995. Addiction protein Phd of plasmid prophage P1 is a substrate of the ClpXP serine protease of Escherichia coli. Proc. Natl. Acad. Sci. USA 92:3274-3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lopilato, J., S. Bortner, and J. Beckwith. 1986. Mutations in a new chromosomal gene of Escherichia coli K-12, pcnB, reduce plasmid copy number of pBR322 and its derivatives. Mol. Gen. Genet. 205:285-290. [DOI] [PubMed] [Google Scholar]
- 43.Meinhart, A., J. C. Alonso, N. Strater, and W. Saenger. 2003. Crystal structure of the plasmid maintenance system epsilon/zeta: functional mechanism of toxin zeta and inactivation by epsilon 2 zeta 2 complex formation. Proc. Natl. Acad. Sci. USA 100:1661-1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nordstrom, K., and S. J. Austin. 1989. Mechanisms that contribute to the stable segregation of plasmids. Annu. Rev. Genet. 23:37-69. [DOI] [PubMed] [Google Scholar]
- 45.Pedersen, K., and K. Gerdes. 1999. Multiple hok genes on the chromosome of Escherichia coli. Mol. Microbiol. 32:1090-1102. [DOI] [PubMed] [Google Scholar]
- 46.Pedersen, K., A. V. Zavialov, M. Pavlov, J. Elf, K. Gerdes, and M. Ehrenberg. 2003. The bacterial toxin RelE displays codon-specific cleavage of mRNAs in the ribosomal A site. Cell 112:131-140. [DOI] [PubMed] [Google Scholar]
- 47.Picardeau, M., C. Le Dantec, G. F. Richard, and I. Saint Girons. 2003. The spirochetal chpK-chromosomal toxin-antitoxin locus induces growth inhibition of yeast and mycobacteria. FEMS Microbiol. Lett. 229:277-281. [DOI] [PubMed] [Google Scholar]
- 48.Rojo, F., and J. C. Alonso. 1994. A novel site-specific recombinase encoded by the Streptococcus pyogenes plasmid pSM19035. J. Mol. Biol. 238:159-172. [DOI] [PubMed] [Google Scholar]
- 49.Rottlander, E., and T. A. Trautner. 1970. Genetic and transfection studies with B. subtilis phage SP 50. I. Phage mutants with restricted growth on B. subtilis strain 168. Mol. Gen. Genet. 108:47-60. [DOI] [PubMed] [Google Scholar]
- 50.Ruiz-Echevarria, M. J., G. Gimenez-Gallego, R. Sabariegos-Jareno, and R. Diaz-Orejas. 1995. Kid, a small protein of the parD stability system of plasmid R1, is an inhibitor of DNA replication acting at the initiation of DNA synthesis. J. Mol. Biol. 247:568-577. [DOI] [PubMed] [Google Scholar]
- 51.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 52.Silhavy, T. J., M. L. Berman, and L. W. Enquist. 1984. Experiments with gene fusions. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 53.Simon, D., and A. Chopin. 1988. Construction of a vector plasmid family and its use for molecular cloning in Streptococcus lactis. Biochimie 70:559-566. [DOI] [PubMed] [Google Scholar]
- 54.Smith, A. S., and D. E. Rawlings. 1997. The poison-antidote stability system of the broad-host-range Thiobacillus ferrooxidans plasmid pTF-FC2. Mol. Microbiol. 26:961-970. [DOI] [PubMed] [Google Scholar]
- 55.Smith, A. S. G., and D. E. Rawlings. 1998. Autoregulation of the pTF-FC2 proteic poison-antidote plasmid addiction system (pas) is essential for plasmid stabilization. J. Bacteriol. 180:5463-5465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Spizizen, J. 1958. Transformation of biochemically deficient strains of Bacillus subtilis by deoxyribonucleate. Proc. Natl. Acad. Sci. USA 44:1072-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Trieu-Cuot, P., C. Carlier, C. Poyart-Salmeron, and P. Courvalin. 1991. Shuttle vectors containing a multiple cloning site and a lacZ alpha gene for conjugal transfer of DNA from Escherichia coli to gram-positive bacteria. Gene 102:99-104. [DOI] [PubMed] [Google Scholar]
- 58.Tsuchimoto, S., Y. Nishimura, and E. Ohtsubo. 1992. The stable maintenance system pem of plasmid R100: degradation of PemI protein may allow PemK protein to inhibit cell growth. J. Bacteriol. 174:4205-4211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tsurimoto, T., T. Hase, H. Matsubara, and K. Matsubara. 1982. Bacteriophage lambda initiators: preparation from a strain that overproduces the O and P proteins. Mol. Gen. Genet. 187:79-86. [DOI] [PubMed] [Google Scholar]
- 60.Van Helvoort, J. M., and C. L. Woldringh. 1994. Nucleoid partitioning in Escherichia coli during steady-state growth and upon recovery from chloramphenicol treatment. Mol. Microbiol. 13:577-583. [DOI] [PubMed] [Google Scholar]
- 61.Van Melderen, L., P. Bernard, and M. Couturier. 1994. Lon-dependent proteolysis of CcdA is the key control for activation of CcdB in plasmid-free segregant bacteria. Mol. Microbiol. 11:1151-1157. [DOI] [PubMed] [Google Scholar]
- 62.Weaver, K. E., K. D. Jensen, A. Colwell, and S. I. Sriram. 1996. Functional analysis of the Enterococcus faecalis plasmid pAD1-encoded stability determinant par. Mol. Microbiol. 20:53-63. [DOI] [PubMed] [Google Scholar]
- 63.Weaver, K. E., D. M. Weaver, C. I. Wells, C. M. Waters, M. E. Gardner, and E. A. Ehli. 2003. Enterococcus faecalis plasmid pAD1-encoded Fst toxin affects membrane permeability and alters cellular responces to lantibiotics. J. Bacteriol. 185:2169-2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Williams, D. R., and M. Thomas. 1992. Active partitioning of bacterial plasmids. J. Gen. Microbiol. 138:1-16. [DOI] [PubMed] [Google Scholar]
- 65.Zhang, Y., J. Zhang, M. Zhu, and M. Inouye. 2004. Interference of mRNA function by sequence-specific endoribonuclease PemK. J. Biol. Chem. 279:20678-20684. [DOI] [PubMed] [Google Scholar]
- 66.Zhang, J., Y. Zhang, K. P. Hoeflich, M. Ikura, G. Qing, and M. Inouye. 2003. MazF cleaves cellular mRNAs specifically at ACA to block protein synthesis in Escherichia coli. Cell 12:913-923. [DOI] [PubMed] [Google Scholar]
- 67.Zielenkiewicz, U., and P. Cegłowski. 2001. Mechanisms of plasmid stable maintenance with special focus on plasmid addiction systems. Acta Biochim. 48:1003-1023. [PubMed] [Google Scholar]