Abstract
Carbapenem antibiotics are used as antibiotics of last resort because they possess a broad spectrum of antimicrobial activity and are not easily hydrolyzed by β-lactamases. Recently, class A enzymes, such as the SME-1, NMC-A, and IMI-1 β-lactamases, have been identified with the capacity to hydrolyze carbapenem antibiotics. Traditional class A β-lactamases, such as TEM-1 and SHV-1, are unable to hydrolyze carbapenem antibiotics and exhibit some differences in sequence from those that are able to hydrolyze carbapenem antibiotics. The positions that differ may contribute to the unique substrate specificity of the class A carbapenemase SME-1. Codons in the SME-1 gene representing residues 104, 105, 132, 167, 237, and 241 were randomized by site-directed mutagenesis, and functional mutants were selected for the ability to hydrolyze imipenem, ampicillin, or cefotaxime. Although several positions are important for hydrolysis of β-lactam antibiotics, no single position was found to uniquely contribute to carbapenem hydrolysis. The results of this study support a model whereby the carbapenemase activity of SME-1 is due to a highly distributed set of interactions that subtly alter the structure of the active-site pocket.
β-Lactamases compromise the efficacy of β-lactam antibiotics by catalyzing the hydrolysis of the β-lactam ring to render the antibiotics ineffective (4, 13, 15). β-Lactamases are grouped into four classes (labeled A to D) based on primary sequence homology (4). Among these, class A enzymes are often on plasmids and are a widespread cause of resistance in gram-negative bacteria. Carbapenem antibiotics such as imipenem are of value because of their broad spectrum of antibacterial activity and their stability to the action of class A β-lactamases (6, 12). Bacterial resistance to carbapenems due to the action of β-lactamases, however, has emerged. Initially, resistance was found in gram-negative bacteria harboring class B metallo-β-lactamases, which utilize a divalent cation to carry out the hydrolysis of the β-lactam ring (13, 20, 24). More recently, however, resistance has also been associated with bacteria harboring class A enzymes (20, 24). Several class A enzymes, including NMC-A (19), IMI-1 (28), SME-1 (18), SME-2 (26), KPC-1 (32), and GES-2 (25), have been shown to hydrolyze carbapenem antibiotics.
Among the class A carbapenemases, the NMC-A and SME-1 enzymes have been extensively characterized. The X-ray structures of NMC-A and SME-1 have been determined to a resolution of 1.64 and 2.13 Å, respectively (29, 31). A striking characteristic of the class A carbapenemases, which is seen in the X-ray structures of NMC-A and SME-1, is the presence of a disulfide bond between positions 69 and 238 (27, 29, 31). Site-directed mutagenesis experiments have shown that the disulfide bond is required for hydrolysis of all β-lactams by SME-1, and therefore, while important, it does not uniquely confer carbapenemase activity (14).
Other potentially important features of the class A carbapenemases have been noted from comparison of the sequences and structures of the NMC-A and SME-1 enzymes with other class A β-lactamases, such as TEM-1 (29, 31). Differences in sequence conservation, such as at positions 104 and 105, and structure, such as at position 132, between the class A carbapenemases and other β-lactamases have been observed for residues 104, 105, 132, 167, 237, and 241 (Fig. 1). This may indicate a role for these positions in the unique substrate specificity of the NMC-A and SME-1 enzymes (29, 31). In this regard, Mourey et al. provided evidence based on the X-ray structure of NMC-A with a 6α substituent similar to that of imipenem that the altered position of Asn132 permits hydrogen bonding to the hydroxethyl substituent (17). The interaction prevents this group from interacting with the hydrolytic water in the active site which is responsible in TEM-1 for the slow turnover of imipenem (17). An additional position that has been shown to play a role in the substrate specificity of SME-1 is Ser237. Sougakoff et al. demonstrated that replacement of this residue with alanine significantly compromises the ability of SME-1 to hydrolyze imipenem (30).
FIG. 1.
Sequence alignment of several class A β-lactamases with conserved motifs highlighted in blue. Residues highlighted in red are conserved among class A carbapenemases. B. lich, Bacillus licheniformis.
Here we examine the amino acid sequence requirements at positions 104, 105, 132, 167, 237, and 241 for the hydrolysis of a carbapenem (imipenem), a penicillin (ampicillin), and an oxyimino-cephalosporin (cefotaxime) (Fig. 2). The goal of this study was to determine if these positions are important for imipenem hydrolysis and to compare the sequence requirements for imipenem hydrolysis with those for other substrates. It was found that the identity of the residue at positions 105, 132, 237, and 241 is critical for imipenem hydrolysis. These positions, however, also contribute to the hydrolysis of ampicillin and cefotaxime. Therefore, a single residue in the active site of SME-1 that uniquely provides for carbapenem hydrolysis was not identified. Rather, the capacity to hydrolyze carbapenems appears to be a more general property of the shape of the active-site pocket, which is determined by many residues via cooperative interactions.
