Abstract
The apical surface of mammalian urothelium is covered by 16-nm protein particles packed hexagonally to form 2D crystals of asymmetric unit membranes (AUM) that contribute to the remarkable permeability barrier function of the urinary bladder. We have shown previously that bovine AUMs contain four major integral membrane proteins, i.e., uroplakins Ia, Ib, II, and IIIa, and that UPIa and Ib (both tetraspanins) form heterodimers with UPII and IIIa, respectively. Using a panel of antibodies recognizing different conformational states of uroplakins, we demonstrate that the UPIa-dependent, furin-mediated cleavage of the prosequence of UPII leads to global conformational changes in mature UPII and that UPIb also induces conformational changes in its partner UPIIIa. We further demonstrate that tetraspanins CD9, CD81, and CD82 can stabilize their partner protein CD4. These results indicate that tetraspanin uroplakins, and some other tetraspanin proteins, can induce conformational changes leading to the ER-exit, stabilization, and cell surface expression of their associated, single-transmembrane-domained partner proteins and thus can function as “maturation-facilitators.” We propose a model of AUM assembly in which conformational changes in integral membrane proteins induced by uroplakin interactions, differentiation-dependent glycosylation, and the removal of the prosequence of UPII play roles in regulating the assembly of uroplakins to form AUM.
INTRODUCTION
Although integral membrane proteins account for almost one-third of all the proteins and they serve essential cellular functions, relatively little is known about how they interact and assemble into multicomponent complexes. A group of mammalian membrane proteins that provides an excellent model system for studying membrane assembly is the uroplakins (UP). There are four major uroplakins, i.e., UPIa (27-kDa), Ib (28-kDa), II (15-kDa), and IIIa (47-kDa). As a model system, uroplakins offer several major advantages: 1) They form 16-nm particles that are naturally assembled into hexagonally packed 2D crystals (the “urothelial plaques”; also known as “asymmetric unit membrane” or AUM) that cover almost the entire apical urothelial surface (Hicks and Ketterer, 1969; Vergara et al., 1969; Brisson and Wade, 1983) forming a remarkable permeability barrier (Negrete et al., 1996; Lewis, 2000; Apodaca, 2004). 2) Because uroplakins are synthesized as the major specialization products of mammalian urothelium, they can be isolated as 2D crystals in milligram quantities, thus facilitating their structural analyses (Wu et al., 1990; Walz et al., 1995; Kachar et al., 1999; Oostergetel et al., 2001; Min et al., 2003). 3) The four uroplakins form two heterodimers each consisting of a tetraspanin member, i.e., UPIa and UPIb (of the “tetraspanin” family; see below), which bind to UPII and UPIIIa, respectively (Wu et al., 1995; Liang et al., 2001). The formation of the UPIa/II and UPIb/IIIa heterodimers is a prerequisite for uroplakins to exit from the ER (Tu et al., 2002), thus facilitating the analysis of uroplakin processing. 4) Cultured urothelial cells can synthesize all uroplakins which, however, do not assemble into 2D crystals (Surya et al., 1990), indicating that the later stages of membrane assembly leading to the formation of 2D crystals can be studied by comparing the processing and assembly of uroplakins in normal urothelium versus cultured urothelial cells. 5) Uroplakins are functionally important as the ablation of uroplakins II or IIIa gene abolishes urothelial plaque formation and compromises the barrier function (Hu et al., 2000, 2002; Kong et al., 2004).
Uroplakins Ia and Ib belong to the tetraspanin family, which contains many cell surface proteins involved in a variety of cellular activities including cell mobility, interactions and fusion, immunological signaling, tumor progression, pathogen invasion, and membrane architecture (Maecker et al., 1997; Boucheix and Rubinstein, 2001; Hemler, 2003). These proteins possess four transmembrane domains, with a small and a large extracellular loop—the latter harboring several highly conserved cysteine residues (Stipp et al., 2003b). Many tetraspanins interact with important single-transmembrane-domained signaling proteins; examples of such tetraspanin/partner interactions include CD9/pro-TGF-α or pro-HB-epidermal growth factor; CD9, CD81, or CD82/CD4; CD82/EGFR; and CD151/integrins (Boucheix and Rubinstein, 2001; Hemler, 2003). It has been suggested that by recruiting the signaling molecules to membrane subdomains called “tetraspanin webs,” tetraspanins can lower the concentration threshold of the ligands that are required to trigger cellular responses (Maecker et al., 1997; Boucheix and Rubinstein, 2001; Hemler, 2003; Tarrant et al., 2003). Studies on tetraspanins are hampered, however, by their usually weak interactions with multiple partners (Hemler, 2001). In contrast, tetraspanin uroplakins interact with their partners in a highly specific, strong, and stoichiometric manner, thus providing unique opportunities for detailed mechanistic studies of their interactions with their partners.
The amino acid sequences of mammalian uroplakins are highly conserved, suggesting that they play an essential function in urinary bladder epithelium (Wu et al., 1994; Sun et al., 1999). Uroplakins Ia (27-kDa) and Ib (28-kDa) are ∼40% identical in their amino acid sequences and are both tetraspanins (Yu et al., 1994). UPII is synthesized as a pre (26 amino acids)–pro (59 amino acids)–mature protein (100 amino acids); the mature UPII (15-kDa) can be divided into a long extracellular domain of 71 amino acids and a transmembrane domain of 25 amino acids, with almost no intracellular domain (Lin et al., 1994). UPIIIa is synthesized as a pre-protein; the mature protein (47-kDa) contains an apoprotein of ∼29-kDa plus ∼18-kDa equivalents of complex glycans, and it is the only uroplakin that has a significant cytoplasmic domain (of ∼52 amino acid residues; Wu and Sun, 1993). How these uroplakins assemble into 2D crystals is unclear.
In this article, we use a panel of antibodies that can distinguish different conformational states of uroplakins to study AUM assembly. We demonstrate that global conformational changes occur during uroplakin interaction and processing. We propose a model in which the differentiation-dependent glycosylation and the removal of UPII prosequence are involved in regulating AUM assembly. Studies on several other nonuroplakin tetraspanins suggest that, possibly as a general rule, members of the tetraspanin family facilitate the ER-exit, stabilization, and surface presentation of their single-transmembrane-domained partner proteins.
MATERIALS AND METHODS
Materials
The reagents and materials were obtained from the following sources. Furin inhibitor 1 (decanoyl-Arg-Val-Lys-Arg-chloromethylketone) and tunicamycin (Calbiochem, La Jolla, CA); disuccinimidyl suberate (DSS; Pierce, Rockford, IL); endoglycosidase H (endo H), N-glycosidase F (endo F) and recombinant human furin (New England Biolabs, Beverly, MA; NEB); clasto-lactacystin β-lactone (CLL), N-acetyl-l-leucinyl-l-leucinyl-norleucinal (ALLN) and carbobenzoxy-l-leucyl-l-leucyl-l-leucinal (MG132; Biomol, Plymouth Meeting, PA); monoclonal antibodies to GST, CD4, and CD82 (Santa Cruz Biotechnology, Santa Cruz, CA); pGEX4T-3 vector (Amersham, Piscataway, NJ); antibodies to CD9 (SYB-1) and CD81 (TS81), and human CD9 and CD81 cDNAs (Eric Rubinstein); human CD82 cDNA (Xin A. Zhang); and human CD4 cDNA (Dan R. Littman). All the cDNAs were subcloned into pcDNA3 (Invitrogen, Carlsbad, CA), a eukaryotic expression vector. For the preparation of urothelial plaques (AUM), bovine urothelium was homogenized in buffer A (10 mM HEPES, pH 7.5; 1 mM EDTA; 1 mM EGTA; 1 mM phenylmethylsulfonyl fluoride), loaded onto a 1.6 M sucrose cushion, and centrifuged at 16,000 rpm for 25 min at 4°C (Beckman, Fullerton, CA; SW28). The crude membranes (CM) concentrated at the interface were isolated, washed with buffer A, treated with 2% Sarkosyl in Buffer A for 10 min at 25°C, and pelleted, resulting in the Sarkosyl-insoluble AUM (Liang et al., 1999).
Cultured Cells
Bovine urothelial cells were cultured in the Dulbecco's modified Eagle's medium (DMEM, Life Technologies, Rockville, MD) containing 15% fetal bovine serum (FBS) in the presence of mitomycin C-pretreated 3T3 feeder cells as described previously (Surya et al., 1990). COS-1 cells (ATCC) and furin-deficient 7.P15 cells (originally selected from COS-7 cells and kindly provided by Joseph F. Sucic; Inocencio et al., 1997) were grown overnight in DMEM containing 10% FBS and in DMEM/F12 (1:1) containing 7.5% FBS, respectively; and transfected using a mixture of cDNA and FuGENE6 reagent (Roche, Indianapolis, IN; 1:3, wt/vol) in 100 μl of serum-free DMEM. The transfectants were analyzed 24–48 h later.
Deglycosylation and Immunoblotting
Deglycosylation was performed according to manufacturer's instructions (New England Biolabs, Beverly, MA). Briefly, cell lysates were denatured in 0.5% SDS and 1% β-mercaptoethanol at 25°C for 10 min, made to contain 50 mM sodium citrate (pH 5.5), and incubated with endo H at 37°C for 16 h (complete deglycosylation) or at 25°C for 6 h (partial deglycosylation). Alternatively, sodium phosphate (pH 7.5) and NP-40 were added to the denatured cell lysates to a final concentration of 50 mM and 1%, respectively, and the mixture was incubated with endo F. SDS-PAGE (17% acrylamide) and immunoblotting were done as described (Liang et al., 2001).
Epitope Mapping
Full-length bovine UPII cDNA (template) and specific PCR primers were used to amplify various truncated UPII fragments, which were subsequently subcloned into the pGEX4T-3 vector. The constructs encoding various GST-(truncated UPII) fusion proteins were sequenced and induced to express in DH5α cells using isopropyl-β-d-thiogalactopyranoside (0.5 mM; Sigma, St. Louis, MO).
