Abstract
The function of Tricornered (Trc), the Drosophila Ndr (Nuclear Dbf2-related) serine/threonine protein kinase, is required for the normal morphogenesis of a variety of polarized outgrowths including epidermal hairs, bristles, arista laterals, and dendrites. In yeast the Trc homolog Cbk1 needs to bind Mob2 to activate the RAM pathway. In this report, we provide genetic and biochemical data that Drosophila Trc also interacts with and is activated by Drosophila Dmob proteins. In addition, Drosophila Mob proteins appear to interact with the related Warts/Lats kinase, which functions as a tumor suppressor in flies and mammals. Interestingly, the overgrowth tumor phenotype that results from mutations in Dmob1 (mats) was only seen in genetic mosaics and not when the entire animal was mutant. We conclude that unlike in yeast, in Drosophila individual Mob proteins interact with multiple kinases and that individual NDR family kinases interact with multiple Mob proteins. We further provide evidence that Mo25, the Drosophila homolog of the RAM pathway hym1 gene does not function along with Trc.
INTRODUCTION
tricornered (trc) encodes the Drosophila Ndr protein kinase (Nuclear Dbf2 related; Geng et al., 2000). The Ndr kinases are members of a subfamily of serine/threonine kinases that includes Sax1 (Caenorhabditis elegans), Cbk1 (Saccharomyces cerevisiae), Dbf2 (S. cerevisiae), Warts/Lats (Drosophila), Orb6 (Schizosaccharomyces pombe) and Cot-1 (Neurospora), which regulate cell growth, cell division and cell morphology (Yarden et al., 1992; Justice et al., 1995; Xu et al., 1995; Verde et al., 1998; Zallen et al., 2000). In S. cerevisiae Cbk1 and Dbf2/Dbf20 play central roles in the RAM (regulation of AceII activity and cellular morphogenesis) and MEN (mitotic exit network) pathways (Mah et al., 2001; Nelson et al., 2003). Mutants of Cbk1 or other RAM pathway genes including its binding partners Mob2 and Tao3 (Pag1) fail to activate the AceII transcription factor in daughter cells and result in rounder than normal cells due to a defect in axial growth of the bud (Colman-Lerner et al., 2001; Du and Novick, 2002; Nelson et al., 2003). Little is known about the in vivo function of the two human Ndr genes but extensive study of their biochemical characteristics has been carried out for nearly 10 years (Millward et al., 1995; Stegert et al., 2004). trc is the Sole Ndr Gene in Drosophila. The function of trc and its partner furry (fry) is required for the development of epidermal hairs, sensory bristles, arista laterals, and dendrite arborization (da) sensory neuron dendrites (Geng et al., 2000; Cong et al., 2001; He and Adler, 2001; Emoto et al., 2004). The morphogenesis of these cell extensions involves the regulated activation of both the actin and microtubule cytoskeletons (Fristrom et al., 1993; Wong and Adler, 1993; Verheyen and Cooley, 1994; Eaton et al., 1996; Hopmann et al., 1996; Turner and Adler, 1998; Tilney et al., 2000; Geng et al., 2000; He and Adler, 2001). Mutations in trc and fry result in split and multipled hairs and laterals, split and deformed bristles and dendrites with extra branching and tiling defects (Geng et al., 2000; Cong et al., 2001; Emoto et al., 2004).
The single warts/large tumor suppressor (wts/lats) gene is the Drosophila kinase most closely related to trc (45% identical and 65% similar over 418 amino acids). Once again there are two wts homologues in mammals. There is no wts ortholog in yeast. wts was first identified in Drosophila as a tumor suppressor (Justice et al., 1995; Xu et al., 1995). Homozygous wts/lats mutant cells display defects in morphogenesis (such as deformed bristles and altered cuticle morphology) and extensive overgrowths (Justice et al., 1995; Xu et al., 1995).
Ndr, like many kinases is regulated by phosphorylation. The phosphorylation of the activation segment site Ser-281 and the hydrophobic motif site Thr-444 of Ndr increase Ndr kinase activity in vitro (Millward et al., 1999). Ser-281 phosphorylation is thought to be due to autophosphorylation, whereas Thr-444 is targeted by an as yet unidentified upstream kinase (Tamaskovic et al., 2003; Stegert et al., 2004). These sites are also important regulatory sites for Trc function in the Drosophila epidermis and nervous system. The mutation of these sites in trc to ala resulted in dominant negative proteins (Emoto et al., 2004; He et al., 2005).
Several Ndr family kinases have been shown to function with members of the Furry protein family, which consists of large conserved proteins that lack informative motifs. The first member of this family to be characterized was the Drosophila fry gene (Cong et al., 2001). Both genetic and biochemical experiments have shown that in flies trc and fry function in a common process, are present together in a complex and that Fry is required for Trc kinase activity (Cong et al., 2001; Emoto et al., 2004; He et al., 2005). Mutations in both result in similar phenotypes in both the epidermis and sensory neurons. In addition, the subcellular localization/accumulation of Trc and Fry is interdependent in pupal wing cells (He et al., 2005). The subcellular localization of Cbk1 and Tao3 in S. cerevisiae and Orb6 and Mor2 in S. pombe (Du and Novick, 2002; Hirata et al., 2002; Nelson et al., 2003) has also been found to be interdependent, although the relationships differ in these systems (Tao3 and Mor2 are the Fry homologues in these systems).
The Trc and Furry family proteins appear to be conserved both in terms of sequence and function in a wide range of eukaryotes. This suggests that homologues of other members of the RAM pathway in S. cerevisiae will also play similar roles in higher eukaryotes. The Mob2 protein of yeast has been shown to bind to Cbk1 and be essential for Cbk1 kinase activity (Weiss et al., 2002). In vivo Mob2 is required along with Cbk1 for both mother/daughter separation after cytokinesis and the maintenance of polarized cell growth (Weiss et al., 2002). Furthermore, Mob2 and Cbk1 show interdependent localization (Nelson et al., 2003). A similar situation exists for the related Dbf2 kinase, which is a component of the mitotic exit network (MEN). Dbf2 binds to Mob1 (which is related to Mob2) and this complex is essential for activity (Mah et al., 2001). Similarly, S. pombe Mob2 interacts physically with the Orb6 protein kinase and is required for Orb6 function in the coordination of cell polarity with the cell cycle (Hou et al., 2003). Multicellular organisms possess multiple mob genes. Recently, it was shown that a basic sequence within the insert in the catalytic domain of Ndr has an autoinhibitory function and that Human Mob1 may stimulate Ndr activity by releasing the autoinhibitory effect of this sequence (Bichsel et al., 2004; Devroe et al., 2004).
There are 4 Drosophila genes related to the yeast mob genes. Evidence for a two-hybrid interaction between CG13852 (Dmob1/mats) and Trc was described in a genome scale experiment; however; no evidence for such interactions were seen between Trc and any of the other Drosophila Mobs (Giot et al., 2003). Nor was there any indication in that paper that any of the Drosophila Mobs function with the related Warts/Lats kinase. In this article we provide evidence that Mob1 interacts with both trc and wts and that at least one additional member of the Dmob gene family CG11711 (Dmob2) can interacts with trc and wts. While this paper was in revision Lai et al. (2005) reported that CG13852 interacted with and activated Wts. They named CG13852 mats, and we will follow their lead and use that name.
MATERIALS AND METHODS
Plasmid Constructs
Mutagenesis. For the Dmob2 mutants generation, the complete Dmob2 EST clone: RE70633 was used as the template. The following oligonucleotides were used to introduce amino acids substitution with the QuickChange Multiplesites Mutagenesis kit (Stratagene, La Jolla, CA): C161W: TGAATGCTAGCGTCACACCGCCTCGCTCTATTTG; C179L: CTATGGAACTATTAGTGAATTTTTGACACAATCCGGTTGCGCTGAC; E151K: CCAGCTGGTCCGGACTACAATGAATGGCCAGAGTCACACACCC; H261Q: CATGTGATAGCGCAACTGTATGCGGCGC; I15V: CGGGCGGCAATCGCATTAGCCGTGCGATC; N280S: CGGCTTGCACACCCATCTAACGCTGACTTTCGCGCACCTC; N65I: CCTTCATGCCTCTTCGGATCACATTCCCGGACGC; Q167R: CCTCGCTCTATTTGAGCAAGTGAATTTGGTCTATGG; T85P: GCTAGTGGGGGACCAGGTGGCACGG; Y193H: GACCGGACCCGGCAACGAAACGTACCCATGGTTCGACGAGAAGG.