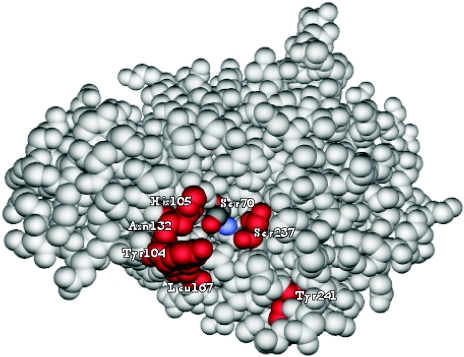
FIG. 2.
Structure of the class A β-lactamase SME-1 with catalytic amino acids Ser70 highlighted along with Tyr104, His105, Asn132, Leu167, Ser237, and Tyr241 in red (PDB 1DY6 [29].
MATERIALS AND METHODS
Site-directed randomization and random library construction.
Site-directed mutagenesis was performed by overlap extension PCR using Pfu polymerase as previously described (14) (Stratagene, La Jolla, CA). Primers were designed such that positions 104, 105, 132, 167, 237, and 241 were randomized using the degenerate NNS codon, where N is any nucleotide and S is either cytosine or guanine. The primer sequences are listed in Table 1. This approach eliminates two of the three stop codons and reduces the number of clones needed to form a complete randomized library. Primers flanking the 5′ and 3′ end of the gene were designed to allow for cloning by restriction digestion into the pTP123 cloning vector (23). In the first step, the flanking primers and the randomizing primers for positions 104, 105, 167, 237, and 241 were utilized to amplify each half of the blaSME gene. The PCR products generated from this first step were then mixed together and amplified using only the flanking primers to reconstitute the entire blaSME genes randomized at positions 104, 105, 132, 167, 237, and 241 (7). The flanking primers contained SacI and BamHI restriction sites for cloning into the pTP123 vector. All primers were obtained from Integrated DNA Technologies, Inc. (Coralville, IA).
TABLE 1.
Oligonucleotides used for randomization and cloning of the blaSME gene
| Primer name | Sequence | Use |
|---|---|---|
| SME-Sac | 5′-GGGGCGGAGCTCAACTCATTCAACACTCGG-3′a | Cloning |
| SME-BamH1 | 5′-GGGGCGGGATCCGCGTCAAGGCCACAGTCAGCTCTAACGC-3′a | Cloning |
| SME-NdeI | 5′-GGGCCGGGACATATGTCAAACAAAGTAAATTT-3′a | Cloning |
| SME-Tyr104-Bot | 5′-TTTGTTGTAATAGGTGAATGSNNTTCTAGATCCCTACTC-3′b | Randomizing |
| SME-Tyr104-Top | 5′-TATGAGAGTAGGGATCTAGAANNSCATTCACCTATTACAA-3′b | Randomizing |
| SME-His105-Bot | 5′-TTTTGTTGTAATAGGTGASNNATATTCTAGATCCCTACTC-3′b | Randomizing |
| SME-His105-Top | 5′-TATGAGAGTAGGGATCTAGAATATNNSTCACCTATTACAA-3′b | Randomizing |
| SME-Asn132-Bot | 5′-ATATTTGTTGCCCCSNNGTCGCTATATTGCAATGCAGC-3′b | Randomizing |
| SME-Asn132-Top | 5′-GCTGCATTGCAATATAGCGACNNSGGGGCAACAAATAT-3′b | Randomizing |
| SME-Leu167-Bot | 5′-GATTGCAGTGTTAAGTTCSNNTTCCCAGCGATCTAACCT-3′b | Randomizing |
| SME-Leu167-Top | 5′-AGGTTAGATCGCTGGGAANNSGAACTTAACACTGCAATC-3′b | Randomizing |
| SME-Ser237-Bot | 5′-CGCAGTACCATATGCCCCACASNNCCCAGTTTGT-3′b | Randomizing |
| SME-Ser237-Top | 5′-ACAAACTGGGNNSTGTGGGGCATATGGTACTGCG-3′b | Randomizing |
| SME-Tyr241-Bot | 5′-CATTCGCAGTACCSNNTGCCCCACAGCTCCCAGTT-3′b | Randomizing |
| SME-Tyr241-Top | 5′-AACTGGGAGCTGTGGGGCANNSGGTACTGCGAATG-3′b | Randomizing |
The SacI, BamHI, and NdeI restriction sites (underlined) were used to clone the PCR products into either pTP123 or pET24a.
The randomizing primers replaced the target codon using NNS randomization, where N represents any nucleotide and S represents guanine or cytosine.
Bacterial strains and construction of random libraries.
All blaSME genes that were randomized at either codon 104, 105, 132, 167, 237, or 241 were inserted as SacI/BamHI fragments into the multicloning site of pTP123. pTP123 is a derivative of the pTrc 99A vector (Amersham Pharmacia Biotech, Piscataway, NJ), which possesses the cat gene for chloramphenicol (cmp) resistance (23). The randomized library was electroporated into the Escherichia coli strain XL-1 Blue [recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac F′::Tn10(Tetr), proAB, δlacIq (lacZ)M15] strain (2). After electroporation, cells were spread on Luria-Bertani (LB) agar plates supplemented with 12.5 μg/ml chloramphenicol (LB/cmp). The colonies were pooled in 1 ml of LB medium. Plasmid DNA was harvested from these bacteria using the QIAGEN Qiaprep Spin Miniprep kit (Valencia, CA). The SacI and BamHI enzymes were obtained from New England Biolabs (Beverley, MA).
