Abstract
We conducted in vitro experiments to evaluate the susceptibility of a clinical isolate of Cryptococcus neoformans to a wide range of concentrations of fluconazole. In vitro susceptibility was tested using broth macrodilution methods modified to provide a numeric count of viable organisms. The association between the quantitative in vitro response and fluconazole drug concentrations was estimated using local nonparametric regression. Regression analysis was used to assess the correspondence between the in vitro fluconazole concentration-response curve and the murine dose-response curve observed in our previously reported murine model. The regression model was then used to predict the murine response. There was a strong correspondence between in vitro measures of response to fluconazole alone and the previously reported biologic effects seen in the mouse. In vitro antifungal drug susceptibility testing can reliably predict the murine response to fluconazole.
The goal of in vitro susceptibility testing is to predict the clinical outcome. Having an in vitro measure that can reliably predict the response following 2 weeks of treatment would permit physicians to select the drug(s) with the greatest activity, extend intensive treatments past 2 weeks, and/or use combination drug therapy. During the last 20 years, considerable effort has been expended to establish a reproducible standard for in vitro testing of yeast susceptibility to antifungal drugs. This standard has been most useful for testing Candida species causing oral thrush (14, 18, 20). Limited data suggest this method might also be useful for Cryptococcus neoformans; however, no interpretive guidelines or breakpoints have been identified (16, 19). The usual approach to identifying breakpoints has been to measure outcome as a categorical response (e.g., sterile cerebrospinal fluid [CSF] versus nonsterile CSF) (1, 11, 24). This approach forces the search for a breakpoint into a search for a drug concentration that predicts an outcome measure that takes on just two values, “sterile CSF” versus “nonsterile CSF.” This search is greatly confounded by the wide range in the severity of meningitis that occurs in clinical practice (21, 24).
In our previously published murine model, we used a clinical isolate of C. neoformans to evaluate the effects of fluconazole (12). The response was the number of CFU per gram of brain recovered at day 16. By using this quantitative response and by testing fluconazole over a wide range of doses, we could use regression methods to estimate the dose-response curve for fluconazole. In the present paper, we report the in vitro susceptibility of this same isolate to fluconazole tested over a wide range of concentrations.
MATERIALS AND METHODS
Test isolate.
The Cryptococcus neoformans var. neoformans isolate (USC 1597) was obtained from a patient with AIDS-associated cryptococcal meningitis that responded promptly to treatment with fluconazole plus flucytosine. The isolate was stored in 20% skim milk at −70°C and subcultured on Sabouraud's dextrose agar before testing.
In vitro antifungal drug susceptibility testing.
The susceptibility of this isolate to fluconazole was measured following the Clinical and Laboratory Standards Institute (CLSI) (formerly the National Committee for Clinical Laboratory Standards [NCCLS]) broth macrodilution method (14), modified to enumerate the numbers of organisms viable after 48 h. Six to 10 colonies from a 4-day growth of isolate USC 1597 grown on Sabouraud's dextrose agar were suspended in 10 ml of buffered RPMI 1640 medium (MediaTech, Herndon, VA) supplemented with l-glutamine and buffered with HEPES. The number of organisms per ml after 24 h of incubation was estimated by a manual hemocytometer count. Further dilutions were prepared to achieve a final inoculum of approximately 500 CFU/ml. This inoculum number was confirmed by quantitative culture. Following 48 h of incubation in drug-containing media, the numbers of viable CFU per ml at each concentration were assessed quantitatively by plating serial dilutions. Fluconazole was tested at concentrations ranging from 0.5 to 20 mg/liter.
Statistical analysis.
Local nonparametric regression as described previously (5-8) was used to estimate the in vitro fluconazole concentration-response curve. Ninety-nine percent confidence intervals (99% CIs) based on the fitted regression were used as described previously (3, 12) to assess the magnitude, variation, and significance of the effects of fluconazole. If the 99% CIs do not overlap, then the difference in response is significant at the 0.01 level. If the CIs do overlap, we use the widths of the CIs to assess the magnitude of the potential difference within the context of the observed variability. The association between the in vitro fluconazole concentration-response curve and the murine fluconazole dose-response curve from our previously reported murine model (12) was assessed using regression analysis (see Appendix) (15, 23). This regression model was then used to predict the murine fluconazole dose-response curve. Descriptive statistics were based on medians and robust CIs (9). All statistical analyses were performed using S-Plus (10, 23).
RESULTS
Observed fluconazole dose-response curves.
