Abstract
Pseudomonas aeruginosa is the most relevant pathogen producing chronic lung infections in patients with chronic underlying diseases such as cystic fibrosis (CF), bronchiectasis, and chronic obstructive pulmonary disease (COPD). Hypermutable (or mutator) P. aeruginosa strains, characterized by increased (up to 1,000-fold) spontaneous mutation rates due to alterations of the DNA mismatch repair (MMR) system have been found at high frequencies in the lungs of CF patients, but their role in other chronic processes is still unknown. Sixty-two P. aeruginosa isolates from 30 patients with underlying non-CF chronic respiratory diseases (22 with bronchiectasis and 8 with COPD) and documented chronic infection were studied. Antibiotic susceptibility profiles and mutation frequencies were determined, and complementation assays using the cloned wild-type mutS gene and molecular epidemiology studies (pulsed-field electrophoresis, [PFGE]) were performed with these strains. Thirty-three (53%) of the isolates were hypermutable, and 17 (57%) of the 30 patients were colonized by hypermutable strains. Strains from 11 of the 17 patients were found to be defective in the MMR mutS gene by complementation assays. Interpatient transmission of strains was ruled out by PFGE. Multiple-antimicrobial resistance was documented in 42% of the hypermutable strains in contrast to 0% resistance in the nonhypermutable strains (P < 0.0001). Hypermutable P. aeruginosa strains are extremely prevalent in chronic infections in contrast to what has been described in acute processes, suggesting a role of hypermutation in bacterial adaptation for long-term persistence. Furthermore, hypermutation is found to be a key factor for the development of multiple-antimicrobial resistance, and therefore these findings are expected to have important consequences for the treatment of chronic infections.
Pseudomonas aeruginosa is a ubiquitous versatile environmental microorganism that is the leading cause of opportunistic human infections (42). This pathogen is one of the major and more severe causes of acute nosocomial infections, especially affecting patients in intensive care units (ICUs) with mechanical ventilation-associated pneumonia or burn wound infections, which are both associated with a high mortality rate (22, 45). Nevertheless, P. aeruginosa is also a major cause of chronic respiratory infections. For instance, this microorganism is the main driver for morbidity and mortality of cystic fibrosis (CF) patients (7, 9, 21); is the leading cause of chronic infection in patients with bronchiectasis, which is associated with more severe lung function deterioration and poorer quality of life (5, 26, 28, 46); and is beginning to be recognized as a marker of intense airway inflammation in patients with chronic obstructive pulmonary disease (COPD) (13). The prevalence of P. aeruginosa in patients with COPD is around 4% but increases to 8 to 13% in patients with advanced airflow obstruction (19).
One of the most striking characteristics of P. aeruginosa is its extraordinary ability for antibiotic resistance acquisition (20). Treatment failure due to the development of resistance is indeed a frequent outcome of Pseudomonas infections (6). This is an especially critical factor in the management of chronic infections such as those occurring in CF patients. After years of intensive antibiotic chemotherapy in an effort to control the negative outcome of chronic colonization, sequential development of resistance to most antibiotics frequently occurs. Resistance rates of P. aeruginosa strains isolated from CF patients are, in fact, substantially higher than those found in other settings, including the strains from patients in ICUs (12, 29). Considerably less information is available for other P. aeruginosa chronic infection settings, such as bronchiectasis or COPD. Nevertheless, a limited number of studies have demonstrated that P. aeruginosa strains isolated from CF and bronchiectasis patients share common features, such as the frequent emergence of mucoid isolates, the intense phenotypic diversification despite unique colonizing clonal lineages, and the frequent emergence over time of antibiotic-resistant variants (9, 11, 14, 33, 38).
A link between the high antibiotic resistance rates in CF patients and the presence of a high proportion of hypermutable (or mutator) P. aeruginosa strains has been previously documented (29). Mutators represented 20% of the total number of P. aeruginosa isolates from CF patients and were found in 37% of the patients in the mentioned study (29). Hypermutable strains are those that have an increased (up to 1,000-fold) spontaneous mutation rate due to defects in genes involved in DNA repair or error avoidance systems (15, 25). In P. aeruginosa strains obtained from CF patients, as well as in other natural bacterial populations (18, 24), the most frequently involved system is the mismatch repair (MMR) system, and mutS is the most frequently affected gene (30). A recent in vitro work has shown that mutS deficiency in P. aeruginosa determines the immediate development of resistance to every single antipseudomonal agent due to the ascent to dominance of resistant mutants in a few hours during drug exposure (31).
