Abstract
Cambodia is located in an area of resistance to multiple antimalarials and has been the first country to implement the systematic use of an artesunate-mefloquine combination as first-line treatment for Plasmodium falciparum malaria. Little is known, however, about the prevalence of resistance mutations within the natural parasite populations, impeding rational drug policy in this context. Using direct sequencing of PCR products, we have analyzed sequence polymorphism of the dihydrofolate reductase-thymidylate synthase, dihydropteroate synthetase, and multidrug resistance 1 genes in a large number of clinical P. falciparum isolates collected in various areas of Cambodia. This highlighted a 100% prevalence of haplotypes with multiple mutations in the target genes of antifolates after more than a decade without use of antifolates for malaria therapy. A high prevalence of mutations in Pfmdr1, including mutations associated with decreased in vitro susceptibility to mefloquine and quinine, was also observed. In addition, novel, low-frequency mutations were detected in Pfmdr1. Our findings show an alarming rate of multilocus resistance genotypes in Cambodia, requiring diligent surveillance and imposing limitations on possible future drug combinations.
Plasmodium falciparum resistance to antimalarials has progressed at a steady rate during the last decades, particularly in Southeast Asia, and nowadays constitutes a major public health concern. Cambodia is one of the most affected regions (26). Chloroquine (CQ) resistance emerged in the early 1960s along the border between Thailand and Cambodia (11). Due to the loss of efficacy of CQ, combinations of diaphenylsulfone or sulfadoxine with pyrimethamine (SP) were gradually introduced in Cambodia from the 1970s onwards. However, treatment failures were rapidly observed, so that a triple therapy combining mefloquine, sulfadoxine, and pyrimethamine was introduced in the early 1980s (27, 29). To date, CQ remains efficacious only in some northeast provinces, but SP has lost efficacy everywhere in the country. Decreased susceptibility to mefloquine has also been reported in patients living near the Cambodia-Thailand border (6, 33). Artesunate in association with mefloquine has been distributed as the first-line treatment throughout the country for the last 5 years. Therapeutic efficacy studies performed by the National Malaria Control Program in 2001-2002 showed treatment failure rates of the artesunate-mefloquine combination of 0%, 3.4%, 5.6%, and 13.3% in Ratanakiri (east), Preah Vihear (north), and Pursat and Pailin (west) provinces, respectively. More recent in vivo monitoring in the Sampovloum area showed a 5.8% recrudescence rate (Yi Poravuth, personal communication).
The present situation with an uneven geographic distribution of drug resistance, with some areas showing quite worrying rates of multiresistant P. falciparum malaria, requires continued monitoring of drug resistance in the country (16). Surveillance for drug-resistant malaria is classically based on assessment of therapeutic efficacy and on measurements of in vitro susceptibility profiles of parasites collected in primary health care facilities. Surveillance can be carried out on a larger scale using molecular markers. Little is known about the evolution of the genes once selection pressures have been withdrawn or drugs have been replaced by other molecules. In view of the various selection pressures exerted on the parasite population in Cambodia over the last 40 years, a descriptive molecular survey of the overall diversity of the identified target genes is warranted in order to help rationalize control policy in the future and design the most appropriate surveillance markers.
Using molecular techniques, we recently described P. falciparum chloroquine resistance transporter (Pfcrt) polymorphism and its association with in vitro chloroquine resistance in Cambodia (8, 17). We now report an analysis of the genetic diversity of the P. falciparum dihydrofolate reductase-thymidylate synthase (Pfdhfr-ts), dihydropteroate synthetase (Pfdhps), and multidrug resistant 1 (Pfmdr1) genes in clinical isolates from Cambodia with documented in vitro sensitivity profiles for pyrimethamine, quinine, chloroquine, artesunate, and mefloquine. The aim of this study was to provide an overall picture of sequence diversity of these genes in several geographic areas from Cambodia with differing patterns of in vitro resistance.
MATERIALS AND METHODS
Sites and blood sampling.