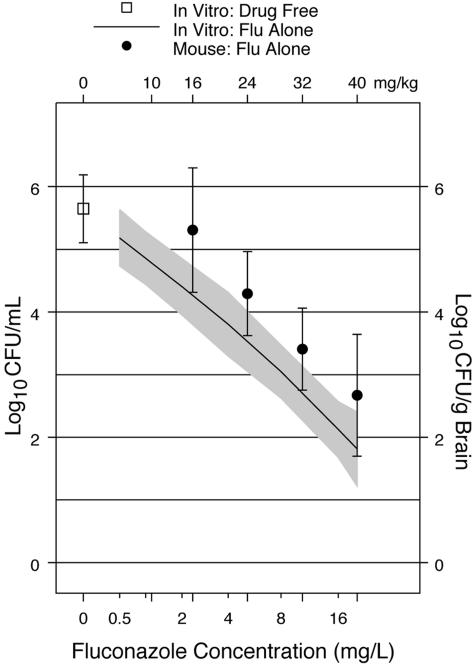
In the in vitro experiments, the loess fit of the association (fit of the association found with LOESS software) between the numbers of log10 CFU/ml viable after 48 h of incubation in fluconazole showed a strong locally linear dependence on the log2 concentration of fluconazole (Fig. 1). In our previously reported murine model, the loess fit of the association between the numbers of log10 CFU/g brain recovered after 14 days of treatment with fluconazole alone also showed a strong linear dependence on the dose of fluconazole (Fig. 1) (12). The range of responses to fluconazole seen in vitro was similar to the range of responses seen in the mouse model. The widths of the CIs for the fitted responses were similar (Fig. 1), showing that the magnitude of the variation in the responses was similar (±0.9 log10 CFU).
FIG. 1.
Loess fits for the in vitro concentration-response curve and murine dose-response curve for fluconazole (Flu) alone. The solid line shows the in vitro susceptibility, and the filled circles show the murine response. The shaded bands and vertical bars show the 99% CIs for the in vitro susceptibility and murine response, respectively. The bottom x axis shows the in vitro concentrations of fluconazole (mg/liter). The top x axis shows the doses of fluconazole (mg/kg/day) administered to the mice. Murine data are from Larsen et al. (12). The animal protocol for our murine model has been described, and the results were reported previously (12). Briefly, male BALB/c mice weighing 23 to 25 g were anesthetized, and meningitidis was induced with approximately 700 CFU of C. neoformans injected directly into the cranial vault. Treatment was initiated 2 days later with the assigned concentrations of fluconazole (Diflucan; Pfizer, Inc.) dissolved in the sole source of drinking water. Treatment was continued for 14 days. The treatment effect was measured by the numbers of CFU of C. neoformans per gram of brain recovered at the end of treatment.
Predicted fluconazole dose-response curve.
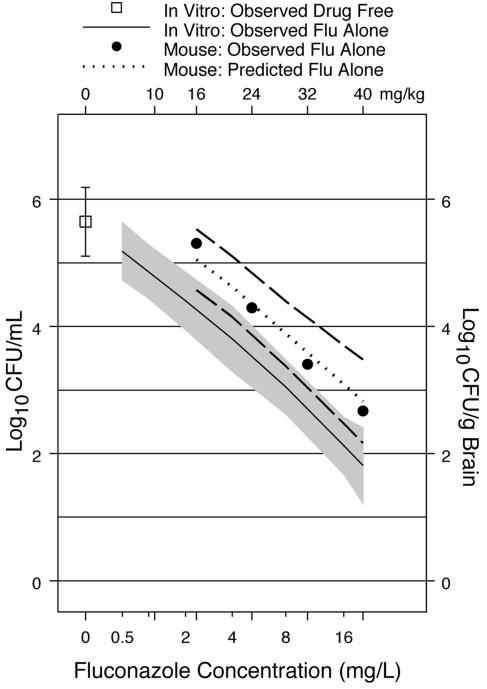
Loess regression analyses showed that both the in vitro and murine fluconazole dose-response curves could be approximated well by global regression models. Therefore, linear regression analysis (see Appendix) was used to evaluate the correspondence between the in vitro fluconazole concentration-response curve and the murine fluconazole dose-response curve previously reported (12). Both the 95% and the 99% CIs for the difference in slopes of the two curves contained zero, indicating that the two curves were parallel. This linear regression model was then used to predict the murine fluconazole dose-response curve (Fig. 2) on the basis of the observed in vitro concentration response. The loess fit for the observed murine dose-response curve lies almost directly on the predicted mouse dose-response curve (Fig. 2). The 95% CIs for the predicted curve overlap the observed murine response curve (Fig. 2). The 99% CIs also overlap the observed murine fluconazole response curve (data not shown).
