Abstract
Arp1p is the only actin-related protein (ARP) known to form actin-like filaments. Unlike actin, Arp1p functions with microtubules, as part of the dynein regulator, dynactin. Arp1p's dissimilar functions imply interactions with a distinct set of proteins. To distinguish surface features relating to Arp1p's core functions and to identify the footprint of protein interactions essential for dynactin function, we performed the first complete charge-cluster-to-alanine scanning mutagenesis of an ARP and compared the results with a similar study of actin. The Arp1p mutations revealed three nonoverlapping surfaces with distinct genetic properties. One of these surfaces encompassed a region unique to Arp1p that is crucial for Jnm1p (dynamitin/p50) and Nip100p (p150Glued) association as well as pointed-end associations. Unlike the actin mutations, none of the ARP1 alleles disrupt filament formation; however, one pointed-end allele delayed the elution of Arp1p on gel filtration, consistent with loss of additional subunits.
INTRODUCTION
Microfilaments and microtubules provide cells with the means of locomotion, intracellular transport, mitosis, and cell division. They are comprised of two core proteins, actin and tubulin, each of which are members of larger families of related proteins. Currently, 11 actin-related proteins (Arps) have been identified. Canonical actin has roles in secretion, endocytosis, mitochondrial movement, spindle positioning, and cytokinesis (Pollard and Cooper, 1986). The ARP family governs a similar number of cellular roles; however, each individual ARP has a more narrowly defined function (Schafer and Schroer, 1999). Arp1 is a subunit of the dynactin complex, a regulator of cytoplasmic dynein. Arp2 and Arp3 contribute to actin filament nucleation and branching. Arps4–9 are nuclear localized, with functions including chromatin remodeling (Goodson and Hawse, 2002). Arp11 is an additional dynactin subunit (Eckley et al., 1999). Although the function of yeast Arp10p is not known, some sequence analyses suggest Arp10 and Arp11 belong to the same group. However, a role for Arp10 in the dynactin complex was discounted based on phenotypic criteria (Eckley and Schroer, 2003). Of the known Arps only actin and Arp1 have the ability to form a polymer (Holleran et al., 1998; Bingham and Schroer, 1999). Moreover, both actin and Arp1 bind and hydrolyze ATP, and the CapZ heterodimer binds their barbed-end. The structural similarity of Arp1 and actin is surprising given that Arp1, as part of dynein's regulatory machinery, is a functional component of the microtubule cytoskeleton, rather than actin cytoskeleton (Lees-Miller et al., 1992).
The metazoan dynactin complex contains a 10 × 40-nm filament of Arp1 that serves as a scaffold for the remaining subunits (Schafer et al., 1994; Schroer, 2004). Using the same terminology developed for conventional actin, the barbedend of the Arp1 filament is bound by the CapZ heterodimer. The pointed-end of the Arp1 filament is bound by a complex consisting of a unique set of proteins identified by their molecular weights: p62, p27, p25, and a second actin-related protein, Arp11. The remaining subunits (p50/dynamitin, p22/24, and p150Glued) form structures referred to as the shoulder and sidearm (Schroer, 2004). Electron microscopy (EM) studies (Schafer et al., 1994) indicated that a portion of the p150Glued subunit projects from the side of the Arp1p filament near the barbed-end. However, recent work suggests that a larger portion of p150Glued also lies along the Arp1 filament (Schroer, 2004). The exact position of the four dynamitin/p50 subunits remains elusive. An ultrastructural analysis of dynactin using antibodies against individual subunits did not include the location of dynamitin/p50 (Schafer et al., 1994). However, the isolation of dynamitin/p50 associated with both p150Glued and p22/24 as well as with the Arp1p filament, suggested it was located near the projecting arm of p150Glued (Eckley et al., 1999). Excess dynamitin/p50 destabilizes dynactin in vitro and in vivo suggesting that it links p150Glued to the Arp1p filament (Schroer, 2004). Recent single-particle EM analysis (Hodgkinson et al., 2005) found additional mass along the Arp1p filament. Although it was proposed that this could represent dynamitin/p50, a definitive assignment awaits further analysis to exclude p150Glued or p22/24.
The dynactin complex is required for two aspects of cytoplasmic dynein activity: cargo selection and processivity. The importance of processivity in the whole organism is revealed by mutations that reduce dynactin's affinity for microtubules. Such mutations are the causative basis of lower motor neuron disease in humans (Puls et al., 2003). Mutations that affect cargo selection are also potential disease locus candidates.
The vertebrate dynactin/dynein complex has distinct roles in different parts of the cell cycle. Throughout interphase, it drives retrograde vesicular trafficking and maintains the microtubule organizing center. During mitosis, it is responsible for focusing spindle microtubules, poleward movement of chromosomes, and rotation of the mitotic spindle via microtubule interactions at the cortex. The budding yeast dynactin complex also is clearly required for the activity of cytoplasmic dynein, but its functions are limited to mitotic roles.
In the course of mitosis in budding yeast, a spindle forms in the mother cell. Half of the spindle must pass through the bud neck to place its complement of chromosomes into the growing bud. This occurs by the coordinated effort of two functionally redundant systems (Cottingham and Hoyt, 1997; DeZwaan et al., 1997; Miller et al., 1998). First, the proteins of the KIP3/KAR9 pathway move the preanaphase spindle to the bud neck, aligning it along the mother bud axis (Korinek et al., 2000; Lee et al., 2000; Miller et al., 2000; Yin et al., 2000). Second, the anaphase spindle moves through the bud neck via the dynein/dynactin pathway, in conjunction with nuclear forces provided by CIN8 and KIP1 (Cottingham and Hoyt, 1997). Normally, either system is capable of completing mitosis alone; however, mitosis cannot be completed in the absence of both systems. Yeast homologues of five of the known subunits of dynactin have been identified (ARP1 encodes Arp1, JNM1 encodes dynamitin/p50, NIP100 encodes p150Glued, and CAP1 and CAP2 encode the capping proteins). Mutations in any of the subunits of yeast dynactin produce a defect in mitosis indistinguishable from that of DYN1. Additional homologues for six proteins of the vertebrate complex have yet to be identified (Holleran et al., 1998; Eckley et al., 1999; Karki and Holzbaur, 1999).
Charge-cluster-to-alanine scanning has been successfully used to define the protein interactions of actin and tubulin as well as their motors myosin II and kinesin (Wertman et al., 1992; Reijo et al., 1994; Woehlke et al., 1997; Sasaki and Sutoh, 1998). For Act1p (actin), alanine scanning clearly defined the binding surfaces of several actin-interacting proteins, including Aip1p, Sac6p/fimbrin, myosin, and the small molecules phalloidin and latrunculin A (Wertman et al., 1992; Holtzman et al., 1994; Amberg et al., 1995; Miller et al., 1995; Belmont et al., 1999b). Furthermore, it has been used to assess actin's functional role in mitochondrial transport and endocytosis (Drubin et al., 1993; Belmont and Drubin, 1998). Finally, alanine scanning has served as a starting point for further mutagenesis (Belmont and Drubin, 1998; Belmont et al., 1999b).
Actin and Arp1p are highly similar, thus, many surface features are conserved; however, several features remain distinct. Here, we investigated the essential differences that allow these two similar proteins to have distinct functions. Presumably, specificity arises from contacts with unique sets of proteins. We reasoned that these Arp1p surfaces could be identified, in vivo, by comparative studies of loss of function through alanine scanning.
MATERIALS AND METHODS
Strains and Yeast Methods
All strains, unless noted, were derived from MY8648 or MY8649. These are derivatives of YPH499 (Sikorski and Hieter, 1989) wherein the ade2-101 allele has been replaced by ade2Δ and ade3Δ has been deleted. Integration was achieved by the TRAFO transformation method (Gietz and Schiestl, 1995). General yeast methods, media, and plasmid transformation were as described previously (Rose et al., 1990).
Isolation of an ARP1 Synthetic Lethal Locus
We deleted ARP1 by replacement with TRP1 (pMR5224) in a strain background (MY8648) harboring mutations in ADE2 and ADE3, to produce strain MY8650. A centromeric plasmid bearing URA3, ADE3, and ARP1 (pMR5222) was introduced to give MY8651. When the centromeric plasmid is lost, white sectors are produced in an otherwise red colony due to the epistasis of ade3. MY8651 was mutagenized with ethyl methane sulfonate. We screened 14,600 colonies for nonsectoring candidates on SC low-adenine media. Plasmid dependence was further examined by sensitivity to 5-fluoroorotic acid (5-FOA). A plasmid substitution test (plasmid shuffle) determined that the synthetic phenotype was specific to ARP1 and not to other markers on the plasmid. The candidates were backcrossed to the parental strain to determine whether a single locus was responsible for the synthetic lethality. Strains resulting from two or three additional backcrosses were used for further analyses.
Cloning and Characterization of KIP3 as the ARP1 Synthetic Lethal Locus
A plasmid library (65-25-1) was prepared by partial Sau3A digestion of genomic DNA from strain MS15 and inserted pMR1865 digested with BamHI as described previously (Rose et al., 1987) with minor modifications. KIP3 was isolated from the library by colony hybridization. A KIP3 minimal open reading frame (ORF) was constructed by subcloning a 4061-base pair EcoRI/BamHI fragment into pMR1869. The KIP3 plasmid was shown to suppress the lethality of an arp1Δ kip3-15 strain.
To delete KIP3, HIS3 was amplified from plasmid pMR1865 with primers that add KIP3 homologous sequences to the ends of the PCR product (Miller et al., 1998). The PCR product was transformed into strain MY8651 to replace KIP3 with kip3Δ::HIS3. The resulting strain is MY8652 (arp1Δ::TRP1 kip3Δ::HIS3). Other strains containing kip3Δ::HIS3 were constructed by standard genetic crosses.
