Abstract
ADP-ribosylation factor (ARF) and ARF-like (ARL) proteins are members of the ARF family, which are critical components of several different vesicular trafficking pathways. ARFs have little or no detectable GTPase activity without the assistance of a GTPase-activating protein (GAP). Here, we demonstrate that yeast Gcs1p exhibits GAP activity toward Arl1p and Arf1p in vitro, and Arl1p can interact with Gcs1p in a GTP-dependent manner. Arl1p was observed both on trans-Golgi and in cytosol and was recruited from cytosol to membranes in a GTP-dependent manner. In gcs1 mutant cells, the fraction of Arl1p in cytosol relative to trans-Golgi was less than it was in wild-type cells. Increasing Gcs1p levels returned the distribution toward that of wild-type cells. Both Arl1p and Gcs1p influenced the distribution of Imh1p, an Arl1p effector. Our data are consistent with the conclusion that Arl1p moves in a dynamic equilibrium between trans-Golgi and cytosol, and the release of Arl1p from membranes in cells requires the hydrolysis of bound GTP, which is accelerated by Gcs1p.
INTRODUCTION
Intracellular membrane trafficking is regulated, in part, by small GTP-binding proteins of the ADP-ribosylation factor (ARF) family. ARFs are critical components of several different vesicular trafficking pathways in all eukaryotic cells and activators of specific phospholipase Ds (reviewed by Moss and Vaughan, 1998). ARF function depends on the controlled binding and hydrolysis of GTP. The conformational changes that accompany the binding of GDP or GTP can lead directly to changes in the affinity of the GTPase for proteins, lipids, and membranes. Binding of GTP activates ARF1, facilitates its association with membranes, and enables it to recruit coat proteins to initiate vesicle budding. Inactivation of ARF by hydrolysis of the bound GTP releases coat proteins (and ARF) from vesicle membranes. Unlike other monomeric small GTP-binding proteins, ARFs do not have detectable GTPase activity (Kahn and Gilman, 1986) and their inactivation requires a GTPase-activating protein (GAP).
In mammalian cells, ARF-GAPs have been found to link aspects of cell signaling and morphogenesis to vesicular transport (Randazzo et al., 2000). Thus, ARF-GAPs may allow temporal as well as spatial coordination of the ARF GTPase cycle (Donaldson, 2000). Several GAPs were identified by their ability to enhance the GTPase activity of ARF proteins in vitro (Cukierman et al., 1995; Poon et al., 1996, 1999; Randazzo, 1997; Brown et al., 1998; Premont et al., 1998; Andreev et al., 1999; Turner et al., 1999). An ARF-GAP localized to the rat liver Golgi complex contains a zinc-finger motif, which was important for GAP activity but had no effect on its subcellular distribution (Cukierman et al., 1995). Structurally, this mammalian GAP, ARFGAP, is very similar to the yeast zinc-finger proteins Gcs1p, Glo3p, and Age2p, which are GAPs for yeast Arf1p in vitro (Poon et al., 1996, 1999, 2001). Gcs1p and Glo3p have overlapping functions in endoplasmic reticulum (ER)-Golgi transport and Glo3p is important for retrieval of proteins from the Golgi to the ER (Andreev et al., 1999; Dogic et al., 1999; Poon et al., 1999). The Gcs1p and Age2p ARF GAPs provide overlapping, essential function in transport from the yeast trans-Golgi network (Poon et al., 2001). Although Gcs1p also was implicated in the regulation of actin cytoskeletal organization (Blader et al., 1999) and mitochondrial morphology (Huang et al., 2002), its multiple functions in cells remain to be established.
ARF-like (ARL) proteins are a subfamily of the ARF branch of the small G protein superfamily. The ARL proteins are highly conserved through evolution and have diverse functions, including regulation of membrane trafficking (Moss and Vaughan, 1998, Pasqualato et al., 2002). In mammalian cells, ARL1 is localized to the TGN and regulates trafficking in TGN-endosomal pathways (Lowe et al., 1996; Lu et al., 2001; Lu and Hong, 2003). ARL2 regulates the interaction of tubulin-folding cofactor D with native tubulin (Bhamidipati et al., 2000), whereas ARL4 and ARL5 may function in the nucleus (Lin et al., 2000, 2002). Saccharomyces cerevisiae has two ARL proteins, Arl1p and Arl3p, both of which seem to play roles in vesicular trafficking (Lee et al., 1997; Huang et al., 1999; Panic et al., 2003). Relatively little is known about biological roles, protein effectors, or the importance of membrane-lipid interactions for the ARLs. Because Arl1p, like Arf1p, lacks detectable GTPase activity (Lee et al., 1997), a GAP should be essential for Arl1p function. To obtain additional clues to the physiological role(s) of Arl1p, we investigated a potential GAP regulator for Arl1p.
Here, we provide both in vivo and in vitro evidence that the yeast Arf1p-GAP, Gcs1p, is also an Arl1p GAP. Arl1p was colocalized with Sft2p at the trans-Golgi apparatus in a GTP-dependent manner. Release of Golgi-bound Arl1p to the cytosol seemed to require hydrolysis of GTP by Gcs1p. Mutant cells lacking Gcs1p failed to redistribute Arl1p from trans-Golgi structures to the cytosol. Thus, our findings indicate that the movement of Arl1p between trans-Golgi and cytosol is regulated in part by Gcs1p acting as its GAP.
MATERIALS AND METHODS
Strains, Media, and Microbiological Techniques
Table 1 lists the yeast strains used in this study. Yeast culture media were prepared as described by Sherman et al. (1986). YPD contained 1% Bacto-yeast extract, 2% Bacto-peptone, and 2% glucose. SD contained 0.17% Difco yeast nitrogen base (without amino acids and ammonium sulfate), 0.5% ammonium sulfate, and 2% glucose. Nutrients essential for auxotrophic strains were supplied at specified concentrations (Sherman et al., 1986). Yeast strains were transformed by the lithium acetate method (Ito et al., 1983). Plasmids were constructed according to standard protocols (Sambrook et al., 1989) (Table 2). Gene disruption was carried out as described by Lee et al. (1994).
Table 1.
Yeast strains used in this study
| Strain | Genotypea | Source |
|---|---|---|
| YPH250 | MATa ade2, his3, leu2, lys2, trp1, ura3-52, ARF1, ARF2, ARF3, ARL1, ARL3, GCS1, GLO3 | Lee et al. (1997) |
| YPH250dr1 | MATa ade2, his3, leu2, lys2, trp1, ura3-52, arf1, ARF2, ARF3, ARL1 ARL3, GCS1, GLO3 | Huang et al. (2003) |
| YPH250dl1 | MATa ade2, his3, leu2, lys2, trp1, ura3-52, ARF1, ARF2, ARF3, arl1, ARL3, GCS1, GLO3 | Huang et al. (1997) |
| YPH250dg1 | MATa ade2, his3, leu2, lys2, trp1, ura3-52, ARF1, ARF2, ARF3, ARL1, ARL3, gcs1, GLO3 | Huang et al. (2002) |
| YPH250dg3 | MATa ade2, his3, leu2, lys2, trp1, ura3-52, ARF1, ARF2, ARF3, ARL1, ARL3, GCS1, glo3 | This work |
| YPH250dgl11 | MATa ade2, his3, leu2, lys2, trp1, ura3-52, ARF1, ARF2, ARF3, arl1, ARL3, gcs1, GLO3 | This work |
| YEM1α | MATα trp1, his3, leu2, 6ops-LEU2, 2ops-LacZ |
ade, adenine-requiring; his, histidine-requiring; trp, tryptophan-requiring; ura, uracil-requiring; leu, leucine-requiring. Arl1 represents arl1::hisG; and arf1 represents arf1::hisG; gcs1 represents gcs1::Leu2 and glo3 represents glo3::Leu2.
Table 2.
