Abstract
The establishment and maintenance of characteristic cellular morphologies is a fundamental property of all cells. Here we describe Schizosaccharomyces pombe Pal1p, a protein important for maintenance of cylindrical cellular morphology. Pal1p is a novel membrane-associated protein that localizes to the growing tips of interphase cells and to the division site in cells undergoing cytokinesis in an F-actin- and microtubule-independent manner. Cells deleted for pal1 display morphological defects, characterized by the occurrence of spherical and pear-shaped cells with an abnormal cell wall. Pal1p physically interacts and displays overlapping localization with the Huntingtin-interacting-protein (Hip1)-related protein Sla2p/End4p, which is also required for establishment of cylindrical cellular morphology. Sla2p is important for efficient localization of Pal1p to the sites of polarized growth and appears to function upstream of Pal1p. Interestingly, spherical pal1Δ mutants polarize to establish a pearlike morphology before mitosis in a manner dependent on the kelch-repeat protein Tea1p and the cell cycle inhibitory kinase Wee1p. Thus, overlapping mechanisms involving Pal1p, Tea1p, and Sla2p contribute to the establishment of cylindrical cellular morphology, which is important for proper spatial regulation of cytokinesis.
INTRODUCTION
The establishment and maintenance of characteristic cellular morphologies is important for proper execution of cell functions. How signaling pathways, the cytoskeleton, and membrane trafficking mechanisms cooperate to control cellular morphogenesis remains a fundamental challenge in understanding cell regulation.
The fission yeast Schizosaccharomyces pombe is an attractive organism for the study of several aspects of cell biology, such as division and morphogenesis (Chang and Verde, 2003). Fission yeast cells are cylindrical in shape with hemispherical caps at the two ends. Cells grow by tip extension with almost no change in cellular diameter (Mitchison and Nurse, 1985). During growth, F-actin is detected in patches at the growing ends (Marks et al., 1986), although microtubule bundles run parallel to the long axis of the cell in a manner such that their minus ends are positioned near the nucleus and the plus ends are oriented toward the cell ends (Brunner and Nurse, 2000; Tran et al., 2001). During cell division, F-actin and associated proteins assemble into a contractile medial actomyosin ring (Le Goff et al., 1999; Feierbach and Chang, 2001; Rajagopalan et al., 2003) and microtubules assemble into a ring structure following anaphase underneath the actomyosin ring (Heitz et al., 2001; Pardo and Nurse, 2003). How growth zones are specified during interphase leading to the localization of F-actin at the cell ends and microtubules along the long axis represents a fundamental question of interest in understanding cell polarity and morphogenesis. To unravel the mechanism of cell polarization and morphogenesis, two major classes of morphological mutants have been isolated and characterized in fission yeast (Chang et al., 1994; Snell and Nurse, 1994; Verde et al., 1995; Arellano et al., 1996).
The first class consists of mutants that are unable to establish and/or maintain a cylindrical morphology. These mutants, referred to collectively as orb mutants, define proteins important for F-actin function and cell wall assembly (Diaz et al., 1993; Chang et al., 1994; Miller and Johnson, 1994; Ottilie et al., 1995; Verde et al., 1998). A second group consists of mutants that are able to establish a cylindrical morphology, but are unable to position and maintain the growth machinery along a straight line leading to the formation of bent and T-shaped cells (Snell and Nurse, 1994; Verde et al., 1995; Beinhauer et al., 1997; Mata and Nurse, 1997; Radcliffe et al., 1998; Browning et al., 2000; Brunner and Nurse, 2000; Behrens and Nurse, 2002; Snaith and Sawin, 2003). These mutants, that include the tea and alp classes, define elements of the microtubule cytoskeleton or those that might link F-actin and microtubules. The functional links between the orb group and tea/alp group have not been fully investigated.
Here we describe a gene, pal1, whose product localizes to the ends of interphase cells and to the division site in cells undergoing cytokinesis. Pal1p appears to contribute to a mechanism that maintains the cylindrical morphology of fission yeast cells in concert with the Hip1-related protein Sla2p/End4p, by modulation of cell wall integrity. We also provide evidence for the existence of at least two pathways important for the maintenance of a cylindrical cellular morphology.
MATERIALS AND METHODS
S. pombe Strains, Media, and Reagents
S. pombe strains used in this study are listed in Table 1. YES medium or Minimal medium with appropriate supplements were used to culture fission yeast cells. Fission yeast strains were constructed by either random spore germination method or by tetrad dissection. For strains with low mating efficiency, diploids were generated by using ade6-210/ade6-216 intragenic complementation and selected on minimal plates lacking adenine. These diploids were then sporulated and tetrads dissected. To disrupt F-actin or microtubules, cells were treated with either 15 μM of latrunculin A (Molecular Probes, Eugene, OR) or 8 μg/ml methyl 1-(butylcarbomyl)-2-benzimidazolecarbamate (MBC; Aldrich Chemical, Milwaukee, WI).
Table 1.
Yeast strains used in this study
| Name | Genotype | Source |
|---|---|---|
| MBY102 | ade6-210 ura4-D18 leu1-32 h+ | Lab collection |
| MBY103 | ade6-216 ura4-D18 leu1-32 h- | Lab collection |
| MBY169 | wee1-50, leu1-32, h- | Lab collection |
| MBY192 | ura4-D18 leu1-32 h- | Lab collection |
| MBY1186 | sph2-3 ade3-58 h90 | Matthias Sipiczki |
| MBY1247 | tea1::ura4+rlc1-GFP::leu1+leu1-32 ura4-D18 ade6-210 h+ | Lab collection |
| MBY2204 | pal1-GFP::ura4+ura4-D18 leu1-32 h- | This study |
| MBY2211 | pal1::ura4+ura4-D18 leu1-32 h- | This study |
| MBY2219 | pal1-GFP::ura4+cdc25-22 | This study |
| MBY2220 | pal1-GFP::ura4+mid1Δ::ura4+ | This study |
| MBY2233 | pal1::ura4+, wee1-50, ura4-D18 | This study |
| MBY2237 | pal1::ura4+ade6-210, h+ | This study |
| MBY2238 | pal1::ura4+ade6-216, h- | This study |
| MBY2239 | pal1::ura4+ade6-216, h+ | This study |
| MBY2240 | pal1-GFP::ura4+h+ | This study |
| MBY2249 | orb2-34 pal1-GFP::ura4+ | This study |
| MBY2250 | orb3-167 pal1-GFP::ura4+ | This study |
| MBY2253 | tea1::ura4+pal1-GFP::ura4+ | This study |
| MBY2260 | pal1::ura4+tea1Δ::ura4+ | This study |
| MBY2262 | pal1::ura4+ expressing α-tubulin GFP plasmid (pCDL646) | This study |
| MBY2264 | pal1::ura4+tea1-YFP::Kanr | This study |
| MBY2327 | sla2-GFP::ura4+ura4-D18 leu1-32 h- | This study |
| MBY2335 | orb6-25, wee1-50 | This study |
| MBY2336 | nda3-KM311 pal1-GFP::ura4+ | This study |
| MBY2338 | pal1-GFP::ura4+ade6-210 leu1-32 ura4-D18 h90 | This study |
| MBY2354 | pal1-13Myc::ura4+ade6-210 ura4-D18 leu1-32 h+ | This study |
| MBY2357 | sla2-GFP::ura4+pal1::ura4+ | This study |
| MBY2360 | sla2-GFP::ura4+pal1-13Myc::ura4+ | This study |
| MBY2451 | sla2-13Myc::ura4+, ade6-210 ura4-D18 leu1-32 h+ | This study |
| MBY2463 | sla2-13Myc::ura4+, pal1-GFP::ura4+ | This study |
| MBY2524 | sla2:: ura4+h+ | This study |
| MBY2532 | pal1-GFP::ura4+, sla2::ura4+ | This study |
| MBY2655 | cdc13-YFP, pal1::ura4+ | This study |
| MBY2784 | pTN-L1-sla2 plasmid in sla2::ura4+ | This study |
| MBY2785 | pTN-L1-pal1 plasmid in sla2::ura4+ | This study |
| MBY2786 | pTN-L1 plasmid in sla2::ura4+ | This study |
Gene Disruption and Epitope Tagging
To create the pal1::ura4+ disruption cassette, a 0.7-kb KpnI/XhoI fragment representing the 5′ UTR of the pal1 open reading frame (ORF) was obtained by PCR from genomic DNA using the primers MOH1532 (CGGGGTACCTTATTGCAGCTCTTTATATC) and MOH1533 (CCGCTCGAGCCCGTATTGCACCCAAACGATAC), and cloned into KpnI/XhoI sites of the pCDL126 vector (contains ura4+ in pBSK+ plasmid). A 0.7-kb XbaI/SacI fragment representing the 3′ UTR of pal1 was also obtained by PCR using the primers MOH1534 (GCTCTAGAGCTTTTCAGGCTTGAAACTTTATTTAC) and MOH1535 (CGAGCTCCCAATTTGGCACCGACTCTAAAAAAC) and cloned into the modified plasmid containing the 5′ UTR and the marker gene ura4+, to generate pCDL863. To generate a pal1 gene deletion strain, a 3.2-kb fragment from pCDL863 was generated by digestion with KpnI and SacI and was introduced into the wild-type haploid strain MBY192 (ura4-D18, leu1-32, h-). A similar strategy was used to create a strain deleted for the sla2 gene. The primers MOH1686 (CGGGGTACCAGAAATGTTAACGAAGACTC) and MOH1687 (CCGCTCGAGGTGATCGGATTGCAGTCGAAACG) were used to amplify the 5′ UTR, and the primers MOH1688 (GCTCTAGATTAAGTCTTTTCGATCC) and MOH1689 (CGAGCTCTCCACCCCGACGGTTTCATTC) were used to amplify the 3′UTR of sla2. The ura4+ gene flanked by the 5′ UTR and 3′ UTR of sla2 was transformed into a diploid strain (ura4-D18/ura4-D18, leu1-32/leu1-32, ade6-210/ade6-216, h+/h-). Yeast transformations were performed by the lithium acetate method (Okazaki et al., 1990). Transformants were selected by growth on supplemented minimal medium lacking uracil. For green fluorescent protein (GFP) tagging of pal1 and sla2, a 1-kb KpnI/SmaI fragment of pal1 and a 1-kb KpnI/SmaI fragment of sla2, encoding the C-termini, were obtained by PCR using the primer pairs MOH1466 (CGGGGTACCTAATCTGCGGAGGCATTAGCCGAACG) and MOH1467 (TCCCCCGGGCGACTTTTTGTGGAATAATCGTC), and MOH1564 (CGGGGTACCTAATCCGTCAACACAAGAAAAATGGATG) and MOH1565 (TCCCCCGGGCTCTTCGGCAACATGATAAGATG), respectively. The fragments were cloned into the KpnI/SmaI sites of pJK210-GFP to yield the plasmids pCDL856 and pCDL916. Plasmids were linearized with SalI or PacI and transformed into the wild-type haploid strain MBY192. Similarly, for tagging pal1 and sla2 with 13-Myc, a 1-kb KpnI/BamHI fragment of pal1 and a 1-kb KpnI/BamHI fragment of sla2, encoding the C-termini, were obtained by PCR using the primers MOH1466, MOH1681 (GCGGATCCGCGACTTTTTGTGGAATAATCGTC) and MOH1564, MOH1685 (GCGGATCCGCTCTTCGGCAACATGATAAGATG), respectively, and cloned into the same sites of pJK210-13Myc to yield the plasmids pCDL919 and pCDL931. The plasmids were linearized with SalI and PmlI, respectively, and transformed into the wild-type haploid strain MBY102. In all cases, correct integration was confirmed by PCR.