Plasmid Construction
For pMR5222, ARP1 was inserted as an XbaI/SalI fragment from pMR5223 into pMR5221. For ARP1 overexpression constructs, all ARP1 alleles were transferred from their respective pBScKS-(XbaI-) ARP1-LEU2 cassettes as an EcoRI fragment into both pMR1865 (CEN, HIS3) and pMR1869 (2μ HIS3). For ARP1“shuffle” plasmid, pMR5225 was created by inserting the XbaI/SalI fragment from pMR5223 bearing ARP1 into pMR1867 a CEN LEU2 plasmid. For arp1Δ1-384::HIS3 integration plasmid (pMR5228), XbaI and SacI of pBScKS-were destroyed by Klenow fill-in (XbaI) or made blunt with T4 DNA polymerase (SacI) followed by religation, producing pMR5230. An EcoRI fragment bearing ARP1 from pMR5223 was recloned into this modified vector at EcoRI, producing pMR5231. This was in turn linearized with SacI, made blunt with T4 DNA polymerase, further cleaved with XbaI, and then filled-in with Klenow. A fragment harboring HIS3 was isolated from pMR1924 by excision with BamHI followed by fill-in with Klenow. This was cloned into pMR5231 (SacI/XbaI), replacing ARP1 with HIS3. HIS3 retains the original orientation of the ARP1 ORF it replaces. This construct was linearized with ClaI and integrated at ARP1 in strain MY8656 producing MY8657. The integration was confirmed by Southern blotting of PstI-cleaved genomic DNA, with a HindIII fragment harboring ARP1 from pMR5223. pMR5467 was created by gap-repair via cotransformation of pMR1869 (cleaved with EcoRI and BamHI) with a PCR product containing the NIP100 ORF and 5′ and 3′ flanking regions. pMR5468 was created in a similar manner, except the JNM1 ORF and surrounding sequence were amplified. Both plasmids were confirmed by sequencing.
Alanine Scanning
Mutagenesis was accomplished using the dut-ung-method (Kunkel, 1985). Primers generally had nine base pairs upstream and 12 base pairs down-stream, with occasional extensions to adjust the GC content. Mutations were screened wherever possible by the gain or loss of a restriction site. All alanine scanning cassettes were sequenced with Sequenase (United States Biochemical, Cleveland, OH) or by the Princeton SYN/SEQ facility. Oligonucleotides were synthesized on an Applied Biosystems 391A DNA synthesizer or obtained from Invitrogen (Carlsbad, CA).
Construction of ARP1
For LEU2 cassette for mutagenesis, an EcoRI fragment harboring ARP1 and FUR1 was cloned from pMR5223 into pMR5229, which had its XbaI site destroyed by fill-in with Klenow, producing pMR5237 wherein the 5′end of ARP1 is adjacent to the KpnI site in the polylinker. An XbaI site downstream of ARP1 was filled by Klenow to accept an XbaI/BamHI (both filled) fragment harboring LEU2 from pMR2254. The final construct (pMR5238) has ARP1 and LEU2 transcribed in the same direction. For gene replacement of the arp1Δ::HIS3 locus, a linear fragment containing ARP1 and LEU2 was released with HindIII.
FUR1-TRP1
arp1Δ1-384::HIS3 results in the truncation of FUR1, leading to 5-FOA sensitivity (Kern et al., 1990). Additionally, a trp1Δ63 strain is inherently cold sensitive. To address these two issues, a plasmid containing both TRP1 and FUR1 was constructed and integrated at trp1Δ63 producing MY8655. Details are available upon request. Probes prepared from TRP1 and FUR1 were used to confirm the integration by Southern blotting. Strains containing the TRP1-FUR1 construct were constructed by standard genetic methods.
Integration of ARP1 Alanine Scanning Alleles
We used the method of (Wertman et al., 1992) to facilitate the screening of integrants directed to the ARP1 locus. Specifically, ARP1 alleles marked with LEU2 were introduced into a strain harboring arp1Δ::HIS3 such that there is no homologous sequence between ARP1 and LEU2 that could direct a double crossover involving LEU2 alone. In this manner, one can quickly screen for correct targeting to the ARP1 locus by screening for transformants that have switched from His+ Leu– to Leu+ His–. By integrating into haploid strains, the likelihood of recombination (gene conversion) from a wild-type source of ARP1 was limited; however, in most cases the presence of the mutation at the ARP1 locus was confirmed by sequencing.
ARP1 under the Tetracycline Promoter
The EUROSCARF dual activation/repression system was used to regulate the level of Arp1p (Belli et al., 1998a, b). Strain MY8660 (kip3Δ::HIS3) was transformed with pMR5240 (pCM244) linearized at EcoRV to introduce CMVp(tetR'-SSN6)::LEU2 at leu2-Δ1, producing MY8661. PwoI polymerase (Roche Diagnostics, Indianapolis, IN) was used to amplify ARP1 with a kanMX4-tetO2 (tetracycline) promoter from NotI-linearized pMR5241 (pCM224) template. This was integrated at ARP1 to produce MY8662. For both solid and liquid media, YPD-doxycycline was prepared by addition from a 5 mg/ml doxycycline (Sigma, St. Louis, MO) stock in 50% ethanol. Then, 1:3 dilutions were made beginning with a 10 μg/ml final concentration. For examination on solid media, strains were pregrown on solid YPD at 23°C, resuspended in distilled H2O, 10-fold serial dilutions were made, and small samples were spotted onto Petri plates with a 48-prong device. For protein extracts, strains were pregrown on solid YPD at 23°C. A single colony was transferred into YPD ± doxycycline and grown overnight at 30°C. Protein extracts were prepared as below.
Protein Methods
Protein extracts were prepared from overnight cultures of exponentially growing cells. For temperature shifts, exponentially growing cells were resuspended in prewarmed media and were harvested after 3 h growth at the specified temperature. Cell pellets were resuspended at 1.2 × 109 cells/ml in freshly prepared ice-cold lysis buffer [50 mM Tris-Cl, pH 8.0, 150 mM NaCl, 1 mM Na2ATP, 1 mM dithiothreitol [DTT], 40 μg/ml phenylmethylsulfonyl fluoride, 5 μg/ml each chymostatin, pepstatin, antipain, arpotinin, and leupeptin). One milliliter of resuspended cells was added to prechilled glass beads (0.5 mm). Cells were lysed with a Beadbeater (Biospec Products, Bartlesville, OK) for 1.5 min. at 4°C. Crude lysates were clarified in a micro-centrifuge for 5 min at 4°C. Cytosols were obtained by centrifugation in a TLA100 rotor at 50,000 rpm for 10 min at 4°C. Protein concentration was determined using the Bio-Rad (Hercules, CA) protein assay with bovine serum albumin as a control. For analysis of steady-state protein levels, an equal quantity of total protein was added to each well, proteins were resolved by SDS-PAGE, and the relative quantity of Arp1p determined by immunoblotting with an Arp1p antibody. Signal strength was examined on appropriate exposures with ImageJ (http://rsb.info.nih.gov/ij/).
Gel Filtration
Cytosols (100 μl) were examined at 4°C on a Superose6 column (Amersham Biosciences, Piscataway, NJ) in 50 mM Tris-Cl, pH 7.4, 150 mM NaCl, 1 mM DTT, and 1 mM ATP at 0.3 ml/min. Strains were shifted to 37°C for 3 h before lysis, where indicated. One-milliliter fractions were collected and trichloroacetic acid (TCA)-precipitated. One-half of the TCA pellet was examined by SDS-PAGE. LOAD corresponds to 5 μl of cytosol. Calibration standards were Void, 6.8 ml; thyroglobulin (669 kDa, Stokes = 85Å), 11.0 ml; ferritin (440 kDa, Stokes = 61Å), 13.0 ml; aldolase (158 kDa, Stokes = 48.1Å), 14.3 ml; ovalbumin (43 kDa, Stokes = 30.5Å), 15.6 ml; and acetone, 19.7 ml.
Modeling of Arp1p
Using the Swiss-Model Server (http://www.expasy.ch/swissmod/), (Peitsch, 1996; Guex and Peitsch, 1997), the Arp1p sequence was modeled and displayed based on a theoretical conformation of an actin subunit in the actin filament (Lorenz et al., 1993). Electrostatic surfaces were calculated and represented with the Swiss-PDBViewer using the Coulomb method.
Bioinformatics
To determine the conservation of Saccharomyces cerevisiae (S.c.) Arp1p charged residues among other Arp1s, other Arps, and actins, we created a custom Perl script. The script took the alignment of Arps and actins from (Poch and Winsor, 1997) and extracted and tabulated the conservation of each S.c. Arp1p charged residue among subgroups of aligned sequences: fungal Arp1s, metazoan Arp1s, all Arp1s, all Arps, all actins, or the total alignment. In each case, the sequence of S.c. Arp1p was removed from the analysis. For each S.c. charged residue, the script tabulated the ARP1 allele it belongs to, the allele's phenotype, and the frequency and type of conservation among a given alignment. Conservation types were 1) exact, 2) conserved change (D/E for E/D) or (R/K for K/R), 3) opposite charge (R/K for D or E, D/E for R or K), or 4) nonconserved (any residue other than D, E, R, or K).
Arp1-(HIS)6p for Bacterial Expression
The N terminus of ARP1 was amplified with primers introducing an NcoI site at the initiator methionine. This PCR product was cloned into pMR5229 at EcoRV modified as a ddT-Vector (Holton and Graham, 1991). The amplified segment was confirmed by sequencing. The cloned fragment was cleaved at StuI within the ARP1 ORF and at XbaI in the pBScKs-polylinker. A StuI/XbaI fragment was isolated from pMR5223 and swapped into the NcoI-modified ARP1 sequence to generate a full-length ARP1 ORF. NcoI-modified ARP1 was then cloned into pMR5248 (pET8c) via NcoI/BamHI (Studier and Moffatt, 1986). The NcoI-modified ARP1 was also cloned into pMR5372 (pGEX-KG) as an NcoI/SacI fragment, producing pMR5245. Antibodies were raised against Arp1-(HIS)6p by CoCalico (Reamstown, PA). Arp1-(HIS)6p and GST-Arp1p were expressed and purified and antibodies were affinity purified on Arp1-(HIS)6p and titrated against either Arp1-(HIS)6p or GST-Arp1p as described previously (Clark and Meyer, 1992). Arp1p was detected by immunoblotting using enhanced chemiluminescence (Amersham Biosciences). A rabbit polyclonal antibody against Jnm1p was the generous gift of K. Tatchell.
