Abstract
Carbapenemases are rare in the United States. This is the first report of a United States nosocomial outbreak of pan-resistant Pseudomonas aeruginosa infections due to an integron-borne metallo-β-lactamase, VIM-2. This emergence of carbapenemases on mobile genetic elements in the United States warrants surveillance.
Βroad-spectrum carbapenems are resistant to hydrolysis by most β-lactamases (4, 9). However, the group 3 metallo-β-lactamases (MBL) that characteristically hydrolyze these agents continue to spread worldwide (7, 13, 20, 25). Despite low amino acid homology (approximately 30%) between the two major MBL families, IMP and VIM, the hydrolytic properties are similar, conferring resistance to all β-lactams except monobactams. They are typically carried on integrons, often in association with other resistance gene cassettes, which commonly include aminoglycoside-modifying enzymes (16).
In this report, we describe the first nosocomial outbreak of MBL-producing Pseudomonas aeruginosa isolates in the United States and characterize the genetic context of blaVIM-2 in the index isolate, P. aeruginosa 7052. Two previous reports of single isolates of P. aeruginosa harboring mobile metalloenzymes in the United States described a patient in Texas, whose isolate produced VIM-7 (24), and a patient in New Mexico with an IMP-like enzyme (N. D. Hanson, A. Hossain, L. L. Buck, E. S. Moland, and K. S. Thomson, Abstr. 44th Intersci. Conf. Antimicrob. Agents Chemother., abstr. C1-291, 2004).
A 71-year-old man with no history of foreign travel was admitted in May of 2003 to our public teaching hospital in Chicago for a perforated duodenal ulcer requiring surgery. He received broad-spectrum antibiotics for peritonitis, including ampicillin-sulbactam and piperacillin-tazobactam. His antibiotic coverage was changed to imipenem for ventilator-associated pneumonia. After two weeks, blood and endotracheal-tube cultures yielded Pseudomonas aeruginosa isolates susceptible only to aztreonam. Colistin was given, but he died from sepsis and multiple-organ failure.
A review of data from our clinical microbiology lab revealed 11 additional P. aeruginosa isolates, which were susceptible only to aztreonam, during the prior 10-month period. Unfortunately, these isolates were no longer available for analysis. A retrospective analysis revealed a clustering of six cases in the trauma intensive care unit. Four isolates were respiratory specimens from patients receiving broad-spectrum β-lactams. Upon the identification of the index case, prospective surveillance continued over the next 17 months, detecting five additional cases. These cases, which constitute the isolates described herein, showed no clustering in space or time.
Two common clinical features were identified among these five cases: all patients were chronically ill, and all had received intravenous antibiotics. Three of them were exposed to β-lactams, one each having received ceftriaxone, piperacillin-tazobactam, and imipenem. Isolates were obtained from diverse sources, including urine (n = 1), wound (n = 2), respiratory secretions (n = 1), and blood (n = 1). The blood isolate likely represented transient bacteremia or contamination, as no targeted antimicrobial therapy was given and the patient was not clinically septic. Most of these patients appeared to be colonized, as only the index case required specific therapy.
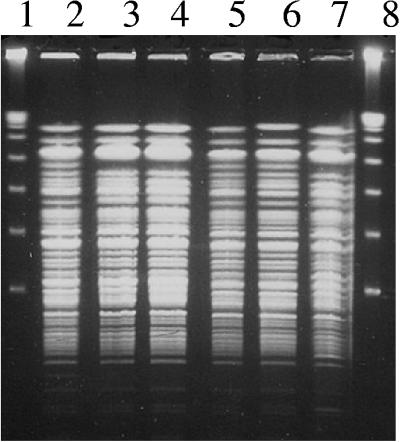
Susceptibilities were determined by use of broth microdilution methodology (12). Four of the six P. aeruginosa case isolates recovered had the unusual antibiogram phenotype of resistance to all β-lactams except aztreonam. The other two case isolates were resistant to aztreonam (Table 1). MBL Etests (AB BIODISK, Solna, Sweden) revealed that imipenem MICs were reduced eightfold in the presence of EDTA, suggesting the presence of an MBL (26). Chromosomal DNA was fingerprinted by pulsed-field gel electrophoresis (PFGE) after digestion with XbaI (Invitrogen, Carlsbad, CA) (11), and isolates revealed similar PFGE banding profiles, indicating clonality (Fig. 1).
TABLE 1.