High-copy Suppressor Screening
To identify genes that act as high-copy suppressors of the colony formation defect of the sla2Δ mutant at 36°C, a genomic DNA library (Nakamura et al., 2001) was transformed into sla2Δ cells. After transformation, cells were incubated at 24°C for 12 h on minimal medium lacking leucine and then shifted to 36°C and incubated for 5 d. The plasmids were recovered from the rescued sla2Δ clones and transformed into Escherichia coli cells. The recovered plasmids were sequenced using T3 and T7 primers. From this screen, we identified sla2 six times and pal1 three times.
Immunoprecipitation and Western Blotting
Immunoprecipitation and Western blotting were performed as described (Wang et al., 2002). Briefly, total cell extracts were prepared from exponentially growing cells by disruption with glass beads. NP-40 buffer, 600 μl, (1% Triton X-100, 150 mM NaCl, 2 mM EDTA, 6 mM Na2HPO4, 4 mM NaH2PO4, 1 mM phenylmethylsulfonyl fluoride [PMSF], 2 mM benzamidine, supplemented with protease inhibitors [Complete, EDTA-free, Roche Diagnostics, Indianapolis, IN]) was used to extract soluble proteins from ∼40 ml early log phase culture. Cell extracts were clarified by centrifugation at 14,000 rpm for 10 min at 4°C. For each immunoprecipitation experiment, 500 μl of soluble protein was incubated with 5 μl of α-GFP antibodies for 1 h at 4°C. Sepharose-Protein A beads (100 μl, Amersham Biosciences, Piscataway, NJ) were added to the antigen-antibody immunocomplex and incubated for 45 min at 4°C. After six washes, beads were resuspended in SDS-PAGE loading buffer and heated at 95°C for 5 min.
To detect Myc or GFP-tagged Pal1p and Sla2p proteins were separated on 8% SDS-polyacrylamide gels (Mini-protein II system; Bio-Rad Laboratories, Richmond, CA) at 120 V for 1 h and transferred (Trans-Blot system; Bio-Rad Laboratories) at 90 V for 1 h to a PVDF membrane (Millipore Co., Bedford, MA). The membrane was blocked with 5% nonfat milk in phosphate-buffered saline-Tween 20 (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4, 0.05% Tween 20, pH 7.4) for 1 h at room temperature or overnight at 4°C. Primary α-GFP (Molecular Probes) and α-Myc (Sigma, St. Louis, MO) antibodies were used at 1:2000 dilutions. Peroxidase-conjugated α-rabbit and α-mouse IgG (Sigma) were used at 1:4000 dilutions, and the chemiluminescent signal was detected using a 1:1 mixture of ECL1 (2.5 mM 3-aminophytaldrazide dissolved in dimethyl sulfoxide (DMSO), 0.4 mM p-coumaric acid, 100 mM Tris-HCl, pH 8.5), and ECL2 (0.02% H2O2, 100 mM Tris-HCl, pH 8.5; Schneppenheim et al., 1991).
Microscopy
Fluorescence microscopy was done as described by Balasubramanian et al. (1997). Cells were fixed with 7% formaldehyde to visualize F-actin structures using Alexa Fluor-488 phalloidin (Molecular Probes). For immunofluorescence studies, cells were fixed with 7% formaldehyde. To detect Sla2p-GFP, Cdc4p, and microtubules, antibodies against GFP, Cdc4p, and β-tubulin (a gift from Dr Keith Gull) were used. α-rabbit and α-mouse IgG-conjugated with either Alexa Fluor-594 or Alexa Fluor-488 (Molecular Probes) were used as the secondary antibodies. Aniline blue and Calcofluor were used to stain cell wall. DNA was stained with 4′,6-diamidino-2-phenylindole (DAPI). Cells were observed using a Leica DMLB microscope (Deerfield, IL) and images were captured using a Photometrics CoolSNAP ES camera (Tucson, AZ). MetaVue software (Universal Imaging, West Chester, PA) was used to acquire images and the images were then assembled in Photoshop 7.0 (Adobe Systems, San Jose, CA). For time-lapse microscopy, cells were concentrated from early log phase culture and spotted on a glass slide containing an agar pad with appropriate medium. The time-lapse analysis was conducted at room temperature (22–24°C) using the Leica DMLB microscope. Assembly of images was done using ImageJ 1.32 (National Institutes of Health, Bethesda, MD). For confocal microscopy, cells were observed under the Zeiss Meta Inverted Laser Scanning Confocal microscope (LSM510; Thornwood, NY). Image acquisition was done at the excitation wavelength of 488 nm with 3–4% of the laser intensity. For electron microscopy, cells were processed as described in Wang et al. (2002).
Density Gradient Centrifugation
S. pombe cells were grown in YES at 30°C to OD595 = 0.5. Cells were harvested and broken using glass beads and total proteins were extracted in 300 μl TNE buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 5 mM EDTA, 1 mM PMSF, supplemented with protease inhibitors [Complete, EDTA-free, Roche Diagnostics]). The lysate was cleared from cell wall debris by low-speed centrifugation (200 × g). To the supernatant, Optiprep (Axis-Shield, Oslo, Norway) was added to a final concentration of 40%, overlaid by 1300 μl of 30% Optiprep in TNE and 100 μl TNE. Samples were centrifuged for 2 h at 55000 rpm at 4°C in a Beckman TLS55 rotor (Fullerton, CA), and six equal fractions were collected from top to bottom of the gradient. Proteins were precipitated using methanol/chloroform (Wessel and Flugge, 1984), and the pellets were dissolved in SDS sample buffer. Protein concentration was determined using amidoblack (Popov et al., 1975) against bovine serum albumin as a standard. For Western blotting, equal volumes of each fraction were separated on 10% SDS-polyacrylamide gels (Laemmli, 1970) and immunoblotted with α-GFP (Molecular Probes), α-Pma1p and α-Cdc8p antibodies.
RESULTS
A Novel Membrane-associated Protein, Pal1p, Localizes to the Sites of Active Growth and Cell Division
We identified a gene, pal1 (pears and lemons; encoded by SPCP1E11.04c), based on its homology to a novel uncharacterized ORF from budding yeast (YDR348c). This gene was chosen because its protein product localizes to the cell division site, as determined from a large-scale effort aimed at cataloging the intracellular localization of all budding yeast proteins (Huh et al., 2003). Proteins related to Pal1p were detected in several ascomycetes. Figure 1A shows the alignment of the conserved part of the amino acid sequence of Pal1p with related members from S. cerevisiae, Magnaporthe grisea, and Neurospora crassa. Proteins related to Pal1p are variable in length and share an ∼160 amino acid region of high similarity, which ends roughly 100 amino acids from the C-termini of these proteins (Figure 1B). Proteins related to Pal1p were not readily identified in plants and metazoans.
Figure 1.
Identification of Pal1p in fission yeast. (A) Schizosaccharomyces pombe Pal1p is aligned with S. cerevisiae YDR348cp, Magnaporthe grisea MG02779.4, Neurospora crassa NCU00687.1. Identical amino acids are shaded in black and similar amino acids are shaded in gray. (B) Schematic representation of the homology between Sp.Pal1p and other related proteins. Identities (%) and the number of amino acids in parentheses are indicated in the conserved region. (C) Localization of Pal1p in wild-type cells. Cells expressing functional GFP-tagged Pal1p grown on YES plates were visualized by fluorescence microscopy. Pal1p localizes to the growing end(s) during monopolar and bipolar growth (cells 1 and 2) and to the medial cell division site during cytokinesis (cells 3–6). Scale bar, 5 μm. (D) Detection of Pal1p. Total cell extracts were prepared from the pal1-GFP (lane 1) and wild-type (lane 2) strains and were subjected to SDS-PAGE and immunoblotted using α-GFP antibody. (E) Pal1p is a membrane associated protein. Total cell extracts prepared from the pal1-GFP strain were fractionated by density gradient centrifugation. The fractions obtained were subjected to SDS-PAGE and immunoblotted using α-GFP, α-PMA1, and α-Cdc8p antibodies. Lanes 1–6 show the different fractions from top to bottom of the density gradient.
To determine the intracellular distribution of Pal1p, we fused the ORF to DNA sequences encoding the GFP. The sole copy of the gene encoding the Pal1p-GFP fusion was expressed under the control of the native promoter sequence. Pal1p showed a cell cycle-dependent intracellular distribution (Figure 1C). Pal1p was detected at one end of the cell in a small proportion of uninucleate cells (Figure 1C; cell 1) and was visualized at both ends of the vast majority of uninucleate cells (Figure 1C; cell 2). In cells undergoing cytokinesis, Pal1p-GFP was present at the medial cell division site (Figure 1C; cells 3–6). Thus Pal1p localizes to the cell tips in interphase and to the division site during mitosis and cytokinesis.
In most cases the Pal1p-GFP signal appeared to be closely associated with the cell periphery, suggesting a possible membrane association of this protein. To test if Pal1p was membrane associated, we carried out membrane preparations from a strain expressing GFP-tagged Pal1p. In denaturing lysates, Pal1p-GFP was detected as a single band of ∼75 kDa, consistent with the predicted size of this fusion protein (Figure 1D). This band was absent in a wild-type strain in which Pal1p was not tagged with the GFP epitope, establishing that the 75-kDa band was the product of the pal1-GFP encoding gene. Interestingly, upon preparation of a membrane fraction by density gradient centrifugation, a significant portion of Pal1p was found to cosediment at the top of the gradient (Figure 1E) in a fraction that contained other known membrane proteins such as plasma membrane ATPase Pma1p (Serrano et al., 1993), whereas a significant fraction also comigrated with Cdc8p, a known soluble protein related to tropomyosins (Balasubramanian et al., 1992). The sedimentation studies suggested that a part of Pal1p was membrane associated.
