Abstract
The “Dsl1p complex” in Saccharomyces cerevisiae, consisting of Dsl1p and Tip20p, is involved in Golgi-ER retrograde transport and it is functionally conserved from yeast to mammalian cells. To further characterize this complex, we analyzed the function of Dsl3p, a protein that interacts with Dsl1p in yeast two hybrids screens. DSL3, recently identified in a genome wide analysis of essential genes as SEC39, encodes a cytosolic protein of 82 kDa that is peripherally associated with membranes derived from the ER. There is strong genetic interaction between DSL3 and other factors required for Golgi-ER retrograde transport. Size exclusion chromatography and affinity purification approaches confirmed that Dsl3p is associated with subunits of the “Dsl1p complex.” The complex also includes the Q/t-SNARE proteins, Use1p, Sec20p, and Ufe1p, integral membrane proteins that constitute the trimeric acceptor for R/v-SNAREs on Golgi-derived vesicles at the ER. Using mutants, we performed a detailed analysis of interactions between subunits of the Dsl1p complex and the ER-localized SNARE proteins. This analysis showed that both Dsl1p and Dsl3p are required for the stable interaction of the SNARE Use1p with a central subcomplex consisting of Tip20p and the SNARE proteins Ufe1p and Sec20p.
INTRODUCTION
The protein trafficking pathway in the yeast S. cerevisiae is composed of several distinct membrane-bounded compartments, which interact via a bidirectional flow of membrane-bounded vesicles in a process referred to as vesicular transport (Kaiser and Schekman, 1990; Rothman and Orci, 1992; Ferro-Novick and Jahn, 1994; Rothman, 1994; Waters and Hughson, 2000). Briefly, secretory proteins destined for transport must be sorted away from resident proteins, packaged into the proper cargo vesicles, and subsequently, delivered to the correct target membrane (Palade, 1975). Each step of this process must be tightly regulated to ensure efficient secretion and maintenance of the distinct cellular compartments. Transport in the retrograde direction ensures further rounds of anterograde transport by recycling components of the transport machinery, recovering wayward proteins and maintaining the balance of lipids between the distinct compartments of the pathway.
The cell makes use of a variety of coated vesicles to transport proteins, whose formation is nucleated by the action of small GTP-binding proteins. Specifically, vesicle budding in the retrograde direction from the Golgi to the ER involves a heptameric coat protein complex called COPI (Waters et al., 1991; Stenbeck et al., 1993; Letourneur et al., 1994; Barlowe, 2000). The COPI coat consists of coatomer, an ∼700–800 kDa protein complex comprised of an equimolar assembly of α-, β-, β'-, γ-, δ-, ε-, and ζ-COP (Cop1[Ret1]p, Sec26p, Sec27p, Sec21p, Ret2p, Sec28p, and Ret3p, respectively, in yeast) and a small ras-like GTPase termed Arf (encoded by ARF1 or ARF2).
Current models of vesicular transport propose that the vesicle coat is removed soon after vesicles are formed (although this has not been directly demonstrated) thereby preparing them for fusion with their specific target membrane (Rothman, 1994). The first contact between vesicle and target membrane is mediated by tethering factors (Barlowe, 1997; Cao et al., 1998). Currently, up to seven tethering factors, either long extended molecules or huge multi subunit complexes, have been identified in yeast that function in the various transport steps (Whyte and Munro, 2002).
Once tethered and uncoated, the vesicle proceeds to the docking stage, which involves SNAREs (for soluble NSF (N-ethylmaleimide-sensitive fusion protein) attachment protein receptor), most of them tail-anchored membrane proteins that reside on the vesicle (v-SNARE) or on the target membrane (t-SNARE) (Söllner et al., 1993a; Söllner et al., 1993b). The formation of trans-SNARE complexes involves the parallel alignment of four helices derived from either three or four different SNARE molecules (Hanson et al., 1997; Sutton et al., 1998; Weber et al., 1998; Antonin et al., 2002). The zippering of the four membrane proximal SNARE motifs results in close apposition of the vesicle and target membranes, perhaps providing the means by which SNAREs directly mediate membrane fusion in vitro. SNAREs implicated in the fusion of Golgi derived vesicles with the ER membrane are Ufe1p, Use1p, Sec20p and Sec22p (Sweet and Pelham, 1992; Lewis and Pelham, 1996; Cosson et al., 1997; Belgareh-Touze et al., 2003; Burri et al., 2003; Dilcher et al., 2003). During trans-SNARE pairing, these four SNARE proteins would form a four helix bundle whose central polar layer is made up by one arginine residue derived from Sec22p, two glutamine residues (Ufe1p, Sec20p) and one aspartate (Use1p). Aspartate can replace the glutamine in some SNARE proteins (Dilcher et al., 2003). Therefore, we will refer to Use1p as a Q-SNARE. Thus, the ER SNARE complex would conform to the rule that a SNARE complex is composed one R/v-SNARE and three Q/t-SNAREs (Fasshauer et al., 1998; Sutton et al., 1998; Antonin et al., 2002).
Two proteins have been described that may act upstream of, or in conjunction with, the retrograde ER-localized SNAREs. Tip20p, which is an essential ∼81-kDa peripheral membrane protein that interacts with the cytosolic domain of Sec20p and is involved in Golgi-ER transport (Sweet and Pelham, 1993; Cosson et al., 1997). The second protein is Dsl1p, which was found to be essential for retrograde traffic from the Golgi to the ER. Dsl1p is an ER-localized peripheral membrane protein that was shown to interact with Tip20p and Sec20p (Andag et al., 2001; Reilly et al., 2001; VanRheenen et al., 2001; Andag and Schmitt, 2003). The complex consisting of Dsl1p, Tip20p, and Sec20p was referred to as the “Dsl1p complex” (Whyte and Munro, 2002; Ungar and Hughson, 2003). Two mammalian proteins that share sequence similarity with Dsl1p and Tip20p (ZW10 and RINT-1) were recently shown to function in ER-Golgi transport (Hirose et al., 2004). Dsl1p and its homologues from fungi may be unique because they carry a central domain, which mediates the interaction with different coatomer subunits (Andag et al., 2001; Reilly et al., 2001; Andag and Schmitt, 2003). Further, we demonstrated that the binding site for Tip20p is at the N-terminus and does not overlap with the binding site for coatomer, suggesting that a simultaneous interaction of Dsl1p, Tip20p, and coatomer could exist (Andag and Schmitt, 2003). This Dsl1p-COPI-Tip20 interaction might help to either tether the properly coated vesicle to the ER, uncoat the vesicle at the appropriate ER docking site, or both.
The approximate mass of the Dsl1p complex is significantly larger than the expected size of the known components, suggesting that other proteins are likely to be present. We therefore searched for additional subunits by utilizing the two-hybrid approach. This method led to the identification of a novel gene, DSL3 (YLR440c) that was identified in a two-hybrid screen with Dsl1p. While this work was in progress Mnaimneh et al. (2004) identified DSL3 as a gene whose product is required for ER-Golgi transport in a genome-wide screen of essential genes using TET-promoter shut-down and named it SEC39. The product of this gene has been localized to the ER (Huh et al., 2003).
We report here that Dsl3p is a novel and stable component of the Dsl1p complex. In an effort to further characterize the Dsl1p complex, we analyzed the effect of mutations in each subunit to define the interactions between components of the Dsl1p ER targeting complex and the ER-localized Q/t-SNARE proteins Use1p, Ufe1p, and Sec20p. Our results show that the Dsl1p complex is required for the stability of the ER-localized SNARE complex, and we discuss implications for its function in ER-Golgi retrograde traffic.
MATERIALS AND METHODS
Media, Strains, Plasmids, and Antibodies
Strains produced by the Saccharomyces Genome Deletion Project (SGDP; Winzeler et al., 1999) were purchased from ResGen (Invitrogen, Carlsbad, CA) and EUROSCARF (Frankfurt, Germany). TAP-tagged strains (Ghaemmaghami et al., 2003) were obtained from Open Biosystems (Huntsville, AL). The diploid strain ylr440cΔ/YLR440c (strain 26044) in the BY4743 background was transformed with pBR11 (a LEU2 CEN plasmid containing DSL3) and then sporulated and dissected in order to obtain the ylr440cΔ haploid strain, GWY600. GWY600A was generated by transforming GWY600 with a 3.3-kb XhoI-EcoRI digest of pMR4266 (trp::HIS3+). To obtain dsl3 mutant plasmids used in this study, DSL3 (YLR440c) was amplified by PCR from pBR11 using primers Bam440 (5′-CGG-GAT-CCG-CAG-AGA-GTA-TTG-ATA-GGT-3′) and Bam440–2 (5′-CTG-TCC-TAA-ATC-CCG-AAT-3′) that placed BamHI sites at each end of the fragment. The PCR product was then digested with BamHI and ligated into a similarly digested plasmid, pRS414 (a TRP1 CEN plasmid) resulting in plasmid pBAK60. BamHI was used for subcloning DSL3 and mutant alleles into vectors pRS313 (CEN, HIS3) and pRS315 (CEN, LEU2; Sikorski and Hieter, 1989). Temperature-sensitive alleles of DSL3 were constructed by PCR mutagenesis (Leung et al., 1989) and selected directly in yeast (Muhlrad et al., 1992) as follows. DSL3 was amplified with TaqDNA polymerase from plasmid pBAK60 using primers Bam440 and Bam440–2 (see above) in the presence of 0.65 mM manganese chloride and lowered levels of dATP (0.2 mM dATP, 1 mM dGTP, 1 mM dCTP, and 1 mM dTTP). pBAK60 was digested with BspEI and NdeI, and the fragment containing the 5′ and 3′ ends of the DSL3 locus (lacking the internal 1.8-kb BspEI/NdeI fragment) was subsequently purified generating a gapped plasmid. The gapped plasmid was isolated and cotransformed with the mutagenized PCR product into strain GWY600A (dsl3Δ + pBR11). After selection on SC lacking tryptophan (SC-TRP) plates at 23°C, transformants were streaked onto 5-FOA to select for cells that had lost the nonmutagenized copy of DSL3 on pBR11. The 5-FOA resistant colonies were streaked onto three YPD plates, which were incubated at 23, 30, and 37°C. Two colonies were identified that were able to grow at 23 and 30°C but not at 37°C. Plasmids bearing the temperature-sensitive alleles of DSL3 were isolated from yeast and retransformed into GWY600A to confirm plasmid linkage to the temperature-sensitive phenotype after removal of the URA3 plasmid harboring wild-type DSL3 by counterselection on 5-FOA. These plasmids were named pBAK4B and pBAK2F. Plasmid pAC5 was constructed by amplification of the complete DSL3 ORF by PCR placing the Bam-HI site upstream of the start codon and the KpnI site downstream of the termination codon with the use of primers 5BamYLR4 (5′-CGC-GGA-TCC-GCG-TTG-GAA-GAG-CAA-CTA-TAT-TTG-TTA-GCT-3′) and 3KpnYLR4(5′-CGG-GGT-ACC-CCG-trichloroacetic acid-GGA-ATT-AGT-CAC-GTT-GTG-GAA-AAT-CTC-3′). The PCR product was then digested with BamHI and KpnI and ligated to a similarly digested vector, pGEM-T easy (Promega, Madison, WI). To generate the plasmid expressing His6-tagged Dsl3p pAC5 was digested with BamHI and KpnI, and the liberated fragment was ligated to a similarly digested pQE30 vector (Qiagen, Santa Clarita, CA) and named pAC7. The plasmid used to express GST-Dsl3p, pAC13, was generated by digesting pAC8 (same construction as pAC5, with the exception of the orientation of the fragment) with BamHI and SalI and ligating the liberated fragment with similarly digested pGEX 4T-1 vector; Amersham Biosciences, Piscataway, NJ).
His6-Dsl3p was expressed from plasmid pAC7, purified according to standard procedure under denaturing conditions (Qiagen) and used to inoculate rabbits by standard procedures (Harlow and Lane, 1988). His6-Dsl3p was also covalently linked to cyanogen bromide-activated Sepharose according to manufacturer's instructions; Amersham Biosciences). The resulting His6-Dsl3p-conjugated Sepharose was used to affinity-purify antibodies against Dsl3p by standard procedures (Harlow and Lane, 1988). GST-Dsl3p was expressed from pAC13 plasmid and purified according to instructions by the manufacturer. The GST-Dsl3p was used to further affinity purify the antibody on nitrocellulose strips according to Smith and Fisher (1984).