All the Dmob2 constructs were subcloned into pGADT7 to test their interaction with trcpGBKT7 using NdeI and EcoRI restriction enzyme sites. The primers used were RE70633NdeI: sense: GGCCTTCCATATGAACTGGGCCATTTC and RE70633EcoRI-antisense: GTGGGAATTCCTACCCCTGTTGCATTC.
trc Constructs Subcloning. Full-length of trc cDNA was subcloned into pGBKT7 from NdeI-BglII. The following primers were used: trc-5′-NdeI: GGGAATTCCATATGCACCATCACCATC; trc-3′-EcoRI: CCGGAATTCTCACTCCAAATTTCGCAC. To get truncated forms of trc cDNA, we used the same trc-5′-NdeI primer and the following 3′ primers (each containing EcoRI site): trcD1: CCGGAATTCTAGCTCACGTATGTGCTCCCAGTC; trcD2: CCGGAATTCTAGGATTGTCCGAGCAGAATGG; trcD3: CCGGAATTCTAGGCACAGTCCGAAGTCGGAGAG; trcD4: CCGGAATTCTAGCGTCATCATATCACCACCAGG; trcD5: CCGGAATTCTAGGTACACATGTCCCGTG; trcD6: CCGGAATTCTAGGCTCTCGTCCTTCAGCTG.
The PCR products were subcloned into pGBKT7 from NdeI-EcoRI sites. To subclone wild-type and mutant forms of trc cDNA into 3xFLAGpCMV7.1, Trc-5′-EcoRI: CCAAGAATTCATGCACCATCACCATCACC; Trc-3′-BglII: GGAAGATCTTCACTCCAAATTTCGCACCTCG.
mats and wts Subcloning for Two-hybrid Assays. The mats cDNA clone LD47553 was used as a template and subcloned into pGADT7 with NdeI and EcoRI after PCR using the following primers. Mats5: GGGAATTCCATATGATGGACTTCTTGTTCGGTTC; Mats3: CCGGAATTCCTATATCTGCCGCTCATC.
The wts cDNA clone SD19495 was used as a template and subcloned into the NdeI and EcoRI sites of pGBKT7 after PCR using the following primers: Wts5: GGGAATTCCATATGAGCAGCAGCATCATGCATCC; Wts3: CCGGAATTCGATGAGCAGCGACATCAG.
Dmob2 and Dmo25 Constructs Subcloning. Dominant negative Dmob2 and Dmo25 constructs were produced by cloning N- or C-terminally truncated subfragments generated by PCR into pUAST. EST clones, GH01659 and GH07469, (Research Genetics, Huntsville, AL) were used as full-length cDNA templates. The primers and restriction sites used are as follows: for truncations of Dmob2 (5′-EcoRI and 3′-BglII): Dmob2-N-5′-EcoRI: CCGGAATTCATGAACTGGGCCATTTC; Dmob2-N-3′-BglII: GGAAGATCTCTACGCCGCATACAGATGC; Dmob2-C-5′- EcoRI: CCGGAATTCATGGTGATAGCGCATC; Dmob2-C-3′-BglII: CGTGGAGATCTCTACCCCTGTTGCATTC. For truncations of Dmo25 (5′-EcoRI and 3′-BglII): Dmo25-N-5′-EcoRI: CCGGAATTCATGCCACTGTTCG; Dmo25-N-3′-BglII: GGAAGATCTCTACTCCACGTAGCGGAAG.
Tagging of Dmob2 (CG11711). Dmob2 (CG11711) was tagged as its C-terminus with CFP or 8xHA by a PCR using both EST clone,GH07469, which expressed the long form of Dmob2 (CG11711), and CFP and 8xHA complete sequence, which was amplified from the ECFPpCMV5 (Clontech, Palo Alto, CA) and 8xHAPag1-N (Provided by Novick lab) as templates. For Dmob2 tagged with 8xHA, GH07469 was modified by insertion of NdeI and HindIII sites before the stop codon and 5′-XhoI and 3′-XbaI sites at both ends. The primers used are as follows: GH07469-C-8× HA (NdeI-HindIII)-XbaI-CTAGTCTAGACTAAAGCTTGAATTCCATATGTGCCGTGGTGGTCG; GH07469-C-8× HA (NdeI-HindIII)-XhoI-CGATTAATTTCTTCTCGAGATGGGCAAGGCCCGTC.
The PCR product was subcloned into pBS vector. Simultaneously, NdeI and HindIII restriction sites were added at both ends of the 8xHA sequence amplified from 8xHAPag1 template and subcloned into GH07469pBS. The resulting XhoI-GH07469–8xHA-XbaI was then cut from pBS and subcloned into pUAST using XhoI-XbaI sites.
For Dmob2 tagged with CFP, PCR products of GH07469 with 5′ sequence of ECFP at 3′ end and ECFP with 3′ sequence at 5′ end were made to be templates to get the final C-terminal tagged Dmob2. 5′ primer of GH07469 and 3′ primer of ECFP also contained a XhoI site and a XbaI site separately for subcloning into pUAST. Primers used are as follows: GH07469-C-ECFP: 5′-GATTGGCTTCTCGAGATGGGCAAG; GH07469-C-ECFP: 3′-CTTGCTCACTGCCGTGGTGGTC; ECFPpCMV5-C: 5′-GACCACCACGGCAGTGAGCAAG; ECFPpCMV5-C: 3′-GACCGGCGCTCAGTTGGAATTC.
Genetics
Flies were grown on standard media at 25°C. Mutant stocks were either obtained from the stock center at Indiana University, generated in Charlottesville or generously supplied by Zhi-Chun Lai (Lai et al., 2005) or Peter Bryant (Justice et al., 1995). Genetic interaction tests were done by individually crossing male ap (or ptc)-Gal4/+; UAS-trcT453A flies to virgin female flies that carried mutations or deficiencies. Clones were generated by FLP/FRT technology (Xu and Rubin, 1993). Deficiency stocks used in the interaction and mapping experiments included the following: Df(3L)vin5, Df(3L)vin4, Df(3L)vin2, Df(3L)vin66, Df(3L)lxd6, Df(3L)vin6, Df(3L)vin7, Df(3L)ED4470, Df(3L)ED4475 and Df(3L)BK9 (for Dmob2), Df(2R)42 and Df(2R)ED1552 (for CG3403), Df(2L)J2 and Df(2L)Exel6026 (for CG4946), Df(3R)hh and Df(3R)Exel6191 (for CG13852). The warts (wts)/large tumor suppressor (lats) gene is known by both names. In line with FlyBase usage, we use warts.
Immunohistochemistry
Immnuostaining was done using standard techniques as described previously (He et al., 2005). Monoclonal anti-FLAG antibody was purchased from Sigma (St. Louis, MO). Anti-GFP polyclonal antibody was purchased from Molecular Probes (Eugene, OR). Secondary antibodies and labeled phalloidin were purchased from Molecular Probes. The anti-Fry antibody has been described previously (He et al., 2005).
Cell Culture and Coimmunoprecipitation
Immunoprecipitations used S2 cells and were done as described previously (Emoto et al., 2004; He et al., 2005).
Microscopy and Photography
Confocal microscopy was done using either a Nikon Laser Scanning confocal microscope (Melville, NY) or an Atto CARV spinning disk confocal attached to a Nikon microscope. Bright field images of wings were obtained using a Spot digital camera (National Diagnostics, Manville, NJ) on a Zeiss Axioskop microscope (Thornwood, NY). Adobe Photoshop (San Jose, CA) was used to compose figures.
Scoring of Mutant Wings
Wings were mounted in Euparal (Asco Labs, Manchester, England) and examined under bright-field microscopy using approached described previously (Wong and Adler, 1993). Because the wing is not effected by the mutants to a uniform level throughout the whole region, the same area within the C region (5 × 20 cells) was chosen for scoring the number of hair per wing cell for ap-GAL4- or ptc-GAL4-driven UAS-mutants.
Scoring of Denticle Phenotypes
We counted the number of split denticles in segments 3–7 of second instar larvae. We used second instar larvae for quantifying the phenotypes because some genotypes of interest yielded very few third instar larvae. The number of denticles varies in different segments, so we only counted larvae in which we could count all of the relevant segments. We found only small differences (nonsignificant) in the number of denticles between most of the relevant genotypes. The one exception was trcP matsPB, which contained ∼15% fewer denticles per segment. To correct for the effect of this on the number of split denticles we did the following normalization. We determined the average number of denticles in abdominal segment 6 for each genotype and normalized the number of split denticles for each by dividing the number observed by the ratio of the number of denticles in the relevant genotype to Oregon R. Strong wts mutants died before the second instar, so we could not compare denticle phenotype to that of the other mutants.
Yeast Two-hybrid Assay
For the yeast two-hybrid screen, we used trc wild-type full-length cDNA and dominant negative form (trcT453A) as a bait subcloned into the Matchmaker GAL4 Two-Hybrid System 3 vector pGBKT7 (Clontech). This was used to screen a cDNA library made from RNA of 0–21h old Drosophila embryos (Clontech). For the analysis of interaction between defined protein partners, yeast strain AH109 was cotransformed with two-hybrid plasmids (pGBKT7 carrying a DBD fusion and a TRP1 marker, and pGADT7 carrying an AD fusion and a LEU2 marker). We followed the directions provided by Clontech. In these experiments we scored three reporter genes (ADE2, HIS3, and MEL1/lacZ), assessing both growth on dropout medium (SD/-Ade/-His/-Leu/-Trp) and the α-galactosidase/β-galactosidase activity. A positive test for all was required for us to score two proteins as interacting.