MICs of various antibiotics for VIM-2-producing P. aeruginosa 7052 and other clinical isolates with similar antibiograms
| Antibiotic(s)a | MIC (μg/ml)
|
|||||
|---|---|---|---|---|---|---|
| P. aeruginosa 7052 | Case no.
|
|||||
| 2 | 3 | 4 | 5 | 6 | ||
| Cefoxitin | >256 | >256 | >256 | >256 | >256 | >256 |
| Ceftriaxone | >256 | >256 | >256 | >256 | >256 | >256 |
| Ceftazidime | 64 | 64 | 128 | 64 | 32 | 128 |
| Cefepime | 128 | 128 | 128 | 64 | 128 | 128 |
| Aztreonam | 8 | 8 | 32 | 8 | 4 | 32 |
| Ampicillin | >128 | >128 | >128 | >128 | >128 | >128 |
| Piperacillin | 256 | 256 | 256 | 256 | 256 | 256 |
| Piperacillin plus TZB | 256 | 256 | 256 | 256 | 256 | 256 |
| Imipenem | 64 | 64 | 64 | 64 | 32 | 64 |
| Meropenem | 32 | 32 | 32 | 32 | 16 | 32 |
| Gentamicin | >128 | >128 | >128 | >128 | >128 | >128 |
| Tobramycin | >128 | >128 | >128 | >128 | >128 | >128 |
TZB, tazobactam at a fixed concentration of 4 μg/ml.
FIG. 1.
PFGE of XbaI-digested chromosomal DNA. Lanes 1 and 8, lambda markers; lane 2, P. aeruginosa 7052; lane 3, case no. 2; lane 4, case no. 3; lane 5, case no. 4; lane 6, case no. 5; lane 7, case no. 6.
PCR analysis of genomic DNA from all case isolates by using VIM primers and PCR conditions as previously described (25) yielded an internal fragment of approximately 770 bp (data not shown). The index case isolate, P. aeruginosa 7052, was studied in depth to further characterize the mechanism of carbapenem resistance, and sequencing of its VIM PCR product was consistent with blaVIM-2.
To examine the entire gene and the surrounding genetic environment, a shotgun cloning approach was used, and Escherichia coli DH5α transformants were selected with ampicillin (50 μg/ml). Freeze-thawed cell extracts were assayed spectrophotometrically against 74 μM imipenem at 297 nm (23). Inhibition of imipenem hydrolysis was tested by preincubating the lysate for 5 min at 25°C by using a final EDTA concentration of 500 μM. β-Lactamases in cell extracts were examined by isoelectric focusing (IEF) (10). Gels were overlaid with 0.3 mM cloxacillin prior to nitrocefin development to identify AmpC-type β-lactamases (21).
All case isolates produced a pI 7.5 β-lactamase. Transformants carrying the pI 7.5 enzyme lacked carbapenemase activity; therefore, this enzyme was not examined further. Isolates from case no. 3 and case no. 6 carried an additional cloxacillin-inhibitable β-lactamase with pI 8.8, consistent with the chromosomal AmpC of P. aeruginosa (4). Extracts of a transformant with imipenem hydrolytic activity inhibitable by EDTA did not reveal a nitrocefin-hydrolyzing band on IEF. According to Poirel et al. (15), the VIM-2 β-lactamase has a pI of 5.6 but is not visible on IEF gels developed with nitrocefin, similar to the group 3b MBL from Aeromonas spp. (19).
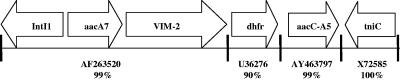
From the imipenem-hydrolyzing transformant, a 4.45-kb plasmid insert was subcloned by using plasmid pBCSK+ (Stratagene, La Jolla, CA) and both strands sequenced. Sequence homologies were determined by using the BLAST program from the National Center for Biotechnology Information (1), revealing the presence of blaVIM-2 in a nontypical integron (Fig. 2). Nucleotides 1 through 2787 had 99% identity to the In58 integron described by Poirel et al. (14) (GenBank accession no. AF263520), which contains the typical class I integron 5′ conserved sequence, intI1, aacA7, and VIM-2 genes. The P. aeruginosa 7052 sequence diverged from In58 after the VIM-2 gene. Instead of the conserved 3′ end with qacEΔ1 and sulI genes, VIM-2 was followed by a dihydrofolate reductase gene for trimethoprim resistance with 90% homology to an E. coli dihydrofolate reductase gene (GenBank accession no. U36276) and an aminoglycoside acetyltransferase gene with 99% identity to Salmonella enterica subsp. enterica serovar Kentucky aacC-A5 (GenBank accession no. AY463797). The 3′ end of the plasmid insert had 100% identity to the tniC portion of the K. aerogenes plasmid pR571, which contains an integron within transposon Tn5090 (18) (GenBank accession no. X72585). The tniC gene, which codes for a recombination protein, has recently been found to be associated with a VIM-2 integron from a French P. aeruginosa isolate (GenBank accession no. AY507153). The unusual structure of this new VIM-2 integron underscores the variety of genetic material in these mobile elements.
FIG. 2.