FIG. 2.
Predicted murine dose-response curve. The dotted line shows the predicted murine dose-response curve for fluconazole (Flu) alone based on the linear regression model. The broken lines show the 95% CIs for the predicted murine dose-response curve. The solid line shows the loess fit for the observed in vitro susceptibility, and the filled circles show the loess fit for the observed murine response. The shaded band shows the 99% CIs for the in vitro susceptibility. The top x axis shows the in vitro fluconazole concentrations (mg/liter). The bottom x axis shows the treatment doses of fluconazole (mg/kg/day). Murine data are from Larsen et al. (12).
Correspondence between in vitro fluconazole concentrations and murine fluconazole doses.
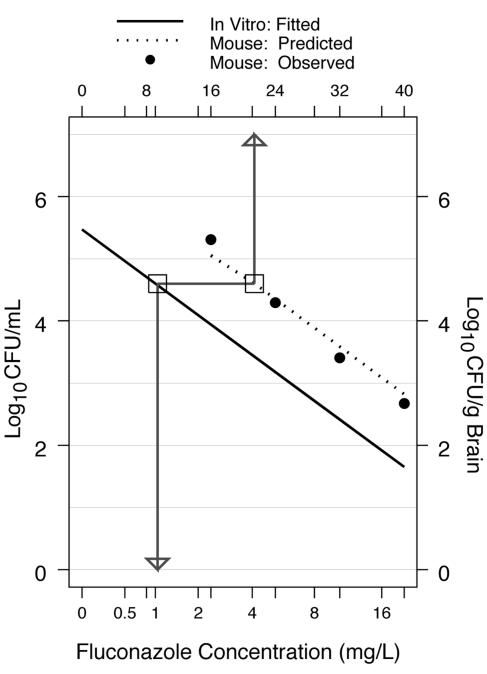
Figure 3 shows how the linear regression model for the in vitro concentration-response curve can be used to estimate the correspondence between the in vitro concentrations and the mouse fluconazole doses (see Appendix). The in vitro response observed at 1 mg/liter of fluconazole was approximately 4.6 log10 CFU/ml (Fig. 3). The global linear regression model predicts that the fluconazole dose administered to mice that would produce this level of response would be approximately 21 mg/kg of body weight (Fig. 3). For comparison, the murine response at 21 mg/kg per day given by the locally linear loess fit of the observed murine response was 4.65 log10 CFU/g (Fig. 3) (12).
FIG. 3.
Correspondence between in vitro fluconazole concentrations and murine fluconazole doses. The dotted line shows the predicted murine fluconazole dose-response curve based on the regression model for the correspondence between the observed in vitro (solid line) and the observed murine response (filled circles). The bottom x axis shows the in vitro fluconazole concentrations (mg/liter). The top x axis shows the treatment doses of fluconazole (mg/kg/day). Murine data are from Larsen et al. (12).
DISCUSSION
In the present report, we evaluated the in vitro susceptibility of a clinical isolate of C. neoformans to a wide range of concentrations of fluconazole. Regression analysis demonstrated a strong correspondence between the observed in vitro response curve and the murine dose-response curve observed in our previously published murine model (12). The in vitro model is a closed static system in which the C. neoformans organisms are exposed continuously to drug for 48 h in a test tube. In contrast, our murine model of cryptococcal meningitis is a dynamic biologic system with intermittent exposure to fluconazole given over 14 days (12). We do not expect to see a mg-for-mg correspondence between in vitro drug concentrations used in the closed static system and the doses administered intermittently to mice in the dynamic biologic system. However, using the same quantitative responses (number of CFU) and testing a wide range of doses using the same level of inoculum of the same isolate, we have demonstrated that the observed in vitro susceptibility of C. neoformans measured at 48 h can reliably predict the murine response to fluconazole when treatment is given for 14 days.
It is not known whether the drug concentrations used in vitro that correspond to the observed murine response represent peak or trough drug concentrations in the brain, CSF, or some other pharmacokinetic compartment or construct. Furthermore, we do not propose that the doses identified in our mouse model can be translated directly to humans. Monitoring the biologic responses in humans is based on CSF cultures, whereas the biologic response in the mouse is based on brain tissue. It is well established that fluconazole metabolism is more rapid in mice than in humans (2, 13, 22). Thus, the corresponding fluconazole concentrations in human subjects will almost certainly differ.