Cross-linking
Ten microliters of disuccinimidyl suberate (50 mM in dimethyl sulfoxide) were added to 100 μl of transfectant lysates in the nuclear extraction buffer (20 mM HEPES, pH 7.6; 20% glycerol; 500 mM NaCl; 1.5 mM MgCl2; 0.2 mM EDTA; 0.1% Triton X-100; 1 mM dithiothreitol) with protease inhibitors (Cocktail Set 1, Calbiochem). The mixture was incubated for 1 h on ice and quenched with 50 mM Tris-HCl (pH 7.5) for 30 min on ice.
Mutagenesis
Mutagenesis was performed according to the instructions provided with the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). After the mutations were confirmed by DNA sequencing, the insert was excised from the old vector and ligated back into the nonamplified pcDNA3 vector to eliminate potential mutations in the plasmid DNA caused by the Pfu amplification reaction.
In Vitro Cleavage of Pro-UPII by Furin
Pro-UPII expressed in the furin-deficient 7.P15 cells was immunopurified using the AE31 antibody attached to protein A/G PLUS-agarose beads (Santa Cruz Biotechnology), suspended in 50 mM sodium acetate (pH 5.4) containing 1 mM CaCl2, and incubated with 10 U of furin in a total volume of 100 μl at 25°C for 16 h.
Pulse-chase Labeling
Cells were starved in the methionine-free DMEM medium containing dialyzed serum (Life Technologies) for 1 h and pulse-labeled for 1 h in the same medium supplemented with 125 μCi/ml [35S]methionine. The cells were then incubated in the DMEM medium for a certain chase period, rinsed with phosphate-buffered saline, lysed in RIPA buffer (10 mM Tris-HCl, pH 7.4; 150 mM NaCl; 1% NP-40; 0.5% sodium deoxycholate; 0.1% SDS; 1 mM EDTA) containing protease inhibitors, dispersed by passing through a 25-gauge needle (15 times), rocked at 4°C for 1 h, centrifuged at 16,000 × g for 10 min at 4°C to pellet the nuclei, and stored at –80°C before further analyses.
Immunoprecipitation
[35S]methionine-labeled cell lysates in RIPA buffer containing protease inhibitors were preincubated with preimmune sera along with Protein A/G-agarose beads at 4°C to reduce the background. The cleared lysate was combined with a primary antibody plus Protein A/G-agarose beads and rotated head-over-head overnight at 4°C. The bead-bound proteins were analyzed by SDS-PAGE. The gel was fixed, impregnated with the Amplify solution (Amersham), dried under vacuum, and fluorographed. The band density was determined by scanning x-ray films using the GS-800 calibrated densitometer (Bio-Rad, Richmond, CA) followed by analyses using the Quantity One program (Bio-Rad).
RESULTS
A Monoclonal Antibody, AE31, Recognized an SDS-resistant Composite Epitope of Pro-UPII
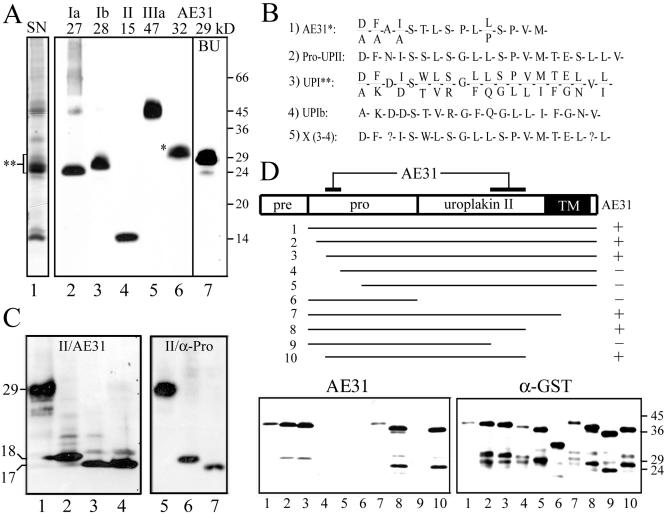
To study the structure and assembly of uroplakins, we generated a panel of monoclonal and polyclonal antibodies monospecific for all four major bovine uroplakins (Figure 1A, lanes 1–5). Mouse monoclonal antibody (mAb), AE31, recognized a 32-kDa bovine AUM protein (Figure 1A, lane 6; Yu et al., 1990). Immunoaffinity-purified AE31 antigen had an N-terminal sequence of D/A-F/A-A-I/A-S-T-L-S... (Figure 1B, sequence 1), which was almost identical to that of the prosequence of UPII (sequence 2; Lin et al., 1994). A similar sequence (sequence 5 or X) was obtained by subtracting the N-terminal sequence of UPIb (sequence 4; Yu et al., 1994) from the mixed N-terminal sequences of the 27–32-kDa “uroplakin I” protein band in AUM (sequence 3). These results raised the possibility that the AE31 antigen was the pro-UPII, instead of being related to uroplakin I as we previously thought (Yu et al., 1990). To test this possibility, we transfected COS-1 cells using cDNA encoding UPII and immunoblotted the total cellular proteins using AE31. We found that AE31 indeed recognized a 29-kDa band (Figure 1C, lane 1), which is also recognized by a rabbit antiserum to the prosequence of UPII (α-Pro; Figure 1C, lane 5). Moreover, the 29-kDa antigen pulled down by the AE31 antibody could be immunoblotted using the α-Pro antibody, and vice versa (see below), thus further establishing the identity of the AE31-antigen as the pro-UPII. The 29-kDa pro-UPII could be converted to the 18- and 17-kDa forms after it was treated with endo H and endo F, respectively (Figure 1C, lanes 2–3), indicating the presence of high mannose glycans (the 1-kDa difference was due to the removal of N-acetylglucosamines). Interestingly, the 29-kDa pro-UPII of cultured bovine urothelial (BU) cells (Figure 1A, lane 7), which are less differentiated than in vivo urothelium (Surya et al., 1990), was ∼3-kDa smaller than the pro-UPII of the in vivo umbrella cells (32-kDa; Figure 1A, lane 6; see below). Finally, we mapped the AE31 epitope on pro-UPII by immunoblotting various recombinant fragments of pro-UPII using the AE31 antibody. We found that the AE31-immunoreactivity required two separate domains of the pro-UPII, one located in the prosequence region (amino acid residues –50 to –40), whereas the other located ∼80 amino acid residues away in the middle of the mature UPII sequence (residues 40–60; Figure 1D). These results established that AE31 recognized a composite pro-UPII epitope consisting of two distant sequences that bound to each other forming presumably a hairpinlike structure that was stable under SDS-PAGE conditions.
Figure 1.
The AE31 mAb recognizes a composite epitope of pro-UPII. (A) Total proteins of purified bovine urothelial plaques (AUM; lanes 1–6) or cultured bovine urothelial (BU) cells (lane 7) were separated by SDS-PAGE and stained with (lane 1) silver nitrate (SN); or immunoblotted using rabbit antibodies to (2) UPIa, (3) UPIb, or (4) UPII, or (5) mouse AU1 mAb to UPIIIa, or (6 and 7) mouse AE31 mAb. Note that AE31 recognized a 32-kDa protein in the AUM and a 29-kDa protein in cultured BU cells. (B) N-terminal sequence of the bovine AE31 antigen. Listed are the N-terminal sequences of (1) the 32-kDa AE31 antigen immunoaffinity-purified from the Triton X-100 solubilized total urothelial plaque proteins; (2) the cDNA-deduced prosequence of pro-UPII (Lin et al., 1994); (3) the SDS-PAGE-purified, 27–32-kDa proteins of the bovine AUM (double-asterisked region in lane 1 of A); (4) the cDNA-deduced UPIb sequence (Yu et al., 1994); and (5) a hypothetical protein (X)— deduced by subtracting the UPIb sequence shown in (4) from the mixed sequence shown in (3) (described as “UPIc” in Yu et al., 1994). Note that the N-terminal sequence of the AE31 antigen was almost identical to that of pro-UPII and X. (C) Immunoblotting of total proteins of UPII cDNA-transfected COS-1 cells using (lanes 1–4) the AE31 mAb or (4–7) a rabbit antiserum against the prosequence portion of pro-UPII. Samples in lanes (2) and (6) were endo H-treated; (3) and (7) endo F-treated; cells of lane (4) were treated with tunicamycin (6.4 μg/ml). Note that both AE31 and the anti-prosequence antibody recognized the same 29-kDa glycoprotein that became 18- and 17-kDa after endo H and endo F treatments, respectively. (D) Mapping of the AE31 epitope in pro-UPII. Lanes 1–10 showed the immunoblotting of 10 pro-UPII fragments (in the form of GST-(truncated pro-UPII) fusion proteins; amino acids –59 to +100, –55 to +100, –50 to +100, –40 to +100, –30 to +100, –59 to –1, –59 to +80, –59 to +60, –59 to +40, and –50 to +60) using the AE31 mAb (lower left) or a mAb against the GST protein (lower right; as a control). The highest MW bands recognized by both AE31 and anti-GST antibodies represent the intact fusion proteins (no. 1–10); lower MW bands are degradation products. Note that AE31-reaction with pro-UPII required the simultaneous presence of two distant domains (amino acid positions –50 to –40 in the prosequence and positions +40 to +60 in the mature UPII), suggesting that pro-UPII assumed a folded, hairpinlike structure that was stable in the SDS-PAGE condition.
