Abstract
Antisense oligodeoxynucleotides (ODNs) and their analogs have been successfully utilized to inhibit gene expression and bacterial growth in vitro or in cell culture. In this study, acpP-targeting antisense peptide nucleic acid (PNA) and its peptide conjugate were tested as potential antibacterial agents in two groups of experiments using a mouse model. In the first group, Escherichia coli mutant strain SM101 with a defective outer membrane was used to induce bacteremia and peritonitis in BALB/c mice by intraperitoneal (i.p.) injection. The resulting bacteremia was fatal within 48 h. A single i.p injection of 5 nmol (or more) of PNA administered 30 min before bacterial challenge significantly reduced the bacterial load in mouse blood. Reductions in serum concentrations of the proinflammatory cytokines tumor necrosis factor alpha, interleukin-1β (IL-1β), IL-6, and IL-12 were also observed. PNA treatment was effective in rescuing 100% of infected animals. In the second group, bacteremia in BALB/c mice was induced by i.p. injection of E. coli wild-type strain K-12. The infected mice were treated by a single intravenous injection of peptide-PNA conjugate 30 min after bacterial challenge. Treatment with the peptide-PNA conjugate significantly reduced the K-12 load, with modest reduction in cytokine concentrations. The conjugate treatment was also able to rescue up to 60% of infected animals. This report is the first demonstration of ODNs' antibacterial efficacy in an animal disease model. The ability of PNA and its peptide conjugate to inhibit bacterial growth and to prevent fatal infection demonstrates the potential for this new class of antibacterial agents.
The use of oligodeoxynucleotides (ODNs) has been successful in a variety of applications to silence gene expression in eukaryotic systems for drug target validation and therapeutic purposes (3-5, 11). With the development of ODN analogs such as phosphorothioate, peptide nucleic acid (PNA), and phosphodiamidate morpholino oligomer (PMO) (for reviews, see references 6 and 19), the ODN gene silencing technology has been increasingly utilized to down-regulate target genes that are critical for bacterial viability, thus inhibiting bacterial growth (14, 15, 31). Antisense ODNs (AS-ODNs) have also been used to interfere with the expression of resistance genes so as to increase the bactericidal action of existing antibiotics (12, 26, 35). A range of expression plasmids have been developed to generate oligonucleotide molecules endogenously in bacterial cells, and these expression systems have been utilized to screen oligonucleotide libraries for identification of essential bacterial genes and to facilitate isolation of bactericidal oligonucleotide agents (18, 32).
A major challenge for the utilization of ODN gene silencing technology is the inefficient entry of sufficient numbers of ODNs into the targeted bacterial pathogen due to restrictions imposed by the outer membrane barrier. Methods have been recently developed to overcome this membrane barrier resistance. For example, some cell-permeabilizing peptides, when covalently attached to ODNs, enable the resulting peptide-ODN conjugate to traverse the outer membrane and have been shown to greatly improve antisense activity (10, 13, 23). Negatively charged liposomes have been successfully used to encapsulate and deliver ODNs into bacterial cells (7).
Given the critical role of fatty acid biosynthesis (FAB) in bacterial survival and the difference between bacterial and mammalian FAB pathways, the exploration of FAB for novel antibacterial agents has increased significantly in the last several years (21, 24). In particular, the FAB protein ACP encoded by the acpP gene has been shown to be a valid target for developing novel antibacterial agents (10, 13). Bacteria such as Escherichia coli normally synthesize fatty acids in three stages: initiation, cyclic elongation, and transfer to the membrane bilayer. The ACP protein acts early in the pathway and is required throughout the process (8, 21). Bacterial ACP protein also has important functions as a donor of activated fatty acyl groups during biosynthesis of phospholipids, lipid A, lipoteichoic acid, and acylated homoserine lactones (1, 2, 16, 17, 25, 27, 28). Therefore, depletion of the ACP pool will interfere with these diverse physiological processes, leading to cell dysfunction and death. It has been shown that acpP-targeting PNA-ODN and PMO-ODN are both bactericidal in vitro (10, 13).