Pal1p Localizes to Growth Zones Independent of F-actin and Microtubules
The fact that Pal1p was detected at one or both ends suggested that Pal1p localizes to the actively growing end(s). In the orb2-34 mutant grown at the permissive temperature (known to grow only at the old end; Verde et al., 1995), Pal1p-GFP was detected at the growing end as determined by costaining with the cell wall stain aniline blue (Figure 2A, a and b). Pal1p-GFP was detected at both ends of heat-arrested cdc25-22 cells that are known to grow in a bipolar manner (Figure 2B). Pal1p-GFP was found to be localized to a larger region of the cell cortex in spherical and isotropically growing mutants such as orb2-34, orb3-167, orb6-25 at the restrictive temperature (Figure 2C; unpublished data for orb2-34 and orb6-25) and was detected at the additional tips generated in a tea1Δ mutant (marked with arrows in Figure 2D). Furthermore, Pal1p-GFP localized to the mispositioned septa in a mid1Δ mutant (Figure 2E). Taken together, these data suggested that Pal1p localizes to sites of polarized growth and secretion.
Figure 2.
Pal1p localizes to the sites of cell growth and division in an F-actin- and microtubule-independent manner. Different strains with functional GFP-tagged Pal1p were grown on YES agar plate at 24°C and then shifted to 36°C for 6 h. (A) orb2-34 at 24°C; a shows Pal1p-GFP; b shows aniline blue staining; (B) cdc25-22 at 36°C; (C) orb3-167 at 36°C; (D) tea1Δ; (E) mid1Δ; (F) Pal1p localizes to the zone of cell fusion in the h90 strain. (G) Role of F-actin in Pal1p localization. cdc25-22 pal1-GFP cells were blocked for 4 h at 36°C in YES medium and then released to 24°C into medium containing 15 μM Lat A or DMSO. Cell samples were taken at different time points (a, 60 min; b, 60 min; c, 120 min; d, 180 min) after release and visualized by fluorescence microscopy. (H) Role of microtubules in Pal1p localization. nda3-KM311 pal1-GFP cells were grown on a YES agar plate at 32°C for 1 d and shifted to 19°C for 6 h. Scale bar, 5 μm.
Fission yeast cells are known to reinitiate polarized growth toward a mating partner of opposing mating type. This growth occurs in random directions, determined by the position of the mating partner (Nielsen and Davey, 1995). During mating, Pal1p-GFP was detected at the zone of cell fusion (Figure 2F). Thus, Pal1p localizes to sites of polarized growth and secretion in vegetative and mating cells.
Given that both the F-actin and microtubule cytoskeletons are important for proper polarized growth and cell division, we studied the role of F-actin and microtubule cytoskeletons in the localization of Pal1p-GFP to the cell tips and the cell division site. To test the role of F-actin in assembly and maintenance of Pal1p at the cell tips and the division site, a cdc25-22 strain was arrested at the G2/M boundary by shift to the restrictive temperature. Cells were then treated with Lat A to cause eventual loss of all F-actin structures and returned to the permissive temperature to allow resumption of the mitotic cycle. Cells were treated with the solvent DMSO as a control. In DMSO-treated cells, Pal1p was detected at the division site in well-formed septa (Figure 2G; cell a). Interestingly, in Lat A-treated cells, Pal1p continued to be present at the cell tips, suggesting that maintenance of Pal1p at the cell tips is F-actin independent (Figure 2G: cells b–d, marked with arrows). In addition, Pal1p-GFP also accumulated at the division site in Lat A-treated cells (Figure 2G: cells b–d, marked with arrowheads), although Pal1p-GFP was not organized into a proper medial ring structure in the absence of an actomyosin ring.
To study the role of the microtubule cytoskeleton in Pal1p-GFP localization, we arrested the cold-sensitive β-tubulin mutant nda3-KM311 at the restrictive temperature and assayed the localization of Pal1p-GFP. Interestingly, Pal1p-GFP was clearly detected at the presumptive cell division site (Figure 2Ha; marked with arrowheads) as well as at the cell tips (Figure 2Hb; marked with arrows). Thus, Pal1p accumulation at the cell division site and its maintenance at the cell tips and the division site are independent of F-actin and microtubule function.
Pal1p Physically Interacts and Shows Overlapping Localization with Sla2p
The budding yeast Pal1p related protein YDR348c has been shown to copurify with Sla2p in genome-scale proteomic analyses in S. cerevisiae (Gavin et al., 2002). We identified a Sla2p related protein encoded by SPAC688.11 in fission yeast (Figure 3A). S. pombe Sla2p is related to the budding yeast Sla2p as well as human proteins HIP1 (Huntingtin-interacting protein) and talin. All four proteins shown in Figure 3A contain the actin-binding I/LWEQ motif. In addition, the AP180-N-terminal homology domain (ANTH domain) is present in Sla2p from budding and fission yeasts as well as in HIP1, but is absent in talin. To test if Pal1p physically interacted with Sla2p, we created S. pombe strains expressing 13-Myc tagged Pal1p, GFP-tagged Sla2p, and both these epitope tagged-proteins. Cell lysates were immunoprecipitated with α-GFP antibodies and the immune complexes were immunoblotted with α-Myc antibodies. Interestingly, Pal1p-Myc was immunoprecipitated by α-GFP antibodies only in cells expressing Sla2p-GFP and Pal1p-Myc (Figure 3B). Furthermore, in strains expressing Sla2p-Myc and Pal1p-GFP, Sla2p-Myc was recovered in immune complexes prepared with GFP antibodies (Figure 3B). Thus, Pal1p physically interacts with Sla2p.
Figure 3.
Pal1p physically interacts with Sla2p, a Hip1-related protein in S. pombe. (A) Schematic representation of the homology between fission yeast Sla2p and other Sla2p/Hip1p family proteins. S. pombe Sla2p (Sp_Sla2p) is aligned with S. cerevisiae Sla2p (Sc_Sla2p), Homo sapiens Hip1p (Hs_Hip1p), Homo sapiens Tln1p (Hs_Tln1p). Identities (%) and the number of amino acids are indicated in two conserved domains, ANTH and I/LWEQ. (B) Coimmunoprecipitation of Pal1p with Sla2p in α-GFP immunoprecipitation from native cell extracts. Total cell extracts were prepared from pal1-Myc, sla2-GFP, pal1-Myc sla2-GFP, pal1-GFP, sla2-Myc, and pal1-GFP sla2-Myc strains. The entire immune complex prepared from 500 μl of lysate and 30 μl of straight-lysate were subjected to SDS-PAGE and immunoblotted with α-GFP or α-Myc antibodies. (C) Localization of Sla2p-GFP in wild-type cells. Cells expressing functional Sla2p-GFP under its native promoter were grown at 30°C and visualized by fluorescence microscopy. (D) Colocalization of Sla2p-GFP and F-actin patches. Cells expressing Sla2p-GFP were grown at 30°C and immunostained with α-GFP antibody and Alexa Fluor-488-conjugated phalloidin. (E) Role of F-actin in Sla2p localization. cdc25-22 sla2-GFP cells were blocked at 36°C for 4 h in YES medium and then released to 24°C into YES medium containing 15 μM Lat A or DMSO. Cell samples were taken at different time points (a, 60 min; b, 60 min; c, 120 min; d, 180 min) after release. (F) Role of microtubules in Sla2p localization. nda3-KM311 sla2-GFP cells were grown in YES liquid medium at 32°C and then shifted to 19°C for 6 h. Scale bar, 5 μm.
We then investigated the localization of Sla2p-GFP, expressed from a sole copy of the gene the under control of native promoter sequences. In interphase cells, Sla2p was detected as patches at the cell ends, whereas in late mitosis, Sla2p was visualized at the medial region of the cell in patches (Figure 3C). To address possible colocalization of Sla2p with F-actin, cells expressing Sla2p-GFP were fixed and stained with antibodies to GFP and Alexa-conjugated phalloidin (Figure 3D). This experiment revealed significant colocalization of F-actin and Sla2p. Some differences were noted as well. First, Sla2p was detected in more patch-like structures compared with F-actin and second, whereas F-actin assembled at the division site early in mitosis, Sla2p assembled at the division site in late anaphase. We conclude that Sla2p colocalizes with a large subset of F-actin structures and displays overlapping localization with Pal1p. As in the case of Pal1p, Sla2p localization was not altered in cells treated with the actin polymerization inhibitor LatA (Figure 3E) and in microtubule mutants (Figure 3F), suggesting that Sla2p localized in an F-actin- and microtubule-independent manner.
Pal1p Is Important for Maintenance of a Cylindrical Shape
To study the phenotypes resulting from the loss of Pal1p function, a strain in which the entire coding region of pal1 was deleted was constructed. The resultant strain, pal1::ura4 (referred to as pal1Δ) was viable, although upon microscopic observation several morphological abnormalities were detected. The morphological phenotypes in pal1Δ cells were classified into three groups; Spherical (short to long axis ratio of 0.80 or higher; Figure 4A; cells marked with arrows), Abnormal (pearlike appearance; Figure 4A; cells marked with arrowheads), and Normal (cylindrical). In spherical cells F-actin and microtubules were disorganized (Figure 4, B and C). F-actin was detected in patches over the entire cortex, as opposed to the pattern of localization of F-actin patches at the cell tips observed in wild-type cells. In spherical cells microtubules were observed to crisscross the cell, as opposed to the presence of properly organized bundles of microtubules observed in wild-type cells (Figure 4C). Thus, the pal1Δ mutant is defective in maintenance of a cylindrical cellular morphology and displays abnormally organized F-actin and microtubules. Staining with calcofluor revealed cell wall abnormalities in pal1Δ cells (Figure 4D). In wild-type cells, intense staining is only detected at hemispherical cell ends during growth and is observed at the division site during cytokinesis. In contrast, in growing interphase pal1Δ cells, the intensity of calcofluor staining was more in the cylindrical part than that in the hemispherical part of the cell, suggesting that cell wall assembly was occurring at improper sites.
Figure 4.