Plasmids used in this study
| Plasmid | Description | Source or reference |
|---|---|---|
| pIG4-5 | 2 μm TRP1 PGAL acid bolb B42 | Gyuris et al. (1993) |
| pEG202 | 2 μm HIS3 PADH LEX A DNA binding domain | Gyuris et al. (1993) |
| pET30a | f1 ori Kan PT7 His.Tag | Novagen (Madison, WI) |
| pGEX4T-1 | f1 ori Amp PT7 GST.Tag | Amersham Biosciences |
| pYEX4T-1 | 2 μm URA3 PCUP1 GST.Tag | BD Biosciences Clontech |
| pRS314 | CEN6 TRP1 | Sikorski and Hieter (1989) |
| pRS315 | CEN6 LEU2 | Sikorski and Hieter (1989) |
| pVT101U | 2 μm URA3 PADH | Verent et al. (1987) |
| YIplac128 | LEU2 | Gietz et al. (1988) |
| GFPSFT2 | GFPSFT2 in pRS406 | Hugh R. B. Pelham |
| EMP47MYC | EMP47MYC in Yiplac128 | Stephan Schroder-Kohne |
| GCS1pJG4-5 | GCS1 in pJG4-5 2 μm TRP1 PGAL | This study |
| GCS1znpJG4-5 | GCS1zn in pJG4-5 2 μm TRP1 PGAL | This study |
| GCS1dPHpJG4-5 | GCS1dPH in pJG4-5 2 μm TRP1 PGAL | This study |
| GCS1NpJG4-5 | GCS1N in pJG4-5 2 μm TRP1 PGAL | This study |
| GCS1pET30a | GCS1 in pET30a kan | Huang et al, 2002 |
| GLO3pJG4-5 | GLO3 in pJG4-52 μm TRP1 PGAL | This study |
| ARL1d17NpEG202 | ARL1d17N in pEG202 2 μm HIS3 PADH | This study |
| ARL1Q/Ld17NpEG202 | ARL1Q72Ld17N in pEG202 2 μm HIS3 PADH | This study |
| ARL1T/Nd17NpEG202 | ARL1T32Nd17N in pEG202 2 μm LEU2 PADH | This study |
| ARF1Q/Ld17NpEG202 | ARF1Q71Ld17N in pEG202 2 μm HIS3 PADH | This study |
| ARF3Q/Ld17NpEG202 | ARF3Q71Ld17N in pEG202 2 μm HIS3 PADH | This study |
| ARL1Q/Ld17NpGEX4T | ARL1Q72Ld17N in pGEX4T-1 Amp | This study |
| ARF1Q/Ld17NpGEX4T | ARF1Q71Ld17N in pGEX4T-1 Amp | This study |
| ARF3Q/Ld17NpGEX4T | ARF3Q71Ld17N in pGEX4T-1 Amp | This study |
| ARL1d17NpYEX | ARL1d17N in pYEX4T-1 2 μm URA3 PCUP1 GST.Tag | This study |
| ARL1Q/Ld17NpYEX | ARL1Q72Ld17N in pYEX4T-1 2 μm URA3 PCUP1 GST.Tag | This study |
| ARL1T32Nd17NpYEX | ARL1T32Nd17N in pYEX4T-1 2 μm URA3 PCUP1 GST.Tag | This study |
| ARL1mRFP | ARL1mRFP in pRS315 CEN6 LEU2 PADH | This study |
| ARF1mRFP | ARF1mRFP in pRS315 CEN6 LEU2 PADH | This study |
| GCS1GFP | GCS1GFP in pVT101U 2 μm URA3 PADH | This study |
| GCS1MYC | GCS1MYC in pVT101U 2 μm URA3 PADH | This study |
| ARL1Yiplac128 | PADH -ARL1 in yYIplac128 LEU2 | This study |
| ARL1Q/LYIplac128 | PADH - ARL1Q72L in YIplac128 LEU2 | This study |
| ARL1T/NYIplac128 | PADH -ARL1T32N in YIplac128 LEU2 | This study |
| GFPIMH1pRS314 | PGAL-GFPIMH1 in pR314 CEN6 TRP1 | This study |
Plasmid Construction
GCS1 mutant expression clones were constructed and subcloned in pJG4-5 for yeast two-hybrid assay, in pET15b for recombinant protein production, or in pVT101U for immunofluorescence microscopy. To introduce site-specific mutations in GCS1, a two-step recombinant PCR procedure was used. In Gcs1zn, two cysteines in the zinc-finger motif were replaced with alanines by primer pairs GCS1.1, GCS1.4 and GCS1.3, GCS1.2 (Table 3). Gcs1dC was generated with GCS1.1 and GCS1.dPH, and Gcs1N was generated with GCS1.1 and GCS1.N. For Myc-tagged Gcs1p expression, primer pairs GCS1.1 and GCS1.myc were used. For construction of Arl1T32N, primer pairs yARL1.1, yL1 T/N.2 and yL1 T/N.1, yARL1.2 (Table 2) were used for the primary PCR. For the construction of Arl1Q72L, primer pairs yARL1.1/yL1 Q/L.2 and yL1 Q/L.1, yARL1.2 were used. For the deletion of N-terminal 17 amino acids, primer yARL1d17N was used. These constructs were subcloned in pEG202 for yeast two-hybrid assay, pGEX4T-1 or pET15b for recombinant protein production, or YIplac128 for immunofluorescence microscopy. For monomeric red fluorescent protein (mRFP)-tagged proteins, template mRFP1 in pRSETb (kindly provided by Dr. Roger Tsien, University of California at San Diego, La Jolla, CA) was used to generate mRFP-fused Arl1p or Arf1p. mRFP-fused proteins were driven by the ADH promoter in a centromeric vector and expression of these proteins was approximately twoto threefold higher than endogenous Arl1p (Supplemental Figure S1A). These expression constructs should be able to minimize the overexpression defect. IMH1 was cloned with IMH1.1 and IMH1.2 primer pairs and subcloned into pRS314, containing the green fluorescent protein (GFP) gene under control of the GAL1 promoter.
Table 3.
Primers used in this study
| Primer | Sequence (5′-3′) |
|---|---|
| GCS1.1 | GAATTCATGTCAGATTGGAAAGTGGACC |
| GCS1.2 | TGAACTTTCCTTGAATGTTGAGAAAA |
| GCS1.3 | CCCTCTATGGATACCGGCAGCTTCAAGGGCAAT |
| GCS1.4 | ATTGCCCTTGAAGCTGCCGGTATCCATAGAGGG |
| GCS1.dPH | TTATGCAGACCGTTCTTGTGGAGG |
| GCS1.N | AAACTGATCCATAGTGATAGATCTTA |
| GCS1.myc | TTACAAGTCTTCTTCAGAAATCAGCTTTTGTTC |
| yARL1.1 | GAATTCATGGGTAACATTTTTAGTTCAATG |
| yARL1.2 | ATTCGGATCCATTTAAAAAGTATGCATCTAC |
| yL1 T/N.1 | GGTGCAGGTAAAAATACCATCTTATATCG |
| yL1 T/N.2 | TAAGATGGTATTTTTACCTGCACCATC |
| yL1 Q/L.1 | GGTGCAGGTAAAAATACCATCTTATATCG |
| yL1 Q/L.2 | TAAGATGGTATTTTTACCTGCACCATC |
| yARL1d17N | GAATTCATGGAATTGCGTATATTGATTTTGGG |
| IMH1.1 | GGATCCATGTTCAAACAGCTGTCACAAATTG |
| IMH1.2 | CGTATCTTCTGCTTTCAGCTAC |
Yeast Two-Hybrid Analysis
Yeast two-hybrid analyses were performed using the “interaction-trap” system (Golemis and Khazak, 1997). In this system, bait (ARF or ARL) was fused with the DNA-binding domain of LexA in pEG202. GCS1, GLO3, AGE2, SAT1/AGE1, SPS18, and GTS1were made in the vector pJG4-5, which uses the inducible yeast GAL1 promoter to express the protein fused to an acidic domain that functions as a transcriptional activation motif. Reporter yeast, YEM1α, containing interacting proteins, can transactivate two reporter genes, LacZ and Leu2, which can express β-galactosidase activity and grow on minimal medium lacking leucine.
In Vitro and In Vivo Binding Analysis
For in vitro binding analyses, 2 μg of purified glutathione S-transferase (GST), GST-Arf1Q71Ld17N, GST-Arf3Q71Ld17N, or GST-Arl1Q72Ld17N bound to glutathione-Sepharose beads were incubated with 2 μg of purified His-tagged Gcs1p in 1 ml of binding buffer (50 mM Tris-Cl, pH 7.5, 150 mM NaCl, 1% Triton X-100, 10 mM MgCl2) for 1 h at 4°C. Beads were then washed three times with binding buffer and boiled in sample buffer (50 μl), and samples of proteins were separated by SDS-PAGE in 10% gel before blotting.
For binding analyses in cells, GST-fused Arl1d17N, Arl1Q72Ld17N, or Arl1T32Nd17N (expressed by pYEX-4T-1) was induced with 0.5 mM Cu2+ in arl1-deleted yeast, which were then lysed with glass beads in phosphate-buffered saline containing 5 mM MgCl2, 10 mM dithiothreitol and protease inhibitor cocktail (1 μg/ml aprotinin, 1 μg/ml leupeptin, 1 μg/ml pepstatin, 1 μM benzamidine, and 1 mM phenylmethylsulfonyl fluoride). After centrifugation (500 × g, 5 min), the cleared lysate (1 ml) was incubated with glutathione-Sepharose (40 μl) at 4°C overnight, and then washed three times with binding buffer. Bound proteins were analyzed as described for in vitro binding assay.
GAP Assays
GAP activity was assessed essentially as described by Huber et al. (2001). GAP activity is assayed by a single-round hydrolysis of ARF-bound GTP by using [γ-32P]GTP and measuring [32P]Pi release after charcoal absorption of the nucleotides. Recombinant myristoylated Arf1p, Arl1p, or Arl3p protein was incubated in 2 mM EDTA, 1 mM MgCl2, 50 mM NaCl, 25 mM 3-(N-morpholino)propanesulfonic acid (MOPS), pH 7.4, and a 1:10 dilution of a dimyristoylphosphatidyl-choline (DMPC)/cholate mixture, with [γ-32P]GTP for 15 min at 30°C, resulting in the association of ∼50% of the labeled nucleotide with Arl1p, Arl3p, and Arf1p proteins. GAP activity was determined at 30°C in 100 μl assay systems at different time points (up to 20 min) containing 5 mM MgCl2, 25 mM MOPS, pH 7.4, and different concentrations (0.1–200 ng) of recombinant Gcs1p or Glo3p. Samples were taken at 0 and 5 min for the typical experiment. Radioactivity is measured by scintillation counter. The GAP assays also show hydrolysis of [γ-32P]GTP in a time-dependent manner.