A deletion ranging from codon 8 to 317 of DSL1 was created by inserting a pair of oligonucleotides encoding one c-myc epitope and fitting into EcoNI-SpeI-cleaved pRS313-DSL1 (HIS3, CEN/ARS). The SpeI site in the polylinker of the pRS313 vector had been removed before. To check the effect of this deletion in strains expressing TAP-tagged strains (His+), we transferred the mutant allele dsl1-ΔN (using flanking XhoI-BamHI sites) into the LEU2 vector pRS315. To modify DSL1 at its chromosomal site an XhoI-EaeI fragment containing the truncated N-terminus of DSL1 (starting at 386 nt upstream of the ATG to 647 downstream of the insertion/deletion site) into the integrating vector pRS305. This plasmid was cut at a unique NsiI site to direct its integration into the DSL1 gene or TAP-tagged version of DSL1. The transformants express a C-terminally truncated as well as an N-terminally truncated DSL1 gene either with or without TAP-tag (strain D1-ΔCΔN-T and D1-ΔCΔN). Plasmids used in this work are listed in Supplementary Table 2.
Synthetic Lethality
GWY601 (MATα dsl3-1) was mated with BY4741, then sporulated and dissected to obtain GWY602. GWY602 (MATa, dsl3-1) was then mated with RSY275 (sec20-1). The resulting diploids were selected for on SD –His –Leu plates and then sporulated and dissected as described previously (Reilly et al., 2001). Segregants of each tetrad were spotted onto YPD, and the plates were incubated at 23, 30, or 35°C for 2 d. Putative tetratypes were identified as sets of tetrads displaying 1:3 viable:inviable ratio at 35°C, as sec20-1 is inviable at 33°C and dsl3-1 is inviable at 35°C. Spore-derived colonies that exhibit G-418 resistance are dsl3Δ bearing dsl3-1 on a plasmid (pBAK4B TRP). Genotypes of all the spore-derived colonies were determined by complementation of temperature sensitivity with plasmids containing either DSL3 or SEC20 (pBR12 or pSTM20, respectively).
Analysis of BiP/Kar2p Secretion, Pulse-chase Experiments, and Sec22-α-factor Retention Assay
Cells were streaked onto YPD plates and grown over night at 25°C. These plates were replica-plated to get a uniform layer of cells. These patches of cells were overlaid with nitrocellulose prewetted in sterile water. After a 24-h incubation at 25 or 30°C, the nitrocellulose was removed and the levels of BiP/Kar2p were analyzed by immunoblotting with a 1:5000 dilution of an anti-BiP/Kar2p antibody and ECL PLUS detection (Amersham Biosicences, Freiburg, Germany). Pulse chase experiments were performed as described previously (Andag et al., 2001).
The retention of Sec22p in early secretory compartments was analyzed using a constitutively expressed Sec22-α-factor fusion protein expressed from vector pWBAcyc-α (Ballensiefen et al., 1998; Ossipov et al., 1999). Transformants were grown overnight in PM medium (2% glucose, 1.7 g/l yeast nitrogen base, 3.3 g/l ammonium sulfate, 0.5% casein hydrolysate, 20 mg/l adenine sulfate). Extracts were prepared as described previously and analyzed by Western blotting with anti-Sec22p, anti-c-myc antibodies, or anti-α-factor antibodies.
Subcellular Fractionation and Membrane Extraction
Subcellular fractionation and membrane extractions were performed as previously described (Reilly et al., 2001) except that supernatant and pellet fractions, derived from equivalent amounts of starting material, were resolved by SDS-12% PAGE and analyzed by immunoblotting with antibodies against Dsl1p, Sed5p, Dpm1p, and phosphoglycerate kinase (PGK).
Extractions were performed on Dsl3p-containing total yeast membranes that had been isolated on a step gradient, as described previously (Reilly et al., 2001). Samples were then resolved by SDS-10% PAGE and analyzed by immunoblotting with affinity-purified anti-Dsl3p and anti-Dpm1p antibodies.
Yeast Two-hybrid Screen
The yeast two-hybrid screen was performed as described by P. James (James et al., 1996) except that PJ69-4A yeast strain carried the bait plasmid pBR2 (Reilly et al., 2001), which encodes a Gal4p-DNA binding domain-Dsl1p fusion protein.
Affinity Purification of TAP-tagged Proteins
To analyze proteins bound to TAP-tagged proteins by Western blotting analysis (single step procedure; see Figure 5 and Supplementary Material, Supplementary Figures S3 and S5–S10) wild-type cells expressing TAP-tagged proteins (Ghaemmaghami et al., 2003) or mutants derived from these strains were lysed in HKET buffer (20 mM HEPES, pH 6.8, 150 mM potassium acetate, 2.5 mM EDTA, 1% Triton X-100, and complete protease inhibitor mixture; Roche Applied Science, Mannheim, Germany) using glass beads. After centrifugation at 30,000 × g for 10 min at 4°C, the supernatant was collected and protein concentration of the extract was determined using a protein assay (Bio-Rad, Richmond, CA). Extract equivalent to 2 mg of protein was added to 20 μl IgG Sepharose (Amersham Biosciences). Extracts and beads were incubated in a total volume of 600 μl of HKET buffer for 1 h at 4°C. After washing four times with HKET buffer, proteins were eluted from the beads using Laemmli buffer and analyzed by immunoblotting. When the proteins analyzed were of similar size as the IgG heavy or light chain, we used Laemmli buffer without 2-mercaptoethanol for elution. One tenth of the input extract was analyzed in parallel to the affinity-purified samples. To analyze the supernatant after the incubation with IgG Sepharose, acetone was used to precipitate proteins from the supernatant after. The obtained pellet was resuspended in Laemmli buffer and equivalent amounts of bound and unbound fraction were analyzed. Proteins immobilized on IgG Sepharose as well as aliquots of unbound supernatants or the input extracts were analyzed by Western blotting. Chemiluminescence was quantified using Lumi-Imager software (Roche Applied Science). A significant amount of Ufe1p bound unspecifically to the IgG Sepharose or IgG agarose under the conditions used for determining Use1p binding to TAP-tagged proteins. TEV protease cleavage of the TAP-tag could be used to elute specifically bound Ufe1p, which depends on the TAP-tagged protein to bind to the resin. A more convenient way to reduce the background was to raise the ratio of cell extract to IgG Sepharose. We either increased the amount of extract to 2.5 mg protein per 10 μl IgG Sepharose. As a control we used extracts from a strain expressing an unrelated TAP-tagged protein (YSC1178–7502847) to show that the binding of Ufe1p to the beads is in fact specific. Notably, Ufe1p that bound to the IgG Sepharose did not result in unspecific binding of Use1p (unpublished data). This suggests that only free, uncomplexed Ufe1p binds in an unspecific way. Extracts of strain YSC1178–7502847 were used as negative control for the experiments shown in Supplementary Figures S3, S5C, and S6.
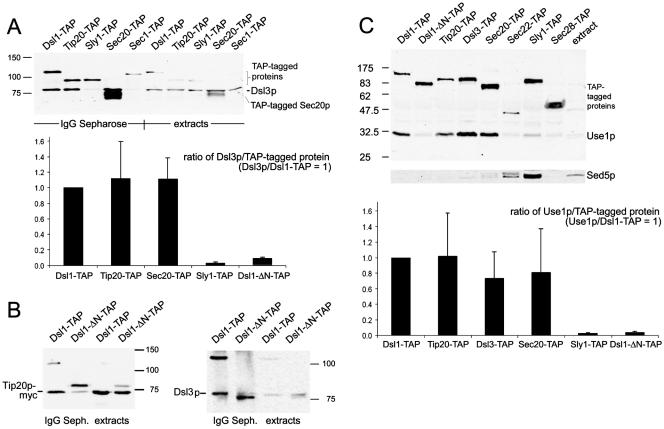
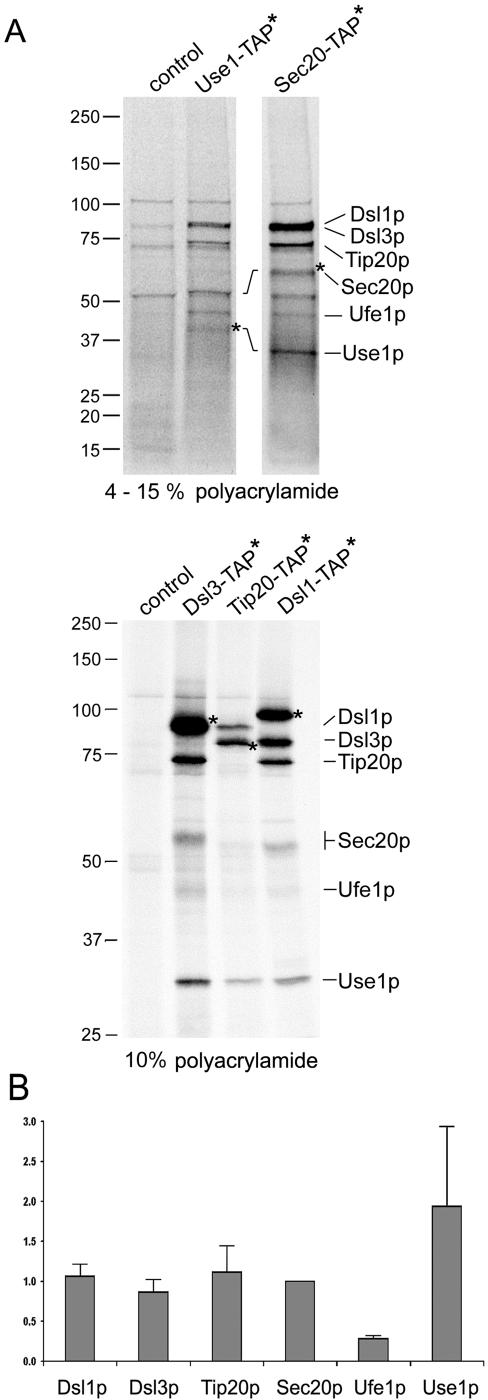
Figure 5.
Dsl3p is part of the Dsl1p complex and binds to the ER-localized SNARE protein Use1p. Extracts of strains expressing various TAP-tagged proteins (YSC1178 stains, Supplementary Table 1; Ghaemmaghami et al., 2003) were prepared as described in Materials and Methods. Proteins bound to IgG Sepharose were eluted with SDS sample buffer and analyzed by SDS-PAGE and immunoblotting with anti-Dsl3p, anti-Use1p, anti-c-myc (9B11, Cell Signaling Technology) and anti-Sed5p antibodies. (A) Coprecipitation of Dsl3p was analyzed in extracts of TAP-tagged Dsl1p, Tip20p, Sly1p, Sec20p, and Sec1p expressing strains. Pulldown fractions and the equivalent of 10% of the input material (extract) were analyzed. The results of several experiments were quantified. The diagram shows a comparison of the ratios of Dsl3p coprecipitating with the various TAP-tagged proteins. The ratio of Dsl3p to Dsl1p-TAP was arbitrarily set to 1. (B) The deletion of residue 8–317 from Dsl1p greatly reduces the binding of myc-tagged Tip20p expressed from plasmid pPMT2 (Sweet and Pelham, 1993) as well as Dsl3p. The TAP-tagged mutant protein was expressed from the DSL1 locus, which was modified by the insertion of plasmid pRS305-ΔNdsl1 into strain YSC1178–7502517, thus creating strain D1-ΔCΔN-T, as described in Materials and Methods. The TAP-tagged proteins are marked (*). (C) Use1p and Sed5p binding to TAP-tagged proteins was analyzed as indicated. The results were quantified as in A to show that Dsl1p, Dsl3p, Tip20p, and Sec20p bind Use1p with equal efficiency. The migration of molecular-weight markers is indicated at the side of each panel.
The tandem affinity purification was performed with the 15 OD600 radiolabeled cells (400 μCi, 90 min, 30°C) using a modified version of the TAP procedure developed by Rigaut et al. (1999). Glass bead lysates were prepared as described above. IgG Sepharose, 200 μl, was added to the cleared lysates. A five times surplus of extracts from unlabeled cells lacking TAP-tagged proteins was added at this point to reduce the unspecific background. Beads and extracts were incubated for 1 h at 4°C. After washing the beads three times with HKET buffer containing 300 mM potassium acetate, beads were washed once with TEV cleavage buffer (10 mM HEPES, pH 8.0, 150 mM NaCl, 0.5 mM EDTA, 1 mM dithiothreitol). TEV protease (Invitrogen) cleavage to remove TAP-tagged proteins from the beads was performed at 16°C for 90 min. Beads were washed with HKET buffer. The collected supernatants were incubated with preimmunserum and protein A Sepharose for 30 min at 4°C. Anti-TAP antibody (3 μl, Open Biosystems) and 10 μl protein A Sepharose was added to the supernatant of this preclearing step. After 1 h at 4°C beads were washed four times with HKET buffer. Proteins were eluted from the beads with Laemmli buffer, and aliquots were either analyzed directly by SDS-PAGE or boiled and subjected to another precipitation with specific antibodies. Gels were immersed in a fluorographic reagent (Amersham Biosciences), dried and analyzed using a Phosphoimager (BAS-2500, Fujifilm, Tokyo, Japan) for quantification (AIDA Biopackage, Raytest, Pittsburgh, PA) and afterward exposed to x-ray films.