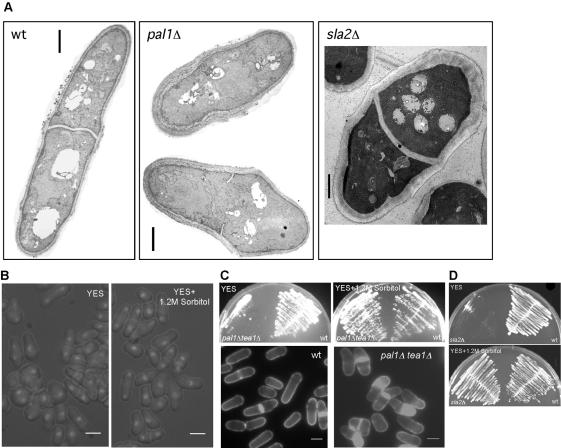
pal1Δ and sla2Δ cells have defects in cell morphology and cell wall. (A) Microscopic analysis of pal1Δ cells. pal1Δ cells were grown in minimal medium at 30°C and stained with DAPI to visualize DNA. Arrows, spherical cell; arrowheads, abnormal shaped cell. (B) pal1Δ cells were grown in minimal medium, fixed, and stained with Alexa Fluor-488-conjugated phalloidin to show the F-actin localization. (C) pal1Δ cells expressing GFP-α-tubulin were used to visualize interphase microtubules. (D) Wild-type and pal1Δ cells were grown in YES liquid medium and stained with Calcofluor. (E) Morphology of sla2Δ cells. sla2Δ cells were grown in YES supplemented with 1.2 M sorbitol, washed, and reinoculated into YES medium for 6 h at 30°C, fixed, and stained with aniline blue and DAPI to visualize division septa and nuclei, respectively. Scale bar, 5 μm.
Given the physical interaction between Pal1p and Sla2p, the phenotype resulting from the loss of Sla2p function was also examined. Strains bearing a deletion of the entire sla2 gene were viable as reported recently by Iwaki et al. (2004) and Castagnetti et al. (2005), although these cells showed a higher incidence of morphological abnormalities (Figure 4E) and were incapable of colony formation at 36°C (Figure 5D). sla2Δ cells were more spherical in nature than pal1Δ cells. Thus, the interacting proteins Pal1p and Sla2p are important for proper cylindrical shape establishment and maintenance.
Figure 5.
pal1Δ and sla2Δ cells have abnormally thick cell wall and are rescued by growth on sorbitol. (A) Exponentially growing wild-type, pal1Δ, and sla2Δ cells were processed for thin section electron microscopy. Electron micrographs of wild-type, pal1Δ cells, and sla2Δ cells are shown in the left, middle, and right panels, respectively. Scale bar, 1 μm. (B) Morphology of pal1Δ mutants is suppressed by sorbitol. pal1Δ cells were grown in YES and YES plus 1.2 M sorbitol medium. Cells were stained with DAPI and visualized by fluorescence microscopy. (C) pal1Δ phenotype can be suppressed by sorbitol in pal1Δ tea1Δ mutant. pal1Δ tea1Δ and wild-type cells were grown on YES plate with or without 1.2 M sorbitol for 2 d at 36°C (top panel). Cells from the plates were stained with aniline blue to visualize the cell shape (bottom panel). (D) Ability of sla2Δ cells to form colonies on YES supplemented with sorbitol. Wild-type and sla2Δ cells were streaked on YES plates with or without sorbitol and incubated at 32°C for 3 d. Scale bar, 5 μm.
Pal1p and Sla2p Are Important for Cell Wall Integrity
In experiments involving calcofluor staining, we found that the cell wall of pal1Δ cells was intensely fluorescent, suggesting that the cell wall in pal1Δ cells might be altered (Figure 4D). To test this, we studied thin sections of wild-type and pal1Δ cells by electron microscopy. Interestingly, whereas the cell wall of wild-type cells was of uniform thickness, the cell wall in pal1Δ cells was significantly thicker (Figure 5A). Thus, pal1Δ cells assemble abnormal cell walls. It remained possible that the spherical morphology might have resulted from improper cell wall integrity. If this were the case, addition of an osmolyte such as sorbitol would be expected to restore cylindrical morphology of pal1Δ cells. This was found to be the case (Figure 5B). An increased number of spherical and abnormal cells was detected when pal1Δ cells were grown in YES compared with those grown in YES medium containing sorbitol.
We have found that Pal1p is essential in cells lacking the kelch-repeat protein Tea1p (discussed in a later section). To more rigorously test the conclusion that cells lacking Pal1p exhibited abnormal cell wall integrity, we assessed the ability of exogenously added sorbitol to restore viability of pal1Δ tea1Δ double mutant. Although pal1Δ tea1Δ double mutants were incapable of colony formation at 36°C on YES plates (Figure 5C; top panel), the double mutants formed healthy colonies on YES plates containing 1.2 M sorbitol (Figure 5C; top panel). Microscopic examination of double mutants grown on sorbitol-containing plates revealed the establishment of a cylindrical morphology in these cells (Figure 5C; left panel). These double mutant cells, however, displayed a tea1Δ phenotype, because the osmolyte only suppressed cell wall-associated defects. Collectively, these studies established that the cell shape defects in pal1Δ cells might result from improper cell wall architecture. sla2Δ cells were also rendered capable of colony formation at 36°C in the presence of 1.2 M sorbitol (Figure 5D). Furthermore, electron microscopic analyses indicated that sla2Δ cells contained abnormally thickened cell walls (Figure 5A), as observed in the pal1Δ cells. These studies established that cells lacking Pal1p and Sla2p contained abnormal cell walls and the morphological phenotypes resulting from the loss of these proteins were suppressed by stabilization of the cell wall.
Pal1p Appears to Function Downstream of Sla2p in Promoting Polarized Growth
Given the similarity in the phenotypes of cells deleted for pal1 and sla2, the intracellular codistribution and the physical interactions, we addressed if Sla2p depended on Pal1p for its localization and vice versa. Sla2p localization was relatively unaffected in cells deleted for pal1 (Figure 6A, a and b). In contrast, the localization of Pal1p was significantly altered in cells deleted for sla2 in two ways (Figure 6Ad). First, a significantly elevated level of Pal1p was detected at the cell periphery in sla2Δ cells (compare cells in Figure 6Ac; wild-type and Figure 6Ad; sla2Δ). Second, ∼14% of sla2Δ cells displayed mislocalization of Pal1p, in that Pal1p was concentrated at the sides of the cell rather than the cell tips or the medial division site (Figure 6Ad, marked with arrowheads). Finally, in spherical sla2Δ cells Pal1p was detected over the entire cortex. Thus, Sla2p appears to be important for optimal localization of Pal1p at the cell tips and the division site and the localization dependencies suggest that Pal1p functions downstream of Sla2p. The localization epistasis was consistent with our isolation of pal1 as a high copy suppressor of the temperature-sensitive growth and morphology defects of sla2Δ cells. Interestingly, although sla2Δ cells expressing empty plasmids were unable to form colonies at 36°C, cells expressing sla2 or pal1 were able to form colonies at 36°C (Figure 6B). Furthermore, cylindrical morphology was largely restored in sla2Δ cells carrying multicopy plasmids expressing pal1 (Figure 6Cc; compared with cells in Figure 6C, a and b). sla2Δ cells suppressed by overproduction of Pal1p were largely incapable of new end growth (Figure 6Cc; marked with arrow), indicating that Sla2p function was essential for new end growth. These experiments suggested that Pal1p might function downstream of Sla2p in the maintenance of a cylindrical morphology and cell wall integrity, by participating in a subset of Sla2p functions.
Figure 6.
Dependency relationships between Sla2p and Pal1p. (A) Cells expressing functional GFP-tagged Sla2p in wild-type (a) and pal1Δ (b) backgrounds were grown in YES liquid medium and imaged by laser scanning confocal microscopy. Cells expressing functional GFP-tagged Pal1p in wild-type (c) and sla2Δ (d) backgrounds were grown in YES liquid medium supplemented with sorbitol and imaged by laser scanning confocal microscopy. Scale bar, 5 μm. (B) Viability of sla2Δ cells at high temperature is suppressed by multiple copies of pal1. sla2Δ cells carrying pal1 or sla2 plasmid were streaked on minimal medium without leucine supplement and were grown at 24°C (a) and 36°C (b) for 3 d. sla2Δ transformed with the empty vector, pTN-L1, was included as a control. (C) Microscopic analysis of sla2Δ cells transformed with vector (a), sla2 (b), and pal1 (c) plasmids. sla2Δ cells carrying vector, sla2, and pal1 plasmids were grown in minimal medium without leucine supplement at 24°C, shifted to 36°C for 8 h, and then stained with Calcofluor. Scale bar, 5 μm.
pal1Δ Cells Exhibit Various Growth Patterns Leading to Different Morphologies
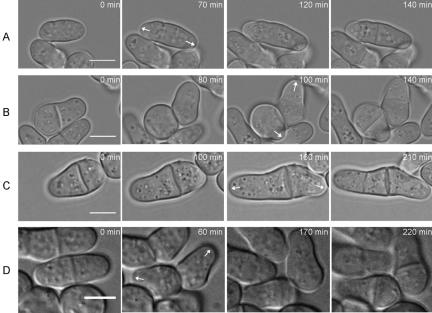
We have described Pal1p, a protein that interacts with Sla2p to regulate cell wall integrity and cellular morphogenesis. We have also described that pal1Δ cells exhibit a variety of morphological phenotypes, such as spherical, abnormal (pear shaped) and normal morphologies. We studied the behavior of cells by time-lapse microscopy to identify growth patterns that might shed light on the mechanism of formation of these various morphologies. Four major growth behaviors were noted. Cells with a cylindrical morphology were capable of bipolar growth as in the case of wild-type cells (Figure 7A). In instances where pear-shaped abnormal cells underwent septation, a spherical and a cylindrical daughter cell were generated (Figure 7B). Interestingly, we found that the spherical daughter cells made a cylindrical outgrowth and generated a tip at or near the new end, whereas the cylindrical daughter cell grew in either a monopolar (growing at the old end) or bipolar manner. In cells incapable of separation after septation, old end growth ensued and new end (branches) growth was not observed (Figure 7C). Finally, in instances where the two daughter cells were born with a pear shape (Figure 7D), both daughters grew from the old ends and did not grow from the new ends. These studies indicated that pal1Δ cells were capable of both old and new end growth, but that old end growth was more common. The exception to this was in spherically born pal1Δ cells that preferentially grew either at or near the new ends. These observations also provided a partial explanation for the mixture of phenotypes observed in pal1Δ cells.
Figure 7.