Deletion of Other Dynactin Subunits and Dynein Heavy Chain
NIP100 was replaced with a kanMX cassette in strain MY8885 using pMR5246 (pFA6akanMX4) as a template for PCR to produce MY8669 (Wach et al., 1994). DYN1 was replaced with URA3 by transformation of strain MY8648 with EcoRI-linearized pMR5247 (pBΔR2U-5), to produce MY8670. Both The nip100Δ and dyn1Δ were confirmed by PCR. URA3 was subsequently replaced at dyn1Δ with HIS3 by transformation with plasmid pMR4269 (PUH7) linearized at XbaI (Cross, 1997), producing MY8671.
Jnm1p-Arp1p Two-Hybrid Interactions
Plasmids with ARP1 mutations fused to the GAL DNA binding domain (GBD) were transformed into two-hybrid tester strains (James et al., 1996) already harboring either ARP1 (MY8883) or JNM1 (MY8884) fused to the GAL activation domain (GAD). Activation of the HIS3 reporter was monitored at 23 and 30°C.
RESULTS
ARP1 Charge-Cluster-to-Alanine Alleles
Charge-cluster-to-alanine scanning mutations target regions with high local charge. Based on the assumption that charge-clusters are located on the protein surface, this method is expected to favor mutations that interfere with protein–protein interactions rather than disturbing tertiary structure. A total of 45 mutations of ARP1 were constructed. ARP1 alanine scanning mutations were chosen along the primary sequence by searching a five-residue window for at least two charged residues (Figure 1 and Tables 1 and S1). To maximize the ability to compare ARP1 with ACT1, we also made six additional mutations. In two cases, the mutants encompass more than five residues to accommodate additional charge (arp1-44 and arp1-45), and in two cases, where the cognate ACT1 charge cluster was of interest, but only a single charge was conserved in Arp1p, only a single residue was mutated (arp1-40 and arp1-42). Finally, because similar mutations in Neurospora crassa Arp1p compromise dynactin function (Minke et al., 2000), a hydrophobic residue (Tyr151) was mutated to alanine (arp1-176) and to serine (arp1-181). All mutations were integrated at single copy at the ARP1 locus (see Supplemental Methods).
Figure 1.
Comparison of ARP1 and ACT1 charge-cluster to alanine mutations. ARP1 allele numbers are indicated. Green indicates pseudo-wild-type, red Ts–, and black lethal. Alleles separated by a slash are overlapping charge clusters where the double underline designates the shared residues. Alanine scanning alleles for actin and their phenotypes are indicated on the lower strand. Actin data are from Wertman et al. (1992).
Table 1.
ARP1 allele position and residues, protein levels, and overexpression rescue
| ARP1 allele | Residue | Pheno | PP | PR | CEN | 2μ |
|---|---|---|---|---|---|---|
| 33 | K222A, E223A, K224A | L | - | - | - | - |
| 35 | R185A, D187A | T | + | + | (+) | + |
| 36 | D321A, R322A | L | - | (-) | - | - |
| 37 | E31A, E32A | W | + | + | ||
| 38 | E374A, D375A | T | (-) | (-) | (-) | + |
| 39 | E84A, D85A | T | (-) | (-) | + | + |
| 40 | D58A | W | + | + | ||
| 41 | D87A, E90A | W | + | + | ||
| 42 | E108A | W | + | + | ||
| 44 | D48A, K49A, E53A | W | (+) | (-) | ||
| 45 | K233A, K234A, E235A, E236A, E237A, K238A | T | (-) | (+) | + | + |
| 46 | K46A, D48A, K49A | W | (+) | (-) | ||
| 85 | R266A, E269A | L | + | - | - | - |
| 86 | R124A, E125A | W | + | + | ||
| 94 | D2A, D6A | T | - | - | + | + |
| 95 | K35A, E38A | W | ||||
| 96 | K6A7, R69A | W | ||||
| 97 | K73A, R75A | W | ||||
| 98 | R75A, K79A | W | ||||
| 99 | K122A, R124A, E125A | W | ||||
| 100 | E133A, D136A | L | - | - | - | (+) |
| 101 | D162A, E165A | W | ||||
| 102 | D192A, E195A | W | ||||
| 103 | R202A, K203A | W | ||||
| 104 | E214A, R215A, E216A | L | - | - | - | (+) |
| 105 | K251A, D254A | L/DC | - | - | - | - |
| 106 | R256A, E259A | W | ||||
| 107 | D263A, R264A | W | ||||
| 108 | D282A, D286A | W | ||||
| 109 | K294A, D296A | W | ||||
| 110 | D298A, R300A, K301A | W | ||||
| 111 | D326A, E328A | T | - | - | + | + |
| 112 | K336A, K338A | W | + | + | ||
| 113 | E344A, R345A, K346A | T | (-) | (-) | - | - |
| 114 | R364A, K368A | W | ||||
| 115 | K369A, D371A | L | - | - | - | + |
| 134 | E31A, E32A, R33A, K35A | W | ||||
| 135 | E214A, R215A, E216A, R219A | L | - | - | ||
| 136 | K230A, K233A, K234A | W | ||||
| 137 | D296A, D298A | W | ||||
| 138 | K317A, D321A | W | ||||
| 139 | K332A, K336A | W | ||||
| 140 | E374A, D375A, R378A | T | (+) | (+) | ||
| 176 | Y151A | W | ||||
| 181 | Y151S | W |
Pheno, phenotype; PP, protein at permissive temperature; PR, protein at restrictive temperature; CEN, rescue by one or two copies of allele; 2μ, rescue by high-copy; L/DC, lethal and dominant cold-sensitive; W, pseudo-wild type; T, temperature sensitive (Ts-); gray highlights exceptions to general trends. Protein levels (PR, PP) or growth (CEN, 2μ) are indicated, in decreasing level, by +, (+), (-), and -.
Phenotypic Spectrum of the ARP1 Alanine Scanning Alleles
Because ARP1 is not essential for life, we conducted a screen for mutations that render ARP1 essential. A mutation in the kinesin-related gene kip3-15, showed synthetic lethality with arp1Δ (see Supplemental Data). “Synthetic lethality” occurs when two pathways, either of which is sufficient to complete a task, are simultaneously deleted resulting in a lethal phenotype. The ARP1 alleles were compared for growth in the kip3-15 background under conditions that counterselect the plasmid carrying the wild-type copy of ARP1 (5-FOA) at four temperatures. The variety of defects ranged from having no discernible phenotype (pseudo-wild type) to as severe as arp1Δ (Figure 2). By definition, an ARP1 pseudo-wild-type mutation produces no synthetic interactions with the KIP3 pathway. Sixteen of the 45 alleles caused a temperature-sensitive (Ts–), Ts–/cold-sensitive (Cs–), or lethal phenotype. Because the phenotype of a copolymer of wild-type and mutant Arp1p could be informative, we also assessed relevant alleles in the heterozygous configuration. One mutation, arp1-105, which maps to a charge cluster on a homotypic interface (Arp1p–Arp1p), exhibited a partially dominant Cs– phenotype (Figure 3). This is similar to the location of dominant lethal clusters in Act1p (Wertman et al., 1992). All other arp1 alleles were recessive (our unpublished data).
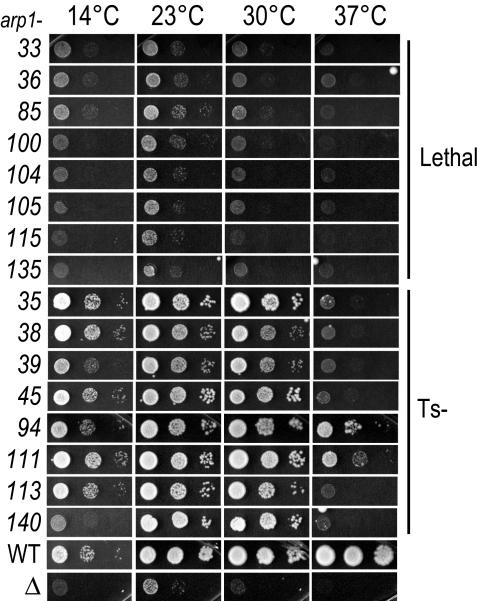
Figure 2.
Phenotypic profiles of lethal and Ts– ARP1 alleles. All 45 ARP1 alleles were examined for growth phenotypes in the kip3-15 background. Ts– and lethal are shown here. Each allele was covered by a plasmid harboring wild-type ARP1 that was counterselected with 5-FOA at four temperatures. The slight growth at 23°C for arp1Δkip3Δ is typical of this strain background. Each spot shows the growth of successive, serial 1:10 dilutions from left to right. WT, ARP1+; Δ, arp1Δ.
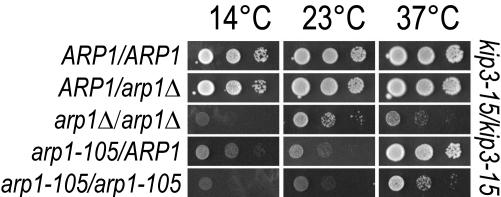
Figure 3.
Hemizygous ARP1 and the partial dominant cold sensitivity of arp1-105. Diploid strains harbored the indicated ARP1 alleles in a homozygous kip3-15 background. Hemizygous ARP1 diploids were compared with the homozygous ARP1 wild-type and homozygous arp1Δ diploids. The hemizygous configuration of ARP1 does not affect growth. These were further compared with arp1-105 in homozygous and heterozygous configurations. The decreased growth of heterozygous arp1-105 at 14 and 23°C relative to heterozygous arp1Δ indicates a dominant cold sensitivity for arp1-105. Growth was examined at three temperatures by 5-FOA counterselection of a plasmid harboring ARP1. Tenfold serial dilutions are shown.