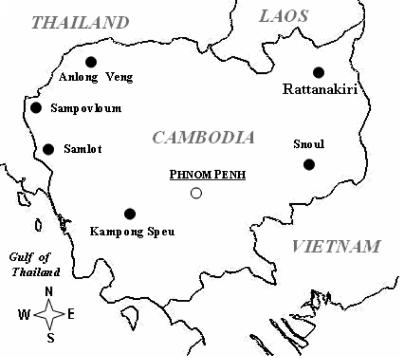
P. falciparum isolates were collected from patients recruited at home or at health centers during in vivo monitoring of the efficacy of the artesunate-plus-mefloquine or alternative artemisinin-based combinations, or during regular control surveys performed in the study areas over the 2001-2002 period. The study was approved by the Ethics Committee of the Cambodian Ministry of Health, and informed consent was obtained from each participant. Samples were collected in six areas where malaria is endemic: Sampovloum and Samlot in western Cambodia, along the Thai border; Anlong Veng in the north; Kampong Speu in the southwest; and finally Rattanakiri and Snoul, in the east, near the Vietnamese border (Fig. 1). Venous blood was collected in EDTA tubes and transported to Phnom Penh at 4°C within 24 to 48 h of collection. Giemsa-stained thin blood smears were examined upon arrival to determine parasite density and identify the single-species infections (with only P. falciparum). The samples were processed for in vitro assays, and the rest was aliquoted and stored at −80°C before genomic DNA isolation
FIG. 1.
Map of Cambodia with location of the sampling sites.
In vitro assays.
In vitro drug sensitivity was assessed using a classical isotopic 48-hour test as fully described elsewhere (16). The final concentrations ranged from 0.05 to 51.2 nM for artesunate, 1 to 1,024 nM for mefloquine, 5 to 5,120 nM for chloroquine, 0.1 to 102.4 μM for pyrimethamine, and 6.2 to 6,400 nM for quinine. Each plate included two drug-free control wells and one control well without parasites. The results were expressed as the 50% inhibitory concentration (IC50), defined as the concentration at which 50% of the incorporation of [3H]hypoxanthine was inhibited, compared to drug-free control wells. The threshold IC50s for in vitro resistance to pyrimethamine, chloroquine, quinine, mefloquine, and artesunate were 2 μM, 100 nM, 100 nM, 20 nM, and 3 nM, respectively. Except for pyrimethamine, chloroquine, and mefloquine, these in vitro thresholds were arbitrarily set and may not reflect in vivo drug susceptibility. Only isolates with acceptable growth during incubation and homogeneous in vitro drug responses characterized by typical sigmoid inhibition curves were included in the study. The validity and reproducibility of the assay were assessed using 3D7 parasites.
PCR amplification and sequencing.
Parasite DNA was extracted directly from 400 to 1,000 μl of infected blood using the phenol-chloroform method. Amplifications were performed in 50 μl reaction buffer containing 2 to 3 μl DNA, 0.5 to 0.8 μM each primer, 200 to 250 μM each deoxynucleoside triphosphate, 2 to 4 mM MgCl2, and 2.5 U Taq polymerase (Promega). The primers used were as follows (sequences in boldface are universal sequences added for sequencing): dhfrQs (5′-CTCGAGGAATTCGGATCCTATGATGGAACAAGTCGTCGAC-3′) and dhfrQas (5′-TCTAGAAAGCTTGGATCCTAAGCAGCCATATCCATTGAAATTT-3′), and dhps1s (5′-CCATTCCTCATGTGTATACAACAC-3′) and dhps2as (5′-GTTTAATCACATGTTTGCACTTTC-3′), designed to amplify the full Pfdhfr-ts coding sequence (1,827 bp, PlasmoDB PFD0830w) and 1,326 bp of the Pfdhps (PlasmoDB PF08_0095) sequence, respectively. Two Pfmdr1 gene regions, namely, mdrA (650 bp) and mdrO (890 bp), located 5′ and 3′ of the gene (PlasmoDB PFE1150w), respectively, were amplified using primers mdrA1s (5′-CTCGAGGAATTCGGATCCTGTTGAAAGATGGGTAAAGAGCAGAAAGAG-3′) and mdrA2as (5′-TCTAGAAAGCTTGGATCCTACTTTCTTATTACATATGACACCACAAACA-3′), and mdrO1s (5′-CTCGAGGAATTCGGATCCAGAAGATTATTTCTGTAATTTGATAGAAAAAGC-3′) and mdrO2as (5′-TCTAGAAAGCTTGGATCCATGATTCGATAAATTCATCTATAGCAGCAA-3′), respectively. These fragments correspond to the polymorphic domains of Pfmdr1 known to contain mutations associated with antimalarial susceptibility. After an initial step of denaturation at 94°C for 3 min, samples were subjected to 35 to 40 cycles of denaturation at 94°C for 30 s to 1 min; hybridization at 60°C (Pfdhps), 65°C (mdrA), or 58°C (mdrO) for 40 s (Pfdhps) or 2 min (mdrO and mdrA); and DNA synthesis at 72°C for 2 to 3 min. For Pfdhfr-ts, amplification, hybridization, and extension were done at the same temperature (66°C); incubation was for 4 min. DNA synthesis was achieved by final extension at 72°C for 10 min.
