Abstract
Sp1 and Sp3 are ubiquitously expressed mammalian transcription factors that activate or repress the expression of a variety of genes and are thought to compete for the same DNA binding site. We used indirect immunofluorescence microscopy and image deconvolution to show that Sp1 and Sp3 are organized into distinct nonoverlapping domains in human breast and ovarian cells. Domains of Sp1 and Sp3 infrequently associate with sites of transcription. Sp3 partitions with the tightly bound nuclear protein fraction of hormone responsive MCF-7 breast cancer cells, whereas only a subpopulation of Sp1 is found in that fraction. Both Sp1 and Sp3 are bound to the nuclear matrix, and the nuclear matrix-associated sites of Sp1 and Sp3 are different. Indirect immunofluorescence studies demonstrate that Sp1 and Sp3 associate with histone deacetylases 1 and 2 and with the estrogen receptor α, albeit at low frequencies in MCF-7 cells. Chromatin immunoprecipitation (ChIP) and re-ChIP assays revealed that although both Sp1 and Sp3 bind to the estrogen-responsive trefoil factor 1 promoter in MCF-7 cells, they do not occupy the same promoter. Our results demonstrate the different features of Sp1 and Sp3, providing further evidence that Sp3 is not a functional equivalent of Sp1.
INTRODUCTION
The structural organization and compartmentalization of the nucleus play an important role in the regulation of gene expression (Zaidi et al., 2005). Many transcription factors are located in foci throughout the nucleus excluding the nucleoli, showing a punctate pattern under high-resolution fluorescence microscopy (Grande et al., 1997). Many of these regulatory factors are associated with the nuclear matrix (Stenoien et al., 2000; Zaidi et al., 2005). Furthermore, visualization studies of nuclear matrix preparations have revealed a punctate distribution of transcription factors (Stenoien et al., 2000; Zaidi et al., 2005), suggesting their targeting to specific sites in the nuclear matrix. Although the function of such a subnuclear compartmentalization of transcription factors is not yet understood, its biological significance has been demonstrated by the occurrence of disease and cancer often linked with mislocalization (Mancini et al., 1999; Choi et al., 2001; Zaidi et al., 2005).
The Sp family of transcription factors binds and acts through GC boxes to regulate gene expression and is recognizable by a DNA binding domain formed of three conserved Cys2His2 zinc fingers (Suske, 1999). Within this family, Sp1 and Sp3 are ubiquitously expressed in mammalian cells and participate in regulating the expression of genes involved in almost all cellular processes (Cawley et al., 2004). Sp3 is structurally similar to Sp1, with similar affinities for Sp1 binding sites, and, as evidenced by a comparison of the phenotypes of Sp1 and Sp3 knockout mice, has many overlapping functions with Sp1 as well as distinct functions (Bouwman et al., 2000). Sp1 is found in both nonmatrix nuclear fractions and nuclear matrix preparations (van Wijnen et al., 1993), but it is not known whether Sp3 is associated with the nuclear matrix. Unlike Sp1 which is usually an activator of transcription, Sp3 functions as an activator or a repressor (Suske, 1999; Bouwman and Philipsen, 2002; Li et al., 2004). Sp3 has four isoforms, two long forms (L1-Sp3 and L2-Sp3) and two short forms (M1-Sp3 and M2-Sp3), that are the products of differential translational initiation (Sapetschnig et al., 2004). The short forms of Sp3 lack the subdomain A of the transactivation domain and act as repressors or weak activators. Sp3 does not have the ability of Sp1 to bind to the DNA as multimers or to activate transcription in a synergistic manner by binding to multiple sites (Yu et al., 2003), but it can antagonize Sp1, blocking its synergistic transactivation function (Yu et al., 2003). Yet, there are promoters for which Sp3 activates rather than represses (Sowa et al., 1999; Gartel et al., 2000). Despite the different roles of Sp1 and Sp3, they are generally thought to compete for the same binding sites, and a number of chromatin immunoprecipitation studies have shown that both Sp1 and Sp3 are associated in situ with the GC box(es) of a variety of promoters (Stoner et al., 2004; Chan et al., 2004; Lee et al., 2004; Liu et al., 2004; Zelko and Folz, 2004; de Leon et al., 2005 Sun et al., 2005). Whether Sp1 and Sp3 occupy these promoters simultaneously is not known. They could associate with the Sp1 element separately or as a multi-protein Sp1/Sp3 complex. Some studies support the existence of a Sp1/Sp3 complex (Choi et al., 2002; Zhang et al., 2003). However, we and others found that Sp1 does not form complexes with Sp3 in human breast cancer MCF-7 or mouse L cells (Sun et al., 2002; Yu et al., 2003). Nonetheless, it is conceivable that although not in complex together, Sp1 and Sp3 are colocalized in the nucleus. A number of imaging studies of Sp1 or Sp3 have been published, most of them showing a diffuse or punctate localization of Sp1 and Sp3 in the nucleus of various cell types (Birnbaum et al., 1995; Pombo et al., 1998; Ross et al., 2002; Sapetschnig et al., 2002, 2004; Spann et al., 2002; Zhang et al., 2003; Spengler et al., 2005). However, no colocalization study of Sp1 and Sp3 in the same image has yet been performed.
In this study, we determined the nuclear organization of Sp1 and Sp3 relative to sites of transcription and their interaction with subnuclear domains, the nuclear matrix, the transcription factor estrogen receptor (ER) α, and chromatin remodeling factors histone deacetylases 1 and 2. Fluorescence microscopy followed by deconvolution analysis in conjunction with biochemical partitioning assays demonstrated that Sp1 and Sp3 are nuclear matrix proteins that are organized into different domains that had minor overlap with transcription sites and interacting proteins, estrogen receptor α, and histone deacetylases 1 and 2. These results show the different features of Sp1 and Sp3.
MATERIALS AND METHODS
Cell Culture and Indirect Immunolocalization
The human breast cancer cell line MCF-7 (T5) (ER positive and estrogen dependent) was grown under estrogen-complete or estrogen-deplete conditions as described previously (Spencer et al., 2000; Sun et al., 2001). HeLa, T47D, and ZR-75 cell lines were grown in the same medium. Cells were plated onto poly-l-lysine-coated coverslips in 24-well tissue culture plates. Cells were fixed in 3.7% formaldehyde in PEM buffer (80 mM piperazine-N,N′-bis(2-ethanesulfonic acid), 5 mM EGTA, and 2 mM MgCl2, pH 6.8) for 30 min on ice. Cells were washed three times 5 min in PEM buffer, quenched with the addition of 75 mM ammonium chloride and 20 mM glycine, and permeabilized with 0.5% Triton X-100 in PEM buffer for 30 min. Coverslips were blocked in 5% dry milk in TBST (20 mM Tris-HCl, pH 7.4, 150 mM NaCl, and 0.1% Tween 20) for 1 h at room temperature and incubated with the appropriate primary antibodies overnight at 4°C. The following antibodies were used at the indicated dilution: mouse monoclonal antibodies against ERα (1:50; Novocastra Laboratories, Newcastle, United Kingdom), Sp1 (1:50; Santa Cruz Biotechnology, Santa Cruz, CA), and HDAC2 (1:250; Santa Cruz Biotechnology) and rabbit polyclonal antibodies against Sp1 (1:500; Upstate Biotechnology, Charlottesville, VA), Sp3 (1:500; Santa Cruz Biotechnology), histone deacetylase (HDAC) 1 (1:5000; Affinity Bioreagents, Golden, CO), and HDAC2 (1:2000; Affinity Bioreagents). Alexa 488-conjugated goat anti-mouse IgG (Molecular Probes, Eugene, OR) or Cy3-conjugated goat anti-rabbit IgG (Sigma, St. Louis, MO) were used as secondary antibodies. Subsequently, DNA was counterstained with 200 ng/ml 4′6-diamidino-2-phenylindole (DAPI), and the coverslips were mounted onto glass slides using the Vectashield mounting medium (Vector Laboratories, Burlingame, CA). Control experiments including epitope peptide blocking or primary antibody omission demonstrated the specificity of the antibodies used. Digital images were captured and processed with fluorescence microscopy and deconvolution analysis.
