Abstract
In the present study we describe the isolation and functional analysis of a sphingolipid biosynthetic gene, IPT1, of Candida albicans. The functional consequence of the disruption of both alleles of IPT1 was confirmed by mass analysis of its sphingolipid composition. The disruption of both alleles or a single allele of IPT1 did not lead to any change in growth phenotype or total sphingolipid, ergosterol, or phospholipid content of the mutant cells. The loss of mannosyl diinositol diphosphoceramide [M(IP)2C] in the ipt1 disruptant, however, resulted in increased sensitivity to drugs like 4-nitroquinoline oxide, terbinafine, o-phenanthroline, fluconazole, itraconazole, and ketoconazole. The increase in drug susceptibilities of ipt1 cells was linked to an altered sphingolipid composition, which appeared to be due to the impaired functionality of Cdr1p, a major drug efflux pump of C. albicans that belongs to the ATP binding cassette superfamily. Our confocal and Western blotting results demonstrated that surface localization of green fluorescent protein-tagged Cdr1p was affected in ipt1 disruptant cells. Poor surface localization of Cdr1p resulted in an impaired ability to efflux fluconazole and rhodamine 6G. The effect of mannosyl inositol phosphoceramide accumulation in the ipt1 mutant and the absence of M(IP)2C from the ipt1 mutant on the efflux of drug substrates was very selective. The efflux of methotrexate, a specific substrate of CaMdr1p, another major efflux pump of major facilitator superfamily, remained unaffected in ipt1 mutant cells. Interestingly, changes in sphingolipid composition affected the ability of mutant cells to form proper hyphae in various media. Taken together, our results demonstrate that an altered composition of sphingolipid, which is among the major constituents of membrane rafts, affects the drug susceptibilities and morphogenesis of C. albicans.
The early steps in mammalian and fungal sphingolipid synthesis are conserved, but finally, they diverge to produce structurally and chemically different types of sphingoid bases, ceramides, and complex sphingolipids (12). Therefore, over the years, the sphingolipid biosynthetic pathway has been exploited as an antifungal drug target in pathogenic yeasts (24, 29). Unlike mammals, fungi do not have phosphatidylcholine as part of their polar head group in the sphingolipids; instead, they have phosphoinositol, which is transferred by Aur1p to the C-1 hydroxyl of ceramide to make inositol phosphoceramide (IPC) (6, 7). IPC is further modified by the addition of mannose by Csg1p, Csg2p, and Vrg4p to make mannosyl inositol phosphoceramide (MIPC) and the addition of a second inositol phosphate group by Ipt1p to make mannosyl diinositol diphosphoceramide [M(IP)2C] (6, 7). The biosynthesis of sphingolipids starts in the endoplasmic reticulum and proceeds up to the formation of ceramides (12). Subsequently, complex fungal sphingolipids [IPC, MIPC, and M(IP)2C] are synthesized in the Golgi apparatus. Recent reports have suggested that the biosynthesis of these sphingolipids is critical to the maintenance of plasma membrane (PM) function; however, the synthesis of M(IP)2C is not critical for viability (8). In Saccharomyces cerevisiae, ipt1 deletion mutants grow normally but display sensitivity to calcium and increased resistance to the polyene antibiotic nystatin (7, 23). Moye-Rowley and his group have observed that the loss of IPT1 has complex effects on drug resistance in S. cerevisiae, mediated through Pdr1p and Pdr3p transcription factors which regulate multidrug resistance genes in S. cerevisiae (15).
Candida albicans is an opportunistic diploid fungus that causes superficial as well as systemic infections in immunocompromised and debilitated patients (5). Unlike in S. cerevisiae, the regulation of synthesis of sphingolipids and their roles in membrane function in C. albicans cells are not well understood. In order to explore the role of sphingolipids, we had earlier used a specific inhibitor, fumonisin B1, which blocks the synthesis of phytoceramide, a precursor of the three major sphingolipid species (28). We observed a close interaction between plasma membrane ergosterol and sphingolipids in C. albicans cells (28), wherein the depletion of either of the two resulted in the impaired functionality of a major drug efflux pump, Cdr1p, and as a consequence turned Candida cells hypersensitive to several drugs. In this study we have specifically blocked the synthesis of M(IP)2C by homozygous disruption of the IPT1 gene, which mediates the conversion of MIPC to M(IP)2C. This provided an opportunity to analyze the role of M(IP)2C, which accounts for the majority of membrane sphingolipids. We observed that the specific accumulation of MIPC in ipt1 disruptants increased sensitivity to several drugs. Interestingly, the ipt1 mutant cells also showed defects in their ability to form hyphae in different media.
MATERIALS AND METHODS
Materials.
Medium chemicals were obtained from Difco (Detroit, Mich.) and HiMedia (Mumbai, India), and lipid N-rhodamine-dioleoyl-phosphatidylethanolamine was purchased from Avanti Polar Lipids Inc. (Albaster, Ala). Sigma Chemical Co. (St. Louis, Mo.) was the source of l-dipalmitoyl-phosphatidylcholine and rhodamine 6G (R6G); the drugs 4-nitroquinoline oxide, terbinafine, o-phenanthroline, nystatin, amphotericin B, cycloheximide, and methotrexate; as well as protease inhibitors (phenylmethylsulfonyl fluoride, leupeptin, pepstatin A, and aprotinin). Nourseothricin was obtained from Werner Bioagents (Jena, Germany). Fluconazole was a kind gift from Ranbaxy Laboratories (New Delhi, India), while sulfomethuron methyl, ketoconazole, and itraconazole were gifts from Du Pont (Wilmington, Del.). [3H]fluconazole ([3H]FLU) was custom prepared by Amersham, United Kingdom. [3H]methotrexate ([3H]MTX) was procured from Amersham. Labeled 6-[(7-nitrobenz-2-oxa-1,3-diazol-4-yl) amino hexanoyl] sphingosylphosphocholine (NBD-sphingomyelin [NBD-SPH]) was purchased from Molecular Probes (Eugene, Oreg.).
Bacterial and yeast strains and growth media.
C. albicans strain SC5314 was procured from the American Type Culture Collection. Escherichia coli was cultured in Luria-Bertani medium (Difco, BD Biosciences), to which ampicillin was added (100 μg/ml). The Candida strains were cultured in YEPD (yeast extract-peptone-dextrose) broth (Bio 101, Vista, CA) or Sabouraud dextrose broth without uracil (SD-URA−, minimal defined media SDU− consisting of 2% glucose supplemented with yeast nitrogen base; Bio 101), as required. For agar plates, 2% (wt/vol) Bacto Agar (Difco, BD Biosciences) was added to the medium. The strains and plasmids used in this study are listed in Table 1. Strains CAF2-1, TPIPT1-1, TPIPT1-3, and TPIPT1-5 were used for all functional assays and are referred to as the wild type (WT), the heterozygous disruptant, the homozygous disruptant, and the heterozygous revertant, respectively. Cdr1p of CAF2-1, TPIPT1-1, TPIPT1-3, and TPIPT1-5 was tagged with green fluorescent protein (GFP); and the respective strains were designated TPWT-GFPCDR1, TPD1-GFPCDR1, TPD2-GFPCDR1, and TPR1-GFPCDR1.