Real Time PCR
We used real time RT-PCR to quantify the amount of Mo25 mRNA. RNA was isolated from either Oregon-R or Dmo25P/Df using the Trizol reagent. Ten- to 20-s instar larvae were ground in a 1.5-ml microfuge tube after freezing in liquid nitrogen. Trizol reagent was added and mixed, and linear acrylamide was added followed by a chloroform extraction and isopropanol precipitation. Before reverse transcriptase the sample was treated with DNase I. We used Oligo dT priming for RT using AMV reverse transcriptase (Roche, Indianapolis, IN). The real time PCR was carried out in a Cepheid Smart Cycler system SC-1600 (Sunnyvale, CA). For Dmo25 we used the following primers: Dmo25-rp: forward: ATGCCACTGTTCGGGAAGTC; Dmo25-rp: reverse: GCAGCTTCAAGCTCTGACGC.
We used rp49 as a control for RNA content. For rp49 amplification we used the following primers: rp49: forward: AAGATCGTGAAGCGCCACCAA; rp49: reverse: CTGTTGTCGGATACCCTTGGGGCTT.
RESULTS
Four mob Genes Are Found in Drosophila
BLAST searches identified 4 Drosophila genes that shared similarity to the yeast mob1 and mob2 genes: CG13852 (mats or Dmob1), CG11711 (Dmob2), CG4946 (Dmob3), and CG3403 (Dmob4). The BLAST analysis and additional clustering analyses did not allow us to identify one or two of the fly genes as being clear orthologues of either yeast mob1 or yeast mob2. CG13852 and CG4946 are the two most closely related to both yeast genes, and CG3403 is the most distantly related. Gene chip analysis of pupal wing RNA showed all four of the fly mob genes are expressed in the pupal wing before, during, and after hair morphogenesis (Adler, unpublished results).
Evidence of Physical Interaction between Trc and Dmob2
The S. cerevisiae Cbk1 and Mob2 proteins are known to interact physically as do the S. pombe homologues (Orb6 and Mob2; Weiss et al., 2002; Hou et al., 2003). In addition the related Dbf2 and Mob1 proteins of S. cerevisiae also interact physically (Komarnitsky et al., 1998). Thus, we expected that Trc and at least one of the Drosophila Mobs would interact physically. Indeed, Trc and Dmob1 (CG13582) were identified as interacting proteins in a genome scale two-hybrid screen (Giot et al., 2003). None of the other Dmobs were reported as being able to interact with Trc, and no Dmob was reported as interacting with Wts in that study (Giot et al., 2003), although a recent article demonstrated an interaction between Wts and Mats (Dmob1; Lai et al., 2005).
We performed a yeast two-hybrid screen of a Drosophila cDNA library using full-length trc cDNA as “bait.” Most of the clear positive clones recovered contained fusions of segments of Dmob2 fused to the GAL4 activation domain. We did not recover clones of any of the other Dmobs. Perhaps they were not present in the library screened. To determine whether Mats and Trc interacted in the yeast two-hybrid system, we obtained a cDNA clone for mats from the BDGP collection and subcloned it into pGADT7. We used this plasmid and confirmed that Trc and Mats interacted in the yeast-two-hybrid system. Thus, Trc appears to be able to interact with at least two different Mob family proteins in Drosophila. We similarly tested and confirmed that Wts was able to interact with both Mats and Dmob2 in the two hybrid system. Thus, we did not see evidence of specificity in the Drosophila NDR/Mob family interactions.
To determine what portion of the Trc protein interacted with Dmobs, we generated a set of plasmids that contained trc C-terminal truncations and assayed them for an interaction with Dmob2 and Mats using the two-hybrid system (Figure 1A). We obtained similar but not identical results with these two mob family members. All of the Trc deleted proteins interacted strongly with Mats and the larger Trc proteins interacted strongly with Dmob2. However, Trc proteins that only contained amino acids 1–60 or 1–119 interacted quite weakly with Dmob2 (slow growth on his- ad- plates and no blue color in the presence of Xgal or X-αGal). The data for the binding of human Ndr1 to hMob1 indicates that important residues are found in the amino terminal region of Ndr1 (Bichsel et al., 2004). In hNdr, Tyr-31, Arg-41, Thr-74, and Arg-78 were found to be absolutely required for interaction, whereas the Lys-24, Arg-44, and Leu-79 mutants displayed reduced interaction (Bichsel et al., 2004). The corresponding residues in Trc are Tyr35, Arg45, Thr78, Arg81, Lys28, Arg48, and Leu82. Hence our data argue that for Mats binding to Trc requires only a subset of the residues needed in human Ndr1, whereas Dmob2 binding is enhanced by additional residues. The significance of these differences is not clear, but each of these examples is consistent with the amino terminal region of the NDR kinase family members being essential for the interaction with Mob family members.
Figure 1.
Trc physically interacts with Dmob2. (A) A diagram of the Trc protein showing important landmarks and the set of truncated proteins tested for an interaction with Dmob2 in the yeast two-hybrid system. The truncated genes were subcloned in pGBKT7 vector and tested for interactions with Dmob2 fused to the GAL4 transcriptional activation domain. The results are shown as “+” (positive) or “-” (negative) using the stringent criteria described in Materials and Methods. The amino acids from 119 to 180 of Trc are important for a strong Trc-Dmob2-interaction as the complementation was quite weak. These experiments showed that neither an intact nor functional Trc kinase domain was essential for the interaction with Dmob2. (B) Trc and Dmob2 coimmunoprecipitate in an S2 cell lysate. Cells were transfected with either UAS-Flag-Trc and/or Dmob2-HA. The top panels show the results from Western blot analysis of the lysate. The bottom panel shows that immunoprecipitation with an anti-HA antibody brought down Flag-Trc protein. The lower band is likely IgG and serves as a loading control. (C) Dmob2 point mutations block its interaction with Trc. Sequence alignment shows the conservation of the Mob family proteins in yeast, frog, fly, mice, and human. The red colored residues are sites that are absolutely conserved, and the blue sites conserved in a majority of the proteins. Arrows indicate the residues mutated and tested. The results from our experiments and the related results from S. cerevisiae mob1 are shown in the table.
Because the protein kinase domain of Trc extends from residue 90–393, these results indicate that the kinase domain does not have to be intact for Trc to interact with Dmob2 or Mats. We further found mutations in the conserved regulatory phosphorylation sites, S292A+T453A did not interfere with the interaction. Thus it appears clear that the kinase activity of Trc is not important for its ability to bind to Dmob2. Our results were similar to those seen between kinase inactive Dbf2 and Mob1 (Komarnitsky et al., 1998).
Mutations in DMob2 Interrupt the Interaction with Trc
In S. cerevisiae Mob1 a number of sites have been identified as being important for the binding of Mob1 and Cbk1 (Luca and Winey, 1998). To test whether these sites are functionally conserved within the Dmob family, we aligned the sequence of the four Drosophila Mobs with yeast, human, and frog to identify the Dmob2 amino acids that corresponded to the important sites in yeast Mob1 for interaction with Dbf2. We generated similar mutants in Dmob2 (RE70633) to confirm the conservation of the Mob-Ndr interaction (Figure 1C). Most of these mutations in Dmob2 disrupted the binding to Trc (Figure 1C), consistent with the conclusion that the mechanism of interaction has been conserved. Most of the residues noted above are also conserved in Mats, Dmob3, and Dmob4, consistent with all Mob family members interacting with Ndr family members in the same manner.
Trc and Mob Interact In Vivo
To assess whether these proteins were capable of associating in vivo in Drosophila cells, immunoprecipitation experiments were carried out in Drosophila S2 cells expressing both Trc and Dmob2. We found Trc in anti-Dmob2–8x HA, but not in control, immunoprecipitates (Figure 1B), consistent with these two proteins interacting in vivo.
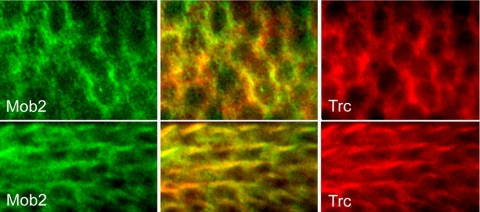
As an additional test of these proteins interacting in vivo we examined the subcellular localization of Trc and Dmob2 protein in wing cells. Trc distribution was examined with an anti-FLAG monoclonal antibody (Sigma) using UAS-trcWT and UAS-trcDN transgenes driven by ptc-GAL4. The proteins encoded by these transgenes carry an amino terminal FLAG epitope. We found that the FLAG staining pattern of overexpressed Trc was the same as the endogenous Trc detected by anti-Trc antibody staining (He et al., 2005). We localized Dmob2 in a similar way using a CFP tag because an anti-Dmob2 antibody was not available. Confocal microscopy demonstrated that before hair formation Trc was cytoplasmic and concentrated at the cell periphery (Figure 2). During hair outgrowth Trc accumulated in the hair, as is the case for the endogenous Trc (He et al., 2005). Both before and after hair initiation Dmob-2 was localized similarly to Trc (Figure 2). The subcellular localization of these proteins was similar in flies that carried a single transgene or both UAS-trc and UAS-Dmob2 transgenes (unpublished data). We concluded that Trc and Mob proteins can interact in vivo in Drosophila cells.
