Abstract
Human anthrax infection cannot always be treated successfully by antibiotics, as highlighted by recent bioterrorist attacks; thus, adjunct therapies are clearly needed for the future. There is a particular need to further develop adjunct therapies that can neutralize secreted toxins, such as antibodies directed towards the 83-kDa protective antigen (PA83). In the absence of human donors, we immunized a macaque (Macaca fascicularis) with PA83 to obtain such antibodies suitable as an adjunct therapy for human anthrax infection. By using bone marrow as a template, we PCR amplified specific Fab-encoding genes and cloned them as an immune library (107 clones). We isolated a high-affinity (equilibrium dissociation constant [KD], 3.4 nM), highly neutralizing (50% inhibitory concentration, 5.6 ± 0.13 nM) Fab (designated 35PA83) from this library by panning. Its epitope was localized by Pepscan analysis between residues 686 and 694 of PA83 and is part of the region which directly interacts with the cell receptor. 35PA83 may thus neutralize the anthrax toxin by competing directly for its receptor. The genes encoding 35PA83 were similar to those of a human immunoglobulin germ line and were assigned to subgroups of human V, (D), or J genes by IMGT/V-QUEST analysis. The 35PA83 framework regions were 92% identical to a representative allele of each subgroup. When compared to framework regions coded by related human germ line genes, only 2 of 74 (VH) or 75 (VK) analyzed amino acids of 35PA83 have different chemical characteristics. A very high degree of identity with human framework regions makes 35PA83 well suited for expression as a whole primatized immunoglobulin G and demonstrates the practicality of using macaque Fabs when immunized human plasma cell donors are not available.
Bacillus anthracis, the causative agent of anthrax, produces a toxin related to the classic A-B family, which plays a major role in disease pathogenesis. The 83-kDa protective antigen (PA83) is the common cell-binding domain (38) that, after proteolytic activation, can interact with two enzymatically active domains that elicit cell damage, the edema factor (EF; 89 kDa) and lethal factor (LF; 90 kDa). LF is a metalloproteinase specific for mitogen-activated protein kinase kinases. EF is a calmodulin-dependent adenylate cyclase that causes dramatic increases in the intracellular concentration of cAMP. These proteins are secreted by the bacterium as nontoxic monomers and assemble on the surface of receptor-bearing cells to form toxic complexes. After binding to its cellular receptor (4, 30, 44), PA83 is cleaved by a furin-like cellular protease(s) (34), causing the release of an amino-terminal 20-kDa fragment and leaving the carboxy-terminal 63-kDa moiety (PA63) bound to its receptor. PA63 then spontaneously oligomerizes to form a ring-shaped homoheptamer that binds LF and/or EF (38) to form the lethal toxin and/or the edema toxin, respectively. Heptamers are internalized by acidic endosomes, and then EF and LF are translocated into the cytosol (32).
This pathogenesis and the prophylactic/therapeutic approaches used to treat anthrax are subjects of great interest due to concerns over intentional or inadvertent exposure to aerosols of Bacillus anthracis spores (47). Five of the 11 patients that inhaled anthrax spores sent intentionally through the United States postal system in 2001 died, despite receiving powerful antibiotic therapy. This confirmed that antibiotics are unable to prevent a fatal outcome in humans once the disease has reached a phase involving toxin production (16). Hence, an adjunct treatment is needed to inactivate the toxins, and this may be achieved by passive immunization with toxin-specific antibodies. This concept was recently tested by Karginov et al. in a mouse model and proved effective (17). In addition, monoclonal antibodies (27, 28) and polyclonal antibodies (Ab) (26) alone can confer passive protection against anthrax toxin in rats and anthrax infection in guinea pigs, respectively. These results emphasize the importance of antibodies directed against PA.
Primate antibodies are more similar in sequence to human, versus murine, antibodies and they are less likely to be immunogenic in humans (25, 36). The V-domains of antibodies of nonhuman primate origin can be fused to the constant regions of human immunoglobulin Gs (IgGs) to produce primatized Ab suitable for clinical use. Three primatized Ab directed against human antigens (IDEC C9.1, IDEC 114, and IDEC 151, directed against CD4, CD80, and CD53, respectively) are currently undergoing clinical trials (Biogen Idec, Inc., Cambridge, MA), and preliminary results show no human anti-primate antibody response (5, 13). Thus, the selection of primate Fab by use of phage display, to be followed by the expression of their variable domains in fusion with constant human domains, could be a valid way of obtaining high-affinity antibodies that neutralize the anthrax toxin and are deemed suitable for medical use. However, this approach has rarely been described in detail (3).
Here, we immunized a macaque with PA83 and subsequently constructed a Fab library from bone marrow. Screening led to the isolation of a high-affinity macaque Fab that neutralizes the anthrax toxin. Due to its similarity with human antibody counterparts, this Fab should be suitable for clinical purposes. Therefore, utilizing nonhuman primate plasma cells and the phage display method is suitable for the production of antimicrobial/anti-toxin reagents, in particular when no human plasma cell donors are available.
MATERIALS AND METHODS
Animal experiments.