Indirect Immunofluorescence
Cells grown to a density of 1–2 × 107 cells/ml in 3 ml of minimal selective medium with 2% glucose or YPD medium were prepared for indirect immunofluorescence essentially as described previously (Lee et al., 1997), except for replacement of the methanol (6-min incubation)-acetone (30-s incubation) step with a 0.2% SDS solution at –20°C before Arl1p staining (Hagan and Ayscough, 1999). Antibodies included affinity-purified anti-Arl1p, commercial monoclonal anti-myc (BD Biosciences Clontech, Palo Alto, CA) and anti-GFP (Sigma, St. Louis, MO) antibodies. Secondary antibodies Alexa FluorTM 488 goat anti-rabbit IgG and Alexa FluorTM 594 goat anti-mouse IgG (Molecular Probes, Eugene, OR) were used at dilutions of 1:1000 and 1:2000, respectively. Nuclei were stained with H33258 (2 μg/ml) included in the mounting solution. Preparations were inspected with a Zeiss Axioskop microscope and photographed on Photometrix coolSNAP fx, and then processed by Image-ProPlus software (Media Cybernetics, Silver Spring, MD).
Cells containing GFP- or mRFP-tag proteins were imaged live in synthetic medium. Except for GFP-Imh1p, which requires galactose for induction, all fluorescence protein-tagged chimera were cultured in glucose-containing medium. After overnight culture, mid-log phase cells were examined, and images were processed as described above. Quantification of those signals was performed using Axio Vision Rel. 4.2 software.
Preparation of Yeast Cell Extracts and Immunoblotting
Whole yeast extracts were prepared by agitating (vortex mixer) yeast cells suspended in TE buffer (10 mM Tris, pH 7.4, 1 mM EDTA) with glass beads for 1 min followed by incubation on ice for 1 min, repeated five times. After brief centrifugation (3300 × g; 5 min) to clarify the lysate, protein was quantified by Coomassie Blue assay (Pierce Chemical, Rockford, IL). Proteins separated by SDS-PAGE were transferred to polyvinylidene diflouride (PVDF) membranes (Millipore, Billerica, MA), which were incubated (60 min; room temperature) with antibodies in Tris-buffered saline, pH 7.4, containing 0.1% Tween 20 and 5% dried skim milk. Bound antibodies were detected with the ECL system (Amersham Biosciences, Piscataway, NJ). Anti-LexA antibody was used in 1:5000 (Invitrogen, Carlsbad, CA), and anti-hemagglutinin (HA) was used in 1:5000 (Roche Diagnostics, Indianapolis, IN); other antibodies used in this study were generated from our laboratory.
RESULTS
Arl1p Interacts with Gcs1p in a Nucleotide-dependent Manner
The yeast genome contains six genes that encode polypeptides with similarity to ARF GAPs. Four of these (Gcs1p, Glo3p, Age2p, and Sat1p/Age1p) are known to be Arf1p GAPs (Antonny et al., 1997; Poon et al., 1999, 2001; Zhang et al., 2003). Among the others, Sps18p is expressed only in sporulating cells, and Gts1p is likely a transcription factor (Coe et al., 1994; Mitsui et al., 1994).
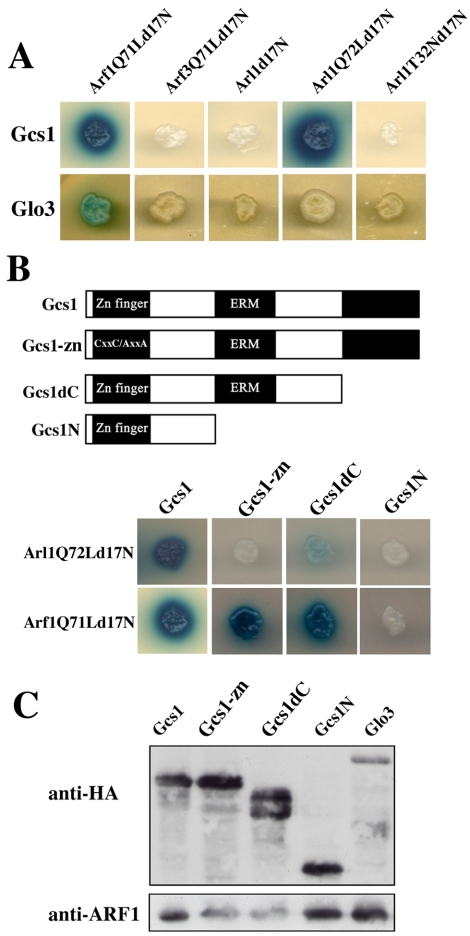
Our initial attempt to identify an Arl1p GAP using full-length, wild-type Arl1 or Arl1Q72L, a putative active form of Arl1p, in a yeast two-hybrid system was unsuccessful (our unpublished data). It was known that deletion of the first 17 N-terminal hydrophobic amino acids of the active GTP-form of Arf1 protein could enhance its interaction with GAP proteins in a two-hybrid system (Eugster et al., 2000; Figure 1A). Therefore, we tested whether Arl1Q72Ld17N could interact with any of the six ARF GAP-related proteins. We found that Arl1Q72Ld17N interacted with Gcs1p but not with Glo3p or other ARF GAP-related proteins (Figure 1A; our unpublished data). The putative GDP-form Arl1T32Nd17N and wild-type Arl1d17N did not interact with Gcs1p (Figure 1A). Thus, Arl1p associated with Gcs1p in a nucleotide-dependent manner. Our data also showed that Gcs1p could interact with Arf1Q71Ld17N but not with Arf3Q71Ld17N and Arl3Q78Ld17N (Figure 1A and Supplemental Figure S2), suggesting that Gcs1p could be a GAP for both Arl1p and Arf1p but not Arf3p or Arl3p. Consistent with previous report (Eugster et al., 2000), Gcs1p seemed to not interact with Arf1d17N in yeast two-hybrid analysis (Supplemental Figure S2).
Figure 1.
Interaction of Arl1Q72Ld17N with Gcs1p in a two-hybrid assay. (A) Arl1Q72Ld17N specifically interacts with Gcs1p but not Glo3p. The yeast reporter strain YEM1α cotransformed with the indicated ARFpEG202 or pEG202-ARL constructs and Gcs1pJG4-5 or Glo3pJG4-5 were plated on synthetic medium lacking histidine and tryptophan (His-Trpplate). Colonies from His-Trpplates were assayed for β-galactosidase activity by spotting on His-Trpplates containing X-gal to test for specificity. (B) Arl1Q72Ld17N and Arf1Q71Ld17N specifically interacts with different domains of Gcs1p. Top, diagram of Gcs1p and deletion constructs. Bottom, yeast reporter strain YEM1α cotransformed with Arf1Q71Ld17NpEG202, or Arl1Q72Ld17NpEG202 and the indicated Gcs1pJG4-5 construct was plated and colonies were assayed for β-galactosidase activity. (C) Expression of HA-tagged fusion proteins. Proteins (∼20 μg) from lysates of the cells expressing Gcs1pJG4-5 or Glo3pJG4-5 constructs, as indicated, were separated by SDS-PAGE in a 10% gel, transferred to PVDF membranes, and reacted with anti-HA antibodies, or anti-Arf1p antibodies as indicated, followed by detection using the ECL system. Because the expression of HA-tagged proteins was not the same, different amounts of lysates were loaded for Western blotting. Arf1p was used as loading control.
Gcs1p binds to phospholipids vesicles and contains a putative zinc-binding motif and ERM-homology domains (Antonny et al., 1997; Blader et al., 1999). To identify the Arl1p-interacting domain of Gcs1p, we generated yeast two-hybrid constructs expressing mutated and truncated Gcs1p: Gcs1-zn with two cysteine residues in the zinc-finger motif replaced by alanine, Gcs1dC lacking 125 amino acids at the C terminus, and Gcs1N lacking both ERM domains and the C terminus (Figure 1B). These constructs were expressed in roughly equal amounts, but Gcs1-zn and Gcs1dC exhibited twofold higher expression (Figure 1C). We detected interactions of Arl1Q72Ld17N with wild-type Gcs1 and Gcs1dC but not Gcs1-zn or Gcs1N (Figure 1B). Thus, structural elements within the zinc-finger and ERM domains and/or intervening sequence may be essential for Gcs1p–Arl1p interaction. The interaction between Arl1Q72Ld17N and Gcs1dC seemed to be much less than that of wild-type Gcs1, indicating a possible role for the C terminus of Gcs1p for this interaction. Arf1Q71Ld17N interacted with wild-type Gcs1, Gcs1dC, and Gcs1-zn, but not Gcs1N (Figure 1B, bottom), consonant with interactions of Arl1Q72Ld17N and Arf1Q71Ld17N with different regions of Gcs1p.