Figure 2.
Trc colocalizes with Dmob2 to the cell periphery in wing cells and wing hairs. Confocal images of ptc-GAL4; UAS-Flag-trcWT/ UAS-Dmob2-CFP pupal wings are shown. Trc (anti-Flag) is shown in red and Dmob2 (anti-CFP) is shown in green. Merged images are also present. The top panels are of a wing before wing hair morphogenesis (∼26 h apf) and the bottom panels of a wing during hair morphogenesis (34 h apf). Note that in the top panels the proteins preferentially accumulated together at the cell periphery and later in development they both accumulated in the hairs.
mats1 Is Required for Both Trc and Wts/Lts Functions In Vivo
As shown previously the directed expression of a dominant negative Trc protein provides a sensitized system for identifying interacting genes (Figure 3A, 1 and 2; He et al., 2005). Deficiencies for each of the fly mob genes enhanced the wing hair phenotype that resulted from driving expression of UAS-trcT453A using either ap-GAL4 or ptc-gal4 (Figure 3 and Table 1). The strongest enhancement was seen with deficiencies for mats and Dmob2 (Table 1). These results suggested the possibility that all 4 Dmobs can redundantly interact with Trc, although it is possible that the interactions could be indirect or due to other genes in deleted regions. It is worth noting that such interactions are not common. When we screened the Drosophila deficiency collection for enhancement or suppression of ap-GAL4 UAS-trcT453A, <10% of the Dfs showed an interaction (unpublished data).
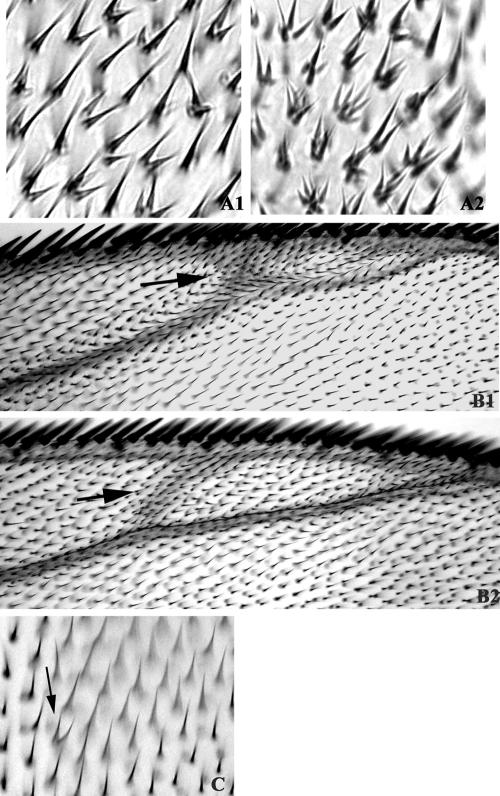
Figure 3.
trc dominant negative phenotype is enhanced by a mutation in mats and the overexpression of a truncated Dmob2 protein caused a very weak trc-like phenotype. (A) Multiple hair cell phenotype of ap-GAL4/+; UAS-trcT453A/+ (A1) and ap-GAL4/+; UAS-UAS-trcT453A/matsPB (A2). Note the dominant enhancement in flies heterozygous for mats. (B) An extra vein phenotype caused by ap-GAL4-driven UAS-trcK122A (arrow) (B1) and ap-GAL4-driven UAS-Dmob2-C (arrow)(B2). (C) Overexpression of a truncated form of Dmob2 (ap-gGAL4-UAS-Dmob2-C) induced a very weak trc-like dominant negative phenotype (arrow).
Table 1.
Genetic interactions of four Dmobs with a trc dominant negative mutant
| Number of wing hairs/cell
|
|||
|---|---|---|---|
| Genotype | Interactiona | Average | Std Error |
| exp 1 | |||
| ap-GAL4/+; UAS-trcT453A2/+ | — | 1.8 | 0.06 |
| ap-GAL4/+;UAS-trcT453A2/Df(Dmob1) | Enhancement | 3.5 | 0.08 |
| ap-GAL4/+;UAS-trcT453A2/Dmob1P | Enhancement | 3.68 | 0.08 |
| ap-GAL4/+;UAS-trcT453A2/Df(Dmob2) | Enhancement | 3.49 | 0.08 |
| ap-GAL4/Df(Dmob3);UAS-trcT453A2/+ | Enhancement | 3.42 | 0.08 |
| ap-GAL4/Df(Dmob4);UAS-trcT453A2/+ | Enhancement | 3.09 | 0.08 |
| exp2 | |||
| ap-GAL4/UAS-trcT453A1 | — | 1.98 | 0.07 |
| ap-GAL4/UAS-trcT453A1;Df(Dmob2)/+ | Enhancement | 3.94 | 0.09 |
| ap-GAL4/UAS-trcT453A1;UAS-Dmob2-C/+ | Enhancement | 2.35 | 0.09 |
| exp 3 | |||
| ptc-Gal4/UAS-trcT453A1/+ | — | 1.8 | 0.11 |
| ptc-Gal4/UAS-trcT453A1/UAS-mats | Suppression | 1.46 | 0.06 |
| exp 4 | |||
| ptc-Gal4/+; UAS-trcT453A2/+ | — | 1.46 | 0.05 |
| ptc-Gal4/+; UAS-trcT453A2/UAS-mob2C | Enhancement | 1.82 | 0.07 |
| ptc-Gal4/UAS-mob2C; UAS-trcT453A2/UAS-mob2C | Enhancement | 2.01 | 0.05 |
| exp 5 | |||
| ptc-Gal4/+; UAS-trcT453A2/+ | — | 1.56 | 0.05 |
| ptc-Gal4/+; UAS-trcT453A2/matsPB | Enhancement | 1.98 | 0.06 |
| ptc-Gal4/+; UAS-trcT453A2/matsPBrv | — | 1.51 | 0.08 |
| ptc-Gal4/+; UAS-trcT453A2/matse235 | Enhancement | 2.02 | 0.03 |
| ptc-Gal4/+; UAS-trcT453A2 +/+ wts3-17 | Enhancement | 1.82 | 0.1 |
Genetic interactions were scored as the effect of loss of one wild-type copy of the genes or transgene co-overexpression on the dominant multiple hair cell phenotype from ap-GAL4 or ptc-Gal4-induced overexpression of TrcT453A2 or TrcT453A1, which are independent lines of UAS-trcT453A. Because the phenotype is sensitive to both genetic background and environmental effects, results are always compared with a control done at the same time.
Interaction: statistical significance was determined by Student's t test (p < 0.001). The only exception to this was the enhancement by wts, which was significant at p < 0.05
To confirm that the interaction between trc and Df (mats) was due to the reduction in mats dose, we utilized two independent alleles. One was the null allele described by Lai et al. (2005) (matse235), which is deleted for almost the entire coding region, and the other was a lethal PiggyBac insertion allele of mats [PBac{RB}CG13852e03077] (we subsequently refer to this allele as matsPB). Because this later allele had not been well characterized, we first determined that the insertion was lethal over a deficiency for the region (Df(3R)Exel6191) and it failed to complement the recessive lethality of matse235 consistent with the lethality being due to the PB insertion. We were able to revert this mutation using a source of PiggyBac transposase (Thibault et al., 2004). We found that both mats alleles dominantly enhanced the trc dominant negative phenotype (Table 1) and this enhancement was lost in the PB revertant. We also found that over expression of mats from a UAS-mats transgene (Lai et al., 2005; driven by ptc-Gal4) partially suppressed the multiple hair cell phenotype that resulted from driving expression of Trc-DN using ptc-Gal4 (Table 1). These dose responses argue that Mats activates Trc. Interestingly, we found that heterozygosity for a wts mutation also enhanced the Trc dominant negative phenotype, although somewhat less strongly (Table 1).
We also obtained evidence for mats functioning with trc and fry using simple loss of function mutations. Wild-type flies or flies heterozygous for either trc, fry, or mats appeared normal and only rarely (on fewer than 5% of wings) was even a single multiple hair cell seen. Flies that were heterozygous for two of these genes showed a slightly higher frequency of wings with one or a couple of multiple hair cells (often ∼10%) but the increase was not routinely significant (Table 2). However, almost half of the wings from flies that were heterozygous for all three genes (e.g., fry2 trc1+/+ + matsPB) showed a weak multiple hair cell phenotype (Table 2), a significant increase. This genetic interaction is further support for the hypothesis that trc, fry, and mats function together in regulating wing hair development. In this assay we did not see an equivalent interaction with wts3–17 (Table 2).
Table 2.