Nascent RNA Labeling
Active transcription sites were labeled by incorporation of 5′-fluorouridine (FU) (Sigma) into nascent RNA, as described previously (Boisvert et al., 2000). Briefly, MCF-7 (T5) cells were grown on coverslips with complete DMEM. To label active transcription sites, 2 mM FU was added to the newly replaced prewarmed medium and then incubated for 5 min at 37°C. The labeled cells were fixed with 3.7% formaldehyde, and immunofluorescence detection was performed as described above. To detect the labeled nascent RNA, the mouse monoclonal anti-bromodeoxyuridine (BrdU) (Sigma) was used as primary antibody and was detected by the secondary antibody Alexa 488 anti-mouse IgG. To verify the specificity of our immunodetection of nascent RNA, the following immunostaining control experiments were satisfactorily performed: 1) no labeling and no primary antibody, 2) no labeling but with anti-BrdU antibody incubation, 3) with labeling but without anti-BrdU incubation, and 4) with labeling followed by RNase A digestion.
Cellular Fractionation
The cellular fractionation of MCF-7 cells by Triton X-100 extraction was performed as described previously (Sun et al., 2001).
Isolation of Nuclear Matrices
Nuclear matrices (NM) were isolated from MCF-7 cells as described previously (Samuel et al., 1997). Briefly, cells were extracted with Triton X-100 to release lipids and soluble proteins. Nuclei were collected, resuspended in DIG buffer (50 mM NaCl, 300 mM sucrose, 3 mM MgCl2, 10 mM Tris-HCl, pH 8.0, 1% thiodiglycol) containing Triton X-100, and digested with DNase I. The pellet was washed with ammonium sulfate at a final concentration of 0.25 M and then with NaCl at a 2 M final concentration to facilitate the removal of chromatin. The resulting pellet contains the nuclear matrix.
Immunoprecipitation and Immunoblotting
Immunoprecipitations and sequential immunoprecipitations were performed as described previously (Sun et al., 2002). The cell lysis included sonication to ensure a complete extraction of the proteins under study, including proteins bound to the nuclear matrix. Immunoblot analyses with the anti-Sp1 and anti-Sp3 polyclonal antibodies were performed as described previously (Sun et al., 2001). The specificity of the anti-Sp1 and anti-Sp3 antibodies used in the imaging experiments was tested by immunoblotting cell lysates from T5 cells.
Chromatin Immunoprecipitation (ChIP) Assay
ZR-75 cells were synchronized by 3 d of culture in DMEM/2% dextrancharcoal-treated fetal calf serum and then cultured with 10–8 M estradiol or ethanol for 30 min. ChIP assays were done as described previously (Sun et al., 2005). Briefly the input formaldehyde cross-linked chromatin, which had been sheared to an average fragment size of 500 base pairs, was diluted fivefold in dilution buffer (1.2 mM EDTA, 167 mM NaCl, 16.7 mM Tris-HCl, pH 8.1, 1.1% Triton X-100, and 0.01% SDS). Immunoprecipitated complexes, which were recovered by an incubation with protein A/G plus-Agarose (Santa Cruz Biotechnology), were serially washed with 1 ml of washing buffer I (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris, pH 8.1, and 150 mM NaCl), washing buffer II (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris, pH 8.1, and 500 mM NaCl), washing buffer III (1 0.25 M LiCl, 1% NP-40, 1% deoxycholate, 1 mM EDTA, and 10 mM Tris, pH 8.0), and then twice with 1 mM EDTA, 10 mM Tris-HCl, pH 8.0. Then, 20 μg of RNase A was added to each ChIP and incubated for 30 min at 37°C. Precipitated chromatin complexes were eluted from the beads through incubation with 100 μl of 1% SDS, 0.1 M NaHCO3. In re-ChIP experiments, after the elution, the supernatant was diluted 10 times with dilution buffer as described above and subjected again to the ChIP procedure. ChIP and input DNA were analyzed by PCR using trefoil factor 1 (TFF1) promoter primers (For-, 5′-GAC GGA ATG GGC TTC ATG AGC-3′; Rev-, 5′-GAT AAC ATT TGC CTA AGG AGG-3′) to amplify a 385-base pairs fragment. The PCR products were electrophoresed on a 1.5% agarose gel, stained with ethidium bromide and quantified using a Kodak Image Station. The linear range of PCR product amplification was determined, and the amount of ChIP-DNA template was optimized.
Nuclear Matrix Immunofluorescence Staining
MCF-7 cells were extracted on poly-l-lysine coated coverslips, using an established procedure (Nickerson et al., 1997). Briefly, soluble proteins were removed by a 5-min incubation in ice-cold CSK buffer (100 mM NaCl, 300 mM sucrose, 3 mM MgCl2, 10 mM PIPES, pH 6.8) containing 0.5% Triton X-100. The remaining nuclear structure was then cross-linked with 3.7% formaldehyde in CSK buffer for 30 min on ice. Chromatin was released by digestion with 0.5 mg/ml DNase I (Sigma) for 50 min at 32°C and two 5-min washes at room temperature. The first wash was done with ammonium sulfate at a final concentration of 0.25 M, and the second wash was done with NaCl at a final concentration of 2 M. The resulting nuclear matrix preparations were then analyzed by immunofluorescence as described above.
Fluorescence Microscopy and Deconvolution Analysis
Fluorescence images were captured on an AxioPhot II microscope (Carl Zeiss, Thornwood, NY) with an AxioHRm camera. The stack images of 100 optical sections with a step size of 200 nm were then deconvolved in three dimensions with the AxioVision 4.1 constrained Iterative Algorithm (Carl Zeiss).
Digital Image Analysis
Quantitative colocalization analysis was performed as described previously (von Mikecz et al., 2000; Zaidi et al., 2003) with some modifications. Briefly, the deconvolved images of each color channel were imported into the Meta-Morph imaging software version 6.1 (Universal Imaging, Downingtown, PA). Subsequently, correlation scatter plots and correlation coefficients were generated to illustrate the concurrence of pixel intensities between the images of different color channels. The scatter plot represents a comparison of pixel coincidence between two channels (i.e., red and green). The intensity of the corresponding pixels in the two images was examined, and the two intensity values were used as the x and y coordinates in the scatter plot. Two completely overlapping images produce a straight diagonal line starting from the origin of the scatter plot. The range of values of r is –1.0 to +1.0. A value of 1.0 shows that the data are perfectly correlated with one another. This will only happen if the two images are identical, resulting in complete overlap. An r of –1.0 is observed when there is an inverse relationship between intensities in the two images. In a linescan, the pixel intensity in each channel is plotted versus the position along a straight line across the dual image. A superposition of red and green peaks indicates image overlap.
RESULTS
Nuclear Distribution of Sp1 and Sp3
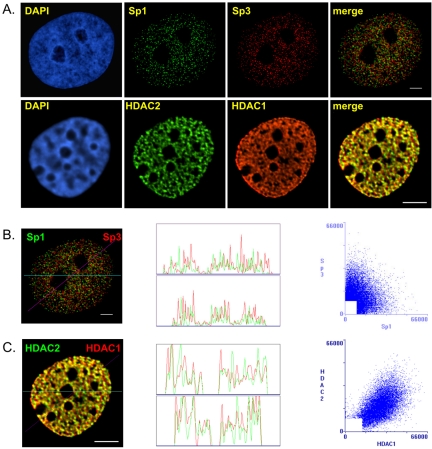
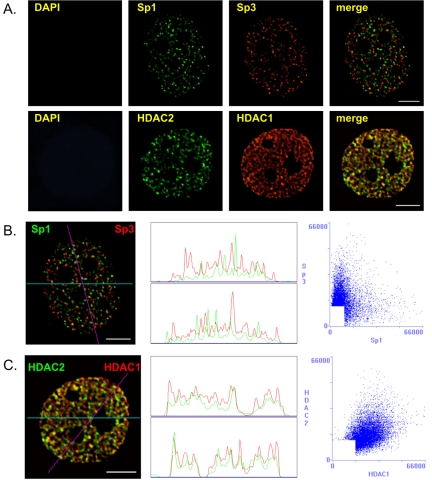
Our previous immunoprecipitation studies demonstrated that Sp1 and Sp3 were in separate complexes (Sun et al., 2002). To determine whether Sp1 and Sp3 resided in different locations in the nucleus, we compared the spatial distributions of Sp1 and Sp3 by fluorescence microscopy and image deconvolution after indirect immunofluorescent labeling of MCF-7 cells grown and fixed on coverslips. Figure 1A shows that Sp1 and Sp3 were distributed in a punctate pattern throughout the nucleus and were excluded from the nucleoli of MCF-7 cells. The merged image shows that Sp1 and Sp3 had distinct subnuclear sites, demonstrating that Sp1 and Sp3 were not colocalized in MCF-7 cells; a result that is consistent with our immunoprecipitation study.