In order to ascertain whether hypermutation is a common feature of chronic lung infections caused by P. aeruginosa, a prospective study on non-CF patients with underlying chronic respiratory diseases was undertaken, finding that the prevalence of hypermutable P. aeruginosa strains is extremely high (more than 50% of the isolates) in these patients. Furthermore, hypermutation is found to be the main driver for the development of multiple-antimicrobial resistance in patients with chronic infections caused by P. aeruginosa.
MATERIALS AND METHODS
Patients and strains.
From March 2003 to June 2004, all P. aeruginosa isolates from respiratory samples submitted to the Microbiology Department from patients with documented underlying chronic respiratory diseases, excluding cystic fibrosis, were prospectively collected. Identification of the isolates was performed with the API 20NE system (Biomérieux, France) and conventional tests. For the definition of chronic P. aeruginosa colonization/infection, only patients with at least 1 year of documented P. aeruginosa colonization prior to the study (according to the Microbiology Department database, with information available up to 1995) or with at least three P. aeruginosa-positive cultures, each separated by more than 1 month, during the study period were considered. All different P. aeruginosa morphological variants isolated from two sputum samples (separated by more than 1 month) from each of the patients with documented chronic colonization were selected for further study. In those patients with only one sample recovered during the study period, all different P. aeruginosa morphological variants from these single samples were included. Strain PAO1 and its hypermutable, mutS-deficient, isogenic derivative, PAOΔmutS (31), were used as controls.
According to a American Thoracic Society/European Respiratory Society consensus document (2), COPD was diagnosed as not fully reversible chronic obstruction to airflow demonstrated by forced postbronchodilator spirometry (forced expiratory volume in 1 s [FEV1]/forced vital capacity [FVC], <0.70) which could not be explained by other pulmonary pathology. The severity of the chronic obstruction to airflow was assessed using the Global Initiative for Chronic Obstructive Lung Disease criteria (2) that classify patients in four categories: mild (decreased FEV1/FVC with normal FEV1), moderate (FEV1 between 50 and 80% of the predicted value), severe (FEV1 between 30 and 50%), and very severe (FEV1 < 30%).
Considering the criteria described by Kang et al. (16), bronchiectasis was diagnosed by computed tomography (CT) chest scan on the basis of an internal bronchial diameter that was greater than that of the adjacent pulmonary artery, lack of tapering of the bronchial lumina, and/or visualization of bronchi within 1 cm of the pleura. Other roentgenologic signs characteristic of bronchiectasis based on the CT classification criteria of Naidich et al. (27) were as follows: bronchial-wall widening or absence of tapering in contiguous axial sections for mild cylindric bronchiectasis, horizontal or vertical course of the bronchi seen as “tramlines” or “signet rings” for cylindric bronchiectasis, dilated bronchi with walls of beaded appearance for varicose bronchiectasis, and dilated bronchi with air fluid levels and/or strings or clusters of cysts for cystic bronchiectasis. A respiratory function test (spirometry) was performed for all patients with bronchiectasis in order to assess the presence of chronic obstruction to airflow and the severity of the obstruction. Obstruction in bronchiectasis was defined by a prebronchodilator FEV1/FVC value of <0.70, and the severity was assessed as for COPD.
Antibiotic susceptibility testing.
MICs were determined for ceftazidime, imipenem, meropenem, ciprofloxacin, and tobramycin in Mueller-Hinton (MH) agar plates by using Etest strips (AB Biodisk, Sweden) following standard procedures, and the resistance breakpoints used were those of the Clinical and Laboratory Standards Institute (formerly NCCLS) document M100-S15 (4). The documentation and interpretation of the antibiotic-resistant mutant subpopulations for the detection and susceptibility testing of hypermutable strains were performed as previously described (23). Briefly, suspensions with a McFarland standard of 0.5 or 1 were used for inoculum standardization of regular or mucoid isolates, respectively. MICs were read after 24 h (36 h for slow-growing strains) of incubation at 37°C. The presence (or absence) of resistant mutant subpopulations within the inhibition zones, as well as their relative numbers (+, ++, and +++ for <10, 10 to 100, and >100 mutants, respectively) and their highest MICs, was recorded after an extra 12 h of incubation. Except when specifically indicated, the MICs considered for the analysis were those of the general bacterial population regardless of the presence of resistant mutant subpopulations.