In this work, the ability of the acpP-targeting antisense PNA and its peptide conjugate to inhibit bacterial growth in vivo and to rescue mice from infection was tested. The results show that treatment of infected mice with PNA and the peptide-PNA conjugate significantly reduced the bacterial load in the blood and effectively prevented fatal infection.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
E. coli strain SM105 [thr-1 araC14 tsx-78 Δ(galK-attλ)99 hisG4 rfbD1 rpsL136 sylA5 mtl-1 thi-1] and its isogenic derivative SM101 (9) with a defective outer membrane were purchased from the E. coli Genetic Stock Center (New Haven, CT). SM101 is deficient in the UDP-N-acetylglucosamine acyltransferase, and its lipid A content is only one-third of that of wild-type SM105. The low lipid A content causes the outer membrane of SM101 to be permeable to high-molecular-weight substances like PNA. SM101 and SM105 cells were grown at 30°C in Luria-Bertani (LB; Becton Dickinson and Company, Sparks, MD) medium containing 50 μg/ml streptomycin. E. coli strain K-12 was obtained from the American Type Culture Collection (ATCC 29425) and grown at 37°C in nutrient broth.
Chemical synthesis of PNA and peptide-PNA conjugate.
PNA and peptide-PNA conjugate were synthesized by Bio-Synthesis (Dallas, TX). Briefly, PNA with the sequence CTCATACTCT and peptide-PNA conjugate with the sequence KFFKFFKFFK-CTCATACTCT were synthesized manually on a solid phase by following the procedures described by Good et al. (13) using PNA monomers from Applied Biosystems (Foster City, CA) and amino acid residues from Novabiochem (San Diego, CA). The crude PNA or conjugate was then purified using high-performance liquid chromatography and identified and confirmed by matrix-assisted laser desorption ionization-time of flight mass spectrometry.
Inhibition of bacterial growth in vitro.
E. coli SM101, SM105, or K-12 isolates were grown as described above (9, 22). Cell cultures were diluted to ∼105 CFU/ml, and PNA or the peptide-PNA conjugate was added to a final concentration of 0, 4, 40, or 400 μM as described by Good and Nielsen (12). After incubation with shaking at 37°C for various times, aliquots of cell cultures were collected for cell growth assay by measuring viable cell counts. Viable cell counting was done by diluting the cultures and plating them in triplicate on LB plates. The plates were then incubated overnight at 37°C, and the colonies were enumerated by visual inspection.
Mouse intraperitoneal model of E. coli infection.
Mice were infected intra-abdominally to induce peritonitis according to the established protocols (20, 29, 30) under University of Texas—Houston Health Science Center animal welfare guidelines (HSC-AWC-02-059). BALB/c mice were used for infection experiments due to genetic control of the endotoxic response linked to lipopolysaccharide and CD14 (33, 34). For each infection experiment, 6- or 8-week-old female BALB/c mice in groups of six to eight were used. Mice were housed four to a cage and kept in a laminar-flow cabinet under specific-pathogen-free conditions for 2 weeks before use and monitored throughout experimentation under Institutional Animal Care and Use Committee-approved guidelines. Bacterial cells of log-phase cultures (optical density at 600 nm = ∼1.0) were collected by centrifugation and then resuspended in sterile phosphate-buffered saline (PBS) at 4°C. This log-phase preparation of bacteria was serially diluted in PBS, and a 70% lethal dose (LD70) of strain SM101 (approximately 1.2 × 109 CFU) or an LD90 of strain K-12 (approximately 8 × 108 CFU) was injected intraperitoneally (i.p.) into mice in 500-μl aliquots. Infected mice were observed for up to 9 days. Blood samples (up to 300 μl) were collected at 24 h after bacterial challenge by puncturing the orbital plexus with a heparinized capillary tube. The collected blood samples were used for bacterial titer analysis and proinflammatory cytokine (tumor necrosis factor alpha [TNF-α], interleukin-1β [IL-1β], IL-6, and IL-12) determination as described below.
PNA or peptide-PNA conjugate treatment.
The therapeutic efficacy of PNA or peptide-PNA conjugate was evaluated using the mouse model above. Groups of mice were challenged by i.p. injection of E. coli SM101 or K-12. SM101-infected mice were treated with a single injection of PNA administered i.p. 30 min before the bacterial challenge. K-12-infected mice were treated with a single injection of peptide-PNA conjugate administered intravenously 30 min after the bacterial challenge. The control groups included mice treated with PBS alone.
