Abstract
Two clinical Streptococcus pneumoniae isolates, identified as resistant to macrolides and chloramphenicol and nonsusceptible to linezolid, were found to contain 6-bp deletions in the gene encoding riboprotein L4. The gene transformed susceptible strain R6 so that it exhibited such resistance, with the transformants also showing a fitness cost. We demonstrate a novel bacterial mechanism of resistance to chloramphenicol and nonsusceptibility to linezolid.
Macrolide resistance in Streptococcus pneumoniae is predominantly caused by acquisition of the erm(B) (target modification) (33) or mef(A) (drug efflux) (28) gene or a combination of these mechanisms (16). Mutations in 23S rRNA and riboproteins L4 and L22 have more recently been found to confer macrolide resistance (6, 29, 30). Nonsusceptibility to linezolid in laboratory-generated resistant Escherichia coli isolates (35) and enterococci (21) as well as in clinical isolates of methicillin-resistant Staphylococcus aureus (17, 32) and enterococci (4, 15) has been found to be conferred by mutations in domain V of 23S rRNA. To date, linezolid-nonsusceptible pneumococcal strains are extremely rare. Chloramphenicol resistance in the pneumococcus occurs by acquisition of the cat gene encoding chloramphenicol acetyltransferase (3, 18). Chloramphenicol acetyltransferase acetylates chloramphenicol, resulting in derivatives that are unable to bind the ribosome (22, 23). In this study, two clinical pneumococcal isolates with elevated macrolide, linezolid, and chloramphenicol MICs were identified and investigated for their resistance mechanisms.
PU1071099 (PROTEKT surveillance study) (7) was isolated in Boston in 2001 from sputum of a 67-year-old, and TN33388 (ABCs program of the Centers for Disease Control) was isolated in Tennessee in 2003 from the blood of a 32-year-old who had been exposed to long-term azithromycin prophylaxis. Unencapsulated laboratory strain S. pneumoniae R6 was used in transformation studies. Pneumococci were routinely cultured at 37°C in 5% CO2 on Mueller-Hinton agar supplemented with 5% horse blood. MICs were determined according to the CLSI (NCCLS) broth microdilution method (19). Serotyping was by the Quellung reaction with antisera from the Statens Serum Institut (Copenhagen, Denmark). Multilocus sequence typing (MLST) was performed as previously described (5) with primers described by Gertz et al. (9). MLST alleles were determined using the Wisconsin version 10.2 package (Genetics Computer Group, Madison, Wisconsin). Sequence types were assigned using the MLST database (http://spneumoniae.mlst.net/). Phenotypic data for both isolates are shown in Table 1. Based on MLST analyses, the isolates were determined to be clonally unrelated.
TABLE 1.
Phenotypic and genotypic data for the isolates, untransformed R6, and the R6 transformants
| Strain | Serotype | Multilocus sequence typea | L4 deletion | MIC (μg/ml) ofb:
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ERY | CLR | AZM | CLI | LZD | S-B | Q-D | CHL | TEL | TET | PEN | ||||
| Isolates | ||||||||||||||
| PU1071099 | 9N | ST66 (2-8-2-4-6-1-1) | 65WR66 | 2 | 1 | 4 | 0.12 | 4 | 4 | 1 | 16 | 0.015 | 0.25 | 0.03 |
| TN33388 | 33F | ST100 (5-12-29-12-9-39-18) | 68KG69 | 2 | 1 | 4 | 0.12 | 4 | 4 | 2 | 16 | 0.015 | 0.25 | 0.03 |
| Transformants | ||||||||||||||
| Untransformed R6 | None | 0.12 | 0.06 | 0.12 | 0.06 | 1 | 4 | 0.5 | 4 | 0.015 | 0.5 | 0.03 | ||
| R6 PU1071099/L4 | 65WR66 | 1 | 1 | 2 | 0.06 | 4 | 8 | 0.5 | 16 | 0.008 | 0.5 | 0.015 | ||
| R6TN33388/L4 | 68KG69 | 2 | 0.5 | 2 | 0.12 | 4 | 8 | 2 | 8 | 0.008 | 0.5 | 0.015 | ||
Numbers in parentheses indicate the allelic profile of each isolate determined by MLST and used to determine the sequence type.
Abbreviations: CLR, clarithromycin; AZM, azithromycin; CLI, clindamycin; LZD, linezolid; S-B, streptogramin B; Q-D, quinupristin-dalfopristin; CHL, chloramphenicol; TEL, telithromycin; TET, tetracycline; PEN, penicillin.