Figure 1.
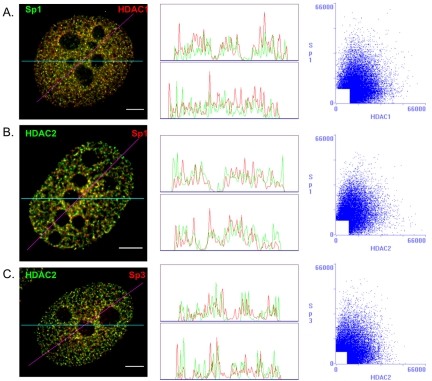
Intranuclear distribution of Sp1 and Sp3 in MCF-7 cells. MCF-7 cells were grown on coverslips in estrogen complete medium, fixed, and double labeled with anti-Sp1 and anti-Sp3 or anti-HDAC1 and anti-HDAC2 antibodies. (A) DNA was stained by DAPI. Sp1 and Sp3 as well as HDAC1 and HDAC2 distributions were visualized by fluorescence microscopy and image deconvolution as described in Materials and Methods. Single optical sections are shown. Yellow in the merged images signifies colocalization. (B and C) The Sp1/Sp3 and HDAC1/HDAC2 merged images were analyzed by the generation of a horizontal linescan (top box) and a diagonal linescan (bottom box) along with a correlation scatter plot. Bar, 5 μm.
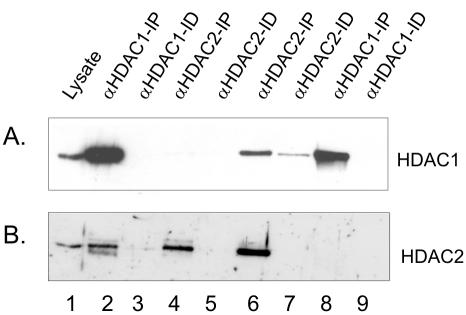
We contrasted our findings on the nuclear organization of Sp1 and Sp3 to that of two chromatin remodeling factors, HDAC 1 and HDAC2, which coexist in multiprotein complexes such as Sin3 and NuRD (De Ruijter et al., 2003). Before visualizing the organization of HDAC1 and HDAC2 in MCF-7 cells, we determined the extent to which HDAC1 and HDAC2 were associated with each other by sequential immunoprecipitations followed by immunoblot detection. The MCF-7 cell lysate was incubated with anti-HDAC1 antibodies under low stringency conditions followed by immunoprecipitation of the HDAC1-immunodepleted supernatant with anti-HDAC2 antibodies. In another experiment the antibodies were added in the reverse order. Immunochemical staining of the blot with anti-HDAC1 antibodies demonstrated that the anti-HDAC1 antibodies efficiently immunoprecipitated most, if not all, of the HDAC1 in the cell lysate (Figure 2A, lanes 2 and 3). This immunoprecipitate also contained most of the HDAC2; however, HDAC2 was detected in the HDAC1 immunodepleted fraction, and this population of HDAC2 was immunoprecipitated with anti-HDAC2 antibodies (Figure 2B, lanes 2–4). Approximately 10% of the HDAC2 was not bound to HDAC1. When the experiment was done with the reverse order of antibodies, the antibody against HDAC2 immunoprecipitated most, if not all, of HDAC2 (Figure 2B, lanes 6 and 7). Although HDAC1 also was immunoprecipitated, a population of HDAC1 (∼20%) was not associated with HDAC2 (Figure 2A, lane 7). When this population of HDAC1 was immunoprecipitated, HDAC2 was not found in the immunoprecipitate (Figure 2B, lane 8). These results demonstrate a considerable level of association between HDAC1 and HDAC2 in MCF-7 cells. In agreement with immunoprecipitation results, substantial colocalization of HDAC1 and HDAC2 in the nucleus of MCF-7 cells was observed (Figure 1A, merged image). The distribution of HDAC1 and to a lesser extent HDAC2 occurred as fibers rather than as the foci observed for Sp1 and Sp3 (Figure 1A, compare HDAC1 with Sp1 and Sp3).
Figure 2.
Coimmunoprecipitation of HDAC1 and HDAC2. Two A260 of MCF-7 cell lysate was incubated with 4 μg of anti-HDAC1 antibodies, and the immunoprecipitated (IP, lane 2) and immunodepleted (ID, lane 3) fractions were collected. The immunodepleted fraction was next incubated with anti-HDAC2 antibodies, yielding IP (lane 4) and ID (lane 5) fractions. Conversely, two A260 of MCF-7 cell lysate were incubated with 4 μg of anti-HDAC2 antibodies, and the immunoprecipitated (IP, lane 6) and immunodepleted (ID, lane 7) fractions were collected. The immunodepleted fraction was next incubated with anti-HDAC1 antibodies, yielding IP (lane 8) and ID (lane 9) fractions. The whole IP fractions and equivalent volumes of lysate (lane 1) and ID fractions, corresponding to 1% of 2 A260 of lysate were loaded onto SDS-10% polyacrylamide gels, transferred to nitrocellulose membranes, and immunochemically stained with anti-HDAC1 (A) or anti-HDAC2 (B) antibodies.
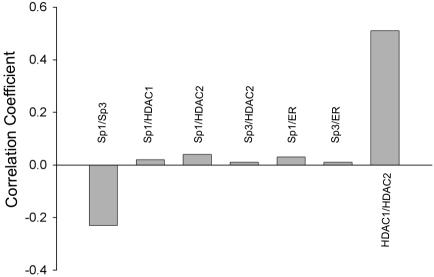
The distribution of Sp1, Sp3, HDAC1, and HDAC2 was analyzed further by the generation of correlation scatter plots and linescans. The scatter plot of Sp1 and Sp3 was diffuse, which indicates an absence of overlap between the two images (Figure 1B). The r was –0.23 (see Materials and Methods), and there was no peak superposition in the linescans (Figure 1B). In contrast, the scatter plot of HDAC1 and HDAC2 immunofluorescence shows a distribution from the origin along the diagonal, which is indicative of a significant pixel coincidence between the two images (Figure 1C). The r was found to be 0.51. The two linescans also show superposition of the two HDAC images (Figure 1C).
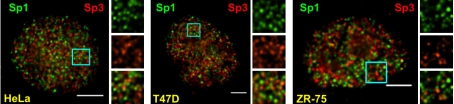
The generality of our observations for Sp1 and Sp3 in MCF-7 cells was investigated in three cell lines: HeLa cells, T47D cells, and ZR-75 cells. The overlays for each of the cell lines showed that the subnuclear position of Sp1 was distinct from that of Sp3 (Figure 3).
Figure 3.
Nuclear distribution of Sp1 and Sp3 in HeLa, T47D and ZR-75 cells. Fixed HeLa, T47D, and ZR-75 cells were double labeled with anti-Sp1 and anti-Sp3 antibodies. Sp1 and Sp3 distributions were visualized by fluorescence microscopy and image deconvolution as described in Materials and Methods. Single optical sections are shown. Yellow in the merged images signifies colocalization. Boxed areas of each image are shown enlarged as green signal only (top insets), red signal only (middle insets), or merged signals (bottom insets). Bar, 5 μm.