Estimation of mutation frequencies and complementation studies.
Mutation frequencies were estimated as previously described (29, 30). Briefly, independent triplicate 10-ml overnight MH broth cultures of the P. aeruginosa isolates were collected by centrifugation and resuspended in 1 ml of saline solution. Serial 10-fold dilutions were plated in MH agar with and without 300 μg/ml of rifampin, and after 36 h of incubation (48 h for slow-growing strains), colonies were counted and the mean fraction of mutants estimated. As previously defined, strains were considered hypermutable when the mutation frequency was at least 20-fold higher than that obtained for control strain PAO1 (28). To explore the genetic basis for the mutator phenotype, complementation studies using the cloned PAO1 wild-type mutS gene were performed with all hypermutable strains. Plasmid pUCPMS harboring the PAO1 mutS gene (31) was electroporated into the different hypermutable P. aeruginosa isolates as previously described (40). Transformant cells harboring plasmid pUCPMS were selected in Luria-Bertani agar plates containing 50 or 250 μg/ml gentamicin. Complementation of the increased mutation frequencies was studied in three independent transformants.
Molecular epidemiology.
The epidemiological relatedness of the strains was studied by pulsed-field gel electrophoresis (PFGE). Bacterial DNA embedded in agarose plugs prepared as previously described (17) was digested with the restriction enzyme SpeI. DNA separation was performed in a CHEF-DRIII apparatus (Bio-Rad, La Jolla, Calif.) under the following conditions: 6 V/cm2 for 26 h with pulse times of 5 to 40 seconds. DNA macrorestriction patterns were interpreted according to the criteria established by Tenover et al. (44)
Statistical analysis.
Fisher's exact test and the Mann-Whitney U test were used for the comparison of resistance rates and distributions of MICs, respectively, between hypermutable and nonhypermutable strains. A P value of <0.05 was considered statistically significant.
RESULTS
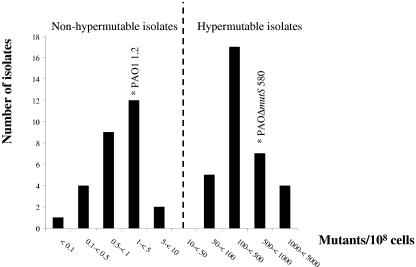
A total of 45 patients with documented underlying chronic lung disease (26 with bronchiectasis and 19 with COPD) had at least one P. aeruginosa-positive sputum culture during the study period. Following the criteria described in Materials and Methods, 30 of them were considered to have documented chronic P. aeruginosa colonization. A total of 62 P. aeruginosa isolates from two sputum samples from each of these 30 patients were studied. Estimations of the mutation frequencies revealed that 33 (53%) of the isolates were hypermutable. The distribution of the mutation frequencies is shown in Fig. 1. The mean mutation frequency of nonhypermutable isolates was 1.8 mutants/108 cells (range, 0.07 to 7.7 mutants/108 cells), and that of hypermutable isolates was 470 mutants/108 cells (range, 52 to 2,100 mutants/108 cells). Seventeen (57%) of the 30 patients were found to be colonized by hypermutable strains. Hypermutable strains from 11 (65%) of the 17 patients were found to be defective in the MMR mutS gene by complementation assays using plasmid pUCPMS. The range of the mutation frequencies of mutS-deficient strains was similar to that of the overall hypermutable strains (from 56 to 1,200 mutants/108 cells).
FIG. 1.
Distribution of mutation frequencies of the 62 P. aeruginosa isolates from chronically infected patients. Mutation frequencies of the control strain PAO1 and its hypermutable derivative PAOΔmutS are also shown.
To evaluate the possibility of the high prevalence of hypermutable strains being a consequence of the dissemination of a single clone (or a few) of hypermutable P. aeruginosa, molecular epidemiology studies were undertaken. The analysis of the PFGE DNA macrorestriction patterns from the collection of 62 P. aeruginosa isolates revealed no interpatient transmission of strains. A total of 32 different P. aeruginosa clones were identified, each found in a single patient. In 28 of the 30 patients, a single P. aeruginosa clone was documented, whereas the remaining 2 patients were found to harbor two different clones. When two or more P. aeruginosa morphological variants were recovered from a single sample, which occurred for 10 of the 30 patients, analysis of PFGE patterns of the different isolates revealed the presence of a single clonal type as is frequently observed in CF patients (38). Twenty-eight (45%) and 13 (21%) of the isolates had the mucoid- or small-colony variant morphotypes, respectively, typical of chronic CF infection (9, 11). Nevertheless, no significant differences in the proportions of these morphotypes between hypermutable and nonhypermutable isolates were observed.