TABLE 1.
Strains and plasmids used in this study
| Plasmid or strain | Description or genotype | Source or reference |
|---|---|---|
| Plasmids | ||
| pBKS | Bluescript II KS+ vector | Stratagene |
| pBSK-S/X | Bluescript II KS+ vector with SalI and XhoI sites removed and then religated | This study |
| pTPIPT1 | 2.614-kb amplicon containing IPT1 inserted into pBKS-S/X at EcoRV | This study |
| pUC19 | Small, high-copy-number plasmid | MBI, Fermentas |
| pMB7 | Plasmid that contains the C. albicans URA3 gene flanked by 1.1-kb direct repeats of the Salmonella enterica serovar Typhimurium hisG DNA | 10 |
| pTPHUH | 4.044-kb BamHI-BglII fragment from pMB7 containing hisG-Ura3-hisG inserted into pUC19 at BamHI | This study |
| pTPIPT1R | 4.061-kb SalI-SmaI fragment from pTPHUH containing hisG-Ura3-hisG inserted into pTPIPT1 at SalI-HpaI | This study |
| pADH1G3 | Plasmid harboring the SAT1 and GFP genes | 26 |
| pCPL51 | Plasmid harboring CDR1-LACZ fusion with 3′ CDR1 | This study |
| pCPG2 | Bluescript II KS+ vector containing a 3.5-kb fragment, GFP fused to the C terminus of CDR1 by mutating stop codon into BamHI, along with CaSAT1 marker gene | This study |
| C. albicans strains | ||
| SC 5314 | URA3/URA3 | 10 |
| CAF2-1 | URA3/Δura3::imm434 | 10 |
| CAI4 | Δura3::imm434/Δura3::imm434 | 10 |
| TPIPT1-1 | Δura3::imm434/Δura3::imm434 IPT1/Δipt1::hisG-URA3-hisG | This study |
| TPIPT1-2 | Δura3::imm434/Δura3::imm434 IPT1/Δipt1::hisG | This study |
| TPIPT1-3 | Δura3::imm434/Δura3::imm434 Δipt1::hisG-URA3-hisG/Δipt1::hisG | This study |
| TPIPT1-4 | Δura3::imm434/Δura3::imm434 Δipt1::hisG/Δipt1::hisG | This study |
| TPIPT1-5 | Δura3::imm434/Δura3::imm434 Δipt1::hisG/Δipt1::hisG::IPT1-URA3 | This study |
| TPWT-GFPCDR1 | URA3/Δura3::imm434 | This study |
| TPD1-GFPCDR1 | Δura3::imm434/Δura3::imm434 IPT1/Δipt1::hisG-URA3-hisG | This study |
| TPD2-GFPCDR1 | Δura3::imm434/Δura3::imm434 Δipt1::hisG-URA3-hisG/Δipt1::hisG | This study |
| TPR1-GFPCDR1 | Δura3::imm434/Δura3::imm434 Δipt1::hisG/Δipt1::hisG::IPT1-URA3 | This study |
Construction of disruption cassette pTPIPT1-HUH.
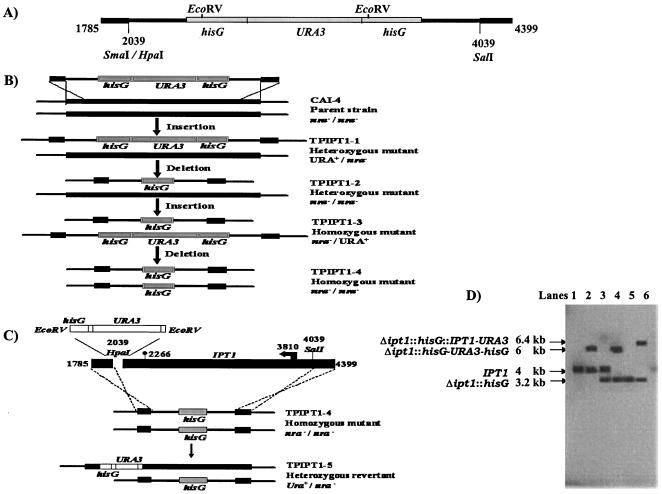
The IPT1 open reading frame (ORF) was amplified from the genomic DNA of C. albicans strain SC5314 by using the primers IPT1F (5′-GTGGTTTACCTTTAGGGTGGTTG-3′) and IPT1R (5′-GGTGGGTGGTGGGTGGTGTTT-3′) and Pfu Turbo (Stratagene). This amplicon was then cloned into the EcoRV site of pBSK-S/X (Table 1), yielding pTPIPT1. For the construction of the URA blaster cassette, plasmid pMB-7 (10), which harbors the C. albicans URA3 gene flanked by Salmonella serovar Typhimurium hisG DNA, was digested with BamHI and BglII, yielding a 4.044-kb fragment which was subsequently cloned into plasmid pUC19 to get the desired restriction sites, yielding pTPHUH. From pTPHUH, a 4.061-kb SalI-SmaI fragment containing hisG-Ura3-hisG was then inserted into pTPIPT1 at SalI-HpaI, yielding the disruption cassette pTPIPT1-HUH. The SacI-KpnI-digested 4.866-kb fragment of pTPIPT1-HUH was used to disrupt IPT1 in Candida wild-type strain CAI-4 (Fig. 1A and B).
FIG. 1.
Schematic representation of the disruption and complementation of IPT1 using the URA3 selection marker. (A) Diagrammatic representation of the disruption cassette derived from pTPIPT1-HUH containing hisG-URA3-hisG with flanking portions (regions between positions 1785 and 2039 and positions 4039 and 4399) on either side for homologous recombination. (B) Sequential targeted disruption of the two IPT1 alleles in C. albicans with the disruption cassette. Strain designations are shown on the right. (C) Schematic representation of the reintegration cassette used for reintroduction of one wild-type IPT1 allele and genomic reintegration of an intact copy into TPIPT1-4 by using the reintegration cassette pTPIPT1R, which led to revertant strain TPIPT1-5. (D) Southern analysis of the genomic DNA digested with SalI. Lanes: 1, CAI4; 2, TPIPT1-1; 3, TPIPT1-2; 4, TPIPT1-3; 5, TPIPT1-4; 6, TPIPT1-5. The hybridization probe was the 1.716-kb SacI-NdeI fragment derived from N-terminal region of pTPIPT1 (Table 1). The exact size and genotype of the expected hybridizing DNA fragment are indicated on the left.
Construction of reintegration cassette pTPIPT1R.
A 2.441-kb fragment obtained by EcoRV digestion of pTPIPT1-HUH was cloned at the HpaI site of pTPIPT1 located downstream of the CaIPT1 ORF, yielding pTPIPT1R (Fig. 1C). A 5.157-kb SacI-KpnI fragment from pTPIPT1R was used to transform the homozygous ipt1 mutant strain, TPIPT1-4, for the construction of heterozygous revertant strain TPIPT1-5, which had one functional IPT1 allele and the URA3 gene as a marker.