The sequencing strategy was as follows. PCR products were purified using a P-100 Gel Fine solution and a Multiscreen MAV N45 kit (Millipore). Sequencing reactions were performed from both ends using ABI Prism BigDye Terminator Cycle Sequencing-Ready Reaction kits and run on a 3700 Genetic Analyzer (Applied Biosystems). Sequencing was done using gene-specific or universal sequences introduced within PCR primers. Internal primers were used for >400-bp PCR fragments. The trace files were base called using phred (9, 10), and a quality file for each sequence was obtained. Sequences with segments of ≥300 bp called with a quality of >20 per base were retained. Sequence assembly was performed using the Phred Phrap consed package (9, 10, 13). Resulting contigs were assembled, visually examined individually, and aligned using Clustalw software. Since the isolates were not preselected for single genotype infections, heterogeneity was detected on chromatograms for a limited number of samples and at a few single nucleotide polymorphism positions. These sequences were rejected for the analysis of the haplotype. They were retained for the analysis of prevalence of individual mutations only if one nucleotide was dominant (i.e., >75% of the signal). Haplotypes for drug resistance markers were reconstituted from full sequences presenting a single allele at all positions and unambiguous signals at all positions.
Statistical analysis.
Statistical analysis was performed using the STATA software (Statacorporation). The nonparametric Mann-Whitney U test was used to evaluate whether differences observed in the in vitro responses to drugs were significant (P < 0.05).
RESULTS
Pfdhfr-ts.
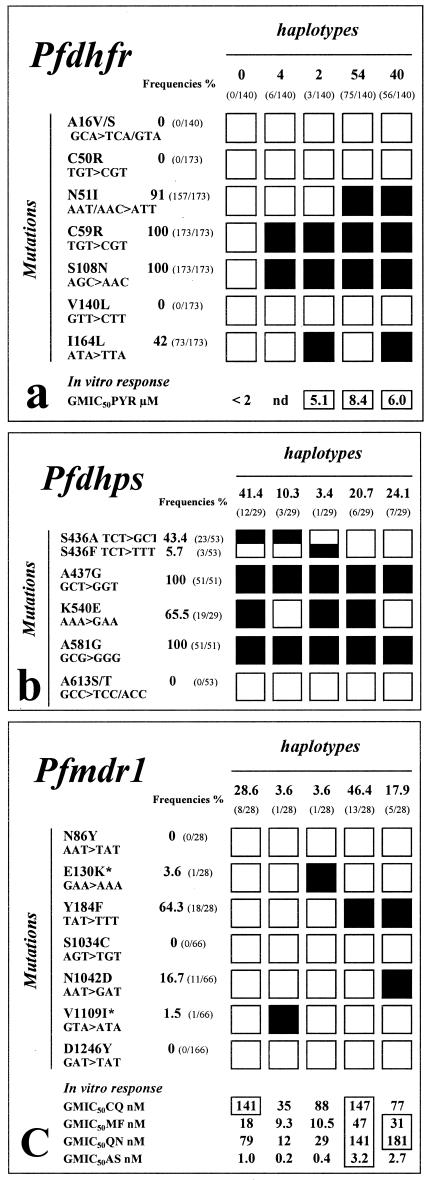
The full-length coding sequence was sequenced in 173 clinical isolates from six different areas. Results are summarized in Fig. 2a. No novel mutation was detected. The C59R and S108N mutations were observed in all isolates (100% frequency), and the N51I mutation was observed in 91% (20). The I164L mutation was present at a 42% frequency. It was less frequent in the east of the country, with only 32% and 19% prevalence in Rattanakiri and Snoul, respectively. The previously reported A16V/S, C50R, and V140L mutations were not detected here (22, 30, 34). Overall, four isoforms were identified. The N51I C59R S108N triple mutant haplotype was observed in 54% of isolates, and the N51I C59R S108N I164L quadruple mutant had an overall 40% frequency. Two percent of the isolates studied carried the C59R S108N I164L triple mutant haplotype. Thus, triple or quadruple mutants accounted for more than 96% of alleles, and the remaining 4% carried two mutations—C59R and S108N—which have invaded the entire local population.
FIG.2.