To evaluate the impact of the high prevalence of hypermutable strains on antibiotic resistance, susceptibility testing for five of the most potent and widely used antipseudomonal agents was performed. Overall antibiotic resistance percentages for the 62 isolates ranged from 3.2% for meropenem to 35.5% for ciprofloxacin (Table 1). When resistance percentages of hypermutable and nonhypermutable strains were analyzed independently, it was found not only that hypermutable strains were much more resistant to all the antibiotics but also that most of the strains resistant to any of the antibiotics were hypermutable. The differences in resistance percentages were statistically significant (Fisher's exact test) for ceftazidime (P = 0.0056), imipenem (P = 0.0019), and ciprofloxacin (P = 0.0048). Differences did not reach statistical significance for meropenem and tobramycin due to the low number of strains resistant to these antibiotics. However, when the MIC distributions were analyzed for these two antibiotics, hypermutable strains were found to be significantly (Mann-Whitney U test) more resistant (higher MICs) than nonhypermutable strains both for meropenem (P = 0.003) and for tobramycin (P = 0.021). When multiple-antimicrobial resistance was considered, an even higher impact of hypermutation was observed. Forty-two percent (14 out of 33) of the hypermutable strains were resistant to at least two of the antibiotics tested, whereas none of the 29 nonhypermutable strains were found to be resistant to more than one antimicrobial agent (P < 0.0001); in other words, all P. aeruginosa strains resistant to two or more antibiotics (22.6%) were hypermutable.
TABLE 1.
Percentages of resistance and geometric means of MICs for P. aeruginosa isolates with and without considering resistant mutant subpopulations
| Antibiotic | Overall isolates (n = 62)
|
Nonhypermutable isolates (n = 29)
|
Hypermutable isolates (n = 33)
|
Statistical significance (P)c | Hypermutable isolates considering RMSd(n = 33)
|
Overall % resistance considering RMS of hypermutable isolatese | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| No. (%) resistanta | Mean MICb | No. (%) resistant | Mean MIC | No. (%) resistant | Mean MIC | No. (%) resistant | Mean MIC | |||
| Ceftazidime | 11 (17.7) | 1.5 | 1 (3.5) | 0.68 | 10 (30.3) | 3.6 | 0.0056 | 23 (69.7) | 37.9 | 38.7 |
| Imipenem | 9 (14.5) | 1.1 | 0 (0) | 0.53 | 9 (27.3) | 1.9 | 0.0019 | 25 (75.8) | 8.5 | 40.3 |
| Meropenem | 2 (3.2) | 0.09 | 0 (0) | 0.04 | 2 (6.1) | 0.18 | 0.27 | 9 (27.3) | 1.4 | 14.5 |
| Ciprofloxacin | 22 (35.5) | 0.65 | 5 (17.2) | 0.31 | 17 (51.5) | 1.2 | 0.0048 | 27 (81.8) | 5.5 | 51.6 |
| Tobramycin | 6 (9.7) | 1.2 | 1 (3.5) | 0.90 | 5 (15.2) | 1.7 | 0.13 | 11 (33.3) | 2.7 | 19.3 |
| ≥2 antibiotics | 14 (22.6) | 0 (0) | 14 (42.4) | <0.0001 | 33 (100) | 53.2 | ||||
The numbers and percentages of resistant isolates are according to the Clinical and Laboratory Standards Institute non-susceptibility breakpoints and therefore include the intermediate and resistant categories.
Geometric mean of the MICs in μg/ml.
P values (Fisher's exact test) resulting from comparison of the numbers of resistant strains among nonhypermutable and hypermutable isolates.
RMS, resistant mutant subpopulations. Results are based on the MICs of the resistant mutant colonies growing within the inhibition zones.
Overall percentages of resistance obtained for the 62 studied P. aeruginosa isolates if the RMS of hypermutable strains were considered in the susceptibility results interpretation.
Hypermutable strains are characterized by the presence of resistant mutant subpopulations within the inhibition zones of the antibiotics due to the high spontaneous mutation rates (23). When these typically resistant mutant subpopulations were considered in susceptibility data interpretation, the rates of resistance for the hypermutable strains were increased much further, reaching values ranging from 27% for meropenem to 82% for ciprofloxacin (Table 1). Then, given the high proportion of hypermutable strains, the consideration of the resistant mutant subpopulations had an important additional effect on the overall resistance rates, as can be observed in Table 1.