A cynomolgus macaque (Macaca fascicularis) was subcutaneously injected with 200 μg of recombinant PA83 (France Biochem, Meudon, France) in complete Freund's adjuvant (Sigma, St. Louis, MO). This was followed by three additional boosts (each of 200 μg in incomplete Freund's adjuvant) given at 2-month intervals. The bone marrow was sampled each week after the first week, at different sites and under general anesthesia (Imalgene; Merial, Lyon, France), using a protocol approved by the local ethics committee for animal care.
Analysis of the macaque immune response.
PA83 was diluted to 5.0 μg ml−1 in phosphate-buffered saline (PBS) to coat microtiter plates (Maxisorp; Nunc, Rosewald, Denmark) overnight at 4°C. Plates were blocked with 5% (wt/vol) skim milk-PBS for 1 h at 37°C, and then washed five times with 0.1% Tween 20-PBS. Serial dilutions of serum (in 3% skim milk-PBS) were placed in the wells and incubated for 2 h at 37°C. After the washing step, peroxidase conjugated to anti-monkey Fab IgG (Tebu, Le Perray, France) was diluted 1/1,000 in 3% skim milk-0.05% Tween 20-PBS, and 100 μl was then incubated in each well for 1 h at 37°C. After washing, ABTS [2,2′-azinobis(3-ethylbenzthiazolinesulfonic acid)] substrate (Roche, Meylan, France) was added, and the absorbance was read at 405 nm.
Construction and screening of γ1/κ antibody phage library.
Total bone marrow RNA was extracted using Tri Reagent (Molecular Research Center, Inc., Cincinnati, OH) according to the manufacturer's instructions. The library was then constructed with the phagemid vector pComb3X and screened, and then phage-enzyme-linked immunosorbent assay (ELISA) was performed as previously described (2, 3).
Fab production, periplasmic extraction, and purification.
Phagemid DNA isolated after the panning process was used to transform the nonsuppressor Escherichia coli strain HB2151 (6) such that it expressed the soluble Fab fragment. Single colonies of randomly chosen transformants were used to inoculate 5 ml of Super Broth (SB) (32 g peptone [Sigma], 20 g yeast extract, 5 g NaCl, 5 ml NaOH [1 M] for 1 liter SB) medium supplemented with carbenicillin (50 μg · ml−1) and 1% glucose. Cultures were incubated overnight at 37°C with vigorous shaking (250 rpm). We then inoculated 500 ml of SB medium supplemented with carbenicillin and 0.1% glucose with 500 μl of each culture. The cultures were grown at 30°C until the optical density at 600 nm reached 1.5. IPTG (isopropyl-β-d-thiogalactopyranoside; 1 mM) was then added overnight to induce gene expression at 22°C. The cells were harvested by centrifugation at 2,500 × g for 15 min at 4°C. Fabs were extracted with polymyxin B sulfate (40) and purified on a nickel column (Ni-NTA spin column; QIAGEN, Valencia, CA) according to the manufacturer's instructions.
ELISA for soluble Fab reactivity.
ELISA was used to assess the specificities of individual soluble Fabs. Microtiter plates were coated with PA83 and blocked. After incubation with periplasmic extract, Fabs were detected by using a peroxidase-conjugated anti-hemagglutinin tag antibody (diluted 1/1,000) (Roche, Meylan, France).
In vitro lethal toxin neutralization assay.
The mouse macrophage cell line J774A.1 was plated overnight at 14,000 cells/well in 96-well dishes. Lethal toxin components (400 ng/ml of PA and 40 ng/ml of LF) were added simultaneously to Fab or medium alone and incubated for 1 h at 37°C and then added to macrophages and incubated at 37°C for 4 h (28). A Cytotox Assay 96 kit (Promega) was used according to the manufacturer's instructions to evaluate the 50% inhibitory concentrations (IC50) for Fab and to neutralize the lethal toxin (48). All assays were performed in triplicate, and results were corrected for the background level.
To evaluate the neutralizing mechanism, PA83 (400 ng/ml) was preincubated with J774A.1 cells for 2 h at 4°C. In parallel, LF (40 ng · ml−1) and Fab (10 times the IC50) were coincubated at 37°C for 1 h. The two components of the assay were then incubated together at 37°C for 4 h, and the assay was continued as described above (29).
Quantification of soluble Fab.
The purity of the Fab was assessed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and its concentration was determined by spectrophotometry (λ = 280 nm). The extinction coefficient (ɛ) was calculated (11) by using the Fab sequence and Vector NTI software (Informax, Frederick, MD) and was found to be equal to 81,850 M−1 cm−1.
Affinity determination using surface plasmon resonance.
Affinity constants were determined by surface plasmon resonance with a Biacore X instrument (Uppsala, Sweden) using a method described in reference 18 and were verified with the internal consistency tests described in reference 43.
Epitope mapping.