Interaction between loss of function alleles at fry, trc, and mats
| Genotype | No. of wings | Fraction with an mhc phenotype | Different from Ore-R |
|---|---|---|---|
| Ore R | 36 | 0 | ND |
| trc1/+ | 33 | 0.03 | ND |
| trcP/+ | 20 | 0 | ND |
| fry1/+ | 20 | 0 | ND |
| fry2/+ | 20 | 0 | ND |
| matsPB/+ | 20 | 0 | ND |
| matse235/+ | 20 | 0 | ND |
| wts3—17/+ | 20 | 0 | ND |
| fry2trc1/+ + | 24 | 0.12 | ND |
| fry1 +/+ matsPB | 23 | 0.09 | ND |
| trcP +/+ matsPB | 20 | 0.05 | ND |
| matPB +/+ wts3—17 | 34 | 0.03 | ND |
| matsPB +/+ wtsP | 16 | 0.06 | ND |
| fry2trc1 +/+ + matse235 | 25 | 0.48 | Differenta |
| fry1trcP +/+ + matsPB | 20 | 0.4 | Differenta |
| fry2trc1 +/+ + matsPB | 22 | 0.41 | Differenta |
| fry2 + trc1 +/+ Df-mob2 + Df-mats | 10 | 0.5 | Differenta |
| fry2trc1+/+ + wts3—17 | 30 | 0 | ND |
A z test was used to compare the fraction showing a multiple hair cell phenotype with that of the control Ore-R wings. mhc, multiple hair cells. ND, no difference at p < 0.05.
Different at p < 0.001
Previous studies established that trc also had a larval denticle phenotype (Geng et al., 2000). In trc mutants the overall pattern of denticles was partly disorganized and many denticles were split (Table 3). Split denticles are infrequent in wild-type larvae (Figure 4A1 and Table 3). The denticle pattern of matsPB/matsPB homozygous larvae was also disorganized and contained many split denticles (Figure 4A2 and Table 3). The number of split denticles was similar in matsPB/Df larvae, suggesting that for this phenotype matsPB is a strong, near phenotypic null allele. The matse235 also showed a similar denticle phenotype (unpublished data). The phenotype of matsPB homozygotes was slightly less severe than that of in trcP/trcP larvae (Figure 4A3 and Table 3). Notably, trcP matsPB/trcP matsPB double mutant larvae did not have a more severe phenotype than the single mutants (Table 3). This lack of additivity argues that trc and mats function in a common pathway during denticle development.
Table 3.
mats and trc function together during denticle development
| Genotype | Mean number of split denticles/segment (2nd instar larvae) | SEM | p vs. trc | p vs. mob |
|---|---|---|---|---|
| Ore R | 0.2 | 0.12 | <0.000001 | <0.000001 |
| trcP | 38.6 | 2.25 | NR | 0.0026 |
| matsP | 27.8 | 2.73 | 0.0026 | NR |
| trcPmatsP | 33.5 | 1.87 | 0.10 | 0.091 |
| matsPB/Df(3)mats | 30.9 | 2.33 | 0.048 | 0.22 |
NR, not relevant. We scored second instar larvae because for several of the genotypes third instar larvae are rare.
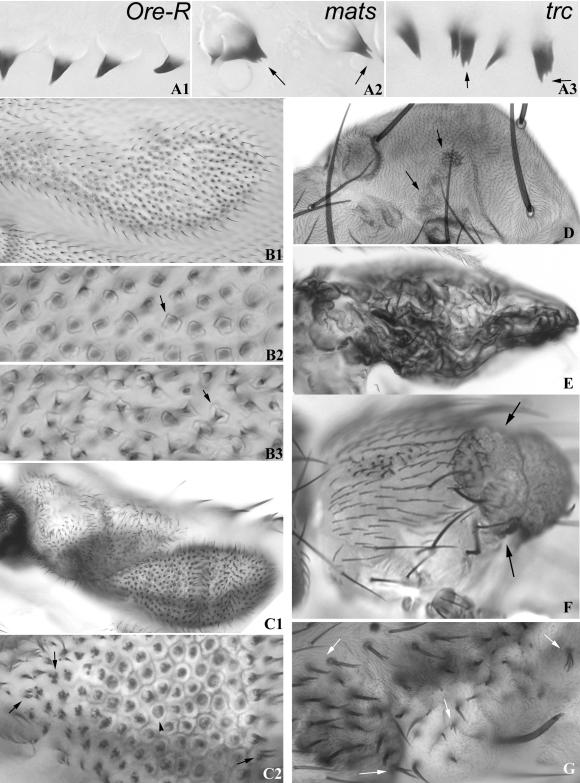
Figure 4.
Cuticular phenotypes of Dmob1. (A1–A3) Larval denticle phenotype of Dmob1 is similar to that of trc. (A1) Oregon R; (A2) matsPB/matsPB (A3) trcP/trcP. Note the split denticles (arrows) in both matsPB/matsPB and trcP/trcP. (B1–B3) Bulging wing cell phenotype of matsPB is similar to that of wts. Unmarked mutant clones were induced using flp/FRT. (B1) A moderate-sized matsPB/matsPB clone results in a dimpling of the wing surface. (B2) A blow up of a small region from B1 showing the cuticle defect in Dmob1 clones. The hair is located on a pedestal like structure (arrows). (B3) Shown is a high magnification image of part of a wts3–17-induced clone on the wing. Note the presence of pedestal like structures that are very similar to those seen in mats clones. (C1-C2) matsPB clones on the wing sometimes resulted in discrete outgrowths. One is shown in C1. Many cells in such outgrowths display strong multiple hair cells. A blow up of C1 is shown in C2. Arrows point to cells with strong multiple hair cell phenotypes that are reminiscent of trc. Cells in the center are elevated (due to the bulge) and here we see the cell outlines (arrowhead) and a darkened center (pedestal and base of the hair). (D) Shown is part of the thorax of a fly showing small matsPB clones. The arrows point to clones that are darkened. (E) A grossly distorted wing due to the presence of a large number of matsPB clones. This fly was vg-GAL4 UAS-flp/+; FRT82/FRT82 matsPB. (F) Shown is a fly with a large mats“tumor” clone (arrows) on the thorax. (G) Shown is the thorax from a fly that was vg-GAL4 UAS-flp/±; FRT82/FRT82 matsPB and contained large regions of matsPB clones. Arrows point to split bristles. Similar defects are seen in trc mutants.
Both mats alleles are larval lethals with death typically in the second or early third instar. To examine the phenotype of mats in wing cells, we generated mosaics using FLP/FRT. mats clones on the wing, leg, thorax, and head displayed two types of phenotype. The most notable was indistinguishable from those produced by clones of wts (Justice et al., 1995; Xu et al., 1995), suggesting that mats also functions with wts as was recently shown by Lai et al. (2005). On the wing small clones produced bulges that could be seen at low magnification with a stereomicroscope. In mounted wings individual cell outlines were visible in the cuticle and the cells appeared to have a bulging apical surface (Figure 4B, 1 and 2). The hairs were located on an elevated pedestal, a phenotype that was indistinguishable from those seen in wts clones ((Figure 4B, 2 and 3). The hairs were often broader than normal. Particularly in other body regions clones were abnormally pigmented (either darker or lighter than normal; Figure 4D) and there were outgrowths of clone tissue (Figure 4F). In highly abnormal wing clones (Figure 4C1) we often saw evidence of clustered and split hairs that were typical of trc mutant clones (Figure 4C2). Some multiple hair cells were seen in very abnormal wts clones but this phenotype appeared less severe (e.g., number of hairs per cell) than that seen with mats or trc. These observations suggest that mats functions with both Trc and Wts.
We also examined matsPB and matse235 clones in pupal wings. Mutant mats cells were able to outcompete their neighbors and ended up comprising most of the wing when clones were induced early (Figure 5). As was expected from the morphology of clones in adult wings the pupal clones produced bulges in the wing and individual cells also often appeared bulged. Clone cells stained more brightly for F-actin (Figure 5). This was true both in developing hairs and in the general apical cortex. This phenotype was clear-cut enough that we could use it as a convenient marker of mats mutant cells. These phenotypes were seen with both mats alleles tested. A similar, increase in actin staining was seen in wts clones (unpublished data). A similar, but perhaps less severe increase in staining, is seen in trc clones (He et al., 2005). In some, but not all mats clones large numbers of multiple hair cells could be seen (Figure 5). At later stages we could see a circular pedestal of actin staining surrounding the base of the hair in mutant cells but not in surrounding wild-type cells (Figure 5). At still later stages the wild-type cells also had a circular pedestal of actin staining, suggesting that the mutant cells might be developmentally more advanced. Consistent with this possibility, in many clones hair initiation and outgrowth appeared to be advanced in mats mutant cells compared with neighbors (Figure 5). This is also the case for wts clones (unpublished data), but it is the opposite of trc clones, in which hair development is often delayed (He et al., 2005). Cells in mats clones had a smaller cross section so that the array of hairs appeared denser (Figure 5), which is also the opposite of what is seen in trc clones, in which there is an increase in cross-sectional area (He et al., 2005). Once again the phenotype of the wts clone cells resembles that seen for mats cells (unpublished data). Thus, for several wing phenotypes mats mutant cells resembled wts and not trc cells. Indeed, the mats phenotype was the opposite of trc for both cell area and the timing of hair morphogenesis.