DISCUSSION
Pseudomonas aeruginosa is the most relevant pathogen producing chronic lung infections in patients with chronic underlying diseases such as CF and bronchiectasis and is beginning to be recognized as a relevant pathogen in COPD patients with advanced airflow obstruction. While long-term treatment with antipseudomonal agents is crucial to avoid a fast decline in the respiratory function of the infected patients, mutants resistant to multiple antimicrobials almost invariably evolve and frequently lead to treatment failure. Unlike many other bacteria, P. aeruginosa can, by changes in single genes, generate mutants that are resistant to clinical concentrations of all antimicrobial agents used for therapy (20).
In vitro and theoretical approaches suggest that the acquisition of a stable mutator phenotype may confer a selective advantage for bacteria, particularly in stressful and fluctuating environments (41, 43). In vivo experiments have also shown a potential benefit of hypermutation for bacterial adaptation to new environments, although once adapted, this advantage tends to disappear, and the transmissibility of the mutator strains is considerably reduced (8). In natural bacterial populations, the presence of hypermutable strains has been linked to resistance to host immunological defenses, such as mutation-dependent phase variation in Neisseria meningitidis (36) or resistance to antibiotics in P. aeruginosa isolates from chronically infected CF patients (29). Up to 37% of the CF patients were colonized by hypermutable strains in the mentioned study (29), whereas not a single hypermutable strain was found in 75 acutely infected patients. Recently, results from another study have suggested that chronicity itself maybe a key factor for the selection of hypermutable P. aeruginosa strains: despite the fact that mutational-antibiotic resistance development was a frequent outcome in a series of 103 acutely infected ICU patients, the prevalence of hypermutable strains was found to be lower than 1% (10). Certainly, the extremely high prevalence of hypermutable strains found in the present work suggests that hypermutation plays a role in the bacterial adaptation to the lung environment required for long-term persistence because hypermutation increases the rates of generation of any mutant phenotype (not only antibiotic resistance), including those involved in the development of resistance to the host immune system. In this sense, it has recently been found in vitro that mismatch repair disruption in P. aeruginosa determines the emergence at a high frequency of multiple morphological variants resembling what naturally occurs in chronic infections (39). Furthermore, the link between hypermutation and chronic infections seems not to be restricted to just P. aeruginosa, since recent works have found a significantly higher prevalence (14%) of hypermutable Staphylococcus aureus and Haemophilus influenzae strains in CF patients than in patients with other infections (1%) (32, 37).
Whether the impressive high frequency of hypermutable P. aeruginosa strains that we have found in chronic infections is a consequence of (i) coselection of the hypermutable variants with the adaptive mutations required for long-term bacterial persistence, (ii) their coselection with antibiotic resistance mutations required for survival during the frequent antibiotic treatments received by these patients, or, more likely, (iii) both selecting forces acting synergistically remains to be elucidated. Nevertheless, results from this work strongly suggest that hypermutation is the main driver for the development of antibiotic resistance in chronic P. aeruginosa infections, since most of the strains resistant to any of the antibiotics and all of the strains resistant to multiple antibiotics were hypermutable.
In the last 2 years, the role of hypermutation in antibiotic resistance development has begun to be acknowledged as a potentially concerning problem (1, 3). A recent in vitro work has shown that mutS deficiency in P. aeruginosa determines the immediate development of resistance to every single antipseudomonal agent, due to the ascent to dominance of resistant mutants in a few hours during drug exposure, and therefore monotherapy should be avoided in the treatment of infections by hypermutable strains (31). These in vitro results, together with those obtained in the present study, certainly demonstrate that hypermutation is a major negative factor for the treatment of chronic infections not only because hypermutable strains are more frequently resistant to the antibiotics, limiting our therapeutic arsenal, but also because development of resistance to additional antibiotics is expected to be extremely fast during therapy.