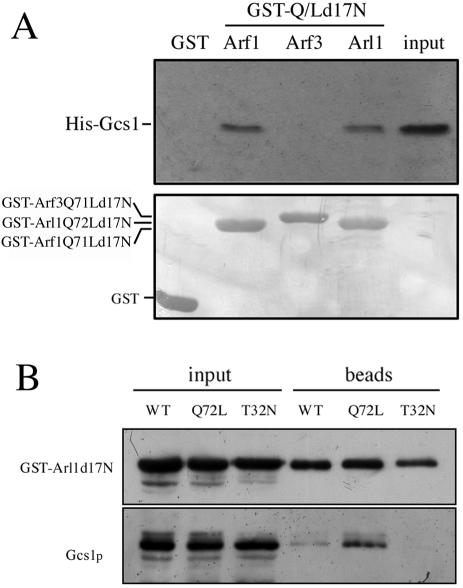
To confirm the direct interactions between Arl1p and Gcs1p, His-tagged Gcs1p was synthesized, purified, and incubated with purified GST, GST-Arl1Q72Ld17N, GST-Arf1Q71Ld17N, or GST-Arf3Q71Ld17N proteins immobilized on glutathione-Sepharose (Figure 2A). Retained His-Gcs1p was detected by immunoblotting (Figure 2A, top). Similar amounts of GST, GST-Arl1Q72Ld17N, GST-Arf1Q71Ld17N, or GST-Arf3Q71Ld17N were present in each reaction, as shown by Coomassie staining (Figure 2A, bottom). GST-Arl1Q72Ld17N and GST-Arf1Q71Ld17N, but not GST or GST-Arf3Q71Ld17N, adsorbed significant amounts of His-Gcs1p. To examine their interaction in vivo, we expressed GST-Arl1d17N, GST-Arl1Q72Ld17N, or GST-Arl1T32Nd17N in arl1 mutant yeast. GST-Arl1 proteins were pulled down on glutathione Sepharose, and the presence of bound endogenous Gcs1p was assessed by immunoblotting (Figure 2B). Arl1Q72Ld17N and to a lesser extent Arl1d17N, but not Arl1T32Nd17N, had bound endogenous Gcs1p, suggesting that Arl1p directly interacted with Gcs1p in a GTP-dependent manner.
Figure 2.
Arl1Q72Ld17N interacts with Gcs1p in vitro and in vivo. (A) In vitro interaction of His-Gcs1 with GST-Arl1Q72Ld17N, GST-Arf1Q71Ld17N, or GST-Arf3Q71Ld17N constructs. Purified recombinant His-Gcs1 (2 μg) was mixed with 2 μg of purified recombinant GST, GST-Arl1Q72Ld17N, GST-Arf1Q71Ld17N, or GST-Arf3Q71Ld17N immobilized on glutathione-Sepharose beads and incubated at 4°C for 1 h. After three times wash, bound proteins were eluted by boiling in 20 μl of 2× sample buffer, and separated by SDS-PAGE in a 12% gel. Input lane contained 1% of the amount added to beads. Proteins were stained with Coomassie Blue (bottom) to assess equal loading, and bound proteins were visualized by Western blotting with anti-Gcs1p antibody (top). (B) In vivo interaction of Arl1Q72Ld17N with endogenous Gcs1p in a GTP-dependent manner. Expression of GST-fused Arl1d17N (WT), Arl1Q72Ld17N (Q72L), or Arl1T32Nd17N (T32N) from pYEX-4T-1 was induced, and yeast were lysed with glass beads. After centrifugation, the cleared lysates were incubated with glutathione-Sepharose beads at 4°C overnight and then washed three times with binding buffer before analysis of bound proteins as described in A. Antibodies used are indicated to the left of the panel.
Gcs1p Possesses GAP Activity for Arl1p and Arf1p In Vitro
To test whether Gcs1p has GAP activity for Arl1p, different amounts of purified recombinant Gcs1p and Glo3p were assayed in vitro for their ability to stimulate the hydrolysis of GTP bound to recombinant-Arl1p, Arf1p, or Arl3p. Figure 3 shows that Gcs1p exhibited GAP activity for both Arl1p and Arf1p. The GAP activity of Glo3 is considerably greater with Arf1p than Arl1p. GAP activity of Arl3p was little affected by Gcs1p or Glo3p. In addition, Gcs1-zn lost GAP activity for both Arl1p and Arf1p (our unpublished data). These data suggest that Gcs1p could be a GAP for Arl1p in vitro.
Figure 3.
GAP activity of Gcs1p for Arl1p in vitro. GAP activity of the different concentrations of purified recombinant Gcs1p or Glo3p (as indicated at bottom) was assayed using Arl1p (square), Arf1p (circle), or Arl3p (triangle) with [γ-32P]GTP bound as substrate as described in Materials and Methods.
Arl1p Localizes to Trans-Golgi Compartment
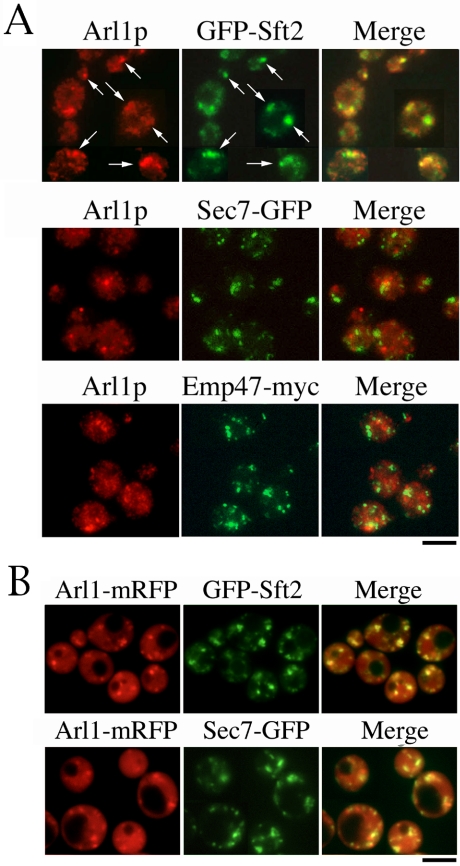
Our previous report suggested that Arl1p is localized to the Golgi (Lee et al., 1997); however, the precise location of Arl1p has yet to be determined. Initially, three Golgi resident proteins, Emp47p (cis-Golgi), Sec7p (late Golgi), and Sft2p (late Golgi), were used to determine the localization of endogenous Arl1p by immunofluorescence microscopy. Constructs encoding GFP-tagged Sec7p and Sft2p, and myc-tagged Emp47p were integrated into wild-type yeast genomic DNA. The coding sequence of GFP was inserted in the chromosome immediately upstream of the translational stop codon for SEC7. Like Emp47p, Sec7p, and Sft2p, Arl1p occurred in typical Golgi-like punctate structures (Figure 4A). It was partially colocalized with Sft2p, but not with Emp47p or Sec7p (Figure 4A, arrows), suggesting that Arl1p is concentrated in a subregion of trans-Golgi, which differs from that occupied by Emp47p or Sec7p. However, our result is inconsistent with a previous report, which suggested that Arl1p was colocalized with Sec7-GFP (Setty et al., 2003). To clarify this difference, we generated Arl1-mRFP to examine localization of Arl1p in live cells. As shown in Figure 4B, Arl1-mRFP colocalized with GFP-Sft2p, but less with Sec7-GFP. Moreover, Arl1-mRFP distributes from cytosol to Golgi, similar to the observation from indirect immunofluorescence staining. Our data suggest that Arl1p is localized to a subdomain of trans-Golgi compartments containing Sft2p.
Figure 4.
Arl1p partially colocalizes with GFP-Sft2p. (A) Endogenous Arl1p partially colocalizes with Sft2p. Myc-tagged Emp47p (cis-Golgi), or GFP-tagged Sec7p (late Golgi), or Sft2p (late Golgi) was used to identify the location of Arl1p by immunofluorescence microscopy, as described in Materials and Methods. Cells were fixed with formaldehyde; spheroplasts were prepared and reacted with purified anti-Arl1p, anti-GFP, or anti-myc antibodies, followed by appropriate secondary antibodies. Arrows indicate colocalization of Arl1p and Sft2p. Observations were replicated in three experiments. (B) Arl1-mRFP on centromeric vector, pRS315, under ADH promoter was transformed into arl1-deleted yeast containing Sec7-GFP or GFP-Sft2. Mid-log phase cells were imaged live with microscopy.