Sequence polymorphism in the Pfdhfr-ts, Pfdhps, and Pfmdr1 genes and resulting amino acid changes in clinical P. falciparum isolates from Cambodia as deduced by a PCR and direct sequencing strategy. Filled boxes show the position of the observed single nucleotide polymorphism for each haplotype. Empty boxes indicate a wild-type sequence at that position. The prevalence of the various mutations and haplotypes is indicated, as well as the in vitro responses of isolates to pyrimethamine (PYR), chloroquine (CQ), mefloquine (MF), quinine (QN), and artesunate (AS). In vitro responses are expressed as the GMIC50. (a) Pfdhfr-ts sequence polymorphism and its association with in vitro pyrimethamine resistance in 173 P. falciparum isolates from six different sites in Cambodia: Rattanakiri (n = 25), Snoul (n = 43), Kampong Speu (n = 21), Samlot (n = 17), Sampovloum (n = 62), and Anlong Veng (n = 5). (b) Pfdhps sequence polymorphism in 53 P. falciparum isolates from Rattanakiri (n = 2), Snoul (n = 26), Kampong Speu (n = 2), Sampovloum (n = 13), Samlot (n = 7), and Anlong Veng (n = 3). (c) Pfmdr1 sequence polymorphism and its association with in vitro susceptibility to a panel of antimalarials in 66 P. falciparum isolates from Rattanakiri (n = 2), Snoul (n = 21), Kampong Speu (n = 6), Sampovloum (n = 20), Samlot (n = 15), and Anlong Veng (n = 2). *, new mutations. The boxed GMIC50s indicate values above the threshold for in vitro resistance (16). In panel a, 95% confidence intervals observed for pyrimethamie were 5.07 to 14.0 μM (n = 26) and 3.06 to 11.8 μM (n = 27) for the ACIRNVI and ACIRNVL Pfdhfr-ts haplotype, respectively. In panel c, confidence interval values observed for the NEYSNVD (wild type), NEFSNVD, and NEFSDVD Pfmdr1 haplotypes were 69.0 to 288.2 nM (n = 6), 82.5 to 263.4 nM (n = 12), and 25.8 to 230.2 (n = 4), respectively, for CQ; 9.6 to 33.2 nM (n = 7), 27.2 to 81.6 nM (n = 13), and 16.9 to 55.6 nM (n = 5), respectively, for mefloquine; 33.0 to 187.8 nM (n = 7), 78.3 to 253.1 nM (n = 11), and 66.5 to 491.7 nM (n = 3), respectively, for quinine; and finally 0.56 to 1.65 nM (n = 7), 2.22 to 4.66 nM (n = 13), and 0.74 to 9.6 nM (n = 5), respectively, for artesunate. nd, no data.
All isolates responded weakly to pyrimethamine in vitro, with IC50s above 5,000 nM. The highest IC50 of pyrimethamine was associated with the N51I C59R S108N haplotype, but differences observed in the in vitro responses of the various haplotypes were below the level of significance (P ≫ 0.05).
Pfdhps.
The full-length Pfdhps could not be amplified with sufficient yield and purity to allow sequencing. We therefore focused on the 3′ region of the gene (1,326 bp), which contains all mutations reported so far (18, 22, 28). This region was fully characterized for 53 P. falciparum isolates from five different sites. No novel mutation was observed. The A437G and A581G mutations were detected at a 100% frequency (Fig. 2b). The S436A/F and K540E mutations were very common, with 49 and 65% prevalence, respectively. Consistent with widespread antifolate resistance in Cambodia, these mutations were homogeneously distributed over the study sites. Triple and quadruple Pfdhps mutants accounted for more than two-thirds of the parasites examined, with the remaining third corresponding to double mutants. The A613S/T mutation was not observed.
Pfmdr1.
Two fragments of 650 and 890 bp in size, located at the 5′ and 3′ ends of the gene, respectively, were amplified and sequenced (Fig. 2c) in 66 samples. Of the five known mutations, only Y184F and N1042D were observed, with a 64 and 17% frequency, respectively (31). Two new mutations, namely, E130K and V1109I, were observed with a 3.5 and 1.5% frequency, respectively. Both were detected in parasites originating from the Snoul region bordering Vietnam. The genotype carrying the single V184F mutation was the most abundant (64.3% frequency). One-third of the isolates carried the wild-type allele. These originated from the eastern and southern regions. The parasites carrying the single Y184F mutation responded poorly to chloroquine, mefloquine, and quinine, whereas the Y184F N1042D double mutant responded satisfactorily to chloroquine while having a geometric mean IC50 (GMIC50) to quinine and mefloquine higher than that of the parasites with a wild-type codon at these positions. The Y184F single and Y184F N1042D double mutants had significant, or nearly significant, elevated GMIC50s to artesunate (P of 0.002 and 0.07, respectively) and mefloquine (P of 0.02 and 0.05, respectively); other differences were below significance (P > 0.05).