Bacterial titer analysis.
Blood samples collected at 24 h postinfection were serially diluted, plated onto LB agar plates, and incubated overnight at 37°C. The colonies on the plates were enumerated by visual inspection.
ELISA evaluation for proinflammatory cytokines.
Blood was collected using heparinized tubes at 24 h postinfection. Cells were removed by centrifugation, and the remaining serum was assessed for production of proinflammatory cytokines TNF-α, IL-1β, IL-6, and IL-12. Evaluation was accomplished using commercially available enzyme-linked immunosorbent assay (ELISA) DuoSet kits (R&D Systems, Minneapolis, MN) according to the manufacturer's instructions. Briefly, Costar 96-well vinyl assay plates were coated with capture antibody overnight (e.g., TNF-α at 0.5 μg/ml, IL-6 at 1 μg/ml). Plates were washed three times with wash buffer (0.05% Tween 20 in PBS). Blocking buffer (1% bovine serum albumin, 5% sucrose, 0.05% NaN3 in PBS) was added for a 3-h incubation. After three washings, 10 μl serum (final dilution of 1:20 in PBS) was added for a 2-h incubation. Biotin-conjugated secondary antibodies were added, incubated for 2 h, washed, and then developed using streptavidin-horseradish peroxidase (Sigma, St. Louis, MO) and TMB Microwell peroxidase substrate (Kirkegaard & Perry Laboratories, Inc., Gaithersburg, MD). Absorbance was read at 570 nm and 450 nm on an ELISA plate reader (Molecular Devices, Sunnyvale, CA). Means of duplicate or triplicate wells were calculated based on a standard curve constructed for each assay, using recombinant murine cytokines (R&D Systems). The limit of sensitivity for these assays is 5 pg/ml.
RESULTS
Inhibition of SM101 growth in vitro by acpP-targeting PNA.
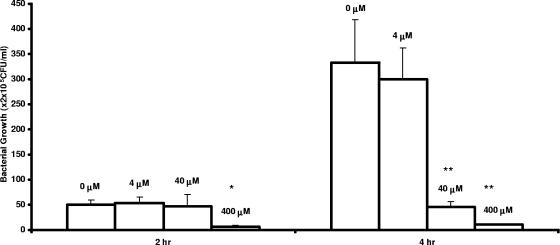
Inhibition of E. coli SM101 growth was evaluated by examining the effects of various PNA concentrations. As shown in Fig. 1, compared with the cell culture without PNA, the viable cells in cultures with PNA were significantly reduced. The inhibition of bacterial growth was both time and concentration dependent. However, this inhibition was not observed in wild-type E. coli SM105 cells (data not shown).
FIG. 1.
Inhibition of E. coli mutant SM101 growth in vitro by acpP-targeting PNA. PNA was added to cell cultures containing 5 × 105 CFU/ml SM101 to a final concentration of 4, 40, or 400 μM as indicated on the graph, with addition of an equal volume of water as a control (0 μM). Cultures were incubated as described in Materials and Methods. Aliquots of each culture were collected at 2 and 4 h, diluted, and plated for viable cell determination. Error bars indicate standard deviations of the results from three experiments (*, P < 0.01 relative to 2 h control; **, P < 0.001 relative to 4-h control [as determined by Student's t test]).
Rescue of SM101-infected mice by acpP-targeting PNA.
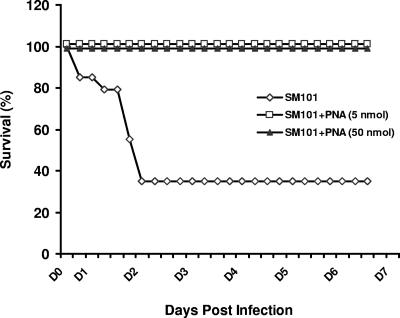
SM101 (LD70) was i.p. introduced into mice, and animals were observed for 1 week. All mice showed evidence of infection soon after bacterial administration (6 h), with characteristics such as lethargy, warmth to the touch, and scruffiness. Of the control (untreated) mice, 62.5% succumbed to infection within 48 h after receipt of SM101 (Fig. 2). Untreated mice (four of six) demonstrated high levels of bacteria in serum at 24 h postinfection, while treatment of mice with 5 nmol or 50 nmol PNA led to complete absence of bacteria at that time (Table 1). Of significance, all of the six mice treated with PNA at either dose level survived (Fig. 2).