Growth patterns of pal1Δ cells. pal1Δ cells (>5 for each morphology) were imaged by time-lapse light microscopy on minimal medium agar pads at room temperature. (A) Bipolar growth pattern of a cylindrical cell. (B) Pattern of growth of the spherical and cylindrical cell generated by division of a pear-shaped cell. Note that the spherical cell generates a tip at or near the new end. (C) Pattern of growth of cells defective in separation. Note the old end growth and no new end (branching) growth. (D) Pattern of growth of pear-shaped cells generated by cell division. Note that both daughters carry out old end growth. Scale bar, 5 μm.
Spherical pal1Δ Cells Polarize in G2 to Establish Pear-shaped Morphology
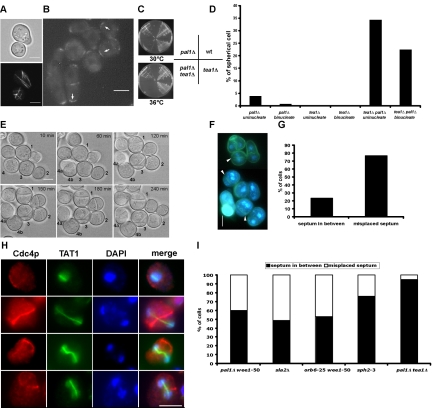
Interestingly, we found that spherical binucleate cells were rarely (<1%) seen, whereas ∼6% of the uninucleate cells were spherical in appearance (Figure 8A). The vast majority of uninucleate and binucleate cells were abnormal (pear shaped) in appearance (Figure 8A; >60% in both cases). The fact that spherical cells were rarely detected with two nuclei suggested that a mechanism might exist to polarize spherically born pal1Δ cells before mitosis. To test if this was the case, spherical pal1Δ cells were imaged by time-lapse microscopy (Figure 8B). Twelve of 15 spherical cells imaged underwent polarized growth, leading to the attainment of a pear-shaped morphology. Cells shown in Figure 8B were found to establish a partially cylindrical morphology and assemble division septa, indicative of successful completion of mitosis. To determine if the polarization of spherical cells took place in interphase (as opposed to during mitosis), we followed the repolarization in pal1Δ cells expressing Cdc13p-YFP (Figure 8C). Cdc13p, the predominant B-type cyclin in fission yeast accumulates in the nucleus in interphase and is transferred to the mitotic spindle and the spindle pole body in metaphase before eventual degradation at anaphase A (Decottignies et al., 2001). We found that the polarization in spherical cells always occurred in cells in which Cdc13p-YFP was detected in the nucleus, suggesting that the polarization event occurred in interphase, and possibly in G2, because this phase represents the majority of interphase in fission yeast. We then addressed if shortening of the G2 phase abrogated polarization of spherical pal1Δ cells. To this end, double mutants of the genotype pal1Δ wee1-50 were constructed. Wee1p regulates timing of entry into mitosis by inhibitory phosphorylation of Cdc2p and wee1 mutants exhibit a shortened G2 phase. Interestingly, we found that pal1Δ cells displayed synthetic lethality in combination with wee1-50 at 36°C (Figure 8D) and nearly 40% of binucleate cells were spherical in shape (Figure 8, E and F), whereas <6% of binucleate wee1-50 single mutants were spherical in morphology (Figure 8F). We therefore conclude that spherical pal1Δ cells polarize and achieve a partially cylindrical morphology before entry into mitosis in a Wee1p-dependent manner.
Figure 8.
Establishment of cylindrical morphology in spherical cells of pal1Δ. (A) Correlation of cell shape and nuclear number in pal1Δ cells. Cells were grown in minimal medium at 30°C to log phase, fixed, and stained with DAPI to visualize DNA. At least 600 cells each were counted for uninucleate and binucleate categories. (B) Time-lapse analysis of the growth of pal1Δ mutant. Cells were imaged by time-lapse light microscopy on minimal medium agar pads at room temperature. Time in minutes is indicated on the top right of each panel at the commencement of observation (t = 0). Cells are numbered for tracking, a and b are used to indicate the products of cell division. (C) Spherical shaped pal1Δ cells repolarize while the Cdc13-YFP signal is present in the nucleus. pal1Δ cells expressing Cdc13-YFP were imaged by time-lapse laser scanning confocal microscopy on YES medium agar pads at room temperature. (D) Wee1p is required for the viability and the repolarization of pal1Δ mutant. Wild-type, pal1Δ, wee1-50, and pal1Δ wee1-50 were streaked on YES agar plate and incubated for 2 d at 36°C. (E) Microscopic analysis of wee1-50 and pal1Δ wee1-50 mutants. Cells were stained with DAPI to show DNA. (F) Quantification of spherical cells with one or two nuclei in pal1Δ, wee1-50, and pal1Δ wee1-50 mutants. Cultures were grown in YES medium at 24°C to log phase and shifted to 36°C for 4 h. Cell samples were stained with DAPI and at least 300 cells were counted for each strain. Scale bar, 5 μm.
Kelch-repeat Protein Tea1p Is Required for Polarization of Spherical pal1Δ Cells
We have shown that spherically shaped pal1Δ cells polarize in G2 and become abnormally shaped with a clearly defined long and short axis before septation. We noticed that microtubules converged into the newly forming cylindrical projections that assemble on the spherical cell bodies (Figure 9A). The microtubule cytoskeleton is important for ensuring proper antipodal growth, although it is not required for establishment of cylindrical morphology (Beinhauer et al., 1997; Mata and Nurse, 1997; Sawin and Nurse, 1998; Browning et al., 2000). It was possible that microtubules were important for the establishment of cylindrical morphology in spherically born pal1Δ cells. Preliminary experiments revealed that pal1Δ cells were hypersensitive to low doses of MBC (8 μg/ml) and that pal1Δ cells were mostly spherical under these conditions (unpublished data). These observations suggested that the microtubule cytoskeleton is important for establishment of a cylindrical morphology in spherical mutants such as pal1Δ.
Figure 9.
Tea1p is essential for the viability and repolarization of pal1Δ cells. (A) Microtubules converge into the cylindrical projection during the repolarization of pal1Δ cells. pal1Δ cells expressing α-tubulin GFP were grown on minimal medium agar pad at room temperature and imaged by confocal microscopy. (B) Tea1p-YFP is spread over the cortex in spherical pal1Δ cells and localizes at tips generated in spherical pal1Δ cells. pal1Δ cells expressing Tea1p-YFP were used to visualize the localization of Tea1p-YFP. (C) Wild-type, pal1Δ, tea1Δ, and pal1Δ tea1Δ were streaked on YES agar plate and incubated for 2 d at 30 or 36°C. (D) Quantification of the percentages of spherical cells with one or two nuclei in pal1Δ, tea1Δ, and pal1Δ tea1Δ mutants. Cultures were grown in YES medium at 24°C to log phase and shifted to 36°C for 6 h. Cell samples were stained with DAPI and at least 300 cells were counted for each strain. (E) The growth of pal1Δ tea1Δ was imaged by time-lapse light microscopy on YES agar pads at room temperature. Time is indicated in minutes from the beginning of observation (t = 0). Cells are numbered for tracking, a and b are used to indicate the daughter cells produced after cell division. (F) Microscopic analysis of pal1Δ tea1Δ cells. Cells were stained with DAPI and aniline blue to visualize DNA and septum. Arrowheads indicate the septum. (G) Quantification of the percentages of spherical cells with properly positioned septum in between two nuclei or misplaced septum. (H) Localization of Cdc4p in pal1Δ tea1Δ double mutant. Exponentially growing cells were fixed with formaldehyde, processed for immunofluorescence, and stained with DAPI to visualize DNA, α-Cdc4p to visualize actomyosin ring, and α-TAT1 to visualize microtubules. Merge shows that the mitotic spindle (green) is not aligned properly with respect to the Cdc4p ring (red). (I) Septum is misplaced in different spherical mutants. pal1Δ wee1-50 and orb6-25 wee1-50 mutants were grown in YES medium at 24°C and shifted to 36°C for 5 h. sph2-3 cells were cultured in YES medium at 30°C. sla2Δ and pal1Δ tea1Δ mutants were grown in YES supplemented with 1.2 M sorbitol at 24°C and shifted to 36°C for 5 h. Scale bar, 5 μm. Except for sorbitol-grown pal1Δ tea1Δ, in which abnormal cells were counted, the phenotype in spherical cells is indicated.
Previous studies have established a strong (but not absolute) requirement for the microtubule cytoskeleton in localizing the polarity factor Tea1p (Mata and Nurse, 1997; Sawin and Nurse, 1998; Behrens and Nurse, 2002). Tea1p was localized in several spots over the cortex in spherical pal1Δ cells and was detected at the ends of abnormal as well as normal pal1Δ cells (Figure 9B, marked with arrows). It was therefore possible that Tea1p was also important in the polarization process in spherical pal1Δ cells. To address this question, pal1Δ tea1Δ double mutants were constructed. Although tea1Δ and pal1Δ cells were capable of growth and colony formation at 36°C, the double mutant was unable to form colonies at 36°C and grew poorly at 30°C (Figure 9C). Furthermore, the percentage of spherical uninucleate and binucleate cells increased dramatically in the double mutants. Although <1% of binucleate pal1Δ cells were spherical, ∼25% of binucleate pal1Δ tea1Δ cells were spherical at 36°C (Figure 9D). Time-lapse studies performed with the pal1Δ tea1Δ double mutants further confirmed the inability of a majority of spherical double mutants to polarize and these cells septated while still spherical (Figure 9E). These experiments led to two conclusions. First, Tea1p is important for polarization in spherical pal1Δ cells. Second, the loss of both Pal1p and Tea1p has a deleterious combinatorial effect, suggesting that the formation of cylindrical morphology is important for maximal cell viability.
Coordination between Mitosis and Cytokinesis Is Altered in Spherical Cells
We have shown a deleterious effect upon combining pal1Δ with tea1Δ. To understand the basis of this lethality, we shifted pal1Δ tea1Δ cells to 36°C, fixed, and stained with DAPI and aniline blue to visualize nuclei and septa, respectively. As controls, pal1Δ and tea1Δ cells were used. We noticed that a high proportion of double mutants, but not either of the single mutants, contained both DNA masses on one side of the division septum, leading to the production of an anucleate and a binucleate compartment. Such abnormally septated cells did not separate to produce anucleate and binucleate daughters (Figure 9, F and G). pal1Δ, tea1Δ, and pal1Δ tea1Δ cells were also fixed and stained with antibodies against Cdc4p and β-tubulin to visualize the actomyosin ring and microtubules. Normally the actomyosin ring and the anaphase B spindle are aligned at right angles to each other. Interestingly, in spherical cells of the double mutant this alignment was severely altered and strikingly some spindles did not intersect the plane of the actomyosin ring at all (Figure 9H), whereas in other cells the spindle was not placed perpendicular to the actomyosin ring. To establish that these defects in spatial regulation of cytokinesis were due to a spherical cell shape and not due to tea1Δ in the background, a variety of spherical mutants were scored for coordination of planes of mitosis and cytokinesis (Figure 9I). The mutants included pal1Δ wee1-50, sph2-3 (Sipiczki et al., 2000), sla2Δ, and orb6-25 wee1-50. We found that the mitotic and cytokinetic planes were not coordinated in spherical cells that were genotypically tea1+ as well as tea1-, although the extent of defects varied in these strains. Finally, defective spatial regulation of cytokinesis in the pal1Δ tea1Δ double mutant was almost completely suppressed by growth in medium containing sorbitol (Figure 9I). These experiments suggested that a spherical morphology does not allow for optimal spatial regulation of cytokinesis.