Reduction in Steady-State Arp1p Levels Is Not the Basis of Their Phenotype
Knowledge of the underlying basis of ARP1 mutant phenotypes is required for analyzing its role in dynactin function. At one extreme, a mutation may destabilize the tertiary structure of Arp1p, leading to degradation of the monomer and destruction of the dynactin complex. At the other extreme, the inherent stability of the Arp1p monomer may be unaffected; but key protein interactions may be disrupted, resulting in loss of function. Monitoring the steady-state level of Arp1p revealed four trends (Table 2 and Figure S2). First, most pseudo-wild-type mutants had wild-type levels of protein. Second, most severe mutants had greatly reduced protein levels. Third, some mutants with greatly reduced levels nevertheless retained considerable function. Fourth, one mutant showed high levels of protein despite a severe growth defect. Together, there is a general correlation between the steady-state level of Arp1p and the severity of the Ts– phenotype. At first blush, one might conclude that the Ts– or lethal phenotype merely reflects steady-state Arp1p levels; however, there are notable exceptions to this rule. First, several alleles caused a strong reduction of protein levels at both 23 and 37°C but retained near-wild-type phenotypes at 23°C (arp1-38, 39, 45, 94, 111, and 113). Second, some alleles caused a markedly reduced steady-state level of Arp1p at 37°C, but exhibited pseudo-wild-type phenotypes (arp1-44 and arp1-46). The issue becomes whether the reduced levels of Arp1p are the cause of the phenotype or a secondary effect of a primary change in Arp1p function or interaction. We took two complementary approaches to identify the functional relationship between ARP1 mutations and steady-state levels of expression. First, we adjusted the levels of the mutant protein to wild-type by overexpression. Second, we directly determined the protein threshold for Arp1p phenotypic defects.
Table 2.
Correlation of phenotype and protein level
| Phenotype class | ARP1 allele | Protein at permissive temperature | Protein at restrictive temperature |
|---|---|---|---|
| Pseudo-wild type | 44 | (+) | (-) |
| 46 | (+) | (-) | |
| 37 | + | + | |
| 40 | + | + | |
| 41 | + | + | |
| 42 | + | + | |
| 86 | + | + | |
| 112 | + | + | |
| Ts- | 45 | (-) | (+) |
| 35 | + | + | |
| 94 | - | - | |
| 111 | - | - | |
| 113 | (-) | (-) | |
| 38 | (-) | (-) | |
| 39 | (-) | (-) | |
| Lethal | 85 | + | - |
| 36 | - | (-) | |
| 33 | - | - | |
| 100 | - | - | |
| 104 | - | - | |
| 105 | - | - | |
| 115 | - | - |
Shaded regions highlight key exceptions to the general pattern of the correlation between phenotype and level. Protein levels were scored at permissive (23°C) and restrictive (37°C) temperature. A qualitative assessment of protein levels (see also Figure S2) are indicated, in decreasing level, by +, (+), (-), and -.
The introduction of just one or two additional copies on a centromeric plasmid (CEN) or higher levels afforded by a high-copy plasmid (2μ) appreciably relieved the phenotypes of several Ts– alleles. The exception was arp1-113 (Table 3 and Figure S3). Conversely, only three of the lethal alleles (arp1-100, 104, and 115) showed rescue when expressed from 2μ. Because overexpression increased steady-state levels for all but a few alleles (Figure S4), this analysis suggests that most lethal alleles produce a direct growth phenotype, and that the reduction in steady-state level of Arp1p is a secondary effect. In contrast, the rescue of most of the Ts– alleles by increased protein levels is consistent with reduced stability. However, many of the Ts– alleles were overexpressed beyond wild-type levels and reduced interactions might be suppressed by mass action. Thus, although there is a gross correlation between the steady-state level of Arp1p and temperature sensitivity, the reduced steady-state level of Arp1p was not the primary basis of the phenotype in many cases.
Table 3.
Remediation of phenotype by overexpression
| Phenotype | ARP1 allele | CEN rescue | 2μ Rescue |
|---|---|---|---|
| Ts- | 113 | - | - |
| 35 | (+) | + | |
| 38 | (-) | + | |
| 39 | + | + | |
| 45 | + | + | |
| 94 | + | + | |
| 111 | + | + | |
| 140 | (+) | (+) | |
| Lethal | 100 | - | (+) |
| 104 | - | (+) | |
| 115 | - | + | |
| 33 | - | - | |
| 36 | - | - | |
| 85 | - | - | |
| 105 | - | - | |
| 135 | - | - |
Shaded regions highlight key exceptions to the remediation of phenotype by overexpression. CEN represents one or two copies per cell; 2μ high-copy. Degree of growth rescue is indicated, in decreasing order, by +, (+), (-), and -.
A complementary approach to assessing the contribution of a mutation's steady-state level to its phenotype is to evaluate the growth threshold for wild-type Arp1p. In the case of actin, cells are very sensitive to protein levels; the hemizygous state (one wild-type allele, one null) confers a growth defect (Wertman et al., 1992), and a Ts– phenotype can result from any mutation that destabilizes the protein. This was not the case for hemizygous ARP1 diploids, which grew indistinguishably from homozygous ARP1 diploids, in a homozygous kip3Δ background (Figure 3). Accordingly, we determined the minimum steady-state level at which wild-type Arp1p supported wild-type growth and compared this with the mutants. This was particularly relevant to Ts– alleles whose mutant phenotype was alleviated by increasing steady-state protein levels. To control Arp1p steady-state expression levels, we integrated a tetracycline operator dual activation/silencing cassette in lieu of the endogenous ARP1 promoter (see Supplemental Material). This configuration constitutively expressed Arp1p in the absence of doxycycline at levels similar to the endogenous ARP1 promoter (Figure 4A). As doxycycline was added, the Arp1p steady-state level gradually decreased, eventually reaching a plateau. Comparison of growth on solid media indicated that the threshold for the growth defect occurred when the corresponding protein level was 10- to 20-fold lower than endogenous Arp1p levels (Figure 4B). Clearly, wild-type cells maintain an excess of Arp1p. Because the cells were viable with greatly reduced Arp1p levels and thus fewer dynactin complexes, an Arp1p mutant need only be expressed above this protein threshold for assembly of sufficient dynactin complexes. The low threshold creates a large window within which mass action can effect rescue. Reconsidering the Ts– alleles, it is evident that alleles arp1-35, 38, 39, and 45, should be pseudo-wild type if protein levels were the sole basis of their phenotype. A similar argument can be made for two lethal alleles, arp1-36 and arp1-85. Many of the remaining alleles have protein levels below the wild-type threshold but produce a phenotype worse than a comparable level of wild-type Arp1p, suggesting that these mutants also cause defects in Arp1p function. Thus, protein levels reflect the severity of the Ts– or lethal phenotype, rather than lying at the root of its cause.
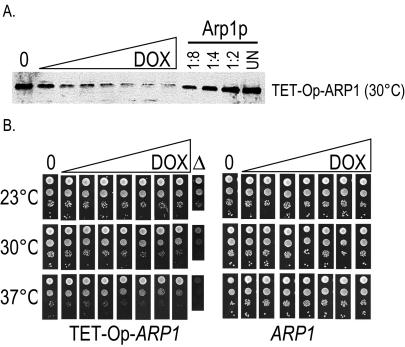
Figure 4.
(A) ARP1 under control of the tetracycline operator allows reduction of the steady-state level of wild-type Arp1p. Immunoblot of ARP1 kip3Δ strains wherein the endogenous ARP1 promoter had been replaced with a tetracycline operator and minimal promoter (TET-Op-ARP1). Cultures were grown with increasing concentrations of the tetracycline analog doxycycline (DOX) (0, no doxycycline). For comparison, serial dilutions of a wild-type ARP1 strain are shown. UN, undiluted. Arp1p was detected with anti-Arp1p antibody. (B) ARP1 under the tetracycline operator defines a growth threshold that can be equated with a steady-state level of wild-type Arp1p. Tenfold serial dilutions of a TET-Op-ARP1 kip3Δ strain and an ARP1 kip3Δ strain were examined for growth at three temperatures on solid media containing increasing concentrations of doxycycline, matching those shown in A. A growth defect is first visible between the third and fourth dilutions of doxycycline. An arp1Δ strain grown in the absence of doxycycline is shown for comparison.
Integrity of the Dynactin Complex in Ts– and Lethal ARP1 Alleles
All of Arp1p's known functions are associated with the dynactin complex and the arp1 mutants could lead to a variety of defects at the molecular level. On one hand, mutant Arp1p could be incapable of forming a filament. Conversely filament formation may be possible, but crucial interactions with other dynactin subunits or extradynactin interactions might be lost. To differentiate between these models we used two approaches to examine the mutant dynactin complexes. First, we used size exclusion chromatography to monitor changes in the apparent elution of the Arp1p filament in ARP1 mutants. Second, we examined the stability of Arp1p in dynactin mutants.
Monitoring the Arp1p filament by size-exclusion chromatography indicated a broad peak of Arp1p (Figure 5). Examination of Arp1p behavior in the absence of Jnm1p or Nip100p resulted in a similar profile (our unpublished data), suggesting that these proteins do not stably associate with the Arp1p filament under these conditions. Although this apparent dissociation limits the usefulness of this assay for determining the effects of ARP1 mutants on specific dynactin subunit associations, we used this behavior to our favor to monitor changes in the Arp1p filament itself. Combining data from sedimentation velocity in sucrose gradients (our unpublished data) with the Stokes radius estimated from the gel filtration, we found the approximate dimensions of the Arp1p filament to be 39 × 7 nm under our conditions (see Supplemental Material). These dimensions agree very well with the expected dimensions of an Arp1p filament lacking side-binding subunits (7 × 40 nm) (Schroer, 2004).
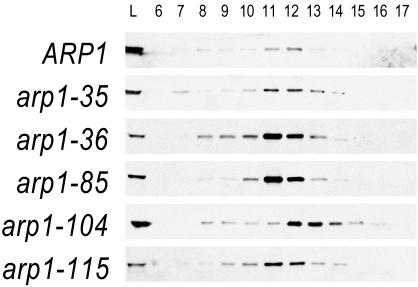
Figure 5.
Elution profiles of Arp1p mutants when overexpressed. Strains harboring mutant Arp1p were examined by gel filtration and Arp1p was detected by immunoblot with anti-Arp1p antibody. Fraction size is 1 ml. Under these conditions, we are likely examining only the Arp1p filament and capping subunits (see main text). The panels compare elution of wild-type Arp1p with that of Ts– and lethal alleles, overexpressed from a 2μ plasmid. For wild-type, 10-fold less protein was examined. The Ts– of arp1-35 and the lethal allele arp1-115 are fully rescued under these conditions, whereas the lethal allele arp1-36 is not rescued by expression to wild-type steady-state levels. Despite these phenotypic differences, all show the same peak elution. Lethal allele arp1-104, which is partially rescued by overexpression, has a notably delayed elution compared with the other mutants. Cytosols were prepared from strains transformed with a 2μ plasmid carrying the same allele as integrated at ARP1. Calibration standards were Void, 6.8 ml, thyroglobulin (669 kDa), 11.0 ml; ferritin (440 kDa), 13.0 ml; aldolase (158 kDa), 14.3 ml; ovalbumin (43 kDa), 15.6 ml; and acetone, 19.7 ml.