Chromosomal DNA was extracted as previously described (24). PCR-based methods were used to screen for the erm(B), mef(A) (27), and cat (34) genes, for which both isolates were negative. The four alleles encoding 23S rRNA were amplified separately according to previously described methods (6, 29). Riboprotein genes rplD (L4) and rplV (L22) were amplified using primer pairs L4F (AAATCAGCAGTTAAAGCTGG) and L4R (GAGCTTTCAGTGATGACAGG) and L22F (GCAGACGACAAGAAAACACG) and L22R (ATTGGATGTACTTTTTGACC), respectively. For each 50-μl reaction mixture, 3 μl of chromosomal DNA was added to a mix containing 2.5 U of Taq DNA polymerase, 1× reaction buffer, 1.5 mM MgCl2, 200 μM (each) dATP, dCTP, dGTP, and dTTP, and 800 nM (each) forward and reverse primers. Cycling parameters were as follows: 94°C for 2 min; 94°C for 1 min, 54°C for 2 min, and 72°C for 3 min for 27 cycles; and 72°C for 10 min. Amplified products were purified from agarose gel with the Geneclean kit (Bio101, Inc., La Jolla, CA). DNA sequencing was performed using the BigDye Terminator cycle sequencing kit (Applied Biosystems, Foster City, CA) and an Applied Biosystems model 310 automated DNA sequencer. Six-base-pair deletions resulting in the deletion of two amino acids from L4 were found in both isolates (Table 1). The mutation in TN33388 (67QKGT70 to 67Q--T70) is a novel mutation in S. pneumoniae. For both isolates, the genes encoding riboprotein L22 and 23S rRNA were found to be of the wild type compared with those in S. pneumoniae R6 and S. pneumoniae ATCC 33400.
The effect of the L4 mutations on susceptibility to protein synthesis-inhibiting antibiotics was investigated. S. pneumoniae R6 was made competent by culture in C-medium (31), and transformation was performed as previously described (25). The L4 gene was used as donor DNA, and transformants (four for each isolate) were selected on Mueller-Hinton agar supplemented with 5% horse blood and containing erythromycin (ERY; 0.25 to 0.5 μg/ml). MICs were determined and mutations confirmed by sequencing. R6PU1071099/L4 and R6PU1071099/L4 transformants showed decreased susceptibility to ERY, clarithromycin, azithromycin, linezolid, and chloramphenicol in comparison with untransformed R6. A one-dilution increase in the streptogramin B MIC was observed for both sets of transformants. R6TN33388/L4 transformants additionally showed reduced susceptibility to clindamycin and quinupristin-dalfopristin (Table 1).
The L4 mutations detected in this study are likely to account for the macrolide resistance of the isolates since L4 mutations, most commonly in a highly conserved region (63KPWRQKGTGRAR74), have been shown to confer macrolide resistance in S. pneumoniae (29, 30). These mutations were also found to be responsible for the nonsusceptibility of the isolates to linezolid, with the MICs for the transformants being equivalent to those for the parent isolates. This represents a novel mechanism of linezolid nonsusceptibility as, in previous reports on gram-positive bacteria, resistance has been attributed to mutations in domain V of 23S rRNA (15, 32). Mutations in the L4 gene confer macrolide resistance in Staphylococcus aureus (20). Should these mutations be shown to confer nonsusceptibility to linezolid in Staphylococcus aureus, they would be of particular significance as linezolid is widely used for the treatment of infection with methicillin-resistant Staphylococcus aureus. The L4 deletions described in this study were also found to confer a novel mechanism of resistance to chloramphenicol. Previous studies have indicated that although macrolides, linezolid, and chloramphenicol have different mechanisms of action, they appear to share a common binding site on the large ribosomal subunit. Suryanarayana (26) showed that extracted E. coli L4 binds to both ERY- and chloramphenicol-coupled affinity columns. In addition, chloramphenicol competes with the binding of the oxazolidinone eperezolid to the 50S ribosomal subunit (14) and mutations in domain V of 23S rRNA of Halobacterium halobium confer resistance to linezolid as well as chloramphenicol (13). From this study, it can be concluded that L4 forms an integral part of this common binding site. The L4 deletion detected in PU1071099 has been previously described in group A streptococci (1, 2). However, susceptibility to linezolid and chloramphenicol was not determined in the previous studies. This mutation is also likely to confer nonsusceptibility to chloramphenicol and linezolid in Streptococcus pyogenes. The clinical significance of riboprotein mutations was emphasized by the death of a patient from an infection with a pneumococcal strain with macrolide resistance conferred by an L22 mutation (D. M. Musher, M. E. Dowell, V. D. Shortridge, R. K. Flamm, J. H. Jorgensen, P. Le Magueres, and K. L. Krause, Letter, N. Engl. J. Med. 346:630-631, 2002).