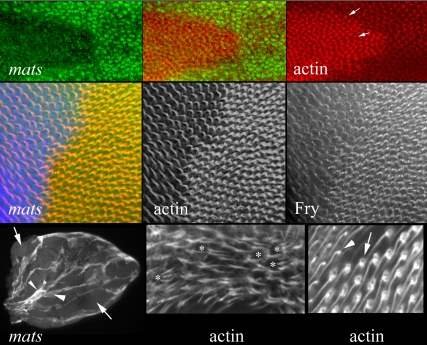
Figure 5.
mats clones in pupal wings. The top panels show a matsPB clone marked by a loss of the Nmyc epitope in a pupal wing. The left panel shows immunostaining for myc (green). The right panel shows F-actin staining (red; Alexa 568 phalloidin). Note that the clone cells stain more brightly than their wild-type neighbors. The arrows point to two hairs. Note that hairs form in the vicinity of the distal most vertex and point distally. Thus, mats function is not required for the function of the frizzled pathway in planar polarity. Also note that the mutant cells appear to be advanced in hair morphogenesis compared with their wild-type neighbors. The middle panels also show a matsPB clone marked by a loss of the Nmyc epitope in a pupal wing. The middle panel in this row shows F-actin staining and the panel on the left anti-Fry antibody staining. The panel on the right is a merge (actin in red, Fry in green and myc in blue). Note once again the increase in actin staining in mats mutant cells. Note also that there is an increase in Fry accumulation in the mutant cells. This is similar to what is seen in trc mutant clones (He et al., 2005) and is consistent with Dmob1 activating Trc. On the left of the bottom row is a low magnification image of a vg gal4 UAS-flp; FRT 82 matsPB/FRT 82 hs-Nmyc pupal wing stained with anti-myc antibody. The arrows point to mats clone cells that lack myc expression. The large arrowhead points to a region with intermediate Myc staining. These should be the heterozygous genotype (no recombination). The small arrowhead points to a region with bright Myc staining. This is the twin spot that has two copies of the myc-expressing transgene. Note how the mats mutant cells have outcompeted the wild-type cells and now comprise most of the wing. If the mob mutation did not alter growth, then the region of mats clones should be the same as that of the two-dose myc twin spots. The middle panel in the lower row shows an example of prominent multiple hair cells (F-actin staining) formed by mats clone cells. This was common only in clones that showed the “outgrowth” phenotype. The right panel on the bottom row shows the boundary between mats and wild-type cells late in hair development stained for F-actin. The arrow points to a Dmob1 cell and its prominent “pedestal.” The arrowhead points to a neighboring wild-type cell.
We previously found that the accumulation of Fry in wing cells is subject to feedback control that is dependent on Trc activity. Hence, in a trc mutant we find increased Fry accumulation (He et al., 2005). Several of the observations described above suggested the hypothesis that mats functioned along with trc and was important for Trc activation. From this we predicted that Fry accumulation would also be elevated in Dmob1 clones. We found increased levels of Fry immunostaining in Dmob1 clone cells, which was consistent with our hypothesis (Figure 5). This was also seen in wts mutant clones (unpublished data), although the increase appeared less dramatic.
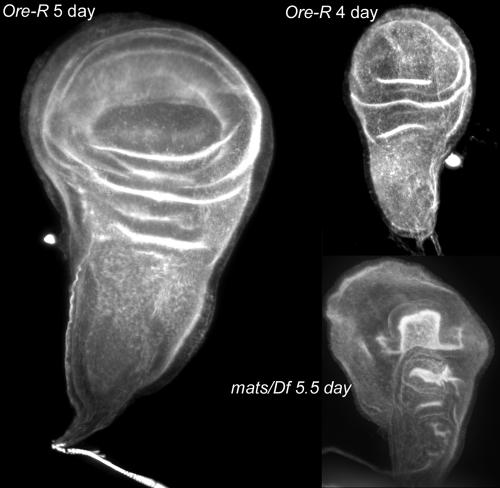
Tumorous overgrowth phenotypes are a consequence of mutations in a number of Drosophila genes. In several cases, such as lethal giant discs overgrown imaginal discs are found in late third instar larvae (Buratovich and Bryant, 1997). To determine whether that was also the case for mats, we examined mats/Df mutant larvae. These larvae grow slowly and after 5 d of growth, when wild-type larvae begin to pupate, mats/Df larvae are the size of early third instar larvae. These larvae routinely die without growing substantially larger. When we dissected 5.5–5-d-old mats larvae, we did not see any evidence of tumors or overgrowth of imaginal or other tissues. Rather, the imaginal discs were approximately the size of those seen in 4-d larvae (Figure 6). However, the mats homozygous discs did not appear normal, as they were abnormally shaped and more folded than normal discs of this size.
Figure 6.
Imaginal wing discs in mats mutants are smaller than normal. Shown are maximum projections of confocal stacks of wing imaginal discs stained with Alexa 568 phalloidin. All micrographs are at the same magnification. Note how much smaller the 5.5-d mats/Df disk is compared with the 5-d Ore-R disk.
In a number of experiments involving mats or wts we observed what appeared to be spontaneous tumors or clones. This was seen most often in flies that also carried reduced doses of Dmob3 and Dmob4. We think that these overgrowths were due to spontaneous mitotic recombination, because when the flies were also mutant for trc or fry, we also saw evidence of trc or fry clones. The trc and fry clones were seen less frequently. This could be due to these genes being located more proximally on the chromosome than mats or wts, but it might also be due to the competitive advantage of mats and wts clones, resulting in these clones being larger and easier to detect. The basis for these clones is unclear but suggests genomic instability in mats and/or wts mutants.
Dmob2 Interacts with trc
We tested 10 deficiencies from the 68C region and further mapped the enhancing region to a small interval (68C11–13) that contained CG11711 (Dmob2). Deficiencies from this region were able to similarly enhance the phenotypes that resulted from the directed expression of other dominant negative Trc proteins (unpublished data). The genome project annotation of Dmob2 suggests it is a complicated gene that encodes at least four variant mRNAs from exons that span >40 kb. These mRNAs encode four proteins with a common c-terminal segment but with different amino terminal regions. There are P insertions in a large intron of Dmob2, but these do not inactivate the gene to produce a mutant phenotype (unpublished data). Attempts to use imprecise excision to produce a deletion that would ensure that no Dmob2 protein could be made were not successful, because we only obtained small deletions that would eliminate one isoform. These did not produce a mutant phenotype. As an alternative approach we generated transgenic flies that carried UAS constructs that encoded either a tagged full-length Dmob2 (GH07469) protein or partial proteins aa 1–157 (Dmob2-N) and aa 148–354 (Dmob2-C) that might act as dominant negative proteins. The directed expression of the wild-type Dmob2 and Dmob2-N proteins by ap-GAL4 did not cause any notable visible phenotype. The interpretation of these results is limited by the fact that we were only expressing one of 4 CG11711 isoforms. On the other hand, overexpression of the common Dmob2-C protein segment resulted in a weak trc-like multiple hair cell phenotype (Figure 3C) and it also enhanced the dominant negative trc wing hair phenotype in a dose-sensitive way (Table 1), consistent with Dmob2-C being a dominant negative and the normal function of Dmob2 being to activate Trc. In addition, overexpression of Dmob2-C caused an extra vein phenotype. This phenotype was enhanced by increasing the number of UAS-mob2c transgenes and it was also enhanced by heterozygosity for a deletion for CG11711 (unpublished data). Thus, Dmob2-C acts as a dominant negative for this phenotype. A similar, but weaker vein phenotype was also seen in when trcDN was overexpressed (Figure 3B, 1 and 2).
Dmo25, the Drosophila Hym1 Homolog Does Not Function with trc
The yeast hym1 gene functions upstream of Cbk1 and is needed for RAM pathway function (Dorland et al., 2000; Nelson et al., 2003). There is a single homolog of hym1 in Drosophila, the intronless Dmo25 gene. Df(3L)81k19, which uncovers Dmo25, did not enhance or suppress the wing hair phenotype obtained from expressing UAS-trcDN using ptc-Gal4. A P insertion mutation in Dmo25 {(PZ)Mo2500274} was obtained from the Bloomington Stock Center. This 17.3-kb insertion is into the 5′ untranslated region of the Dmo25 mRNA. We confirmed that the insertion was lethal over Df(3L)81k19 and that the lethal mutation could be induced to revert in the presence of a source of P transposase. Thus, the P insertion in Dmo2500274 is responsible for the recessive lethal mutation. For simplicity we refer to Dmo2500274 as Dmo25P. Homozygous (or hemizygous) Dmo25P mutant animals died as larvae without any obvious morphological defect. There was no difference in the lethal stage between homozygous Dmo25P and Dmo25P/Df animals arguing that the P insertion is a strong phenotypic null allele. We compared the amount of Dmo25 mRNA in wild-type and Dmo25P/Df second instar larvae using real time RT-PCR. The relative abundance of Dmo25 mRNA (i.e., compared with the mRNA for the rp49 ribosomal protein) in the mutant was <1% of that seen for Oregon R. Because there is no intron in Dmo25, there was no way to distinguish between mRNA and contaminating DNA in these samples. Further, maternally derived Dmo25 mRNA could also be contributing to our amplification. Hence the <1% estimate of Dmo25 mRNA in the mutant is likely to be an underestimate. The molecular and phenotypic tests indicate that Dmo25P is a strong if not null allele.