Primer design and cloning of SME-1 for protein expression.
Primers were designed to insert SME-1 and the SME variants into the pET24a expression vector (Novagen, Madison, WI). The restriction sites used for inserting SME genes into pET24a differed from those used for constructing libraries in the pTP123 vector. The PCR primers used for amplifying the SME genes for insertion into pET24a contained sequences recognized by the NdeI and BamHI restriction enzymes (Table 1).
The PCR products resulting from amplification of the SME genes were digested with the NdeI and BamHI restriction enzymes and inserted into the pET24a vector that had also been digested with the NdeI and BamHI restriction enzymes. The ligation reactions were used to transform E. coli BL21(DE3) [F− ompT hsdβ (rB−, mB−) gal dcm (DE3)] cells.
Antibiotic selection, MICs, and DNA sequencing.
Three antibiotics, ampicillin, imipenem, and cefotaxime, were used to select functional clones from the SME random libraries. For this purpose, E. coli XL-1 Blue cells were electroporated with random library plasmid DNA, and transformed cells were spread on LB plates containing 12.5 μg/ml chloramphenicol. To identify functional clones from the libraries, a sterile blank paper disk (Becton Dickinson, Cockeysville, MD) was soaked in either 10 mg/ml ampicillin (Sigma Chemical Co., St. Louis, MO), 4 mg/ml imipenem (Merck Research Laboratories, Rahway, NJ), or 2 mg/ml cefotaxime (Sigma Chemical Co., St. Louis, MO) and placed in the center of an agar plate on which the transformed E. coli cells had been spread, and the bacteria were allowed to grow overnight at 37°C. Colonies were picked from the zone of clearing around the disk and replated on LB/cmp plates. The blaSME gene was amplified from each colony by PCR for subsequent DNA sequencing. The DNA sequence of the blaSME gene from several selected clones was determined by automated DNA sequencing using an Applied Biosystems Instruments 3100 automated sequencer.
MICs of ampicillin, imipenem, and cefotaxime were determined for SME mutants using the twofold dilution method, with intermediate concentrations included, in a 96-well format in LB media containing 12.5 μg/ml chloramphenicol. The 96-well plates were inoculated with 1 × 105 cells/ml, incubated overnight at 37°C, and scored for growth.
Enzyme purification.
The SME-1, SME H105R, SME H105W, and SME H105Y enzymes were purified for biochemical characterization. For this purpose, 10-ml overnight cultures were diluted into 1 liter of LB medium supplemented with 12.5 μg/ml kanamycin and grown at 37°C until an optical density at 600 nm of 0.5 was reached. Protein expression was then induced with 0.5 mM isopropyl-β-d-thiogalactopyranoside, and the cultures were grown an additional 12 to 16 h. Cells were harvested by centrifugation at 3,000 × g for 20 min at 4°C in a Sorvall RC50 centrifuge and frozen at −80°C. Cells were thawed and treated with 20% glucose, 1 mM EDTA, and 20 mM Tris-HCl, pH 8.0, to release the periplasmic contents. The insoluble fraction was removed by centrifugation in a Sorvall RC50 centrifuge at 20,000 × g for 15 min at 4°C, and the soluble fraction was concentrated to a volume of 10 ml using an Amicon Centriprep-10 column (Millipore, Bedford, MA). The concentrated soluble fraction was dialyzed into 50 mM HEPES buffer, pH 7.5, using a Slide-A-Lyzer Dialysis Cassette with a 10-kDa cutoff (Pierce, Rockford, IL). The dialyzed soluble fraction was separated on a HiTrap SP FF ion exchange column (Amersham Biosciences, Piscataway, NJ). Purified protein was collected as 2-ml fractions, and fractions containing active enzyme were identified using nitrocefin (Becton Dickinson and Co., Sparks, MD). The active fractions were then concentrated using the Amicon Centriprep-10 column and separated on a HiLoad 16/60 XK16 Superdex 200 gel filtration column (Amersham Biosciences, Piscataway, NJ). Active fractions were then concentrated using the Amicon Centriprep-10 column, and protein concentrations were determined using the Bradford method (Bio-Rad Bradford reagent kit; Hercules, CA). Protein purity was assessed via sodium dodecyl sulfate-polyacrylamide gel electrophoresis.
Enzyme kinetic analysis.
Kinetic parameters were determined for the SME-1, SME H105R, SME H105W, and SME H105Y enzymes to characterize the contribution of changes at position 105 towards the hydrolysis of several β-lactam antibiotics. Kinetic parameters were determined for ampicillin (Sigma Chemical Co., St. Louis, MO), cefotaxime (Sigma Chemical Co., St. Louis, MO), and imipenem (Merck Research Laboratories, Rahway, NJ) using concentrations ranging from 10 μM to 3,200 μM depending on the substrate. The change in absorbance of substrate upon hydrolysis was measured using a Beckman DU 640 spectrophotometer (Fullerton, CA) to determine initial velocities. Km and Vmax values were calculated using Enzyme Kinetics Pro (SynexChem/ChemSW, Fairfield, CA). kcat was calculated using the known enzyme concentration and the calculated Vmax value. The measurements were repeated two to four times, and the standard deviation is reported with the kinetic parameters.