FIG. 2.
Survival of SM101-infected mice due to PNA treatment. Six-week-old BALB/c were infected and treated as described in the text. The infected mice (LD70) were monitored every 4 to 6 h for survival through 7 days. Six to eight mice were infected per group, and the experiments were repeated with nearly identical results.
TABLE 1.
Reduction of SM101 inoculum in mouse blood by acpP-targeting PNAa
| Treatment | No. of mice bled | No. of CFU/ml |
|---|---|---|
| SM101 + no PNA | 6 | 0, 0, 6 × 103, 1.6 × 103, 4.6 × 103, >105 |
| SM101 + 5-nmol PNA | 4 | 0, 0, 0, 0 |
| SM101 + 50-nmol PNA | 4 | 0, 0, 0, 0 |
Mice were infected with an LD70 of E. coli SM101. Treatment with acpP-targeting PNA occurred in vivo 30 min prior to infection. Two-hundred-microliter blood samples were collected at 24 h after SM101 challenge, and bacteria were enumerated. Samples were diluted and plated on LB plates and incubated overnight at 37°C, and colonies were assessed by visual inspection.
As shown in Table 2, untreated mice exhibited marked increases in IL-6 and IL-12 and minor elevations in TNF-α and IL-1β. Treatment with PNA at a 5-nmol level led to significant reductions (P < 0.05) in serum IL-6 and IL-12. At a dose of 50 nmol, significant and substantial reductions were seen in TNF-α, IL-6, and IL-12.
TABLE 2.
Reduction of proinflammatory cytokines in serum by acpP-targeting PNAa
| Treatment | TNF-α | IL-1β | IL-6 | IL-12 |
|---|---|---|---|---|
| SM101 + no PNA | 0.86 ± 0.08 | 0.43 ± 0.33 | 12.16 ± 3.08 | 4.58 ± 0.01 |
| SM101 + 5-nmol PNA | 0.58 ± 0.18 | 0.50 ± 0.12 | 0.05 ± 0.01b | 0.62 ± 0.07b |
| SM101 + 50-nmol PNA | 0.54 ± 0.03b | 0.31 ± 0.07 | 0.04 ± 0.03b | 0.44 ± 0.06b |
Mice were infected and treated as described in the footnote to Table 1. Serum collected 24 h post SM101 challenge was examined for proinflammatory mediators TNF-α, IL-1β, IL-6, and IL-12 by standard ELISA. Treated mice demonstrated marked reductions in TNF-α, IL-6, and IL-12. Data are presented as μg/ml (± standard deviation) of mediator in serum. ELISA values were assessed in duplicate. Similar results were obtained in repeat experiments.
P < 0.05 relative to controls as determined by Student's t test.
Inhibition of K-12 growth in vitro by peptide-PNA conjugate.
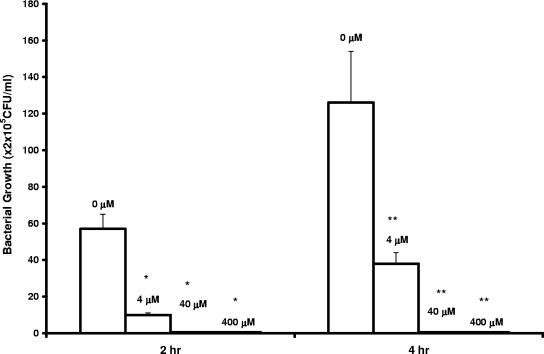
The cationic peptide with the sequence KFFKFFKFFK was previously shown to be effective in carrying PNA (as peptide-PNA conjugate) across the membrane barrier (13). The acpP-targeting peptide-PNA conjugate was produced to examine antibacterial efficacy against wild-type E. coli (strain K-12). As shown in Fig. 3, compared with the cell cultures in the absence of the conjugate, the viable cells in culture were significantly reduced in the presence of the conjugate; no viable K-12 cell was detected in the cultures with 40 μM or 400 μM conjugate. This inhibition was not observed in cell cultures with addition of peptide alone; after incubation for 4 h, 2.1 × 107 CFU/ml and 1.4 × 107 CFU/ml K-12 cells were detected in the cultures with 40 μM and 400 μM peptide, respectively (data not shown in Fig. 3).