RESULTS
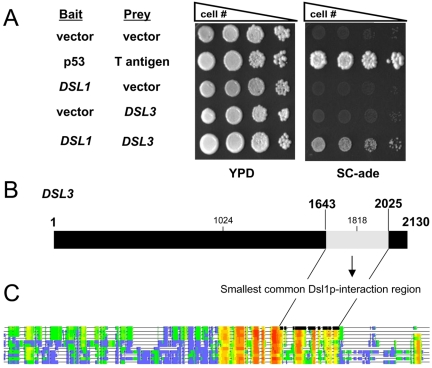
In our previous work (Reilly et al., 2001), we performed a yeast two-hybrid screen with Dsl1p fused to the Gal4p-DNA binding domain (BD) as “bait” (James et al., 1996) in order to identify proteins that physically interact with Dsl1p. This system utilizes a strain in which transcription of three reporter genes ADE2, HIS3, and MET2 are under control of the Gal4p transcriptional activator (James et al., 1996). We isolated three different Dsl1p-interacting components in the two-hybrid screen, each one multiple times. Two were represented by uncharacterized open reading frames (YLR440c and YKR022c) and the third was the Ret2p/δ-COP component of the retrograde COPI coat (Reilly et al., 2001).
We initially obtained 13 isolates verified by sequencing and then another 53 isolates were confirmed by PCR to be YLR440c (a total of 66 YLR440c isolates), whereas only four isolates of YKR022c were obtained from the screen. In addition, a recent genome wide two-hybrid screen has been carried out that also identified YLR440c as a potential Dsl1p interacting protein (Ito et al., 2001). We therefore concentrated our efforts on YLR440c, which will be referred to as DSL3. As shown in Figure 1A, only strains expressing Dsl1p-Gal4p-BD and Dsl3p-Gal4p-activation domain (AD) are able to grow on media lacking adenine, indicating that Dsl1p was able to recruit the Dsl3p-Gal4p-AD fusion protein to the ADE2 promoter, thus activating transcription of the reporter gene. Figure 1B shows that the 382-base pair region (bases 1643–2025) common to all the YLR440c isolates found in the screen (light shading) lies near the C-terminal portion of Dsl3p. We cannot rule out that additional factors are involved in the Dsl1p/Dsl3p interaction, because the two-hybrid screen was performed in yeast cells. We were unable to observe mutual binding of GST-Dsl1p or his6-Dsl3p proteins produced in E. coli. However, both recombinant proteins interacted very efficiently with proteins overexpressed in yeast (Supplementary Figure S1). We observed that the simultaneous overproduction of Dsl3p and Tip20-myc did not improve binding to GST-Dsl1p. Thus additional factors present in yeast are likely required for the Dsl1p-Dsl3p interaction. On the other hand, Dsl1p, Dsl3p, and Tip20p are rather large proteins, and the proper folding of these proteins depends very much on Hsp70 chaperones (Mayer and Bukau, 2005). This factor may be limiting in E. coli where the average protein is smaller than in eukaryotes. Binding of Dsl3p to Dsl1p may require that at least Dsl3p is properly folded.
Figure 1.
Dsl3p interacts with Dsl1p in a two-hybrid system. (A) A log phase two-hybrid strain (PJ69-4A) bearing bait (DNA-binding domain) and prey (activation domain) vectors as indicated was serially diluted and spotted onto YPD medium to assess viability or onto synthetic complete medium lacking adenine to assay the two-hybrid interaction. Coexpression of p53 fused to the Gal4p-BD and T-antigen fused to the Gal4p-AD was used as a positive control. (B) The Dsl1p-interacting domain of Dsl3p. The 3′ portion (bases 1643–2025) of DSL3 encodes the smallest common Dsl1p-interacting region found in the isolates obtained from the screen. The region deleted in the dsl3-1 mutant (see below; nt 1024–1818) is also marked. The Dsl1p-binding region is part of a conserved domain as indicated by the schematic alignment shown in C, and the two lines connecting the two drawings. Regions conserved within fungal homologues of Dsl3p are highlighted red and orange; nonconserved regions are blue. The sequences were from different yeast species including S. pombe, filamentous fungi, and basidiomycetes fungi. T-COFFEE software was used for the alignment (Notredame et al., 2000).
DSL3 is predicted to encode a 709-amino acid residue protein with a molecular weight of 82.4 kDa. The sequence of DSL3 does not reveal a signal sequence, transmembrane domain, or any other significant motifs. BLAST searches identified homologues of unknown function in many fungi including Schizosaccharomyces pombe and Neurospora crassa as well as basidiomycetes species (Figure 1C). The schematic sequence comparison shows that the Dsl1p-binding domain is part of the conserved region in the Dsl3p-like proteins from fungi. Human and plant proteins with a similar domain could be identified using PSI-BLAST searches (human neoblastoma amplified protein, GenBank NP_056993; unnamed protein from Arabidopsis thaliana, GenBank BAB11228).
Genetic and Functional Characterization of dsl3 Mutants
Because DSL3/SEC39 is an essential gene (Winzeler et al., 1999), we generated two conditional lethal mutants (see Materials and Methods), pBAK4B from strain GWY601 bearing dsl3-1 and pBAK2F in strain GWY603 bearing dsl3-2. dsl3-1 (pBAK4B) contains along with two point mutations a 794-base pair deletion between nt 1024 and 1818, thus removing large parts of the most conserved domain within Dsl3p (Figure 1B). Both mutants grow at 23 and 30°C but do not grow at 37°C (Figure 2A).
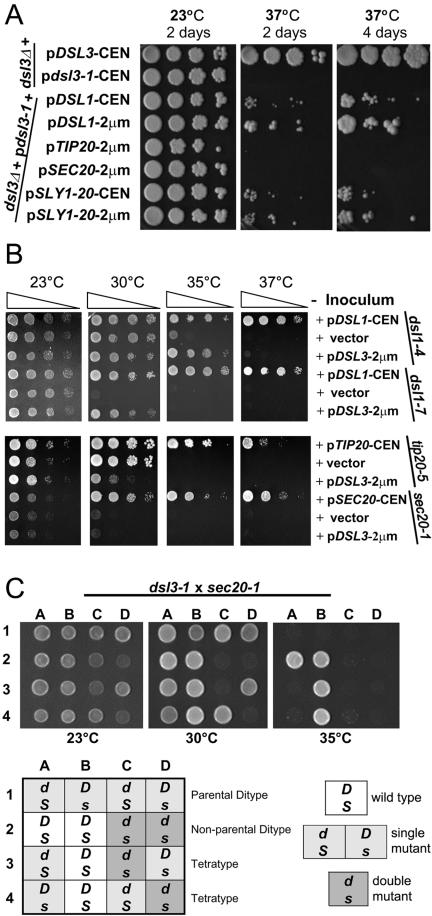
Figure 2.
Genetic interactions between DSL3 and other genes required for transport between ER to Golgi. (A) Suppression of dsl3-1 by DSL1 and SLY1-20. Log phase dsl3Δ (GWY601), bearing pBR12 (CEN DSL3), or pBAK4B (CEN dsl3-1), and log phase dsl3Δ (GWY601) bearing pBAK4B (CEN dsl3-1) and pSC2 (CEN DSL1), pSC10 (2 μm DSL1), pTM2 (2 μm TIP20), pSTM20 (2 μm SEC20), pSV11 (CEN SLY1-20), or pSK54 (2 μm SLY1-20) were subjected to 10-fold serial dilutions, spotted onto rich medium, and incubated at 23 and 37°C for 2 d, or at 37°C for 4 d. (B) The effect of overexpression of DSL3 in dsl1, tip20, and sec20 mutants. Log phase dsl1-4 (GWY230), and dsl1-7 (GWY233), tip20-5 (PC137), and sec20-1 (RSY275), strains bearing pYES2 (vector) or the indicated vector [pDSL1 (pSC2), pTIP20-CEN (pBRT20), pSEC20-CEN (pBRS20), pDSL3-2 μm (pBAK70)] were grown to stationary phase and subjected to 10-fold serial dilutions that were then spotted onto GAL media and incubated for 2 d at 23, 30, 35 or 37°C. (C) DSL3 and SEC20 exhibit genetic interaction. dsl3-1 cells (GWY602) was mated to sec20-1 cells (RSY275) and the resulting diploid strains were sporulated, dissected onto YPD plates, and incubated at 23°C for 2 d. Spores were then spotted onto YPD and incubated at the indicated temperatures for 2 d. Representative sets of spores are shown as indicated: row 1, parental ditype; row 2, nonparental ditype; rows 3 and 4, tetratypes. Genotypes (indicated in the chart, D, DSL1; d, dsl3-1; S, SEC20; s, sec20-1) were determined by G418R and complementation of the temperature sensitivity with plasmid-borne SEC20.
Because previous analysis of DSL1 suggested a possible role in retrograde traffic (Andag et al., 2001; Reilly et al., 2001), we performed high-copy suppression analyses to determine if DSL3 displayed genetic interactions with known retrograde factors. We tested whether the temperature-sensitive growth defect of dsl3-1 could be suppressed by overexpression of the following retrograde transport components: Dsl1p, Sec20p, and Tip20p, the COPI coat components α-COP, β′-COP, γ-COP, and δ-COP, the retrograde t-SNARE Ufe1p, and the Sec1/munc18-related (SM protein) Sly1p, whose Sly1–20p mutant version suppresses mutations in DSL1 (Reilly et al., 2001; VanRheenen et al., 2001). Among these genes only DSL1 and the dominant SLY1-20 allele were able to weakly suppress dsl3-1 (Figure 2A). The suppression was dose-dependent in that expression of either DSL1 or SLY1-20 from high-copy plasmids suppressed the temperature-sensitive growth defect of dsl3-1 better than expression from low-copy vectors (Figure 2A).
SLY1-20 has been shown to weakly suppress components of the retrograde pathway (Ossig et al., 1991) and strongly suppress mutant alleles of many proteins involved in anterograde tethering and docking in the early secretory pathway (Sapperstein et al., 1996; Sacher et al., 1998; VanRheenen et al., 1998, 1999). We therefore examined whether overexpression of anterograde tethering and docking components could suppress dsl3-1 as well: the Rab protein Ypt1p; the tethering factors Uso1p, Cog3p, and Cog2p; the TRAPP component Bet3p (Gallwitz et al., 1983; Sapperstein et al., 1996; Sacher et al., 1998; VanRheenen et al., 1998, 1999); the SNAREs Bet1p, Bos1p, Sec22p, Ykt6p, and Sed5p (Søgaard et al., 1994). In addition we tested the cargo selector Erv14p for its ability to suppress dsl3-1 because it suppressed dsl1 defects as well (VanRheenen et al., 2001). None of those genes required for anterograde docking, with the exception of SLY1-20 (as demonstrated above), could suppress the dsl3-1 mutant phenotype (our unpublished results).
We next tested whether high-copy DSL3 could suppress the temperature sensitivity of the dsl1-4, dsl1-7, sec20-1, and tip20-5 mutations. A plasmid bearing DSL3 under the control of the GAL promoter was introduced into these strains. We found that GAL-dependent overexpression of DSL3 was able to partially suppress both the dsl1-4 and dsl1-7 strains (Figure 2B). Thus, high levels of Dsl1p can compensate for a defect in the essential function of Dsl3p and vice versa. We also found that overexpression of DSL3 was toxic to cells that harbor the tip20-5 mutant allele, suggesting that the two genes show a genetic interaction (Figure 2B). One possible interpretation is that overexpression of the Dsl3p can compete for components required for proper retrograde function (such as Dsl1p, see below). No effect of overexpression of DSL3 was observed on sec20-1. However, we did find strong genetic interactions when we combined the dsl3-1 mutation with the sec20-1 defect. Representative tetrads from a dissection of the dsl3-1/DSL3 sec20-1/SEC20 diploids grown at 23, 30, and 35°C are shown in Figure 2C. The dsl3-1 sec20-1 double mutant is inviable at 30°C, whereas the dsl3-1 and sec20-1 single mutants are viable (Figure 2C). These genetic data, suppression analysis as well as synthetic lethality, provides further support for a retrograde role for Dsl3p and suggests that it may act in conjunction with Dsl1p, Sec20p, and Tip20p at the ER target site.
