Abstract
Plasmid-mediated resistance to quinolones is increasingly reported in studies of Enterobacteriaceae. Using a PCR-based strategy, a series of gram-negative species were screened for qnrA-like genes. Shewanella algae, an environmental species from marine and fresh water, was identified as its reservoir. This is a one of the very few examples of progenitor identification of an acquired antibiotic resistance gene.
Multidrug resistance in Enterobacteriaceae, including resistance to quinolones, is currently among the top antibiotic resistance problems in the United States and is rising worldwide (5, 13). Quinolone resistance in Enterobacteriaceae results mostly from chromosomal mutations in genes coding for DNA gyrase (topoisomerase II), for efflux and outer membrane proteins, or for their regulatory elements (11). However, Qnr (later termed QnrA), a plasmid-mediated quinolone resistance determinant (G. A. Jacoby, K. Walsh, D. Mills, V. Wolker, A. Robicsek, H. Oh, and D. C. Hooper, Abstr. 44th Intersci. Conf. Antimicrob. Agents Chemother., Abstr. C2-1898a, 2004), had been reported in 1998 from Klebsiella pneumoniae first from the United States (16). It has been reported since then in Citrobacter freundii, Escherichia coli, Enterobacter cloacae, Enterobacter sakazakii, K. pneumoniae, and Klebsiella oxytoca from Asia and Europe (8, 15, 18, 26, 32).
The 218-amino-acid protein QnrA, which belongs to the pentapeptide repeat family, protects DNA gyrase and topoisomerase IV from the inhibitory activity of quinolones (29, 30). QnrA confers resistance to nalidixic acid and increases MICs of fluoroquinolones up to 32-fold (7, 15, 16).
The recent emergence of plasmid-mediated quinolone resistance led us to search for its natural reservoir. Our working hypothesis was that this novel resistance determinant could derive from an environmental, human, or animal gram-negative species, taking into account our knowledge on horizontal gene transfer in bacteria. A total of 48 gram-negative bacterial species were screened that included clinically significant bacterial species such as representatives of the Enterobacteriaceae, Aeromonadaceae, Pseudomonadaceae, Xanthomonadaceae, Moraxellaceae, and Shewanellaceae families.
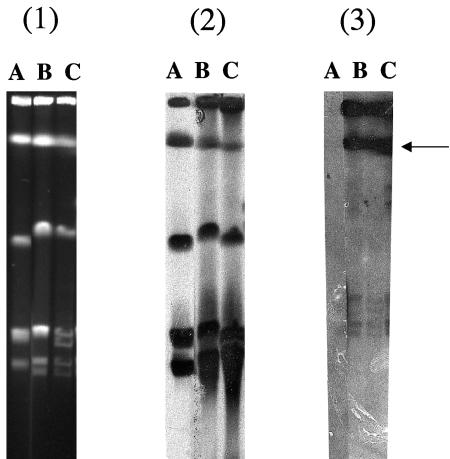
PCR-based experiments using specific primers for the qnrA gene (15) identified a positive signal for three Shewanella algae clinical isolates (KB-1 to KB-3) (10) and reference strain S. algae CIP106454T (Institut Pasteur strain collection, Paris, France). The locations of the qnrA genes were determined precisely by using the endonuclease I-Ceu-I technique (14). Pulsed-field gel electrophoresis (28) gave six DNA fragments from Shewanella sp. strains (Fig. 1). The DNA probe for rRNA consisting of a 1,504-bp PCR fragment for 16S and 23S rRNA genes (6) hybridized with all the fragments from the Shewanella strains. Hybridization with a DNA probe internal to qnrA gene (15) gave a single signal for the S. algae strains only (Fig. 1).
FIG. 1.
(Panel 1) Pulsed-field gel electrophoresis profiles of I-Ceu-I digested whole-cell DNAs of Shewanella sp. strains. Lane A, S. putrefaciens reference strain CIP8040 (Institut Pasteur, Paris); lane B, S. algae clinical isolate KB-1; lane C, S. algae reference strain CIP106454T. (Panels 2 and 3) Southern hybridization was performed with a specific probe for the 16S-23S rRNA gene (6) and an internal probe for the qnrA gene (15). The horizontal arrow indicates hybridization position with the qnrA probe. Negative hybridization results with the qnrA probe were obtained for the S. putrefaciens reference strain.