DISCUSSION
Together with our previous reports highlighting associations of some specific Pfcrt haplotypes with the level of in vitro CQ susceptibility in Cambodia (8, 17), the work presented here constitutes the first multilocus inventory of sequence polymorphism in Cambodia and even in the Mekong region, the epicenter of multidrug resistance in Southeast Asia.
Pyrimethamine and cycloguanil target the bifunctional enzyme dihydrofolate reductase-thymidylate synthase. Dapsone and, particularly, sulfadoxine in association with pyrimethamine (Fansidar) inhibit the dihydropteroate synthetase. For both enzymes, specific polymorphisms have been associated with therapeutic failures (14). Consistent with a complete loss of efficacy of SP throughout the country and very high IC50s for pyrimethamine in vitro, triple or quadruple Pfdhfr-ts mutants were observed in nearly all isolates examined. The N51I, C59R, and S108N mutations have reached fixation. No wild types and single Pfdhfr-ts mutants were detected. Similarly, there was no wild-type or single mutant allele for Pfdhps. Triple and quadruple mutants accounted for more than two-thirds of the parasites examined with the remaining third corresponding to double mutants. To our knowledge, no similar situation has been reported elsewhere, including in Thailand, Laos, and Vietnam, where the inefficacy of antifolate drugs has long been established. In all three countries, resistance-associated mutations have a lower frequency (1, 2, 18, 30).
The P-glycoprotein homologue 1 modulates susceptibility to several antimalarials including mefloquine, quinine, chloroquine, and possibly artemisinins (5, 7, 24). Pfmdr1 showed considerable polymorphism in Cambodian isolates, although there was no picture of complete invasion of the population by specific mutations as observed for the antifolate targets. Two new mutations were detected at low frequency, and interestingly out of the five mutations previously described only Y184F and N1042D were observed (12). The N86Y, S1034C, and D1246Y codons were absent. This again differs from the situation reported for Thailand, where 86Y and 1034C mutants are present at a significant level (21, 23). In contrast, the observed high frequency of the Y184F mutation in the set of Cambodian samples studied here is in line with data from Thailand (21, 23). Interestingly, parasites with a wild-type Pfmdr1 haplotype originated from the eastern and southern regions, where the selection pressures exerted by quinine and mefloquine are also the weakest. An association of the Y184F mutation with mefloquine resistance in Asia was reported by Pickard et al. (21), who also noticed an increased susceptibility to chloroquine in isolates with a wild-type Pfmdr1 allele, which was not observed here. This discrepancy might reflect different geographic prevalences of Pfcrt haplotypes associated with chloroquine susceptibility and/or of Pfmdr1 copy number and allelic form of the amplified copy, a parameter that was not investigated here (3, 8, 17, 23). An estimate of Pfmdr1 copy number in our collection of Cambodian isolates is needed to help clarify the association of this locus with susceptibility to mefloquine and possibly other antimalarials in the country. The characterization of the various Pfmdr1 alleles present in Cambodia will help in the design of such studies.
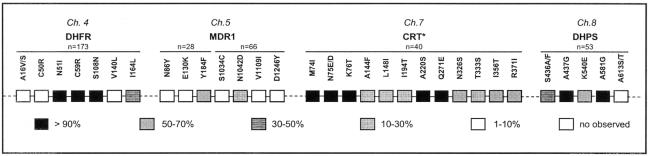
Our previous work showed an unusually high rate of Pfcrt mutations associated with resistance to chloroquine. The K76T mutation was observed in 93% of Cambodian isolates together with specific multiple mutant haplotypes (8, 17). Actually, the synopsis presented in Fig. 3 shows that Cambodian isolates have now accumulated numerous mutations at multiple loci. These data point to an alarmingly high rate of resistance mutations in key target genes of several antimalarials. Such a high frequency of multiresistant parasite genotypes a couple of decades after withdrawing use of antifolates and chloroquine for P. falciparum malaria indicates fixation, a situation that differs from the reversion to wild-type Pfcrt alleles after chloroquine withdrawal in some African settings (15). These differences may have arisen from differing patterns of drug policies and pressures after withdrawal.
FIG. 3.