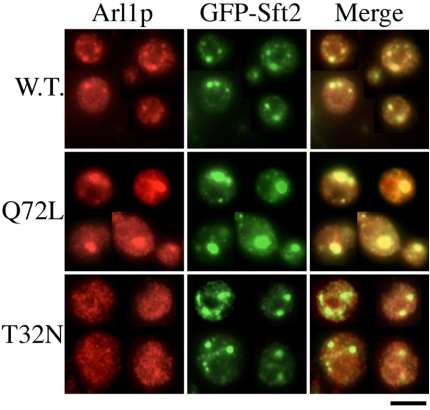
Localization of Arl1p on Trans-Golgi Is Nucleotide Dependent
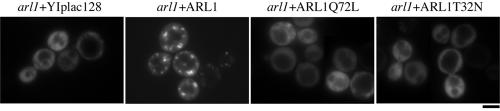
To determine whether localization of Arl1p was guanine nucleotide dependent, we constructed nontagged wild-type Arl1p, GTP binding-defective Arl1T32N, and GTP hydrolysis-deficient Arl1Q72L mutants under the control of the ADH promoter in YIplac128 (Giets and Sugino, 1988). After linearization with EcoRV, these plasmids were integrated into the leu2 locus of the arl1-deleted yeast genome. The expression levels of these integrated wild-type and mutated Arl1p were approximately threeto fivefold higher than the level of endogenous Arl1p (Supplemental Figure S1B). The integrated wild-type Arl1p could rescue the arl1deleted phenotype, suggesting that it possesses at least some of the properties of endogenous Arl1p (our unpublished data). Figure 5 shows that Arl1Q72L and some Arl1p wild-type were colocalized with the late Golgi marker GFP-Sft2p, whereas Arl1T32N staining was, for the most part, distributed through the cytoplasm. In addition, expression of Arl1Q72L seemed to cause the enlargement of trans-Golgi structures. Although the enlarged Golgi is adjacent to the vacuole, the morphology of the vacuole seems normal (Supplemental Figure S3). Our results indicate that the distributions of the GTP-bound and GDP-bound forms of Arl1p were distinct from each other and resembled, in part, those of the corresponding ARL1 in mammalian cells (Lu et al., 2001).
Figure 5.
Localization of Arl1p on trans-Golgi is nucleotide dependent. Wild-type Arl1, Arl1Q72L, or Arl1T32N from integrated plasmid YIplac128 under the control of the ADH promoter was transformed into arl1 (YPH250dl1) cells expressing GFP-Sft2p from pRS314 vector. Logarithmically growing cells were fixed and spheroplasts were prepared for reaction with affinity-purified anti-Arl1p antibody and monoclonal anti-GFP antibody. Indirect immunofluorescence staining was done as described in Materials and Methods. Observations were replicated in at least three experiments.
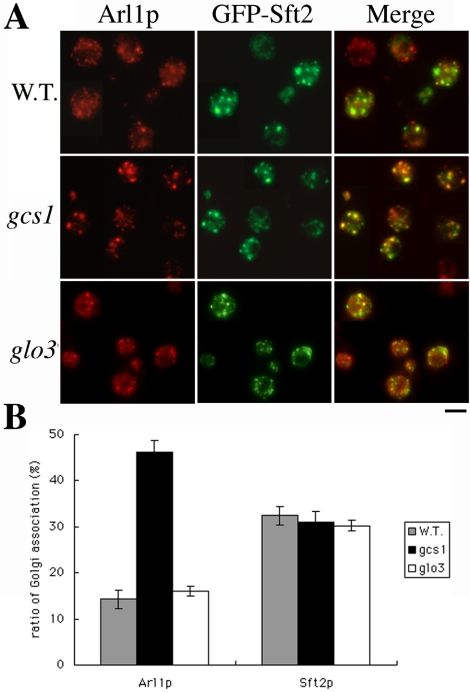
Gcs1p Affects Localization of Arl1p
It is thought GTP-bound active ARF associates with membrane and disassociates when GTP is hydrolyzed to GDP. Thus, the localization of ARF may reflect the identity of the bound guanine nucleotide. If Gcs1p has GAP activity for Arl1p in cells, the amount of Gcs1p in a cell might affect the GTP-binding equilibrium of Arl1p. To investigate whether absence of Gcs1p in a cell affects the intracellular localization of Arl1p, endogenous Arl1p in wild-type, gcs1-deleted, or glo3-deleted yeast was examined (Figure 6). In the gcs1-null mutant, Arl1p was colocalized with GFP-Sft2p to a much greater extent than it was in cells containing Gcs1p, 45 and 13%, respectively. This effect on Arl1p localization was not observed in the glo3 mutant. This phenomenon was also observed when we examined Arl1-mRFP in arl1- or arl1gcs1- deleted yeasts (Supplemental Figure S4). Our findings suggest that failure to hydrolyze GTP on Arl1p in the gcs1 mutant could result in accumulation of Arl1p-GTP on trans-Golgi membranes, thus inhibiting vesicular trafficking.
Figure 6.
More Arl1p is present in trans-Golgi of gcs1 mutant than wild-type yeast. (A) Wild-type, gcs1-, or glo3-deleted yeast were stained with anti-GFP and affinity-purified anti-Arl1p antibodies to monitor the Golgi association of Arl1p. GFP-Sft2p was the late-Golgi marker. (B) Quantification of Golgi association ratio of Arl1p. Signals in A were quantified by Axio Vision Rel. 4.2 software. Punctate signals were summed and divided with whole cell signals (n = 50 and p < 0.05).
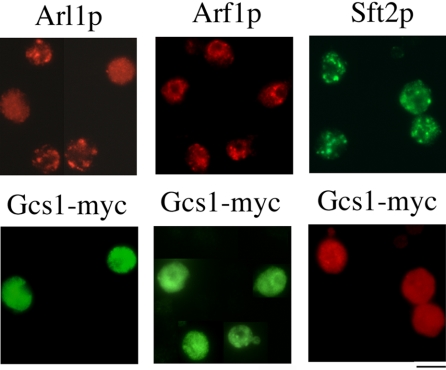
Next, we determined whether overexpression of Gcs1p altered the intracellular localization of Arl1p. We used indirect immunofluorescence staining to observe the effect of Gcs1-myc overexpression on endogenous Arl1p. Figure 7 shows that, in gcs1 cells overexpressing Gcs1p-myc, Arl1p, but not Arf1p, was no longer in a punctate pattern, whereas localization displayed a cytoplasmic pattern. In contrast, localization of GFP-Sft2p seemed normal, suggesting that the Golgi structure is intact. Thus, this effect of Gcs1p seemed to be specific for Arl1p. Similar results also were observed when plasmids encoding C-terminal GFP-tagged Gcs1p and Arl1-mRFP or Arf1-mRFP were cotransformed into gcs1 mutants (Supplemental Figure S5A). We also used Gcs1-zn to demonstrate that GAP activity is important for Gcs1p in regulation of Arl1p at trans-Golgi compartments. The overexpression of Gcs1-zn-GFP seemed normal like Gcs1p-GFP, but it showed no effect on Arl1p localization (Supplemental Figure S5A). Overexpression of Gcs1-zn-myc protein also was attempted; however, it seemed lumpy (possibly due to aggregation of the recombinant protein) and could not be used in this experiment (our unpublished data). In addition, Arl1p was not dissociated from its punctate patterns in glo3 cells overexpressing Glo3p-myc (Supplemental Figure S5B). Age2p, a GAP protein, which provides overlapping function with Gcs1p (Poon et al., 2001), also had no effect on Arl1p distribution (Supplemental Figure S5B). Our findings suggest that the effect of Gcs1p on intracellular localization of Arl1p might be anticipated from its acceleration of GTP hydrolysis on Arl1p.
Figure 7.
Overexpression of Gcs1-myc, but not Gcs1-zn-myc, caused dissociation of endogenous Arl1p from trans-Golgi. Gcs1-myc overexpression affected the distribution of Arl1p. Gcs1-mycpVT101U was transformed into gcs1 cells with or without GFP-Sft2pRS314. Mid-log phase cells overexpressing Gcs1-myc were stained with anti-myc, anti-GFP, affinity-purified anti-Arl1p, or affinity-purified anti-Arf1p antibody.
Gcs1p and Arl1p Interact Functionally
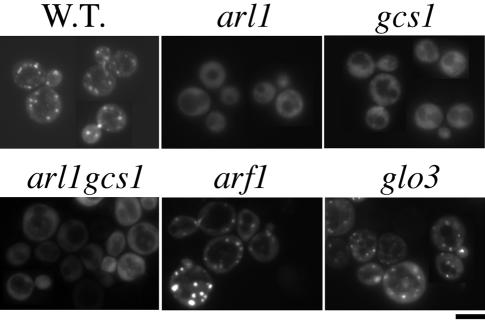
Previous report showed that GFP-Imh1, an effector of Arl1p, lost its Golgi localization signals in arl1 null cells (Panic et al., 2003; Setty et al., 2003). We next examined whether the localization of Imh1p also was affected in the gcs1 mutant. We constructed a GFP-IMH1 that was driven by a GAL1 promoter in a centromeric plasmid. The expression level of GFP-Imh1p was similar to that of endogenous Imh1p (Supplemental Figure 1C). Figure 8 shows that GFP-Imh1p displays a Golgi-like distribution in wild-type cells and a diffuse pattern in arl1-deleted yeast. Unexpectedly, substantial amounts of GFP-Imh1p became diffuse in gcs1-null mutants (Figure 8). The diffuse GFP-Imh1p in the gcs1-null mutant is specific because GFP-Imh1p was concentrated at punctate Golgi-structures in arf1- or glo3-null mutants.
Figure 8.