The linearity of the epitope was evaluated by testing the reactivity of the Fab by Western blotting, after migration of PA83 in denaturing conditions. Pepscan analysis (Pepscan Systems, Lelystad, The Netherlands) was carried out as described in reference 15, with overlapping 15-mer peptides spanning domain four (between residues 595 and 735), which is involved in cell receptor binding (37). Surface accessibility was calculated using the ASA View program (1), available at http://netasa.org. Synthetic peptides were ordered from NeoMPS (Strasbourg, France), and inhibition of 35PA83 binding to PA83 by these peptides was tested as described previously (45).
Nucleic acid analysis of PA83-specific Fab clones.
Sequences of the light and heavy Fab chains from selected clones were determined by Genome Express (Meylan, France) using the primers OMPSEQ/KPEL and RevKPEL/GBACK, respectively (RevKPEL being the reverse complementary sequence of KPEL) (2). The sequences were analyzed online, using the International ImMunoGeneTics information system (IMGT) (http://imgt.cines.fr) and compared with the sequences of the human germ line immunoglobulin genes using IMGT/V-QUEST and IMGT/JunctionAnalysis software (19-21, 23).
Nucleotide sequence accession numbers.
The M. fascicularis 35PA83H and 35PA83L sequences (VH and VK domain sequences, respectively) are accessible in the GenBank/EMBL data banks under the accession numbers AJ810486 and AJ810487, respectively.
RESULTS
Library construction and isolation of Fab specific for PA83.
We monitored humoral responses after immunization with PA83 by ELISA, using a series of dilutions of both preimmune and immune sera. PA-specific Ab were detectable after the first injection, and the titer increased to 100,000 after three subsequent immunizations. Total RNA was extracted from the bone marrow (5 ml), and the κ- and γ1-chain genes were best amplified 35 days after the last boost. These PCR products were purified and cloned into pComb3X DNA to yield a library containing approximately 107 independent clones. The primate Fab-phage library was then subjected to five rounds of enrichment against PA83. The increasing number of phages eluted after the fourth round (4 × 105 clones, a 40-fold increase compared to the previous round) and fifth round (107 clones, a 25-fold increase) indicated that the panning had successfully enriched the library in antigen-specific clones. Phages from the various panning rounds were screened for reactivity with PA83 by ELISA, and those eluted after the fourth round gave a signal three- to fourfold higher than the background level, thus reacting specifically with the antigen. The library eluted after the fifth round was expressed as soluble Fabs. We tested 50 isolated clones for their reactivity with PA83 and for in vitro neutralization. Five Fab clones were then selected for both their high reactivity and neutralization properties. Sequencing showed that the five clones were identical, and Fab 35PA83 was used for the rest of the study. Systematic sequencing of the 50 clones later showed that Fab 35PA83 represented half of them.
After expression and purification, sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Coomassie blue staining demonstrated that the heavy and light chains were equally represented, with a purity exceeding 90%. Expression levels of the purified Fab 35PA83 were about 150 μg per liter of culture per optical density of 1.
In vitro neutralizing activity of the macaque Fab 35PA83.
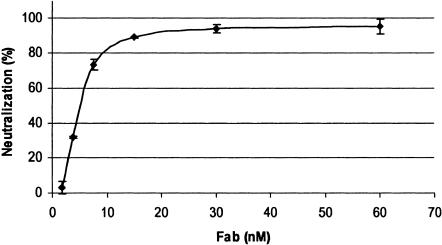
After selection and sequencing, we assayed 35PA83 for neutralization activity using serial dilutions of the purified Fab. When preincubated with toxin, this anti-PA83 Fab has a 50% neutralization value of 5.6 ± 0.13 nM (mean ± standard deviation) and a 80% neutralization value of 11 ± 0.16 nM (Fig. 1). Similar results were obtained on numerous occasions and were also obtained with the use of Alamar Blue, as opposed to the Cytotox assay, to assess cell viability (data not shown).
FIG. 1.
Toxin neutralization assay. The assay was performed in triplicate as described in Materials and Methods.
To evaluate the mechanism of neutralization of 35PA83, we examined whether the Fab could neutralize toxin after PA83 had bound to the receptor. Even at high concentrations (70 nM; 10 IC50), the Fab completely failed to show any neutralizing properties. This suggests that the 35PA83 Fab mediates neutralization by preventing PA from interacting with the cellular receptors, so that its epitope was expected to be located in domain 4 (37).
Affinity determination.
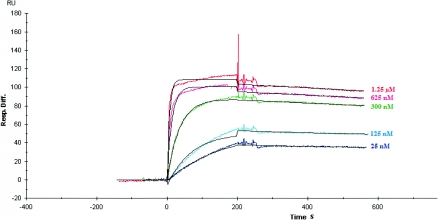
The affinity of the 35PA83 was measured by surface plasmon resonance using Biacore. Association and dissociation kinetics were measured for binding of Fab 35PA83 to the protective antigen (Fig. 2). The kon and koff rate constants were determined directly from the sensorgram in the Biacore analysis, and KD was deduced. The Fab 35PA83 has an affinity for PA83 of 3.4 nM. The low off rate (3.23 × 10−4 s−1) may confer significant physiological advantages for toxin neutralization in vivo.
FIG. 2.