One practical application of this approach is the design of in vitro and animal experiments to determine whether these results apply to the many different strains of C. neoformans. However, the ultimate goal of in vitro susceptibility testing is not to cure cryptococcal meningitis in mice. Thus, a more important application would be to use our approach to identify quantitative measures of in vitro susceptibility that can reliably predict clinical outcome. The usual approach to identifying clinical correlations with in vitro susceptibility has been to use a response that takes on just two values, “sterile CSF” versus “nonsterile CSF.” Two recently reported clinical trials have used quantitative counts based on the patient's CSF as a more informative measure of treatment response (4, 17). Employing quantitative microbiologic measures of response to drug exposure in both in vitro testing and in the clinic will greatly facilitate evaluating associations between in vitro susceptibility and the patient's response to drug treatment. This approach holds the greatest promise for identifying in vitro measures of drug susceptibility that can be used to select the best therapy for each patient.
Having an in vitro measure that can reliably predict response following 2 weeks of treatment would permit physicians to select the drug(s) with the greatest activity, extend intensive treatments past 2 weeks, and/or use combination drug therapy. Continued investigation of in vitro drug susceptibility testing for C. neoformans is warranted and urgently needed given the large numbers of individuals with AIDS-associated cryptococcal meningitis resulting from the global AIDS pandemic.
Acknowledgments
This work was supported in part by the University of California University-wide AIDS Research Program (UARP CC99-SD-0030) through the California Collaborative Treatment Group and unrestricted educational grants from Pfizer, Inc. The mouse experiments were supported in part by the National Institute of Allergy and Infectious Diseases (N01-AI-85351).
We gratefully acknowledge the students who participated in these experiments: David Holtom, Karen Holtom, Ryan Larsen, Scott Larsen, Nicole Leonard, Meredith Pitts, Nazia Siddiqui, and Elissa Singer.
APPENDIX
Linear regression.The regression model for the global linear association between the in vitro response (log10 CFU/ml) and fluconazole concentration (log2) is given by
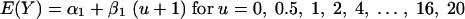 |
(A1) |
where E(Y) is the expected in vitro response at the fluconazole concentration u. The quantity 1 is added to each concentration before the log2 transformation, since log2 of zero is undefined. α1 is the intercept − the value of the response at u = 0, and β1 is the slope − the change in response as the concentration increases 1 unit.
Similarly, the regression model for the linear association between the murine response (log10 CFU/g brain) and fluconazole dose is given by
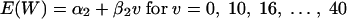 |
(A2) |
where E(W) is the expected murine response at the fluconazole dose v.
Note that α2, the intercept for the murine regression curve in equation A2, can be rewritten in terms of α1, the intercept for the in vitro regression curve in equation A1, that is: α2 = α1 + α.
Similarly, β2, the slope for the murine regression curve in equation A2, can be rewritten in terms of β1, the slope for the in vitro regression curve in equation A1, that is: β2 = β1 + β.
Thus, equation A2 can be rewritten as
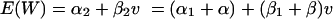 |
Evaluating the association between the two curves.
In order to evaluate the association between the two curves, we define an indicator variable, δ, where δ = 0 for the in vitro data and δ = 1 for the mouse data. Then, we can use a single regression model to encompass both regression curves:
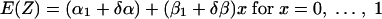 |
(A3) |
where E(Z) is the expected response at the scaled dose (concentration) x:
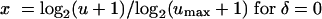 |
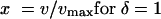 |
and where umax is the maximum in vitro fluconazole concentration tested and vmax is the maximum murine fluconazole dose tested. In this single regression model, α is the difference between the intercept for the in vitro dose-response curve and the intercept for the murine dose-response curve. Similarly, β is the difference between the slopes for the two curves. If the in vitro concentration-response curve and the murine dose-response curve have the same slope (i.e., the curves are parallel), then β will be zero.
To evaluate β, we fit the regression model (equation A3) and use the fitted model to estimate the CI for β. If the CI for β contains zero, then we conclude that the curves have similar slopes.
Predicting the murine dose-response curve.
When the two response curves have the same slope, β will be zero so the regression model shown in equation A3 becomes:
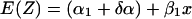 |
(A4) |
We fit this regression model and use it to predict the murine response curve on the basis of the in vitro response curve. Thus, the expected murine response is given by:
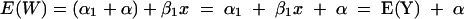 |
That is, the expected murine response can be predicted by shifting the expected in vitro response by α, which is the difference between the intercepts for the two curves.