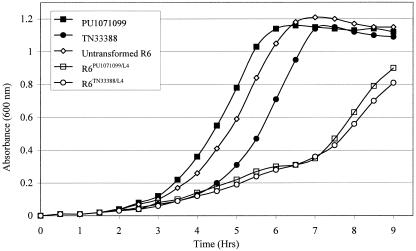
Growth studies were performed in duplicate by inoculating glycerol stocks of pneumococci into tryptone soy broth (1:100 dilution) and monitoring turbidity at 600 nm every 30 min for 9 h (Fig. 1). Mass doubling times (in minutes) during the exponential phase of growth were as follows: PU1071099, 53.9; TN33388, 53.4; untransformed R6, 59.6; R6PU1071099/L4, 88.1; and R6TN33388/L4, 102.6. The reduced growth rates of the transformants suggest that the L4 mutations are associated with a fitness cost. The rplD gene is essential and is regarded as one of the minimal set of genes necessary for bacterial life (10). L4 forms a part of the exit tunnel of the large ribosomal subunit and is thought to be involved in processing of the nascent polypeptide chains (8). Mutations may inhibit antibiotic binding; however, as a consequence protein synthesis may be affected. Decreased growth rates may also be due to the fact that L4 mutations perturb the three-dimensional structure of 23S rRNA (12). In contrast, the mass doubling times of the clinical isolates were shorter than that for R6. Bacteria adapt to a decrease in fitness as a result of resistance mutations by developing compensatory mutations that restore their fitness without affecting resistance (11). Our data suggest that the isolates may have acquired such compensatory mutations.
FIG. 1.
Growth curves of the isolates, untransformed R6, and R6 transformants carrying L4 deletion mutations. Bacteria were grown at 37°C.
In conclusion, we have for the first time described mutations in pneumococcal isolates conferring nonsusceptibility to linezolid together with a novel mechanism of simultaneous resistance to macrolides, oxazolidinones, and chloramphenicol.
Acknowledgments
This research was supported by grants from the Medical Research Council, the National Institute for Communicable Diseases, and the University of the Witwatersrand, South Africa. DNA sequencing was performed with an automated DNA sequencer funded by the Wellcome Trust (grant 061017). The PROTEKT study is financially supported by Aventis.
We thank Lesley McGee of Emory University and Bernie Beall of the CDC, Atlanta, Ga., for the molecular epidemiology of the isolates and for critical review of the manuscript. We thank Jemma Shackcloth of GR Micro Ltd. for her assistance. We thank André Bryskier of Sanofi Aventis, Paris, France, for providing streptogramin B and Steven Brown of the Clinical Microbiology Institute, Wilsonville, Oreg., for providing demographic data on the patient from Boston, Mass. We are grateful to Brenda Barnes and Samir Hanna for their assistance in gathering data on the isolate from Tennessee.
REFERENCES
- 1.Bingen, E., R. Leclercq, F. Fitoussi, N. Brahimi, B. Malbruny, D. Deforche, and R. Cohen. 2002. Emergence of group A streptococcus strains with different mechanisms of macrolide resistance. Antimicrob. Agents Chemother. 46:1199-1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bozdogan, B., P. C. Appelbaum, L. Ednie, I. N. Grivea, and G. A. Syrogiannopoulos. 2003. Development of macrolide resistance by ribosomal protein L4 mutation in Streptococcus pyogenes during miocamycin treatment of an eight-year-old Greek child with tonsillopharyngitis. Clin. Microbiol. Infect. 9:966-969. [DOI] [PubMed] [Google Scholar]
- 3.Dang-Van, A., G. Tiraby, J. F. Acar, W. V. Shaw, and D. H. Bouanchaud. 1978. Chloramphenicol resistance in Streptococcus pneumoniae: enzymatic acetylation and possible plasmid linkage. Antimicrob. Agents Chemother. 13:557-583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dibo, I., S. K. Pillai, H. S. Gold, M. R. Baer, M. Wetzler, J. L. Slack, P. A. Hazamy, D. Ball, C. B. Hsiao, P. L. McCarthy, and B. H. Segal. 2004. Linezolid-resistant Enterococcus faecalis isolated from a cord blood transplant recipient. J. Clin. Microbiol. 42:1843-1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Enright, M. C., and B. G. Spratt. 1998. A multilocus sequence typing scheme for Streptococcus pneumoniae: identification of clones associated with serious invasive disease. Microbiology 144:3049-3060. [DOI] [PubMed] [Google Scholar]