Construction of pCPG2.
Plasmid vector pCPG2 was constructed to fuse GFP with the CDR1 ORF at its own locus by mutating its stop codon into a BamHI site. This 250 bp of the C-terminal CDR1 ORF was PCR amplified by using primers CDR29F (5′-ATTTGGTACCATACATTAAATTTGCTGGTGGG-3′) and CDR30R (5′-GTTTGGATCCTTTCTTATTTTTTTTCTCTCTGTTACCC-3′). The PCR amplicon was digested with KpnI-BamHI (the sites were introduced into the forward and the reverse primers during synthesis). GFP, along with ACT1 termination sequence and dominant selection marker gene SAT1, was taken out by digesting vector pADH1G3 with BamHI-PstI. The KpnI-PstI-digested vector backbone of pCPL51 (2.9 kb) was ligated with the KpnI-BamHI-digested C terminus of CDR1 along with the BamHI-PstI-digested GFP-SAT1 fragment to generate the final clone, pCPG2 (6.5 kb). The KpnI-SacII restriction sites were used to take out the linearized cassette (CT-CDR1-GFP-SAT1-3′ untranslated region) from plasmid pCPG2 and transformed into the C. albicans wild-type and the IPT1 disruptant and heterozygous revertant strains to produce strains with GFP-tagged Cdr1p.
DNA and RNA isolation and hybridization.
Yeast genomic DNA was isolated as described elsewhere (1). A total of 5 μg of genomic DNA from each transformant and the WT cells was digested with SalI and electrophoresed for Southern analyses. Southern hybridizations were carried out with the 1.716-kb SacI-NdeI fragment, which was used as the probe and which was derived from N-terminal region of pTPIPT1. Total RNA were isolated and Northern blot analyses were done essentially by standard protocols, as described before (1, 22, 31). Equal loading of RNA was checked by detection of equal intensity of the rRNA bands, and the relative intensities of the CDR1 mRNA signals in Northern hybridizations were quantitated in an FLA5000 Fuji phosphoimager.
Candida transformation.
The lithium acetate method was used for the transformation of C. albicans cells, as described previously (13). Transformants were selected on synthetic minimal medium (Sabouraud dextrose medium) to obtain Ura+ transformants, and these Ura+ transformants were cured of Ura by the use of 5-fluoroorotic acid (1 mg/ml) and 25 μg/ml uridine.
Strains CAF2-1, TPIPT1-1, TPIPT1-3, and TPIPT1-5 were transformed with a linearized 3.5-kb KpnI-SacII fragment from plasmid pCPG2 to produce strains TPWT-GFPCDR1, TPD1-GFPCDR1, TPD2-GFPCDR1, and TPR1-GFPCDR1 with GFP-tagged Cdr1p, respectively. The transformants were selected on YEPD plates containing 200 μg/ml of nourseothricin.
Detection of sphingolipids by mass spectrometry.
Sphingolipids were extracted by following a protocol used for S. cerevisiae described earlier by Takemoto and colleagues (33). Negative-ion mass electrospray spectra were recorded on a liquid chromatography-time of flight mass spectrometer (Micromass-Waters). The spectral data were analyzed with MassLynx software. The m/z range was recorded to be from 500 to 1,500 Da. A constant flow of the solvent chloroform-methanol-water (16:16:5 vol/vol/vol) was maintained with a pump connected to the spraying capillary. Solutions of extracted sphingolipids in the solvent described above were injected into the solvent flow of the above liquid chromatograph-mass spectrometer for analysis.
Drug efflux assay.
The glucose-induced efflux of fluorescent R6G and radiolabeled [3H]FLU (specific activity, 19 Ci/mmol) was carried out essentially as described in our earlier publications (21, 28). R6G and [3H]FLU were used to final concentrations of 10 μM and 100 nM, respectively. MTX transport was determined as described earlier (20). [3H]MTX (specific activity, 5.7 Ci/mmol) was added to a final concentration of 25 μM.
Labeling of cells with NBD-SPH.
Labeling of cells with NBD-SPH was carried out essentially as described earlier (28).
Morphogenetic studies on solid and liquid media.
Studies of hyphal induction by wild-type and mutant cells of C. albicans were carried out on solid Spider (1% nutrient broth, 1% mannitol, 0.2% K2HPO4, 2% Bacto Agar) and nitrogen starvation SLAD (0.17% yeast nitrogen base without amino acids and ammonium sulfate, 2% glucose, 50 μM ammonium sulfate, 2% Bacto Agar) medium plates and with 10% bovine calf serum in liquid YEPD and 2.5 mM N-acetyl-d-glucosamine in liquid salt base medium (containing 0.45% NaCl and 0.335% yeast nitrogen base without amino acids), as described elsewhere (2, 3, 19, 35).
Nucleotide sequence accession number.
The IPT1 ORF amplified from the genomic DNA of C. albicans SC5314 with primers IPT1F and IPT1R has been submitted to GenBank and has been given accession number AY884203.
RESULTS
Identification and homozygous disruption of CaIPT1.
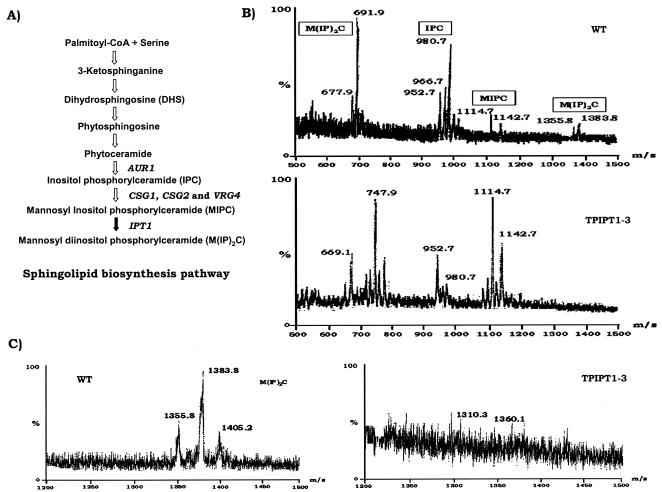
BLAST analysis revealed one gene in the C. albicans genome that bore significant homology (expected value = 3e-36) with S. cerevisiae ScIPT1, which encodes inositol phosphoryl transferase, and was designated CaIPT1 (orf 6.927). CaIPT1 also shows similarity to another sphingolipid biosynthetic gene of S. cerevisiae, AUR1 (expected value = 1e-24) and CaAUR1 of C. albicans (expected value = 1e-24) (Fig. 2A), which performs a similar function of adding an inositol phosphate group, as predicted by homology to ScAUR1 (18). The 1,545-bp CaIPT1 gene is located on chromosome I. The Stanford contig 6-1754 has the CaIPT1 sequence from positions 2266 to 3810. The estimated molecular mass of the putative membrane protein CaIpt1p (514 amino acids) is 59.43 kDa.
FIG. 2.