DISCUSSION
In this study we describe a mechanism of cellular polarization leading to the formation of a cylindrical cellular morphology in fission yeast. We describe the role of a novel protein Pal1p and its binding partner, Sla2p/End4p in cellular polarization, by modulation of cell wall integrity. Second, from the study of division patterns in pal1Δ cells, we describe an alternate pathway leading to cellular polarization in the absence of Pal1p. Finally, we show that a cylindrical morphology is important for spatial coordination of cytokinesis. We discuss these points in the subsequent sections.
Pal1p, a Novel Protein that Localizes to the Growing Cell Ends and the Division Site
We identified Pal1p as a relative of the budding yeast uncharacterized ORF, YDR348c, which has been shown to localize to the bud neck (Huh et al., 2003). Pal1p-related proteins are found in fungi, but not in plants or metazoans. As with the budding yeast YDR348c gene product, Pal1p is also detected at the cell division site. Interestingly, Pal1p is in addition detected at the growing ends of the cell. The localization of Pal1p is independent of F-actin and microtubule functions. The localization of Pal1p to the growing ends was established by its restriction to the growing end in cells growing in a monopolar manner and to both ends in G2 arrested cdc25-22 cells that grow in a bipolar manner. The localization of Pal1p to the ectopic cell ends generated in a tea1Δ mutant and the localization of Pal1p to a larger region of the cell cortex in spherically shaped cells with no clear long and short axis further established that Pal1p was associated with the growth zones. Thus, Pal1p might either mark the growth sites or be a part of the growth machinery itself in addition to marking the growth zones. Pal1p also localizes to the zone of cell fusion in mating cells. Thus, Pal1p function is associated with polarized growth during the vegetative life cycle and during cell fusion before meiosis.
Although the Pal1p sequence does not contain obvious signal sequences or prenylation motifs, Pal1p-GFP localization experiments suggested that Pal1p might be closely associated with the plasma membrane at the cell ends and the division site. Consistent with this, Pal1p was found in the membrane fraction in centrifugation experiments. The intracellular localization of Pal1p is very similar to the distribution of sterols in fission yeast (Wachtler et al., 2003; Takeda et al., 2004). Sterol-rich membrane-domains, termed lipid-rafts, are insoluble when extracted with buffers containing Triton X-100. We have been unable to ascertain the molecular mechanism of membrane association of Pal1p and if Pal1p is a component of lipid-rafts, given that Pal1p is only partially solubilized in buffers containing urea, bicarbonate, and salt (our unpublished observations) and is largely solubilized by treatment with Triton X-100.
Pal1p and Sla2p Are Important for the Maintenance of a Cylindrical Shape and Control Cell Wall Integrity
Cells deleted for pal1 display morphological abnormalities, suggesting a role for Pal1p in the maintenance of cylindrical cell morphology. Cells deleted for pal1 are largely pear-shaped, although spherical morphology is also commonly observed. What role might Pal1p play in cellular morphogenesis? The cell wall of budding and fission yeast cells plays a key role in the establishment of proper cell morphology. Furthermore, several spherical mutants define proteins important for cell wall metabolism in fission yeast (Ribas et al., 1991; Arellano et al., 1996; Hochstenbach et al., 1998; Katayama et al., 1999; Martin et al., 2003). We have shown abnormalities in the cell wall of pal1Δ cells by electron microscopy. Excessive deposition of cell wall material was revealed when thin sections were examined by electron microscopy. Although excessive cell wall deposition is observed in pal1Δ cells, it is likely that the cell wall is structurally defective. This is due to our observation that the deleterious consequences resulting from loss of Pal1p are effectively rescued by exogenous addition of sorbitol.
We have shown that Pal1p interacts with the Huntingtin-interacting-protein (Hip1; Wanker et al., 1997) -related molecule Sla2p/End4p in coimmunoprecipitation experiments. We roughly estimate that 5–10% of soluble pools of Sla2p and Pal1p associate physically based on the coimmunoprecipitation assay, although it is unclear if the antibodies or buffer conditions destabilize the interaction. Sla2p localizes to the regions of cell growth and division and is required for establishment and maintenance of a cylindrical morphology (this study and Castagnetti et al., 2005). We have also shown that cells deleted for sla2 display reduced cell wall integrity and the growth and morphological phenotype of sla2Δ is suppressed by growth on sorbitol medium. Furthermore, cell walls of sla2Δ cells, like pal1Δ cells, were thicker in appearance when studied by electron microscopy. Interestingly, the budding yeast sla2Δ also displays a thickened cell wall phenotype similar to S. pombe pal1Δ (Mulholland et al., 1997; Gourlay et al., 2003). Proteins related to Sla2p have been proposed to recruit additional molecules such as Sla1p, leading to Arp2/3-dependent polarized actin assembly (Li, 1997; Warren et al., 2002; Gourlay et al., 2003). It is therefore possible that Sla2p and Pal1p play a role in recruiting other proteins to the cell tips and the cell equator to aid F-actin assembly and to establish and maintain cylindrical morphology by remodeling of the cell wall. Alternatively, it is possible that Pal1p and Sla2p might be important for positioning the growth machinery, loss of which leads to targeting of the growth and the cell wall synthesizing machineries to improper sites and might not directly influence F-actin function. In both cases, Pal1p might provide a link between cellular membranes and cell polarization, given that part of Pal1p is membrane associated. In this context, it is interesting to note that the ANTH domain of budding yeast Sla2p interacts with membrane lipids such as phosphoinositide (4,5) bis-phosphate (Sun et al., 2005). Future studies should examine the regions of Pal1p and Sla2p that interact with each other as well as with cellular membranes.
Pal1p Appears to Function Downstream of Sla2p
We have shown that Pal1p and Sla2p localize to the sites of polarized growth and division in an actin and microtubule independent manner (this study for Pal1p and this study and Castagnetti et al., 2005, for Sla2p). We have also shown that although Sla2p localizes normally in pal1Δ cells, Pal1p localization is altered in cylindrical and abnormal sla2Δ cells. Thus, Pal1p localization depends on Sla2p function and might function downstream of Sla2p. Consistent with this idea, we have shown that overproduction of Pal1p suppresses the colony formation defect of sla2Δ cells at 36°C. The fission yeast and budding yeast Sla2 proteins are required for endocytosis (Gourlay et al., 2003; Holtzman et al., 1993; Wesp et al., 1997; Iwaki et al., 2004). Furthermore, Hip1R has been shown to interact with the endocytic protein clathrin in mammalian cells (Engqvist-Goldstein et al., 1999). Cells deleted for pal1, however, are not defective for endocytosis as assayed by the uptake of the lipophilic dye FM4-64 (2 μg/ml FM4-64 treatment for 30 min; unpublished data). Additionally, overproduction of Pal1p suppresses the colony formation and morphogenetic defects of sla2Δ cells, but not its endocytosis defects (unpublished data). Thus, it is likely that Pal1p is downstream of Sla2p only for a subset of functions of Sla2p. We have also shown that sla2Δ cells overproducing Pal1p are defective in new end growth as suggested recently by Castagnetti et al, (2005). Additional studies are required to ascertain if the Sla2p-Pal1p module functions largely to regulate old end growth.
Polarization of Spherical pal1Δ Cells Involves the Kelch-repeat Protein Tea1p
Although Pal1p is important for establishment of a cylindrical morphology, the proportion of pal1Δ cells that are spherical is low (∼5%). Interestingly, spherical pal1Δ cells with two nuclei are only rarely observed. These observations suggested that upon loss of Pal1p function, a second pathway might compensate for its loss and allow polarization. Consistent with this, the majority of pal1Δ cells are pearlike in appearance. The shape change from spherical to pearlike depends on Tea1p and occurs by formation of a “tip” at or near the new end generated by the previous septation event. In the absence of Tea1p, which is known to be important for new end growth, spherical pal1Δ cells are unable to carry out tip growth and over several generations the vast majority of cells accumulate and die with a spherical morphology. A similar mechanism also operates to alter the morphology of spherical cells of other orb mutants such as orb3-167, ras1Δ, and sph2-3 (TGC, WG, and MKB, unpublished observations), suggesting that the shape correction is not unique to pal1Δ. Several orb mutants also exhibit synthetic lethal interactions with tea1Δ mutants (orb3-167; TGC, WG, and MKB, unpublished; scd1Δ and ras1Δ; described by Papadaki et al., 2002). Thus, we propose that Tea1p becomes essential in spherical cells and that “tip” formation is the equivalent of new end growth in such spherical cells. Our studies are consistent with the proposal of multiple pathways of cell polarization (Feierbach et al., 2004) and with recent studies of Sawin and Snaith (2004), suggesting a role for Tea1p in resetting polarity.
Previous studies have shown that Tea1p and microtubules are important for proper antipodal growth of fission yeast cells (Snell and Nurse, 1994; Verde et al., 1995; Mata and Nurse, 1997; Sawin and Nurse, 1998), although they are not required for establishment of the cylindrical morphology per se (Sawin and Snaith, 2004). It is likely that Tea1p plays a major role in targeting the growth machinery to the tips generated in spherically born pal1Δ cells. An attractive possibility is that stochastic accumulation of Tea1p at the cell cortex to “critical” levels might allow F-actin and cell wall assembly, leading to tip growth. Although it seems likely that other molecules involved in microtubule-based polarization, such as Tip1p, Mal3p, Tea2p, Pom1p, Tea3p, and Tea4p (Bahler et al., 1998; Brunner and Nurse, 2000; Arellano et al., 2002; Browning et al., 2003; Busch and Brunner, 2004; Martin et al., 2005) should become essential when cylindrical morphology is compromised, future studies should establish if this is indeed the case.