Early treatment of P. aeruginosa lung colonization in patients with underlying chronic respiratory diseases before the typical markers of chronic P. aeruginosa infection, among them hypermutation, are selected should be the main therapeutic strategy (35). Once the chronic infection is established and the hypermutable strains selected, eradication is expected to be almost impossible, and avoidance of treatment failure due to multiple-antimicrobial resistance development should be the main objective. Maintenance inhaled-tobramycin treatment has been found to have a positive effect on the pulmonary function of CF patients, whereas resistance selection has been found to be relatively low due to the higher (up to 100-fold) antibiotic concentrations reached in the colonization site than by the systemic route (34). The high antibiotic concentrations may prevent resistance development even in the hypermutable strains, since the accumulation of multiple resistance mutations is required for reaching such high-level resistance.
In summary, in this work we find that hypermutable P. aeruginosa strains are extremely frequent in chronic lung infections, in contrast to what is observed in acute processes. Furthermore, hypermutation is found to be the main driver for the development of multiple-antimicrobial resistance in patients with chronic P. aeruginosa infections. In order to avoid this negative outcome, treatment strategies should be directed to the avoidance of the selection of hypermutable variants.
Acknowledgments
This work was partially financed by grant SAF2003-02851 from the Ministerio de Ciencia y Tecnología of Spain to A.O. and was supported in part by the networks “Red Española de Investigación en Patología Infecciosa (REIPI)” and “Red Española de Investigación en Patología Respiratoria (RESPIRA)” from the Ministerio de Sanidad of Spain.
REFERENCES
- 1.Blazquez, J. 2003. Hypermutation as a factor contributing to the acquisition of antimicrobial resistance. Clin. Infect. Dis. 37:1201-1209. [DOI] [PubMed] [Google Scholar]
- 2.Celli, B. R., and W. MacNee. 2004. Standards for the diagnosis and treatment of patients with COPD: a summary of the ATS/ERS position paper. Eur. Respir. J. 23:932-946. [DOI] [PubMed] [Google Scholar]
- 3.Chopra, I., A. J. O'Neill, and K. Miller. 2003. The role of mutators in the emergence of antibiotic-resistant bacteria. Drug. Resist. Updates 6:137-145. [DOI] [PubMed] [Google Scholar]
- 4.CLSI. 2005. Performance standards for antimicrobial susceptibility testing. Fifteenth informational supplement, M100-S15. CLSI, Wayne, Pa.
- 5.Evans, S. A., S. M. Turner, B. J. Bosch, C. C. Hardy, and M. A. Woodhead. 1996. Lung function in bronchiectasis: the influence of Pseudomonas aeruginosa. Eur. Respir. J. 9:1601-1604. [DOI] [PubMed] [Google Scholar]
- 6.Fish, D. N., S. C. Piscitelli, and L. H. Danziger. 1995. Development of resistance during antimicrobial therapy: a review of antibiotic classes and patient characteristics in 173 studies. Pharmacotherapy 15:279-291. [PubMed] [Google Scholar]
- 7.Gibson, R. L., J. L. Burns, and B. W. Ramsey. 2003. Pathophysiology and management of pulmonary infections in cystic fibrosis. Am. J. Respir. Crit. Care Med. 168:918-951. [DOI] [PubMed] [Google Scholar]
- 8.Giraud, A., I. Matic, O. Tenaillon, A. Clara, M. Radman, M. Fons, and F. Taddei. 2001. Costs and benefits of high mutation rates: adaptive evolution of bacteria in the mouse gut. Science 291:2606-2608. [DOI] [PubMed] [Google Scholar]
- 9.Govan, J. R. W., and V. Deretic. 1996. Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol. Rev. 60:539-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gutiérrez, O., C. Juan, J. L. Pérez, and A. Oliver. 2004. Lack of association between hypermutation and antibiotic resistance development in Pseudomonas aeruginosa isolates from intensive care unit patients. Antimicrob. Agents Chemother. 48:3573-3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haussler, S., B. Tummler, H. Weissbrodt, M. Rohde, and I. Steinmetz. 1999. Small-colony variants of Pseudomonas aeruginosa in cystic fibrosis. Clin. Infect. Dis. 29:621-625. [DOI] [PubMed] [Google Scholar]