Antibodies Recognizing Selectively the Mature Uroplakin II
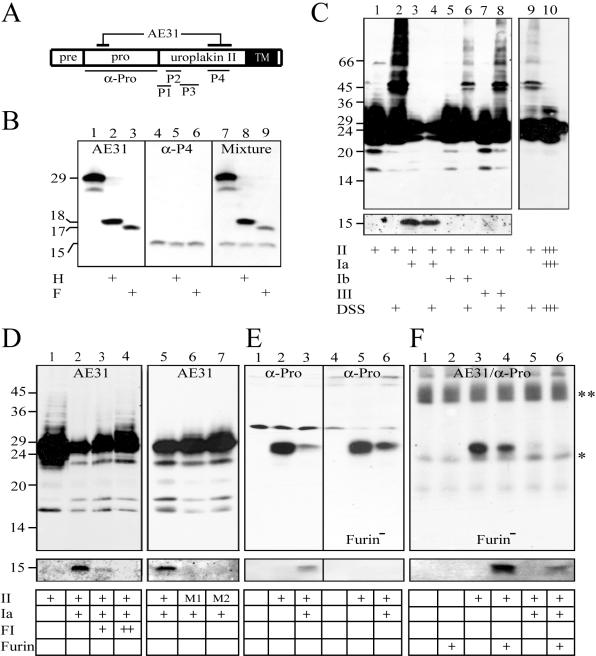
In addition, we generated rabbit antisera against synthetic peptides (designated P1–P4 in Figure 2A) corresponding to four separate regions of the mature UPII (Wu et al., 1994; Liang et al., 2001). Although the AE31 antibody selectively recognized pro-UPII (Figure 2B, lanes 1–3), all four rabbit antisera recognized only the 15-kDa mature UPII, in cultured BU cells (Figure 2B, lanes 4–6 for anti-P4; and unpublished data). Because pro-UPII obviously contained all four linear sequences against which the rabbit antisera were raised, these findings indicated that such epitopes were somehow masked or conformationally altered in the tightly packed pro-UPII so that they were not recognized by the rabbit antisera, that the removal of the prosequence led to the unmasking/conformational changes of P1-P4 epitopes, and that the immunological distinction between pro-UPII and mature UPII was maintained under SDS-PAGE conditions (Figure 2B).
Figure 2.
Immunological distinction between pro-UPII and mature UPII and the processing of pro-UPII by furin. (A) A diagram showing the epitopes of various UPII antibodies used in this study. (AE31) The composite epitope of the AE31 mAb; (α-Pro) the prosequence of UPII used to generate the rabbit “α-Pro” antiserum; (P1–P4) synthetic peptides used to generate rabbit antisera against amino acid positions +1 to +11 (P1); +7 to +19 (P2); +18 to +33 (P3); and +40 to +58 (P4) of the mature UPII. (B) Immunological distinction between pro-UPII and mature UPII. Total proteins of cultured bovine urothelial (BU) cells were immunoblotted using (1–3) AE31 mAb, (4–6) anti-P4 (similar results were obtained using anti-P1 to P3), and (7–9) a mixture of AE31 and anti-P4. Proteins were incubated with buffer (lanes 1, 4, and 7), endo H (lanes 2, 5, and 8), or endo F (lanes 3, 6, and 9). Note that AE31 and anti-UPII antibodies recognized the 29-kDa pro-UPII and the 15-kDa mature UPII, respectively (with extremely weak cross-reactivity; cf. Tu et al., 2002). (C) Tetraspanin UPIa blocked the aggregation and facilitated the maturation of pro-UPII. Total lysates of COS-1 cells transfected with cDNAs encoding UPII (all lanes), plus UPIa (lanes 3, 4, and 10), UPIb (lanes 5 and 6), or UPIIIa (lanes 7 and 8) were immunoblotted using AE31 (top panel) or the anti-P4 antibody (bottom panel). Samples of lanes 2, 4, 6, 8, 9, and 10 were cross-linked with disuccinimidyl suberate (DSS). The sample in lane 10 was identical to that in lane 4 except it was fivefold overloaded to illustrate the absence of pro-UPII aggregates. Note the cleavage of pro-UPII, in the presence of UPIa, to yield the 15-kDa mature UPII. (D) Furin is involved in cleaving the prosequence of pro-UPII in the presence of UPIa. Total lysates of COS-1 cells transfected with cDNA of UPII (lanes 1–5), or UPII mutant 1 (Arg(-1)Ala; lane 6) and mutant 2 (Arg(-4)Ala; lane 7), both inactivating the furin cutting site, were immunoblotted using AE31 (top panel) or the anti-P4 antibody (bottom panel). Some cells were double-transfected with UPIa (lanes 2–7), or were, in addition, treated with decanoyl-Arg-Val-Lys-Arg-chloromethylketone, a furin inhibitor (FI; Stieneke-Grober et al., 1992), at 25 μM (lane 3) and 50 μM (lane 4). Note the complete blockage of pro-UPII processing by FI and by furin-site mutations. (E) Pro-UPII processing is blocked in furin-deficient cells. COS-1 (lanes 1–3) and furin-deficient 7.P15 (lanes 4–6) cells were transfected with an empty pcDNA3 plasmid (lanes 1 and 4; control), UPII cDNA (lanes 2 and 5), or UPII plus UPIa cDNAs (lanes 3 and 6). The lysates were immunoblotted using the α-Pro (top panel) or anti-P4 antibody (bottom panel). Note the absence of the 15-kDa UPII in the furin-deficient cells. (F) Pro-UPII processing in the furin-deficient cells can be restored by exogenous furin. The furindeficient 7.P15 cells were transfected with an empty pcDNA3 plasmid (lanes 1 and 2), UPII cDNA (lanes 3 and 4), or UPII plus UPIa cDNAs (lanes 5 and 6). The pro-UPII was immunopurified using the AE31 antibody, incubated in vitro with recombinant human furin (lanes 2, 4, and 6), and analyzed by immunoblotting using the α-Pro (top panel) or anti-P4 antibody (bottom panel). Note that pro-UPII produced even in singly transfected 7.P15 cells could be processed by exogenous furin (lane 4) because of disruptions of cellular compartments in RIPA buffer. The asterisked and double-asterisked bands represent light chains and heavy chains of immunoglobulins.
Interactions between UPIa and UPII
The availability of these two sets of conformation-dependent antibodies, which can distinguish pro-UPII from mature UPII (Figure 2B), allowed us to study how pro-UPII interacted with its tetraspanin partner, UPIa. Transfecting COS-1 cells with the full-length UPII cDNA alone resulted in the formation of pro-UPII aggregates, which could best be visualized after cross-linking (Figure 2C, lanes 1 and 2). Coexpression of UPII with its partner UPIa (Figure 2C, lanes 3–4), but not with UPIb or UPIIIa (lanes 5–8), led to the processing of pro-UPII giving rise to the 15-kDa UPII recognized by rabbit antisera to mature UPII; in such cells the pro-UPII aggregation became undetectable even in overloaded gels (Figure 2C, lanes 9 and 10).
Four lines of evidence established that furin, a trans-Golgi network (TGN)-associated endopeptidase, was responsible for pro-UPII processing. First, the UPIa-dependent processing of pro-UPII could be blocked by decanoyl-Arg-Val-Lys-Arg-chloromethylketone, a furin inhibitor (Figure 2D, lanes 2–4). Second, pro-UPII processing was completely blocked by two mutations (Arg(-1)Ala and Arg(-4)Ala) in its RGRR furin cleavage motif (Figure 2D, lanes 5–7). Third, pro-UPII processing was blocked in two furin-deficient cell lines, 7.P15 (Figure 2E, lane 6) and RPE.40 (unpublished data). Finally, the processing of the pro-UPII expressed in furin-deficient 7.P15 cells could be restored in vitro using an exogenous recombinant human furin (Figure 2F, lanes 4 and 6).
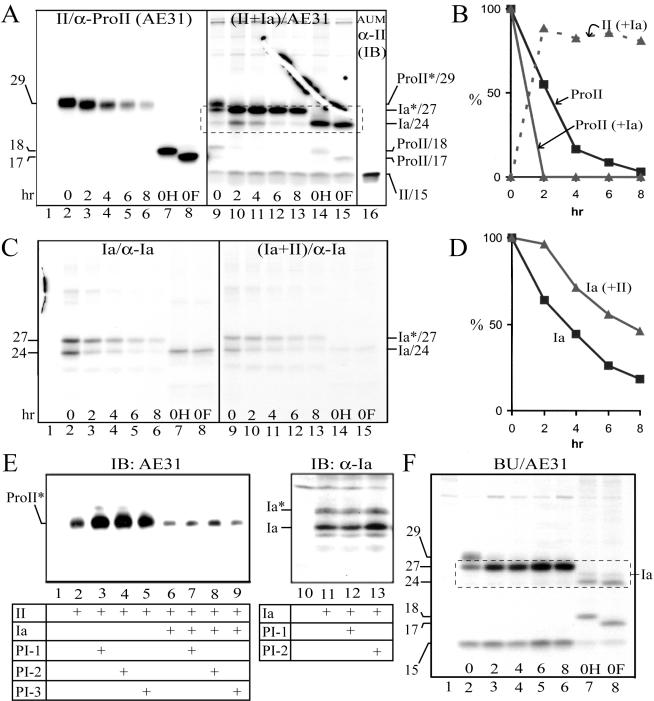
To analyze the effects of UPIa on pro-UPII processing, we [35S]methionine-labeled COS-1 cells that had been transfected with cDNA of either UPII or UPIa alone, or both, and performed immunoprecipitations after various chase intervals to assess the turnover rate of these uroplakins (Figure 3). Pro-UPII expressed alone turned over with a half-life of ∼2.2 h (Figure 3, A, lanes 2–6, and B); treatment of the cells with proteasome inhibitors led to a tremendous accumulation of pro-UPII (Figure 3E, lanes 2–5), suggesting that the unpaired pro-UPII was trapped in the ER and was degraded via the proteasome pathway. As shown earlier (Figure 2C), pro-UPII coexpressed with UPIa was rapidly processed (T1/2 < 1 h) into mature UPII (Figure 3A, lanes 9–15), which was stable and could be recognized by the rabbit antisera (Figure 3A, lane 16); AE31 (which did not recognize mature UPII; Figures 1A and 2B) could pull down in these cotransfected cells small amounts of mature UPII, most likely because of binding of the mature UPII to the coprecipitated UPIa (Figure 3A). A somewhat different situation existed for UPIa, which when expressed alone turned over with a half-life of ∼3.5 h (Figure 3, C and D), probably also via the proteasome pathway (Figure 3E, lanes 11–13). UPIa was, however, stabilized when coexpressed with UPII (Figure 3, C and D). Similar results were obtained by pulse-chase studies of the uroplakins synthesized by cultured BU cells (Figure 3F), indicating that our transfection data obtained using COS-1 cells were applicable to diploid urothelial cells. Taken together, these results indicated that UPIa enabled pro-UPII to exit from the ER in the form of UPIa/pro-UPII heterodimer and to be proteolytically processed by furin in the TGN to yield a conformationally distinct and stable, mature UPII (see Discussion).