Analysis of the 35PA83 macaque Fab by surface plasmon resonance. Fab 35PA83 binding responses at concentrations of 1.25 μM, 625 nM, 312 nM, 125 nM, 25 nM, and 0 nM. The black lines represent a global fit for a 1:1 interaction model.
Epitope mapping.
The epitope was found to be linear, and the following four sequences were selected by Pepscan analysis: PLYISNPNY (P1), EVINSS (P2), RKILSG (P3), and MLNISS (P4). P1 was selected because four overlapping peptides centered on this sequence reacted with 35PA83, whereas only one peptide centered on each of the other three sequences reacted with 35PA83. The peptide centered on PLYISN (this sequence is designated P1′) reacted at least twice as strongly with 35PA83 as with any other peptide selected by the Pepscan analysis. It was seen that its fifth residue is a serine, which was also shared by P2, P3, and P4. Taken together, these results suggested that P2, P3, and P4 were mimics of P1′ and P1, which were regarded as the best candidates. Accessible surface area was evaluated as 39% for P1 (and 39% for P1′), 43% for P2, 17% for P3, and 21% for P4; therefore, peptides P3 and P4 were not considered the best candidates in this respect. Two soluble peptides, representing P1 and P2, were synthesized, and P1 was able to strongly compete with PA83 for its binding to 35PA83 (50% inhibition at 2 × 10−5 M). P2 did not compete, nor did a peptide having an irrelevant sequence (representing a tetanus toxin epitope), which was used as a negative control. These data suggested that most of the residues that react with 35PA83 are contained within P1, which represents the fragment of PA limited by residues 686 and 694 (PA686-694).
Computational analysis.
The M. fascicularis 35H and 35L sequences (VH and V-KAPPA domain sequences, respectively) were automatically analyzed with IMGT/V-QUEST software (12, 19), which identifies the immunoglobulin germ line V, (D), and J genes from which a specific immunoglobulin chain is derived, and with the IMGT/JunctionAnalysis tool (21, 35), which analyzes in detail the V-D-J and V-J junctions. For the 35H VH domain (V-D-J-REGION), the M. fascicularis IGHV gene clearly belongs to the IGHV4 subgroup. An initial analysis with the IMGT/V-QUEST software showed that there were two additional amino acids compared to the closest human IGHV genes, one in the CDR1-IMGT (at position “31A”), the other in the CDR2-IMGT at position 59A. In order to identify the closest human IGHV gene for the M. fascicularis 35H sequence and to calculate the percentage of similarity, the amino acids “31A” and “59A” were omitted in a second analysis with the IMGT/V-QUEST tool. The V-REGION was recognized as originating from a M. fascicularis IGHV gene similar to the human germ line IGHV4-59*01 allele (22), with a 92% nucleotide sequence identity in the V-REGION frameworks. The 35H J-REGION gene was recognized by the IMGT/V-QUEST tool as originating from a M. fascicularis IGHJ gene close to the human germ line IGHJ5*01 gene (22). The D-REGION was recognized as originating from a M. fascicularis IGHD gene similar to the human germ line IGHD3-3*01 gene (22) with three mutations in the D-REGION, zero in the 5′ V-REGION and two in the 3′ J-REGION.
For the V-KAPPA (V-J-REGION) domain, the 35L V-REGION was recognized as originating from a M. fascicularis IGKV gene similar to the human germ line IGKV1-13*02 (22), with a 92% nucleotide sequence identity in the V-REGION frameworks. The J-REGION gene was recognized by IMGT/V-QUEST as originating from a M. fascicularis IGKJ gene similar to the IGKJ3*01 human germ line gene.
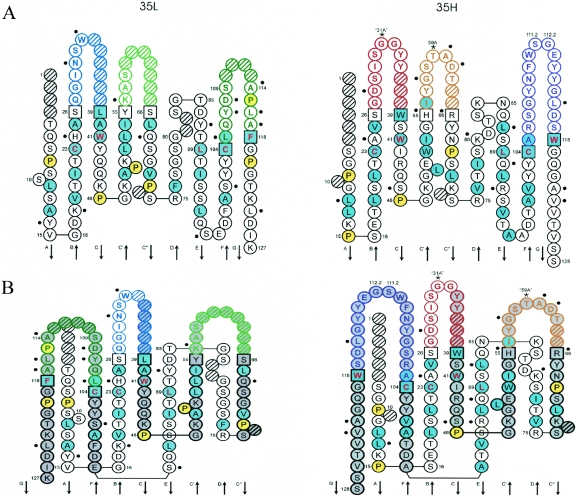
Only two framework (FR) positions in the 35H domain (24 and 87) and two others in the 35L domain (14 and 87) have different chemical characteristics (see Plate 3 in reference 39) compared to those encoded by the most similar human germ line genes. The IMGT Collier de Perles graphical two-dimensional representation (23, 41) of the Fab is shown in Fig. 3.
FIG. 3.