- 12.Henwood, C. J., D. M. Livermore, D. James, M. Warner, and the Pseudomonas Study Group. 2001. Antimicrobial susceptibility of Pseudomonas aeruginosa: results of a UK survey and evaluation of the British society for antimicrobial chemotherapy disc susceptibility test. J. Antimicrob. Chemother. 47:789-799. [DOI] [PubMed] [Google Scholar]
- 13.Hill, A. T., E. J. Campbell, S. L. Hill, D. L. Bayley, and R. A. Stockley. 2000. Markers of airway inflammation in patients with stable chronic bronchitis. Am. J. Med. 109:288-295. [DOI] [PubMed] [Google Scholar]
- 14.Hla, S. W., K. P. Hui, W. C. Tan, and B. Ho. 1996. Genome macrorestriction analysis of sequential Pseudomonas aeruginosa isolates from bronchiectasis patients without cystic fibrosis. J. Clin. Microbiol. 34:575-578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Horst, J. P., T. H. Wu, and M. G. Marinus. 1999. Escherichia coli mutator genes. Trends Microbiol. 7:29-36. [DOI] [PubMed] [Google Scholar]
- 16.Kang, E. Y., R. R. Miller, and N. L. Müller. 1995. Bronchiectasis: comparison of preoperative thin-section CT and pathologic findings in resected specimens. Radiology 195:649-654. [DOI] [PubMed] [Google Scholar]
- 17.Kaufmann, M. E. 1998. Pulsed-field gel electrophoresis. Methods Mol. Med. 15:17-31. [DOI] [PubMed] [Google Scholar]
- 18.LeClerc, J. E., B. Li, W. L. Payne, and T. A. Cebula. 1996. High mutation frequencies among Escherichia coli and Salmonella pathogens. Science 274:1208-1211. [DOI] [PubMed] [Google Scholar]
- 19.Lieberman, D., and D. Lieberman. 2003. Pseudomonal infections in patients with COPD: epidemiology and management. Am. J. Respir. Med. 2:459-468. [DOI] [PubMed] [Google Scholar]
- 20.Livermore, D. M. 2002. Multiple mechanisms of antimicrobial resistance in Pseudomonas aeruginosa: our worst nightmare? Clin. Infect. Dis. 34:634-640. [DOI] [PubMed] [Google Scholar]
- 21.Lyczak, J. B., C. L. Cannon, and G. B. Pier. 2002. Lung infections associated with cystic fibrosis. Clin. Microbiol. Rev. 15:194-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lynch, J. P. 2001. Hospital-acquired pneumonia: risk factors, microbiology, and treatment. Chest 119(Suppl. 2):373-384. [DOI] [PubMed] [Google Scholar]
- 23.Maciá, M. D., N. Borrell, J. L. Pérez, and A. Oliver. 2004. Detection and susceptibility testing of hypermutable Pseudomonas aeruginosa strains with the Etest and disk diffusion. Antimicrob. Agents Chemother. 48:2665-2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matic, I., M. Radman, F. Taddei, B. Picard, C. Doit, E. Bingen, E. Denamur, and J. Elion. 1997. High variable mutation rates in commensal and pathogenic Escherichia coli. Science 277:1833-1834. [DOI] [PubMed] [Google Scholar]
- 25.Miller, J. H. 1996. Spontaneous mutators in bacteria: insights into pathways of mutagenesis and repair. Annu. Rev. Microbiol. 50:625-643. [DOI] [PubMed] [Google Scholar]
- 26.Nagaki, M., S. Shimura, Y. Tanno, T. Ishibashi, H. Sasaki, and T. Takishima. 1992. Role of chronic Pseudomonas aeruginosa infection in the development of bronchiectasis. Chest 102:1464-1469. [DOI] [PubMed] [Google Scholar]
- 27.Naidich, D. P., D. I. McCauley, H. F. Khouri, F. P. Stitik, and S. S. Siegelman. 1982. Computed tomography of bronchiectasis. J. Comput. Assisted Tomogr. 6:437-444. [DOI] [PubMed] [Google Scholar]
- 28.Nicotra, M. B., M. Rivera, A. M. Dale, R. Shepherd, and R. Carter. 1995. Clinical, pathophysiologic, and microbiologic characterization of bronchiectasis in an aging cohort. Chest 108:955-961. [DOI] [PubMed] [Google Scholar]
- 29.Oliver, A., R. Cantón, P. Campo, F. Baquero, and J. Blázquez. 2000. High frequency of hypermutable Pseudomonas aeruginosa in cystic fibrosis lung infection. Science 288:1251-1253. [DOI] [PubMed] [Google Scholar]