Dual Labeling of Active Transcription Sites and Sp1 or Sp3
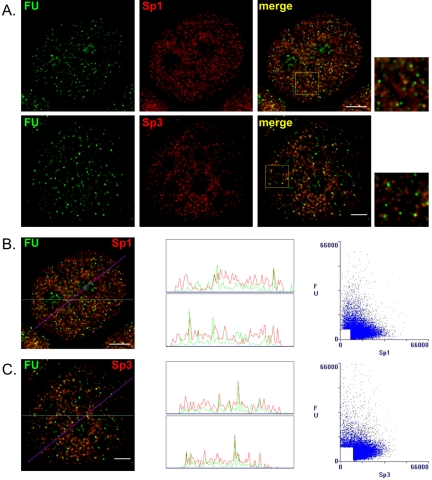
Transcription sites containing RNA polymerase holoenzyme complexes and nascent RNA transcripts have a nuclear punctate distribution similar to that of many transcription factors. Several transcription factors such as BRG1, TFIIH, AML-1B, p53, AhR, and ARNT were found to be partially colocalized with active transcription sites (Grande et al., 1997; Zeng et al., 1998; Rubbi and Milner, 2000; Elbi et al., 2002). On the other hand, the colocalization of transcription factors such as Oct1, E2F-1, the glucocorticoid receptor, and ERα with transcription sites was minimal (Grande et al., 1997; Stenoien et al., 2000). To determine whether Sp1 and Sp3 were colocalized with active transcription sites, fluorescence microscopy followed by deconvolution analysis was performed. 5′-Fluorouridine was incorporated for 5 or 30 min into the nascent RNA transcripts of MCF-7 cells. Nascent RNA was detected with anti-BrdU antibodies. Figure 4A shows hundreds of active transcription sites spread throughout the nucleus in a punctate pattern. Very few Sp1 or Sp3 foci were colocalized with nascent RNA. This observation was emphasized by the correlation scatter plots displaying very diffuse patterns and the linescans where peak superposition was rare (Figure 4, B and C). Correlation coefficients for the Sp1/FU and Sp3/FU pairs were –0.13 and –0.14, respectively. A 30-min labeling of the nascent RNA transcripts resulted in the same punctate pattern throughout the nucleoplasm and in bright clusters located in nucleoli representing RNA polymerase I transcript. Nonetheless, foci showing colocalization of Sp1 or Sp3 with the longer nascent RNA transcripts were rare (our unpublished data). Thus, the majority of Sp1 and Sp3 was not associated with active transcription sites.
Figure 4.
Sp1 and Sp3 distribution relative to active transcription sites. MCF-7 cells were grown on coverslips in estrogen complete medium, fixed, and double labeled with anti-Sp1 or anti-Sp3 antibodies and anti-BrdU antibodies that detect nascent RNA after in situ incorporation of FU. (A) Spatial distribution was visualized by fluorescence microscopy and image deconvolution as described in Materials and Methods. Single optical sections are shown. Yellow in the merged images signifies colocalization. The boxed area in each merged image is shown enlarged. (B and C) The Sp1/FU and Sp3/FU merged images were analyzed by the generation of a horizontal linescan (top box) and a diagonal linescan (bottom box) along with a correlation scatter plot. Bar, 5 μm.
Comparison of the Sp1/Sp3 Distribution with the HDAC1/HDAC2 Distribution
Sp1 and Sp3 are associated with HDAC1 and HDAC2 in MCF-7 cells (Sun et al., 2002). The spatial relationship between HDAC1/HDAC2 and Sp1 or Sp3 was determined by fluorescence microscopy and deconvolution analysis. Figure 5 shows a slight colocalization between Sp1 and HDAC1, Sp1 and HDAC2, or Sp3 and HDAC2. The correlation scatter plots show patterns that neither followed the diagonal, nor were totally diffuse, with the correlation coefficients for the Sp1/HDAC1, Sp1/HDAC2, and Sp3/HDAC2 pairs being 0.02, 0.04, and 0.01, respectively (Figure 7). The linescans also show limited colocalization, with only a few peaks being superposed. Mostly, Sp1 and Sp3 occupied sites different from those of HDAC1 and HDAC2.
Figure 5.
Sp1 and Sp3 distribution relative to HDAC1 and HDAC2. MCF-7 cells were grown on coverslips in estrogen complete medium, fixed, and double labeled with anti-Sp1 or anti-Sp3 and anti-HDAC1 or anti-HDAC2 antibodies. Spatial distribution was visualized by fluorescence microscopy and image deconvolution as described in Materials and Methods. Single optical sections are shown. Yellow in the merged images signifies colocalization. The merged images were analyzed by the generation of a horizontal linescan (top box) and a diagonal linescan (bottom box) along with a correlation scatter plot. Relative distributions of Sp1 and HDAC1 (A), Sp1 and HDAC2 (B), and Sp3 and HDAC2 (C). Bar, 5 μm.
Figure 7.
Comparison of correlation coefficients. Correlation coefficients, obtained as described in Materials and Methods, measure the degree of colocalization for each pair, as indicated. Correlation coefficient values theoretically range from 1.0 to –1.0, with 1.0 representing a complete colocalization (two identical images).
Comparison of the Sp1/Sp3 Distribution with the Estrogen Receptor α Distribution
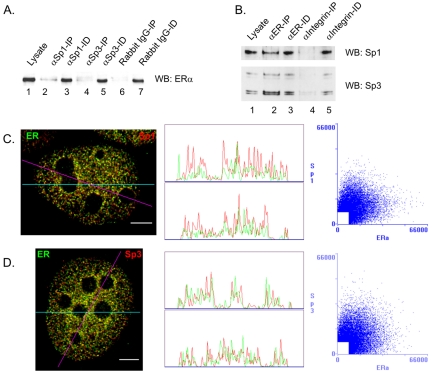
ERα is a ligand activated transcription factor that has a critical role in the proliferation and differentiation of breast cancer cells. ERα forms complexes with Sp1 or Sp3 to regulate the expression of several estrogen-responsive genes (Kim et al., 2005). To confirm the association of Sp1 or Sp3 with ERα, immunoprecipitations followed by immunoblot detection were performed with MCF-7 cell extracts. Figure 6A shows that either anti-Sp1 (lanes 1–3) or anti-Sp3 (lanes 1, 4, and 5) antibodies coimmunoprecipitated a small fraction of the ERα population, as opposed to the normal rabbit IgG negative control (lanes 6 and 7, which did not precipitate ERα. Inversely, Figure 6B shows that the anti-ERα antibody coprecipitated a small fraction of the Sp1 or Sp3 populations. These data demonstrate that, under estrogencomplete conditions, some of the ERα molecules were associated with Sp1 and Sp3. We compared their relative spatial distributions in MCF-7 cells by fluorescence microscopy and image deconvolution. Figure 6C shows that, in the presence of estrogen, ERα was located throughout the nucleus in a punctate pattern and was excluded from the nucleoli, as reported previously (Htun et al., 1999; Stenoien et al., 2000). There was a limited colocalization between ERα and Sp1 or Sp3, evidenced by a small number of yellow dots, as well as by the correlation scatter plots that show patterns that were neither close to the diagonal nor totally diffuse (Figure 6, C and D). The correlation coefficients were 0.03 and 0.01 for the Sp1/ERα and Sp3/ERα pairs, respectively. The linescans were in agreement with the scatter plots, showing a superposition of only a few peaks. As shown in Figure 7 summarizing our colocalization data in MCF-7 cells, the degrees of colocalization between Sp1 or Sp3 and ERα were similar to those between Sp1 or Sp3 and HDAC1 or HDAC2. They were minor compared with the substantial level of colocalization found between HDAC1 and HDAC2, but not negligible compared with the absence of colocalization between Sp1 and Sp3.
Figure 6.
Sp1 and Sp3 distribution relative to ERα. (A) Seven A260 of lysate from MCF-7 cells grown in complete medium were incubated with 5 μg of anti-Sp1 or anti-Sp3 antibodies or with normal rabbit IgG nonspecific antibodies as negative control, and the immunoprecipitated (IP, lanes 2, 4, and 6) and immunodepleted (ID, lanes 3, 5, and 7) fractions were collected. The whole IP fractions and equivalent volumes of lysate (lane 1) and ID fractions, corresponding to 1% of 7 A260 of lysate were loaded onto a SDS-10% polyacryl-amide gel, transferred to nitrocellulose membranes, and immunochemically stained with anti-ERα antibodies. (B) Nine A260 of lysate from MCF-7 cells grown in complete medium were incubated with 4 μg of anti-ERα or antiintegrin (control) antibodies, and the immunoprecipitated (IP, lanes 2 and 4) and immunodepleted (ID, lanes 3 and 5) fractions were collected. The whole IP fractions and equivalent volumes of lysate (lane 1) and ID fractions, corresponding to 0.5% of 9 A260 of lysate were loaded onto a SDS-10% polyacryl-amide gel, transferred to a nitrocellulose membrane, and immunochemically stained with anti-Sp1 antibodies. After stripping of the anti-Sp1 antibodies, the membrane was restained with anti-Sp3 antibodies. (C and D) MCF-7 cells were grown on coverslips in complete medium, fixed, and double labeled with anti-Sp1 or anti-Sp3 and anti-ERα antibodies. Spatial distribution was visualized by fluorescence microscopy and image deconvolution as described in Materials and Methods. Single optical sections are shown. Yellow in the merged images signifies colocalization. The merged images were analyzed by the generation of a horizontal linescan (top box) and a diagonal linescan (bottom box) along with a correlation scatter plot. Bar, 5 μm.