IMGT Colliers de Perles of the VH and VK domains of the M. fascicularis Fab 35PA83 on one layer (A) and on two layers (B). IMGT Colliers de Perles representations are displayed according to the IMGT unique numbering (23). Dots indicate differences from the human genes most similar to 35PA83, and 35PA83. The positions of hydrophobic amino acids (hydropathy index with positive value, i.e., I, V, L, F, C, M, A) and tryptophan (W) are shown in blue. All proline (P) residues are shown in yellow. The CDR-IMGT sequences are limited by amino acids shown in squares (anchor positions), which belong to the neighboring FR-IMGT. Hatched circles correspond to missing positions according to the IMGT unique numbering. Arrows indicate the direction of the beta strands (from IMGT Repertoire, http://imgt.cines.fr). For the VH domain, CDR1-IMGT is shown in red, CDR2-IMGT in orange, and CDR3-IMGT in purple. For the V-KAPPA domain, CDR1-IMGT is shown in blue, CDR2-IMGT in green, and CDR3-IMGT in turquoise (21).
DISCUSSION
The recent use of B. anthracis spores as a terrorist weapon has rekindled interest in the mechanisms of protective immunity towards B. anthracis, and the partial failure of antibiotic therapy for treatment of anthrax (5 out of 11 patients died) (16) has highlighted the importance of developing new or complementary therapeutic approaches. As anthrax pathogenicity involves both bacterial replication and toxin production, a combination of approaches inhibiting B. anthracis replication (by antibiotic therapy) and neutralizing the anthrax toxin could be of great value. The interest of neutralizing antibodies directed against PA has long been demonstrated, and the protection conferred by the anthrax vaccine is mediated largely by antibodies specific for this toxin component (24). In addition, passive immunization with anti-PA Ab that neutralize anthrax toxin has been shown to be an effective adjunct treatment with antibiotics (17). Accordingly, the main goal of this work was to select neutralizing fragments of antibodies (Fab) directed against PA.
Although human monoclonal antibodies may represent the best option for developing immunotherapeutics meant for human use, these reagents are not always available, as few humans are immunized against the antigen of interest. Nonhuman sources can also be restricted by financial considerations, as is the case with the patented “xenomice,” in which the native immunoglobulin genes have been replaced by some of their human counterparts (14). In France, no anti-anthrax vaccine is currently authorized for human use, and natural anthrax cases are rare. We therefore had no human source for the generation of anti-anthrax antibodies, unlike other investigators (42, 48), even though anthrax toxin-neutralizing antibodies would be particularly useful precisely because of the absence of a vaccine. Our alternative approach has been to use phage display to select a primate antibody fragment against PA. This approach, which has rarely been described in detail despite its advantages (3), yielded the high-affinity (KD = 3.4 nM) and highly neutralizing (IC50 = 5.36 nM; IC80 = 11 nM) 35PA83 Fab. The Fab 35PA83 generated here has affinity and neutralization properties that are equivalent to those of a human anti-PA Fab recently obtained by Wild et al., who noted that its IC50 value was close to equimolar with the concentration of PA83 used in the assay (4.8 nM) (48). These results are also consistent with a previous study showing that the antibody-mediated neutralization of PA correlates with affinity (31). 35PA83 will be further tested with a rat model using PA and LF and with animal models of the disease itself.
Computer analysis with the IMGT/V-QUEST software and the IMGT/JunctionAnalysis tool revealed that the amino acid sequences of the 35PA83H and 35PA83L chains are quite similar; their homologs are encoded by the most similar human germ line sequences, at 92% for the framework regions and 80% and 84% for the entire VH and VK sequences, respectively. Moreover, compared to several human sequences coded by VH or VK germ line genes, only 2 of 74 (VH) or 75 (VK) analyzed amino acids located in the 35PA83 framework region differ significantly in their chemical characteristics. Only these four differences would be retained in the primatization process, and this high level of sequence similarity and a number of other observations suggest that it should be possible to administer primate antibody fragments or primatized Ab directly to humans without the need for further modification (9, 10).
Our epitope mapping suggests that 35PA83 interacts with residues located in the segment of PA limited by residues 686 and 694 (PA686-694). This is consistent with previous data, as PA686-694 is included in the broader region (PA671-721) previously known to comprise neutralizing epitopes (29) and is part of the small loop (PA679-693) shown by mutagenesis techniques to interact with the cell receptor (46). Thus, 35PA83 may neutralize PA83 by direct competition for its receptor. According to a recent report describing a population of anti-PA monoclonal antibodies capable of enhancing the cellular cytotoxicity of the anthrax toxin (33), blocking the binding of PA to its cell receptor, such as with 35PA83, eliminates the possibility of enhancement. 35PA83 Fab may thus be expressed as a whole primatized IgG for therapeutic use or be pegylated to prolong its in vivo half-life (7, 8).
The identification and analysis of 35PA83 suggests that it is possible to obtain macaque Fabs that are equivalent to their human counterparts in affinity, functionality, and safety. The generation of macaque Fabs against a range of antigens may lead to medical reagents of interest and would be especially valuable given the difficulties and ethical problems associated with immunizing humans against target antigens. Macaques may also be useful for the production of antibodies against epitopes or antigens that do not elicit a humoral response in humans, for example, those of human origin. More generally, nonhuman primates may be regarded as a rich source of largely untapped therapeutic molecules that include anti-toxin/antimicrobial agents.