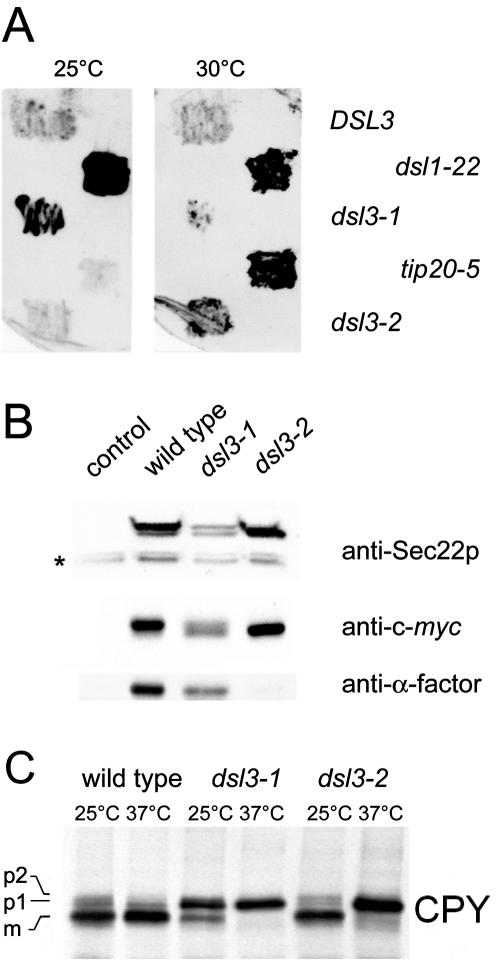
In an effort to provide further support that Dsl3p functions at the retrograde target site, we sought to determine if Dsl3p played a role in retrograde traffic, as has been shown for Dsl1p. One of the many tasks of retrograde traffic is to recycle ER-resident proteins, such as the lumenal chaperone BiP/Kar2p, that have escaped from the ER. A consequence of mutations in components required for proper functioning of the retrograde pathway is the failure to accurately retrieve ER-resident proteins, such as BiP/Kar2p, which instead are secreted outside the cell. This can be assayed by growing cells in contact with a nitrocellulose membrane, to which the secreted BiP/Kar2p will bind, allowing for subsequent immunodetection. To examine whether dsl3 mutants exhibit this phenotype, a dsl3Δ strain containing a control low-copy plasmid bearing DSL3 or the mutant alleles dsl3-1 or dsl3-2, were grown at 25 or 30°C for 24 h in contact with nitrocellulose. As shown in Figure 3A, the dsl3-1 and dsl3-2 mutants secrete BiP/Kar2p at the permissive temperatures like other mutants involved in retrograde transport from Golgi to ER (dsl1-22 and tip20-5). Control experiments indicate that BiP/Kar2p is not released because of cell lysis (our unpublished results). A defect in retrograde transport is also indicated by an assay utilizing a Sec22p-derived fusion protein whose cleavage in the late Golgi can be conveniently followed by Western blotting (Ballensiefen et al., 1998). The C-terminal reporter fused to Sec22p consists of a c-myc epitope and one α-factor pheromone repeat separated by a cleavage site that is recognized by the late Golgi protease Kex2p. Figure 3B shows the steady-state level of processing of the Sec22-α-factor chimera in wild-type and dsl3 mutant strains incubated at 25°C. Wild-type cells contained only the full-length Sec22-α-factor. This protein can be detected with anti-Sec22p, anti-myc and anti-α-factor antibodies. In dsl3-2 mutants only the Kex2p-processed form was found indicating that Sec22-α-factor is mislocalized. The apparently stronger effect of the dsl3-2 mutation compared with dsl3-1 is due to the more prominent anterograde transport defect in this mutant as shown below.
Figure 3.
dsl3ts strains secrete the normally ER-resident BiP/Kar2p and block the ER-Golgi transport of the vacuolar hydrolyze CPY. (A) Log phase dsl3Δ cells, bearing pBR12 (CEN, DSL3), (CEN, pRS315-dsl3-1), or pBAK2F (CEN pRS313-dsl3-2) as well as dsl1-22 and tip20-5 mutants (strains BY-D3WT, BY-d3/1, BY-d3/2, YUA1–9C, BY-t20/5) were streaked onto rich medium, overlaid with nitrocellulose, and allowed to grow at 25 or 30°C for 24 h. BiP/Kar2p that had been secreted and bound to the nitrocellulose was detected by immunoblotting with anti-BiP/Kar2p antibody followed by ECL-PLUS detection. (B) Sec22-α-factor mislocalization assay. Cells harboring plasmid pWBAcyc-α were grown at 25°C overnight. The equivalent of 2 OD600 units of cells were analyzed by Western blotting using antibodies to detect either the whole protein (anti-Sec22p), the c-myc-tag linking Sec22p, and the α-factor moiety (9B11, Cell Signaling Technology, Beverly, MA), or anti-α-factor, which is cleaved off from the hybrid protein after reaching the late Golgi. An asterisk marks endogenous Sec22p. (C) CPY transport assay. Wild-type and mutant cells were shifted to the indicated temperatures and labeled with 35S-methionine and -cysteine for 10 min. After a 30-min chase, samples were removed and the extracts were analyzed by immunoprecipitation using a serum against the vacuolar hydrolase CPY. (p1), core-glycosylated ER form; (p2), Golgi form; (m), mature/vacuolar form.
Pulse-chase experiments were used to analyze the effect of the dsl3 mutations on forward transport between the ER and Golgi. A block in the exit of CPY from the ER was observed in dsl3-1 mutants at permissive as well as restrictive temperature, whereas dsl3-2 mutants showed a CPY transport defect only at the restrictive temperature. A strong block in forward traffic is consistent with the results obtained by Mnaimneh et al. (2004), who used a TET promoter shut down approach to stop expression of DSL3. It is conceivable that a strong block in retrograde transport in dsl3-1 can lead to a concomitant block in anterograde transport (Cosson et al., 1997; Kamena and Spang, 2004).
Dsl3p Is a Peripheral Membrane Protein of the ER
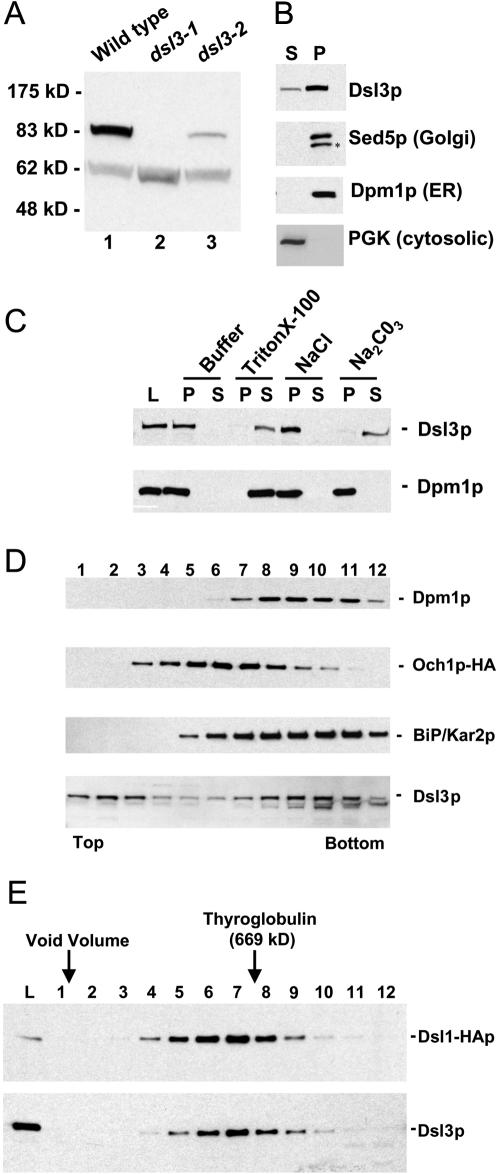
To perform a biochemical analysis of the Dsl3 protein, we generated polyclonal anti-Dsl3p antisera in rabbits. After affinity purification, the antibodies recognized a single protein in crude yeast extracts that migrated at ∼83 kDa (Figure 4A, lane 1), the predicted molecular mass of Dsl3p, and a cross-reactive protein at ∼63 kDa. Examination of the sequence of the temperature-sensitive allele dsl3-1 revealed that this mutant encodes a truncated protein of ∼62 kDa. Western blotting of dsl3-1 cell lysates with the anti-Dsl3p antisera revealed a band of ∼62 kDa, migrating just slightly under the cross-reactive band at 63 kDa (Figure 4A, lane 2). There was no residual immunoreactivity at 83 kDa, indicating that immunoreactivity at 83 kDa in wild-type cells is due solely to Dsl3p. In addition, the immunoreactivity at 83 kDa is increased in a strain containing DSL3 on a multicopy plasmid, whereas the expression level of the cross-reactive band is unaffected (our unpublished data). Interestingly, the truncated dsl3-1 mutant protein as well as the dsl3-2 protein were less abundant than wild-type Dsl3p (Figure 4A, lanes 2 and 3). These experiments show that the 83-kDa protein recognized by our antibody is Dsl3p.
Figure 4.
Dsl3p is a peripheral membrane protein that cofractionates with ER proteins and is in the same large complex as Dsl1p. (A) Characterization of the anti-Dsl3p antibody. Total yeast lysates prepared from a wild-type strain (RSY255) or a dsl3Δ strain bearing dsl3-1 (GWY601) or the dsl3-2 (GWY603) were separated by SDS-12% PAGE and immunoblotted with antibodies against Dsl3p. The migration of molecular mass standards (New England Biolabs, Beverly, MA) is shown on the left. (B) Subcellular fractionation of Dsl3p. A yeast lysate was centrifuged at 175,000 × g and separated into supernatant (S) and pellet (P) fractionation, which were resolved by SDS-12% PAGE and immunoblotted with antibodies against Dsl3p, Sed5p, Dpm1p, or PGK, as indicated. The asterisk indicates a proteolytic product of Sed5p. (C) Dsl3p is a tightly bound peripheral membrane protein. Total yeast membranes were isolated on a step gradient and incubated for 45 min in either buffer, 1% Triton X-100, 1 M NaCl, or 100 mM Na2CO3, pH 11. Reaction mixtures were separated into supernatant (S) and pellet (P) fractions by centrifugation at 175,000 × g for 60 min and analyzed by SDS-12% PAGE and immunoblotting with antibodies against Dsl3p (top panel) or Dpm1p (bottom panel). (D) Dsl3p cofractionates with BiP/Kar2p and Dpm1p and does not significantly overlap with Och1p-HA by buoyant density centrifugation. Membranes from an Och1p-HA-expressing strain (RSY255 bearing pSV66) were separated by buoyant density centrifugation. Gradient fractions were analyzed by immunoblotting with anti-Dsl3p, anti-BiP/Kar2p, anti-Dpm1p, and anti-HA antibodies as indicated. Fractions obtained from the top and bottom of the gradient are indicated. (E) Dsl3p is present in a large complex with Dsl1p. Size exclusion chromatography of Triton X-100-extracted membranes was followed by immunoblotting fractions with anti-Dsl1p and anti-Dsl3p antibodies.
Because Dsl1p was found to associate with the ER membrane, we investigated whether Dsl3p was also associated with the ER membrane. We centrifuged yeast lysate at 175,000 × g to separate cytosolic from membrane-associated proteins and probed the supernatant (S) and pellet (P) fractions for Dsl3p by immunoblotting. A significant portion of Dsl3p was contained in the pellet fraction, which contains a majority of the membrane-associated proteins, as were the integral membrane proteins Sed5p and Dpm1p (Figure 4B); a small fraction of Dsl3p was found in the supernatant with the cytosolic marker protein phosphoglycerate kinase (PGK). This suggests that the majority of the Dsl3 protein is associated with a membrane.
To investigate the nature of Dsl3p membrane association, we determined whether we could extract Dsl3p from crude yeast membranes that were isolated on a buoyant density step gradient. We treated the membranes with reagents that either solubilize integral membrane proteins (1% Triton X-100), or extract many (but not all) peripherally associated proteins (1 M NaCl or pH 11, sodium carbonate). After centrifugation at 175,000 × g the supernatant (S) and pellet (P) fractions were analyzed by immunoblotting with antibodies against Dsl3p (Figure 4C). Buffer alone did not remove Dsl3p, again indicating that it is associated with a membrane. On incubation with Triton X-100, Dsl3p was almost completely released into the supernatant fraction. As expected, the integral membrane protein, Dpm1p, was also completely solubilized after incubation with buffer containing Triton X-100. Dsl3p was not extracted from the membrane by high-salt treatment, similar to the extraction behavior of Dsl1p. Lastly, Dsl3p was efficiently removed to the supernatant fraction upon exposure to pH 11, sodium carbonate, whereas the integral membrane protein Dpm1p remained associated with the pellet. These data suggest that Dsl3p behaves like a tightly associated peripheral membrane protein, as would be expected from the lack of a transmembrane domain in the primary sequence of the protein. It is noteworthy that the inability to extract Dsl3p with high salt is unusual and identical to the behavior of Dsl1p (Reilly et al., 2001).