Solubility Partitioning and Association with the Nuclear Matrix of Sp1 and Sp3
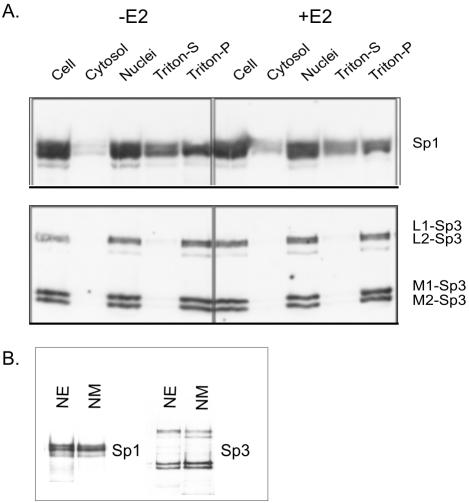
After the addition of estradiol to MCF-7 cells, ERα goes from being loosely bound in the nucleus to a tightly bound form, which parallels the increased affinity of ERα for the nuclear matrix (Stenoien et al., 2000). This prompted the analysis of the subcellular distribution of Sp1 and Sp3 in MCF-7 cells grown under estrogen deplete or replete conditions. MCF-7 cells were lysed in TNM buffer (100 mM NaCl, 300 mM sucrose, 2 mM MgCl2, 10 mM Tris-HCl, pH 8.0, 1% thiodiglycol) without detergent to prevent loosely bound nuclear proteins from leaking out of the nuclei. The nuclei were resuspended in TNM buffer with 0.5% Triton X-100 and incubated on ice to release loosely bound nuclear proteins (Triton-S fraction). The resulting pellet contained the tightly bound nuclear proteins, which includes proteins associated with the nuclear matrix (Triton-P). Immunoblot analyses showed that both Sp1 and Sp3 were predominantly in the nuclear fraction. Sp1 was present in the Triton-S and Triton-P fractions, with ∼60% of the nuclear Sp1 being associated with the latter fraction (Figure 8A). The presence of estradiol had no effect on the distribution of Sp1 among these fractions. Figure 8A shows that the distribution of Sp3 was distinct from that of Sp1 in that Sp3 was present in the Triton-P, but not in the Triton-S, fraction. Moreover, the four isoforms of Sp3 partitioned into the Triton-P fraction. As with Sp1, the presence of estradiol did not influence the fractionation of Sp3 isoforms.
Figure 8.
Subcellular distribution of Sp1 and Sp3 in MCF-7 cells. (A) MCF-7 cells incubated with (+E2) or without (–E2) 10 nM estradiol at 37°C for 20 min were fractionated by Triton X-100 extraction. Equal volumes of all fractions were analyzed by SDS- 10% PAGE, transferred to a membrane, and immunochemically stained with anti-Sp1 (top) or anti-Sp3 antibodies (bottom). The long isoforms of Sp3 are shown as L1 and L2, whereas the short isoforms are denoted by M1 and M2. (B) Association of Sp1 and Sp3 with nuclear matrix: 20 μg of protein of nuclear extract (NE) and nuclear matrix (NM) from MCF-7 cells, grown in estrogen complete medium, were electrophoresed on a SDS-10% polyacrylamide gel, and immunochemically stained with anti-Sp1 (left) or anti-Sp3 (right) antibodies.
The partitioning of Sp1 and Sp3 with the Triton-P fraction suggested that these transcription factors were associated with chromatin and/or the nuclear matrix. Nuclear matrices were isolated from MCF-7 cells grown under estrogen-complete conditions and the presence of Sp1 and Sp3 determined by immunoblot analyses. The results demonstrated that Sp1 and the four isoforms of Sp3 were associated with the nuclear matrix of MCF-7 cells (Figure 8B).
The association of Sp1 and Sp3 with the nuclear matrix was further analyzed by determining the location of Sp1 and Sp3 on the nuclear matrix by using fluorescence microscopy and deconvolution analysis of in situ nuclear matrix preparations from MCF-7 cells. Figure 9A shows that the residual nuclear matrix from MCF-7 cells grown in presence of estrogen was devoid of chromatin (lack of DAPI staining), but retained Sp1 and Sp3. Furthermore, both Sp1 and Sp3 had a punctate distribution similar to that seen in intact cells. The merging of Sp1 and Sp3 images shows that these regulatory factors were not colocalized in the nuclear matrix (Figure 9A). In contrast, HDAC1 and HDAC2, which are nuclear matrix proteins, had a substantial degree of colocalization (Figure 9A). These results were confirmed by correlation scatter plots and linescans shown in Figure 9, B and C, as well as by the r values of –0.16 and 0.24 for the Sp1/Sp3 and HDAC1/HDAC2 pairs, respectively.
Figure 9.
Nuclear matrix distribution of Sp1 and Sp3. MCF-7 cells were grown on coverslips in estrogen complete medium, extracted as described in Materials and Methods, and the remaining nuclear matrix was double labeled with anti-Sp1 and anti-Sp3 or anti-HDAC1 and anti-HDAC2 antibodies. (A) DNA was stained by DAPI. Sp1 and Sp3 as well as HADC1 and HDAC2 distributions were visualized by fluorescence microscopy and image deconvolution as described in Materials and Methods. Single optical sections are shown. Yellow in the merged images signifies colocalization. (B-C) The Sp1/Sp3 and HDAC1/HDAC2 merged images were analyzed by the generation of a horizontal linescan (top box) and a diagonal linescan (bottom box) along with a correlation scatter plot. Bar, 5 μm.
The ERα distribution in nuclear matrix preparations from MCF-7 cells was determined and compared with that of Sp1 or Sp3. ERα had a punctate distribution within the nuclear matrix, either without estradiol (Figure 10, A and B) or with estradiol (Figure 10, C and D). Note in the absence of estradiol a much longer exposure was needed to obtain the ERα fluorescent image; a longer exposure was necessary as only a low level of ERα was bound to the nuclear matrix in the absence of ligand. There was a low level of colocalization on the nuclear matrix between ERα and Sp1 or Sp3 in the absence of estradiol. The presence of estradiol and the resulting affinity of ERα for the nuclear matrix did not lead to a significant increase of colocalization between Sp1 or Sp3 and ERα within the nuclear matrix. Indeed, the Sp1/ERα r values were 0.01 and 0.02, without and with estradiol, respectively, whereas for the Sp3/ERα pair, the values were 0.02 without estradiol and 0.06 with estradiol. Thus, when relocated to the nuclear matrix upon addition of estradiol, ERα mostly binds to sites that were different from the Sp1 and Sp3 sites of residence.
Figure 10.
Nuclear matrix distribution of Sp1 and Sp3 relatively to ERα with or without estradiol treatment. MCF-7 cells were grown on coverslips for 4 d with phenol red free DMEM complemented with 7% 2× calf serum-fetal bovine serum. After ethanol (A and B) or 10 nM estradiol (C and D) treatment for 60 min, the nuclear matrices were prepared and double labeled with anti-Sp1 or anti-Sp3 and anti-ERα antibodies. Spatial distribution was visualized by fluorescence microscopy and image deconvolution as described in Materials and Methods. Single optical sections are shown. Yellow in the merged images signifies colocalization. The merged images were analyzed by the generation of a horizontal linescan (top box) and a diagonal linescan (bottom box) along with a correlation scatter plot. Bar, 5 μm.