Figure 3.
Tetraspanin UPIa facilitates the processing of pro-UPII. (A) The processing of pro-UPII is facilitated by UPIa. COS-1 cells were transfected with the control pcDNA3 plasmid (lane 1; control), UPII cDNA (lanes 2–8), or UPII plus UPIa cDNAs (lanes 9–15); the cells were [35S]methionine-labeled for 1 h and chased. The cell lysates were immunoprecipitated using the AE31 mAb. Immunoprecipitated proteins combined from the 0- and 2-h chase samples were used for endo H (lanes 7 and 14) or endo F (lanes 8 and 15) deglycosylation. Lane 16 showed the mature UPII in the AUM (immunoblotted using anti-P4; see Figure 2A). Note that UPIa facilitated the processing of pro-UPII yielding the stable, 15-kDa mature UPII; and that large amounts of UPIa (broken-lined box) could be coimmunoprecipitated by the AE31 mAb in the form of pro-UPII/UPIa complex (cf. panel C, lanes 9–13, using identical lysates and same exposure time). (B) Bands in A were quantified by densitometry (see Materials and Methods). ProII and ProII (+Ia) denote pro-UPII in the UPII transfectants and in the UPII/UPIa double transfectants, respectively; II (+Ia) denotes the mature UPII in the double transfectants. The amount of UPII that was present at time 0 (at the end of a 60-min labeling) was subtracted as a background for subsequent time points. Similar results were obtained from two additional experiments. (C) UPIa can be stabilized by pro-UPII. COS-1 cells were transfected with the pcDNA3 plasmid (lane 1), UPIa cDNA (lanes 2–8), or UPIa plus UPII cDNAs (lanes 9–15), radiolabeled for 1 h, chased, and analyzed by immunoprecipitation using the anti-UPIa antibody. Immunoprecipitated proteins from the 0-h chase sample were used for endo H (lanes 7 and 14) or endo F (lanes 8 and 15) deglycosylation. Ia* and Ia denote the glycosylated, mature UPIa (27-kDa), and the unglycosylated, immature UPIa (24-kDa), respectively. (D) Bands in C were quantified by densitometry and the sum of the glycosylated and unglycosylated UPIa bands was plotted. Ia indicates UPIa in the UPIa single transfectants, and Ia (+II) represents UPIa in the UPIa/UPII double transfectants. Similar results were obtained from two additional experiments. Note the stabilization of UPIa by UPII. (E) Pro-UPII in singly transfected cells is degraded by proteasomes. COS-1 cells were transfected with the pcDNA3 plasmid (lanes 1 and 10), UPII cDNA (lanes 2–5), UPII plus UPIa cDNAs (lanes 6–9), or UPIa cDNA (lanes 11–13). After 24 h, the cells were treated for 16 h with proteasome inhibitors, clasto-lactacystin β-lactone (CLL or PI-1; 10 μM; lanes 3, 7, and 12), N-acetyl-l-leucinyl-l-leucinyl-norleucinal (ALLN or PI-2; 20 μM; lanes 4, 8, and 13), or carbobenzoxy-l-leucyl-l-leucyl-l-leucinal (MG132 or PI-3; 50 μM; lanes 5 and 9; Xiong et al., 1999). The lysates were immunoblotted by AE31 or the anti-UPIa antibody. Note that the proteasome inhibitors resulted in >10-fold accumulation of pro-UPII (lanes 2–5), and about twofold accumulation of UPIa (lanes 11–13) in single transfectants; and a less significant accumulation of pro-UPII in double transfectants (lanes 6–9). (F) The processing of pro-UPII in cultured bovine urothelial (BU) cells. Cultured BU cells were radiolabeled for 1 h and analyzed by immunoprecipitation using a control antibody (lane 1) or the AE31 mAb (lanes 2–8). Immunoprecipitated proteins from the 0-h chase sample were deglycosylated using endo H (lane 7) or endo F (lane 8).
Interactions between UPIb and UPIIIa
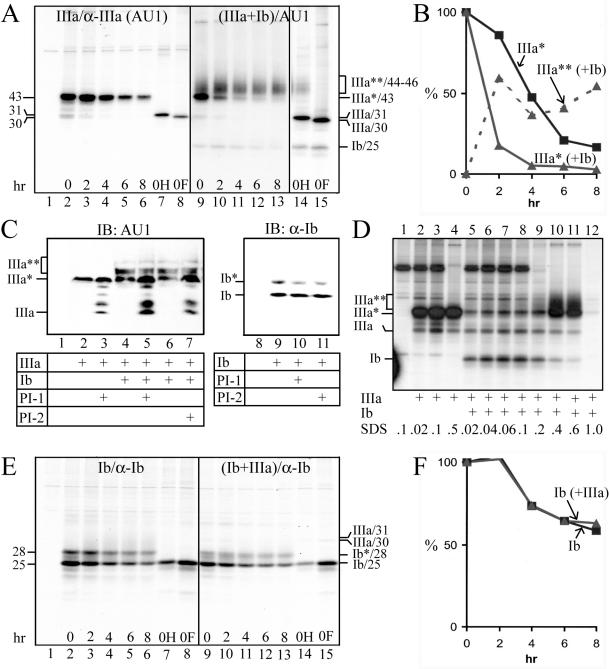
Similar pulse-chase experiments were conducted to study the interactions between UPIb and UPIIIa (Figure 4). UPIIIa expressed alone could be pulled down by all our antibodies to UPIIIa, including the AU1 mAb (Figure 4A, lanes 2–8) and rabbit antisera to synthetic peptides corresponding to four separate regions of the UPIIIa molecule (amino acid residues 87–101, 179–191, 243–257, and 261–276; unpublished data). The singly expressed UPIIIa had a half-life of ∼3.8 h (Figure 4B) and was degraded by proteasomes (Figure 4C, lanes 2 and 3).
Figure 4.
Tetraspanin UPIb facilitates the processing of UPIIIa. (A) The processing of UPIIIa is facilitated by UPIb. COS-1 cells were transfected with the control pcDNA3 plasmid (lane 1), UPIIIa cDNA (lanes 2–8), or UPIIIa plus UPIb cDNAs (lanes 9–15), [35S]methionine-labeled for 1 h, chased, and analyzed by immunoprecipitation using the AU1 anti-UPIIIa mAb. Immunoprecipitated proteins from the 0-h chase sample were used for endo H (lanes 7 and 14) or endo F (lanes 8 and 15) deglycosylation. Note the conversion of the unstable and endo H-sensitive UPIIIa (IIIa*), in the presence of UPIb, to the stable and endo H-resistant, mature UPIIIa (III**, lanes 9–14). (B) Bands in A were quantified by densitometry. IIIa* indicates UPIIIa in the UPIIIa single transfectants; IIIa* (+Ib) and IIIa** (+Ib) denote the immature and mature UPIIIa, respectively, in the UPIIIa/UPIb double transfectants. Note that the stable, mature UPIIIa [IIIa** (+Ib)] was immunoprecipitated in the RIPA buffer containing 0.5% SDS instead of the usual 0.1% SDS (see D). Similar results were obtained from two additional experiments. (C) UPIIIa is degraded by proteasomes. COS-1 cells were transfected with the pcDNA3 plasmid (lanes 1 and 8), UPIIIa cDNA (lanes 2 and 3), UPIIIa plus UPIb cDNAs (lanes 4–7), or UPIb cDNA (lanes 9–11). After 24 h, the cells were treated for 16 h with proteasome inhibitors, CLL (PI-1; 10 μM; lanes 3, 5, and 10) or ALLN (PI-2; 20 μM; lanes 7 and 11). The lysates were immunoblotted using the AU1 anti-UPIIIa mAb or an anti-UPIb antibody. Note that proteasome inhibitor treatments led to the accumulation of UPIIIa (and some partially deglycosylated intermediates targeting to proteasome degradation; Hirsch et al., 2003) in the UPIIIa transfectants (lanes 2 and 3), but did not lead to the accumulation of UPIb in the UPIb transfectants (lanes 9–11). Also note that in the UPIIIa/UPIb double transfectants, the immature (ER-form) UPIIIa (IIIa*) increased in the presence of proteasome inhibitors, but the mature (Golgi-form) UPIIIa (IIIa**) was unaffected by the inhibitors (lanes 4–7), suggesting that UPIIIa acquired stability upon maturation. (D) Tetraspanin UPIb induces a conformational change in UPIIIa. COS-1 cells were transfected with the pcDNA3 plasmid (lane 1), UPIIIa cDNA (lanes 2–4), or UPIIIa plus UPIb cDNAs (lanes 5–12), radiolabeled for 1 h, lysed in RIPA buffer containing 0.1% SDS, and analyzed by immunoprecipitation using the AU1 mAb. The SDS concentrations were adjusted, as indicated, by adding SDS or by diluting the lysates with RIPA buffer containing no SDS. Note that immunoprecipitation of even the immature UPIIIa (IIIa*) in the UPIIIa/UPIb double transfectants required the presence of 0.4–0.6% SDS, suggesting that the immature UPIIIa, when binding to UPIb in the ER, has already undergone conformational changes. (E) UPIb is stable by itself. COS-1 cells were transfected with the pcDNA3 plasmid (lane 1), UPIb cDNA (lanes 2–8), or UPIb plus UPIIIa cDNAs (lanes 9–15). The transfectants were radiolabeled for 1 h and analyzed after indicated chase intervals by immunoprecipitation using the anti-UPIb antibody. Immunoprecipitated proteins from the 0-h chase sample were deglycosylated using endo H (lanes 7 and 14) or endo F (lanes 8 and 15). Ib*/28-kDa and Ib/25-kDa denote the glycosylated, mature UPIb and the unglycosylated, immature UPIb, respectively. (F) Bands in E were quantified by densitometry. Ib denotes UPIb in the UPIb single transfectants; Ib (+IIIa) denotes UPIb in the UPIb/UPIIIa double transfectants. Similar results were obtained from two additional experiments.