We next investigated the subcellular distribution of Dsl3p to determine which membrane population Dsl3p primarily associates with. Membranes from a wild-type strain bearing the Golgi resident protein Och1p tagged with an HA-epitope (Harris and Waters, 1996) were subjected to sucrose density centrifugation utilizing a magnesium-containing gradient method (Antebi and Fink, 1992) followed by fractionation and immunoblotting with anti-Dsl3p, anti-BiP/Kar2p, anti-Dpm1p, and anti-HA antibodies (Figure 4D). This method resolves a variety of yeast organelles including ER, Golgi, and vacuolar membranes. Dsl3p comigrated with the ER-resident BiP/Kar2p, and with Dpm1p, an ER integral membrane protein. Further, the Dsl3p peak was resolved from the Golgi Och1p-HA peak, with only a slight overlap. A small fraction of Dsl3p failed to enter the gradient and remained in the initial fractions, suggesting that either some Dsl3p may have dissociated from the membrane during the isolation procedure or that there is a small cytoplasmic pool of Dsl3p that is not associated with the membrane. Because the magnesium-containing sucrose gradient does not resolve ER membrane fractions from the plasma membrane, we also utilized a method that uses EDTA and a harsh lysis procedure to remove ribosomes from the ER, thus lowering its buoyant density (Roberg et al., 1997). Again, Dsl3p comigrated with the ER marker BiP/Kar2p and was resolved from the plasma membrane marker Pma1p (our unpublished results).
These fractionation profiles suggest that Dsl3p primarily associates with ER membranes, a result that is entirely consistent with, and further supports, our findings that Dsl3p may interact with Dsl1p at the retrograde target site on the ER membrane.
Dsl1p and Dsl3p Are Found in the Same Complex at the Retrograde Target Site
Dsl1p is an ER-localized peripheral membrane protein found in a large complex that contains proteins of ∼80 and ∼55 kDa, which we have identified as Tip20p and Sec20p (Reilly et al., 2001). To examine whether the Dsl3p may associate with Dsl1p in this complex, we extracted Dsl3p from microsomal membranes with Triton X-100. We found that Dsl3p was extracted fairly efficiently (∼50%) from enriched microsomal membranes (our unpublished data). The detergent extracts were subjected to size exclusion chromatography followed by immunoblotting with anti-Dsl1p and anti-Dsl3p antibodies (Figure 4E). We found that Dsl3p precisely cochromatographed with Dsl1p eluting from the column just ahead of thyroglobulin at a size comparable to Dsl1p complex of ∼700 kDa.
To corroborate these findings we first performed affinity purification assays followed by Western blotting analysis. We used strains from a fusion library carrying the tandem affinity tag (TAP) fused to the chromosomal copies of the genes of interest (Ghaemmaghami et al., 2003). Extracts from yeast strains expressing TAP-tagged proteins functionally implicated with Dsl1p were incubated with the IgG Sepharose affinity matrix. After extensive washing, SDS-PAGE and blotting, Dsl3p bound to the tagged proteins was visualized using specific antibodies. The proteins carrying the affinity tag were also detected since the two protein A moieties within the TAP tag are recognized by primary as well as secondary antibodies (Rigaut et al., 1999). Figure 5A shows that Dsl3p interacts with Dsl1-TAP, Tip20-TAP, and Sec20-TAP equally well. A small and varying amount of Dsl3p consistently associated with the Sec1/munc18(SM)-like protein Sly1p (our unpublished results). This result was expected because Sly1p was shown to interact with the ER SNARE Ufe1p (Yamaguchi et al., 2002) and because the mammalian Sly1 protein copurifies with proteins homologous to Dsl1p and Tip20p (Hirose et al., 2004). Moreover, Li et al. (2005) provided direct evidence for Sly1p being involved in Golgi–ER retrograde transport (see pp. 3951–3962 of this issue). In contrast, no Dsl3p binds to the TAP-tagged Sec1p, a SM protein involved in fusion of vesicles at the plasma membrane (Figure 5A, fifth lane).
The results of the two-hybrid experiments indicated that Dsl1p may engage in pairwise interactions with Dsl3p and Tip20p. The domain in Dsl1p that mediates the binding to Tip20p was mapped to the N-terminal 200 residues (Andag and Schmitt, 2003). To determine if the N-terminus of Dsl1p is also involved in the binding of Dsl3p, we created a mutant carrying a split DSL1 gene. This mutant strain produced two truncated versions of Dsl1p, a nonfunctional N-terminal fragment lacking 221 C-terminal amino acid residues and a TAP-tagged version in which most of the N-terminal half of the protein (residue 8–317) was replaced by a c-myc epitope. The mutant was viable but showed a growth defect at temperatures above 30°C and a BiP/Kar2p secretion defect. The latter phenotype, however, could be attributed to the presence of the TAP-tag at the end of the C-terminal Dsl1p fragment (see Supplementary Figure S2 for a detailed characterization of the different dsl1-ΔN mutants). We expected this mutant to be viable because a recent multicopy suppressor screen using dsl1-22 mutants had yielded a DSL1 fragment encoding a 58-kDa C-terminal fragment that was able to complement the deletion of DSL1. Figure 5B shows that the TAP-tagged version of this truncated protein is equally affected in its ability to bind myc-tagged Tip20p and Dsl3p. This result together with the two-hybrid data suggested that the N-terminal part of Dsl1p mediates the interaction with both Tip20p and Dsl3p.
Mutants expressing the Δ8-317 deletion allele alone (dsl1-ΔN) were also viable and were included in the experiments described below. This deletion mutant is only slightly temperature sensitive. It shows normal CPY processing even at 37°C, secretes little BiP/Kar2p, and mislocalizes Sec22-α-factor only at higher temperatures (Supplementary Figure S2). Thus the presence of a separate N-terminal Tip20p/Dsl3p-interacting domain has a negative effect on cells expressing the essential C-terminal fragment of DSL1. The absence of this domain has only weak effects.
Interaction of Dsl Proteins and Tip20p with SNARE Proteins
Next we tested whether Dsl3p is involved in the interaction with SNARE proteins. Figure 5C shows that Use1p, the yeast homolog of mammalian p31 (Burri et al., 2003; Dilcher et al., 2003; Hirose et al., 2004), is associated with the Dsl1p complex. Binding was equally strong to TAP-tagged Dsl3p, Dsl1p, Tip20p, or Sec20p (Figure 5C). Very little Use1p was associated with Sly1-TAP, and no significant interaction was observed in case of Sec22-TAP, Sec28(ε-COP)-TAP. As a control we incubated the same blot with anti-Sed5p antibodies. As expected, Sed5p interacts with Sly1-TAP and Sec22-TAP. Because Sed5p is specifically engaged in forward traffic, this indicated that at steady state conditions only a very small fraction of Sly1p and Sec22p is active in retrograde transport. In addition to Sec22p and Sed5p, we were not able to detect the SNARE proteins Bet1p and Bos1p in pulldown experiments with TAP-tagged Dsl1p or Dsl3p (Supplementary Figure S3). Taken together, these biochemical and genetic data suggest that Dsl3p is a stable component of the Dsl1p retrograde complex. Like Tip20p, Dsl1p, and Sec20p, Dsl3p is associated with the ER SNARE Use1p.
Tandem Affinity Purification of the Dsl1p Complex
To assess the stoichiometry of the Dsl1p complex and the SNARE proteins associated with it, we purified the complex using a modified tandem affinity protocol (Rigaut et al., 1999; see Materials and Methods). To analyze several samples in parallel and to aid protein detection, cells expressing TAP tagged Dsl1p complex subunits as well as Sec20p and Use1p were radiolabeled and purified utilizing a quick protocol (see Materials and Methods) to minimize protein degradation and prevent the loss of subunits from the complex. Fluorographs showing proteins associated with the purified proteins are shown in Figure 6 and Supplementary Figure S4. Asterisks mark the proteins that carried the tag. Note that the remaining TAP-tag after TEV cleavage increases the size of the protein by 6 kDa. Individual proteins were identified by boiling aliquots of the preparations and reprecipitating proteins either with anti-TAP-tag antibodies or specific antibodies (anti-Dsl3p, anti-Use1p, anti-Ufe1p, or anti-c-myc; Supplementary Figure S4A). Extracts from cells not expressing any tagged protein were used to identify unspecific bands. Figure 6 shows that a similar set of proteins was isolated regardless of what subunit carried the affinity tag. Six proteins corresponding to Dsl1p (89 k Da), Dsl3p (82 kDa), Tip20p (81 kDa), Sec20p (∼ 50 kDa, O-glycosylated), Ufe1p (40 kDa), and Use1p (29 kDa) represented the most prominent bands. Dsl1p, Dsl3p, and Tip20p were present in equal amounts when copurified with Sec20-TAP (see also Supplementary Figure S4B). A quantification of the results is shown in Figure 6B. When the data were corrected for differences in the content of sulfur-containing amino acids, it appeared that nearly equal amounts of Dsl1p, Dsl3p, Tip20p, and Sec20p were present in the preparations. The apparently lower and higher yield of Ufe1p and Use1p, respectively, may either be due to different labeling efficiencies or, in case of Ufe1p, a weaker association with the complex. The results also indicated that any additional proteins that may be specifically associated with the Dsl1p complex are present at substoichiometic amounts, for instance, the weak band above Sec20p.
Figure 6.
Tandem affinity purification of the Dsl1p complex and associated SNARE proteins. The composition of the Dsl1p complex was analyzed by applying a modified tandem affinity purification protocol (Rigaut et al., 1999) as described in Materials and Methods. Cells expressing the TAP-tagged SNARE proteins Use1p and Sec20p as well as the Dsl1p complex subunits Dsl1p, Dsl3p, and Tip20p were labeled with 35S-methionine and -cysteine to facilitate the detection of purified proteins. The wild-type strain BY4741 was also analyzed to visualize proteins that bind unspecifically to IgG Sepharose and protein A Sepharose carrying anti-TAP antibodies. Proteins were identified by an additional immunoprecipitation. Before the reprecipitation, these samples had been incubated at 95°C for 5 min to disassemble protein complexes (Supplementary Figure S4A). Samples were loaded either on a 4–15% gradient SDS polyacrylamide gel (Bio-Rad; top panel) to distinguish small proteins or on a one-percentage 10% gel to better resolve the larger proteins (bottom panel). For comparison, Supplementary Figure S4B shows a one-percentage gel with a preparation using a Sec20-TAP expressing strain and a 4–15% gradient gel with samples from Dsl3-TAP, Tip20-TAP, and Dsl1-TAP expressing strains. The migration of molecular-weight markers is indicated on the left. (B) Phosphoimager pictures from two independent Sec20-TAP preparations as well as one Dsl3-TAP, Tip20-TAP, and Dsl1-TAP preparation were quantified (AIDA Biopackage, Raytest). Data were corrected for different contents of methionine and cysteine of the proteins. The intensity values for each protein were normalized to the number of methionines and cysteines in Sec20p (minus the initiator methionine). Sec20p with eight methionines and cysteines was assigned a normalization factor of 1 because it was discernable as a single band in all preparations.
All Known Subunits of the Dsl1 Complex Are Required for the Stability and Integrity of the ER-localized Retrograde SNARE Complex
Finally we explored the possibility to draw an interaction map linking all known subunits of the Dsl1p complex and the associated SNARE proteins by using extracts from various mutants for pulldown experiments. In short, we repeated pulldown experiments like that shown in Figure 5C with cells expressing a combination of the TAP-tagged versions of Dsl1p, Dsl3p, Tip20p, and Sec20p and the following mutations: dsl3-1 (this work), two truncated versions of DSL1, dsl1-ΔN, whose gene product lacks the Tip20p and Dsl3p interacting domain (this work), dsl1-22, a deletion of 30 codons at the conserved C-terminus of DSL1 (Andag et al., 2001), a TIP20 mutant allele, tip20-5 (Cosson et al., 1997), or a mutation affecting the SNARE motif of Sec20p, sec20-1 (Sweet and Pelham, 1992; Lewis et al., 1997). We were not able to analyze all possible combinations of TAP-tagged proteins with mutations in the Dsl1 complex. Ufe1-TAP expression was not observed (Ghaemmaghami et al., 2003) and recombinant spores expressing a TAP-tagged Use1p were either not viable or formed very small colonies. Sec20-TAP, may be exceptional because it is one of the few SNAREs that carry a long luminal domain. In fact, Sec20-TAP-expressing strains carrying the dsl1-22, dsl3-1, tip20-5, and dsl1-ΔN were obtained and analyzed (Supplementary Figure S6B). To facilitate the detection of Sec20p, a series of mutant strains was created that express a c-myc tagged Sec20p version (Sweet and Pelham, 1993).