Synopsis of multilocus genotypes in P. falciparum isolates from Cambodia. *, mutations in the Pfcrt gene have been reported elsewhere (8, 17).
The countrywide distribution of numerous resistance mutations, including those associated with decreased susceptibility to mefloquine, is of concern for the future of mefloquine-based combinations. Fortunately enough, artesunate and mefloquine combinations were efficacious despite a significant rate of mefloquine resistance in the population (19). Furthermore, deployment of the combination halted the progression of mefloquine resistance and possibly reduced it (4, 19). Since our data indicate a higher prevalence of mutations in Cambodia than Thailand, accurate and regular monitoring of drug resistance in Cambodia is needed to adapt guidelines for better management of malaria cases. It is worth noting that Cambodia was the first country to introduce the artesunate-mefloquine combination as first-line treatment for uncomplicated P. falciparum malaria. The current levels of multidrug resistance in Cambodia have therefore important implications for the design of epidemiologic surveillance of drug resistance using molecular markers. This is particularly important in view of recent evidence showing for both antifolates and chloroquine that resistance alleles have arisen in Southeast Asia and subsequently invaded Africa (25, 32). Thus, the present situation in Cambodia may well prefigure future situations in Africa and in other areas of endemicity where resistance is presently less prevalent. Our data also indicate that any future drug combinations and development of novel antimalarial compounds should target such multiresistant genetic backgrounds.
Acknowledgments
We are grateful to the staff of the National Malaria Center (Cambodian Ministry of Health) and to the Institut Pasteur du Cambodge, especially Pheaktra Chim, Rithy Sem, and Sina Nehm, who actively participated in this study. We also thank Chansuda Wongsrichanalai, Frédéric Ariey, and Reiko Tsuyuoka for fruitful discussions.
This work was supported by the French Academy of Sciences and by a grant from the European Commission (contract QLK2-CT20021-1503-RESMALCHIP).
REFERENCES
- 1.Berens, N., B. Schwoebel, S. Jordan, V. Vanisaveth, R. Phetsouvanh, E.-M. Christophel, S. Phompida, and T. Jelinek. 2003. Plasmodium falciparum: correlation of in vitro resistance to chloroquine and antifolates with genetic polymorphisms in isolates from the south of Lao PDR. Trop. Med. Int. Health 8:775-782. [DOI] [PubMed] [Google Scholar]
- 2.Biswas, S., A. Escalante, S. Chaiyaroj, P. Angkasekwinai, and A. A. Lal. 2000. Prevalence of point mutations in the dihydrofolate reductase and dihydropteroate synthetase genes of Plasmodium falciparum isolates from India and Thailand: a molecular epidemiologic study. Trop. Med. Int. Health 5:737-743. [DOI] [PubMed] [Google Scholar]
- 3.Bray, P. G., R. E. Martin, L. Tilley, S. A. Ward, K. Kirk, and D. A. Fidock. 2005. Defining the role of PfCRT in Plasmodium falciparum chloroquine resistance. Mol. Microbiol. 56:323-333. [DOI] [PubMed] [Google Scholar]
- 4.Brockman, A., R. Price, M. Van Gugt, D. G. Heppner, D. Walsh, P. Sookto, S. Wimonwattrawatee, S. Looareesuwan, N. J. White, and F. Nosten. 2000. Plasmodium falciparum antimalarial drug susceptibility on the north-western border of Thailand during five years of extensive use of artesunate-mefloquine. Trans. R. Soc. Trop. Med. Hyg. 94:537-544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cowman, A. F., D. Galatis, and J. K. Thompson. 1994. Selection for mefloquine resistance in Plasmodium falciparum is linked to amplification of the Pfmdr1 gene and cross-resistance to halofantrine and quinine. Proc. Natl. Acad. Sci. USA 91:1143-1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Denis, M. B., R. L. Kouznetsov, and M. Gibboda. 1991. In vivo response of multi-resistant Plasmodium falciparum infections to mefloquine and its combination with sulfadoxine/pyrimethamine in Cambodia. Folia Parasitol. 38:187-188. [PubMed] [Google Scholar]