- 30.Oliver, A., F. Baquero, and J. Blazquez. 2002. The mismatch repair system (mutS, mutL and uvrD genes) in Pseudomonas aeruginosa: molecular characterization of naturally occurring mutants. Mol. Microbiol. 43:1641-1650. [DOI] [PubMed] [Google Scholar]
- 31.Oliver, A., B. R. Levin, C. Juan, F. Baquero, and J. Blázquez. 2004. Hypermutation and the preexistence of antibiotic-resistant Pseudomonas aeruginosa mutants: implications for susceptibility testing and treatment of chronic infections. Antimicrob. Agents Chemother. 48:4226-4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Prunier, A. L., B. Malbruny, M. Laurans, J. Brouard, J. F. Duhamel, and R. Leclerc. 2003. High rate of macrolide resistance in Staphylococcus aureus strains from patients with cystic fibrosis reveals high proportions of hypermutable strains. J. Infect. Dis. 187:1709-1716. [DOI] [PubMed] [Google Scholar]
- 33.Pujana, I., L. Gallego, G. Martín, F. López, J. Canduela, and R. Cisterna. 1999. Epidemiological analysis of sequential Pseudomonas aeruginosa isolates from chronic bronchiectasis patients without cystic fibrosis. J. Clin. Microbiol. 37:2071-2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ramsey, B. W., H. L. Dorkin, J. D. Eisenberg, R. L. Gibson, I. R. Harwood, R. M. Kravitz, D. V. Schidlow, R. W. Wilmott, S. J. Astley, M. A. McBurney, K. Wentz, and A. L. Smith. 1993. Efficacy of aerosolized tobramycin in patients with cystic fibrosis. N. Engl. J. Med. 328:1740-1746. [DOI] [PubMed] [Google Scholar]
- 35.Ratjen, F., G. Doring, and W. Nikolaizik. 2001. Effect of inhaled tobramycin on early Pseudomonas aeruginosa colonisation in patients with cystic fibrosis. Lancet 358:983-984. [DOI] [PubMed] [Google Scholar]
- 36.Richardson, A., R. Z. Yu, T. Popovic, and I. Stojiljkovic. 2002. Mutator clones of Neisseria meningitidis in epidemic serogroup A disease. Proc. Natl. Acad. Sci. USA 99:6103-6107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Román, F., R. Cantón, M. Pérez-Vázquez, F. Baquero, and J. Campos. 2004. Dynamics of long-term colonization of respiratory tract by Haemophilus influenzae in cystic fibrosis patients shows a marked increase in hypermutable strains. J. Clin. Microbiol. 42:1450-1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Romling, U., B. Feidler, J. Bobhammer, D. Grothues, J. Greipel, H. Von der Hardt, and B. Tummler. 1994. Epidemiology of chronic Pseudomonas aeruginosa infections in cystic fibrosis. J. Infect. Dis. 170:1616-1621. [DOI] [PubMed] [Google Scholar]
- 39.Smania, A. M., I. Segura, R. J. Pezza, C. Becerra, I. Albesa, and C. E. Argaraña. 2004. Emergence of phenotypic variants upon mismatch repair disruption in Pseudomonas aeruginosa. Microbiology 150:1327-1338. [DOI] [PubMed] [Google Scholar]
- 40.Smith, A. W., and B. H. Iglewski. 1989. Transformation of Pseudomonas aeruginosa by electroporation. Nucleic Acids Res. 17:10509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sniegowski, P. D., P. J. Gerrish, and R. E. Lenski. 1997. Evolution of high mutation rates in experimental populations of E. coli. Nature 387:703-705. [DOI] [PubMed] [Google Scholar]
- 42.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrock-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. Lim, K. Smith, D. Spencer, G. K. Wong, Z. Wu, I. T. Paulsen, J. Reizer, M. H. Saier, R. E. Hancock, S. Lory, and M. V. Olson. 2000. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406:959-964. [DOI] [PubMed] [Google Scholar]
- 43.Taddei, F., M. Radman, J. Maynard-Smith, B. Toupance, P. H. Gouyon, and B. Godelle. 1997. Role of mutator alleles in adaptive evolution. Nature 19:700-702. [DOI] [PubMed] [Google Scholar]
- 44.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vincent, J. L. 2003. Nosocomial infections in adult intensive-care units. Lancet 361:2068-2077. [DOI] [PubMed] [Google Scholar]
- 46.Wilson, C. B., P. W. Jones, C. J. O'Leary, D. M. Hansell, P. J. Cole, and R. Wilson. 1997. Effect of sputum bacteriology on the quality of life of patients with bronchiectasis. Eur. Respir. J. 10:1754-1760. [DOI] [PubMed] [Google Scholar]