RESULTS AND DISCUSSION
Selection of SME randomized libraries on antibiotics.
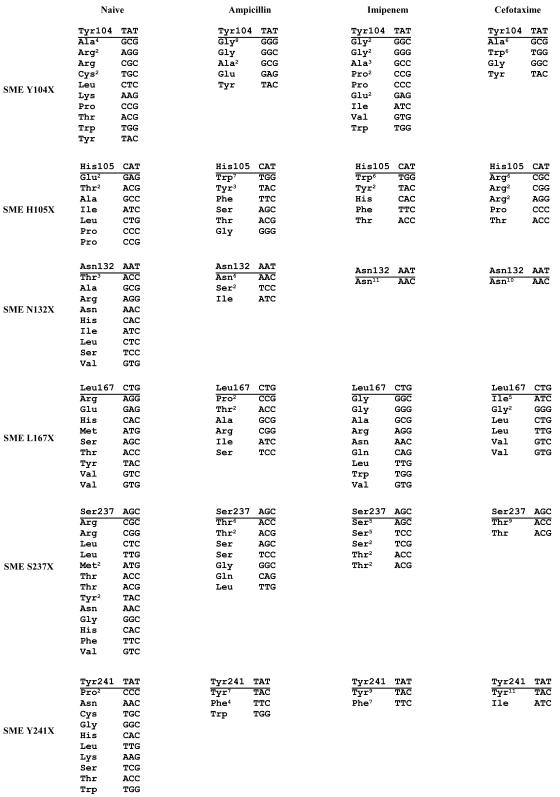
The codons for residues 104, 105, 132, 167, 237, and 241 in the blaSME gene were randomized to create libraries containing all possible substitutions at each position. In order to assess the importance of each position for the structure and function of the SME-1 β-lactamase, functional clones were selected from each library based on the ability to confer antibiotic resistance. The DNA sequence of several functional clones was then determined to identify amino acids consistent with hydrolysis of the antibiotic. Sequencing of library clones that had not been subjected to selection was performed to test the extent of randomization of the SME libraries. The sequences shown in Fig. 3 demonstrate that the libraries are diverse and not obviously biased towards any particular codon. Comparison of the set of sequences obtained before and after antibiotic selection reveals the information content of the residue position (1, 8). If, upon selection of functional clones for resistance to a β-lactam antibiotic, sequence analysis reveals codons for only one or a few amino acids, it indicates the position contains information important for the structure and function of the enzyme. In contrast, if many different amino acids are encoded in the population of clones after selection, it indicates the position contains little information for the structure and function of the enzyme (1, 8). Three different antibiotic selections were performed to determine if the information content of a position is dependent on the substrate used. For example, a position may encode information critical for the hydrolysis of imipenem while encoding essentially no information for the hydrolysis of ampicillin or cefotaxime (16).
FIG. 3.
DNA sequences of clones from the SME-1 random libraries. Sequencing results of clones picked from the naïve, ampicillin-selected, imipenem-selected, and cefotaxime-selected SME-1 random libraries are shown. Superscripts indicate the number of times a sequence occurred.
There is not strong sequence conservation at position 104 among class A β-lactamases; however, aromatic amino acids are conserved at this position among the class A carbapenemases (Fig. 1). Sequence analysis of clones selected from the SME Y104X library on the basis of imipenem resistance revealed many amino acid types and thus indicated position 104 does not encode information important for imipenem hydrolysis (Fig. 3). In contrast, sequence analysis of Y104X clones selected for ampicillin or cefotaxime resistance revealed a bias for clones with either small amino acids such as glycine and alanine or the large aromatic amino acids tyrosine and tryptophan. MIC measurements confirmed the sequencing results in that the Y104A, Y104G, and Y104W mutants retained high levels of resistance to all three antibiotics while the Y104P and Y104V mutants only retained high levels of resistance to imipenem (Table 2). The fact that the alanine and glycine as well as tyrosine and tryptophan but not other types of residues are consistent with ampicillin and cefotaxime hydrolysis suggests two different versions of the active site pocket in the vicinity of residue 104 are consistent with binding and hydrolysis of these drugs.
TABLE 2.