FIG. 3.
Inhibition of E. coli wild-type K-12 growth in vitro by acpP-targeting peptide-PNA conjugate. Peptide-PNA conjugate was added to cell cultures containing 5 × 105 CFU/ml E. coli strain K-12 to a final concentration of 0, 4, 40, or 400 μM as indicated on the graph. Cell cultures were grown, and viable cells were determined as described in the legend to Fig. 1. Error bars indicate standard deviations of the results from two experiments (*, P < 0.001 relative to 2-h control; **, P < 0.005 relative to 4-h control [as determined by Student's t test]).
Rescue of K-12-infected mice by the peptide-PNA conjugate.
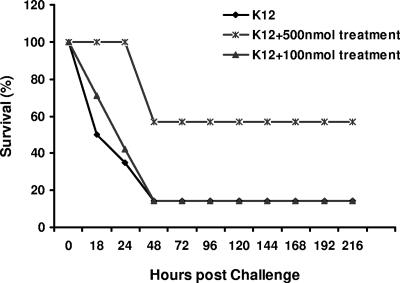
As shown in Fig. 4, untreated mice (90%) succumbed to infection within 48 h. Mice treated with the low PNA conjugate dose (100 nmol) succumbed to infection in a manner similar to controls. However, all mice treated with 500 nmol peptide-PNA conjugate exhibited a survival benefit. Treatment with peptide-PNA conjugate also significantly reduced the bacterial load; serum collected at 24 h postinfection showed significantly reduced levels of bacteria for the groups treated with 100 or 500 nmol (P < 0.05) (Table 3).
FIG. 4.
Survival of K-12-infected mice due to treatment with acpP-targeting peptide-PNA conjugate. Six-week-old BALB/c mice were i.p. infected with 8 × 108 CFU E. coli strain K-12 in 500 μl PBS (LD90). Infected mice were treated with the peptide-PNA conjugate (100 or 500 nmol) 30 min postinfection; comparisons are made to untreated mice. Survival and general health of these animals was monitored every 6 to 12 h through 9 days.
TABLE 3.
Reduction of E. coli K-12 bacteremia in mice following treatment with peptide-PNA conjugatea
| Treatment | Avg log CFU/ml ± SD | P value |
|---|---|---|
| K-12 + no conjugate | 4.27 ± 0.05 | |
| K-12 + 100-nmol conjugate | 3.45 ± 0.18 | <0.05 |
| K-12 + 500-nmol conjugate | 3.89 ± 0.16 | <0.05 |
Mice were infected with an LD90 of E. coli K-12, followed by treatment with the acpP-targeting peptide-PNA conjugate in vivo at 30 min postinfection. Serum was collected 24 h after bacterial challenge and assessed for organisms. Dilutions were plated on LB agar, and CFU were enumerated by visual inspection. Significance shown was determined by Student's t test.
Although the level of bacteremia correlated well with survival, there was only a modest reduction in the concentrations of the proinflammatory cytokines; TNF-α, IL-1β, and IL-12 were all marginally reduced, but not significantly (Table 4).
TABLE 4.
Reduction of proinflammatory mediators in serum by peptide-PNA conjugatea
| Treatment | TNF-α | IL-1β | IL-6 | IL-12 |
|---|---|---|---|---|
| K-12 + no conjugate | 2.41 ± 0.18 | 2.14 ± 0.42 | 8.36 ± 3.65 | 3.54 ± 1.38 |
| K-12 + 100-nmol conjugate | 1.68 ± 0.09 | 1.63 ± 0.50 | 7.71 ± 6.88 | 1.13 ± 0.02 |
| K-12 + 500-nmol conjugate | 1.55 ± 0.11 | 1.82 ± 0.43 | 8.83 ± 4.97 | 3.31 ± 1.02 |
Mice were infected and treated as described in the footnote to Table 3. Serum collected at 24 h post K-12 challenge was examined for proinflammatory cytokines TNF-α, IL-1β, IL-6, and IL-12 by standard ELISA. Only a modest (not statistically significant) reduction in mediators was observed. Data are presented as μg/ml (± standard deviation) of mediator in serum; ELISA values were assessed in duplicate. Results were comparable in repeated experiments.