Genetic context of blaVIM-2-containing integron fragment from P. aeruginosa 7052. Schematic structure (not to scale) of the 4.45-kb BamHI plasmid insert. Open reading frames of the various resistance genes and the transcriptional orientation are represented by arrows. Percent homologies are to the GenBank sequences listed in the text.
Attempts to transfer P. aeruginosa 7052 plasmid DNA by electroporation using E. coli DH5α as the recipient failed. Thus, this VIM-containing integron is most likely chromosomal, although insertion into a large nontransferable plasmid remains a possibility.
The broad resistance of organisms harboring metalloenzymes represents a major threat compromising treatment options. Although metalloenzymes do not hydrolyze aztreonam, it is not clear that aztreonam is therapeutically useful. One animal model study has shown aztreonam to be effective at high doses in treating experimental pneumonia due to a VIM-2-producing P. aeruginosa isolate (3). It is noteworthy that two of our isolates were resistant to aztreonam, probably due to an AmpC type β-lactamase. The utility of older agents, like colistimethate, against such resistant strains has led to a resurgence in their use.
The arrival of carbapenemases on mobile genetic elements in clinical isolates of P. aeruginosa in the United States may pose an infection control hazard, as reported in several countries with outbreaks, including Italy, Colombia, Canada, Greece, and Japan (5, 6, 7, 17, 22). Although these MBLs are rare thus far, a very real threat to patients exists. Many, although not all, MBL producers show high-level imipenem resistance. Detection may be problematic, as automated susceptibility systems report carbapenem susceptibility only at the NCCLS breakpoint of ≥16 μg/ml. One strategy is to subject isolates that are identified as resistant to all β-lactams except aztreonam to further testing with Etest MBL strips (26) or with a double-disk synergy test (2, 8). Resistance to aztreonam remains a possibility in these strains due to other resistance mechanisms, such as porin mutations or chromosomal AmpC cephalosporinase hyperproduction.
Nucleotide sequence accession number.
The sequence of the integron obtained from P. aeruginosa 7052 was assigned GenBank accession no. AY943084.
Acknowledgments
This work was funded in part by Merck Research Laboratories, Whitehouse Station, N.J., and by the Chicago Infectious Disease Research Institute, Chicago, Ill.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arakawa, Y., N. Shibata, K. Shibayama, H. Kurokawa, T. Yagi, H. Fujiwara, and M. Goto. 2000. Convenient test for screening metallo-β-lactamase-producing gram-negative bacteria by using thiol compounds. J. Clin. Microbiol. 38:40-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bellais, S., O. Mimoz, S. Leotard, A. Jacolet, O. Petitjean, and P. Nordmann. 2002. Efficacy of β-lactams for treating experimentally induced pneumonia due to a carbapenem-hydrolyzing metallo-β-lactamase-producing strain of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 46:2032-2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bush, K., G. A. Jacoby, and A. A. Medeiros. 1995. A functional classification scheme for β-lactamases and its correlation with molecular structure. Antimicrob. Agents Chemother. 39:1211-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cornaglia, G., A. Mazzariol, L. Lauretti, G. M. Rossolini, and R. Fontana. 2000. Hospital outbreak of carbapenem-resistant Pseudomonas aeruginosa producing VIM-1, a novel transferable metallo-β-lactamase. Clin. Infect. Dis. 31:1119-1125. [DOI] [PubMed] [Google Scholar]
- 6.Crespo, M. P., N. Woodford, A. Sinclair, M. E. Kaufmann, J. Turton, J. Glover, J. D. Velez, C. R. Castaneda, M. Recalde, and D. M. Livermore. 2004. Outbreak of carbapenem-resistant Pseudomonas aeruginosa producing VIM-8, a novel metallo-β-lactamase, in a tertiary care center in Cali, Colombia. J. Clin. Microbiol. 42:5094-5101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gibb, A. P., C. Tribuddharat, R. A. Moore, T. J. Louie, W. Krulicki, D. M. Livermore, M. F. Palepou, and N. Woodford. 2002. Nosocomial outbreak of carbapenem-resistant Pseudomonas aeruginosa with a new bla(IMP) allele, bla(IMP-7). Antimicrob. Agents Chemother. 46:255-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee, K., Y. S. Lim, D. Yong, J. H. Yum, and Y. Chong. 2003. Evaluation of the Hodge test and the imipenem-EDTA double-disk synergy test for differentiating metallo-β-lactamase-producing isolates of Pseudomonas spp. and Acinetobacter spp. J. Clin. Microbiol. 41:4623-4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Livermore, D. M., A. M. Sefton, and G. M. Scott. 2003. Properties and potential of ertapenem. J. Antimicrob. Chemother. 52:331-344. [DOI] [PubMed] [Google Scholar]