MICs for E. coli XL-1 Blue cells harboring SME-1, SME variants, TEM-1, and pTP123 vector
| Mutant | MIC (μg/ml) for drug:
|
||
|---|---|---|---|
| Ampicillin | Imipenem | Cefotaxime | |
| SME-1 | 1,750 | 50 | 0.0625 |
| SME Y104A | 2,000 | 70 | 0.125 |
| SME Y104G | 1,750 | 64 | 0.0625 |
| SME Y104P | 62.5 | 32 | 0.03125 |
| SME Y104V | 62.5 | 32 | 0.03125 |
| SME Y104W | 500 | 70 | 0.125 |
| SME H105R | 125 | 4 | 1.0 |
| SME H105W | 2,000 | 80 | 0.0625 |
| SME H105Y | 2,000 | 80 | 0.0625 |
| SME N132I | 62.5 | 0.5 | 0.0625 |
| SME N132S | 62.5 | 0.5 | 0.03125 |
| SME S237G | 31.25 | 0.5 | 0.03125 |
| SME S237Q | 62.5 | 0.5 | 0.03125 |
| SME S237T | 2,000 | 32 | 0.250 |
| SME Y241F | 1,750 | 32 | 0.0625 |
| SME Y241W | 1,500 | 4 | 0.0625 |
| TEM-1 | 2,000 | 1 | 0.03125 |
| pTP123 | 2 | 0.0625 | 0.03125 |
A clear preference for the aromatic amino acids tryptophan and tyrosine at position 105 was observed when clones from the SME H105X library were selected for imipenem or ampicillin resistance despite the fact that histidine is the wild-type amino acid (Fig. 3). Eight out of the 11 imipenem-resistant clones sequenced from this selection replaced histidine with either tryptophan or tyrosine, while 10 of the 14 ampicillin-resistant clones sequenced contained tryptophan or tyrosine. Consistent with the sequencing results, MIC measurements indicated that the H105W and H105Y mutants possess slightly higher levels of imipenem and ampicillin resistance than wild-type SME-1 containing histidine at this position (Table 2). It is of interest that a similar preference for aromatic residues was observed for penicillin and cephalosporin substrates in site saturation mutagenesis studies of the analogous Tyr105 position in TEM-1 β-lactamase (3).
A strong preference for a single amino acid type was observed at position 105 among clones selected for cefotaxime resistance. However, the preference was for arginine rather than an aromatic residue or histidine. Ten of the 12 cefotaxime-resistant clones sequenced possessed an arginine codon at position 105. There are six arginine codons present in nature, but only three are represented in this library due to primer design (see Materials and Methods). All three arginine codons are represented among the cefotaxime-resistant clones, indicating the strong preference for arginine when the substrate is cefotaxime (Fig. 3). MIC measurements confirmed the selection and sequencing results in that the H105R mutant is 10-fold more resistant to cefotaxime than wild-type SME-1 (Table 2). The H105R mutant also exhibits significantly reduced resistance to both imipenem and ampicillin, which is consistent with the strong preference for aromatic residues at position 105 for these substrates.
Because of the clear difference in sequence requirements at position 105 depending on the antibiotic used for selection, the H105W, H105Y, H105R, and wild-type SME-1 enzymes were purified, and kinetic parameters were determined for imipenem, ampicillin, and cefotaxime hydrolysis (Table 3). As expected based on the MIC data, the catalytic efficiency of the H105W and H105Y enzymes for ampicillin and imipenem hydrolysis is similar to that of the wild-type enzyme. Also consistent with the MIC and sequencing results is the fact that the H105R enzyme exhibits increased catalytic efficiency for cefotaxime hydrolysis but reduced efficiency for imipenem and ampicillin hydrolysis (Table 3).
TABLE 3.
Kinetic parameters of SME β-lactamasesc
| Enzyme |
kcat (s−1)
|
Km (μM)
|
kcat/Km (μM−1 s−1)
|
Relative kcat/km
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AMP | IMP | CTX | AMP | IMP | CTX | AMP | IMP | CTX | AMP | IMP | CTX | |
| SME-1 | 58 ± 4 | 185 ± 5 | NDa | 590 ± 17 | 535 ± 27 | ND | 0.10 | 0.35 | 0.0064b | 1 | 1 | 1 |
| SME H105R | 5.7 ± 0.8 | 1.1 ± 0.3 | ND | 185 ± 45 | 159 ± 52 | ND | 0.031 | 0.007 | 0.014b | 0.3 | 0.02 | 2.2 |
| SME H105W | 45 ± 2 | 230 ± 7 | ND | 132 ± 17 | 442 ± 23 | ND | 0.34 | 0.52 | 0.0093b | 3.4 | 1.5 | 1.5 |
| SME H105Y | 25 ± 9 | 566 ± 54 | ND | 150 ± 113 | 1801 ± 165 | ND | 0.17 | 0.31 | 0.0038b | 1.7 | 0.9 | 0.6 |
kcat and Km not determined (ND) directly because it is assumed that Km ≫ [S].
kcat/Km determined using v0 = kcat/Km [E][So], where vo is velocity, E is enzyme concentration, S0 is initial substrate concentration.
AMP, ampicillin; IMP, imipenem; CTX, cefotaxime.
It is apparent from the sequencing, MIC, and kinetic data that position 105 has high information content and therefore is important for the structure and function of the enzyme. Interestingly, the sequence requirements at position 105 are strongly dependent on the antibiotic used for the selection. Therefore, substitutions at position 105 can directly influence the substrate specificity of SME-1 β-lactamase. It is of interest that TEM-1 β-lactamase exhibits poor catalytic efficiency for cefotaxime hydrolysis, but mutants with a lysine at position 104 in TEM-1 have been identified in clinical isolates of bacteria resistant to cefotaxime. It is curious that no such mutants were selected from the SME Y104X library but that the arginine substitutions with increased cefotaxime resistance were selected from the H105X library. No substitutions that result in increased cefotaxime hydrolysis have been identified at position 105 in TEM-1 β-lactamase despite saturation mutagenesis experiments directed at this position (3, 22). The TEM-1 and SME-1 wild-type enzymes are similar in that they both poorly hydrolyze cefotaxime. It appears, however, that the mutational pathway leading to increase cefotaxime hydrolysis is different for these enzymes, presumably due to differences in the conformation of the 104 to 105 region.