DISCUSSION
AS-ODNs and their analogs have been demonstrated to be effective in inhibiting bacterial gene expression and cell growth (10, 12-15, 26, 31), making this technology attractive for developing highly specific and efficacious antibacterial agents. Inhibition of bacterial growth by AS-ODN analogs was well established in vitro or in cell culture, but evidence of their biological effect in vivo is lacking. In this work, we tested the acpP-targeting antisense PNA and its peptide conjugate for their abilities to inhibit bacterial growth in vivo, using a mouse model of bacterial infection. Previous studies have shown that acpP-targeting AS-PMO and AS-PNA can significantly inhibit bacterial growth by specifically silencing acpP expression (10, 13). Our results further confirm the inhibition of bacterial growth in vitro by acpP-targeting AS-ODN. More importantly, treatment of the bacterium-infected mice with the acpP-targeting PNA or its peptide conjugate can significantly reduce the bacterial load in mouse blood (Tables 1 and 3), with concomitant reductions in the concentrations of the proinflammatory cytokines in serum (Tables 2 and 4). acpP-targeting PNA and its peptide conjugate were also able to rescue more than 60% (up to 100%) of the infected animals (Fig. 2 and 4).
Although the therapeutic application of the acpP-targeting AS-ODN was examined, it should be feasible to utilize AS-ODNs targeted toward any critical bacterial gene. Due to the genetic nature of antisense technology, it holds promise for the development of both broad-spectrum and specific-spectrum antibacterial agents. AS-ODNs designed to target the conserved regions of highly conserved genes are more likely to be useful as broad-spectrum agents. A cocktail of AS-ODNs targeting many disparate genes is another strategy to pursue broad-spectrum activity, as it has been demonstrated that the effects of AS-ODNs against different genes are cumulative (15). In contrast, AS-ODNs targeting genes unique to certain organisms are less likely to cross-react with others.
In conclusion, the results presented here demonstrate that AS-ODNs, when conjugated to membrane-permeabilizing peptides, can effectively inhibit bacterial growth in vivo and may be useful as therapeutic agents. Furthermore, additional research should be accomplished so that the carrier peptide and its cross-linker to ODN may be optimized for clinical application.
Acknowledgments
We thank Harilyn McMicken and Shen-An Hwang for technical support and Samuel Kaplan, Malcolm Skolnick, and Kurt Berens for reviewing the manuscript and consultation on data interpretation.
REFERENCES
- 1.Anderson, M. S., and C. R. Raetz. 1987. Biosynthesis of lipid A precursors in Escherichia coli. A cytoplasmic acyltransferase that converts UDP-N-acetylglucosamine to UDP-3-O-(R-3-hydroxymyristoyl)-N-acetylglucosamine. J. Biol. Chem. 262:5159-5169. [PubMed] [Google Scholar]
- 2.Cao, J. G., and E. A. Meighen. 1993. Biosynthesis and stereochemistry of the autoinducer controlling luminescence in Vibrio harveyi. J. Bacteriol. 175:3856-3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen, Y., Y. J. Ji, R. Roxby, and C. Conrad. 2000. In vivo expression of single-stranded DNA in mammalian cells with DNA enzyme sequences targeted to C-raf. Antisense Nucleic Acid Drug Dev. 10:415-422. [DOI] [PubMed] [Google Scholar]
- 4.Chen, Y., and H. W. McMicken. 2003. Intracellular production of DNA enzyme by a novel single-stranded DNA expression vector. Gene Ther. 10:1776-1780. [DOI] [PubMed] [Google Scholar]