Spherical Morphology Leads to Loss of Spatial Regulation of Cytokinesis
We have shown that pal1Δ tea1Δ double mutants are unable to form colonies under conditions in which both parents are capable of colony formation. Double mutants of the genotypes scd1Δ tea1Δ and ras1Δ tea1Δ have also been shown to display a synthetic lethal defect, although the basis of this lethality has not been fully characterized (Papadaki et al., 2002). We have shown that the spherical pal1Δ tea1Δ cells are unable to spatially coordinate mitosis and cytokinesis frequently, leading to the formation of a septated-binucleate cell in which both nuclei are placed on one side of the septum. Such defects are observed in spherical cells that are genotypically tea1+ (pal1Δ wee1-50, sla2Δ, sph2-3, orb6-25 wee1-50) as well as tea1- (pal1Δ tea1Δ and orb3-167 tea1Δ; unpublished data for orb3-167 tea1Δ). Furthermore, <5% of cytokinetic events are spatially impaired in pal1Δ tea1Δ cells, whose morphology is suppressed in medium containing the osmolyte sorbitol. We propose that it is the spherical shape, rather than the absence of Tea1p that leads to the observed defects in spatial regulation of cytokinesis, although a role for Tea1p in ring positioning in certain genetic backgrounds cannot be ruled out, given the low percentage of cells with spatial defects in cytokinesis in sorbitol suppressed pal1Δ tea1Δ mutants. It is likely that the lethality of spherical mutants defining essential genes (such as mor2, orb6, mob2, bgs3, etc.) might in part result from defective spatial coordination of cytokinesis, in addition to possible defects in cell wall assembly (Verde et al., 1998; Hirata et al., 2002; Hou et al., 2003; Martin et al., 2003). Interestingly, mutants defining genes important for division site placement, such as mid1Δ and pom1Δ (Chang et al., 1996; Sohrmann et al., 1996; Bahler and Pringle, 1998) are viable. It is possible that the cylindrical morphology of cells of the division site selection mutants (such as mid1Δ and pom1Δ) might largely ensure that mitotic events lead to the production of daughters with one nucleus each.
Spherical Morphology and the Cell Cycle
We have shown that spherical pal1Δ cells polarize in interphase, because polarizing cells contain an interphase array of microtubules and cyclin B (Cdc13p) is detected in the nucleus, as opposed to its presence on the spindle (metaphase) or lack of detectable localization (anaphase). We have also shown that shortening of G2 utilizing a wee1 mutation, leads to an increase in the number of spherical mitotic cells and lethality. Previous studies have shown that spherical S. pombe mutants, such as mor2, and orb6 (Hirata et al., 2002; Verde et al., 1998) are delayed in G2 in a Wee1p-dependent manner. This has been proposed to signify a morphogenesis checkpoint, although the functional end point of this checkpoint mechanism has not been fully explored. An attractive possibility is that some aspect of morphogenesis inactivates Wee1p, leading to entry into mitosis as has been proposed from studies in budding yeast (Lew, 2003). The formation of a tip might represent such a morphogenetic event that relieves the G2 delay. Future studies should assess if pal1Δ cells are delayed in G2 until tip formation and if tip formation in turn represents the functional end point upon morphogenetic checkpoint activation.
Acknowledgments
We thank Drs. V. Boulton, K. Gull, Yasushi Hiraoka, P. Nurse, R. Serrano, M. Sipiczki, and M. Yamamoto for strains and antibodies. We acknowledge the contribution of Ms. A. Bimbo, who constructed the tea1Δ strain. Special thanks are due to A. Bimbo, M. Mishra, V. Rajagopalan, S. Oliferenko, and all other members of the yeast and fungal laboratories for discussions and Drs. J. Karagiannis, D. McCollum, and Ms. A. Bimbo for critical reading of the manuscript. This work was supported by research funds from the Temasek Life Sciences Laboratory. V.W. and S.N.N. were supported by the Singapore Millennium Foundation.
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E04-11-0976) on June 22, 2005.
References
- Arellano, M., Duran, A., and Perez, P. (1996). Rho 1 GTPase activates the (1–3)beta-D-glucan synthase and is involved in Schizosaccharomyces pombe morphogenesis. EMBO J. 15, 4584-4591. [PMC free article] [PubMed] [Google Scholar]
- Arellano, M., Niccoli, T., and Nurse, P. (2002). Tea3p is a cell end marker activating polarized growth in Schizosaccharomyces pombe. Curr. Biol. 12, 751-756. [DOI] [PubMed] [Google Scholar]
- Bahler, J., and Pringle, J. R. (1998). Pom1p, a fission yeast protein kinase that provides positional information for both polarized growth and cytokinesis. Genes Dev. 12, 1356-1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian, M. K., Helfman, D. M., and Hemmingsen, S. M. (1992). A new tropomyosin essential for cytokinesis in the fission yeast S. pombe. Nature 360, 84-87. [DOI] [PubMed] [Google Scholar]
- Balasubramanian, M. K., McCollum, D., and Gould, K. L. (1997). Cytokinesis in fission yeast Schizosaccharomyces pombe. Methods Enzymol. 283, 494-506. [DOI] [PubMed] [Google Scholar]
- Behrens, R., and Nurse, P. (2002). Roles of fission yeast tea1p in the localization of polarity factors and in organizing the microtubular cytoskeleton. J. Cell Biol. 157, 783-793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beinhauer, J. D., Hagan, I. M., Hegemann, J. H., and Fleig, U. (1997). Mal3, the fission yeast homologue of the human APC-interacting protein EB-1 is required for microtubule integrity and the maintenance of cell form. J. Cell Biol. 139, 717-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning, H., Hayles, J., Mata, J., Aveline, L., Nurse, P., and McIntosh, J. R. (2000). Tea2p is a kinesin-like protein required to generate polarized growth in fission yeast. J. Cell Biol. 151, 15-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning, H., Hackney, D. D., and Nurse, P. (2003). Targeted movement of cell end factors in fission yeast. Nat. Cell Biol. 5, 812-818. [DOI] [PubMed] [Google Scholar]
- Brunner, D., and Nurse, P. (2000). CLIP170-like tip1p spatially organizes microtubular dynamics in fission yeast. Cell 102, 695-704. [DOI] [PubMed] [Google Scholar]
- Busch, K. E., and Brunner, D. (2004). The microtubule plus end-tracking proteins mal3p and tip1p cooperate for cell-end targeting of interphase microtubules. Curr. Biol. 14, 548-559. [DOI] [PubMed] [Google Scholar]
- Castagnetti, S., Behrens, R., and Nurse, P. (2005). End4/Sla2 is involved in establishment of a new growth zone in Schizosaccharomyces pombe. J. Cell Sci. 118, 1843-1850. [DOI] [PubMed] [Google Scholar]
- Chang, E. C., Barr, M., Wang, Y., Jung, V., Xu, H. P., and Wigler, M. H. (1994). Cooperative interaction of S. pombe proteins required for mating and morphogenesis. Cell 79, 131-141. [DOI] [PubMed] [Google Scholar]
- Chang, F., and Verde, F. (2003). Control of cell polarity and morphogenesis in fission yeast. In: The Molecular Biology of Schizosaccharomyces pombe: Genetics, Genomics and beyond, ed. R. Egel, Springer, 255-268.
- Chang, F., Woollard, A., and Nurse, P. (1996). Isolation and characterization of fission yeast mutants defective in the assembly and placement of the contractile actin ring. J. Cell Sci. 109, 131-142. [DOI] [PubMed] [Google Scholar]
- Decottignies, A., Zarzov, P., and Nurse, P. (2001). In vivo localisation of fission yeast cyclin-dependent kinase cdc2p and cyclin B cdc13p during mitosis and meiosis. J. Cell Sci. 114, 2627-2640. [DOI] [PubMed] [Google Scholar]
- Diaz, M., Sanchez, Y., Bennett, T., Sun, C. R., Godoy, C., Tamanoi, F., Duran, A., and Perez, P. (1993). The Schizosaccharomyces pombe cwg2+ gene codes for the beta subunit of a geranylgeranyltransferase type I required for beta-glucan synthesis. EMBO J. 12, 5245-5254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engqvist-Goldstein, A. E., Kessels, M. M., Chopra, V. S., Hayden, M. R., and Drubin, D. G. (1999). An actin-binding protein of the Sla2/Huntingtin interacting protein 1 family is a novel component of clathrin-coated pits and vesicles. J. Cell Biol. 147, 1503-1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feierbach, B., and Chang, F. (2001). Cytokinesis and the contractile ring in fission yeast. Curr. Opin. Microbiol. 4, 713-719. [DOI] [PubMed] [Google Scholar]
- Feierbach, B., Verde, F., and Chang, F. (2004). Regulation of a formin complex by the microtubule plus end protein tea1p. J. Cell Biol. 7, 697-707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin, A. C. et al. (2002). Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415, 141-147. [DOI] [PubMed] [Google Scholar]
- Gourlay, C. W., Dewar, H., Warren, D. T., Costa, R., Satish, N., and Ayscough, K. R. (2003). An interaction between Sla1p and Sla2p plays a role in regulating actin dynamics and endocytosis in budding yeast. J. Cell Sci. 116, 2551-2564. [DOI] [PubMed] [Google Scholar]
- Heitz, M. J., Petersen, J., Valovin, S., and Hagan, I. M. (2001). MTOC formation during mitotic exit in fission yeast. J. Cell Sci. 114, 4521-4532. [DOI] [PubMed] [Google Scholar]
- Hirata, D. et al. (2002). Fission yeast Mor2/Cps12, a protein similar to Drosophila Furry, is essential for cell morphogenesis and its mutation induces Wee1-dependent G(2) delay. EMBO J. 21, 4863-4874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochstenbach, F., Klis, F. M., van den Ende, H., van Donselaar, E., Peters, P. J., and Klausner, R. D. (1998). Identification of a putative alpha-glucan synthase essential for cell wall construction and morphogenesis in fission yeast. Proc. Natl. Acad. Sci. USA 95, 9161-9166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtzman, D. A., Yang, S., and Drubin, D. G. (1993). Synthetic-lethal interactions identify two novel genes, SLA1 and SLA2, that control membrane cytoskeleton assembly in Saccharomyces cerevisiae. J. Cell Biol. 122, 635-644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou, M. C., Wiley, D. J., Verde, F., and McCollum, D. (2003). Mob2p interacts with the protein kinase Orb6p to promote coordination of cell polarity with cell cycle progression. J. Cell Sci. 116, 125-135. [DOI] [PubMed] [Google Scholar]