Position 132 is part of the highly conserved S130-X131-N132 motif found in all class A β-lactamases, where it forms a critical hydrogen bond with Lys73 in the active site and also interacts with the acylamide side chain common to penicillins and cephalosporins (13, 15). Consistent with a critical role in substrate binding and catalysis, residue 132 has high information content in that only the wild-type asparagine codon was present in clones selected for imipenem or cefotaxime resistance. Asparagine also predominates among the ampicillin-resistant clones, but clones encoding serine or isoleucine were also detected. MIC measurements demonstrate that the N132S and N132I mutants do not exhibit significant resistance to ampicillin, imipenem, or cefotaxime (Table 2). Therefore, these clones may represent false positives in the selection. This result is not surprising in that Asn132 is highly conserved among class A β-lactamases, and both saturation and site-directed mutagenesis studies of position 132 in class A β-lactamases indicate the asparagine is important for β-lactam antibiotic hydrolysis (10, 21). The result is also consistent with the suggestion of Mourey et al. that Asn132 positions the imipenem hydroxyethyl group away from the hydrolytic water and thereby permits imipenem hydrolysis in the related NMC-A enzyme (17). The randomization and selection experiments will indicate whether a residue is essential, but, as may be the case with Asn132, that residue could be essential for different reasons depending on the substrate.
Position 167 is not strongly conserved among class A β-lactamases; however, leucine is conserved at this position among the subset of carbapenem hydrolyzing class A enzymes (Fig. 1). Sequencing of resistant clones indicated that the information content of position 167 is low regardless of the antibiotic used for the selection (Fig. 3). Therefore, the identity of the amino acid at position 167 is not critical for β-lactam antibiotic hydrolysis by the SME-1 enzyme.
The importance of serine at position 237 for SME-1 substrate specificity has been previously demonstrated (30). Substitution of Ser237 with alanine results in an enzyme that is unable to efficiently hydrolyze imipenem as well as cephalosporin substrates such as cephaloridine and cephalothin (30). Although the alanine substitution diminishes the ability of SME-1 to hydrolyze imipenem, data are not available as to whether other substitutions can support carbapenemase activity. Sequencing of resistant clones indicated that serine or threonine is strongly conserved among imipenem- and ampicillin-resistant clones. Consistent with this result, the S237T mutant exhibits MICs similar to those of the wild type for ampicillin and cefotaxime (Table 2). In contrast, only threonine was observed at position 237 when clones were selected for cefotaxime resistance. In addition, the S237T mutant exhibits a significant increase in the MIC for cefotaxime. These results indicate that serine or threonine at position 237 is important for imipenem hydrolysis and that threonine may result in increased hydrolysis of the oxyimino-cephalosporin cefotaxime. Interestingly, several extended spectrum TEM-1 β-lactamases have a threonine in place of an alanine at position 237 (5).
Position 241 is also conserved in class A β-lactamases with the ability to hydrolyze carbapenems (Fig. 1). In the structure of SME-1, position 241 is occupied by a tyrosine that points away from the protein towards the solvent (29). The main chain nitrogen of position 241 makes hydrogen bond interactions with the main chain carboxyl group of Lys270. The main chain carbonyl group of position 241 also makes hydrogen bond interactions with a side chain hydroxyl group and main chain nitrogen of Thr243 and with the main chain nitrogen of Arg267. Sequencing of resistant clones indicated position 241 has high information content with a strong preference for tyrosine or other aromatic amino acids regardless of the antibiotic used for selection. Despite its importance for the structure and function of SME-1, it is unlikely that the tyrosine residue directly influences substrate binding and catalysis. Rather, this position may stabilize the enzyme or influence the structure of the active-site pocket.
One model for how the substrate specificity of a class A β-lactamase is controlled is that there is a canonical class A β-lactamase fold, and onto this constant fold are grafted amino acids whose side chains provide interactions with substrates such as imipenem that favor hydrolysis of the compound. An alternative hypothesis is that, although the class A folds are similar, many interactions throughout the structure of each class A enzyme influence the structure of the active site, and subtle differences in the active site can have profound effects on the substrate specificity of the enzymes. Comparisons of the X-ray structures of carbapenemases such as NMC-A (31), extended-spectrum cephalosporinases such as Toho-1 (9), and the common class A enzyme, TEM-1 (11), support the latter hypothesis in that multiple changes distributed widely in the enzymes result in altered positioning of key groups such as Asn132 and the omega loop structure in these enzymes.
In this study, several residues in SME-1 have been identified that are important for imipenem hydrolysis. It is of interest that these positions are not uniquely important for hydrolysis of imipenem but also affect the ability of SME-1 to hydrolyze ampicillin and cefotaxime. Previous studies have sought to identify single structural determinants for the carbapenemase activity in SME-1, such as the disulfide bond between positions 69 and 238 or the role of serine at position 237, but have found the structures to be important for the hydrolysis of many β-lactam antibiotics (14, 30). Taken together, these results suggest that single residues grafted into the SME-1 active site are not uniquely responsible for imipenem hydrolysis. Instead, the results are consistent with the hypothesis that multiple residue positions influence the structure of the active site and thereby affect substrate specificity. The possibility that the catalytic properties of the SME-1 enzyme are due to a highly cooperative set of interactions suggests it will be difficult to precisely define substrate specificity determinants by site-directed mutagenesis approaches.