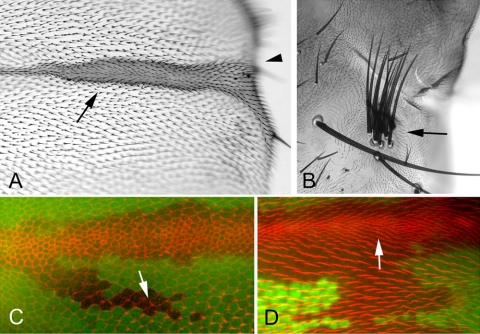
The denticle pattern in the Dmo25P and Dmo25P/Df mutants was normal and we did not see a large number of split denticles as we did in trc and Dmob1. To assess the possible function of Dmo25 for wing hair and bristle development, we used FLP/FRT-mediated recombination to generate clones of mutant cells. We did not see any trc-like multiple hair cells or split bristles, although several phenotypes were seen. When clones were located close to or overlapping wing veins there was a “swelling” of the veins (Figure 7, A and D). Clones some distance from a normal vein did not show any phenotype and differentiated normal looking hairs (Figure 7, C and D). We did not see any evidence for a delay in hair initiation (Figure 7C), as is often the case for cells in trc mutant clones (He et al., 2005). When we examined adult wings bearing clones, there was often a loss of wing margin (wing notching), suggesting cell death in this region (Figure 7A). In adult flies bearing unmarked clones we also saw examples of “multipled bristles” (Figure 7B). These were not split, but rather appeared to be due to extra bristles. All three of these phenotypes are reminiscent of mutant phenotypes associated with defects in Notch signal transduction, suggesting that Dmo25 might function in this pathway. To further test this possibility, we examined the ability of the Dmo25P allele and a Df for the region to interact genetically with Notch pathway mutations. In one set of crosses, we found that a reduction in Dmo25 dose enhanced the weak wing notching phenotype seen in male flies that carried the weak Nno1 allele. The fraction of the wing margin lost increased from 0.31 in Nno1 wings to 0.47 (p = 0.14) in Nno1; Dmo25P/+ and to 0.59 (p = 2 × 10-6) in Nno1; Df(3L)81k19/+. Similarly we found that a reduction in Dmo25 dose enhanced the expanded wing vein phenotype of flies that carried a single copy of the weak DlP05151 allele. The ratio of the vein area to length for Vein 2 distal to the distal cross vein increased from 16.2–21.7 (p = 0.00016) for Dmo25P +/+ DlP05151 and 20.2 (p = 0.0002) for Df(3L)81k19 +/DlP05151. These data are consistent with the possibility that Dmo25 functions in the Notch pathway, but more extensive experiments will be needed to get definitive evidence for this.
Figure 7.
Dmo25, the Drosophila Homolog of Yeast Hym1, does not function along with Trc. (A) The induction of unmarked Dmo25 P homozygous clones results in thickened wing veins (arrow) and loss of the wing margin (arrowhead). Phenotypes similar to these are often seen in Notch pathway gene mutations. (B) Flies with Dmo25P clones often showed a multiple bristle phenotype. An example of an unmarked notum clone is shown (arrow). In control experiments (i.e., when Dmo25P were not induced) such multipled bristles were not seen. (C) A confocal image of a 32-h pupal wing stained with Phalloidin (red) and anti-GFP (green) antibody. The wing contains a large Dmo25P clone marked by the loss of GFP. Note that some cells in the region are just starting to elaborate hairs. The genotype with regard to Dmo25 does not appear to influence the timing of hair initiation. The arrow points to a newly formed hair inside the clone. (D) A confocal image of a 36-h pupal wing stained with phalloidin (red) and anti-GFP (green) antibody. The wing contains a large Dmo25P clone marked by the loss of GFP. Within the clone the vein region was enlarged, whereas wing hair development is not altered.
We also generated flies that carried UAS-Dmo25-N, a C-terminal half truncated form of Dmo25 (aa 1–173). Driving expression of this transgene by ap-GAL4 resulted in swollen wing veins that were similar to the phenotype seen in Dmo25P mutant clones (unpublished data). This suggests that the C-terminal half of Dmo25 is essential for the protein's function.
DISCUSSION
Trc, Dmob, and Fry Function in a Common Signaling Pathway
Previous genetic data pointed out the importance of trc and fry for the morphogenesis of polarized cellular extensions (Geng et al., 2000; Cong et al., 2001; He and Adler, 2001; Emoto et al., 2004). Based on homology to the RAM, MEN, and SIN pathways (Mah et al., 2001; Hou et al., 2003; Nelson et al., 2003), it seemed likely that one or more of the Drosophila mob genes would function along with trc and fry. We found evidence supporting this hypothesis but the results were complicated by both pleiotropy and redundancy. This was illustrated most clearly in our experiments with mats. Mutations in mats displayed phenotypes that were typical of both trc (split denticles and multiple hair cells) and of wts/lats (tumors, bulged cells, advanced hair differentiation). We also found that Mats could interact with both Trc and Wts using the yeast two-hybrid system. These observations stand in contrast to the situation in yeast, in which individual mob genes show specificity for individual Ndr family members. Further evidence for redundancy came from the gene dosage interactions seen between trc and the other Dmobs.
Evidence for a direct physical interaction has been reported for Ndr and Mob family members from yeast, flies and mammals (Colman-Lerner et al., 2001; Mah et al., 2001; Weiss et al., 2002; Hou et al., 2003; Bichsel et al., 2004; Lai et al., 2005). Previous yeast two-hybrid experiments showed evidence for a physical interaction between Trc and Mats (Giot et al., 2003) and Mats and Wts (Lai et al., 2005). Our results extended this by showing a similar interaction between Trc and Dmob2 by both 2 hybrid and coimmunoprecipitation experiments and that Wts and Dmob2 could interact in the 2 hybrid system. We further showed that residues known to be important for the interaction between yeast Mob1 and Dbf2 were also important for the interaction between Trc and Dmob2. The conservation of many of these residues in Dmob3 and Dmob4 suggests that these proteins will also interact with Trc. The genetic interactions seen between trc and Dmobs suggest that the binding of Dmobs to Trc is essential for in vivo function and activation of the protein. Consistent with this hypothesis we found that Dmob2 and Trc colocalized to growing hairs in pupal wing cells.
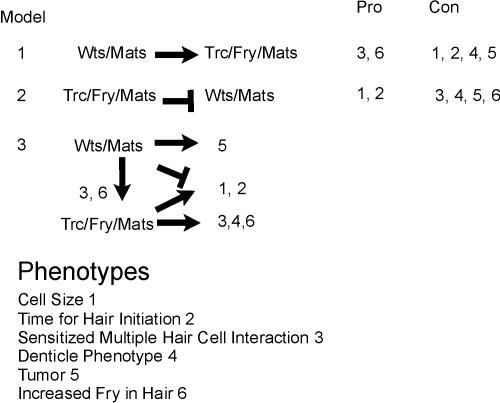
The observation that for two phenotypes (pupal wing cell cross section and time of hair initiation) mats and wts clone cells share a similar phenotype that is the opposite of trc is intriguing and needs to be reconciled with the positive gene dose interactions seen between mats and trc for the sensitized multiple wing hair cell assay and the similar denticle phenotype. Given this complexity it seems unlikely that a single simple mechanism is involved. Because Mats appears to function along with both Trc and Wts, some of the complexity may reflect interactions between these two kinase modules. Cells mutant for mats could have both modules inactive, although it is possible that the degree of possible mats redundancy might not be equivalent for the two modules. In principle these two modules could function in parallel or one could be upstream of the other (Figure 8). The observation that mats and wts clones have increased Fry accumulation in hairs is consistent with mats/wts being upstream of trc/fry/mats(mob). Because increased Fry accumulation in hairs is also seen in trc mutant cells, this hypothesis is also consistent with the positive gene dosage interactions. However, a different explanation is needed to explain the observation that with regard to cell size and the timing of hair initiation the mats/wts phenotype is opposite to that of trc/fry. If we consider both modules to be equally inactivated in a mats cell, then these latter observations suggest that trc/fry/mats could function antagonistically and upstream of mats/wts. In this situation a lack of trc function would result in increased wts function (and increased cell size and delayed hair formation). A lack of wts function would result in the reduced cross section and advanced hair morphogenesis. In a mats mutant we expect a reduction in both trc and wts function. This would result in a wts-like phenotype because a lack of trc inhibition of wts would be of no consequence in cells that already lack wts activity. However, this model does not explain the multiple hair cell interactions. Given the difficulties in any single model we suggest that the interactions are context dependent and/or the two modules function entirely in parallel (Figure 8).
Figure 8.
Models to explain the interaction between the trc and wts modules. Listed are phenotypes that need to be explained and three possible genetic models. Two of the models are only able to explain a subset of the phenotypes. The third model can explain all of the phenotypic interactions.