We next examined whether remediation by overexpression was manifest as a change in Arp1p's elution properties. First, we noted that the Ts– allele arp1-35 showed an elution profile similar to wild-type at the restrictive temperature (Figure 5), consistent with full rescue of the Ts– phenotype when overexpressed. Next, we examined overexpressed lethal alleles from both rescued (arp1-115) and nonrescued (arp1-36) classes. For all, a very similar elution profile was evident, indicating that these mutants do not effect their phenotype by gross disassembly of the Arp1p filament. Finally, we examined arp1-104, a lethal allele that is partially rescued by overexpression. This allele is of interest because its actin cognate exhibits a dominant lethal phenotype and it is positioned at a homotypic interaction surface, exposed at the pointed-end. Arp1-104p exhibited a delayed elution profile, peaking between fractions 12 and 13, consistent with a smaller complex or shorter filament. The position of arp1-104 suggests that it could affect either Arp1p homotypic interactions or proteins binding to the pointed end of the Arp1p filament (Figure 6A). Because most ARP1 mutants assembled an Arp1p filament with the same apparent elution profile, we conclude that the mutant Arp1p monomers were competent for assembly and that a modest alteration of the Arp1p filament can render it nonfunctional.
Figure 6.
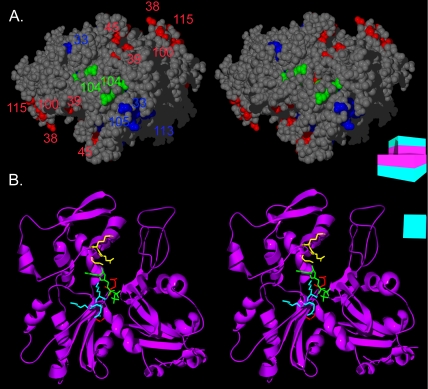
The pointed-end interface and the ATP binding pocket of Arp1p. (A) A stereoview of the pointed-end of an Arp1p pentamer. Ts– and lethal residues are colored by mass-action rescue (red, rescued; blue, not rescued), and numbered by allele. arp1-104 (a rescued lethal allele) is highlighted in green. (B) Three of the ARP1 mutations line the ATP binding pocket of Arp1p. A stereoview is shown of the Arp1p ribbon structure. The ATP is represented in green. The backbone and side chains of arp1-33 are shown in yellow, arp1-101 in red, and arp1-113 in blue. A schematic representation of the filament consisting of boxes representing each monomer, each with a magenta inner face and teal outer face, serves as an orienting guide to the relative rotation of each filament.
Because our gel filtration data suggested that the Arp1p filament was stable in the absence of the remaining dynactin complex, we directly examined the effects of the absence of other dynactin subunits and dynein. Arp1p was not destabilized in the absence of Jnm1p, Nip100p, or Dyn1p (Figure S5), and neither Jnm1p nor Nip100p was unstable in the absence of Arp1p (our unpublished data). Although some ARP1 alleles may disrupt the association of these dynactin subunits, their absence does not lead to Arp1p destabilization.
A Structural Model for Interpretation of Arp1p Mutations
The positions of individual alleles in the primary sequence of Arp1p is less informative than their positions in the tertiary and quaternary structure of the Arp1p filament. The high level of sequence similarity between actin and Arp1p allows one to make a reasonable model of the three-dimensional structure of Arp1p (Figures 6, 7, 8). Among the Arps, crystal structures are known only for Arp2 and Arp3 (Robinson et al., 2001). Although less closely related to actin than Arp1, Arp2, and Arp3 have very similar structures to conventional actin. Their peptide insertions form extended loops that do not perturb the overall actin fold. As a group, the Arp1 class has greatest similarity to conventional actin (Poch and Winsor, 1997). Consequently, Arp1's similarity to actin, the lack of large insertions, and the structural similarity of more distantly related proteins (Arp2 and Arp3) to actin justifies modeling ARP1 on the conventional actin structure.
Figure 7.
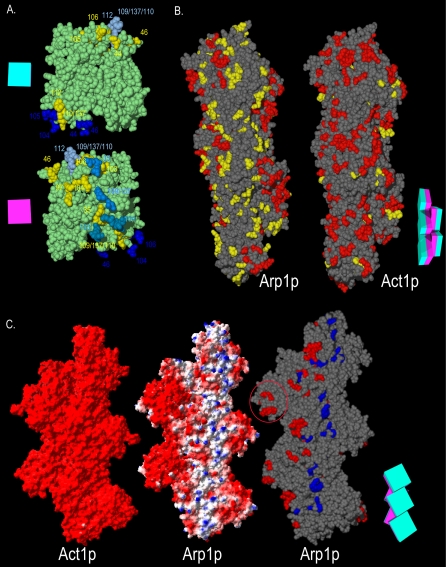
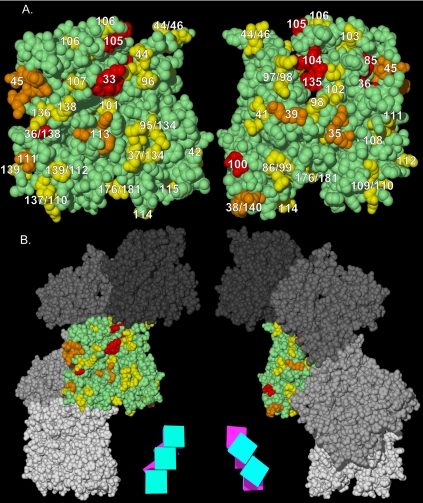
The functional surfaces of the Arp1p filament. (A) Homotypic contact regions in the Arp1p filament. A single Arp1p monomer (green) makes contacts with four surrounding monomers. The charge clusters belonging to the polypeptide chain of the central monomer that make contact with surrounding monomers are shown in yellow. The side chains of charge clusters from the four adjacent monomers that contact the central monomer are shown in four shades of blue (only the relevant side chains are shown, the rest of the polypeptide is not shown for clarity). Opposite faces of the central monomer are shown. The schematic magenta box represents the monomer's inner facing surface; the teal box represents the outer face. (B) The Arp1p filament pseudo-wild-type surface. ARP1 and ACT1 alleles are mapped onto a section of their respective filaments. The “pointed-end” is up and the “barbed-end” is down in all images. Although likely shorter than the actual Arp1p filament, a pentamer is shown to allow visualization of trends, while still providing sufficient detail. Red indicates Ts– and lethal alleles; yellow indicates pseudo-wild type. (C) The positive-charge and mass-action rescue surfaces of Arp1p. We compared the electrostatic potential of Act1p and Arp1p pentamers. Arp1 has a net positive charge surface that encompasses the bulk of Ts– and lethal alleles. Red, –3.5; blue, 3.5 (bottom, left and middle filaments). For mass-action rescue, red, rescued, and blue, nonrescued (bottom, right filament). Note the coincident position of the positive-charge surface and the nonrescued alleles. For clarity, pseudo-wild-type alleles are not shown; they would be found along the three projections to the right. The red circle indicates the Arp1-specific projection encompassing arp-38, 100, and 115. A schematic representation of the Arp1p filament consisting of five boxes, each with a magenta inner face and teal outer face, serves as an orienting guide to the relative position of monomers in each filament.
Figure 8.
ARP1 alleles mapped on a predicted structure of the Arp1p filament. (a) Alleles are represented by number. arp1-94 is not indicated because it resides in the extreme N terminus, not represented in the structure. The “pointed-end” is up and the “barbed-end” is down in all images. Yellow indicates pseudo-wild type, orange indicates Ts–, and red indicates Lethal. (b) The monomers in (a) are shown relative to the Arp1p filament. A schematic representation of the filament consisting of five boxes, each with a magenta inner face and teal outer face, serves as an orienting guide to the relative rotation of each filament.
Alleles Affecting Common Core Functions: Polymerization
Analysis of the environment of charge clusters in the tertiary and quaternary structure of the Arp1p filament suggests the basis of several mutant defects. Based on the model of Arp1p, we considered two subgroups of alleles expected to affect the core functions of polymerization and nucleotide binding: those lying on homotypic interaction surfaces and those likely to contact nucleotide. Each subunit in the filament would make homotypic contacts with four other subunits: one each from the pointed and barbed directions and two lateral subunits. Nearly one-fifth of the ARP1 charge clusters would be predicted to lie, at least in part, along surfaces making homotypic contacts (Figure 7A). From the complementary perspective, in actin, about half of the hydrogen bonds that are engaged in homotypic interactions involve atoms represented in the charge clusters. All seven of these charge clusters are conserved in ARP1, as well as five others lying elsewhere on the homotypic contact interface. In a few cases, the homotypic interaction involves two charge clusters, one on each polypeptide. In Arp1p, at least one of these charge cluster mutations is pseudo-wild type in every case. Particularly notable is a group of pseudo-wild-type alleles (arp1-109, arp1-110, and arp1-137) all of whose ACT1 cognates are lethal.
Looking at all of the homotypic contact charge clusters in Arp1p, we find that most (9/12) are pseudo-wild type. In contrast, in actin, most (8/12) are Ts– or lethal. The excess of pseudo-wild-type clusters among Arp1p homotypic-binding surfaces is indicative of two key differences in the assembly of Arp1p and actin. The first of these involves the context of the Arp1p filament. Unlike actin, Arp1p always functions within a larger complex. Thus, the association of other dynactin subunits may stabilize the Arp1p filament. The second factor involves the critical concentration for polymer assembly. Arp1p assembles at concentrations two orders of magnitude lower than actin (Bingham and Schroer, 1999), indicating that subunit associations in the Arp1p filament are stronger than actin.