Acknowledgments
We thank Patrice Nordmann for providing the Serratia marcescens S6 strain containing the SME-1 β-lactamase gene and Merck Research Laboratories for providing imipenem.
This work was supported by NIH grant AI32956 to T.P.
REFERENCES
- 1.Bowie, J. U., J. F. Reidhaar-Olson, W. A. Lim, and R. T. Sauer. 1990. Deciphering the message in protein sequences: tolerance to amino acid substitutions. Science 27:1306-1310. [DOI] [PubMed] [Google Scholar]
- 2.Bullock, W. O., J. M. Fernandez, and J. M. Short. 1987. XL-1 Blue: a high efficiency plasmid transforming recA Eschericia coli strain with β-galactosidase selection. BioTechniques 5:376-379. [Google Scholar]
- 3.Doucet, N., P.-Y. De Wals, and J. N. Pelletier. 2004. Site-saturation mutagenesis of Tyr-105 reveals its importance in substrate stabilization and discrimination in TEM-1 β-lactamase. J. Biol. Chem. 279:46295-46303. [DOI] [PubMed] [Google Scholar]
- 4.Fisher, J. F., S. O. Meroueh, and S. Mobashery. 2005. Bacterial resistance to β-lactam antibiotics: compelling opportunism, compelling opportunity. Chem. Rev. 105:395-424. [DOI] [PubMed] [Google Scholar]
- 5.Giakkoupi, P., A. M. Hujer, V. Miriagou, E. Tzelepi, R. A. Bonomo, and L. S. Tzouvelekis. 2001. Substitution of Thr for Ala-237 in TEM-17, TEM-12 and TEM-26: alterations in beta-lactam resistance conferred on Escherichia coli. FEMS Microbiol. Lett. 201:37-40. [DOI] [PubMed] [Google Scholar]
- 6.Hellinger, W. C., and N. S. Brewer. 1999. Carbapenems and monobactams: imipenem, meropenem, and aztreonam. Mayo Clin. Proc. 74:420-434. [DOI] [PubMed] [Google Scholar]
- 7.Ho, S. N., H. D. Hunt, R. M. Horton, J. K. Pullen, and L. R. Pease. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51-59. [DOI] [PubMed] [Google Scholar]
- 8.Huang, W., J. Petrosino, M. Hirsch, P. S. Shenkin, and T. Palzkill. 1996. Amino acid sequence determinants of β-lactamase structure and activity. J. Mol. Biol. 258:688-703. [DOI] [PubMed] [Google Scholar]
- 9.Ibuka, A. S., Y. Ishii, M. Galleni, M. Ishiguro, K. Yamaguchi, J. M. Frere, H. Matsuzawa, and H. Sakai. 2003. Crystal structure of extended-spectrum beta-lactamase Toho-1: insights into the molecular mechanism for catalytic reaction and substrate specificity expansion. Biochemistry 42:10634-10643. [DOI] [PubMed] [Google Scholar]
- 10.Jacob, F., B. Joris, S. Lepage, J. Dusart, and J.-M. Frere. 1990. Role of the conserved amino acids of the 'SDN′ loop (Ser130, Asp131, and Asn132) in class A β-lactamse studied by site-directed mutagenesis. Biochem. J. 271:399-406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jelsch, C., M. Lionel, J.-M. Masson, and J.-P. Samama. 1993. Crystal structure of Escherichia coli TEM-1 β-lactamase at 1.8 Å resolution. Proteins Struct. Funct. Genet. 16:364-383. [DOI] [PubMed] [Google Scholar]
- 12.Livermore, D. M. 1996. Are all beta-lactams created equal? Scand. J. Infect. Dis. Suppl. 101:33-43. [PubMed] [Google Scholar]
- 13.Majiduddin, F. K., I. C. Materon, and T. G. Palzkill. 2002. Molecular analysis of beta-lactamase structure and function. Int. J. Med. Microbiol. 292:127-137. [DOI] [PubMed] [Google Scholar]
- 14.Majiduddin, F. K., and T. Palzkill. 2003. Amino acid sequence requirements at residues 69 and 238 for the SME-1 beta-lactamase to confer resistance to beta-lactam antibiotics. Antimicrob. Agents Chemother. 47:1062-1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matagne, A., J. Lamotte-Brasseur, and J.-M. Frere. 1998. Catalytic properties of class A β-lactamases: efficiency and diversity. Biochem. J. 330:581-598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Materon, I. C., Z. Beharry, W. Huang, C. Perez, and T. Palzkill. 2004. Analysis of the context dependent sequence requirements of active site residues in the metallo-beta-lactamase IMP-1. J. Mol. Biol. 344:653-663. [DOI] [PubMed] [Google Scholar]
- 17.Mourey, L., K. Miyashita, P. Swaren, A. Bulychev, J.-P. Samama, and S. Mobashery. 1998. Inhibition of the NMC-A β-lactamase by a penicillanic acid derivative and the structural bases for the increase in substrate profile of this antibiotic resistance enzyme. J. Am. Chem. Soc. 120:9382-9383. [Google Scholar]