(A) Schematic representation of the sphingolipid biosynthetic pathway in fungi. AUR1 encodes IPC synthase; CSG1, CSG2, and VRG4 add mannose to IPC (possible mannosyltransferases); and IPT1 encodes inositol phosphotransferase. (B) Mass spectrometric analysis of sphingolipids. Negative-ion mass spectra of the m/z range of 500 to 1,500 Da of the sphingolipids extracted from the WT and homozygous IPT1 mutant strain TPIPT1-3. The positions of the respective three prominent sphingolipid species are indicated as follows: for IPC, 952.7 [M-H]−, 966.7 [M-H]−, and 980.7 [M-H]−; for MIPC, 1,114.7 [M-H]− and 1,142.7 [M-H]−; and for M(IP)2C, 677.9 [M-2H]2− and 691.9 [M-2H]2− for doubly charged ions and 1,355.8 [M-H]− and 1,383.8 [M-H]− for ions with a single charge. (C) Enlarged negative-ion mass spectra of the m/z range of 1,300 to 1,500 Da of the sphingolipids extracted from the WT and homozygous IPT1 mutant strain TPIPT1-3 to show the absence of the M(IP)2C peak at the indicated position for TPIPT1-3, unlike the WT strain.
The IPT1 ORF was amplified from the genomic DNA of C. albicans SC5314 with primers IPT1F and IPT1R (see Materials and Methods) and sequenced (GenBank accession number AY884203). As described in Materials and Methods, the URA blaster cassette was used for homozygous disruption of CaIPT1 (10). Both IPT1 alleles were sequentially disrupted by using the disruption cassette (Fig. 1A). Strains TPIPT1-2 and TPIPT1-4 were obtained by looping out the URA3 gene from TPIPT1-1 and TPIPT1-3, respectively (Fig. 1B). To obtain heterozygous revertant strain TPIPT1-5, which has one functional IPT1 allele, homozygous disruptant mutant TPIPT1-4 was transformed with the SacI-KpnI fragment of pTPIPT1R (Table 1; Fig. 1C). Strains CAF2-1 (wild type), TPIPT1-1 (heterozygous disruptant), TPIPT1-3 (homozygous disruptant), and TPIPT1-5 (heterozygous revertant) were used for all functional assays (Table 1). Correct insertion of the disruption cassette was checked by Southern analysis (Fig. 1D). Two independent clones were picked and analyzed simultaneously. Pairs of isogenic mutant strains were identical in all phenotypes (data not shown).
Sphingolipid compositions of ipt1 mutants.
The disruption of CaIPT1 was further confirmed by sphingolipid analysis of the mutant and WT cells by using mass spectrometric analysis. The mass analysis of the extracted sphingolipids of WT Candida cells indicated three major peaks for sphingolipid species: for IPC, singly charged species [M-H]− at m/z = 952.7, 966.7, and 980.7 Da; for MIPC, singly charged molecular ions [M-H]− at m/z = 1,114.7 and 1,142.7 Da, and for M(IP)2C, mass spectrum showed signals at m/z = 677.9 and 691.9 Da for doubly charged molecular species [M-2H]2− and at m/z = 1,355.8 and 1,383.8 Da for singly charged molecular ions [M-H]− (Fig. 2B and C). The observed mass difference of 162 Da between IPC and MIPC confirmed the expected difference due to the additional mannose group in MIPC. The observed difference in mass of 241.1 Da between MIPC and M(IP)2C is due to the additional inositolphosphate group in M(IP)2C. The mass spectra of the three sphingolipids showed signals for two to four species of the sphingolipids which could be attributed to heterogeneity in the number of —CH2 groups, since IPC, MIPC, and M(IP)2C have different fatty acid chain lengths that vary from 22 to 26 and the presence of the abundant long chain bases as C18 or C20 phytosphingosines (17, 36) (Fig. 2). Unlike the wild-type Candida cells, homozygous mutant TPIPT1-3 cells revealed a total absence of M(IP)2C, indicated by the absence of the mass spectrum peak at m/z = 691.5 [M-2H]2− as well as at 1,383.8 [M-H]− and 1,355 [M-H]− and showed, instead, an increased area of the peak for MIPC.
Of note, the variations in the sphingolipid compositions of the TPIPT1-3 mutant and its heterozygous counterpart (TPIPT1-1) resulted in no significant changes in cell growth when they were grown on rich medium like YEPD compared to that of the WT cells (data not shown). There was no major change in the total phospholipid, sphingolipid, or ergosterol contents of TPIPT1-1 and TPIPT1-3 cells (data not shown).
IPT1 mutants of C. albicans are susceptible to various drugs.
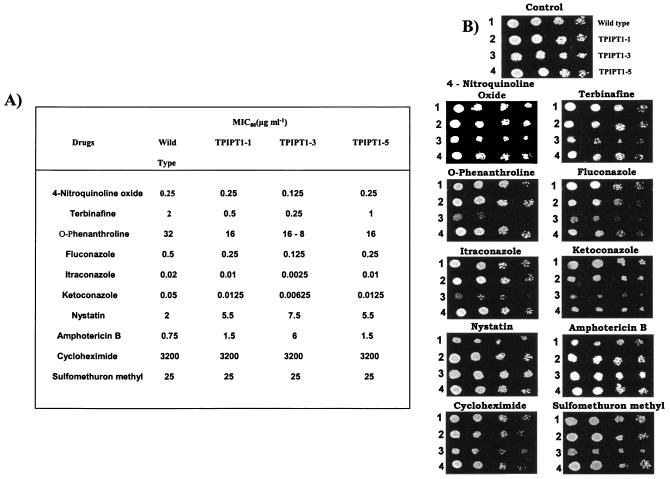
As mentioned above, since the homozygous disruption of IPT1 led to the specific accumulation of MIPC without significantly affecting the total contents of sphingolipids and other phospholipids, we examined if the loss of M(IP)2C could in any way affect the drug susceptibilities of ipt1 mutants. The results of the drug susceptibility tests by two independent methods, MIC (MIC80) and spot assays, demonstrated that the ipt1 mutants were susceptible to 4-nitroquinoline oxide, terbinafine, and o-phenanthroline and to the azoles tested, which included fluconazole, itraconazole, and ketoconazole (Fig. 3A and B). Although the spot assay revealed that these mutants were additionally sensitive to cycloheximide and sulfomethuron methyl, there was no change in the MIC80 values between the mutant and WT cells for these drugs (Fig. 3A and B). The discrepancy between the results of the MIC assay and that of the spot assay is not surprising, since in many instances such differences between various drug susceptibility assays have been reported previously (9, 27, 28). We also checked the susceptibilities of the ipt1 mutant cells to two well-known polyenes, namely, nystatin and amphotericin B. Interestingly, both spot assay and MIC80 values revealed that ipt1 mutants were resistant to polyenes (Fig. 3A and B).
FIG. 3.