Acknowledgments
We thank Vincent Negre for the IMGT Colliers de Perles representations; Martha Wild, Bradle Stiles, and Thibaut Pelat for helpful discussions; and the “biologie appliquée” team for excellent technical support.
This work was sponsored by grants from the DGA (PEA no. 803) and EMA (service125: op3-c/LFR).
REFERENCES
- 1.Ahmad, S., M. Gromiha, H. Fawareh, and A. Sarai. 2004. ASAView: database and tool for solvent accessibility representation in proteins. BMC Bioinformatics 5:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andris-Widhopf, J., C. Rader, P. Steinberger, R. Fuller, and C. F. Barbas III. 2000. Methods for the generation of chicken monoclonal antibody fragments by phage display. J. Immunol. Methods 242:159-181. [DOI] [PubMed] [Google Scholar]
- 3.Andris-Widhopf, J., P. Steinberger, R. Fuller, C. Rader, and C. F. Barbas III. 2001. Generation of antibody libraries: PCR amplification and assembly of light- and heavy-chain coding sequences. In C. F. Barbas III, D. R. Burton, J. K. Scott, and G. J. Silverman (ed.), Phage display: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York.
- 4.Bradley, K. A., J. Mogridge, M. Mourez, R. J. Collier, and J. A. Young. 2001. Identification of the cellular receptor for anthrax toxin. Nature 414:225-229. [DOI] [PubMed] [Google Scholar]
- 5.Bugelski, P. J., D. J. Herzyk, S. Rehm, A. G. Harmsen, E. V. Gore, D. M. Williams, B. E. Maleeff, A. M. Badger, A. Truneh, S. R. O'Brien, R. A. Macia, P. J. Wier, D. G. Morgan, and T. K. Hart. 2000. Preclinical development of keliximab, a Primatized anti-CD4 monoclonal antibody, in human CD4 transgenic mice: characterization of the model and safety studies. Hum. Exp. Toxicol. 19:230-243. [DOI] [PubMed] [Google Scholar]
- 6.Carter, P., H. Bedouelle, and G. Winter. 1985. Improved oligonucleotide site-directed mutagenesis using M13 vectors. Nucleic Acids Res. 13:4431-4443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chapman, A. P. 2002. PEGylated antibodies and antibody fragments for improved therapy: a review. Adv. Drug Deliv. Rev. 54:531-545. [DOI] [PubMed] [Google Scholar]
- 8.Chapman, A. P., P. Antoniw, M. Spitali, S. West, S. Stephens, and D. J. King. 1999. Therapeutic antibody fragments with prolonged in vivo half-lives. Nat. Biotechnol. 17:780-783. [DOI] [PubMed] [Google Scholar]
- 9.Chassagne, S., E. Laffly, E. Drouet, F. Herodin, M. P. Lefranc, and P. Thullier. 2004. A high-affinity macaque antibody Fab with human-like framework regions obtained from a small phage display immune library. Mol. Immunol. 41:539-546. [DOI] [PubMed] [Google Scholar]
- 10.Ehrlich, P. H., Z. A. Moustafa, K. E. Harfeldt, C. Isaacson, and L. Ostberg. 1990. Potential of primate monoclonal antibodies to substitute for human antibodies: nucleotide sequence of chimpanzee Fab fragments. Hum. Antibodies Hybridomas 1:23-26. [PubMed] [Google Scholar]
- 11.Gill, S. C., and P. H. von Hippel. 1989. Calculation of protein extinction coefficients from amino acid sequence data. Anal. Biochem. 182:319-326. [DOI] [PubMed] [Google Scholar]
- 12.Giudicelli, V., D. Chaume, and M. P. Lefranc. 2004. IMGT/V-QUEST, an integrated software program for immunoglobulin and T cell receptor V-J and V-D-J rearrangement analysis. Nucleic Acids Res. 32:W435-W440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gottlieb, A. B., S. Kang, K. G. Linden, M. Lebwohl, A. Menter, A. A. Abdulghani, M. Goldfarb, N. Chieffo, and M. C. Totoritis. 2004. Evaluation of safety and clinical activity of multiple doses of the anti-CD80 monoclonal antibody, galiximab, in patients with moderate to severe plaque psoriasis. Clin. Immunol. 111:28-37. [DOI] [PubMed] [Google Scholar]
- 14.Green, L. L. 1999. Antibody engineering via genetic engineering of the mouse: XenoMouse strains are a vehicle for the facile generation of therapeutic human monoclonal antibodies. J. Immunol. Methods 231:11-23. [DOI] [PubMed] [Google Scholar]
- 15.Hijnen, M., F. R. Mooi, P. G. van Gageldonk, P. Hoogerhout, A. J. King, and G. A. Berbers. 2004. Epitope structure of the Bordetella pertussis protein P.69 pertactin, a major vaccine component and protective antigen. Infect. Immun. 72:3716-3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Inglesby, T. V., T. O'Toole, D. A. Henderson, J. G. Bartlett, M. S. Ascher, E. Eitzen, A. M. Friedlander, J. Gerberding, J. Hauer, J. Hughes, J. McDade, M. T. Osterholm, G. Parker, T. M. Perl, P. K. Russell, and K. Tonat. 2002. Anthrax as a biological weapon, 2002: updated recommendations for management. JAMA 287:2236-2252. [DOI] [PubMed] [Google Scholar]