Sp1 and Sp3 Association with the TFF1 Promoter
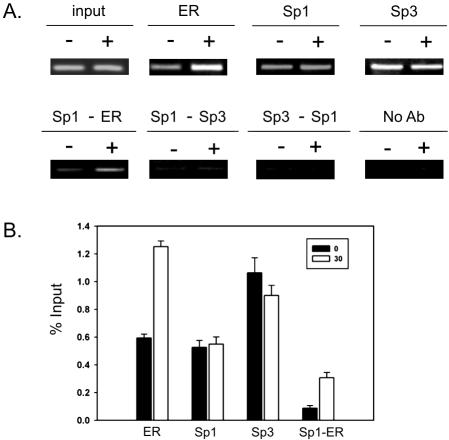
Whether in the nucleus or nuclear matrix our results show that Sp1 and Sp3 have distinct locations. However, there are a multitude of studies using the ChIP assay, indicating that a variety of promoters are loaded with Sp1 and Sp3. Considering the dynamic environment at a promoter to which several transcription factors, coactivators and chromatin remodeling complexes are recruited, it is possible that Sp1 and Sp3 could transiently colocalize at a promoter. An association of Sp1 and Sp3 at a specific promoter would fall below the limit of detection by fluorescence microscopy and deconvolution analysis. Consequently, we carried out ChIP and re-ChIP assays to investigate the binding of Sp1, Sp3, and ERα on the TFF1 promoter in ZR-75 breast cancer cells. The estrogen-responsive TFF1 gene has an estrogen-responsive element (ERE) positioned next to a Sp1 binding site. We had previously showed in a ChIP experiment that both Sp1 and Sp3 were bound to the TFF1 promoter in MCF-7 breast cancer cells (Sun et al., 2005). Sp1 and Sp3 were associated with the TFF-1 promoter in ZR-75 cells cultured in the presence (30 min) and absence of estradiol (Figure 11). Furthermore, ERα association with the TFF1 promoter increased (∼2.1-fold) after estradiol addition (Figure 11B). The loading of Sp1 and Sp3 onto the TFF-1 promoter in ZR-75 cells versus MCF-7 cells differed, with Sp1 loading being reduced and Sp3 loading remaining constant on the TFF1 promoter in MCF-7 cells after 30 min of estradiol treatment. In ZR-75 cells a decline in the loading of Sp1 after estradiol treatment was not observed. This result is consistent with observations of others, showing that Sp1 loading onto an estrogen-responsive gene is different in MCF-7 and ZR-75 cells (Castro-Rivera et al., 2001).
Figure 11.
Association of Sp1 and Sp3 with the TFF1 promoter. (A) ChIP and re-ChIP experiments were performed on chromatin prepared from ZR-75 cells treated with ethanol (–) or 10 nM estradiol (+) for 30 min. The antibodies were used as indicated. No antibody: primary antibody was omitted. Control immunoprecipitations with normal rabbit IgG nonspecific antibodies did not produce a PCR product (our unpublished data). Input, ChIP, and re-ChIP DNAs were analyzed by PCR using TFF1 promoter specific primers. This is a representative result of an experiment repeated three times. (B) Quantitation of ChIP and re-ChIP assay PCR products corrected with the input data were determined to evaluate the binding of ERα, Sp1, Sp3, and Sp1-ERα to the TFF1 promoter region. Error bars show the SD (n = 3).
To determine whether Sp1 and Sp3 were bound together onto the same DNA fragment, we performed a re-CHIP assay. Cross-linked chromatin preparations from ZR-75 cells treated or not with estradiol were subjected to the ChIP procedure with anti-Sp1 antibodies and again precipitated using anti-Sp3 or anti-ERα antibodies. The TFF1 promoter sequence was not detected in the immunoprecipitate after the Sp1-Sp3 re-ChIP assay. Similarly, the TFF1 DNA fragment was not precipitated when the chromatin was first immunoprecipitated by anti-Sp3 antibodies and then by anti-Sp1 antibodies (Figure 11). Conversely, Figure 11 shows that the Sp1-ERα re-ChIP assay resulted in the precipitation of the TFF1 promoter DNA fragment, with the amount of precipitated DNA being markedly enhanced (∼3.7-fold) when cells were treated with estradiol. These data demonstrate that although Sp1 and ERα were bound together on the same TFF1 promoter, Sp1 and Sp3 were bound to different TFF1 promoters located either on different alleles or in different cells.
DISCUSSION
Sp1 and Sp3 are often considered as being functionally equivalent, based upon their structural similarity. Both regulatory factors bind to oligonucleotides with an Sp binding site in in vitro binding assays, and in ChIP assays, it is common to find Sp1 and Sp3 loaded onto a promoter bearing an Sp binding site. However, these observations do not tell us whether Sp1 and Sp3 occupy the promoter at the same time. There are significant differences between these two factors as illustrated in knockout studies and studies investigating their transcriptional roles, isoforms, and modifications (Sapetschnig et al., 2004). Sp1 typically functions as an activator. In contrast the function of the long versus short Sp3 isoforms as transcriptional activators or repressors is dependent upon several parameters, including promoter context, state of modification, and chromatin structure (Sapetschnig et al., 2004). Sp1 and Sp3 also differ in their ability to form multimers (Yu et al., 2003). Our results add to the list of features that distinguish Sp1 from Sp3. We demonstrate that Sp1 and Sp3 occupy different subnuclear sites and exhibit different associations with the nuclear matrix.
The lack of colocalization of Sp1 and Sp3 in four different cell lines and the mutually exclusive binding of Sp1 and Sp3 to the TFF1 promoter argue strongly against the existence of complexes containing both Sp1 and Sp3. Our imaging results concur with the data obtained by others and us in coimmunoprecipitation studies that demonstrated that Sp1 and Sp3 are in separate complexes (Sun et al., 2002; Yu et al., 2003). In contrast, Zhang et al. (2003) observed that Sp1 and Sp3 coimmunoprecipitated in mouse renal epithelial mIMCD-3 cell extracts; however, from the results presented it is difficult to interpret the extent to which Sp1 and Sp3 are in complex together. Nevertheless, in human breast cancer, human cervical carcinoma, and mouse L cells, Sp1 and Sp3 are not in the same complex (Sun et al., 2002; Yu et al., 2003).
The subnuclear foci containing Sp1 or Sp3 were infrequently associated with sites of transcription. This observation is consistent with reports for other transcription factors (e.g., ERα and Oct1) (Grande et al., 1997; Stenoien et al., 2000). However, there are exceptions with Runx2 subnuclear foci colocalizing with sites of transcription (Zaidi et al., 2002). It is possible that the Sp1 and Sp3 foci represent storage sites, allowing the sequestration of Sp1 and Sp3 and regulating their concentration in the nucleus. The distribution of transcription factors into foci may play a role in the regulation of nucleoplasmic concentrations of factors and/or the assembly of multiprotein complexes (Hendzel et al., 2001). Because Sp1 can bind its DNA binding site in nucleosomes, it is possible that without high-affinity nuclear matrix binding sites to temporally retain these transcription factors that Sp1 and Sp3 would associate with Sp binding sites in chromatin, leading to the aberrant remodeling of chromatin and dysfunction of the genome.
Results shown here and our previous studies show a small percentage of the total Sp1 and Sp3 population is associated with HDAC1/2 or ERα (Sun et al., 2002). The interaction of Sp1 with ERα is well documented (Stoner et al., 2004) and was recently demonstrated in living MCF-7 cells (Kim et al., 2005). However, using fluorescence resonance energy transfer, Kim et al. (2005) detected 50% of colocalization between Sp1 and ERα in the absence of estradiol, and a further 10% increase in colocalization in the presence of estradiol (Kim et al., 2005). Our data are not consistent with these numbers. It is possible that the high level of association of Sp1 and ERα observed by Kim et al. (2005) is a consequence of overexpression of the transfected proteins.
Our biochemical and morphological studies show that Sp3 and a subpopulation of Sp1 are bound to the nuclear matrix. Our finding that ∼40% of Sp1 is not in the tightly bound nuclear protein fraction is in agreement with results published previously (van Wijnen et al., 1993). The fact that Sp1 and Sp3 have different solubility properties may contribute to their difference in functionality. Furthermore, even though Sp1 and Sp3 have a strong affinity for the nuclear matrix, they do not colocalize. This indicates that Sp1 and Sp3 are associated with different nuclear matrix proteins.
In contrast to Sp1 and Sp3, most HDAC1 and HDAC2 are in complex together and are colocalized in the nucleus. Both HDAC1 and HDAC2 are associated with the nuclear matrix. Interestingly, HDAC1 occurred as fibers rather than as the foci. Because HDAC1 has the ability to homo-oligomerize, we may be observing HDAC1 oligomers that are decorating the nuclear matrix filament network (Taplick et al., 2001). Our results suggest that high-affinity nuclear matrix proteins that are involved in the nuclear organization of HDAC1 and HDAC2 are different from those binding to Sp1, Sp3, and ERα. The high-affinity nuclear matrix proteins specifically binding these transcription factors and chromatin remodeling enzymes would assign different nuclear domains to these proteins. Thus, the nuclear matrix may have an architectural role in the nuclear organization of proteins regulating gene expression.