- 5.Datta, H. J., and P. M. Glazer. 2001. Intracellular generation of single-stranded DNA for chromosomal triplex formation and induced recombination. Nucleic Acids Res. 29:5140-5147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dias, N., and C. A. Stein. 2002. Antisense oligonucleotides: basic concepts and mechanisms. Mol. Cancer Ther. 1:347-355. [PubMed] [Google Scholar]
- 7.Fillion, P., A. Desjardins, K. Sayasith, and J. Lagace. 2001. Encapsulation of DNA in negatively charged liposomes and inhibition of bacterial gene expression with fluid liposome-encapsulated antisense oligonucleotides. Biochim. Biophys. Acta 1515:44-54. [DOI] [PubMed] [Google Scholar]
- 8.Flaman, A. S., J. M. Chen, S. C. van Iderstine, and D. M. Byers. 2001. Site-directed mutagenesis of acyl carrier protein (ACP) reveals amino acid residues involved in ACP structure and acyl-ACP synthetase activity. J. Biol. Chem. 276:35934-35939. [DOI] [PubMed] [Google Scholar]
- 9.Galloway, S. M., and C. R. Raetz. 1990. A mutant of Escherichia coli defective in the first step of endotoxin biosynthesis. J. Biol. Chem. 265:6394-6402. [PubMed] [Google Scholar]
- 10.Geller, B. L., J. D. Deere, D. A. Stein, A. D. Kroeker, H. M. Moulton, and P. L. Iversen. 2003. Inhibition of gene expression in Escherichia coli by antisense phosphodiamidate morpholino oligomers. Antimicrob. Agents Chemother. 47:3233-3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gewirtz, A. M., D. L. Sokol, and M. Z. Ratajczak. 1998. Nucleic acid therapeutics: state of the art and future prospects. Blood 92:712-736. [PubMed] [Google Scholar]
- 12.Good, L., and P. E. Nielsen. 1998. Antisense inhibition of gene expression in bacteria by PNA targeted to mRNA. Nat. Biotechnol. 16:355-358. [DOI] [PubMed] [Google Scholar]
- 13.Good, L., S. K. Awasthi, R. Dryselius, O. Larsson, and P. E. Nielsen. 2001. Bactericidal antisense effects of peptide-PNA conjugates. Nat. Biotechnol. 19:360-364. [DOI] [PubMed] [Google Scholar]
- 14.Harth, G., P. C. Zamecnik, J. Y. Tang, D. Tabatadze, and M. A. Horwitz. 2000. Treatment of Mycobacterium tuberculosis with antisense oligonucleotides to glutamine synthetase mRNA inhibits glutamine synthetase activity, formation of the poly-l-glutamate/glutamine cell wall structure and bacterial replication. Proc. Natl. Acad. Sci. USA 97:418-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harth, G., M. A. Horwitz, D. Tabatadze, and P. C. Zamecnik. 2002. Targeting the Mycobacterium tuberculosis 30/32-kDa mycolyl transferase complex as therapeutic strategy against tuberculosis: proof of principle by using antisense technology. Proc. Natl. Acad. Sci. USA 99:15614-15619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heaton, M. P., and F. C. Neuhaus. 1994. Role of the d-alanyl carrier protein in the biosynthesis of d-alanyl-lipoteichoic acid. J. Bacteriol. 176:681-690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Issartel, J. P., V. Koronakis, and C. Hughes. 1991. Activation of Escherichia coli prohaemolysin to the mature toxin by acyl carrier protein-dependent fatty acylation. Nature 351:759-761. [DOI] [PubMed] [Google Scholar]
- 18.Ji, Y., B. Zhang, S. F. van Horn, P. Warren, G. Woodnutt, M. K. Burnham, and M. Rosenberg. 2001. Identification of critical staphylococcal genes using conditional phenotypes generated by antisense RNA. Science 293:2266-2269. [DOI] [PubMed] [Google Scholar]
- 19.Kurreck, J. 2003. Antisense Technologies: improvement through novel chemical modifications. Eur. J. Biochem. 270:1628-1644. [DOI] [PubMed] [Google Scholar]