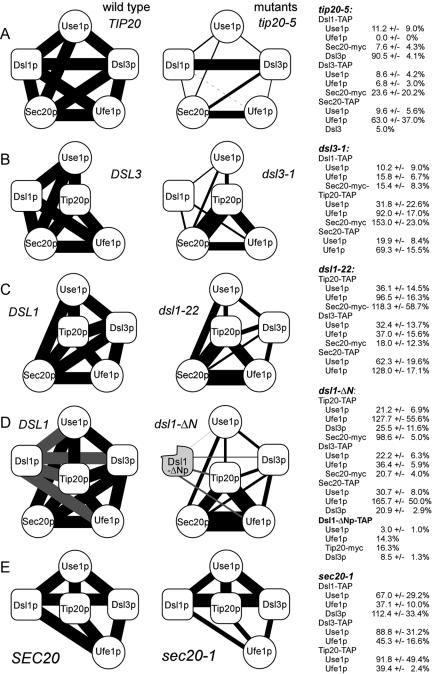
Those mutants that carried point mutations or small deletions (dsl1-22, tip20-5, and sec20-1) were shifted to the restrictive temperature of 35°C for 1 h before the preparation of lysates. Mutant strains carrying large deletions (dsl3-1, dsl1-ΔN) were kept at the permissive temperature of 25°C to avoid artifacts of the temperature shift (see below). Extracts were analyzed as described in Materials and Methods. TAP-tagged proteins served as a convenient internal standard because they were detected along with the protein against which the antibodies were directed (Rigaut et al., 1999; see also Figure 5). This allowed us to determine the ratio of protein copurifying with the tagged protein and the protein carrying the affinity tag (Figures S5 and S6 for a representation of such experiments). A quantification and schematic overview of the results is presented in Figure 7. The schemes on the left side of Figure 7 show the interactions that were analyzed and indicate the wild-type level of interaction. The width of lines between the schematic proteins depicted in Figure 7 illustrates the extent of coprecipitations. They do not indicate pairwise interactions.
Figure 7.
Summary of the interaction data. The results if experiments like those shown in Supplementary Figures S5, S6, and S9 were quantitated using Lumi-Imager software (Roche Applied Science). The wild-type interaction was set to 100% and the level of interaction observed in mutants was expressed as average percentage ± SD. The wild-type column illustrates the coprecipitations that were actually analyzed for each wild-type/mutant combination. The results obtained with mutants are shown on the right side together with the quantification data. SNARE-proteins are shown as white spheres, and the Dsl1p complex subunits as oval boxes. (A–E) Five different pairs of wild-type and mutant alleles that were analyzed. The width of the lines connecting the symbol is proportional to the binding strength. Results obtained with TAP-tagged Dsl1-ΔN protein (D) expressed from the split dsl1-ΔCΔN-T allele are represented as gray lines.
We found that the binding of Use1p to the Dsl1p complex is sensitive to mutations in subunits of this complex. To rule out that this effect is due to a general block in vesicular transport and a subsequent disassembly of protein complexes involved in transport, we analyzed the effects of three other mutations affecting membrane traffic: sec18-1, a mutation, which shows a very rapid arrest in all transport steps at restrictive temperature (Graham and Emr, 1991), sec22-3 and ret2-1 (δ-COP defect). But even growth at restrictive temperature did not reduce the binding of Use1p from these three mutants to TAP-tagged Dsl1p or Dsl3p (Supplementary Figures S5B, lanes 1–10, and S6B, lane 5).
Additional controls addressed the stability of Use1p during the pulldown experiments or the variations in the amount of Use1p in the lysates. In dsl1-22 and tip20-5 mutants the decrease in binding efficiency was not due to instability of the Use1 protein because normal amounts of this protein were recovered from the supernatants after the incubation of extracts with IgG Sepharose (Supplementary Figure S5B, lane 11–15). In dsl3-1 mutant cells, however, the amount of Use1p in the cell lysate was lower than in wild-type cells (63 ± 17%; Supplementary Figure S5A, lane 14 and 16). Even less Use1p was present in dsl3-1 mutants shifted to 35°C (Supplementary Figure S5A, lane 6). We tested the supernatants after pulldown experiments with extracts from dsl3-1 mutants grown at 25°C and found no indication for Use1p being less stable in extracts from dsl3-1 mutants than that of wild type (Supplementary Figure S7). The dsl3-1 mutation consistently decreased the rate at which Use1p copurified with Dsl1-TAP, Tip20-TAP, and Sec20-TAP to 21% of wild-type level (Figure 7B).
Among the mutants tested, tip20-5 had the strongest effect on the binding of Use1p to the TAP-tagged proteins (Figure 7A). It reduced the binding of Use1p to Dsl1-TAP, Dsl3-TAP to 12% of wild-type level or less. To observe this decrease in binding of Use1p to Dsl3-TAP, it was necessary to shift tip20-5 mutant cells to the restrictive temperature before the preparation of lysates. In contrast, the tip20-8 mutation reduced the binding of Use1p to Dsl3-TAP even when cells were kept at 25°C (Supplementary Figure S8). As shown recently, tip20-5 and tip20-8 have quite different effects when analyzed in in vitro experiments (Kamena and Spang, 2004). The tip20-5 mutation did not reduce the binding of Dsl3p to TAP-tagged Dsl1p (Figure 7A; Supplementary Figure S9), thus ruling out the possibility that lower Use1p-binding efficiency in the tip20-5 is due to a decreased stability of the two Dsl proteins.
The comparison of the interaction data obtained for the different mutants shows that the whole complex consists of a core that is resistant to mutations within DSL1 and DSL3 (Figure 7). Another subcomplex consists of Dsl1p and Dsl3p, which remain associated in the presence of the tip20-5 mutation, a mutation with the most dramatic effects on the stability of the complex. The binding of the SNARE Use1p to Dsl proteins is equally sensitive to mutations in Dsl1p, Dsl3p, and Tip20p. Thus, Use1p depends on three other proteins for efficient binding to the other Q-SNAREs, whereas Ufe1p/Sec20p interaction on average never drops below 60% of wild-type level.
In contrast to the mutations in large subunits, a mutation in one of the SNARE proteins, sec20-1, showed a different effect. The sec20-1 mutation represents a leucine to serine substitution in the -1 layer of the SNARE motif (Lewis et al., 1997) and may therefore mainly interfere with interaction mediated by this motif. In fact, sec20-1 reduces the binding efficiency of Ufe1p to the other subunits, whereas the interaction of Use1p with the Dsl proteins and Tip20p is less strongly affected. Taken together, there are two types of interactions involving SNARE proteins, a Dsl-dependent binding of Use1p to Sec20p and Ufe1p and a Dsl-independent interaction between Sec20p and Ufe1p. Tip20p may play a central role at the interface between the SNAREs because it is also required for recruiting Use1p and the association of Tip20p with the Sec20p/Ufe1p does not require Dsl proteins.
DISCUSSION
Dsl3p and Dsl1p Have Related Functions
In this work we identified a new component of the Dsl1 ER targeting complex, Dsl3p. Dsl3p and Dsl1p as well as the third component of the complex, Tip20p, are required for the stability of the ER-localized SNARE complex consisting of the Q-SNAREs Ufe1p, Sec20p, and Use1p. All proteins, Dsl1p, Tip20p, and the newly identified Dsl3p, are essential and have a similar size (88, 82, and 81 kDa, respectively). Interactions between Dsl1p and Tip20p as well as Dsl1p and Dsl3p were observed in screens for yeast two-hybrid interactions and were confirmed using immunoprecipitation or affinity purification (Ito et al., 2001; Reilly et al., 2001; Andag and Schmitt, 2003; this work). Dsl3p, upon extraction from the membrane with detergents, chromatographed in a manner nearly identical to that of Dsl1p; both proteins are found in a large complex of approximately ∼700 kDa. A significant proportion of both proteins is associated with the ER as shown by sucrose gradient centrifugation (Reilly et al., 2001; this work). Tip20p and Dsl1p were implicated in retrograde transport from Golgi to ER (Cosson et al., 1997; Frigerio, 1998; Andag et al., 2001; Reilly et al., 2001; VanRheenen et al., 2001; Andag and Schmitt, 2003). Evidence for a related function of Dsl1p and Dsl3p came from genetic experiments. Mutations in DSL3 and DSL1 were partially suppressed by overexpressing DSL1 and DSL3, respectively. This reciprocal interaction suggests that the gene products act in parallel. Further, we found that overexpression of DSL3 was toxic to cells that harbor the tip20-5 mutant allele. Synthetic negative effects were also observed when dsl3-1 mutation was combined with the sec20--1 defect. Thus the genetic evidence suggests that Dsl3p may act in conjunction with Dsl1p, Sec20p, and Tip20p at the ER target site. Some phenotypes of dsl3ts mutants confirmed this notion. Like many other mutants affected in retrograde traffic from Golgi to ER dsl3ts mutants secreted BiP/Kar2p, mislocalized a Sec22p-α-factor reporter protein and showed a very strict block in anterograde transport at room temperature (Semenza et al., 1991; Ballensiefen et al., 1998; Cosson et al., 1997). Tet-promoter shut-down experiments, which had been used to identify Dsl3p in a genome-wide screen, had already indicated a role of Dsl3p in forward traffic. A block in forward traffic can be either an indirect effect of a block in recycling, which leads to a depletion of essential components at the ER or it can be the immediate result of a defect in the ER targeting machinery, which cannot differentiate between forward and backward moving vesicles (Kamena and Spang, 2004).
The Dsl Proteins Are Required for the Binding of Use1p to the Targeting Complex
It was shown before that Tip20p and Dsl1p interact with SNARE proteins at the ER (Sweet and Pelham, 1993; Lewis et al., 1997; Reilly et al., 2001; Dilcher et al., 2003). We confirmed that Dsl3p is also part of this complex by Western blotting. The results of our attempts to purify the whole complex showed that the complex is rather stable and that equivalent amounts of Dsl1p, Dsl3p, Tip20p, Sec20p, and Use1p are copurified regardless of which subunit carried the TAP-tag. Quantification of the data suggested that Ufe1p is present in lower amounts than the other subunits of the complex (Figure 6B). The quantification of single-step binding experiments like those shown in Figure 5C indicated that similar fractions of Ufe1p and Use1p bind to Dsl3p or Sec20p (Supplementary Figure S10), suggesting that all subunits including Ufe1p are present in comparable amounts at the beginning tandem affinity purification. More work is required to determine whether the different amounts of Ufe1p and Use1p visible in Figure 6 simply reflect varying labeling efficiencies or loss subunits during the lengthy preparation. As shown in Figure 7E, the binding of Ufe1p to the complex is affected by a mutation in the SNARE motif of Sec20p. Unlike Use1p, Ufe1p does not depend on Dsl proteins for its binding to Tip20p and Sec20p. The possibly easy release of Ufe1p from the complex may reflect the need to make Ufe1p available for other membrane-fusion events because it is involved in homotypic ER membrane fusion as well (Patel et al., 1998).
The strongest evidence for a related and comparable function of Dsl1p and Dsl3p came from experiments that tested the integrity of the ER-targeting complex in the presence of mutations in various members of the complex. There is a mutual requirement for Dsl1p and Dsl3p to bind to the complex as well as for the binding of the SNARE protein Use1p to the core of the complex. Almost all mutations within the complex had an effect on the interaction between other partner proteins, suggesting that their primary function lies in the assembly of this complex. Dsl3p may also be required to stabilize Use1p. The summary of the pulldown experiments presented in Figure 7 suggests that Tip20p has a central role within this complex of SNARE proteins and putative tethering factors. Mutations within TIP20 had the most dramatic effect on the integrity of the whole complex, whereas mutations outside of this central part of the complex had little effect on the interaction between Tip20p, Sec20p, and Ufe1p. Conversely, a mutation that changes the SNARE motif of Sec20p had a strong effect on the core of the whole complex as demonstrated by the weaker interaction of Ufe1p with the Dsl proteins and Tip20p. This mutation in the SNARE motif of Sec20p had little effect on the binding of Use1p to the complex (Figure 7E). Therefore, we think that it is possible to draw a map of interactions that reflects their sensitivity to a set of different mutations.
Very tight interaction between the Tip20p, Sec20p, and Ufe1p was suggested by the previous finding that the targeting of overproduced Tip20p to the ER is restored by excess Sec20p as well as by data suggesting that the transmembrane helices of Sec20p and Ufe1p are closely packed in the membrane (Sweet and Pelham, 1993; Lewis et al., 1997). The latter finding is the only experimental evidence so far that Sec20p occupies the Qb position within the putative four helix bundle in the core of the ER-SNARE complex, the position next to the Qa-SNARE Ufe1p. The classification of SNAREs as R-, Qa-, Qb-, and Qc-SNARE is based on their position within the four helix bundle (Sutton et al., 1998; Bock et al., 2001; Antonin et al., 2002). The SNARE domain of syntaxin-like proteins occupies the Qa-position next to the R-SNARE. The very tight interaction between Ufe1p and Sec20p implies that Sec20p represents the other direct neighbor of Ufe1p in the membrane and thus contributes the Qb-helix to the complex. According to this model Use1p must be the Qc-SNARE in the acceptor complex at the ER. The somewhat loose association of Use1p with the rest of the acceptor complex is reminiscent of other Qc-SNAREs or those parts of SNAP-25 homologues that are equivalent to the Qc-SNARE helix (Ungermann et al., 2000; Boeddinghaus et al., 2002; An and Almers, 2004; Fasshauer and Margittai, 2004; Thorngren et al., 2004).