Although our results failed to show a significant change in the colocalization of Sp1 or Sp3 with ERα in cells cultured with and without estradiol, we did observe an increase in the cooccupation of Sp1 and ERα on the same TFF1 promoter after addition of estradiol to ZR-75 breast cancer cells. The results of the re-ChIP assay also demonstrated that Sp1 and Sp3 did not occupy the TFF1 promoter at the same time. This result demonstrates that it is either Sp1 or Sp3 that resides at any particular time on a TFF1 promoter. If both alleles are active, it is possible that Sp1 is on the TFF1 promoter of one allele, whereas Sp3 is on the TFF1 promoter of the other, but this could vary from cell to cell. ChIP assays will present an average of these interactions. Pending the Sp3 isoform and its modification state associated with a TFF1 promoter, the transcriptional activity of an Sp3-charged promoter may be very different from that of a Sp1 occupied promoter. We propose that the temporal and dynamic occupancy of Sp1 and Sp3 onto a promoter and the activation capacity of the Sp family member will confer heterogeneity in the activation state of that promoter. Possibly the position of a promoter next to Sp1 or Sp3 foci will influence which of the two factors occupies the Sp binding site. The relative position of a promoter with Sp1/Sp3 binding sites next to nuclear matrix sites enriched in Sp1 or Sp3 in three-dimensional nuclear space may decide which transcription factor will reside on the promoter. As with splicing factors, which dynamically move from a nuclear speckle storage site to nearby transcripts engaged in the splicing process (Lamond and Spector, 2003; Moen et al., 2004), we envisage that a promoter positioned next to a nuclear matrix site rich in Sp3 will tend to have Sp3 and not Sp1 dynamically residing on that promoter. Thus, spatial organization of genes next to Sp1- or Sp3-rich domains would influence gene regulation. Future studies will need to address the nuclear positioning of Sp1/Sp3-regulated genes next to nuclear matrix sites storing Sp1 or Sp3.
Acknowledgments
We thank Dr. Genevieve Delcuve for preparation of the manuscript. This research was supported by Canadian Institute of Health Research Grant MOP-15183, a Canada Research Chair (to J.R.D.), and a U.S. Army Medical and Materiel Command Breast Cancer Research Program (W81XWH-05-1-0284) studentship (to L. L.).
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05–05–0388) on June 29, 2005.
Abbreviations used: ERα, estrogen receptor α; FU, 5′-fluorouridine; HDAC, histone deacetylase.
References
- Birnbaum, M. J., van Wijnen, A. J., Odgren, P. R., Last, T. J., Suske, G., Stein, G. S., and Stein, J. L. (1995). Sp1 trans-activation of cell cycle regulated promoters is selectively repressed by Sp3. Biochemistry 34, 16503–16508. [DOI] [PubMed] [Google Scholar]
- Boisvert, F. M., Hendzel, M. J., and Bazett-Jones, D. P. (2000). Promyelocytic leukemia (PML) nuclear bodies are protein structures that do not accumulate RNA. J. Cell Biol. 148, 283–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouwman, P., Gollner, H., Elsasser, H. P., Eckhoff, G., Karis, A., Grosveld, F., Philipsen, S., and Suske, G. (2000). Transcription factor Sp3 is essential for post-natal survival and late tooth development. EMBO J. 19, 655–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouwman, P., and Philipsen, S. (2002). Regulation of the activity of Sp1-related transcription factors. Mol. Cell. Endocrinol. 195, 27–38. [DOI] [PubMed] [Google Scholar]
- Castro-Rivera, E., Samudio, I., and Safe, S. (2001). Estrogen regulation of cyclin D1 gene expression in ZR-75 breast cancer cells involves multiple enhancer elements. J. Biol. Chem. 276, 30853–30861. [DOI] [PubMed] [Google Scholar]
- Cawley, S., et al. (2004). Unbiased mapping of transcription factor binding sites along human chromosomes 21 and 22 points to widespread regulation of noncoding RNAs. Cell 116, 499–509. [DOI] [PubMed] [Google Scholar]
- Chan, Y., Fish, J. E., D'Abreo, C., Lin, S., Robb, G. B., Teichert, A. M., Karantzoulis-Fegaras, F., Keightley, A., Steer, B. M., and Marsden, P. A. (2004). The cell-specific expression of endothelial nitric-oxide synthase-A role for DNA methylation. J. Biol. Chem. 279, 35087–35100. [DOI] [PubMed] [Google Scholar]
- Choi, H. S., Lee, J. H., Park, J. G., and Lee, Y. I. (2002). Trichostatin A, a histone deacetylase inhibitor, activates the IGFBP-3 promoter by upregulating Sp1 activity in hepatoma cells: alteration of the Sp1/Sp3/HDAC1 multiprotein complex. Biochem. Biophys. Res. Commun. 296, 1005–1012. [DOI] [PubMed] [Google Scholar]
- Choi, J. Y., et al. (2001). Subnuclear targeting of Runx/Cbfa/AML factors is essential for tissue-specific differentiation during embryonic development. Proc. Natl. Acad. Sci. USA 98, 8650–8655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Leon, M. B., Montanez, C., Gomez, P., Morales-Lazaro, S. L., Tapia-Ramirez, V., Valadez-Graham, V., Recillas-Targa, F., Yaffe, D., Nudel, U., and Cisneros, B. (2005). Dystrophin Dp71 expression is down-regulated during myogenesis - role of Sp1 and Sp3 on the Dp71 promoter activity. J. Biol. Chem. 280, 5290–5299. [DOI] [PubMed] [Google Scholar]
- De Ruijter, A. J., Van Gennip, A. H., Caron, H. N., Kemp, S., and Van Kuilenburg, A. B. (2003). Histone deacetylases (HDACs): characterization of the classical HDAC family. Biochem. J. 370, 737–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbi, C., Misteli, T., and Hager, G. L. (2002). Recruitment of dioxin receptor to active transcription sites. Mol. Biol. Cell 13, 2001–2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gartel, A. L., Goufman, E., Najmabadi, F., and Tyner, A. L. (2000). Sp1 and Sp3 activate p21 (WAF1/CIP1) gene transcription in the Caco-2 colon adenocarcinoma cell line. Oncogene 19, 5182–5188. [DOI] [PubMed] [Google Scholar]
- Grande, M. A., Van der Kraan, I., de Jong, L., and van Driel, R. (1997). Nuclear distribution of transcription factors in relation to sites of transcription and RNA polymerase II. J. Cell Sci. 110, 1781–1791. [DOI] [PubMed] [Google Scholar]
- Hendzel, M. J., Kruhlak, M. J., MacLean, N. A., Boisvert, F., Lever, M. A., and Bazett-Jones, D. P. (2001). Compartmentalization of regulatory proteins in the cell nucleus. J. Steroid Biochem. Mol. Biol. 76, 9–21. [DOI] [PubMed] [Google Scholar]
- Htun, H., Holth, L. T., Walker, D., Davie, J. R., and Hager, G. L. (1999). Direct visualization of the human estrogen receptor α reveals a role for ligand in the nuclear distribution of the receptor. Mol. Biol. Cell 10, 471–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, K., Barhoumi, R., Burghardt, R., and Safe, S. (2005). Analysis of estrogen receptor {α}-Sp1 interactions in breast cancer cells by fluorescence resonance energy transfer. Mol. Endocrinol. 19, 843–854. [DOI] [PubMed] [Google Scholar]
- Lamond, A. I., and Spector, D. L. (2003). Nuclear speckles: a model for nuclear organelles. Nat. Rev. Mol. Cell. Biol. 4, 605–612. [DOI] [PubMed] [Google Scholar]
- Lee, L.T.O., Tan-Un, K. C., Pang, R.T.K., Lam, D.T.W., and Chow, B.K.C. (2004). Regulation of the human secretin gene is controlled by the combined effects of CpG methylation, Sp1/Sp3 ratio, and the E-box element. Mol. Endocrinol. 18, 1740–1755. [DOI] [PubMed] [Google Scholar]
- Li, L., Sun, J.-M., and Davie, J. R. (2004). Gene regulation by Sp1 and Sp3. Biochem. Cell Biol. 82, 460–471. [DOI] [PubMed] [Google Scholar]