- 17.Karginov, V. A., T. M. Robinson, J. Riemenschneider, B. Golding, M. Kennedy, J. Shiloach, and K. Alibek. 2004. Treatment of anthrax infection with combination of ciprofloxacin and antibodies to protective antigen of Bacillus anthracis. FEMS Immunol. Med. Microbiol. 40:71-74. [DOI] [PubMed] [Google Scholar]
- 18.Karlsson, R., A. Michaelsson, and L. Mattsson. 1991. Kinetic analysis of monoclonal antibody-antigen interactions with a new biosensor based analytical system. J. Immunol. Methods 145:229-240. [DOI] [PubMed] [Google Scholar]
- 19.Lefranc, M. P. 2003. IMGT, the international ImMunoGeneTics database. Nucleic Acids Res. 31:307-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lefranc, M. P., V. Giudicelli, C. Ginestoux, N. Bosc, G. Folch, D. Guiraudou, J. Jabado-Michaloud, S. Magris, D. Scaviner, V. Thouvenin, K. Combres, D. Girod, S. Jeanjean, C. Protat, M. Yousfi-Monod, E. Duprat, Q. Kaas, C. Pommie, D. Chaume, and G. Lefranc. 2004. IMGT-ONTOLOGY for immunogenetics and immunoinformatics. In Silico Biol. 4:17-29. [PubMed] [Google Scholar]
- 21.Lefranc, M. P., V. Giudicelli, C. Ginestoux, and D. Chaume. 2003. IMGT, the international ImMunoGeneTics information system, http://imgt.cines.fr: the reference in immunoinformatics. Stud. Health Technol. Inform. 95:74-79. [PubMed] [Google Scholar]
- 22.Lefranc, M. P., and G. Lefranc. 2001. The Immunoglobulin FactsBook. Academic Press, London, United Kingdom.
- 23.Lefranc, M. P., C. Pommie, M. Ruiz, V. Giudicelli, E. Foulquier, L. Truong, V. Thouvenin-Contet, and G. Lefranc. 2003. IMGT unique numbering for immunoglobulin and T cell receptor variable domains and Ig superfamily V-like domains. Dev. Comp. Immunol. 27:55-77. [DOI] [PubMed] [Google Scholar]
- 24.Leppla, S. H., J. B. Robbins, R. Schneerson, and J. Shiloach. 2002. Development of an improved vaccine for anthrax. J. Clin. Investig. 110:141-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lewis, A. P., K. A. Barber, H. J. Cooper, M. J. Sims, J. Worden, and J. S. Crowe. 1993. Cloning and sequence analysis of kappa and gamma cynomolgus monkey immunoglobulin cDNAs. Dev. Comp. Immunol. 17:549-560. [DOI] [PubMed] [Google Scholar]
- 26.Little, S. F., B. E. Ivins, P. F. Fellows, and A. M. Friedlander. 1997. Passive protection by polyclonal antibodies against Bacillus anthracis infection in guinea pigs. Infect. Immun. 65:5171-5175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Little, S. F., S. H. Leppla, and E. Cora. 1988. Production and characterization of monoclonal antibodies to the protective antigen component of Bacillus anthracis toxin. Infect. Immun. 56:1807-1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Little, S. F., S. H. Leppla, and A. M. Friedlander. 1990. Production and characterization of monoclonal antibodies against the lethal factor component of Bacillus anthracis lethal toxin. Infect. Immun. 58:1606-1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Little, S. F., J. M. Novak, J. R. Lowe, S. H. Leppla, Y. Singh, K. R. Klimpel, B. C. Lidgerding, and A. M. Friedlander. 1996. Characterization of lethal factor binding and cell receptor binding domains of protective antigen of Bacillus anthracis using monoclonal antibodies. Microbiology 142:707-715. [DOI] [PubMed] [Google Scholar]
- 30.Liu, S., and S. H. Leppla. 2003. Cell surface tumor endothelium marker 8 cytoplasmic tail-independent anthrax toxin binding, proteolytic processing, oligomer formation, and internalization. J. Biol. Chem. 278:5227-5234. [DOI] [PubMed] [Google Scholar]
- 31.Maynard, J. A., C. B. Maassen, S. H. Leppla, K. Brasky, J. L. Patterson, B. L. Iverson, and G. Georgiou. 2002. Protection against anthrax toxin by recombinant antibody fragments correlates with antigen affinity. Nat. Biotechnol. 20:597-601. [DOI] [PubMed] [Google Scholar]
- 32.Milne, J. C., S. R. Blanke, P. C. Hanna, and R. J. Collier. 1995. Protective antigen-binding domain of anthrax lethal factor mediates translocation of a heterologous protein fused to its amino- or carboxy-terminus. Mol. Microbiol. 15:661-666. [DOI] [PubMed] [Google Scholar]