Our results indicate that Dsl1p and Dsl3p help to recruit Use1p to the ternary complex consisting of the Ufe1p/Sec20p Q-SNARE dimer and Tip20p (Sweet and Pelham, 1993; Lewis et al., 1997). Tip20p plays a central role because Dsl1p and Dsl3p depend on functional Tip20p for their interaction with Use1p. The Dsls and Tip20p may stabilize the Q-SNARE helices and make them ready for the binding of vesicular R-SNARE. The involvement of auxiliary factors adds an additional level of control and may confer an element of selectivity during the Q-SNARE nucleation process. They may leave before or during the formation of the quaternary complex. This may explain why we found very little if any Sec22p, the R-SNARE thought to be involved in Golgi-ER retrograde transport. It is possible that the Dsl1p complex is a place marker for the R-SNARE because Dsl1p carries a partial R-SNARE motif at the C-terminus. This part of the molecule is the best conserved part within Dsl1p-like proteins, including the ZW10 proteins from higher eukaryotes (Andag and Schmitt, 2003). To test the significance of our failure to detect Sec22p interaction with the Dsl1p complex, we wanted to reproduce the interaction between Sec22p and the other SNAREs by the same approaches used above and employing TAP-tagged Sec22p and Sec20p. We also used sec18-1 mutants to block the disassembly of SNARE complexes. We could detect only very low amounts of Sec22p or Sec20p-myc. However, the interaction may be hampered by the mislocalization of Sec22-TAP to the vacuole because of the presence of a bulky tag (Ballensiefen et al., 1998) or the masking of the HDEL ER retrieval signal of Sec20p in Sec20-TAP (Sweet and Pelham, 1992). Moreover, dsl1 and dsl3 mutants mislocalize a Sec22p-derived reporter protein (Andag et al., 2001; this work). Sec22p mislocalization would make the analysis of effects of dsl mutants on the ternary SNARE complex even more complicated.
Instead of acting one after another, our results might also indicate that the Dsl1p complex and Sec22p act in parallel. Sec22p is not essential and some sec22 deletion strains are not even temperature sensitive (Ossig et al., 1991). It is known that the R-SNARE Ykt6p can take over the function of Sec22p in forward traffic (Liu and Barlowe, 2002). It is unlikely that Ykt6p as a lipid-anchored SNARE can replace Sec22p as a v/R-SNARE in retrograde transport as well (McNew et al., 2000), although we found that overproduced Ykt6p can alleviate the need for Sec22p in dsl1-22 mutants (Andag et al., 2001). The fact that dsl1 mutations can make the cells Sec22p-dependent (Andag et al., 2001) indicates that the Dsl1p complex is involved in this Sec22p-independent retrograde transport. Likewise, it is possible that retrograde transport can become Dsl-independent, because the dsl1-ΔN mutation, which has dramatic effects on the stability of the Dsl1p complex, has only slight effects on growth, BiP/Kar2p retention, or Sec22-α-factor localization (Supplementary Figure S2). Similarly, the tip20-8 mutation affects the Use1p/Dsl1p complex interaction at a permissive temperature. As discussed below in more detail, more work is necessary to determine whether the essential function of the Dsl1p complex is in fact its role in vesicle fusion as indicated by in vitro experiments (Kamena and Spang, 2004) or a regulatory role.
The ER-targeting Complex Is Conserved during Evolution
As a general rule, all members of the Dsl1p complex as well as the ER SNAREs Ufe1p, Use1p, and Sec20p exhibit an unusually low degree of sequence conservation compared with proteins acting later in the secretory pathway. The functionally equivalent pairs of yeast and mammalian proteins are Dsl1p/ZW10, Tip20p/RINT-1, Ufe1p/syntaxin 18, Use1p/p31, and Sec20p/BNip1 (Hatsuzawa et al., 2000; Andag and Schmitt, 2003; Hirose et al., 2004; Nakajima et al., 2004). We now identified a new member of the complex, Dsl3p, that has no apparent equivalent among the syntaxin 18-associated proteins (Hirose et al., 2004). There are two properties of the two multiprotein complexes that are conserved despite the low sequence similarities: In both Dsl1p and ZW10 the N-terminal part mediates the interaction with Tip20p/RINT-1 (Andag and Schmitt, 2003; Hirose et al., 2004). Thus, an interaction conserved from yeast to mammals involves a domain, which shows no sequence conservation (Supplementary Data in Andag and Schmitt, 2003; Hirose et al., 2004). The second well-conserved property is the tight functional or physical association of Use1p with the Dsl1p complex (this work) and p31 with ZW10 and RINT-1 (Hirose et al., 2004). This latter subcomplex persists the disassembly of syntaxin 18-associated proteins in the presence of ATP, NSF, and α-SNAP. Maybe the biggest difference between the yeast and the mammalian complex is the rather tight association of mammalian rSly1 and Sec22b with the syntaxin 18-associated proteins, whereas in our experiments very little or no Sly1p and Sec22p was bound to the Dsl1p complex.
Three mammalian syntaxin 18-associated proteins have been known before to be involved in cell cycle checkpoint control (ZW10 and RINT-1) or apoptosis (BNip1; Chan et al., 2000; Xiao et al., 2001; Zhang et al., 2003). In mammalian cells ZW10, the putative equivalent of Dsl1p, helps to recruit dynactin and dynein to the kinetochore (Starr et al., 1998). Interestingly, a new cell cycle checkpoint was recently identified in yeast that monitors cell wall synthesis and involves dynactin (Suzuki et al., 2004). Like ZW10 and RINT-1, Dsl1p, Dsl3p, and Tip20p may have additional roles besides their function in vesicular transport. It will be important to identify these roles and to determine whether the two possible functions lead to crosstalk between these processes.
Supplementary Material
Acknowledgments
We thank Gabi Fischer von Mollard, Mike Lewis, and Uwe Andag for their gift of antibodies, plasmids, and proteins; Hanneget Frahm and Heike Behr for excellent technical assistance; and Anne Spang and Carmen Graf for comments on the manuscript. We thank D. Ungar, N. Erdeniz, and D. Matheos for their assistance with chromatography and synthetic lethality. Additionally, we thank M. Rose for his gift of antibodies, plasmids, and bench space utilized during these studies and Dieter Gallwitz for his support. This work was supported by the Max-Planck Society.
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05–01–0056) on June 15, 2005.
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
References
- An, S. J., and Almers, W. (2004). Tracking SNARE complex formation in live endocrine cells. Science 306, 1042–1046. [DOI] [PubMed] [Google Scholar]
- Andag, U., Neumann, T., and Schmitt, H. D. (2001). The coatomer-interacting protein Dsl1p is required for Golgi-to-endoplasmic reticulum retrieval in yeast. J. Biol. Chem. 276, 39150–39160. [DOI] [PubMed] [Google Scholar]
- Andag, U., and Schmitt, H. D. (2003). Dsl1p, an essential component of the Golgi-endoplasmic reticulum retrieval system in yeast, uses the same sequence motif to interact with different subunits of the COPI vesicle coat. J. Biol. Chem. 278, 51722–51734. [DOI] [PubMed] [Google Scholar]
- Antebi, A., and Fink, G. R. (1992). The yeast Ca(2+)-ATPase homologue, PMR1, is required for normal Golgi function and localizes in a novel Golgi-like distribution. Mol. Biol. Cell 3, 633–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonin, W., Fasshauer, D., Becker, S., Jahn, R., and Schneider, T. R. (2002). Crystal structure of the endosomal SNARE complex reveals common structural principles of all SNAREs. Nat. Struct. Biol. 9, 107–111. [DOI] [PubMed] [Google Scholar]
- Ballensiefen, W., Ossipov, D., and Schmitt, H. D. (1998). Recycling of the yeast v-SNARE Sec22p involves COPI-proteins and the ER transmembrane proteins Ufe1p and Sec20p. J. Cell Sci. 111, 1507–1520. [DOI] [PubMed] [Google Scholar]
- Barlowe, C. (1997). Coupled ER to Golgi transport reconstituted with purified cytosolic components. J. Cell Biol. 139, 1097–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlowe, C. (2000). Traffic COPs of the early secretory pathway. Traffic 1, 371–377. [DOI] [PubMed] [Google Scholar]
- Belgareh-Touze, N., Corral-Debrinski, M., Launhardt, H., Galan, J. M., Munder, T., Le Panse, S., and Haguenauer-Tsapis, R. (2003). Yeast functional analysis: identification of two essential genes involved in ER to Golgi trafficking. Traffic 4, 607–617. [DOI] [PubMed] [Google Scholar]
- Bock, J. B., Matern, H. T., Peden, A. A., and Scheller, R. H. (2001). A genomic perspective on membrane compartment organization. Nature 409, 839–841. [DOI] [PubMed] [Google Scholar]
- Boeddinghaus, C., Merz, A. J., Laage, R., and Ungermann, C. (2002). A cycle of Vam7p release from and PtdIns 3-P-dependent rebinding to the yeast vacuole is required for homotypic vacuole fusion. J. Cell Biol. 157, 79–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burri, L., Varlamov, O., Doege, C.A., Hofmann, K., Beilharz, T., Rothman, J. E., Sollner, T. H., and Lithgow, T. (2003). A SNARE required for retrograde transport to the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA 100, 9873–9877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, X., Ballew, N., and Barlowe, C. (1998). Initial docking of ER-derived vesicles requires Uso1p and Ypt1p but is independent of SNARE proteins. EMBO J. 17, 2156–2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, G. K., Jablonski, S. A., Starr, D. A., Goldberg, M. L., and Yen, T. J. (2000). Human Zw10 and ROD are mitotic checkpoint proteins that bind to kinetochores. Nat. Cell Biol. 2, 944–947. [DOI] [PubMed] [Google Scholar]
- Cosson, P., Schroder-Kohne, S., Sweet, D. S., Demolliere, C., Hennecke, S., Frigerio, G., and Letourneur, F. (1997). The Sec20/Tip20p complex is involved in ER retrieval of dilysine-tagged proteins. Eur. J. Cell Biol. 73, 93–97. [PubMed] [Google Scholar]
- Dilcher, M., Veith, B., Chidambaram, S., Hartmann, E., Schmitt, H. D., and Fischer Von Mollard, G. (2003). Use1p is a yeast SNARE protein required for retrograde traffic to the ER. EMBO J. 22, 3664–3674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasshauer, D., and Margittai, M. (2004). A transient N-terminal interaction of SNAP-25 and syntaxin nucleates SNARE assembly. J. Biol. Chem. 279, 7613–7621. [DOI] [PubMed] [Google Scholar]
- Fasshauer, D., Sutton, R. B., Brunger, A. T., and Jahn, R. (1998). Conserved structural features of the synaptic fusion complex: SNARE proteins reclassified as Q- and R-SNAREs. Proc. Natl. Acad. Sci. USA 95, 15781–15786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferro-Novick, S., and Jahn, R. (1994). Vesicle fusion from yeast to man. Nature 370, 191–193. [DOI] [PubMed] [Google Scholar]
- Frigerio, G. (1998). The Saccharomyces cerevisiae early secretion mutant tip20 is synthetic lethal with mutants in yeast coatomer and the SNARE proteins Sec22p and Ufe1p. Yeast 14, 633–646. [DOI] [PubMed] [Google Scholar]
- Gallwitz, D., Donath, C., and Sander, C. (1983). A yeast gene encoding a protein homologous to the human c-has/bas proto-oncogene product. Nature 306, 704–707. [DOI] [PubMed] [Google Scholar]
- Ghaemmaghami, S., Huh, W. K., Bower, K., Howson, R. W., Belle, A., Dephoure, N., O'Shea, E. K., and Weissman, J. S. (2003). Global analysis of protein expression in yeast. Nature 425, 737–741. [DOI] [PubMed] [Google Scholar]
- Graham, T. R., and Emr, S. D. (1991). Compartmental organization of Golgi-specific protein modification and vacuolar protein sorting events defined in a yeast sec18 (NSF) mutant. J. Cell Biol. 114, 207–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson, P. I., Roth, R., Morisaki, H., Jahn, R., and Heuser, J. E. (1997). Structure and conformational changes in NSF and its membrane receptor complexes visualized by quick-freeze/deep-etch electron microscopy. Cell 90, 523–535. [DOI] [PubMed] [Google Scholar]
- Harlow, E., and Lane, D. (1988). Antibodies. A Laboratory Manual, Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
- Harris, S. L., and Waters, M. G. (1996). Localization of a yeast early Golgi mannosyltransferase, Och1p, involves retrograde transport. J. Cell Biol 132, 985–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatsuzawa, K., Hirose, H., Tani, K., Yamamoto, A., Scheller, R. H., and Tagaya, M. (2000). Syntaxin 18, a SNAP receptor that functions in the endoplasmic reticulum, intermediate compartment, and cis-Golgi vesicle trafficking. J. Biol. Chem. 275, 13713–13720. [DOI] [PubMed] [Google Scholar]