Proteins associating into filamentous structures can be poisoned by incorporation of subunits that do not participate in further associations. Alanine scanning of ACT1 produced two such dominant alleles (Wertman et al., 1992), both of which have conserved cognates in ARP1 (arp1-104 and arp1-112). For the lethal allele, arp1-104, we expected that coassembly with wild-type would block monomer addition at the pointed end. Surprisingly, this allele is not dominant even when expressed at wild-type levels. This result is inconsistent with arp1-104 acting as a poison subunit for polymerization. The cognate of the second allele, arp1-112, a pseudo-wild type in Arp1p, resides on the opposite face of the Arp1p monomer from arp1-104 (Figure 8). Structural refinements place arp1-112 on both a solvent-exposed and homotypic interface. That the actin cognate of arp1-112 may be more important for heterotypic (rather than homotypic) protein association is supported by two-hybrid data that shows interference with actin binding to Aip1p, but not to actin itself (Amberg et al., 1995).
Alleles Affecting Common Core Functions: Nucleotide Hydrolysis
The hydrolysis of nucleotide in actin is a key element of filament dynamics. The subsequent ejection of Pi leads to a conformational change favoring depolymerization (Belmont et al., 1999a). Although Arp1p has been shown to bind and hydrolyze ATP (Bingham and Schroer, 1999), the extreme stability of the Arp1 filament suggests a lesser role for hydrolysis and depolymerization. Although it is beyond the scope of this study to examine nucleotide binding and hydrolysis directly, we can make predictions based on the mutant analysis. Only a few of the nucleotide-binding residues are represented as charge clusters (arp1-33, 101, and 113) (Figure 6B). The ACT1 cognate of lethal mutation arp1-33 is known to contact the purine moiety of ATP and causes a severe Ts–/Cs– phenotype in actin. Other lethal ACT1 charge-cluster mutations with well-conserved cognates in ARP1, contact ATP (arp1-101) or line the nucleotide pocket (arp1-113). Surprisingly, arp1-101 is pseudo-wild type and arp1-113 is merely Ts–. Examination of other ARPs may provide an explanation for the reduced severity of these mutations in Arp1p. Although the aspartate of arp1-101 is required for ATPase activity in ParM, a bacterial relative of actin, (Jensen and Gerdes, 1997), the cognate mutations in ARP7 and ARP9 are pseudo-wild type. These and other ATP-contact mutations were used to argue that Arp7p and Arp9p do not bind nucleotide (Cairns et al., 1998). In addition, the putative ATP-contacting lysine residue mutated in arp1-113 is lacking in vertebrate Arp1 (Supplemental Figure 3), casting doubt on its importance for nucleotide contact in Arp1. We conclude that either budding yeast Arp1p has compensatory changes that allow arp1-101 and arp1-113 to bind and hydrolyze nucleotide or that ATP turnover is not necessary for dynactin function. The reduced need for ATP turnover in Arp1p may be a reflection of a fundamental difference between Arp1p and actin: dynactin functions as a stable complex with a short filament, whereas actin filaments are long and dynamic.
Alleles Affecting Arp1p-specific Functions: Binding of Jnm1p and Nip100p
We tested several arp1 mutants for their interaction with Arp1p and Jnm1p by two-hybrid. Most Ts– and lethal alleles reduced the association of both Arp1p and Jnm1p; however, we could make a distinction between those that strongly affected Jnm1p at elevated temperature (arp1-35, 36, 38, 39, 45, 46, 100, 111, 112, 113, and 115) and those that did not (arp1-42, 44, 85, and 104) (Figure 9A). The fact that even pseudo-wild-type alleles affect the interaction in this assay suggests that it is more sensitive to perturbation than are interactions in the native filament. We mapped the outcome of this analysis on the Arp1p filament. The residues that affect the Jnm1p–Arp1p interaction map to a distinct surface.
Figure 9.
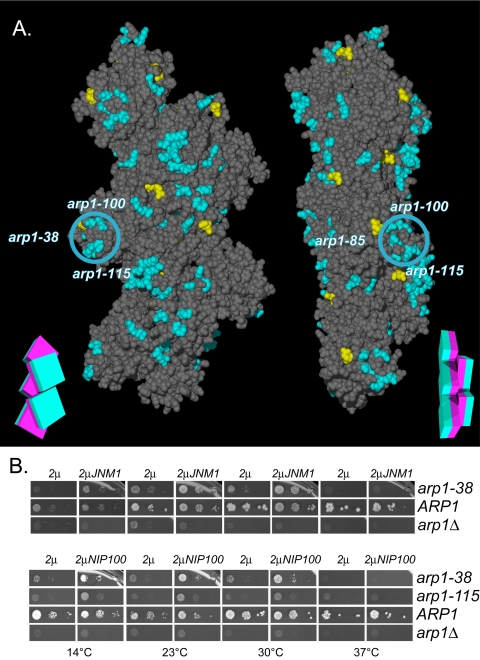
Jnm1–Arp1p 2-hybrid interaction and overexpression-rescue of the Arp1p-specific arp1-100/115 projection by Jnm1p and Nip100p. (A) The results of two-hybrid interaction between Jnm1p and mutant Arp1p. Mutation of the Arp1p residues shown in blue decreased the interaction with Jnm1p, those in yellow had no effect. The blue circle indicates the position of the Arp1p-specific projection, encompassing arp1-38, 100, and 115. A schematic representation of the filament consisting of five boxes, each with a magenta inner face and teal outer face, serves as an orienting guide to the relative rotation of each filament. (B) Plasmids overexpressing (2μ His+) either JNM1 or NIP100 were introduced into all Ts– and lethal ARP1 mutant strains, ARP1 wild-type, or arp1Δ. Their ability to rescue the ARP1 mutation was accessed by growth under conditions (5-FOA-HIS) that counterselect the wild-type ARP1 covering plasmid. Though all Ts– and lethal alleles were examined, only the ARP1 alleles demonstrating rescue are shown.
The rescue of ARP1 alleles by their overexpression (Figure S3) suggested mass-action as the mechanism of suppression. If true, then it follows that overexpression of the dislodged subunits should provide a complementary rescue of ARP1 mutations that are important to that subunit's association with the Arp1p filament. We examined the effect of overexpression of Jnm1p or Nip100p on the growth of all Ts– and lethal ARP1 mutants. Overexpression of Jnm1p clearly suppressed arp1-38, and Nip100p weakly suppressed both arp1-38 and arp1-115 (Figure 9B), indicative of these residues' importance to Jnm1p and Nip100p binding to the Arp1p filament. Because there is a known interdependence of subunit binding to Arp1p (Kahana et al., 1998), overexpression of individual subunits is not expected to restore growth as well as overexpression of the Arp1p filament scaffold.
DISCUSSION
Actin-specific Regions
Overall, mutation of 10 conserved charge clusters produced a lethal or Ts–/Cs– phenotype in ACT1 but a pseudo-wild-type phenotype in ARP1. We propose that these clusters represent regions unique to the function of actin. Among this group, the ACT1 cognates of arp1-40 and arp1-42 eliminate binding of Sac6p (fimbrin) (Holtzman et al., 1994; Whitacre et al., 2001), and the cognates of arp1-37, 42, 44, 46, 114, and 134 would lie under the footprint of myosin (Rayment et al., 1993; Schroder et al., 1993; Miller et al., 1995; Milligan, 1996). Two-hybrid analysis with the ACT1 alanine-scanning alleles indicated a well defined footprint for the actin binding protein Aip1p (Amberg et al., 1995). The Aip1p interaction surface is defined by six ACT1 alleles, all but one of which are lethal. In contrast, only one of the ARP1 cognates is lethal. Together, these results suggest that these residues on the Arp1p surface are not required for the same protein interactions as actin, consistent with a presumed lack of interaction with myosin, fimbrin, and Aip1p.
Alanine Scanning Defines the Unique Interfaces of Arp1p
This work provides the first tools to delineate the surfaces on Arp1p where subunits of the dynactin complex and other proteins are likely to bind. In contrast to the actin-specific regions, ARP1 possesses a group of charge clusters that are either not present in ACT1 or, if conserved, the effect of mutations is more severe in ARP1 than ACT1 (arp1-39, 100, and 115). It is likely that these alleles affect a surface unique to Arp1p function (Figures 7B and 9A). All of the mutations are hypomorphs (having reduced function, rather than null); arp1-39 is rescued by only one or two copies additional copies, arp1-115 is the only lethal allele fully rescued by overexpression, and arp1-100 can be partially suppressed at low temperatures by overexpression (Figure S3). Although they lie at opposite ends of the Arp1p primary sequence, arp1-100 and arp1-115 are adjacent on the tertiary structure and arp1-39 is nearby. arp1-100 and arp1-115 map to an exposed projection, which may be a site of Arp1p-specific protein interactions (Figure 9A).
Overall, the mutations can be grouped into five key interfaces. To simplify the discussion of their properties, we have assigned the following names: 1) the pseudo-wild-type surface, 2) the Arp1p-specific arp1-100/115 projection, 3) the arp1-104 pointed-end region, 4) the mass-action rescued surface, and 5) the positive-charge surface, which cannot be rescued by mass action.
Surface 1: The Pseudo-wild-Type Interface Differentiates Arp1p and Actin
Mapped on the Arp1p filament according to their phenotypes, the pseudo-wild-type alleles (yellow) were found to lie on an uninterrupted surface extending along the length of the filament, separate from the Ts– and lethal clusters (red) (Figure 7B). The equivalent surface of actin includes the regions where fimbrin and myosin interact. The pseudo-wild-type surface also covers the barbed end of Arp1p. By definition, the pseudo-wild type mutations do not affect functions of dynactin redundant with the KIP3 pathway. However, alternative synthetic lethal backgrounds might reveal functions for these surfaces.
Surface 2: Unique ARP1 Mutations on a Filament Projection Affect Jnm1p Interaction
The arp1-100 and arp1-115 clusters are unique to Arp1p and reside on a distinct filament projection that also includes arp1-38 (Figure 9A). In actin, the corresponding mutations represent a pseudo-wild-type surface. These clusters are among the residues important for Jnm1p association with Arp1p in the two-hybrid assay. The severity of their defect and their ability to be rescued by overexpression of Jnm1p, suggest that they play a role in the binding of Jnm1p to the Arp1p filament.
Surface 3: A Mutant at the Arp1p Pointed End Implies a Cargo-binding Subcomplex in Budding Yeast
Two dominant lethal mutations in ACT1, affecting residues conserved in both human and budding yeast Arp1 (arp1-104, arp1-112), have been interpreted as filament capping mutants (Wertman et al., 1992). These alleles would be expected to behave as dominant inhibitors of polymerization. Due to the lower critical concentration for Arp1p polymerization relative to actin filaments (Bingham and Schroer, 1999), such mutations may have a lesser effect on filament formation. However, they may have more impact on the incorporation of other dynactin subunits.