Drug resistance profiles of the C. albicans wild type and IPT1 mutants determined by the broth microdilution assay (A) and spot assay (B), which were done as described previously (27, 28). The MIC80 (defined as the lowest drug concentration that gave >80% inhibition of growth compared to the growth of the drug-free controls) was determined by the broth microdilution method and was evaluated both visually and by reading the absorbance at 620 nm in a microplate reader. In the spot assay, 5 μl of fivefold serial dilutions of each yeast culture (A600 = 0.1) was spotted onto YEPD plates in the absence (control) and the presence of the following drugs: 4-nitroquinoline oxide (0.1 μg/ml), terbinafine (0.2 μg/ml), o-phenanthroline (8 μg/ml), fluconazole (0.1 μg/ml), itraconazole (0.01 μg/ml), ketoconazole (0.01 μg/ml), nystatin (1.5 μg/ml), amphotericin B (0.5 μg/ml), cycloheximide (4 mg/ml), sulfomethuron methyl (20 μg/ml), and methotrexate (4 μg/ml). Growth differences were evaluated by using drug-free controls following incubation of the plates for 48 h at 30°C. 4-Nitroquinoline oxide, terbinafine, o-phenanthroline, fluconazole, itraconazole, ketoconazole, nystatin, cycloheximide, and sulfomethuron methyl are known substrates of CDR1; and methotrexate is a well-known substrate of CaMDR1 (20, 21, 27, 32).
Enhanced drug sensitivity of IPT1 mutants is associated with reduced efflux of substrates.
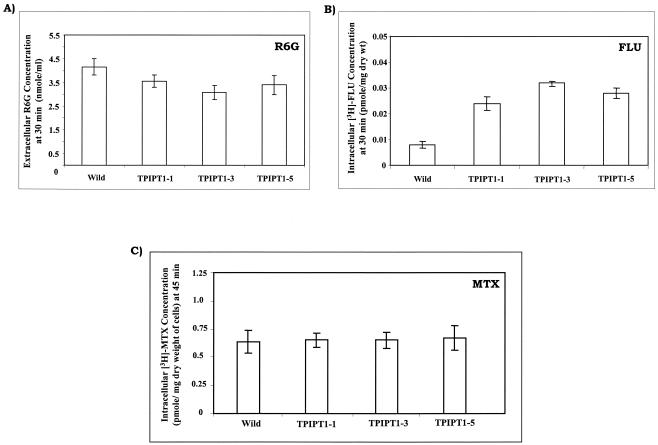
Since the ipt1 mutant cells were found to be susceptible to drugs which are also substrates of Cdr1p, a major drug efflux pump belonging to the ATP binding cassette (ABC) superfamily of transporters, we checked the functionality of this pump. R6G and FLU are well-known substrates of Cdr1p and were earlier used to monitor the drug efflux activity of Cdr1p (28). Fluorescent R6G and [3H]FLU were allowed to equilibrate in deenergized Candida cells by passive diffusion, and the energy-dependent extrusion of R6G was then initiated by the addition of glucose. As depicted in Fig. 4A, after glucose addition, a decrease in the extracellular concentration (due to low efflux) of R6G was observed in TPIPT1-1 and TPIPT1-3 mutant cells compared to that in the WT cells, which clearly indicated impaired efflux activity by the mutant cells. That the efflux ability of TPIPT1-3 mutant cells was reduced was further confirmed by using another substrate, [3H]FLU. The efflux of equilibrated [3H]FLU in deenergized cells also was initiated by the addition of glucose. As shown in Fig. 4B, the intracellular levels of accumulation of [3H]FLU were higher (due to low levels of efflux) in the mutant cells than in the WT cells. Of note, deenergized ipt1 cells also showed increased passive diffusion of drugs (data not shown). This would mean that enhanced passive entry could also increase the intracellular concentration of the drug, which may contribute to the observed changes in the drug susceptibilities of ipt1 cells.
FIG. 4.
(A) Glucose-induced R6G efflux from C. albicans cells. The protocol of the efflux assay has been described in our earlier publication (28). To achieve deenergization of exponentially grown Candida cells for depletion of the intracellular ATP, cells were resuspended in deenergization buffer (phosphate-buffered saline without glucose and with 5 mM deoxyglucose and 5 mM dinitrophenol) at a cell density of 108 cells ml−1. R6G was then added to the cell suspension to a final concentration of 10 μM, and the mixture was incubated for 2 h at 30°C. Efflux was initiated by addition of 2% glucose, and the absorption of the extruded dye in the supernatant was measured at 527 nm. Open bars, extracellular concentration of R6G at 30 min after the addition of glucose. The values are the means ± standard deviations (indicated by the error bars) of three independent experiments. (B) Intracellular concentration of [3H]FLU in deenergized C. albicans WT and IPT1 mutant strains 30 min after glucose-induced drug efflux. The values indicated by the bars represent the means ± standard deviations (indicated by the error bars) of three independent experiments. (C) Intracellular concentration of [3H]MTX in C. albicans WT and IPT1 mutant strains at 45 min. The values indicated by the bars represent the means ± standard deviations (indicated by the error bars) of three independent experiments.
Variations in sphingolipid composition selectively affect Cdr1p functioning.
CaMdr1p, which is a member of major facilitator superfamily (MFS) of proteins, is another drug extrusion pump whose upregulation has been linked to azole resistance in C. albicans (11, 14, 37, 38). We had earlier observed that among the ABC and MFS transporters of C. albicans, the former are more sensitive to changes in the lipid composition (27). To verify this observation, we monitored the efflux of MTX, which is a specific substrate for CaMdr1p. Similar intracellular levels of [3H]MTX in ipt1 mutant and WT cells indicated no change in efflux of this substrate (Fig. 4C). Of note, the MIC80 values of the IPT1 mutant cells for MTX were also not different between WT and mutant cells (data not shown).
The levels of Cdr1p in PM of IPT1 mutant cells are reduced.
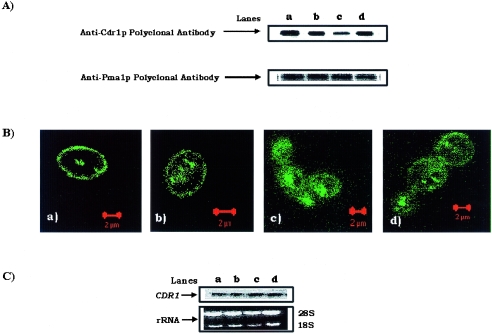
Since the functioning of Cdr1p appeared to be selectively affected in TPIPT1-1 and TPIPT1-3 mutant cells, we explored if it was due to the poor functioning of the protein or was due to improper surface localization of the protein. For this, we checked the expression of Cdr1p in the PM fractions of the mutant cells. Interestingly, Western blotting results revealed low levels of Cdr1p protein in PM of TPIPT1-3 (Fig. 5A, lane c), while Northern analysis showed no difference in CDR1 transcript (Fig. 5C). In order to verify if the low level of efflux protein in PM is indeed due to its poor localization, we examined the surface localization of Cdr1p in ipt1 mutant cells. For this, we expressed Cdr1p as a GFP-tagged protein by integrating the CDR1-GFP cassette into the CDR1 locus by using the SAT1 marker in both wild-type and ipt1 mutant cells (30). Of note, SAT1 gene is a dominant selection marker derived from the bacterial transposon Tn1825, which encodes streptothricin acetyltransferase, which confers resistance to nourseothricin by inactivating the antibiotic (30). It is evident from confocal pictures (Fig. 5B, panel a) that while Cdr1p is properly localized on the surface (rimmed appearance of the fluorescence) in the WT Candida cells, the GFP fluorescence, in contrast, appeared to be concentrated inside the ipt1 mutant cells (Fig. 5B, panels b and c). The rimmed fluorescence of Cdr1p-GFP appeared to be partly restored in revertant strain TPIPT1-5 (Fig. 5B, panel d).