GFP-Imh1p substantially loses its Golgi association in arl1- and gcs1-null cells. GFP-IMH1 under the control of the Gal1 promoter (centromeric plasmid) was transformed into different deletion yeast as indicated. After overnight induction (∼12 h) with 2% galactose and 0.1% glucose, GFP-Imh1p was observed in live cells and photographed.
Because more Arl1p proteins were retained in Golgi structures in gcs1-null cells, we speculated that GTP-hydrolysisdefective Arl1p in gcs1-null cells might cause diffusion of the GFP-Imh1p from Golgi. Therefore, we examined the localization of GFP-Imh1p in arl1-deleted cells containing integrated wild-type, constitutively active Arl1 (Arl1Q72L) or inactive Arl1 (Arl1T32N) (Figure 9). Interestingly, wild-type Arl1p, but not Arl1Q72L or Arl1T32N, could rescue the Golgi-association of GFP-Imh1p. These data suggest that proper GTP-GDP recycling of Arl1p is required for its function on effector recruitment.
Figure 9.
Constitutive active and inactive mutant forms of Arl1p did not complement the loss of Arl1p. GFP-IMH1pRS314 was transformed into arl1-deleted cells containing integrated wild-type, constitutively active (Q72L), constitutively inactive (T32N), Arl1p, or empty vector control (YIplac128). After induction with 2% galactose and 0.1% glucose medium, live cells were observed with microscopy.
DISCUSSION
Yeast cells contain six structurally related proteins with the potential to provide GAP activity for the yeast Arf family proteins and to participate in vesicular transport (Poon et al., 1996, 1999; Zhang et al., 2003). Three of these proteins, Gcs1p, Glo3p, and Age2p have been shown to be Arf GAPs capable of stimulating the GTPase activity of yeast Arf1p (Poon et al., 1996, 1999, 2001). In this study, we demonstrate that Gcs1p is also a GAP for Arl1p. Several types of evidence indicate that in yeast Gcs1p may play a crucial role in regulating Arl1p at trans-Golgi compartments.
The extensive sequence identity of Arl1p and Arf1p led us to hypothesize that they would interact similarly with the Gcs1p molecule. Surprisingly, our data showed that although all three Gcs1p domains seemed to be important for Arl1p interaction (Figure 1B), the zinc-binding domain and C terminus were not essential for Arf1Q71Ld17N interaction (Figure 1B). The zinc-finger mutation eliminates Arl1p binding; therefore, the general structure for interaction must contain these two cysteine residues. Other elements may contribute to establish a stronger interaction between the two proteins. Alternatively, the deletions or truncations may alter the structure of the zinc-finger domain sufficiently to disrupt interaction. In the crystal structure of the Arf1-ARFGAP1 complex determined by Goldberg (1999), the zinc-finger residues do not make direct contacts with Arf1, consistent with the interaction we observed between Arf1Q71Ld17N and Gcs1-zn. Moreover, a recent report (Lee et al., 2005) also has shown that ARFGAP1R50K, a mutant ARFGAP1 without GAP activity, could interact with ARF1 in vitro.
From many studies, it has been suggested that for ARFGAP1 to exert its GAP activity on Arf1 participation of coatomer, GGA and membrane curvature are necessary (Goldberg, 1999; Szafer et al., 2000; Jacques et al., 2002; Bigay et al., 2003). Our data showed that Gcs1p has GAP activity for both Arl1p and Arf1p. Ding et al. (1996) also reported that purified spleen ARF GAP accelerated hydrolysis of GTP bound to recombinant mammalian ARL1. From the crystal structure study, it is thought that the N-terminal helix and myristic acid of Arf1p should leave the core of the protein and insert into the membrane when it is activated (GTP-bound). Therefore, it is reasonable that the interaction between active Arf1p or Arl1p and Gcs1p should occur on the membrane structure. Because the interaction of these proteins in the yeast two-hybrid experiments does not occur on a membrane structure, an N-terminal deletion seems to be required. In contrast, the in vitro GAP assay is performed in reaction buffer containing micelles (DMPC/cholate); thus, Gcs1p could exhibit activity with both the full-length Arf1p and Arl1p. Bigay et al. (2003) reported that the GAP activity of ARFGAP1, the mammalian homologue of Gcs1p, is greatly increased by highly curved liposomes and that Gcs1p shares with ARFGAP1 a preference for binding to conical lipids. Their data showed that ARFGAP1 stimulates the GTPase reaction of ARF1, and the rate constant of GTP hydrolysis on small liposome was in the subsecond range (kcat > 1 s–1). Randazzo (1997) also showed that the GAP activity of rat liver GAP2 and GAP1 is phospholipids dependent; and, when using ARF1 as a substrate, GAP activity was 7.2 and 1.92 s–1 for GAP2 and GAP1, respectively. Our study showed that the average GAP activity of Gcs1p was 0.18 and 0.22 s–1 for Arl1p and Arf1p, respectively, suggesting that membrane curvature or different phospholipids may be required to increase GAP activity. Additional studies of Arl1p and Gcs1p with their interacting adaptor protein(s) or phospholipids are needed to characterize the activity Gcs1p for Arl1p and Arf1p. Thus, we suspect that limitation of those factors may reduce the efficiency of Gcs1p GAP activity for GTP hydrolysis by Arf1p relative to that by Arl1p.
We reported previously in 1997 that Arl1p is associated in part with the Golgi complex (Lee et al., 1997). Moreover, a recent study (Setty et al., 2003) showed that an Arl1p-GFP fusion protein colocalized with RFP-Imh1p-GRIP, and RFP-Imh1p-GRIP colocalized with Sec7p-GFP. Based on these observations, it was suggested that Arl1p colocalized with Sec7p. Here, we demonstrate that Arl1 partially colocalizes with the trans-Golgi integral membrane protein Sft2p, but not with Sec7p. The differences between these two experiments perhaps result from the fact that the overexpression of RFP-Imh1p-GRIP may affect the subcellular distribution of the trans-Golgi network. In Setty's report (Setty et al., 2003) and our unpublished observation, the Golgi punctate structures in cells expressing RFP-Imh1p-GRIP or GFP-Imh1p were larger than those in normal cells. Moreover, overexpression of RFP-GRIP results in the recruitment of Arl1-GFP to Golgi (Setty et al., 2003), which differs from distribution of wild-type Arl1p (Huh et al., 2003). Thus, our data provide evidence that Arl1p localizes at one subdomain of the trans-Golgi compartment containing Sft2p.
From rescue experiments involving fragmented vacuole and localization of GFP-Imh1p observed with arl1-null mutant, we found that C-terminal-tagged Arl1p, with either GFP or mRFP, would lose part of its function, although its localization seems normal (our unpublished observation). This phenomenon also was observed by Trautwein et al. (2004) who reported that cyan fluorescent proteinor yellow fluorescent protein (YFP)-tagged Arf1 was partly defective in its function. Thus, the use of a GFPor mRFP-tag may be a convenient approach to track Arfp and Arlp location, but it is not an appropriate way to identify their cellular function.
We also found that the Golgi association of Arl1p is nucleotide dependent. The GTP-bound form, Arl1Q72L, seems to colocalize with Sft2p and is associated with the aggregation of the trans-Golgi apparatus. This phenomenon is reminiscent of the Golgi expansion caused by overexpression of active human Arl1p (Lu et al., 2001). These results suggest that the GTPase cycle of Arl1p determines its distribution between Golgi and cytosol in a manner that is fundamentally similar to that seen for Arf1p. Moreover, Arl1Q72L seemed not to alter vacuole morphology and vacuole enzyme trafficking was not substantially affected in arl1and gcs1-deleted cells (our unpublished observation), suggesting that Arl1p and Gcs1p are not directly involved in Golgi-to-vacuole transport.
The GTPase cycle of ARF proteins is regulated by their GAPs, which, in turn, leads to the dissociation of ARFs from membranes. When we examined the localization of Arl1p in cells containing different amount of Gcs1p, we found that eliminating Gcs1p from cells prevented Arl1p release from the trans-Golgi to the cytosol. We also found that increasing amounts of Gcs1-myc facilitated the release of Golgi-bound Arl1p to the cytosol. However, no phenotypic differences were observed in cells overexpressing the zinc-finger mutant, Gcs1-zn-GFP. The latter observations support the idea that the pattern of Arl1p distribution upon elimination (or overexpression) of Gcs1p resulted from decreased (or increased) GAP catalytic activities. These data also fit the general model of ARF proteins, namely, that the GTP-bound active ARF localizes to membranes and subsequently disassociate from membranes when the GTP is hydrolyzed to GDP. Again, although Gcs1p possesses GAP activity for both Arl1p and Arf1p in vitro, distribution of Arf1p was not affected by the level of Gcs1p. These results suggest that Gcs1p GAP activity toward Arf1p in cells needs other cofactors, or perhaps equally, Arl1p and Arf1p interact with membranes in different ways.