Correspondence between the in vitro concentrations and murine doses.
We can also use the regression model (equation A4) to estimate the correspondence between the in vitro fluconazole concentration and the murine fluconazole dose that would produce the same level of response.
Let Y be the in vitro response at the in vitro concentration u. To find v, the murine dose that produces the same level of response, we start with the murine response:
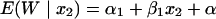 |
(A5) |
where E(W | x2) is the expected murine response at the scaled murine dose
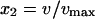 |
and the in vitro response is
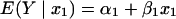 |
(A6) |
where E(Y | x1) is the expected in vitro response at the scaled concentration
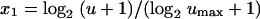 |
Then, to find v, the murine dose at which the murine response equals the in vitro response at u, we use the scaled murine dose x2 and the scaled in vitro concentration x1.
By assumption,
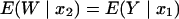 |
and, substituting equations A5 and A6, we get
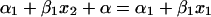 |
(A7) |
Solving equation A7 for x2, the scaled murine dose, we get
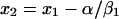 |
(A8) |
Finally, to get the murine dose in the original fluconazole units, multiply x2 by vmax = 40.
Example.
In the example shown in Fig. 3, the in vitro response at u = 1 mg/liter is 4.6 log10 CFU/ml. For u = 1, the scaled in vitro concentration x1 equals log2 (1 + 1)/log2 (20 + 1) or 0.23. In the fitted linear regression, α is 1.17 and β1 is −3.8. Using equation A8, the scaled murine dose is given by
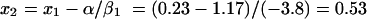 |
Then, v, the murine dose at which the expected murine response equals 4.6 log10 CFU is given by 0.53 × 40 = 21.2 mg/kg per day.
REFERENCES
- 1.Aller, A. I., E. Martin-Mazuelos, F. Lozano, J. Gomez-Mateos, L. Steele-Moore, W. J. Holloway, M. J. Gutierrez, F. J. Recio, and A. Espinel-Ingroff. 2000. Correlation of fluconazole MICs with clinical outcome in cryptococcal infection. Antimicrob. Agents Chemother. 44:1544-1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andes, D., and M. van Ogtrop. 1999. Characterization and quantitation of the pharmacodynamics of fluconazole in a neutropenic murine disseminated candidiasis infection model. Antimicrob. Agents Chemother. 43:2116-2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bauer, M., A. M. Thomas, J. R. Graybill, and R. A. Larsen. 1998. Display of confidence intervals for response surfaces in combination drug experiments, p. 100-105. In Proceedings of statistical graphics, 1997 JSM. American Statistical Association, Alexandria, Va.
- 4.Brouwer, A. E., A. Rajanuwong, W. Chierakul, G. E. Griffin, R. A. Larsen, N. J. White, and T. S. Harrison. 2004. Combination antifungal therapies for HIV-associated cryptococcal meningitis: a randomised trial. Lancet 363:1764-1767. [DOI] [PubMed] [Google Scholar]
- 5.Cleveland, W. S. 1993. Visualizing data. Hobart, Summit, N.J.
- 6.Cleveland, W. S., E. Grosse, and W. M. Shyu. 1991. Local regression models, p. 309-376. In J. M. Chambers and T. Hastie (ed.), Statistical models in S. Chapman and Hall, New York, N.Y.
- 7.Diamond, D. M., M. Bauer, B. E. Daniel, M. E. Leal, D. Johnson, B. K. Williams, A. M. Thomas, J. C. Ding, L. Navjar, J. R. Graybill, and R. A. Larsen. 1998. Fungicidal activity of amphotericin B colloidal dispersion combined with flucytosine with or without fluconazole in murine cryptococcal meningitis. Antimicrob. Agents Chemother. 42:528-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ding, J. C., M. Bauer, D. M. Diamond, M. E. Leal, D. Johnson, A. M. Thomas, L. Navjar, J. R. Graybill, and R. A. Larsen. 1997. Effects of delayed treatment on the fungicidal activity of flucytosine combined with fluconazole in murine cryptococcal meningitis. Antimicrob. Agents Chemother. 41:1589-1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoaglin, D. C., F. Mosteller, and J. W. Tukey. 1983. Understanding robust and exploratory data analysis. J. Wiley, New York, N.Y.
- 10.Insightful, Inc. 2000. S-PLUS guide to statistical and mathematical analysis, S-Plus 2000 for Windows. Insightful, Inc., Seattle, Wash.