FIG. 5.
(A) Immunodetection of Cdr1p in plasma membrane of CAF2-1 (lane a), TPIPT1-1 (lane b), TPIPT1-3 (lane c), and TPIPT1-5 (lane d). The PMs were prepared from C. albicans cells as described earlier (32). The Western blot analyses were done with anti-CDR1p polyclonal antibody (1:500 dilution). The purity of the PM fraction was assessed by using anti-Pma1p polyclonal antibody (1:10,000 dilution). Pma1p is a marker of the PM fraction (32). Immunoblots were visualized by using a chemiluminescence assay system (ECL kit; Amersham Biosciences). (B) Fluorescence imaging by confocal microscope of strains TPWT-GFPCDR1 (a), TPD1-GFPCDR1 (b), TPD2-GFPCDR1 (c), and TPR1-GFPCDR1 (d). Cells were grown overnight and were directly viewed on a glass slide under a ×100 oil-immersion objective on a Bio-Rad confocal microscope. The fluorescence signal from strain TPWT-GFPCDR1 showed localization of Cdr1p on the plasma membrane. On depletion of the sphingolipids, the GFP fluorescence from strains TPD1-GFPCDR1, TPD2-GFPCDR1, and TPR1-GFPCDR1 appeared to be concentrated inside the cells, indicating poor localization of Cdr1p on the plasma membrane; the concentration was maximum inside TPD2-GFPCDR1 cells and in the heterozygous mutant TPD1-GFPCDR1 was intermediate between those in WT and TPD2-GFPCDR1. This defect is reduced in the revertant strain, TPR1-GFPCDR1, compared to that in TPD2-GFPCDR1. (C) Northern blot analyses of CDR1. Upper panel, equal CDR1 transcript levels in strains TPWT-GFPCDR1 (lane a), TPD1-GFPCDR1 (lane b), TPD2-GFPCDR1 (lane c), and TPR1-GFPCDR1 (lane d); lower panel, loading control to indicate equal gel loading of total RNA.
IPT1 mutants show enhanced NBD-SPH exchange.
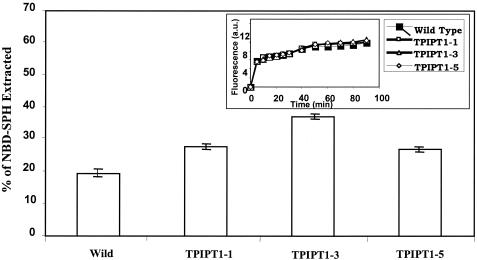
We had earlier observed that in erg mutants of C. albicans larger amounts of fluorescent NBD-SPH could be exchanged compared to that in WT cells (28). In order to explore if changes in sphingolipid composition had any effect within its local environment, we labeled Candida WT and IPT1 mutant cells with fluorescent NBD-SPH, as described earlier (28). At 90 min postlabeling, when NBD-SPH incorporation into Candida membrane was found to be maximum (Fig. 6, inset), the cells were washed and the NBD-SPH of the labeled cells was backextracted by using bovine serum albumin (28). The amount of NBD-SPH backextracted from IPT1 mutant cells was much larger in comparison to that backextracted from the WT cells (Fig. 6). This would mean that the depletion of M(IP)2C and the accumulation of MIPC in ipt1 mutant cells results in the disruption of the interactions between M(IP)2C, MIPC, and IPC and the other components of microdomains (particularly ergosterol). Interestingly, the lower exchange of NBD-SPH in the WT cells evidently indicates that M(IP)2C interacts with ergosterol with a propensity higher than those of the other two species of sphingolipids, namely, IPC and MIPC. Taken together, our earlier observations (28) as well as those from our present study suggest that the changes in the composition of either ergosterol or sphingolipids, which are raft constituents, affect their close interactions.
FIG. 6.
Postlabeling transbilayer exchange of NBD-SPH in the C. albicans wild type and IPT1 mutants. Cells were labeled with NBD-SPH (inset) and then backexchanged with 2% bovine serum albumin, as described in Materials and Methods. The graph depicts data for the 90-min time point, at which the maximum backexchanged fluorescence in the supernatant was recovered. The values are the means (indicated by the error bars) ± standard deviations of three independent experiments.
Sphingolipid composition affects morphogenesis.
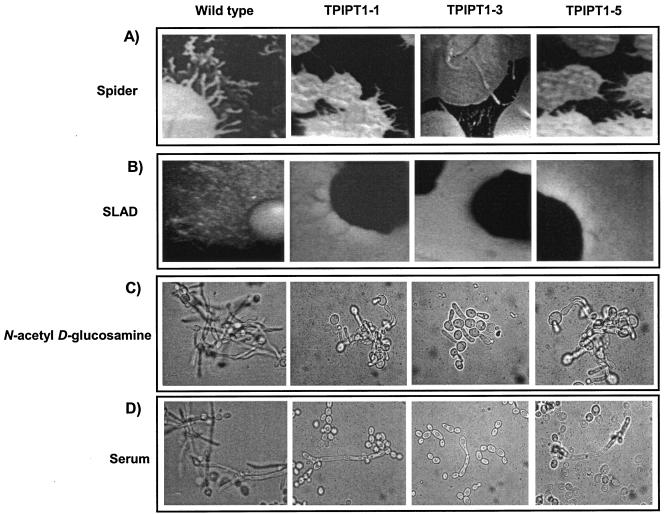
Compared to the wild-type Candida cells, the TPIPT1-1 and TPIPT1-3 mutants were unable to make long hyphal filaments in the presence of serum or N-acetyl d-glucosamine (Fig. 7). The filamentous growth from mature colony borders of cells plated on solid Spider and SLAD medium plates was also reduced in the TPIPT1-1 and TPIPT1-3 mutants. The morphogenesis defect could partially be restored in TPIPT1-5 when it was reconstituted with a single functional copy of the gene (Fig. 7).
FIG. 7.
Phenotypic responses of the IPT1 mutant strains under different hypha-inducing conditions. (A and B) Colony morphologies on solid Spider and SLAD plates, respectively; (C and D) the induction of filamentation in liquid media in response to N-acetyl-d-glucosamine and serum, respectively, in the WT and strains TPIPT1-1, TPIPT1-3, and TPIPT1-5.