- 6.Farrell, D. J., S. Douthwaite, I. Morrissey, S. Bakker, J. Poehlsgaard, L. Jakobsen, and D. Felmingham. 2003. Macrolide resistance by ribosomal mutation in clinical isolates of Streptococcus pneumoniae from the PROTEKT 1999-2000 study. Antimicrob. Agents Chemother. 47:1777-1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Farrell, D. J., I. Morrissey, S. Bakker, S. Buckridge, and D. Felmingham. 2004. In vitro activities of telithromycin, linezolid, and quinupristin-dalfopristin against Streptococcus pneumoniae with macrolide resistance due to ribosomal mutations. Antimicrob. Agents Chemother. 48:3169-3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gabashvili, I. S., S. T. Gregory, M. Valle, R. Grassucci, M. Worbs, M. C. Wahl, A. E. Dahlberg, and J. Frank. 2001. The polypeptide tunnel system in the ribosome and its gating in erythromycin resistance mutants of L4 and L22. Mol. Cell 8:181-188. [DOI] [PubMed] [Google Scholar]
- 9.Gertz, R. E., Jr., M. C. McEllistrem, D. J. Boxrud, L. Zhongya, V. Sakota, T. A. Thompson, R. R. Facklam, J. M. Besser, L. H. Harrison, C. G. Whitney, and B. Beall. 2003. Clonal distribution of invasive pneumococcal isolates from children and selected adults in the United States prior to 7-valent conjugate vaccine introduction. J. Clin. Microbiol. 41:4194-4216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gil, R., F. J. Silva, J. Peretó, and A. Moya. 2004. Determination of the core of a minimal bacterial gene set. Microbiol. Mol. Biol. Rev. 68:518-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gillespie, S. H. 2001. Antibiotic resistance in the absence of selective pressure. Int. J. Antimicrob. Agents 17:171-176. [DOI] [PubMed] [Google Scholar]
- 12.Gregory, S. T., and A. E. Dahlberg. 1999. Erythromycin resistance mutations in ribosomal proteins L22 and L4 perturb the higher order structure of 23S ribosomal RNA. J. Mol. Biol. 289:827-834. [DOI] [PubMed] [Google Scholar]
- 13.Kloss, P., L. Xiong, D. L. Shinabarger, and A. S. Mankin. 1999. Resistance mutations in 23S rRNA identify the site of action of the protein synthesis inhibitor linezolid in the ribosomal peptidyl transferase center. J. Mol. Biol. 294:93-101. [DOI] [PubMed] [Google Scholar]
- 14.Lin, A. H., R. W. Murray, T. J. Vidmar, and K. R. Marotti. 1997. The oxazolidinone eperezolid binds to the 50S ribosomal subunit and competes with binding of chloramphenicol and lincomycin. Antimicrob. Agents Chemother. 41:2127-2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marshall, S. H., C. J. Donskey, R. Hutton-Thomas, R. A. Salata, and L. B. Rice. 2002. Gene dosage and linezolid resistance in Enterococcus faecium and Enterococcus faecalis. Antimicrob. Agents Chemother. 46:3334-3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McGee, L., K. P. Klugman, A. Wasas, T. Capper, A. Brink, and the Antibiotics Surveillance Forum of South Africa. 2001. Serotype 19F multiresistant pneumococcal clone harboring two erythromycin resistance determinants [erm(B) and mef(A)] in South Africa. Antimicrob. Agents Chemother. 45:1595-1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meka, V. G., S. K. Pillai, G. Sakoulas, C. Wennersten, L. Venkataraman, P. C. DeGirolami, G. M. Eliopoulos, R. C. Moellering, and H. S. Gold. 2004. Linezolid resistance in sequential Staphylococcus aureus isolates associated with a T2500A mutation in the 23S rRNA gene and a loss of a single copy of rRNA. J. Infect. Dis. 190:311-317. [DOI] [PubMed] [Google Scholar]
- 18.Miyamura, S., H. Ochiai, Y. Nitahara, Y. Nakagawa, and M. Terao. 1977. Resistance mechanism of chloramphenicol in Streptococcus haemolyticus, Streptococcus pneumoniae and Streptococcus faecalis. Microbiol. Immunol. 21:69-76. [DOI] [PubMed] [Google Scholar]
- 19.National Committee for Clinical Laboratory Standards. 2003. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 6th ed. Approved standard. NCCLS document M7-A6. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 20.Prunier, A., B. Malbruny, M. Laurans, J. Brouard, J. Duhamel, and R. Leclercq. 2003. High rate of macrolide resistance in Staphylococcus aureus strains from patients with cystic fibrosis reveals high proportions of hypermutable strains. J. Infect. Dis. 187:1709-1716. [DOI] [PubMed] [Google Scholar]