Mutation arp1-104, which resides at the pointed end of the Arp1p monomer, could affect either polymerization or association with pointed-end subunits (Figures 6A and 8A). The wild-type ARP1 filament is short relative to actin. Therefore, mutations interfering with polymerization might not show a dominant phenotype, because sufficient wild-type polymers might form by chance, especially if there is a low ratio of Arp1-104p to wild-type Arp1p. However, increasing the ratio of mutant to wild-type protein should then produce a dominant negative effect, rather than rescue as was actually observed for arp1-104. This lack of a dominant phenotype argues against a role for arp1-104 in polymerization per se.
The role of the arp1-104 lethal charge cluster has been examined in the vertebrate dynactin complex (Garces et al., 1999). Residing at the pointed end of the Arp1 filament, this mutation specifically disrupted association with pointed-end protein, p62, a subunit implicated in cargo binding (Eckley et al., 1999; Muresan et al., 2001; Salina et al., 2002). It has been proposed that budding yeast dynactin lacks a pointed-end complex; however, the related ascomycete, N. crassa, possesses a dynactin complex that contains all key subunits of the vertebrate dynactin complex but exhibits a reduced sedimentation coefficient similar to that of budding yeast (Minke et al., 1999; Lee et al., 2001). Our genetic findings argue against a reduction in polymerization in arp1-104 mutants, yet the Arp1p filament is reduced in mass. This phenotype of arp1-104 may be more consistent with loss of a pointed-end complex (our unpublished data).
Surfaces 4 and 5: Mass-Action and Protein Interaction
Suppression by mass-action defines the fourth and fifth surfaces (Figure 7C). The mass-action-rescued surface includes both the arp1-104 pointed-end mutation and the arp1-100/115 cluster important for Jnm1p binding. The introduction of a mutation on a protein interaction surface may decrease the bimolecular association, shifting the apparent equilibrium toward dissociation. By mass action, the overexpression of one of these proteins may then be sufficient to drive the equilibrium toward stable association. Remarkably, the location of the charge cluster on the filament surface is the strongest predictor of suppression and supersedes effects of phenotypic severity. We propose that for Arp1p, the mass-action-rescued surface represents a region of additional protein interactions. More generally, it is likely that the ability of a mutation to be suppressed by mass action is a hallmark of a defect in protein interaction.
Arp1p and actin differ in their distribution of charged residues, and Act1p has a greater proportion of acidic residues. Whereas yeast and vertebrate actin have uniform negative surface electropotentials, the arrangement of charges in yeast and vertebrate Arp1 produces a narrow region of positive surface electropotential (Figure 7C). This positive-charge surface lies adjacent to the pseudo-wild-type surface and encompasses many of the surface exposed Ts– and lethal alleles (arp1-33, 36, 45, 113, and 105). Many alleles not rescued by overexpression coincide with this positive-charge surface. On the one hand, the lack of overexpression suppression on the positive-charge surface could reflect the severity of the Arp1p mutation with respect to protein interaction. If so, then the surface we have identified for Jmm1p and Nip100p binding to Arp1p, may extend to the adjacent positive-charge surface. Alternatively, because neither of the obvious candidates (Jmm1p or Nip100p) gave a positive result for overexpression rescue, this surface may involve functions or contacts other than protein–protein interactions.
A recently published single particle EM analysis of bovine dynactin (Hodgkinson et al., 2005) gives good agreement with our model of the yeast Arp1p filament (Figure S7). The bovine Arp1 filament model could not account for all the mass found in the averaged EM images. A portion of this mass at the barbed end was attributed to the CapZ heterodimer and a portion to p150Glued. Extra mass also was found along the Arp1 filament lying atop the third and fifth Arp1 subunits (counting from the barbed-end). It was suggested this could represent dynamitin/p50/Jnm1p, but could also be a portion of the large C-terminal region of p150Glued/Nip100p or p22/24 (no homolog known in budding yeast). The extra mass maps to the Arp1p positive-charge surface, where mutations were not rescued by overexpression. This is adjacent to but distinct from the residues we identified as being important for Jnm1p and Nip100p association (Figure S7, yellow residues).
In regard to the positive charge surface, it is important to consider that 1) there are eight to 10 Arp1p subunits in the filament, and 2) that the Arp1p filament has a twofold symmetry (the long axis surfaces are equivalent on opposite sides). Because the surfaces extend the entire length of the filament, but the extra mass observed by EM was mostly located near the barbed end, any given Arp1p mutation's effects may be different near the barbed end versus the pointed-end. Accordingly, suppression by overexpression of Arp1p, Jnm1p, or Nip100p may reflect subunit associations only near the barbed-end, whereas the lack of rescue may reflect mutations with more global effects or which disrupt other associations near the pointed-end. In addition, Nip100p/p150Glued projects only from one face of the filament, and it is assumed that Jnm1p/dynamitin/p50 also binds along this face. The opposite face of the Arp1p filament may be associated with the C-terminal 2/3rd of Nip100p/p150Glued (Schroer, 2004) or may be bare of subunit associations. In that regard, we note that the C terminus of the N. crassa Nip100p homolog seems to regulate the association of dynactin with membranes (Kumar et al., 2001). Although speculative, this raises the possibility that the Arp1p positive-charge surface interacts directly with membranes in budding yeast.
In summary, we have created the first complete set of alanine-scanning alleles in an actin-related protein to delineate both common and distinct surfaces of actin and Arp1p filaments. Furthermore, these mutants allowed us to foot-print protein interactions essential to dynactin function, including Jnm1p and Nip100p, while suggesting the existence of subunits binding at the pointed-end.
Supplementary Material
Acknowledgments
This work is dedicated to K.L.L.C. We thank M. Lorenz for the actin structure and helix.c; B. Winsor, D. Meyer, K. Tatchell, D. Eshel, and D. Drubin for strains, plasmids, and antibodies; and E. Muller and L. Schneper for critical comments. This work was funded by National Institutes of Health Grant GM-52526. S.W.C. was supported by the American Cancer Society (PF4404) and National Institutes of Health (CA09528).
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05–02–0093) on June 22, 2005.
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
References
- Amberg, D. C., Basart, E., and Botstein, D. (1995). Defining protein interactions with yeast actin in vivo. Nat. Struct. Biol. 2, 28–35. [DOI] [PubMed] [Google Scholar]
- Belli, G., Gari, E., Aldea, M., and Herrero, E. (1998a). Functional analysis of yeast essential genes using a promoter-substitution cassette and the tetracycline-regulatable dual expression system. Yeast 14, 1127–1138. [DOI] [PubMed] [Google Scholar]
- Belli, G., Gari, E., Piedrafita, L., Aldea, M., and Herrero, E. (1998b). An activator/repressor dual system allows tight tetracycline-regulated gene expression in budding yeast. Nucleic Acids Res. 26, 942–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belmont, L. D., and Drubin, D. G. (1998). The yeast V159N actin mutant reveals roles for actin dynamics in vivo. J. Cell Biol. 142, 1289–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belmont, L. D., Orlova, A., Drubin, D. G., and Egelman, E. H. (1999a). A change in actin conformation associated with filament instability after Pi release. Proc. Natl. Acad. Sci. USA 96, 29–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belmont, L. D., Patterson, G. M., and Drubin, D. G. (1999b). New actin mutants allow further characterization of the nucleotide binding cleft and drug binding sites. J. Cell Sci. 112, 1325–1336. [DOI] [PubMed] [Google Scholar]
- Bingham, J. B., and Schroer, T. A. (1999). Self-regulated polymerization of the actin-related protein Arp1. Curr. Biol. 9, 223–226. [DOI] [PubMed] [Google Scholar]
- Cairns, B. R., Erdjument-Bromage, H., Tempst, P., Winston, F., and Kornberg, R. D. (1998). Two actin-related proteins are shared functional components of the chromatin-remodeling complexes RSC and SWI/SNF. Mol. Cell 2, 639–651. [DOI] [PubMed] [Google Scholar]
- Clark, S. W., and Meyer, D. I. (1992). Centractin is an actin homologue associated with the centrosome. Nature 359, 246–250. [DOI] [PubMed] [Google Scholar]
- Cottingham, F. R., and Hoyt, M. A. (1997). Mitotic spindle positioning in Saccharomyces cerevisiae is accomplished by antagonistically acting microtubule motor proteins. J. Cell Biol. 138, 1041–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross, F. R. (1997). `Marker swap' plasmids: convenient tools for budding yeast molecular genetics. Yeast 13, 647–653. [DOI] [PubMed] [Google Scholar]
- DeZwaan, T. M., Ellingson, E., Pellman, D., and Roof, D. M. (1997). Kinesin-related KIP3 of Saccharomyces cerevisiae is required for a distinct step in nuclear migration. J. Cell Biol. 138, 1023–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drubin, D. G., Jones, H. D., and Wertman, K. F. (1993). Actin structure and function: roles in mitochondrial organization and morphogenesis in budding yeast and identification of the phalloidin-binding site. Mol. Biol. Cell 4, 1277–1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckley, D. M., Gill, S. R., Melkonian, K. A., Bingham, J. B., Goodson, H. V., Heuser, J. E., and Schroer, T. A. (1999). Analysis of dynactin subcomplexes reveals a novel actin-related protein associated with the arp1 minifilament pointed end. J. Cell Biol. 147, 307–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckley, D. M., and Schroer, T. A. (2003). Interactions between the evolutionarily conserved, actin-related protein, Arp11, actin, and Arp1. Mol. Biol. Cell 14, 2645–2654. Epub 2003 Mar 2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garces, J. A., Clark, I. B., Meyer, D. I., and Vallee, R. B. (1999). Interaction of the p62 subunit of dynactin with Arp1 and the cortical actin cytoskeleton. Curr. Biol. 9, 1497–1500. [DOI] [PubMed] [Google Scholar]