Unexpectedly, when coexpressed with UPIb, UPIIIa could no longer be immunoprecipitated by any of these UPIIIa antibodies (unpublished data), suggesting conformational changes. Increasing the SDS concentration in the immunoprecipitation reaction (from 0.1 to 0.5%), however, led to the unmasking of all the UPIIIa epitopes (defined by AU1 and rabbit sera) in both the 43-kDa immature UPIIIa and the 44–46-kDa mature UPIIIa (that harbored endo H-resistant complex sugars) in the double transfectants (Figure 4D, lanes 10 and 11). These results were analogous to those of UPII and suggested that when UPIIIa bound to its partner UPIb, it underwent considerable conformational changes so that its multiple epitopes became masked (see Discussion). By repeating the immunoprecipitations of the pulse-chase experiments at 0.5% SDS instead of the usual 0.1% SDS, we found that UPIIIa, when coexpressed with UPIb, rapidly passed through the ER and the Golgi apparatus as evidenced by its size increase and its acquisition of endo H-resistance (Figure 4A, lanes 9–14); the endo H-resistant, mature UPIIIa became very stable (Figure 4, A and B) and the proteasome inhibitor treatments did not cause its accumulation (Figure 4C, lanes 4–7). Similar results were obtained from studying cultured BU cells (unpublished data).
Interestingly, UPIb was the only major uroplakin that was stable by itself with a half-life (>8 h) not affected by its partner UPIIIa (Figure 4, E and F), and UPIb did not accumulate in response to proteasome inhibitor treatments (Figure 4C, lanes 9–11). These results are consistent with the facts that UPIb can by itself exit from the ER (Tu et al., 2002) and that UPIb is singly expressed in several nonurothelial epithelia such as the corneal epithelium (Adachi et al., 2000).
Differentiation-dependent Glycosylation of Uroplakins
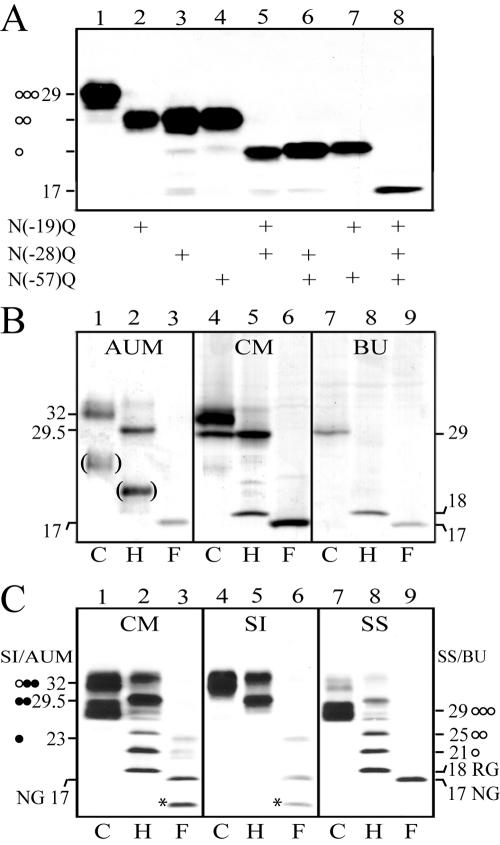
An interesting feature of the pro-UPII, which contained three potential N-glycosylation sites in its prosequence, was that its size was differentiation-dependent. Thus it appeared as a 32-kDa protein in the AUM purified from in vivo bovine urothelium, but as a 29-kDa protein in cultured BU cells (Figure 1A, lanes 6 and 7). Transfection studies using UPII cDNA with the three N-glycosylation sites mutated, singly or in various combinations, showed that all these sites were glycosylated and harbored about equal amounts of sugar (Figure 5A). Complete deglycosylation of the 32- and 29-kDa glycoproteins by endo F yielded an identical protein core of 17-kDa (Figure 5B, lanes 1–3 and 7–9). Both the 32- and 29-kDa species were present in the crude membrane (CM) fraction isolated from in vivo urothelium (Figure 5B, lanes 4–6). Studies of the partial deglycosylation intermediates established that the 29-kDa band harbored three high-mannose glycans, whereas the AUM-associated 32-kDa band harbored one high-mannose glycan and two complex glycans (Figure 5C). Interestingly, the 32- and 29-kDa bands were exclusively associated with the Sarkosyl-insoluble (SI) and Sarkosyl-soluble (SS) fractions, respectively (Figure 5C), suggesting that only the former was present in the fully assembled AUM.
Figure 5.
Differentiation-dependent glycosylation of pro-UPII. (A) COS-1 cells were transfected with wild-type UPII cDNA (lane 1) or with UPII cDNAs mutating one (lanes 2–4), two (lanes 5–7), or all three (lane 8) of the potential N-glycosylation sites (Asn at positions –19, –28 and –57 mutated to Gln) in the prosequence region, and the cell lysates were immunoblotted using the AE31 mAb. Note that the mutation of each site led to the loss of an almost identical size reduction suggesting that all three sites harbored about the same amounts of sugar. (B) Proteins of bovine urothelial plaques (AUM), crude membranes (CM) of normal bovine urothelium, and cultured BU cells (total lysates) were incubated with the deglycosylation buffer (C or control; lanes 1, 4, and 7), endo H (H; lanes 2, 5, and 8), or endo F (F; lanes 3, 6, and 9) at 37°C for 16 h (for complete deglycosylation), and immunoblotted using the AE31 mAb. Note the presence of the 32-kDa AE31 antigen in the AUM, the 29-kDa antigen in cultured BU cells, and both of the two bands in the crude membranes (CM) of normal bovine urothelium. (C) Crude membranes (CM) of bovine urothelium were separated into the Sarkosylinsoluble (SI) AUM and Sarkosyl-soluble (SS) membrane protein fractions. All these proteins were incubated with the deglycosylation buffer (C; lanes 1, 4, and 7), endo H (H; lanes 2, 5, and 8), or endo F (F; lanes 3, 6, and 9) at 25°C for 6 h (for partial deglycosylation), and immunoblotted using the AE31 mAb. Note that the 32-kDa pro-UPII of the CM was Sarkosyl-insoluble (lane 4), whereas the 29-kDa species was Sarkosyl-soluble (lane 7). The numbers indicate molecular weights; the open and closed circles denote high-mannose and complex glycans, respectively; and RG/18-kDa and NG/17-kDa denote the pro-UPII harboring the residual glycan (N-acetylglucosamines) and no glycan, respectively. The bracketed bands in B (showing samples that have been subjected to prolonged deglycosylation reactions) mostly likely represented degradation products; the asterisked ∼14-kDa degradation product in C was only occasionally observed. These results indicate that the 29-kDa pro-UPII harbors three high-mannose glycans (as indicated by the 25-, 21-, and 18-kDa intermediates), whereas the 32-kDa pro-UPII harbors one high-mannose and two complex glycans (29.5-, 23-, and 17-kDa intermediates; it is unknown, however, which two sites harbor the complex glycans).
Ability of Other Tetraspanins to Stabilize Their Single-transmembrane-domained Partner
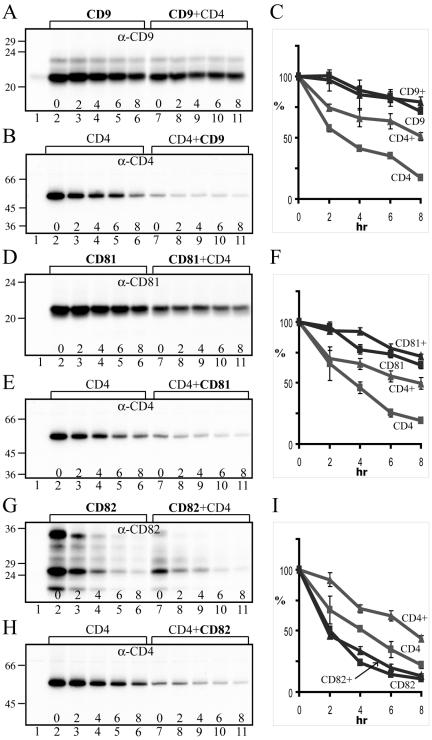
Our results thus indicated that UPIa and UPIb, two tetraspanins, had profound influence on the processing and stabilization of their single-transmembrane-domained partner proteins. Because many other tetraspanin proteins are also known to interact with partner proteins containing a single transmembrane domain, we assessed the effects of CD9, CD81, and CD82 on the stability of their partner protein, CD4, in parallel cotransfection experiments. We chose to do these experiments in COS-1 cells because preliminary surveys of multiple cell types showed that this particular cell line had no detectable, endogenous CD4, CD81, and CD82, and had only minute amounts of CD9 (Figure 6, lane 1 in each immunoblot panel; and unpublished data). Our results indicated that, under our experimental conditions, CD9 and CD81 were by themselves quite stable, with half-lives of ≫8 h, which were not significantly affected by their partner protein, CD4 (Figure 6, A and C for CD9; Figure 6, D and F for CD81). In contrast, CD4 was relatively unstable (T1/2 = ∼3.5 h), but could be significantly stabilized by tetraspanins CD9 and CD81 (T1/2 increased to ∼8 h; see Figure 6, B and C for the effect of CD9, and Figure 6, E and F for the effect of CD81, on the stability of CD4). Tetraspanin CD82 was intrinsically quite unstable (T1/2 =∼2 h; Figure 6, G and I) but could still exert a significant stabilizing effect on CD4 (T1/2 increased from ∼4 h to ∼7.3 h; Figure 6, H and I). These findings indicated that nonuroplakin tetraspanins could also stabilize their partner protein, but not vice versa.
Figure 6.