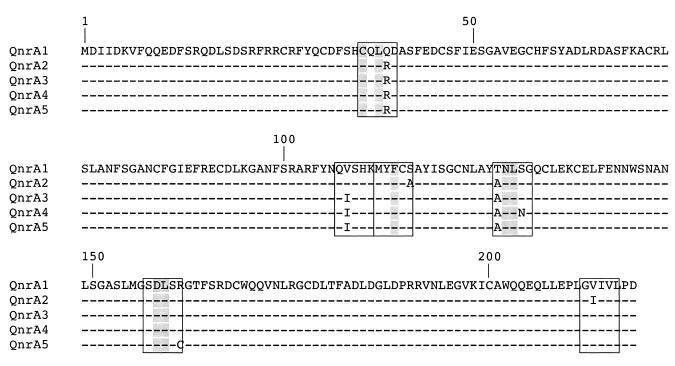
The four corresponding QnrA-like proteins of S. algae reference strains and isolates KB-1 to KB-3 had only two to four amino acid substitutions compared to QnrA (termed now QnrA1) (Fig. 2). Note that the G+C content (52%) of the qnrA-like genes of S. algae matched exactly that of the genome of S. algae (4). S. algae is a gram-negative species belonging to the Shewanellaceae family that is widely distributed in marine and freshwater environments (3, 19, 21). Whereas S. putrefaciens is also a human pathogen (2), it seems now that most of the S. algae isolates have been misidentified for S. putrefaciens (4) and most of the S. algae infections are related to seawater exposure (3). The MIC of nalidixic acid was 2 μg/ml, and those of fluoroquinolones ciprofloxacin, ofloxacin, sparfloxacin, and norfloxacin were 0.12, 0.5, 0.5, and 0.5 μg/ml, respectively, being identical for the four S. algae strains and remaining in the susceptibility range (17). Based on analysis of an antibiotic resistance phenotype, the presence of a QnrA-like determinant in S. algae could not be suspected. However, these MIC levels were four- to eightfold higher than those of the closely related species Shewanella putrefaciens that was qnrA negative.
FIG. 2.
Sequence comparison of plasmid-mediated QnrA-like determinants to those of S. algae strains. The plasmid-mediated QnrA1 and QnrA2 determinants are from K. pneumoniae from the United States (16) and from Klebsiella oxytoca from China (GenBank accession number AY675584), respectively. QnrA3 is from S. algae reference strain CIP106454T and S. algae clinical isolate KB-1, whereas QnrA4 and QnrA5 are from S. algae clinical isolates KB-2 and KB-3, respectively (10). DQ058661, DQ058662, and DQ058663 are GenBank accession numbers for QnrA3, QnrA4, and QnrA5, respectively. Dashes are for identical amino acid residues. Pentapeptide motifs in which amino acid substitutions have been identified compared to the QnrA1 sequence are boxed. In these motifs, the conserved residues identified by Tran and Jacoby (29) are shaded.
Since the plasmid-encoded qnrA gene had been found in a sul1-type integron downstream of the open reading frame orf513 coding for a recombinase (7, 15, 26, 29, 32), we also used a PCR-based strategy with primers ORF513D3 and ORF513D5(15) for detecting this gene in S. algae. These experiments failed as well as those designed to amplify the ampR gene that has been identified in In36 and In37 just downstream of the qnrA gene (data not shown) (32). These results indicated that the CR1 element that provides promoter sequences for high-level expression of the plasmid-mediated QnrA gene in Enterobacteriaceae (15) was not associated with qnrA in S. algae.
Further work may identify in other psychrophilic species the reservoir of the two novel plasmid-mediated quinolone resistance determinants, i.e., QnrB identified from the United States and South India (Jacoby et al., abstr. C2-1898a, 2004) and QnrS identified from Japan (9), which share only 40 and 59% amino acid identity with QnrA, respectively. Notably, it has been shown recently that Vibrio parahaemolyticus possesses a QnrA homologue (58% identity) (27).
This report indicates that gram-negative bacterial species not only of the veterinary world but also of the environment may be a reservoir for emerging antibiotic resistance genes spreading in human pathogens, as suspected (33, 34). Our finding emphasizes the possible role of the aquatic environment as a reservoir of antibiotic resistance genes. In addition, we had determined recently that another Shewanella species, Shewanella oneidensis, is the natural reservoir of OXA-48, a plasmid-encoded carbapenem-hydrolyzing β-lactamase gene that was identified in K. pneumoniae, further indicating gene exchange between Shewanella spp. and Enterobacteriaceae (24). The present report adds knowledge on the origin of clinically significant antibiotic resistance genes that has been established without ambiguity in very few cases, such as for the SHV-, CTX-M-, and AmpC-type β-lactamase genes originating in enterobacterial species (12, 23, 25) and for tetracycline resistance determinants identified in mycobacteria and originating in Streptomyces rimosus (22).
The quinolones prescribed in human therapy are also extensively used in aquaculture (20) as synthetic molecules stable in a water environment (31). Thus, it is tempting to speculate that subinhibitory concentrations of quinolones in water may select for waterborne S. algae strains and therefore enhance transfer of this naturally occurring quinolone resistance determinant to Enterobacteriaceae. The role of quinolones for inducing this antibiotic resistance gene transfer may be related to induction of the SOS bacterial repair system, as shown previously (1).