Our data showed that GFP-Imh1p became diffuse in gcs1-null mutants (Figure 8), suggesting that GTP-hydrolysis defective Arl1p in gcs1-null cells might cause diffusion of the GFP-Imh1p from Golgi. Although the current model for recruiting Imh1p to Golgi is in favor of having an active form of Arl1p in Golgi (Panic et al., 2003), our data showed that constitutively active Arl1p not only could not complement Arl1p function but also seemed to cause the disassociation of GFP-Imh1p from Golgi. These data suggest that a proper GTP-GDP recycling of Arl1p as well as other factors in Golgi membrane may be required for Imh1p recruitment.
Gcs1p has been implicated in mitochondrial function, vesicular trafficking, and actin cytoskeletal organization (Filipak et al., 1992; Wang et al., 1996; Poon et al., 1996; Blader et al., 1999). Overexpressed Gcs1p was concentrated in patches in yeast and colocalized partially with actin structures, suggesting that its overexpression affects mitochondrial morphology as a consequence of perturbing actin structures (Huang et al., 2002). The effect of Gcs1p overexpression on mitochondrial structure does not require Arl1p or Arf1p, because the effect on mitochondria can be observed in the arl1 or arf1 mutants (our unpublished data). The zinc-finger motif of Gcs1p is required for its functional interaction with Arl1p but not for its effects on mitochondria (Huang et al., 2002).
Consistent with our findings, the studies of Eugster et al. (2000) indicate Glo3p to be the major Arf1 GAP in vivo. They reported that Arf1Q71Ld17N could interact with Glo3p, Gcs1p, and COPI subunits and only Glo3p, but not Gcs1p, could interact with COPI subunits. In addition, gcs1 is synthetic lethal with arf1 or glo3 (Zhang et al., 2003) but not with arl1 (this work). Analysis of knockout strains of all six GLO3 related proteins in yeast revealed that the glo3 mutant alone results in a defect in KKXX retrieval from the Golgi (Dogic et al., 1999). The effectiveness of vesicular transport in glo3 mutant cells is not limited by the amount of Gcs1 protein. The glo3 mutant, but not the gcs1 mutant, showed strong synthetic lethality with sec21-1, a mutant of γ-COP (Poon et al., 1999). These observations are consistent with the notion that the Gcs1p may normally supply only a minor portion of the ARF GAP activity for transport from the Golgi to the ER in the cells.
The Gcs1p and Age2p ARF GAPs provide overlapping functions for transport from the trans-Golgi network (Poon et al., 2001). Consistent with our results, it seems that Gcs1p may provide the major GAP activity for Arl1p in trans-Golgi vesicular trafficking. Thus, distinct but partially overlapping functions of the Gcs1p, Glo3p, and Age2p suggest that each of these GAPs may serve in several vesicular transport pathways and interact with distinct sets of proteins characteristic of the different loci.
Identification of the Gcs1p interaction with Arl1p, as well as with Arf1p, led us to infer that Gcs1p is a multifunctional molecule, which may act at different intracellular membranes to regulate intracellular membrane dynamics and/or actin cytoskeletal organization. Consistent with the previous model for ARF function, our findings also suggest that Arl1p-GTP is tethered in a relatively immobile complex on the surface of the Golgi membrane and does not come off until after the hydrolysis of GTP has occurred (Rothman, 1994). Integration of our unexpected findings into an understanding of the role of Gcs1p and Arl1p in the overall intracellular membrane trafficking remains a challenge for future studies.
Supplementary Material
Acknowledgments
We thank Drs. Stephan Schroder-Kohne, Olivia W. Rossanese, Roger Tsien, and Hugh R. B. Pelham for providing us with antibodies and plasmids. We also thank Drs. Randy Haun, Martha Vaughan, and Joel Moss for critical reading of this manuscript. This work was supported by grants from the National Science Council, R.O.C. (NSC-91-2314-B-002-136, NSC-92-2320-B-002-176) and Yung-Shin Biomedical Research Funds (to F.-J.S.L.).
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05–01–0023) on June 22, 2005.
Abbreviations used: ARF, ADP-ribosylation factor; ER, endoplasmic reticulum; GAP, GTPase-activating protein.
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
References
- Antonny, B., Huber, I., Paris, S., Chabre, M., and Cassel, D. (1997). Activation of ADP-ribosylation factor 1 GTPase-activating protein by phosphatidylcholine-derived diacylglycerols. J. Biol. Chem. 272, 30848–30851. [DOI] [PubMed] [Google Scholar]
- Andreev, J., Simon, J. P., Sabatini, D. D., Kam, J., Plowman, G., Randazzo, P. A., and Schlessinger, J. (1999). Identification of a new Pyk2 target protein with Arf-GAP activity. Mol. Cell. Biol. 19, 2338–2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhamidipati, A., Lewis, S. A., and Cowan, N. J. (2000). ADP ribosylation factor-like protein 2 (Arl2) regulates the interaction of tubulin-folding cofactor D with native tubulin. J. Cell Biol. 149, 1087–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigay, J., Pierre, G., Robineau, S., and Antonny, B. (2003). Lipid packing sensed by ArfGAP1 couples COPI coat disassembly to membrane bilayer curvature. Nature 426, 563–566. [DOI] [PubMed] [Google Scholar]
- Blader, I. J., Cope, M.J.T.V., Jackson, T. R., Profit, A. A., Greenwood, A. F., Drubin, D. G., Prestwich, G. D., and Theibert, A. B. (1999). GCS1, an Arf guanosine triphosphatase-activating protein in Saccharomyces cerevisiae, is required for normal actin cytoskeletal organization in vivo and stimulates actin polymerization in vitro. Mol. Biol. Cell 10, 581–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, M. T., Andrade, J., Radhakrishna, H., Donaldson, J. G., Cooper, J. A., and Randazzo, P. A. (1998). ASAP1, a phospholipid-dependent ARF GTPase-activating protein that associates with and is phosphorylated by Src. Mol. Cell. Biol. 18, 7038–7051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coe, J. G., Murray, L. E., and Dawes, I. W. (1994). Identification of a sporulation-specific promoter regulating divergent transcription of two novel sporulation genes in Saccharomyces cerevisiae. Mol. Gen. Genet. 244, 661–672. [DOI] [PubMed] [Google Scholar]
- Cukierman, E., Huber, I., Rotman, M., and Cassel, D. (1995). The ARF1 GTPase-activating protein: zinc finger motif and Golgi complex localization. Science 270, 1999–2002. [DOI] [PubMed] [Google Scholar]
- Ding, M., Vitale, N., Tsai, S. C., Adamik, R., Moss, J., and Vaughan, M. (1996). Characterization of a GTPase-activating protein that stimulates GTP hydrolysis by both ADP-ribosylation factor (ARF) and ARF-like proteins comparison to the ARD1 GAP domain. J. Biol. Chem. 271, 24005–24009. [DOI] [PubMed] [Google Scholar]
- Dogic, D., de Chassey, B., Pick, E., Cassel, D., Lefkir, Y., Hennecke, S., Cosson, P., and Letourneur, F. (1999). The ADP-ribosylation factor GTPase-activating protein Glo3p is involved in ER retrieval. Eur. J. Cell Biol. 78, 305–310. [DOI] [PubMed] [Google Scholar]
- Donaldson, J. G. (2000). Filling in the GAPs in the ADP-ribosylation factor story. Proc. Natl. Acad. Sci. USA 97, 3792–3794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eugster, A., Frigerio, G., Dale, M., and Rainer, D. (2000). COP I domains required for coatomer integrity, and novel interactions with ARF and ARF-GAP. EMBO J. 19, 3905–3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipak, M., Drebot, M. A., Ireland, L. S., Singer, R. A., and Johnston, G. C. (1992). Mitochondrial DNA loss by yeast reentry-mutant cells conditionally unable to proliferate from stationary phase. Curr. Genet. 22, 471–477. [DOI] [PubMed] [Google Scholar]
- Giets, R. D., and Sugino, A. (1988). New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 74, 527–534. [DOI] [PubMed] [Google Scholar]
- Goldberg, J. (1999). Structural and functional analysis of the ARF1-ARFGAP1 complex reveals a role for coatomer in GTP hydrolysis. Cell 96, 893–902. [DOI] [PubMed] [Google Scholar]
- Golemis, E. A., and Khazak, V. (1997). Alternative yeast two-hybrid systems. The interaction trap and interaction mating. Methods Mol. Biol. 63, 197–218. [DOI] [PubMed] [Google Scholar]
- Gyuris, J., Golemis, E., Chertkov, H., and Brent, R. (1993). Cdi1, a human G1 and S phase protein phosphatase that associates with Cdk2. Cell 75, 791–803. [DOI] [PubMed] [Google Scholar]
- Hagan, I. M., and Ayscough, K. R. (1999). Fluorescence microscopy in yeast. In: Protein Localization by Fluorescence Microscopy, ed. V. J. Allan, Oxford: Oxford University Press, 194–196.