- Liu, A. G., Hoffman, P. W., Lu, W. W., and Bai, G. (2004). NF-kappa B site interacts with Sp factors and up-regulates the NR1 promoter during neuronal differentiation. J. Biol. Chem. 279, 17449–17458. [DOI] [PubMed] [Google Scholar]
- Mancini, M. G., Liu, B., Sharp, Z. D., and Mancini, M. A. (1999). Subnuclear partitioning and functional regulation of the Pit-1 transcription factor. J. Cell. Biochem. 72, 322–338. [PubMed] [Google Scholar]
- Moen, P. T., Jr., Johnson, C. V., Byron, M., Shopland, L. S., de, l. S., I, Imbalzano, A. N., and Lawrence, J. B. (2004). Repositioning of muscle-specific genes relative to the periphery of SC-35 domains during skeletal myogenesis. Mol. Biol. Cell 15, 197–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickerson, J. A., Krockmalnic, G., Wan, K. M., and Penman, S. (1997). The nuclear matrix revealed by eluting chromatin from a cross-linked nucleus. Proc. Natl. Acad. Sci. USA 94, 4446–4450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pombo, A., Cuello, P., Schul, W., Yoon, J. B., Roeder, R. G., Cook, P. R., and Murphy, S. (1998). Regional and temporal specialization in the nucleus: a transcriptionally-active nuclear domain rich in PTF, Oct1 and PIKA antigens associates with specific chromosomes early in the cell cycle. EMBO J. 17, 1768–1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross, S., Best, J. L., Zon, L. I., and Gill, G. (2002). SUMO-1 modification represses Sp3 transcriptional activation and modulates its subnuclear localization. Mol. Cell 10, 831–842. [DOI] [PubMed] [Google Scholar]
- Rubbi, C. P., and Milner, J. (2000). Non-activated p53 co-localizes with sites of transcription within both the nucleoplasm and the nucleolus. Oncogene 19, 85–96. [DOI] [PubMed] [Google Scholar]
- Samuel, S. K., Minish, M. M., and Davie, J. R. (1997). Nuclear matrix proteins in well and poorly differentiated human breast cancer cell lines. J. Cell. Biochem. 66, 9–15. [PubMed] [Google Scholar]
- Sapetschnig, A., Koch, F., Rischitor, G., Mennenga, T., and Suske, G. (2004). Complexity of translationally controlled transcription factor Sp3 isoform expression. J. Biol. Chem. 279, 42095–42105. [DOI] [PubMed] [Google Scholar]
- Sapetschnig, A., Rischitor, G., Braun, H., Doll, A., Schergaut, M., Melchior, F., and Suske, G. (2002). Transcription factor Sp3 is silenced through SUMO modification by PIAS1. EMBO J. 21, 5206–5215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowa, Y., Orita, T., Minamikawa-Hiranabe, S., Mizuno, T., Nomura, H., and Sakai, T. (1999). Sp3, but not Sp1, mediates the transcriptional activation of the p21/WAF1/Cip1 gene promoter by histone deacetylase inhibitor. Cancer Res. 59, 4266–4270. [PubMed] [Google Scholar]
- Spann, T. P., Goldman, A. E., Wang, C., Huang, S., and Goldman, R. D. (2002). Alteration of nuclear lamin organization inhibits RNA polymerase II-dependent transcription. J. Cell Biol. 156, 603–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer, V. A., Samuel, S., and Davie, J. R. (2000). Nuclear matrix proteins associated with DNA in situ in hormone-dependent and hormone-independent human breast cancer cell lines. Cancer Res. 60, 288–292. [PubMed] [Google Scholar]
- Spengler, M. L., Kennett, S. B., Moorefield, K. S., Simmons, S. O., Brattain, M. G., and Horowitz, J. M. (2005). Sumoylation of internally initiated Sp3 isoforms regulates transcriptional repression via a trichostatin A-insensitive mechanism. Cell Signal. 17, 153–166. [DOI] [PubMed] [Google Scholar]
- Stenoien, D. L., Mancini, M. G., Patel, K., Allegretto, E. A., Smith, C. L., and Mancini, M. A. (2000). Subnuclear trafficking of estrogen receptor-α and steroid receptor coactivator-1.Mol. Endocrinol. 14, 518–534. [DOI] [PubMed] [Google Scholar]
- Stoner, M., Wormke, M., Saville, B., Samudio, I., Qin, C., Abdelrahim, M., and Safe, S. (2004). Estrogen regulation of vascular endothelial growth factor gene expression in ZR-75 breast cancer cells through interaction of estrogen receptor α and SP proteins. Oncogene 23, 1052–1063. [DOI] [PubMed] [Google Scholar]
- Sun, J. M., Chen, H. Y., Moniwa, M., Litchfield, D. W., Seto, E., and Davie, J. R. (2002). The transcriptional repressor Sp3 is associated with CK2 phosphorylated histone deacetylase 2. J. Biol. Chem. 277, 35783–35786. [DOI] [PubMed] [Google Scholar]
- Sun, J.-M., Chen, H. Y., and Davie, J. R. (2001). Effect of estradiol on histone acetylation dynamics in human breast cancer cells. J. Biol. Chem. 276, 49435–49442. [DOI] [PubMed] [Google Scholar]
- Sun, J.-M., Spencer, V. A., Li, L., Chen, H. Y., Yu, J., and Davie, J. R. (2005). Estrogen-regulation of trefoil factor 1 expression by estrogen receptor α and Sp proteins. Exp. Cell Res. 302, 96–107. [DOI] [PubMed] [Google Scholar]
- Suske, G. (1999). The Sp-family of transcription factors. Gene 238, 291–300. [DOI] [PubMed] [Google Scholar]
- Taplick, J., Kurtev, V., Kroboth, K., Posch, M., Lechner, T., and Seiser, C. (2001). Homo-oligomerisation and nuclear localisation of mouse histone deacetylase 1. J. Mol. Biol. 308, 27–38. [DOI] [PubMed] [Google Scholar]
- van Wijnen, A. J., Bidwell, J. P., Fey, E. G., Penman, S., Lian, J. B., Stein, J. L., and Stein, G. S. (1993). Nuclear matrix association of multiple sequencespecific DNA binding activities related to SP-1,ATF, CCAAT, C/EBP, OCT-1, and AP-1.Biochemistry 32, 8397–8402. [DOI] [PubMed] [Google Scholar]
- von Mikecz, A., Zhang, S., Montminy, M., Tan, E. M., and Hemmerich, P. (2000). CREB-binding protein (CBP)/p300 and RNA polymerase II colocalize in transcriptionally active domains in the nucleus. J. Cell Biol. 150, 265–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, B., Datta, P. K., and Bagchi, S. (2003). Stability of the Sp3-DNA complex is promoter-specific: Sp3 efficiently competes with Sp1 for binding to promoters containing multiple Sp-sites. Nucleic Acids Res. 31, 5368–5376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidi, S. K., Sullivan, A. J., van Wijnen, A. J., Stein, J. L., Stein, G. S., and Lian, J. B. (2002). Integration of Runx and Smad regulatory signals at transcriptionally active subnuclear sites. Proc. Natl. Acad. Sci. USA 99, 8048–8053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidi, S. K., Young, D. W., Choi, J. Y., Pratap, J., Javed, A., Montecino, M., Stein, J. L., van Wijnen, A. J., Lian, J. B., and Stein, G. S. (2005). The dynamic organization of gene-regulatory machinery in nuclear microenvironments. EMBO Rep. 6, 128–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidi, S. K., Young, D. W., Pockwinse, S. M., Javed, A., Lian, J. B., Stein, J. L., van Wijnen, A. J., and Stein, G. S. (2003). Mitotic partitioning and selective reorganization of tissue-specific transcription factors in progeny cells. Proc. Natl. Acad. Sci. USA 100, 14852–14857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelko, I. N., and Folz, R. J. (2004). Sp1 and Sp3 transcription factors mediate trichostatin A-induced and basal expression of extracellular superoxide dismutase. Free Radic. Biol. Med. 37, 1256–1271. [DOI] [PubMed] [Google Scholar]
- Zeng, C., et al. (1998). Intranuclear targeting of AML/CBFα regulatory factors to nuclear matrix-associated transcriptional domains. Proc. Natl. Acad. Sci. USA 95, 1585–1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X., Li, Y., Dai, C., Yang, J., Mundel, P., and Liu, Y. (2003). Sp1 and Sp3 transcription factors synergistically regulate HGF receptor gene expression in kidney. Am. J. Physiol. 284, F82–F94. [DOI] [PubMed] [Google Scholar]