- 33.Mohamed, N., J. Li, C. S. Ferreira, S. F. Little, A. M. Friedlander, G. L. Spitalny, and L. S. Casey. 2004. Enhancement of anthrax lethal toxin cytotoxicity: a subset of monoclonal antibodies against protective antigen increases lethal toxin-mediated killing of murine macrophages. Infect. Immun. 72:3276-3283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Molloy, S. S., P. A. Bresnahan, S. H. Leppla, K. R. Klimpel, and G. Thomas. 1992. Human furin is a calcium-dependent serine endoprotease that recognizes the sequence Arg-X-X-Arg and efficiently cleaves anthrax toxin protective antigen. J. Biol. Chem. 267:16396-16402. [PubMed] [Google Scholar]
- 35.Monod, M. Y., V. Giudicelli, D. Chaume, and M. P. Lefranc. 2004. IMGT/JunctionAnalysis: the first tool for the analysis of the immunoglobulin and T cell receptor complex V-J and V-D-J JUNCTIONs. Bioinformatics 20(Suppl. 1):I379-I385. [DOI] [PubMed] [Google Scholar]
- 36.Newman, R., J. Alberts, D. Anderson, K. Carner, C. Heard, F. Norton, R. Raab, M. Reff, S. Shuey, and N. Hanna. 1992. “Primatization” of recombinant antibodies for immunotherapy of human diseases: a macaque/human chimeric antibody against human CD4. Biotechnology 10:1455-1460. [DOI] [PubMed] [Google Scholar]
- 37.Noskov, A. N., T. B. Kravchenko, and V. P. Noskova. 1996. Detection of the functionally active domains in the molecule of protective antigen of the anthrax exotoxin. Mol. Gen. Mikrobiol. Virusol. 1996:16-20. [PubMed] [Google Scholar]
- 38.Petosa, C., R. J. Collier, K. R. Klimpel, S. H. Leppla, and R. C. Liddington. 1997. Crystal structure of the anthrax toxin protective antigen. Nature 385:833-838. [DOI] [PubMed] [Google Scholar]
- 39.Pommie, C., S. Levadoux, R. Sabatier, G. Lefranc, and M. P. Lefranc. 2004. IMGT standardized criteria for statistical analysis of immunoglobulin V-REGION amino acid properties. J. Mol. Recognit. 17:17-32. [DOI] [PubMed] [Google Scholar]
- 40.Renard, M., L. Belkadi, N. Hugo, P. England, D. Altschuh, and H. Bedouelle. 2002. Knowledge-based design of reagentless fluorescent biosensors from recombinant antibodies. J. Mol. Biol. 318:429-442. [DOI] [PubMed] [Google Scholar]
- 41.Ruiz, M., and M. P. Lefranc. 2002. IMGT gene identification and Colliers de Perles of human immunoglobulins with known 3D structures. Immunogenetics 53:857-883. [DOI] [PubMed] [Google Scholar]
- 42.Sawada-Hirai, R., I. Jiang, F. Wang, S. M. Sun, R. Nedellec, P. Ruther, A. Alvarez, D. Millis, P. R. Morrow, and A. S. Kang. 2004. Human anti-anthrax protective antigen neutralizing monoclonal antibodies derived from donors vaccinated with anthrax vaccine adsorbed. J. Immune Based Ther. Vaccines 2:5-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schuck, P., and A. P. Minton. 1996. Analysis of mass transport-limited binding kinetics in evanescent wave biosensors. Anal. Biochem. 240:262-272. [DOI] [PubMed] [Google Scholar]
- 44.Scobie, H. M., G. J. Rainey, K. A. Bradley, and J. A. Young. 2003. Human capillary morphogenesis protein 2 functions as an anthrax toxin receptor. Proc. Natl. Acad. Sci. USA 100:5170-5174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thullier, P., C. Demangel, H. Bedouelle, F. Megret, A. Jouan, V. Deubel, J. C. Mazie, and P. Lafaye. 2001. Mapping of a dengue virus neutralizing epitope critical for the infectivity of all serotypes: insight into the neutralization mechanism. J. Gen. Virol. 82:1885-1892. [DOI] [PubMed] [Google Scholar]
- 46.Varughese, M., A. V. Teixeira, S. Liu, and S. H. Leppla. 1999. Identification of a receptor-binding region within domain 4 of the protective antigen component of anthrax toxin. Infect. Immun. 67:1860-1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wein, L. M., D. L. Craft, and E. H. Kaplan. 2003. Emergency response to an anthrax attack. Proc. Natl. Acad. Sci. USA 100:4346-4351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wild, M. A., H. Xin, T. Maruyama, M. J. Nolan, P. M. Calveley, J. D. Malone, M. R. Wallace, and K. S. Bowdish. 2003. Human antibodies from immunized donors are protective against anthrax toxin in vivo. Nat. Biotechnol. 21:1305-1306. [DOI] [PubMed] [Google Scholar]