- 21.Prystowsky, J., F. Siddiqui, J. Chosay, D. L. Shinabarger, J. Millichap, L. R. Peterson, and G. A. Noskin. 2001. Resistance to linezolid: characterization of mutations in rRNA and comparison of their occurrences in vancomycin-resistant enterococci. Antimicrob. Agents Chemother. 45:2154-2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shaw, W. V. 1967. The enzymatic acetylation of chloramphenicol by extracts of R factor-resistant Escherichia coli. J. Biol. Chem. 242:687-693. [PubMed] [Google Scholar]
- 23.Shaw, W. V., and J. Unowsky. 1968. Mechanism of R factor-mediated chloramphenicol resistance. J. Bacteriol. 95:1976-1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith, A. M., K. P. Klugman, T. J. Coffey, and B. G. Spratt. 1993. Genetic diversity of penicillin-binding protein 2B and 2X genes from Streptococcus pneumoniae in South Africa. Antimicrob. Agents Chemother. 37:1938-1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smith, A. M., and K. P. Klugman. 2000. Non-penicillin-binding protein mediated high-level penicillin and cephalosporin resistance in a Hungarian clone of Streptococcus pneumoniae. Microb. Drug Resist. 6:105-110. [DOI] [PubMed] [Google Scholar]
- 26.Suryanarayana, T. 1983. Identification by affinity chromatography of Escherichia coli ribosomal proteins that bind erythromycin and chloramphenicol. Biochem. Int. 7:719-725. [PubMed] [Google Scholar]
- 27.Sutcliffe, J., T. Grebe, A. Tait-Kamradt, and L. Wondrack. 1996. Detection of erythromycin-resistant determinants by PCR. Antimicrob. Agents Chemother. 40:2562-2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sutcliffe, J., A. Tait-Kamradt, and L. Wondrack. 1996. Streptococcus pneumoniae and Streptococcus pyogenes resistant to macrolides but sensitive to clindamycin: a common resistance pattern mediated by an efflux system. Antimicrob. Agents Chemother. 40:1817-1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tait-Kamradt, A., T. Davies, M. Cronan, M. R. Jacobs, P. C. Appelbaum, and J. Sutcliffe. 2000. Mutations in 23S rRNA and ribosomal protein L4 account for resistance in pneumococcal strains selected in vitro by macrolide passage. Antimicrob. Agents Chemother. 44:2118-2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tait-Kamradt, A., T. Davies, P. C. Appelbaum, F. Depardieu, P. Courvalin, J. Petitpas, L. Wondrack, A. Walker, M. R. Jacobs, and J. Sutcliffe. 2000. Two new mechanisms of macrolide resistance in clinical strains of Streptococcus pneumoniae from Eastern Europe and North America. Antimicrob. Agents Chemother. 44:3395-3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tomasz, A., and R. D. Hotchkiss. 1964. Regulation of the transformability of pneumococcal cultures by macromolecular cell products. Proc. Natl. Acad. Sci. USA 51:480-487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsiodras, S., H. S. Gold, G. Sakoulas, G. M. Eliopoulos, C. Wennersten, L. Venkataraman, R. C. Moellering, and M. J. Ferraro. 2001. Linezolid resistance in a clinical isolate of Staphylococcus aureus. Lancet 358:207-208. [DOI] [PubMed] [Google Scholar]
- 33.Weisblum, B. 1995. Erythromycin resistance by ribosome modification. Antimicrob. Agents Chemother. 39:577-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Widdowson, C. A., P. V. Adrian, and K. P. Klugman. 2000. Acquisition of chloramphenicol resistance by the linearization and integration of the entire staphylococcal plasmid pC194 into the chromosome of Streptococcus pneumoniae. Antimicrob. Agents Chemother. 44:393-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xiong, L., P. Kloss, S. Douthwaite, N. M. Andersen, S. M. Swaney, D. L. Shinabarger, and A. S. Mankin. 2000. Oxazolidinone resistance mutations in 23S rRNA of Escherichia coli reveal the central region of domain V as the primary site of drug action. J. Bacteriol. 182:5325-5331. [DOI] [PMC free article] [PubMed] [Google Scholar]