DISCUSSION
The present study exploits the sphingolipid biosynthetic pathway to show that any change in sphingolipid composition leads to increased drug sensitivity and morphological defects in C. albicans cells. We show that the change in sphingolipid composition caused by disruption of its biosynthetic gene, IPT1, led to the improper surface localization of a major ABC drug efflux protein, Cdr1p. The inability of ipt1 mutant cells to efflux drugs appeared to be the main cause of the increased drug sensitivities of these cells. However, the impaired plasma membrane barrier function (increased passive diffusion) due to the altered sphingolipid composition may also contribute to enhanced sensitivity to drugs (data not shown). Similar to Saccharomyces cerevisiae, homozygous IPT1 disruptants of Candida albicans had increased resistance to calcium chloride (data not shown). It seems that sphingolipid metabolism in yeast cells is linked to calcium homeostasis or is regulated by the calcium ion concentration (8). Interestingly, changes in the compositions of sphingolipids had a selective effect on the functioning of the efflux pump proteins. For example, the efflux of MTX, which is a specific substrate of an MFS drug efflux protein, CaMdr1p (20), remained largely unaffected in IPT1 mutant cells.
Membrane sphingolipids and ergosterol are important subsets of lipids and are part of distinct domains known as membrane rafts (4, 16). Of note, since the total sphingolipid content in ipt1 mutant cells remained unchanged, it can be speculated that of the three species of sphingolipids, namely, IPC, MIPC, and M(IP)2C, M(IP)2C probably has a greater propensity to interact with ergosterol within microdomains of the plasma membrane. That ipt1 mutants lacking M(IP)2C had larger amounts of exchangeable NBD-tagged sphingolipid available than the WT supports the contention presented above (Fig. 6). It appears that the depletion of M(IP)2C and the accumulation of MIPC in the ipt1 mutant cells result in the disruption of their interactions with ergosterol, which leads to the greater exchangeability of NBD-SPH. This supports our earlier observation, in which we found that depletion of ergosterol also resulted in enhanced exchangeability of NBD-SPH (28).
While the changes in sphingolipid composition did not cause a severe growth defect in IPT1 mutant cells, it did affect their ability to form hyphae. Recent reports suggest the involvement of sterol- and sphingolipid-enriched microdomains in hyphal morphogenesis in C. albicans, wherein membrane lipid polarization appears to contribute to the ability of this pathogen to grow in a highly polarized manner to form hyphae (25). Our results are in agreement with those of such reports, since we also observed that disruption of ergosterol- and M(IP)2C-rich domains in the plasma membranes of the IPT1 mutants results in defective hyphal morphogenesis. It appears that not only is there a close interaction between membrane raft constituents and the drug susceptibilities of yeast cells but there is also a well-coordinated control of their synthesis (27, 28, 34, 39). In this context, it is pertinent to mention that Pdr1p and Pdr3p, well-known Zn(II)-Cys6 transcription factors which regulate pleiotropic drug resistance in Saccharomyces cerevisiae, also target IPT1 (15). Whether such coordination of raft constituents and MDR genes exists in C. albicans cells remains to be established. Nonetheless, our results do point toward a close interaction between raft lipid constituents and drug susceptibilities as well as morphogenesis in C. albicans.
Acknowledgments
The work presented in this paper has been supported in parts by grants to R.P. from the Department of Biotechnology, India (BT/PR1110/MED/09/186/98); Volkswagen Foundation, Germany (1/76 798); and the European Commission, Brussels (QLK-CT-2001-02377). T.P., P.S., and N.A.G. thank the Council of Scientific and Industrial Research for the award of senior research fellowships.
We thank R. L. Lester, University of Kentucky College of Medicine, Lexington, for providing us the yeast sphingolipid standards. We thank Amitabha Mukhopadhyay, National Institute of Immunology, New Delhi, India, for providing us with confocal facilities. We also thank Archana for excellent assistance in mass spectrometry and Ali Abdul Lattif for help with microscopy. Our special thanks go to Sneha Sudha Komath for her valuable suggestions during the preparation of the manuscript.
Our thanks to Joachim Morschhauser, University of Würzburg, Germany, for allowing us to use the plasmid with the CDR1-GFP cassette and SAT1 marker constructed in his laboratory.
REFERENCES
- 1.Adams, A., D. E. Gottschling, C. A. Kaiser, and T. Stearns. 1997. Methods in yeast genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 2.Alonso-Monge, R., F. Navarro-Garcia, G. Molero, R. Diez-Orejas, M. Gustin, J. Pla, M. Sanchez, and C. Nombela. 1999. Role of the mitogen-activated protein kinase Hog1p in morphogenesis and virulence of Candida albicans. J. Bacteriol. 181:3058-3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Augsten, M., C. Hubner, M. Nguyen, W. Kunkel, A. Hartl, and R. Eck. 2002. Defective hyphal induction of a Candida albicans phosphatidylinositol 3-phosphate 5-kinase null mutant on solid media does not lead to decreased virulence. Infect. Immun. 70:4462-4470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bagnat, M., S. Keranen, A. Shevchenko, A. Shevchenko, and K. Simons. 2000. Lipid rafts function in biosynthetic delivery of proteins to the cell surface in yeast. Proc. Natl. Acad. Sci. USA 97:3254-3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bossche, H. V., P. Marichal, and F. C. Odds. 1994. Molecular mechanisms of drug resistance in fungi. Trends Microbiol. 2:393-400. [DOI] [PubMed] [Google Scholar]
- 6.Dickson, R. C., and R. L. Lester. 1999. Metabolism and selected functions of sphingolipids in the yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta 1438:305-321. [DOI] [PubMed] [Google Scholar]
- 7.Dickson, R. C., and R. L. Lester. 1999. Yeast sphingolipids. Biochim. Biophys. Acta 1426:347-357. [DOI] [PubMed] [Google Scholar]
- 8.Dickson, R. C., E. E. Nagiec, G. B. Wells, M. M. Nagiec, and R. L. Lester. 1997. Synthesis of mannose-(inositol-P)2-ceramide, the major sphingolipid in Saccharomyces cerevisiae, requires the IPT1 (YDR072c) gene. J. Biol. Chem. 272:29620-29625. [DOI] [PubMed] [Google Scholar]