- 10.Matthew, A., A. M. Harris, M. J. Marshall, and G. W. Ross. 1975. The use of analytical isoelectric focusing for detection and identification of beta-lactamases. J. Gen. Microbiol. 88:169-178. [DOI] [PubMed] [Google Scholar]
- 11.Matushek, M. G., M. J. M. Bonten, and M. K. Hayden. 1996. Rapid preparation of bacterial DNA for pulsed-field gel electrophoresis. J. Clin. Microbiol. 34:2598-2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.National Committee for Clinical Laboratory Standards. 2003. Performance standards for antimicrobial disk susceptibility tests, 8th ed. Approved standard. NCCLS document M2-A8. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 13.Nordmann, P., and L. Poirel. 2002. Emerging carbapenemases in gram-negative aerobes. Clin. Microbiol. Infect. 8:321-331. [DOI] [PubMed] [Google Scholar]
- 14.Poirel, L., T. Lambert, S. Turkoglu, E. Ronco, J. Gaillard, and P. Nordmann. 2001. Characterization of class 1 integrons from Pseudomonas aeruginosa that contain the bla(VIM-2) carbapenem-hydrolyzing β-lactamase gene and of two novel aminoglycoside resistance gene cassettes. Antimicrob. Agents Chemother. 45:546-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Poirel, L., T. Naas, D. Nicolas, L. Collet, S. Bellais, J. D. Cavallo, and P. Nordmann. 2000. Characterization of VIM-2, a carbapenem-hydrolyzing metallo-β-lactamase and its plasmid- and integron-borne gene from a Pseudomonas aeruginosa clinical isolate in France. Antimicrob. Agents Chemother. 44:891-897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Poirel, L., and P. Nordmann. 2002. Acquired carbapenem-hydrolyzing beta-lactamases and their genetic support. Curr. Pharm. Biotechnol. 3:117-127. [DOI] [PubMed] [Google Scholar]
- 17.Pournaras, S., M. Maniati, E. Petinaki, L. S. Tzouvelekis, A. Tsakris, N. J. Legakis, and A. N. Maniatis. 2003. Hospital outbreak of multiple clones of Pseudomonas aeruginosa carrying the unrelated metallo-beta-lactamase gene variants blaVIM-2 and blaVIM-4. J. Antimicrob. Chemother. 5:1409-1414. [DOI] [PubMed] [Google Scholar]
- 18.Radstrom, P., O. Skold, G. Swedberg, J. Flensburg, P. H. Roy, and L. Sundstrom. 1994. Transposon Tn5090 of plasmid R751, which carries an integron, is related to Tn7, Mu, and the retroelements. J. Bacteriol. 176:3257-3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rasmussen, B. A., and K. Bush. 1997. Carbapenem-hydrolyzing β-lactamases. Antimicrob. Agents Chemother. 41:223-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sader, H. S., M. Castanheira, R. E. Mendes, M. Toleman, T. R. Walsh, and R. N. Jones. 2005. Dissemination and diversity of metallo-beta-lactamases in Latin America: report from the SENTRY Antimicrobial Surveillance Program. Int. J. Antimicrob. Agents 25:57-61. [DOI] [PubMed] [Google Scholar]
- 21.Sanders, C. C., W. E. Sanders, Jr., and E. S. Moland. 1986. Characterization of β-lactamases in situ on polyacrylamide gels. Antimicrob. Agents Chemother. 30:951-952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Senda, K., Y. Arakawa, K. Nakashima, H. Ito, S. Ichiyama, K. Shimokata, N. Kato, and M. Ohta. 1996. Multifocal outbreaks of metallo-β-lactamase-producing Pseudomonas aeruginosa resistant to broad-spectrum β-lactams, including carbapenems. Antimicrob. Agents Chemother. 40:349-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sykes, R. B., D. P. Bonner, K. Bush, and N. H. Georgopapadakou. 1982. Azthreonam (SQ 26,776), a synthetic monobactam specifically active against aerobic gram-negative bacteria. Antimicrob. Agents Chemother. 21:85-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Toleman, M. A., K. Rolston, R. N. Jones, and T. R. Walsh. 2004. blaVIM-7, an evolutionarily distinct metallo-β-lactamase gene in a Pseudomonas aeruginosa isolate from the United States. Antimicrob. Agents Chemother. 48:329-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Toleman, M. A., A. M. Simm, T. A. Murphy, A. C. Gales, D. J. Biedenbach, R. N. Jones, and T. R. Walsh. 2002. Molecular characterization of SPM-1, a novel metallo-beta-lactamase isolated in Latin America: report from the SENTRY antimicrobial surveillance programme. J. Antimicrob. Chemother. 50:673-679. [DOI] [PubMed] [Google Scholar]
- 26.Walsh, T. R., A. Bolmstrom, A. Qwarnstrom, and A. Gales. 2002. Evaluation of a new Etest for detecting metallo-β-lactamases in routine clinical testing. J. Clin. Microbiol. 40:2755-2759. [DOI] [PMC free article] [PubMed] [Google Scholar]