- Huang, C. F., Buu, L. M., Yu, W. L., and Lee, F. J. (1999). Characterization of a novel ADP-ribosylation factor-like protein (yARL3) in Saccharomyces cerevisiae. J. Biol. Chem. 274, 3819–3827. [DOI] [PubMed] [Google Scholar]
- Huang, C. F., Chen, C. C., Tung, L., Buu, L. M., and Lee, F. J. (2002). The yeast ADP-ribosylation factor GAP, Gcs1p, is involved in maintenance of mitochondrial morphology. J. Cell Sci. 115, 275–282. [DOI] [PubMed] [Google Scholar]
- Huang, C. F., Liu, Y. W., Tung, L., Lin, C. H., and Lee, F. J. (2003). Role for Arf3p in development of polarity, but not endocytosis, in Saccharomyces cerevisiae. Mol. Biol. Cell 14, 3834–3847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber, I., Rotman, M., Pick, E., Makler, V., Rothem, L., Cukierman, E., and Cassel, D. (2001). Expression, purification, and properties of ADP-ribosylation factor (ARF) GTPase activating protein-1. Methods Enzymol. 329, 307–316. [DOI] [PubMed] [Google Scholar]
- Huh, W. K., Falvo, J. V., Gerke, L. C., Carroll, A. S., Howson, R. W., Weissman, J. S., and O'Shea, E. K. (2003). Global analysis of protein localization in budding yeast. Nature 425, 686–691. [DOI] [PubMed] [Google Scholar]
- Ito, H., Fukuda, Y., Murata, K., and Kimura, A. (1983). Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 153, 163–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacques, K. M., Nie, Z., Stauffer, S., Hirsch, D. S., Chen, L. X., Stanley, K. T., and Randazzo, P. A. (2002). Arf1 disassociates from the clathrin adaptor GGA prior to being inactivated by Arf GTPase-activating proteins. J. Biol. Chem. 277, 47235–47241. [DOI] [PubMed] [Google Scholar]
- Kahn, R. A., and Gilman, A. G. (1986). The protein cofactor necessary for ADP-ribosylation of Gs by cholera toxin is itself a GTP binding protein. J. Biol. Chem. 261, 7906–7911. [PubMed] [Google Scholar]
- Lee, F. J., Huang, C. F., Yu, W. L., Buu, L. M., Lin, C. Y., Huang, M. C., Moss, J., and Vaughan, M. (1997). Characterization of an ADP-ribosylation factor-like 1 protein in Saccharomyces cerevisiae. J. Biol. Chem. 272, 30998–31005. [DOI] [PubMed] [Google Scholar]
- Lee, J. G., Cho, S. P., Lee, H. S., Lee, C. H., Bae, K. S., and Maeng, P. J. (1994). Identification of a cryptic N-terminal signal in Saccharomyces cerevisiae peroxisomal citrate synthase that functions in both peroxisomal and mitochondrial targeting. J. Biochem. 128, 1059–1072. [DOI] [PubMed] [Google Scholar]
- Lee, S. Y., Yang, J. S., Hong, W., Premont, R. T., and Hsu, V. W. (2005). ARFGAP1 plays a central role in coupling COPI cargo sorting with vesicle formation. J. Cell Biol. 168, 281–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, C. Y., Huang, P. H., Liao, W. L., Cheng, H. J., Huang, C. F., Kuo, J. C., Patton, W. A., Massenburg, D., Moss, J., and Lee, F. J. (2000). ARL4, an ARF-like protein that is developmentally regulated and localized to nuclei and nucleoli. J. Biol. Chem. 275, 37815–37823. [DOI] [PubMed] [Google Scholar]
- Lin, C. Y., Li, C. C., Huang, P. H., and Lee, F. J. (2002). A developmentally regulated ARF-like 5 protein (ARL5), localized to nuclei and nucleoli, interacts with heterochromatin protein 1. J. Cell Sci. 115, 4433–4445. [DOI] [PubMed] [Google Scholar]
- Lowe, S. L., Wong, S. H., and Hong, W. (1996). The mammalian ARF-like protein 1 (Arl1) is associated with the Golgi complex. J. Cell Sci. 109, 209–220. [DOI] [PubMed] [Google Scholar]
- Lu, L., Horstmann, H., Ng, C., and Hong, W. (2001). Regulation of Golgi structure and function by ARF-like protein 1 (Arl1). J. Cell Sci. 114, 4543–4555. [DOI] [PubMed] [Google Scholar]
- Lu, L., and Hong, W. (2003). Interaction of Arl1-GTP with GRIP domains recruits autoantigens Golgin-97 and Golgin-245/p230 onto the Golgi. Mol. Biol. Cell 14, 3767–3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsui, K., Yaguchi, S., and Tsurugi, K. (1994). The GTS1 gene, which contains a Gly-Thr repeat, affects the timing of budding and cell size of the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 14, 5569–5578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss, J., and Vaughan, M. (1998). Molecules in the ARF orbit. J. Biol. Chem. 273, 21431–21434. [DOI] [PubMed] [Google Scholar]
- Panic, B., Whyte, J. R., and Munro, S. (2003). The ARF-like GTPases Arl1p and Arl3p act in a pathway that interacts with vesicle-tethering factors at the Golgi apparatus. Curr. Biol. 13, 405–410. [DOI] [PubMed] [Google Scholar]
- Pasqualato, S., Renault, L., and Cherfils, J. (2002). Arf, Arl, Arp and Sar proteins: a family of GTP-binding proteins with a structural device for `front-back' communication. EMBO Rep. 3, 1035–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon, P. P., Wang, X., Rotman, M., Huber, I., Cukierman, E., Cassel, D., Singer, R. A., and Johnston, G. C. (1996). Saccharomyces cerevisiae Gcs1 is an ADP-ribosylation factor GTPase-activating protein. Proc. Natl. Acad. Sci. USA 93, 10074–10077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon, P. P., Cassel, D., Spang, A., Rotman, M., Pick, E., Singer, R. A., and Johnston, G. C. (1999). Retrograde transport from the yeast Golgi is mediated by two ARF GAP proteins with overlapping function. EMBO J. 18, 555–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon, P. P., Nothwehr, S. F., Singer, R. A., and Johnston, G. C. (2001). The Gcs1 and Age2 ArfGAP proteins provide overlapping essential function for transport from the yeast trans-Golgi network. J. Cell Biol. 155, 1239–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Premont, R. T., Claing, A., Vitale, N., Freeman, J. L., Pitcher, J. A., Patton, W. A., Moss, J., Vaughan, M., and Lefkowitz, R. J. (1998). Beta2-Adrenergic receptor regulation by GIT1, a G protein-coupled receptor kinase-associated ADP ribosylation factor GTPase-activating protein. Proc. Natl. Acad. Sci. USA 95, 14082–14087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randazzo, P. A. (1997). Resolution of two ADP-ribosylation factor 1 GTPase-activating proteins from rat liver. Biochem. J. 324, 413–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randazzo, P. A., Andrade, J., Miura, K., Brown, M. T., Long, Y. Q., Stauffer, S., Roller, P., and Cooper, J. A. (2000). The Arf GTPase-activating protein ASAP1 regulates the actin cytoskeleton. Proc. Natl. Acad. Sci. USA 97, 4011–4016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothman, J. E. (1994). Mechanisms of intracellular protein transport. Nature 372, 55–63. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E. F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual, Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
- Setty, S. R., Shin, M. E., Yoshino, A., Marks, M. S., and Burd, C. G. (2003). Golgi recruitment of GRIP domain proteins by Arf-like GTPase 1 is regulated by Arf-like GTPase 3. Curr. Biol. 13, 401–404. [DOI] [PubMed] [Google Scholar]
- Sherman, F., Fink, G. R., and Hicks, J. B. (1986). Methods in Yeast Genetics, Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
- Sikorski, R. S., and Hieter, P. (1989). A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122, 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szafer, E., Pick, E., Rotman, M., Zuck, S. Huber, I., and Cassel, D. (2000). Role of coatomer and phospholipids in GTPase-activating protein-dependent hydrolysis of GTP by ADP-ribosylation factor-1. J. Biol. Chem. 275, 23615–23619. [DOI] [PubMed] [Google Scholar]
- Trautwein, M., Dengjel, J., Schirle, M., and Spang, A. (2004). Arf1p provides an unexpected link between COPI vesicles and mRNA in Saccharomyces cerevisiae. Mol. Biol. Cell 15, 5021–5037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner, C. E., Brown, M. C., Perrotta, J. A., Riedy, M. C., Nikolopoulos, S. N., McDonald, A. R., Bagrodia, S., Thomas, S., and Leventhal, P. S. (1999). Paxillin LD4 motif binds PAK and PIX through a novel 95-kD ankyrin repeat, ARF-GAP protein: a role in cytoskeletal remodeling. J. Cell Biol. 145, 851–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verent, T., Dignard, D., and Thomas, D. Y. (1987). A family of yeast expression vectors containing the phage f1 intergenic region. Gene. 52, 225–233. [DOI] [PubMed] [Google Scholar]
- Wang, X., Hoekstra, M. F., DeMaggio, A. J., Dhillon, N., Vancura, A., Kuret, J., Johnston, G. C., and Singer, R. A. (1996). Prenylated isoforms of yeast casein kinase I, including the novel Yck3p, suppress the gcs1 blockage of cell proliferation from stationary phase. Mol. Cell. Biol. 16, 5375–5385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, C. J., Bowzard, J. B., Anido, A., and Kahn, R. A. (2003). Four ARF GAPs in Saccharomyces cerevisiae have both overlapping and distinct functions. Yeast 20, 315–330. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.