- Huh, W. K., Falvo, J. V., Gerke, L. C., Carroll, A. S., Howson, R. W., Weissman, J. S., and O'Shea, E. K. (2003). Global analysis of protein localization in budding yeast. Nature 425, 686-691. [DOI] [PubMed] [Google Scholar]
- Iwaki, T., Tanaka, N., Takagi, H., Giga-Hama, Y., and Takegawa, K. (2004). Characterization of end4+, a gene required for endocytosis in Schizosaccharomyces pombe. Yeast 21, 867-881. [DOI] [PubMed] [Google Scholar]
- Katayama, S., Hirata, D., Arellano, M., Perez, P., and Toda, T. (1999). Fission yeast alpha-glucan synthase Mok1 requires the actin cytoskeleton to localize the sites of growth and plays an essential role in cell morphogenesis downstream of protein kinase C function. J. Cell Biol. 144, 1173-1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli, U. K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680-685. [DOI] [PubMed] [Google Scholar]
- Le Goff, X., Utzig, S., and Simanis, V. (1999). Controlling septation in fission yeast: finding the middle, and timing it right. Curr. Genet. 35, 571-584. [DOI] [PubMed] [Google Scholar]
- Lew, D. J. (2003). The morphogenesis checkpoint: how yeast cells watch their figures. Curr. Opin. Cell Biol. 15, 648-653. [DOI] [PubMed] [Google Scholar]
- Li, R. (1997). Bee1, a yeast protein with homology to Wiscott-Aldrich syndrome protein, is critical for the assembly of cortical actin cytoskeleton. J. Cell Biol. 136, 649-658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks, J., Hagan, I. M., and Hyams, J. S. (1986). Growth polarity and cytokinesis in fission yeast: the role of the cytoskeleton. J. Cell Sci. 5, 229-241. [DOI] [PubMed] [Google Scholar]
- Martin S. G., McDonald W. H., Yates J. R., 3rd., and Chang, F. (2005). Tea4p links microtubule plus ends with the formin for3p in the establishment of cell polarity. Dev. Cell 8, 479-491. [DOI] [PubMed] [Google Scholar]
- Martin, V., Garcia, B., Carnero, E., Duran, A., and Sanchez, Y. (2003). Bgs3p, a putative 1,3-beta-glucan synthase subunit, is required for cell wall assembly in Schizosaccharomyces pombe. Eukaryot. Cell 2, 159-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mata, J., and Nurse, P. (1997). Tea1 and the microtubular cytoskeleton are important for generating global spatial order within the fission yeast cell. Cell 89, 939-949. [DOI] [PubMed] [Google Scholar]
- Miller, P. J., and Johnson, D. I. (1994). Cdc42p GTPase is involved in controlling polarized cell growth in Schizosaccharomyces pombe. Mol. Cell Biol. 14, 1075-1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison, J. M., and Nurse, P. (1985). Growth in cell length in the fission yeast Schizosaccharomyces pombe. J. Cell Sci. 75, 357-376. [DOI] [PubMed] [Google Scholar]
- Mulholland, J., Wesp, A., Riezman, H., and Botstein, D. (1997). Yeast actin cytoskeleton mutants accumulate a new class of Golgi-derived secretary vesicle. Mol. Biol. Cell 8, 1481-1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura, T., Nakamura-Kubo, M., Hirata, A., and Shimoda, C. (2001). The Schizosaccharomyces pombe spo3+ gene is required for assembly of the fore-spore membrane and genetically interacts with psy1+-encoding Syntaxin-like protein. Mol. Biol. Cell 12, 3955-3972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen, O., and Davey, J. (1995). Pheromone communication in the fission yeast Schizosaccharomyces pombe. Semin. Cell Biol. 6, 95-104. [DOI] [PubMed] [Google Scholar]
- Okazaki, K., Okazaki, N., Kume, K., Jinno, S., Tanaka, K., and Okayama, H. (1990). High-frequency transformation method and library transducing vectors for cloning mammalian cDNAs by trans-complementation of Schizosaccharomyces pombe. Nucleic Acids Res. 18, 6485-6489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottilie, S., Miller, P. J., Johnson, D. I., Creasy, C. L., Sells, M. A., Bagrodia, S., Forsburg, S. L., and Chernoff, J. (1995). Fission yeast pak1+ encodes a protein kinase that interacts with Cdc42p and is involved in the control of cell polarity and mating. EMBO J. 14, 5908-5919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardo, M., and Nurse, P. (2003). Equatorial retention of the contractile actin ring by microtubules during cytokinesis. Science 300, 1569-1574. [DOI] [PubMed] [Google Scholar]
- Rajagopalan, S., Wachtler, V., and Balasubramanian, M. (2003). Cytokinesis in fission yeast: a story of rings, rafts and walls. Trends Genet. 19, 403-408. [DOI] [PubMed] [Google Scholar]
- Radcliffe, P., Hirata, D., Childs, D., Vardy, L., and Toda, T. (1998). Identification of novel temperature-sensitive lethal alleles in essential β-tubulin and nonessential α2-tubulin genes as fission yeast polarity mutants. Mol. Biol. Cell 9, 1757-1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribas, J. C., Diaz, M., Duran, A., and Perez, P. (1991). Isolation and characterization of Schizosaccharomyces pombe mutants defective in cell wall (1–3)beta-D-glucan. J. Bacteriol. 173, 3456-3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadaki, P., Pizon, V., Onken, B., and Chang, E. C. (2002). Two ras pathways in fission yeast are differentially regulated by two ras guanine nucleotide exchange factors. Mol. Cell Biol. 22, 4598-4606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popov, N., Schmitt, M., Schulzeck, S., and Matthies, H. (1975). Reliable micromethod for determination of the protein content in tissue homogenates. Acta Biol. Med. Ger. 34, 1441-1446. [PubMed] [Google Scholar]
- Sawin, K. E., and Nurse, P. (1998). Regulation of cell polarity by microtubules in fission yeast. J. Cell Biol. 142, 457-471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawin, K. E., and Snaith, H. A. (2004). Role of microtubules and tea1p in establishment and maintenance of fission yeast cell polarity. J. Cell Sci. 117, 689-700. [DOI] [PubMed] [Google Scholar]
- Schneppenheim, R., Budde, U., Dahlmann, N., and Rautenberg, P. (1991). Luminography—a new, highly sensitive visualization method for electrophoresis. Electrophoresis 12, 367-372. [DOI] [PubMed] [Google Scholar]
- Serrano, R., Monk, B. C., Villalba, J. M., Montesinos, C., and Weiler, E. W. (1993). Epitope mapping and accessibility of immunodominant regions of yeast plasma membrane H(+)-ATPase. Eur. J. Biochem. 212, 737-744. [DOI] [PubMed] [Google Scholar]
- Sipiczki, M., Yamaguchi, M., Grallert, A., Takeo, K., Zilahi, E., Bozsik, A., and Miklos, I. (2000). Role of cell shape in determination of the division plane in Schizosaccharomyces pombe: random orientation of septa in spherical cells. J. Bacteriol. 182, 1693-1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snaith, H. A., and Sawin, K. E. (2003). Fission yeast mod5p regulates polarized growth through anchoring of tea1p at cell tips. Nature 423, 647-651. [DOI] [PubMed] [Google Scholar]
- Snell, V., and Nurse, P. (1994). Genetic analysis of cell morphogenesis in fission yeast—a role for casein kinase II in the establishment of polarized growth. EMBO J. 13, 2066-2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohrmann, M., Fankhauser, C., Brodbeck, C., and Simanis, V. (1996). The dmf1/mid1 gene is essential for correct positioning of the division septum in fission yeast. Genes Dev. 10, 2707-2719. [DOI] [PubMed] [Google Scholar]
- Sun, Y., Kaksonen, M., Madden, D. T., Schekman, R., and Drubin, D. G. (2005). Interaction of Sla2p's ANTH domain with PtdIns(4,5)P2 is important for actin-dependent endocytic internalization. Mol. Biol. Cell 16, 717-730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda, T., Kawate, T., and Chang, F. (2004). Organization of a sterol-rich membrane domain by cdc15p during cytokinesis in fission yeast. Nat. Cell Biol. 6, 1142-1144. [DOI] [PubMed] [Google Scholar]
- Tran, P. T., Marsh, L., Doye, V., Inoue, S., and Chang, F. (2001). A mechanism for nuclear positioning in fission yeast based on microtubule pushing. J. Cell Biol. 153, 397-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verde, F., Mata, J., and Nurse, P. (1995). Fission yeast cell morphogenesis: identification of new genes and analysis of their role during the cell cycle. J. Cell Biol. 131, 1529-1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verde, F., Wiley, D. J., and Nurse, P. (1998). Fission yeast orb6, a ser/thr protein kinase related to mammalian rho kinase and myotonic dystrophy kinase, is required for maintenance of cell polarity and coordinates cell morphogenesis with the cell cycle. Proc. Natl. Acad. Sci. USA 95, 7526-7531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wachtler, V., Rajagopalan, S., and Balasubramanian, M. K. (2003). Sterol-rich plasma membrane domains in the fission yeast Schizosaccharomyces pombe. J. Cell Sci. 116, 867-874. [DOI] [PubMed] [Google Scholar]
- Wang, H., Tang, X., Liu, J., Trautmann, S., Balasundaram, D., McCollum, D., and Balasubramanian, M. K. (2002). The multiprotein exocyst complex is essential for cell separation in Schizosaccharomyces pombe. Mol. Biol. Cell 13, 515-529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanker, E. E., Rovira, C., Scherzinger, E., Hasenbank, R., Walter, S., Tait, D., Colicelli, J., and Lehrach, H. (1997). HIP-I: a huntingtin interacting protein isolated by the yeast two-hybrid system. Hum. Mol. Genet. 6, 487-495. [DOI] [PubMed] [Google Scholar]
- Warren, D. T., Andrews, P. D., Gourlay, C. G., and Ayscough, K. R. (2002). Sla1p couples the yeast endocytic machinery to proteins regulating actin dynamics. J. Cell Sci. 115, 1703-1715. [DOI] [PubMed] [Google Scholar]
- Wessel, D., and Flugge, U. I. (1984). A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal. Biochem. 138, 141-143. [DOI] [PubMed] [Google Scholar]
- Wesp, A., Hicke, L., Palecek, J., Lombardi, R., Aust, T., Munn, A., and Riezman, H. (1997). End4p/S1a2p interacts with actin-associated proteins for endocytosis in Saccharomyces cerevisiae. Mol. Biol. Cell 8, 2291-2306. [DOI] [PMC free article] [PubMed] [Google Scholar]