Tetraspanins CD9, CD81, and CD82 can stabilize CD4. (A) CD9 is relatively stable. COS-1 cells transfected with the control pcDNA3 plasmid (lane 1), CD9 cDNA (lanes 2–6), or CD9 plus CD4 cDNAs (lanes 7–11) were labeled with [35S]methionine for 1 h, chased, and analyzed by immunoprecipitation using an anti-CD9 antibody. (B) CD4 can be partially stabilized by CD9. COS-1 cells transfected with pcDNA3 (lane 1), CD4 cDNA (lanes 2–6), or CD4 plus CD9 cDNAs (lanes 7–11) were radio-labeled, chased, and analyzed by immunoprecipitation using an anti-CD4 antibody. (C) The bands in A and B were quantified by densitometry. Data from three independent experiments were presented (mean ± SD). (D) CD81 is relatively stable. COS-1 cells transfected with pcDNA3 (lane 1), CD81 cDNA (lanes 2–6), or CD81 plus CD4 cDNAs (lanes 7–11) were radiolabeled, chased, and analyzed by immunoprecipitation using an anti-CD81 antibody. (E) CD4 can be partially stabilized by CD81. COS-1 cells transfected with pcDNA3 (lane 1), CD4 cDNA (lanes 2–6), or CD4 plus CD81 cDNAs (lanes 7–11) were radiolabeled, chased, and analyzed by immunoprecipitation using the anti-CD4 antibody. (F) The bands in D and E were quantified. Data from three independent experiments were presented (mean ± SD). (G) CD82 is quite unstable. COS-1 cells transfected with pcDNA3 (lane 1), CD82 cDNA (lanes 2–6), or CD82 plus CD4 cDNAs (lanes 7–11) were radiolabeled, chased, and analyzed by immunoprecipitation using an anti-CD82 antibody. (H) CD4 can be partially stabilized by CD82. COS-1 cells transfected with pcDNA3 (lane 1), CD4 cDNA (lanes 2–6), or CD4 plus CD82 cDNAs (lanes 7–11) were radiolabeled, chased, and analyzed by immunoprecipitation using the anti-CD4 antibody. (I) The bands in G and H were quantified. Data from three independent experiments were presented (mean ± SD).
DISCUSSION
Using a panel of conformation-dependent antibodies, we demonstrated that major conformational changes occur during multiple steps of urothelial plaque (AUM) assembly and that secondary modifications of uroplakin subunits are likely to play roles in regulating AUM assembly. We present a model demonstrating the intermediate steps and the possible regulations of AUM assembly (Figure 7). Our data on uroplakins (Figures 1, 2, 3, 4, 5), coupled with our studies on three other tetraspanins (Figure 6), revealed a pivotal function of tetraspanin family proteins as “maturation-facilitators” that can promote the ER-exit, stabilization, and surface expression of their partner proteins.
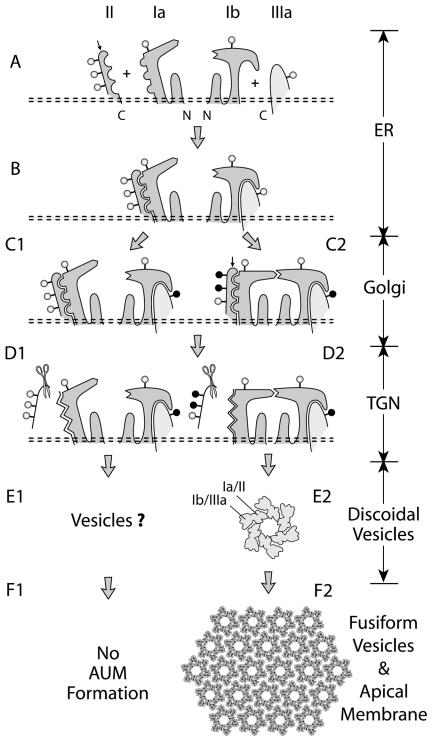
Figure 7.
A model depicting how the four major uroplakins (UPIa, Ib, II, and IIIa) assemble into 2D crystals of urothelial particles and how this process may be regulated. Symbols and abbreviations: open and closed circles (high-mannose and complex glycans); small arrows on UPII (the furin-cutting site at the end of the prosequence); scissors (furin); ER (endoplasmic reticulum); and TGN (trans-Golgi network). The four uroplakins acquire high-mannose glycans and form two heterodimers (UPIa/II and UPIb/IIIa) in the ER (stages A and B). Major conformational changes occur during dimerization (not depicted) leading to a presumably more stable protein complex that can exit from the ER. In normal urothelium, the glycans on two of the three N-glycosylation sites on the prosequence of UPII become complex glycans in the Golgi apparatus (C2); we hypothesize that this can cause conformational changes of Pro-UPII and its partner UPIa thus allowing heterotetramer formation (C2). Furin-mediated removal of the prosequence in the TGN (D2) then triggers oligomerization to form a 16-nm particle (E2) in which UPIa/II and UPIb/IIIa are associated with the inner and outer subdomains of a dumbbell-shaped subunit, respectively (Min et al., 2002), and to later form 2D crystals (F2). In cultured BU cells, the differentiation-dependent glycosylation of pro-UPII does not occur, thus hampering the formation of the uroplakin heterotetramer and the 16-nm particle. For details see the text.
The Enigmatic Nature of the AE31 Antigen
Although AE31 antibody played a key role in enabling us to initially identify and isolate uroplakins (Yu et al., 1990), the precise nature of the AE31 antigen has remained unclear for more than a decade. The 27- to 32-kDa urothelial membrane protein recognized by the AE31 mAb, previously known as “uroplakin I” (Yu et al., 1990), has several puzzling properties, which can now be understood in light of our current data. 1) Although the N-terminal sequence of the AE31 antigen was identical to that of pro-UPII (Figure 1B; sequences 1 and 2), it was puzzling that this antigen was not recognized by antibodies against four separate UPII epitopes (Figure 2B). This can now be explained by the prosequence-induced conformational changes of UPII (Figure 7, C and D). 2) Previous pulse-chase experiments using cultured BU cells yielded data, suggesting that the AE31 antigen was synthesized as a 32-kDa precursor that was processed, via a 30-kDa intermediate, to a mature 27-kDa uroplakin I, which in turn coimmunoprecipitated the 15-kDa mature UPII (Yu et al., 1990). Our current data have made it clear that the previously described 32-, 30-, and 15-kDa bands are actually prepro-UPII, pro-UPII, and mature UPII, respectively, and that the 27-kDa species is actually the coimmunoprecipitated, mature UPIa (cf. Figure 3F). 3) The AE31 antigen of cultured BU cells (29-kDa; Figure 1A, lane 7) is smaller than that of the mature bovine AUM (32-kDa; Figure 1A, lane 6). This can now be explained by the differentiation-dependent glycosylation of the prosequence (see below).
Conformational Changes in Tetraspanins and Their Associated Proteins
The use of a large panel of antibodies to multiple epitopes of uroplakins enabled us to study the accessibility of these epitopes in individual uroplakins, most likely as a function of protein folding/conformation, in multiple steps of AUM assembly. For example, tetraspanin UPIa, once bound to its partner pro-UPII (forming the UPIa/pro-UPII complex), becomes much less available for antibody recognition (compare the band intensities in lanes 9–13 of Figure 3, A vs. C). The fact that two separate anti-UPIa antibodies pulled down only UPIa without immunoprecipitating the UPIa/pro-UPII complex (Figure 3C, lanes 9–13) suggests masking or conformational changes of UPIa epitopes in the complex. Similar conformational changes were detected in UPIIIa, which almost completely loses its immunoreactivities with five independent antibodies when it binds with tetraspanin UPIb (Figure 7, A and B); however, UPIIIa can be recognized by all these antibodies when the UPIb/IIIa dimer is loosened by increasing the SDS concentration from 0.1 to 0.5% (Figure 4D). The fact that multiple UPIIIa epitopes become unavailable upon UPIb binding argues against the masking of only a small portion of the UPIIIa polypeptide by UPIb; rather, it supports the idea of global conformational changes of UPIIIa. Perhaps most striking is the complete switch in the immunoreactivities of UPII when pro-UPII is proteolytically processed to become the mature UPII (Figure 2, B and C). These findings are significant in two accounts. 1) This type of conformational changes, if unrecognized, can lead to misinterpretation of data generated using certain antibodies to tetraspanins and their associated proteins. 2) It is possible that these conformational changes can “lock” the membrane protein complex into an energetically more favorable and stable configuration and/or can expose new binding sites for the complex to further oligomerize.
Heterodimer Formation
Our cotransfection studies showed that UPIa and UPIb bind to pro-UPII and UPIIIa, respectively, forming two heterodimers, UPIa/pro-UPII and UPIb/UPIIIa, which can then exit from the ER (Figures 3, 4, and 7B; Tu et al., 2002). The ER-trapped, singly expressed pro-UPII and UPIIIa are degraded via the proteosome pathway (Figures 3 and 4); the unpaired UPIa and UPIb are, however, more stable than their partners. The trapped pro-UPII undergoes aggregation, which can be blocked by its correct partner UPIa, but not by UPIb or UPIIIa (Figure 2C). Genetic ablation of mouse UPII gene results in the ER-trapping of UPIa, whereas the remaining UPIb/UPIIIa pair is still targeted to the apical surface although its amount is vastly reduced and no 2D crystal is formed (Kong et al., 2004). Taken together, these data established the heterodimerization of UPIa/pro-UPII and UPIb/UPIIIa as an initial step of AUM assembly (Figure 7B) and suggested that uroplakin heterodimers are stabilized upon heterotetramer and/or particle formation (Figure 7, C2 to F2).
Differentiation-dependent Glycosylation of Pro-UPII and Possible Regulation of Heterotetramer Formation
The prosequence of UPII has three potential N-glycosylation sites (Lin et al., 1994), and the glycosylation reaction appears to be differentiation-dependent (Figures 1A and 5B). All three sites of the 29-kDa, Sarkosyl-soluble pro-UPII from cultured BU cells harbor high-mannose glycans; however, two of the three sites of the 32-kDa pro-UPII from purified, Sarkosyl-insoluble AUM harbor complex glycans (Figure 5). The fact that cultured BU cells lack the AUM-associated 32-kDa pro-UPII is interesting, because such cells, although synthesizing all four uroplakins, lack fusiform vesicles and fail to assemble plaques (Surya et al., 1990). This raises the possibility that the addition of complex glycans to the prosequence of pro-UPII plays a role in uroplakin assembly. In this regard, it is important to note that glycosylation can affect protein conformation (Imperiali and O'Connor, 1999; Helenius and Aebi, 2004) and that the expression and subcellular localization of some glycosyltransferases are known to be differentiation-dependent (Berger, 2002). We therefore hypothesize that a special, complex glycan-forming glycosyltransferase in superficial umbrella cells may be involved in producing the AUM-associated 32-kDa pro-UPII, which can then promote the binding between two heterodimers to form a heterotetramer (Figure 7C2). Because the extracellular large loop of tetraspanin CD81 can mediate homo- and heterodimer formation through its conserved stalk domains (Kitadokoro et al., 2001), it is possible that the heterotetramerization of the two uroplakin pairs (UPIa/pro-UPII and UPIb/UPIIIa) is mediated through UPIa:UPIb binding (Figure 7C2).