- 20.Laine V. J., D. S. Grass, and T. J. Nevalainen. 2000. Resistance of transgenic mice expressing human group II phospholipase A2 to Escherichia coli infection. Infect. Immun. 68:87-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Magnuson, K., S. Jackowski, C. O. Rock, and J. E. Cronan. 1993. Regulation of fatty acid biosynthesis in Escherichia coli. Microbiol. Rev. 57:522-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mohan, S., T. M. Kelly, S. S. Eveland, C. R. Raetz, and M. S. Anderson. 1994. An Escherichia coli gene (fabZ) encoding (3R)-hydroxymyristoyl acyl carrier protein dehydrase. Relation to fabA and suppression of mutations in lipid A biosynthesis. J. Biol. Chem. 269:32896-32903. [PubMed] [Google Scholar]
- 23.Nekhotiaeva, N., S. K. Awasthi, P. E. Nielsen, and L. Good. 2004. Inhibition of Staphylococcus aureus gene expression and growth using antisense peptide nucleic acids. Mol. Ther. 10:652-659. [DOI] [PubMed] [Google Scholar]
- 24.Rock, C. O., and J. E. Cronan. 1996. Escherichia coli as a model for the regulation of dissociable (type II) fatty acid biosynthesis. Biochim. Biophys. Acta 1302:1-16. [DOI] [PubMed] [Google Scholar]
- 25.Rumley, M. K., H. Therisod, A. C. Weissborn, and E. P. Kennedy. 1992. Mechanisms of regulation of the biosynthesis of membrane-derived oligosaccharides in Escherichia coli. J. Biol. Chem. 267:11806-11810. [PubMed] [Google Scholar]
- 26.Sarno, R., H. Ha, N. Weinsetel, and M. E. Tolmasky. 2003. Inhibition of aminoglycoside 6′-N-acetyltransferase type Ib-mediated amikacin resistance by antisense oligodeoxynucleotides. Antimicrob. Agents Chemother. 47:3296-3304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schaefer, A. L., D. L. Val, B. L. Hanzelka, J. E. Cronan, Jr., and E. P. Greenberg. 1996. Generation of cell-to-cell signals in quorum sensing: acyl homoserine lactone synthase activity of a purified Vibrio fischeri Luxl protein. Proc. Natl. Acad. Sci. USA 93:9505-9509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shen, B., R. G. Summers, H. Gramajo, M. J. Bibb, and C. R. Hutchinson. 1992. Purification and characterization of the acyl carrier protein of the Streptomyces glaucescens tetracenomycin C polyketide synthase. J. Bacteriol. 174:3818-3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Somerville, J. E., Jr., L. Cassiano, and R. P. Darveau. 1999. Escherichia coli msbB gene as a virulence factor and a therapeutic target. Infect. Immun. 67:6583-6590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sugita-Konishi, Y., S. Shimura, T. Nishikawa, F. Sunaga, H. Naito, and Y. Suzuki. 2003. Effect of bisphenol A on non-specific immunodefenses against non-pathogenic Escherichia coli. Toxicol. Lett. 136:217-227. [DOI] [PubMed] [Google Scholar]
- 31.Tan, X.-X., K. Rose, W. Margolin, and Y. Chen. 2004. DNA enzyme generated by a novel single-stranded DNA expression vector inhibits expression of the essential bacterial cell division gene ftsZ. Biochemistry 43:1111-1117. [DOI] [PubMed] [Google Scholar]
- 32.Tan, X.-X., and Y. Chen. 2005. A novel genomic approach identifies bacterial DNA-dependent RNA polymerase as the target of an antibacterial oligodeoxynucleotide, RBL-1. Biochemistry 44:6708-6714. [DOI] [PubMed] [Google Scholar]
- 33.Watson, J., M. Largen, and K. P. McAdam. 1978. Genetic control of endotoxic responses in mice. J. Exp. Med. 147:39-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Watson, J., R. Riblet, and B. A. Taylor. 1977. The response of recombination inbred strains of mice to bacterial lipopolysaccharides. J. Immunol. 118:2088-2093. [PubMed] [Google Scholar]
- 35.White, D. G., K. Maneewannakul, E. von Hofe, M. Zillman, W. Eisenberg, A. K. Field, and S. B. Levy. 1997. Inhibition of the multiple antibiotic resistance (mar) operon in Escherichia coli by antisense DNA analogs. Antimicrob. Agents Chemother. 41:2699-2704. [DOI] [PMC free article] [PubMed] [Google Scholar]