- 7.Duraisingh, M. T., C. Roper, D. Walliker, and D. C. Warhurst. 2000. Increased sensitivity to the antimalarials mefloquine and artemisinin is conferred by mutations in the Pfmdr1 gene of Plasmodium falciparum. Mol. Microbiol. 36:955-961. [DOI] [PubMed] [Google Scholar]
- 8.Durrand, V., A. Berry, R. Sem, P. Glaziou, J. Beaudou, and T. Fandeur. 2004. Variations in the sequence and expression of the Plasmodium falciparum chloroquine resistance transporter (Pfcrt) and their relationship to chloroquine resistance in vitro. Mol. Biochem. Parasitol. 136:273-285. [DOI] [PubMed] [Google Scholar]
- 9.Ewing, B., and P. Green. 1998. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 8:186-194. [PubMed] [Google Scholar]
- 10.Ewing, B., L. Hillier, M. C. Wendl, and P. Green. 1998. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8:175-185. [DOI] [PubMed] [Google Scholar]
- 11.Eyles, D. E., C. C. Hoo, M. C. Warren, and A. A. Sandosham. 1963. Plasmodium falciparum resistant to chloroquine in Cambodia. Am. J. Trop. Med. Hyg. 12:840-843. [DOI] [PubMed] [Google Scholar]
- 12.Foote, S. J., D. E. Kyle, R. K. Martin, A. M. J. Oduola, D. J. Forsyth, D. J. Kemp, and A. F. Cowman. 1990. Several alleles of the multidrug resistance gene are closely linked to chloroquine resistance in Plasmodium falciparum. Nature 345:225-258. [DOI] [PubMed] [Google Scholar]
- 13.Gordon, D., C. Abajian, and P. Green. 1998. Consed: a graphical tool for sequence finishing. Genome Res. 8:195-202. [DOI] [PubMed] [Google Scholar]
- 14.Gregson, A., and C. V. Plowe. 2005. Mechanisms of resistance of malaria parasites to antifolates. Pharmacol. Rev. 57:117-145. [DOI] [PubMed] [Google Scholar]
- 15.Kublin, J. G., J. F. Cortese, E. M. Njunju, A. G. Mukadam, J. J. Wirima, P. N. Kazembe, A. A. Djimde, B. Kouriba, T. E. Taylor, and C. V. Plowe. 2003. Reemergence of chloroquine-sensitive Plasmodium falciparum malaria after cessation of chloroquine use in Malawi. J. Infect. Dis. 187:1870-1875. [DOI] [PubMed] [Google Scholar]
- 16.Lim, P., P. Chim, R. Sem, S. Nemh, Y. Poravuth, C. Lim, S. Seila, R. Tsuyuoka, M. B. Denis, D. Socheat, and T. Fandeur. 2005. In vitro monitoring of Plasmodium falciparum susceptibility to artesunate, mefloquine, quinine and chloroquine in Cambodia: 2001-2002. Acta Trop. 93:31-40. [DOI] [PubMed] [Google Scholar]
- 17.Lim, P., S. Chy, F. Ariey, S. Incardona, P. Chim, R. Sem, M. B. Denis, S. Hewit, S. Hoyer, D. Socheat, O. Puijalon, and T. Fandeur. 2003. The Pfcrt polymorphism and chloroquine resistance in Plasmodium falciparum isolates from Cambodia. Antimicrob. Agents Chemother. 47:87-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Masimirembwa, C. M., N. Phuong-dung, B. Q. Phuc, L. Duc-Dao, N. D. Sy, O. Skold, and G. Swedberg. 1999. Molecular epidemiology of Plasmodium falciparum antifolate resistance in Vietnam: genotyping for resistance variants of dihydropteroate synthase and dihydrofolate reductase. Int. J. Antimicrob. Agents 12:203-211. [DOI] [PubMed] [Google Scholar]
- 19.Nosten, F., M. van Vugt, R. Price, C. Luxemburger, K. L. Thway, A. Brockman, R. McGready, F. ter Kuile, S. Looareesuwan, and N. J. White. 2000. Effects of artesunate-mefloquine combination on incidence of Plasmodium falciparum malaria and mefloquine resistance in western Thailand: a prospective study. Lancet 356:297-302. [DOI] [PubMed] [Google Scholar]