According to this hypothesis, the failure of the cultured BU cells to form the properly glycosylated 32-kDa pro-UPII (Figure 5B, lane 7) blocks heterotetramerization (Figure 7C1). Consistent with this, we can readily detect heterodimers in cultured BU cells by coimmunoprecipitation (Figures 3 and 4); however, we have not been able to detect heterotetramers. Because the 29-kDa pro-UPII is also produced by in vivo bovine urothelium (Figure 5, B and C), this protein is clearly not an artifact of cultured cells. In bovine urothelium, the superficial umbrella cells exhibit numerous discoidal and fusiform vesicles that deliver urothelial plaques to the apical surface. In contrast, cells located below the umbrella cells are less differentiated in that they contain much less uroplakins, exhibit almost no discoidal or fusiform vesicles, and lack cell surface-associated plaques (Surya et al., 1990; Wu et al., 1990; Yu et al., 1990; Kachar et al., 1999); these features are similar to those of cultured BU cells (Surya et al., 1990). It is possible that these relatively undifferentiated intermediate cells, like cultured cells, produce the 29-kDa pro-UPII (Figure 5B, lanes 4 and 7), which prevents uroplakins from assembling precociously into particles and plaques.
UPII Prosequence and Plaque Assembly
Our data indicate that antibodies to four distinct epitopes of the mature UPII (Figure 2A) fail to recognize pro-UPII (Figure 2B), and that the furin-mediated removal of the prosequence leads to the unmasking of these epitopes (Figure 2, B and D–F). Given the fact that these four epitopes are located throughout the mature sequence (Figure 2A), it seems unlikely that they are all “masked” by the prosequence per se; rather, our data favor a global conformational change. Although the function of the UPII prosequence is unknown, there are several examples in which the furin-mediated removal of a prosequence leads to protein oligomerization; these include Clostridium septicum α-toxin (Ballard et al., 1993), anthrax toxin (Petosa et al., 1997; Sellman et al., 2001), and proaerolysin (Abrami et al., 1998). It is therefore possible that the furin-mediated removal of the UPII prosequence can facilitate the oligomerization of uroplakin heterotetramers (each constitutes one of the 6 dumbbell-shaped “subunits” in the 16-nm particle; Figure 7E2) to form the 16-nm urothelial particles. These particles further assemble into small 2D crystals in discoidal vesicles and later into large crystals in fusiform vesicles (Figure 7F2).
Mechanism by Which Tetraspanin CDs Promote the Surface Expression of Their Partner Proteins
Although emerging data indicate that tetraspanins can associate with other tetraspanins, growth factor receptors, integrins, and other membrane proteins (Boucheix and Rubinstein, 2001; Hemler, 2001, 2003), relatively little is known about the subcellular compartments in which these interactions occur and the effects of such interactions on the structure and processing of their interacting partners. Our data indicate that tetraspanins UPIa and UPIb are required for the ER-exit, processing, stabilization, and cell surface presentation of their respective partner proteins, UPII and UPIIIa (Figures 2, 3, 4; Tu et al., 2002). In addition, we showed that tetraspanins CD9, CD81, and CD82 can stabilize their partner CD4 (Figure 6); most likely this stabilization reflects CD9, CD81, and CD82's abilities to facilitate CD4's escape from the ER-associated degradation. Unlike uroplakins where the “magnitude” of interdependence between the tetraspanins (UPIa and Ib) and their single-transmembrane-domained associated proteins (UPII and UPIIIa) is practically absolute, the magnitude of stabilization between nonuroplakin tetraspanin CDs and their partners is relatively small. Possible reasons for this include: 1) Unlike uroplakins Ia and Ib, which interact selectively with UPII and UPIIIa, respectively, forming highly specific heterodimers, tetraspanin CDs can form complexes promiscuously with multiple partners; thus the stabilization effects within a given “pair” may be smaller. But this may be compensated by the fact that such partners can interact with, and can be stabilized by, multiple tetraspanins. 2) Although we have carefully selected the cell lines for transfection, these cells do contain some endogenous tetraspanin CDs and their associated proteins, which may reduce the observed degree of stabilization.
Taken together, our data suggest that tetraspanins serve the novel function as “maturation-facilitators” to enhance the ER-exit and surface presentation of their partner proteins. This model sheds light on some existing data. For example, Shi et al. (2000) showed that tetraspanin CD9 interacts with, and suppresses the proteolytic maturation of, the transmembrane form of TGF-α (pro-TGF-α) and leads to the accumulation and surface expression of pro-TGF-α; the mechanism of pro-TGF-α stabilization was, however, unclear. Our model raises the possibility that, in this system, tetraspanin CD9 facilitates the processing of pro-TGF-α, resulting in its stabilization. Shoham et al. (2003) showed that although the ablation of tetraspanin CD81 gene has no effect on the mRNA level of CD19 (a single-transmembrane-domained, CD81-binding protein involved in B-cell receptor signaling), it diminishes the protein level and surface expression of CD19. Based on the finding that CD81 deficiency leads to an increase in the amounts of the endo H-sensitive form of CD19 (and a corresponding decrease in the more mature, endo H-resistant form), it was suggested that CD81 regulates the expression of CD19 in an undefined post-ER compartment. Our model implies that this regulation occurs in the ER, instead of post-ER. Finally, Stipp et al. (2003a) studied the interactions between tetraspanin CD81 and its partner EWI-2 (an immunoglobulin superfamily member with a single-transmembrane domain) and found that, in the absence of CD81, EWI-2 is somewhat decreased in quantity, is shifted to a slightly lower size most likely representing an immature, underglycosylated form, and fails to reach cell surface. Based on these data, it was concluded that CD81 controlled EWI-2 maturation and cell surface localization. Such results are totally consistent with our findings and again suggest that tetraspanin CD81 facilitates the ER-exit of EWI-2. Overall, the results from the above studies are in concert with ours (Table 1) and support the general idea that tetraspanins not only serve as “molecular facilitators” to recruit many cell surface-associated signaling molecules to specialized membrane subdomains, as having been suggested earlier (Maecker et al., 1997; Boucheix and Rubinstein, 2001; Hemler, 2003), but also function as “maturation-facilitators.”
Table 1.
Facilitated processing and/or stabilization of their single-transmembrane-domained partners by tetraspanins
Acknowledgments
We thank Eric Rubinstein (INSERM U268, Villejuif, France) for antibodies and cDNAs of CD9 and CD81; Xin Zhang (University of Tennessee Health Science Center) for human CD82 cDNA; Dan Littman (NYU School of Medicine) for human CD4 cDNA; and Joseph Sucic (University of Michigan, Flint) for the 7.P15 and RPE.40 cells. We are indebted to Fang-Ming Deng, Xue-Ru Wu, Xiang-Peng Kong, Jan Sap, Richard Margolis, Stevan Hubbard, and Moosa Mohammadi for useful discussions; Cristina Villagra for her assistance in densitometry; Gordon Cook for the illustration; and Irwin Freedberg and Herbert Lepor for their support. These studies were supported by National Institutes of Health Grants DK39753, DK52206, DK66491, and P30 CA16087.
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05–02–0136) on June 15, 2005.