- Hirose, H., Arasaki, K., Dohmae, N., Takio, K., Hatsuzawa, K., Nagahama, M., Tani, K., Yamamoto, A., Tohyama, M., and Tagaya, M. (2004). Implication of ZW10 in membrane trafficking between the endoplasmic reticulum and Golgi. EMBO J. 23, 1267–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh, W. K., Falvo, J. V., Gerke, L. C., Carroll, A. S., Howson, R. W., Weissman, J. S., and O'Shea, E. K. (2003). Global analysis of protein localization in budding yeast. Nature 425, 686–691. [DOI] [PubMed] [Google Scholar]
- Ito, T., Chiba, T., Ozawa, R., Yoshida, M., Hattori, M., and Sakaki, Y. (2001). A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl. Acad. Sci. USA 98, 4569–4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James, P., Halladay, J., and Craig, E. A. (1996). Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144, 1425–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser, C. A., and Schekman, R. (1990). Distinct sets of SEC genes govern transport vesicle formation and fusion early in the secretory pathway. Cell 61, 723–733. [DOI] [PubMed] [Google Scholar]
- Kamena, F., and Spang, A. (2004). Tip20p prohibits back-fusion of COPII vesicles with the endoplasmic reticulum. Science 304, 286–289. [DOI] [PubMed] [Google Scholar]
- Letourneur, F., Gaynor, E. C., Hennecke, S., Demolliere, C., Duden, R., Emr, S. D., Riezman, H., and Cosson, P. (1994). Coatomer is essential for retrieval of dilysine-tagged proteins to the endoplasmic reticulum. Cell 79, 1199–1207. [DOI] [PubMed] [Google Scholar]
- Leung, D. W., Chen, E., and Goeddel, D. V. (1989). A method for random mutagenesis of a defined DNA segment using a modified polymerase chain reaction. Technique 1, 11–15. [Google Scholar]
- Lewis, M. J., and Pelham, H.R.B. (1996). SNARE-mediated retrograde traffic from the Golgi complex to the endoplasmic reticulum. Cell 85, 205–215. [DOI] [PubMed] [Google Scholar]
- Lewis, M. J., Rayner, J. C., and Pelham, H.R.B. (1997). A novel SNARE complex implicated in vesicle fusion with the endoplasmic reticulum. EMBO J. 16, 3017–3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y., Gallwitz, D., and Peng, R. (2005). Structure-based functional analysis reveals a role for the SM protein Sly1p in retrograde transport to the endoplasmic reticulum. Mol. Biol. Cell 16, 3951–3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y., and Barlowe, C. (2002). Analysis of Sec22p in endoplasmic reticulum/Golgi transport reveals cellular redundancy in SNARE protein function. Mol. Biol. Cell 13, 3314–3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer, M. P., and Bukau, B. (2005). Hsp70 chaperones: cellular functions and molecular mechanism. Cell Mol. Life Sci. 62, 670–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNew, J. A., Weber, T., Parlati, F., Johnston, R. J., Melia, T. J., Sollner, T. H., and Rothman, J. E. (2000). Close is not enough: SNARE-dependent membrane fusion requires an active mechanism that transduces force to membrane anchors. J. Cell Biol. 150, 105–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mnaimneh, S. et al. (2004). Exploration of essential gene functions via titratable promoter alleles. Cell 118, 31–44. [DOI] [PubMed] [Google Scholar]
- Muhlrad, D., Hunter, R., and Parker, R. (1992). A rapid method for localized mutagenesis of yeast genes. Yeast 8, 79–82. [DOI] [PubMed] [Google Scholar]
- Nakajima, K., Hirose, H., Taniguchi, M., Kurashina, H., Arasaki, K., Nagahama, M., Tani, K., Yamamoto, A., and Tagaya, M. (2004). Involvement of BNIP1 in apoptosis and endoplasmic reticulum membrane fusion. EMBO J. 23, 3216–3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notredame, C., Higgins, D. G., and Heringa, J. (2000). T-Coffee: a novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 302, 205–217. [DOI] [PubMed] [Google Scholar]
- Ossig, R., Dascher, C., Trepte, H. H., Schmitt, H. D., and Gallwitz, D. (1991). The yeast SLY gene products, suppressors of defects in the essential GTP-binding Ypt1 protein, may act in endoplasmic reticulum-to-Golgi transport. Mol. Cell. Biol. 11, 2980–2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossipov, D., Schroder-Kohne, S., and Schmitt, H. D. (1999). Yeast ER-Golgi v-SNAREs Bos1p and Bet1p differ in steady-state localization and targeting. J. Cell Sci. 112, 4135–4142. [DOI] [PubMed] [Google Scholar]
- Palade, G. (1975). Intracellular aspects of the process of protein synthesis (secretion). Science 189, 347–358. [DOI] [PubMed] [Google Scholar]
- Patel, S. K., Indig, F. E., Olivieri, N., Levine, N. D., and Latterich, M. (1998). Organelle membrane fusion: a novel function for the syntaxin homolog Ufe1p in ER membrane fusion. Cell 92, 611–620. [DOI] [PubMed] [Google Scholar]
- Reilly, B. A., Kraynack, B. A., VanRheenen, S. M., and Waters, M. G. (2001). Golgi-to-endoplasmic reticulum (ER) retrograde traffic in yeast requires Dsl1p, a component of the ER target site that interacts with a COPI coat subunit. Mol. Biol. Cell 12, 3783–3796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigaut, G., Shevchenko, A., Rutz, B., Wilm, M., Mann, M., and Seraphin, B. (1999). A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17, 1030–1032. [DOI] [PubMed] [Google Scholar]
- Roberg, K. J., Rowley, N., and Kaiser, C.A. (1997). Physiological regulation of membrane protein sorting late in the secretory pathway of Saccharomyces cerevisiae. J. Cell Biol. 137, 1469–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothman, J. E. (1994). Mechanisms of intracellular protein transport. Nature 372, 55–63. [DOI] [PubMed] [Google Scholar]
- Rothman, J. E., and Orci, L. (1992). Molecular dissection of the secretory pathway. Nature 355, 409–415. [DOI] [PubMed] [Google Scholar]
- Sacher, M., Jiang, Y., Barrowman, J., Scarpa, A., Burston, J., Zhang, L., Schieltz, D., Yates, I. J., Abeliovich, H., and Ferro-Novick, S. (1998). TRAPP, a highly conserved novel complex on the cis-Golgi that mediates vesicle docking and fusion. EMBO J. 17, 2494–2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapperstein, S. K., Lupashin, V. V., Schmitt, H. D., and Waters, M. G. (1996). Assembly of the ER to Golgi SNARE complex requires Uso1p. J. Cell Biol. 132, 755–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenza, J. C., Hardwick, K. G., Dean, N., and Pelham, H.R.B. (1990). ERD2, a yeast gene required for the receptor-mediated retrieval of luminal ER proteins from the secretory pathway. Cell 61, 1349–1357. [DOI] [PubMed] [Google Scholar]
- Sikorski, R. S., and Hieter, P. (1989). A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122, 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, D. E., and Fisher, P. A. (1984). Identification, developmental regulation, and response to heat shock of two antigenically related forms of a major nuclear envelope protein in Drosophila embryos: application of an improved method for affinity purification of antibodies using polypeptides immobilized on nitrocellulose blots. J. Cell Biol. 99, 20–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Søgaard, M., Tani, K., Ye, R. R., Geromanos, S., Tempst, P., Kirchhausen, T., Rothman, J. E., and Söllner, T. (1994). A Rab protein is required for the assembly of SNARE complexes in the docking of transport vesicles. Cell 78, 937–948. [DOI] [PubMed] [Google Scholar]
- Söllner, T., Bennett, M. K., Whiteheart, S. W., Scheller, R. H., and Rothman, J. E. (1993a). A protein assembly-disassembly pathway in vitro that may correspond to sequential steps of synaptic vesicle docking, activation, and fusion. Cell 75, 409–418. [DOI] [PubMed] [Google Scholar]
- Söllner, T., Whiteheart, S. W., Brunner, M., Erdjument-Bromage, H., Geromanos, S., Tempst, P., and Rothman, J. E. (1993b). SNAP receptors implicated in vesicle targeting and fusion. Nature 362, 318–324. [DOI] [PubMed] [Google Scholar]
- Starr, D. A., Williams, B. C., Hays, T. S., and Goldberg, M. L. (1998). ZW10 helps recruit dynactin and dynein to the kinetochore. J. Cell Biol. 142, 763–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenbeck, G., Harter, C., Brecht, A., Herrmann, D., Lottspeich, F., Orci, L., and Wieland, F. T. (1993). β′-COP, a novel subunit of coatomer. EMBO J. 12, 2841–2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton, R. B., Fasshauer, D., Jahn, R., and Brunger, A. T. (1998). Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4 A resolution. Nature 395, 347–353. [DOI] [PubMed] [Google Scholar]
- Suzuki, M., Igarashi, R., Sekiya, M., Utsugi, T., Morishita, S., Yukawa, M., and Ohya, Y. (2004). Dynactin is involved in a checkpoint to monitor cell wall synthesis in Saccharomyces cerevisiae. Nat. Cell Biol. 6, 861–871. [DOI] [PubMed] [Google Scholar]
- Sweet, D. J., and Pelham, H. R. (1992). The Saccharomyces cerevisiae SEC20 gene encodes a membrane glycoprotein which is sorted by the HDEL retrieval system. EMBO J. 11, 423–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweet, D. J., and Pelham, H. R. (1993). The TIP1 gene of Saccharomyces cerevisiae encodes an 80 kDa cytoplasmic protein that interacts with the cytoplasmic domain of Sec20p. EMBO J. 12, 2831–2840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorngren, N., Collins, K. M., Fratti, R. A., Wickner, W., and Merz, A. J. (2004). A soluble SNARE drives rapid docking, bypassing ATP and Sec17/18p for vacuole fusion. EMBO J. 23, 2765–2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungar, D., and Hughson, F. M. (2003). SNARE protein structure and function. Annu. Rev. Cell Dev. Biol. 19, 493–517. [DOI] [PubMed] [Google Scholar]
- Ungermann, C., Price, A., and Wickner, W. (2000). A new role for a SNARE protein as a regulator of the Ypt7/Rab-dependent stage of docking. Proc. Natl. Acad. Sci. USA 97, 8889–8891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanRheenen, S. M., Cao, X., Lupashin, V. V., Barlowe, C., and Waters, M. G. (1998). Sec35p, a novel peripheral membrane protein, is required for ER to Golgi vesicle docking. J. Cell Biol. 141, 1107–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanRheenen, S. M., Cao, X., Sapperstein, S. K., Chiang, E.C., Lupashin, V. V., Barlowe, C., and Waters, M. G. (1999). Sec34p, a protein required for vesicle tethering to the yeast Golgi apparatus, is in a complex with Sec35p. J. Cell Biol. 147, 729–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanRheenen, S. M., Reilly, B. A., Chamberlain, S. J., and Waters, M. G. (2001). Dsl1p, an essential protein required for membrane traffic at the endoplasmic reticulum/Golgi interface in yeast. Traffic 2, 212–231. [DOI] [PubMed] [Google Scholar]
- Waters, M. G., and Hughson, F. M. (2000). Membrane tethering and fusion in the secretory and endocytic pathways. Traffic 1, 588–597. [DOI] [PubMed] [Google Scholar]
- Waters, M. G., Serafini, T., and Rothman, J. E. (1991). `Coatomer': a cytosolic protein complex containing subunits of non-clathrin-coated Golgi transport vesicles. Nature 349, 248–251. [DOI] [PubMed] [Google Scholar]
- Weber, T., Zemelman, B. V., McNew, J. A., Westermann, B., Gmachl, M., Parlati, F., Sollner, T. H., and Rothman, J. E. (1998). SNAREpins: minimal machinery for membrane fusion. Cell 92, 759–772. [DOI] [PubMed] [Google Scholar]
- Whyte, J. R., and Munro, S. (2002). Vesicle tethering complexes in membrane traffic. J. Cell Sci. 115, 2627–2637. [DOI] [PubMed] [Google Scholar]
- Winzeler, E. A. et al. (1999). Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285, 901–906. [DOI] [PubMed] [Google Scholar]
- Xiao, J., Liu, C. C., Chen, P. L., and Lee, W. H. (2001). RINT-1, a novel Rad50-interacting protein, participates in radiation-induced G(2)/M checkpoint control. J. Biol. Chem. 276, 6105–6111. [DOI] [PubMed] [Google Scholar]
- Yamaguchi, T., Dulubova, I., Min, S. W., Chen, X., Rizo, J., and Südhof, T. C. (2002). Sly1 binds to Golgi and ER syntaxins via a conserved N-terminal peptide motif. Dev. Cell 2, 295–305. [DOI] [PubMed] [Google Scholar]
- Zhang, H. M., Cheung, P., Yanagawa, B., McManus, B. M., and Yang, D. C. (2003). BNips: a group of pro-apoptotic proteins in the Bcl-2 family. Apoptosis 8, 229–236. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.