- 20.Peterson, D. S., D. Walliker, and T. E. Wellems. 1988. Evidence that a point mutation in dihydrofolate reductase-thymidylate synthase confers resistance to pyrimethamine in falciparum malaria. Proc. Natl. Acad. Sci. USA 85:9114-9118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pickard, A. L., C. Wongsrichanalai, A. Purfield, D. Kamwendo, K. Emery, C. Zalewski, F. Kawamoto, R. S. Miller, and S. R. Meshnick. 2003. Resistance to antimalarials in Southeast Asia and genetic polymorphisms in Pfmdr1. Antimicrob. Agents Chemother. 47:2418-2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Plowe, C. V., J. F. Cortese, A. Djimde, O. C. Nwanyanwu, W. M. Watkins, P. A. Winstanley, J. G. Estrada-Franco, R. E. Mollinedo, J. L. Avila, J. L. Cespedes, D. Carter, and O. K. Doumbo. 1997. Mutations in Plasmodium falciparum dihydrofolate reductase and dihydropteroate synthase and epidemiologic patterns of pyrimethamine-sulfadoxine use and resistance. J. Infect. Dis. 176:1590-1596. [DOI] [PubMed] [Google Scholar]
- 23.Price, R. N., A.-C. Uhlemann, A. Brockman, R. McGready, E. Ashley, L. Phaipun, R. Patel, K. Laing, S. Looareesuwan, N. J. White, F. Nosten, and S. Krishna. 2004. Mefloquine resistance in Plasmodium falciparum increased Pfmdr1 gene copy number. Lancet 364:438-447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reed, M. B., K. J. Saliba, S. R. Caruana, K. Kirk, and A. F. Cowman. 2000. Pgh1 modulates sensitivity and resistance to multiple antimalarials in Plasmodium falciparum. Nature 403:906-909. [DOI] [PubMed] [Google Scholar]
- 25.Roper, C., R. Pearce, S. Nair, B. Sharp, F. Nosten, and T. Anderson. 2004. Intercontinental spread of pyrimethamine-resistant malaria. Science 30:1124. [DOI] [PubMed] [Google Scholar]
- 26.Singhasivanon, P. 1999. Mekong malaria. Malaria, multi-drug resistance and economic development in the greater Mekong subregion of Southeast Asia. Southeast Asian J. Trop. Med. Public Health 30(Suppl. 4):1-101. [PubMed] [Google Scholar]
- 27.Thimasarn, K., J. Sirichaisinthop, S. Vijaykadga, S. Tansophalaks, P. Yamokgul, A. Laomiphol, C. Palananth, U. Thamewat, S. Thaitong, and W. Rooney. 1995. In vivo study of the response of Plasmodium falciparum to standard mefloquine/sulfadoxine/pyrimethamine (MSP) treatment among gem miners returning from Cambodia. Southeast Asian J. Trop. Med. Public Health 26:204-212. [PubMed] [Google Scholar]
- 28.Triglia, T., J. G. T. Menting, C. Wilson, and A. F. Cowman. 1997. Mutations of dihydropteroate synthase are responsible for sulfone and sulfonamide resistance in Plasmodium falciparum. Proc. Natl. Acad. Sci. USA 94:13944-13949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Verdrager. J. 1986. Epidemiology of emergence and spread of drug-resistant falciparum malaria in South-East Asia and Australasia. J. Trop. Med. Hyg. 89:277-289. [PubMed] [Google Scholar]
- 30.Wang, P., C.-S. Lee, R. Bayoumi, A. Djimbe, O. Doumbo, G. Swedberg, L. D. Duc-Dao, H. Mshinda, M. Tanner, W. M. Watkins, P. F. G. Sims, and J. E. Hyde. 1997. Resistance to antifolate in Plasmodium falciparum monitored by sequence analysis of dihydropteroate synthetase and dihydrofolate reductase alleles in a large number of field samples of diverse origins. Mol. Biochem. Parasitol. 89:161-177. [DOI] [PubMed] [Google Scholar]
- 31.Wongsrichanalai, C., A. L. Pickard, H. Wernsdorfer, and S. R. Meshnick. 2002. Epidemiology of drug-resistant malaria. Lancet Infect. Dis. 2:209-218. [DOI] [PubMed] [Google Scholar]
- 32.Wootton, J. C., X. Feng, M. T. Ferdig, R. A. Cooper, J. Mu, D. I. Baruch, A. J. Magill, and X. Z. Su. 2002. Genetic diversity and chloroquine selective sweeps in Plasmodium falciparum. Nature 418:320-332. [DOI] [PubMed] [Google Scholar]
- 33.World Health Organization. 2000. Report on the informal consultation on monitoring resistance to antimalarial drugs in the Mekong Region. World Health Organization, Geneva, Switzerland.
- 34.Zindrou, S., N. P. Dung, N. D. Sy, O. Sköld, and G. Swedberg. 1996. Plasmodium falciparum: mutation pattern in the dihydrofolate reductase thymidylate synthase genes of Vietnamese isolates, a novel mutation, and coexistence of two clones in a Thai patient. Exp. Parasitol. 84:56-64. [DOI] [PubMed] [Google Scholar]