References
- Abrami, L., Fivaz, M., Decroly, E., Seidah, N. G., Jean, F., Thomas, G., Leppla, S. H., Buckley, J. T., and van der Goot, F. G. (1998). The pore-forming toxin proaerolysin is activated by furin. J. Biol. Chem. 273, 32656–32661. [DOI] [PubMed] [Google Scholar]
- Adachi, W., Okubo, K., and Kinoshita, S. (2000). Human uroplakin Ib in ocular surface epithelium. Invest. Ophthalmol. Vis. Sci. 41, 2900–2905. [PubMed] [Google Scholar]
- Apodaca, G. (2004). The uroepithelium: not just a passive barrier. Traffic 5, 117–128. [DOI] [PubMed] [Google Scholar]
- Ballard, J., Sokolov, Y., Yuan, W. L., Kagan, B. L., and Tweten, R. K. (1993). Activation and mechanism of Clostridium septicum alpha toxin. Mol. Microbiol. 10, 627–634. [DOI] [PubMed] [Google Scholar]
- Berger, E. G. (2002). Ectopic localizations of Golgi glycosyltransferases. Glycobiology 12, 29R–36R. [DOI] [PubMed] [Google Scholar]
- Boucheix, C., and Rubinstein, E. (2001). Tetraspanins. Cell Mol. Life Sci. 58, 1189–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brisson, A., and Wade, R. H. (1983). Three-dimensional structure of luminal plasma membrane protein from urinary bladder. J. Mol. Biol. 166, 21–36. [DOI] [PubMed] [Google Scholar]
- Helenius, A., and Aebi, M. (2004). Roles of N-linked glycans in the endoplasmic reticulum. Annu. Rev. Biochem. 73, 1019–1049. [DOI] [PubMed] [Google Scholar]
- Hemler, M. E. (2001). Specific tetraspanin functions. J. Cell Biol. 155, 1103–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemler, M. E. (2003). Tetraspanin proteins mediate cellular penetration, invasion, and fusion events and define a novel type of membrane microdomain. Annu. Rev. Cell Dev. Biol. 19, 397–422. [DOI] [PubMed] [Google Scholar]
- Hicks, R. M., and Ketterer, B. (1969). Hexagonal lattice of subunits in the thick luminal membrane of the rat urinary bladder. Nature 224, 1304–1305. [DOI] [PubMed] [Google Scholar]
- Hirsch, C., Blom, D., and Ploegh, H. L. (2003). A role for N-glycanase in the cytosolic turnover of glycoproteins. EMBO J. 22, 1036–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, P., Deng, F. M., Liang, F. X., Hu, C. M., Auerbach, A. B., Shapiro, E., Wu, X. R., Kachar, B., and Sun, T. T. (2000). Ablation of uroplakin III gene results in small urothelial plaques, urothelial leakage, and vesicoureteral reflux. J. Cell Biol. 151, 961–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, P., Meyers, S., Liang, F. X., Deng, F. M., Kachar, B., Zeidel, M. L., and Sun, T. T. (2002). Role of membrane proteins in permeability barrier function: uroplakin ablation elevates urothelial permeability. Am. J. Physiol. Renal. Physiol. 283, F1200–F1207. [DOI] [PubMed] [Google Scholar]
- Imperiali, B., and O'Connor, S. E. (1999). Effect of N-linked glycosylation on glycopeptide and glycoprotein structure. Curr. Opin. Chem. Biol. 3, 643–649. [DOI] [PubMed] [Google Scholar]
- Inocencio, N. M., Sucic, J. F., Moehring, J. M., Spence, M. J., and Moehring, T. J. (1997). Endoprotease activities other than furin and PACE4 with a role in processing of HIV-I gp160 glycoproteins in chinese hamster ovary-K1 cells. J. Biol. Chem. 272, 1344–1348. [DOI] [PubMed] [Google Scholar]
- Kachar, B., Liang, F., Lins, U., Ding, M., Wu, X. R., Stoffler, D., Aebi, U., and Sun, T.-T. (1999). Three-dimensional analysis of the 16 nm urothelial plaque particle: luminal surface exposure, preferential head-to-head interaction, and hinge formation. J. Mol. Biol. 285, 595–608. [DOI] [PubMed] [Google Scholar]
- Kitadokoro, K., Bordo, D., Galli, G., Petracca, R., Falugi, F., Abrignani, S., Grandi, G., and Bolognesi, M. (2001). CD81 extracellular domain 3D structure: insight into the tetraspanin superfamily structural motifs. EMBO J. 20, 12–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong, X. T. et al. (2004). Roles of uroplakins in plaque formation, umbrella cell enlargement and urinary tract diseases. J. Cell Biol. 167, 1195–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis, S. A. (2000). Everything you wanted to know about the bladder epithelium but were afraid to ask. Am. J. Physiol. Renal. Physiol. 278, F867–F874. [DOI] [PubMed] [Google Scholar]
- Liang, F., Kachar, B., Ding, M., Zhai, Z., Wu, X. R., and Sun, T.-T. (1999). Urothelial hinge as a highly specialized membrane: detergent-insolubility, urohingin association, and in vitro formation. Differentiation 65, 59–69. [DOI] [PubMed] [Google Scholar]
- Liang, F. X., Riedel, I., Deng, F. M., Zhou, G., Xu, C., Wu, X. R., Kong, X. P., Moll, R., and Sun, T. T. (2001). Organization of uroplakin subunits: transmembrane topology, pair formation and plaque composition. Biochem. J. 355, 13–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, J. H., Wu, X. R., Kreibich, G., and Sun, T. T. (1994). Precursor sequence, processing, and urothelium-specific expression of a major 15-kDa protein subunit of asymmetric unit membrane. J. Biol. Chem. 269, 1775–1784. [PubMed] [Google Scholar]
- Maecker, H. T., Todd, S. C., and Levy, S. (1997). The tetraspanin superfamily: molecular facilitators. FASEB J. 11, 428–442. [PubMed] [Google Scholar]
- Min, G., Stolz, M., Zhou, G., Liang, F., Sebbel, P., Stoffler, D., Glockshuber, R., Sun, T. T., Aebi, U., and Kong, X. P. (2002). Localization of uroplakin Ia, the urothelial receptor for bacterial adhesin FimH, on the six inner domains of the 16 nm urothelial plaque particle. J. Mol. Biol. 317, 697–706. [DOI] [PubMed] [Google Scholar]
- Min, G., Zhou, G., Schapira, M., Sun, T. T., and Kong, X. P. (2003). Structural basis of urothelial permeability barrier function as revealed by cryo-EM studies of the 16 nm uroplakin particle. J. Cell Sci. 116, 4087–4094. [DOI] [PubMed] [Google Scholar]
- Negrete, H. O., Lavelle, J. P., Berg, J., Lewis, S. A., and Zeidel, M. L. (1996). Permeability properties of the intact mammalian bladder epithelium. Am. J. Physiol. 271, F886–F894. [DOI] [PubMed] [Google Scholar]
- Oostergetel, G. T., Keegstra, W., and Brisson, A. (2001). Structure of the major membrane protein complex from urinary bladder epithelial cells by cryoelectron crystallography. J. Mol. Biol. 314, 245–252. [DOI] [PubMed] [Google Scholar]
- Petosa, C., Collier, R. J., Klimpel, K. R., Leppla, S. H., and Liddington, R. C. (1997). Crystal structure of the anthrax toxin protective antigen. Nature 385, 833–838. [DOI] [PubMed] [Google Scholar]
- Sellman, B. R., Mourez, M., and Collier, R. J. (2001). Dominant-negative mutants of a toxin subunit: an approach to therapy of anthrax. Science 292, 695–697. [DOI] [PubMed] [Google Scholar]
- Shi, W., Fan, H., Shum, L., and Derynck, R. (2000). The tetraspanin CD9 associates with transmembrane TGF-alpha and regulates TGF-alpha-induced epidermal growth factor receptor activation and cell proliferation. J. Cell Biol. 148, 591–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoham, T., Rajapaksa, R., Boucheix, C., Rubinstein, E., Poe, J. C., Tedder, T. F., and Levy, S. (2003). The tetraspanin CD81 regulates the expression of CD19 during B cell development in a postendoplasmic reticulum compartment. J. Immunol. 171, 4062–4072. [DOI] [PubMed] [Google Scholar]
- Stieneke-Grober, A., Vey, M., Angliker, H., Shaw, E., Thomas, G., Roberts, C., Klenk, H. D., and Garten, W. (1992). Influenza virus hemagglutinin with multibasic cleavage site is activated by furin, a subtilisin-like endoprotease. EMBO J. 11, 2407–2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stipp, C. S., Kolesnikova, T. V., and Hemler, M. E. (2003a). EWI-2 regulates alpha3beta1 integrin-dependent cell functions on laminin-5. J. Cell Biol. 163, 1167–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stipp, C. S., Kolesnikova, T. V., and Hemler, M. E. (2003b). Functional domains in tetraspanin proteins. Trends Biochem. Sci. 28, 106–112. [DOI] [PubMed] [Google Scholar]
- Sun, T.-T., Liang, F. X., and Wu, X. R. (1999). Uroplakins as markers of urothelial differentiation. Adv. Exp. Med. Biol. 462, 7–18. [DOI] [PubMed] [Google Scholar]
- Surya, B., Yu, J., Manabe, M., and Sun, T. T. (1990). Assessing the differentiation state of cultured bovine urothelial cells: elevated synthesis of stratification-related K5 and K6 keratins and persistent expression of uroplakin I. J. Cell Sci. 97(Pt 3), 419–432. [DOI] [PubMed] [Google Scholar]
- Tarrant, J. M., Robb, L., van Spriel, A. B., and Wright, M. D. (2003). Tetraspanins: molecular organisers of the leukocyte surface. Trends Immunol. 24, 610–617. [DOI] [PubMed] [Google Scholar]
- Tu, L., Sun, T. T., and Kreibich, G. (2002). Specific heterodimer formation is a prerequisite for uroplakins to exit from the endoplasmic reticulum. Mol. Biol. Cell 13, 4221–4230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergara, J. A., Longley, W., and Robertson, J. D. (1969). A hexagonal arrangement of subunits in membrane of mouse urinary bladder. J. Mol. Biol. 46, 593–596. [DOI] [PubMed] [Google Scholar]
- Walz, T., Haner, M., Wu, X. R., Henn, C., Engel, A., Sun, T. T., and Aebi, U. (1995). Towards the molecular architecture of the asymmetric unit membrane of the mammalian urinary bladder epithelium: a closed “twisted ribbon” structure. J. Mol. Biol. 248, 887–900. [DOI] [PubMed] [Google Scholar]
- Wu, X. R., Lin, J. H., Walz, T., Haner, M., Yu, J., Aebi, U., and Sun, T. T. (1994). Mammalian uroplakins. A group of highly conserved urothelial differentiation-related membrane proteins. J. Biol. Chem. 269, 13716–13724. [PubMed] [Google Scholar]
- Wu, X. R., Manabe, M., Yu, J., and Sun, T. T. (1990). Large scale purification and immunolocalization of bovine uroplakins I, II, and III. Molecular markers of urothelial differentiation. J. Biol. Chem. 265, 19170–19179. [PubMed] [Google Scholar]
- Wu, X. R., Medina, J. J., and Sun, T. T. (1995). Selective interactions of UPIa and UPIb, two members of the transmembrane 4 superfamily, with distinct single transmembrane-domained proteins in differentiated urothelial cells. J. Biol. Chem. 270, 29752–29759. [DOI] [PubMed] [Google Scholar]
- Wu, X. R., and Sun, T.-T. (1993). Molecular cloning of a 47 kDa tissue-specific and differentiation-dependent urothelial cell surface glycoprotein. J. Cell Sci. 106, 31–43. [DOI] [PubMed] [Google Scholar]
- Xiong, X., Chong, E., and Skach, W. R. (1999). Evidence that endoplasmic reticulum (ER)-associated degradation of cystic fibrosis transmembrane conductance regulator is linked to retrograde translocation from the ER membrane. J. Biol. Chem. 274, 2616–2624. [DOI] [PubMed] [Google Scholar]
- Yu, J., Lin, J. H., Wu, X. R., and Sun, T. T. (1994). Uroplakins Ia and Ib, two major differentiation products of bladder epithelium, belong to a family of four transmembrane domain (4TM) proteins. J. Cell Biol. 125, 171–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, J., Manabe, M., Wu, X. R., Xu, C., Surya, B., and Sun, T. T. (1990). Uroplakin I: a 27-kD protein associated with the asymmetric unit membrane of mammalian urothelium. J. Cell Biol. 111, 1207–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]