- Gietz, R. D., and Schiestl, R. H. (1995). Transforming yeast cells with DNA. Mol. Cell. Biol. 5, 255–269. [Google Scholar]
- Goodson, H. V., and Hawse, W. F. (2002). Molecular evolution of the actin family. J. Cell Sci. 115, 2619–2622. [DOI] [PubMed] [Google Scholar]
- Guex, N., and Peitsch, M. C. (1997). SWISS-MODEL and the Swiss-Pdb-Viewer: an environment for comparative protein modeling. Electrophoresis 18, 2714–2723. [DOI] [PubMed] [Google Scholar]
- Hodgkinson, J. L., Peters, C., Kuznetsov, S. A., and Steffen, W. (2005). Three-dimensional reconstruction of the dynactin complex by single-particle image analysis. Proc. Natl. Acad. Sci. USA 102, 3667–3672. Epub 2005 Feb 3628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holleran, E. A., Karki, S., and Holzbaur, E. L. (1998). The role of the dynactin complex in intracellular motility. Int. Rev. Cytol. 182, 69–109. [DOI] [PubMed] [Google Scholar]
- Holton, T. A., and Graham, M. W. (1991). A simple and efficient method for direct cloning of PCR products using ddT-tailed vectors. Nucleic Acids Res. 19, 1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtzman, D. A., Wertman, K. F., and Drubin, D. G. (1994). Mapping actin surfaces required for functional interactions in vivo. J. Cell Biol. 126, 423–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James, P., Halladay, J., and Craig, E. A. (1996). Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144, 1425–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen, R. B., and Gerdes, K. (1997). Partitioning of plasmid R1. The ParM protein exhibits ATPase activity and interacts with the centromere-like ParR-parC complex. J. Mol. Biol. 269, 505–513. [DOI] [PubMed] [Google Scholar]
- Kahana, J. A., Schlenstedt, G., Evanchuk, D. M., Geiser, J. R., Hoyt, M. A., and Silver, P. A. (1998). The yeast dynactin complex is involved in partitioning the mitotic spindle between mother and daughter cells during anaphase B. Mol. Biol. Cell 9, 1741–1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karki, S., and Holzbaur, E. L. (1999). Cytoplasmic dynein and dynactin in cell division and intracellular transport. Curr. Opin. Cell Biol. 11, 45–53. [DOI] [PubMed] [Google Scholar]
- Kern, L., de Montigny, J., Jund, R., and Lacroute, F. (1990). The FUR1 gene of Saccharomyces cerevisiae: cloning, structure and expression of wild-type and mutant alleles. Gene 88, 149–157. [DOI] [PubMed] [Google Scholar]
- Korinek, W. S., Copeland, M. J., Chaudhuri, A., and Chant, J. (2000). Molecular linkage underlying microtubule orientation toward cortical sites in yeast. Science 287, 2257–2259. [DOI] [PubMed] [Google Scholar]
- Kumar, S., Zhou, Y., and Plamann, M. (2001). Dynactin-membrane interaction is regulated by the C-terminal domains of p150(Glued). EMBO Rep. 2, 939–944. Epub 2001 Sep 2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel, T. A. (1985). Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc. Natl. Acad. Sci. USA 82, 488–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, I. H., Kumar, S., and Plamann, M. (2001). Null mutants of the Neurospora actin-related protein 1 pointed-end complex show distinct phenotypes. Mol. Biol. Cell 12, 2195–2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, L., Tirnauer, J. S., Li, J., Schuyler, S. C., Liu, J. Y., and Pellman, D. (2000). Positioning of the mitotic spindle by a cortical-microtubule capture mechanism. Science 287, 2260–2262. [DOI] [PubMed] [Google Scholar]
- Lees-Miller, J. P., Helfman, D. M., and Schroer, T. A. (1992). A vertebrate actin-related protein is a component of a multisubunit complex involved in microtubule-based vesicle motility. Nature 359, 244–246. [DOI] [PubMed] [Google Scholar]
- Lorenz, M., Popp, D., and Holmes, K. C. (1993). Refinement of the F-actin model against X-ray fiber diffraction data by the use of a directed mutation algorithm. J. Mol. Biol. 234, 826–836. [DOI] [PubMed] [Google Scholar]
- Miller, C. J., Cheung, P., White, P., and Reisler, E. (1995). Actin's view of actomyosin interface. Biophys. J. 68, 50S–54S. [PMC free article] [PubMed] [Google Scholar]
- Miller, R. K., Cheng, S. C., and Rose, M. D. (2000). Bim1p/Yeb1p mediates the Kar9p-dependent cortical attachment of cytoplasmic microtubules. Mol. Biol. Cell 11, 2949–2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, R. K., Heller, K. K., Frisen, L., Wallack, D. L., Loayza, D., Gammie, A. E., and Rose, M. D. (1998). The kinesin-related proteins, Kip2p and Kip3p, function differently in nuclear migration in yeast. Mol. Biol. Cell 9, 2051–2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milligan, R. A. (1996). Protein-protein interactions in the rigor actomyosin complex. Proc. Natl. Acad. Sci. USA 93, 21–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minke, P. F., Lee, I. H., Tinsley, J. H., Bruno, K. S., and Plamann, M. (1999). Neurospora crassa ro-10 and ro-11 genes encode novel proteins required for nuclear distribution. Mol. Microbiol. 32, 1065–1076. [DOI] [PubMed] [Google Scholar]
- Minke, P. F., Lee, I. H., Tinsley, J. H., and Plamann, M. (2000). A Neurospora crassa Arp1 mutation affecting cytoplasmic dynein and dynactin localization. Mol. Gen. Genet. 264, 433–440. [DOI] [PubMed] [Google Scholar]
- Muresan, V., Stankewich, M. C., Steffen, W., Morrow, J. S., Holzbaur, E. L., and Schnapp, B. J. (2001). Dynactin-dependent, dynein-driven vesicle transport in the absence of membrane proteins: a role for spectrin and acidic phospholipids. Mol. Cell 7, 173–183. [DOI] [PubMed] [Google Scholar]
- Peitsch, M. C. (1996). ProMod and Swiss-Model: Internet-based tools for automated comparative protein modelling. Biochem. Soc. Trans. 24, 274–279. [DOI] [PubMed] [Google Scholar]
- Poch, O., and Winsor, B. (1997). Who's who among the Saccharomyces cerevisiae actin-related proteins? A classification and nomenclature proposal for a large family. Yeast 13, 1053–1058. [DOI] [PubMed] [Google Scholar]
- Pollard, T. D., and Cooper, J. A. (1986). Actin and actin-binding proteins. A critical evaluation of mechanisms and functions. Annu. Rev. Biochem. 55, 987–1035. [DOI] [PubMed] [Google Scholar]
- Puls, I., et al. (2003). Mutant dynactin in motor neuron disease. Nat. Genet. 33, 455–456. [DOI] [PubMed] [Google Scholar]
- Rayment, I., Holden, H. M., Whittaker, M., Yohn, C. B., Lorenz, M., Holmes, K. C., and Milligan, R. A. (1993). Structure of the actin-myosin complex and its implications for muscle contraction. Science 261, 58–65. [DOI] [PubMed] [Google Scholar]
- Reijo, R. A., Cooper, E. M., Beagle, G. J., and Huffaker, T. C. (1994). Systematic mutational analysis of the yeast beta-tubulin gene. Mol. Biol. Cell 5, 29–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson, R. C., Turbedsky, K., Kaiser, D. A., Marchand, J. B., Higgs, H. N., Choe, S., and Pollard, T. D. (2001). Crystal structure of arp2/3 complex. Science 294, 1679–1684. [DOI] [PubMed] [Google Scholar]
- Rose, M. D., Novick, P., Thomas, J. H., Botstein, D., and Fink, G. R. (1987). A Saccharomyces cerevisiae genomic plasmid bank based on a centromere-containing shuttle vector. Gene 60, 237–243. [DOI] [PubMed] [Google Scholar]
- Rose, M. D., Winston, F., and Hieter, P. (1990). Methods in Yeast Genetics, Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
- Salina, D., Bodoor, K., Eckley, D. M., Schroer, T. A., Rattner, J. B., and Burke, B. (2002). Cytoplasmic dynein as a facilitator of nuclear envelope breakdown. Cell 108, 97–107. [DOI] [PubMed] [Google Scholar]
- Sasaki, N., and Sutoh, K. (1998). Structure-mutation analysis of the ATPase site of Dictyostelium discoideum myosin II. Adv. Biophys. 35, 1–24. [PubMed] [Google Scholar]
- Schafer, D. A., Gill, S. R., Cooper, J. A., Heuser, J. E., and Schroer, T. A. (1994). Ultrastructural analysis of the dynactin complex: an actin-related protein is a component of a filament that resembles F-actin. J. Cell Biol. 126, 403–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer, D. A., and Schroer, T. A. (1999). Actin-related proteins. Annu. Rev. Cell Dev. Biol. 15, 341–363. [DOI] [PubMed] [Google Scholar]
- Schroder, R. R., Manstein, D. J., Jahn, W., Holden, H., Rayment, I., Holmes, K. C., and Spudich, J. A. (1993). Three-dimensional atomic model of F-actin decorated with Dictyostelium myosin S1. Nature 364, 171–174. [DOI] [PubMed] [Google Scholar]
- Schroer, T. A. (2004). Dynactin. Annu. Rev. Cell Dev. Biol. 20, 759–779. [DOI] [PubMed] [Google Scholar]
- Sikorski, R. S., and Hieter, P. (1989). A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122, 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier, F. W., and Moffatt, B. A. (1986). Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 189, 113–130. [DOI] [PubMed] [Google Scholar]
- Wach, A., Brachat, A., Pohlmann, R., and Philippsen, P. (1994). New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 10, 1793–1808. [DOI] [PubMed] [Google Scholar]
- Wertman, K. F., Drubin, D. G., and Botstein, D. (1992). Systematic mutational analysis of the yeast ACT1 gene. Genetics 132, 337–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitacre, J., Davis, D., Toenjes, K., Brower, S., and Adams, A. (2001). Generation of an isogenic collection of yeast actin mutants and identification of three interrelated phenotypes. Genetics 157, 533–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woehlke, G., Ruby, A. K., Hart, C. L., Ly, B., Hom-Booher, N., and Vale, R. D. (1997). Microtubule interaction site of the kinesin motor. Cell 90, 207–216. [DOI] [PubMed] [Google Scholar]
- Yin, H., Pruyne, D., Huffaker, T. C., and Bretscher, A. (2000). Myosin V orientates the mitotic spindle in yeast. Nature 406, 1013–1015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.