- 11.Jessup, C. J., M. A. Pfaller, S. A. Messer, J. Zhang, M. Tumberland, E. K. Mbidde, and M. A. Ghannoum. 1998. Fluconazole susceptibility testing of Cryptococcus neoformans: comparison of two broth microdilution methods and clinical correlates among isolates from Ugandan AIDS patients. J. Clin. Microbiol. 36:2874-2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Larsen, R. A., M. Bauer, A. M. Thomas, and J. R. Graybill. 2004. Amphotericin B and fluconazole, a potent combination therapy for cryptococcal meningitis. Antimicrob. Agents Chemother. 48:985-991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lortholary, O., M. Nicolas, S. Soreda, L. Improvisi, O. Ronin, O. Petitjean, B. Dupont, M. Tod, and F. Dromer. 1999. Fluconazole, with or without dexamethasone for experimental cryptococcosis: impact of treatment timing. J. Antimicrob. Chemother. 43:817-824. [DOI] [PubMed] [Google Scholar]
- 14.National Committee for Clinical Laboratory Standards. 2002. Reference method for broth dilution antifungal susceptibility testing of yeasts: approved standard, 2nd ed. M27-A2. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 15.Neter, J., W. Wasserman, and M. Kutner. 1989. Applied linear regression models, 2nd ed. Irwin, Homewood, Ill.
- 16.Pfaller, M. A., and W. L. Yu. 2001. Antifungal susceptibility testing. New technology and clinical applications. Infect. Dis. Clinics N. Am. 15:1227-1261. [DOI] [PubMed] [Google Scholar]
- 17.Pitisuttithum, P., S. Tansuphasawadikul, J. Simpson, P. A. Howe, and N. J. White. 2001. A prospective study of AIDS-associated cryptococcal meningitis in Thailand treated with high-dose amphotericin B. J. Infect. 43:226-233. [DOI] [PubMed] [Google Scholar]
- 18.Rex, J. H., and M. A. Pfaller. 2002. Has antifungal susceptibility testing come of age? Clin. Infect. Dis. 35:982-989. [DOI] [PubMed] [Google Scholar]
- 19.Rex, J. H., M. A. Pfaller, J. N. Galgiani, M. S. Bartlett, A. Espinel-Ingroff, M. A. Ghannoum, M. Lancaster, F. C. Odds, M. G. Rinaldi, T. J. Walsh, A. L. Barry, and the Subcommittee on Antifungal Susceptibility Testing of the National Committee for Clinical Laboratory Standards. 1997. Development of interpretive breakpoints for antifungal susceptibility testing: conceptual framework and analysis of in vitro-in vivo correlation data for fluconazole, itraconazole, and Candida infections. Clin. Infect. Dis. 24:235-247. [DOI] [PubMed] [Google Scholar]
- 20.Rex, J. H., M. A. Pfaller, T. J. Walsh, V. Chaturvedi, A. Espinel-Ingroff, M. A. Ghannoum, L. L. Gosey, F. C. Odds, M. G. Rinaldi, D. J. Sheehan, and D. W. Warnock. 2001. Antifungal susceptibility testing: practical aspects and current challenges. Clin. Microbiol. Rev. 14:643-658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Robinson, P. A., M. Bauer, M. A. Leal, S. G. Evans, P. D. Holtom, D. A. Diamond, J. M. Leedom, and R. A. Larsen. 1999. Early mycological treatment failure in AIDS-associated cryptococcal meningitis. Clin. Infect. Dis. 28:82-92. [DOI] [PubMed] [Google Scholar]
- 22.Thaler, F., B. Bernard, M. Tod, C. P. Jedynak, O. Petitjean, P. Derome, and P. Loirat. 1995. Fluconazole penetration in cerebral parenchyma in humans at steady state. Antimicrob. Agents Chemother. 39:1154-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Venables, W. N., and B. D. Ripley. 2002. Modern applied statistics with S-PLUS, 4th ed. Springer-Verlag, Inc., New York, N.Y.
- 24.Witt, M. D., R. J. Lewis, R. A. Larsen, E. N. Milefchik, M. E. Leal, R. H. Haubrich, J. A. Richie, J. E. Edwards, and M. A. Ghannoum. 1996. Identification of patients with acute AIDS-associated cryptococcal meningitis who can be effectively treated with fluconazole: the role of antifungal susceptibility testing. Clin. Infect. Dis. 22:322-328. [DOI] [PubMed] [Google Scholar]