Acknowledgments
This work was funded by a grant from the Ministère de l'Education Nationale et de la Recherche (UPRES-EA3539), Université Paris XI, France, and by a grant from the European Community (6th PCRD, LSHM-CT-2003-503335). L.P. is a researcher from the INSERM (Paris, France), and J.-M.R.-M. was a recipient of a travel grant from the Spanish Society for Clinical Microbiology and Infectious Diseases in 2004. We also thank A. Pascual for constant support of J.-M.R.-M.
REFERENCES
- 1.Beaber, J. W., B. Hochhut, and M. K. Waldor. 2004. SOS response promotes horizontal dissemination of antibiotic resistance genes. Nature 427:72-74. [DOI] [PubMed] [Google Scholar]
- 2.Brink, A. J., A. Van Straten, and A. J. Rensburg. 1995. Shewanella (Pseudomonas) putrefaciens bacteremia. Clin. Infect. Dis. 20:1327-1332. [DOI] [PubMed] [Google Scholar]
- 3.Fonnesbech-Vogel, B., H. M. Holt, P. Gerner-Smidt, A. Bundvad, P. Sogaard, and L. Gram. 2000. Homogeneity of Danish environmental and clinical isolates of Shewanella algae. Appl. Environ. Microbiol. 66:443-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fonnesbech-Vogel, B., K. Jorgensen, H. Christensen, J. Elmerdhahl-Olsen, and L. Gram. 1997. Differentiation of Shewanella putrefaciens and Shewanella alga on the basis of whole-cell protein profiles, ribotyping, phenotypic characterization, and 16S rRNA gene sequence analysis. Appl. Environ. Microbiol. 63:2189-2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Garau, J., M. Xercavins, M. Rodriguez-Carballeira, J. R. Gomez-Vera, I. Coll, D. Vidal, T. Llovet, and A. Ruiz-Bremon. 1999. Emergence and dissemination of quinolone-resistant Escherichia coli in the community. Antimicrob. Agents Chemother. 43:2736-2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Girlich, D., L. Poirel, A. Leelaporn, A. Karim, C. Tribuddharat, M. Fennewald, and P. Nordmann. 2001. Molecular epidemiology of the integron-located VEB-1 extended-spectrum β-lactamase in nosocomial enterobacterial isolates in Bangkok, Thailand. J. Clin. Microbiol. 39:175-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jacoby, G. A., N. Chow, and K. B. Waites. 2003. Prevalence of plasmid-mediated quinolone resistance. Antimicrob. Agents Chemother. 47:559-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jonas, D., K. Biehler, D. Hartung, B. Spitzmuller, and F. D. Daschner. 2005. Plasmid-mediated quinolone resistance in isolates obtained in German intensive care units. Antimicrob. Agents Chemother. 49:773-775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hata, M., M. Suzuki, M. Matsumoto, M. Takahashi, K. Sato, S. Ibe, and K. Sakae. 2005. Cloning of a novel quinolone resistance from a transferable plasmid in Shigella flexneri 2b. Antimicrob. Agents Chemother. 49:801-803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Héritier, C., L. Poirel, and P. Nordmann. 2004. Genetic and biochemical characterization of a chromosome-encoded carbapenem-hydrolyzing Ambler class D β-lactamase from Shewanella algae. Antimicrob. Agents Chemother. 48:1670-1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hooper, D. C. 2001. Emerging mechanisms of fluoroquinolone resistance. Emerg. Infect. Dis. 7:337-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huemeniuk, C., G. Arlet, V. Gautier, P. Grimont, R. Labia, and A. Philippon. 2002. β-Lactamases of Kluyvera ascorbata, probable progenitors of some plasmid-mediated CTX-M types. Antimicrob. Agents Chemother. 46:3045-3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lautenbach, E., B. L. Strom, I. Nachamkin, W. B. Bilker, A. M. Marr, L. A. Larosa, and N. O. Fishman. 2004. Longitudinal trends in fluoroquinolone resistance among Enterobacteriaceae isolates from inpatients and outpatients, 1989-2000; differences in the emergence and epidemiology of resistance across organisms. Clin. Infect. Dis. 38:655-662. [DOI] [PubMed] [Google Scholar]
- 14.Liu, S. L., A. Hessel, and K. E. Sanderson. 1993. Genomic mapping with I-Ceu-I, an intron-encoded endonuclease specific for genes for ribosomal RNA in Salmonella spp., Escherichia coli, and other bacteria. Proc. Natl. Acad. Sci. USA 90:6874-6878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mammeri, H., M. Van De Loo, L. Poirel, L. Martinez-Martinez, and P. Nordmann. 2005. Emergence of plasmid-mediated quinolone resistance in Escherichia coli in Europe. Antimicrob. Agents Chemother. 49:71-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martinez-Martinez, L., A. Pascual, and G. A. Jacoby. 1998. Quinolone resistance from a transferable plasmid. Lancet 351:797-799. [DOI] [PubMed] [Google Scholar]
- 17.National Committee for Clinical Laboratory Standards. 2004. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically: approved standard M100-S14. NCCLS, Wayne, Pa.