- 9.Espinel-Ingroff, A. 2001. In vitro fungicidal activities of voriconazole, itraconazole, and amphotericin B against opportunistic moniliaceous and dematiaceous fungi. J. Clin. Microbiol. 39:954-958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fonzi, W. A., and M. Y. Irwin. 1993. Isogenic strain construction and gene mapping in Candida albicans. Genetics 134:717-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Franz, R., S. L. Kelly, D. C. Lamb, D. E. Kelly, M. Ruhnke, and J. Morschhauser. 1998. Multiple molecular mechanisms contribute to a stepwise development of fluconazole resistance in clinical Candida albicans strains. Antimicrob. Agents Chemother. 42:3065-3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Funato, K., B. Vallee, and H. Riezman. 2002. Biosynthesis and trafficking of sphingolipids in the yeast Saccharomyces cerevisiae. Biochemistry 41:15105-15114. [DOI] [PubMed] [Google Scholar]
- 13.Gietz, D., A. St. Jean, R. A. Woods, and R. H. Schiestl. 1992. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 20:1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Graybill, J. R., E. Montalbo, W. R. Kirkpatrick, M. F. Luther, S. G. Revankar, and T. F. Patterson. 1998. Fluconazole versus Candida albicans: a complex relationship. Antimicrob. Agents Chemother. 42:2938-2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hallstrom, T. C., L. Lambert, S. Schorling, E. Balzi, A. Goffeau, and W. S. Moye-Rowley. 2001. Coordinate control of sphingolipid biosynthesis and multidrug resistance in Saccharomyces cerevisiae. J. Biol. Chem. 276:23674-23680. [DOI] [PubMed] [Google Scholar]
- 16.Hearn, J. D., R. L. Lester, and R. C. Dickson. 2003. The uracil transporter Fur4p associates with lipid rafts. J. Biol. Chem. 278:3679-3686. [DOI] [PubMed] [Google Scholar]
- 17.Hechtberger, P., E. Zinser, R. Saf, K. Hummel, F. Paltauf, and G. Daum. 1994. Characterization, quantification and subcellular localization of inositol-containing sphingolipids of the yeast, Saccharomyces cerevisiae. Eur. J. Biochem. 225:641-649. [DOI] [PubMed] [Google Scholar]
- 18.Heidler, S. A., and J. A. Radding. 2000. Inositol phosphoryl transferases from human pathogenic fungi. Biochim. Biophys. Acta 1500:147-152. [DOI] [PubMed] [Google Scholar]
- 19.Jung, W. H., and L. I. Stateva. 2003. The cAMP phosphodiesterase encoded by CaPDE2 is required for hyphal development in Candida albicans. Microbiology 149:2961-2976. [DOI] [PubMed] [Google Scholar]
- 20.Kohli, A. K., V. Gupta, S. Krishnamurthy, S. E. Hasnain, and R. Prasad. 2001. Specificity of drug transport mediated by CaMDR1: a major facilitator of Candida albicans. J. Biosci. 26:101-107. [DOI] [PubMed] [Google Scholar]
- 21.Kohli, A. K., M. Smriti, K. Mukhopadhyay, and R. Prasad. 2002. In vitro low-level resistance to azoles in Candida albicans is associated with changes in membrane lipid fluidity and asymmetry. Antimicrob. Agents Chemother. 46:1046-1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krishnamurthy, S., V. Gupta, R. Prasad, S. L. Panwar, and R. Prasad. 1998. Expression of CDR1, a multidrug resistance gene of Candida albicans: in vitro transcriptional activation by heat shock, drugs and human steroid hormones. FEMS Microbiol. Lett. 160:191-197. [DOI] [PubMed] [Google Scholar]
- 23.Leber, A., P. Fischer, R. Schneiter, S. D. Kohlwein, and G. Daum. 1997. The yeast mic2 mutant is defective in the formation of mannosyl-diinositolphosphorylceramide. FEBS Lett. 411:211-214. [DOI] [PubMed] [Google Scholar]
- 24.Mandala, S. M., R. A. Thornton, M. Rosenbach, J. Milligan, M. Garcia-Calvo, H. G. Bull, and M. B. Kurtz. 2002. Khafrefungin, a novel inhibitor of sphingolipid synthesis. J. Biol. Chem. 272:32709-32714. [DOI] [PubMed] [Google Scholar]
- 25.Martin, S. W., and J. B. Konopka. 2004. Lipid rafts polarization contributes to hyphal growth in Candida albicans. Eukaryot. Cell 3:675-684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morschhauser, J., S. Michel, and J. Hacker. 1998. Expression of a chromosomally integrated, single-copy GFP gene in Candida albicans, and its use as a reporter of gene regulation. Mol. Gen. Genet. 257:420. [DOI] [PubMed] [Google Scholar]
- 27.Mukhopadhyay, K., A. K. Kohli, and R. Prasad. 2002. Drug susceptibilities of yeast cells are affected by membrane lipid composition. Antimicrob. Agents Chemother. 46:3695-3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mukhopadhyay, K., T. Prasad, P. Saini, T. J. Pucadyil, A. Chattopadhyay, and R. Prasad. 2004. Membrane sphingolipid-ergosterol interactions are important determinants of multidrug resistance in Candida albicans. Antimicrob. Agents Chemother. 48:1778-1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nagiec, M. M., E. E. Nagiec, J. A. Baltisberger, G. B. Wells, R. L. Lester, and R. C. Dickson. 1997. Sphingolipid synthesis as a target for antifungal drugs. J. Biol. Chem. 272:9809-9817. [DOI] [PubMed] [Google Scholar]
- 30.Reuss, O., A. Vik, R. Kolter, and J. Morschhauser. 2004. The SAT1 flipper, an optimized tool for gene disruption in Candida albicans. Gene 341:119-127. [DOI] [PubMed] [Google Scholar]
- 31.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 32.Shukla, S., P. Saini, Smriti, S. Jha, S. V. Ambudkar, and R. Prasad. 2003. Functional characterization of the ABC drug transporter Cdr1p from Candida albicans. Eukaryot. Cell 2:1361-1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stock, S. D., H. Hama, D. B. DeWald, and J. Y. Takemoto. 1999. SEC14-dependent secretion in Saccharomyces cerevisiae. Nondependence on sphingolipid synthesis-coupled diacylglycerol production. J. Biol. Chem. 274:12979-12983. [DOI] [PubMed] [Google Scholar]
- 34.Swain, E., K. Baudry, J. Stukey, V. McDonough, M. Germann, and J. T. Nickels, Jr. 2002. Sterol-dependent regulation of sphingolipid metabolism in Saccharomyces cerevisiae. J. Biol. Chem. 277:26177-26184. [DOI] [PubMed] [Google Scholar]
- 35.Timpel, C., S. Zink, S. Strahl-Bolsinger, K. Schroppel, and J. Ernst. 2000. Morphogenesis, adhesive properties, and antifungal resistance depend on the Pmt6 protein mannosyltransferase in the fungal pathogen Candida albicans. J. Bacteriol. 182:3063-3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wells, G. B., R. C. Dickson, and R. L. Lester. 1996. Isolation and composition of inositolphosphorylceramide-type sphingolipids of hyphal forms of Candida albicans. J. Bacteriol. 178:6223-6226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.White, T. C. 1997. Increased mRNA levels of ERG16, CDR, and MDR1 correlate with increased azole resistance in Candida albicans isolates from a patient infected with human immunodeficiency virus. Antimicrob. Agents Chemother. 41:1482-1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.White, T. C., K. A. Marr, and R. A. Bowden. 1998. Clinical, cellular, and molecular factors that contribute to antifungal drug resistance. Clin. Microbiol. Rev. 11:382-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Worgall, T. S., R. A. Johnson, T. Seo, H. Gierens, and R. J. Deckelbaum. 2002. Unsaturated fatty acid-mediated decreases in sterol regulatory element-mediated gene transcription are linked to cellular sphingolipid metabolism. J. Biol. Chem. 277:3878-3885. [DOI] [PubMed] [Google Scholar]