- 18.Naas, T., L. Vandel, W. Sougakoff, D. M. Livermore, and P. Nordmann. 1994. Cloning and sequencing of the gene for a carbapenem-hydrolyzing class A β-lactamase, Sme-1, from Serratia marcescens S6. Antimicrob. Agents Chemother. 38:1262-1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nordmann, P., S. Mariotte, T. Naas, R. Labia, and M. H. Nicolas. 1993. Biochemical properties of a carbapenem-hydrolyzing beta-lactamase from Enterobacter cloacae and cloning of the gene into Escherichia coli. Antimicrob. Agents Chemother. 37:939-946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nordmann, P., and L. Poirel. 2002. Emerging carbapenemases in gram-negative aerobes. Clin. Microbiol. Infect. 8:321-331. [DOI] [PubMed] [Google Scholar]
- 21.Osuna, J., H. Viadiu, A. L. Fink, and X. Soberon. 1995. Substitution of Asp for Asn at position 132 in the active site of TEM β-lactamase. J. Biol. Chem. 270:775-780. [PubMed] [Google Scholar]
- 22.Palzkill, T., and D. Botstein. 1992. Identification of amino acid substitutions that alter the substrate specificity of TEM-1 β-lactamase. J. Bacteriol. 174:5237-5243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Petrosino, J., G. W. Rudger, H. Gilbert, and T. Palzkill. 1999. Contribution of aspartate 49 and phenylalanine 142 residues of a tight binding inhibitory protein of β-lactamases. J. Biol. Chem. 274:2394-2400. [DOI] [PubMed] [Google Scholar]
- 24.Poirel, L., and P. Nordmann. 2002. Acquired carbapenem-hydrolyzing beta-lactamases and their genetic support. Curr. Pharm. Biotechnol. 3:117-127. [DOI] [PubMed] [Google Scholar]
- 25.Poirel, L., G. F. Weldhagen, T. Naas, C. De Champs, M. G. Dove, and P. Nordmann. 2001. GES-2, a class A beta-lactamase from Pseudomonas aeruginosa with increased hydrolysis of imipenem. Antimicrob. Agents Chemother. 45:2598-2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Queenan, A. M., C. Torres-Viera, H. S. Gold, Y. Carmeli, G. M. Eliopoulos, R. C. Moellering, Jr., J. P. Quinn, J. Hindler, A. A. Medeiros, and K. Bush. 2000. SME-type carbapenem-hydrolyzing class A beta-lactamases from geographically diverse Serratia marcescens strains. Antimicrob. Agents Chemother. 44:3035-3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Raquet, X., J. Lamotte-Brasseur, F. Bouillenne, and J.-M. Frere. 1997. A disulfide bridge near the active site of carbapenem hydrolyzing class A β-lactamases might explain their unusual substrate profile. Proteins 27:47-58. [DOI] [PubMed] [Google Scholar]
- 28.Rasmussen, B. A., K. Bush, D. Keeney, Y. Yang, R. Hare, C. O'Gara, and A. A. Medeiros. 1996. Characterization of IMI-1 beta-lactamase, a class A carbapenem-hydrolyzing enzyme from Enterobacter cloacae. Antimicrob. Agents Chemother. 40:2080-2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sougakoff, W., G. L'Hermite, L. Pernot, T. Naas, V. Guillet, P. Nordmann, V. Jarlier, and J. Delettre. 2002. Structure of the imipenem-hydrolyzing class A beta-lactamase SME-1 from Serratia marcescens. Acta Crystallogr. D Biol. Crystallogr. 58:267-274. [DOI] [PubMed] [Google Scholar]
- 30.Sougakoff, W., T. Naas, P. Nordmann, E. Collatz, and V. Jarlier. 1999. Role of Ser-237 in the substrate specificity of the carbapenem-hydrolyzing class A beta-lactamase Sme-1. Biochim. Biophys. Acta. 1433:153-158. [DOI] [PubMed] [Google Scholar]
- 31.Swaren, P., L. Maveyraud, X. Raquet, S. Cabantous, C. Duez, J.-D. Pedelacq, S. Mariotte-Boyer, L. Mourey, R. Labia, M.-H. Nicolas-Chanoine, P. Nordmann, J.-M. Frere, and J.-P. Samama. 1998. X-ray analysis of the NMC-A β-lactamase at 1.64-Å resolution, a class A carbapenemase with broad spectrum specificity. J. Biol. Chem. 273:26714-26721. [DOI] [PubMed] [Google Scholar]
- 32.Yigit, H., A. M. Queenan, G. J. Anderson, A. Domenech-Sanchez, J. W. Biddle, C. D. Steward, S. Alberti, K. Bush, and F. C. Tenover. 2001. Novel carbapenem-hydrolyzing beta-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob. Agents Chemother. 45:1151-1161. [DOI] [PMC free article] [PubMed] [Google Scholar]