- 18.Nazik, A., L. Poirel, and P. Nordmann. 2005. Further identification of plasmid-mediated quinolone resistance determinant in Enterobacteriaceae, in Turkey. Antimicrob. Agents Chemother. 49:4050-4053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nedoluha, P. C., S. Owens, E. Russek-Cohen, and D. C. Westhoff. 2001. Effect of sampling method on the representative recovery of microorganisms from the surfaces of aquaculture finfish. J. Food Prot. 10:1515-1520. [DOI] [PubMed] [Google Scholar]
- 20.NORM/NORM-VET2002. 2003. Consumption of antimicrobial agents and occurrence of antimicrobial resistance in Norway. Tromso/Oslo 2003. ISSN:1502-2307, p. 13.
- 21.Nozue, H., T. Hayashi, Y. Hashimoto, T. Ezaki, K. Hamasaki, K. Ohwada, Y., and Y. Terawaki. 1992. Isolation and characterization of Shewanella alga from human clinical specimens and emendation of the description of S. alga Simidu et al., 1990, 335. Int. J. Syst. Bacteriol. 42:628-634. [DOI] [PubMed] [Google Scholar]
- 22.Pang, Y., B. A. Brown, V. A. Steingrube, R. J. Wallace, Jr., and M. C. Roberts. 1994. Tetracycline resistance determinants in Mycobacterium and Streptomyces. Antimicrob. Agents Chemother. 38:1408-1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Philippon, A., G. Arlet, and G. A. Jacoby. 2002. Plasmid-determined AmpC-type β-lactamases. Antimicrob. Agents Chemother. 46:1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Poirel, L., C. Héritier, and P. Nordmann. 2004. Chromosome-encoded Ambler class D β-lactamase of Shewanella oneidensis as a progenitor of carbapenem-hydrolyzing oxacillinase. Antimicrob. Agents Chemother. 48:348-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Poirel, L., P. Kämpfer, and P. Nordmann. 2002. Chromosome-encoded Ambler class A β-lactamase of Kluyvera georgiana as a probable progenitor of a subgroup of CTX-M extended-spectrum β-lactamases. Antimicrob. Agents Chemother. 46:4038-4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Poirel, L., M. Van De Loo, H. Mammeri, and P. Nordmann. 2005. Association of plasmid-mediated resistance with extended-spectrum β-lactamase VEB-1. Antimicrob. Agents Chemother. 49:3091-3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saga, T., M. Kaku, Y. Onodera, S. Yamachika, K. Sato, and H. Takase. 2005. Vibrio parahaemolyticus chromosomal qnr homologue VPA0095: demonstration by transformation with a mutated gene of its potential to reduce quinolone susceptibility in Escherichia coli. Antimicrob. Agents Chemother. 49:2144-2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 29.Tran, J. H., and G. A. Jacoby. 2002. Mechanism of plasmid-mediated quinolone resistance. Proc. Natl. Acad. Sci. USA 99:5638-5642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tran, J. H., G. A. Jacoby, and D. C. Hooper. 2005. Interaction of plasmid-encoded quinolone resistance protein Qnr with Escherichia coli DNA gyrase. Antimicrob. Agents Chemother. 49:118-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Turiel, E., A. Martin-Esteban, G. Bordin, and A. R. Rodriguez. 2004. Stability of fluoroquinolones antibibiotics in river water samples and in octadecyl silica solid-phase extraction cartridges. Anal. Bioanal. Chem. 380:123-128. [DOI] [PubMed] [Google Scholar]
- 32.Wang, M., J. H. Tran, G. A. Jacoby, Y. Zhang, F. Wang, and D. C. Hooper. 2003. Plasmid-mediated quinolone resistance in clinical isolates of Escherichia coli from Shanghai, China. Antimicrob. Agents Chemother. 47:2242-2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Witte, W. 1998. Medical consequences of antibiotic use in agriculture. Science 279:996-997. [DOI] [PubMed] [Google Scholar]
- 34.Young, H. K. 1993. Antimicrobial resistance spread in aquatic environments. J. Antimicrob. Chemother. 31:627-635. [DOI] [PubMed] [Google Scholar]