The Tumor Phenotype of mats
The “tumors” produced by mats clones were characterized by altered cell shape and proliferation. The altered cell shape could be seen in the cuticle of clone cells. The altered proliferation of clone cells was associated with them outcompeting neighboring cells so that, in wings in which recombination was induced at a high level using vg-Gal4 and UAS-flp, most cells in the wing were mutant and the wing was grossly larger than normal. These observations are very similar to those seen in wts/lats mutant clones (Justice et al., 1995; Xu et al., 1995). It is unclear how the altered cell shape and proliferation are related. Excess proliferation of a clone of cells that is surrounded by slower growing normal cells is expected to lead to compression of the faster growing clone cells and their immediate neighbors (Shraiman, 2005). This could be responsible for the decreased cross sectional area of mats and wts cells and their bulged apical surface. However, if compression was responsible for the change in cell shape, we would expect that this change would smoothly spread into the surrounding wild-type cells and would be more severe in the center of clones than near their periphery. This did not appear to be the case although this issue deserves further study. The cells in entirely mutant discs also appeared to have a bulged shape, which further argues against excess growth-mediated compression being responsible for the cell shape changes.
The tumors produced by mats clones and their ability to outcompete neighbors suggests the possibility that mats mutant cells grow and/or divide more rapidly than normal. Thus, it was surprising when we found that discs in mats homozygous and hemizygous mutants were smaller than normal. This could be due to the mutant larvae being impaired in feeding, digestion, or absorption of nutrients. This could lead the disk cells to be effectively starved reducing their growth. Alternatively it could be due to the mutation resulting in a defect in the secretion of a growth factor. It remains possible, however, that the overgrowth requires the direct contact of mats mutant and wild-type cells.
All RAM Pathway Components Do not Function along with trc in Drosophila
Hym1 is an essential upstream component of the RAM pathway in yeast (Nelson et al., 2003). Dmo25 is the single hym1 homolog in flies. Our analysis of Dmo25 mutant clones in the wing argued that it did not function in the Trc pathway in the adult epidermis of Drosophila. Hym1 is not thought to bind directly to Cbk1 and this may have facilitated it evolving a different function in Drosophila. We did however see three distinct mutant phenotypes associated with Dmo25 mutant clones. There was an enlargement of wing veins when clones were close to the vein, a loss of wing margin, and multipled bristles. Similar phenotypes can result from disruption of a number of pathways including the Notch/Delta signaling pathway. Consistent with this possibility, we saw genetic interactions between Dmo25 and N and Dl. The mammalian Mo25 protein has been shown to be present in a complex with the Lkb tumor suppressor and the Stradα adapter protein (Boudeau et al., 2003). There is no evidence for homologues of Lkb or Strad functioning with Trc or in N signaling. The Drosophila lkb homolog has been shown to function in oocyte polarization, but little is known about its function in the epidermis. fray is the fly gene most closely related to strad. fray has been principally studied with respect to its role in nerve development (Leiserson et al., 2000). In mammalian cells the Lkb, Strad, Mo25 complex activated the AMP-activated protein kinase (Baas et al., 2004). The Mamk homolog in Drosophila is CG3051 and little is known about the in vivo function of this gene. No deficiency that removed this gene showed an interaction with Trc (unpublished data). It will be interesting to determine whether Mo25 functions with Lkb and Strad in Drosophila and if Mamk is a downstream target.
Acknowledgments
We thank Bloomington Stock Center for fly stocks. We also thank Zhi-Chun Lai and Peter Bryant for sending mutant fly lines. We thank Mario Stegert and Brian Hemmings for communicating information before publication. This work was supported by grants from the National Institutes of Health to P.N.A. (GM53498) and J.Y.N. (R01NS40929). K.E. is a research associate and Y.N.J. is an investigator of the Howard Hughes Medical Institute.
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05-01-0018) on June 22, 2005.
References
- Baas, A. F., Kuipers J., van der Wel, N. N., Batlle, E., Koerten, H. K., Peters, P. J., Clevers, H. C. (2004). Complete polarization of single intestinal epithelial cells upon activation of LKB1 by STRAD. Cell 116, 457-466. [DOI] [PubMed] [Google Scholar]
- Bichsel, S. J., Tamaskovic, R., Stegert, M. R., and Hemmings, B. A. (2004). Mechanism of activation of NDR (nuclear Dbf2-related) protein kinase by the hMOB1 protein. J. Biol. Chem. 279, 35228-35235. [DOI] [PubMed] [Google Scholar]
- Boudeau, J., Baas, A. F., Deak, M., Morrice, N. A., Kieloch, A., Schutkowski, M., Prescott, A. R., Clevers, H. C., Alessi, D. R. (2003). MO25alpha/beta interact with STRADalpha/beta enhancing their ability to bind, activate and localize LKB1 in the cytoplasm. EMBO J. 22, 5102-5114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buratovich, M. A., and Bryant, P. J. (1997). Enhancement of overgrowth by gene interactions in Lethal (2)giant discs imaginal discs from Drosophila melanogaster. Genetics 147, 657-670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colman-Lerner, A., Chin, T. E., and Brent, R. (2001). Yeast Cbk1 and Mob2 activate daughter-specific genetic programs to induce asymmetric cell fates. Cell 107, 739-750. [DOI] [PubMed] [Google Scholar]
- Cong, J., Geng, W., He, B., Liu, J., Charlton, J., and Adler, P.N. (2001). The furry gene of Drosophila is important for maintaining the integrity of cellular extensions during morphogenesis. Development 128, 2793-2802. [DOI] [PubMed] [Google Scholar]
- Devroe, E., Erdjument-Bromage, H., Tempst, P., and Silver, P.A. (2004). Human Mob proteins regulate the NDR1 and NDR2 serine-threonine kinases. J. Biol. Chem. 279, 24444-24451. [DOI] [PubMed] [Google Scholar]
- Dorland, S., Deegenaars, M. L., and Stillman, D. J. (2000). Roles for the Saccharomyces cerevisiae SDS3, CBK1 and HYM1 genes in transcriptional repression by SIN3. Genetics 154, 573-586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du, L. L., and Novick, P. (2002). Pag1p, a novel protein associated with protein kinase Cbk1p, is required for cell morphogenesis and proliferation in Saccharomyces cerevisiae. Mol. Biol. Cell 13, 503-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton, S., Wepf, R., and Simons, K. (1996). Roles for Rac1 and CDC42 in planar polarization and hair outgrowth in the wing of Drosophila. J. Cell Biol. 135, 1277-1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emoto, K., He, Y., Ye, B., Grueber, W. B., Adler, P. N., Jan, L. Y., and Jan, Y. N. (2004). Control of dendritic branching and tiling by the Tricornered-kinase/Furry signaling pathway in Drosophila sensory neurons. Cell. 119, 245-256. [DOI] [PubMed] [Google Scholar]
- Fristrom, D., Wilcox, M., and Fristrom, J. (1993). The distribution of PS integrins, laminin A and F actin during key stages in Drosophila wing development. Development 117, 509-523. [DOI] [PubMed] [Google Scholar]
- Geng, W., He, B., Wang, M., and Adler, P. N. (2000). The tricornered gene, which is required for the integrity of epidermal cell extensions, encodes the Drosophila nuclear DBF2-related kinase. Genetics 156, 1817-1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giot, L. et al. (2003). A protein interaction map of Drosophila melanogaster. Science 302, 1727-1736. [DOI] [PubMed] [Google Scholar]
- He, B., and Adler, P. N. (2001). Cellular mechanisms in the development of the Drosophila arista. Mech. Dev. 104, 69-78. [DOI] [PubMed] [Google Scholar]
- He, Y., Fang, X., Emoto, K., Jan, Y. N., and Adler, P. N. (2005). The tricornered Ser/Thr protein kinase is regulated by phosphorylation and interacts with Furry during, Drosophila wing hair development. Mol. Biol. Cell 16, 689-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirata D. et al. (2002). Fission yeast Mor2/Cps12, a protein similar to Drosophila Furry, is essential for cell morphogenesis and its mutation induces Wee1-depdendent G(2) delay. EMBO J. 21, 4863-4874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopmann, R., Cooper, J. A., and Miller, K. G. (1996). Actin organization, bristle morphology, and viability are affected by actin capping protein mutations in Drosophila. J. Cell Biol. 133, 1293-1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou, M. C., Wiley, D. J., Verde, F., and McCollum, D. (2003). Mob2p interacts with the protein kinase Orb6p to promote coordination of cell polarity with cell cycle progression. J. Cell Sci. 116, 125-135. [DOI] [PubMed] [Google Scholar]
- Justice, R. W., Zilian, O., Woods, D. F., Noll, M., and Bryant, P. J. (1995). The Drosophila tumor suppressor gene warts encodes a homolog of human myotonic dystrophy kinase and is required for the control of cell shape and proliferation. Genes Dev. 9, 534-546. [DOI] [PubMed] [Google Scholar]
- Komarnitsky, S. I., Chiang, Y. C., Luca, F. C., Chen, J., Toyn, J. H., Winey, M., Johnston, L. H., and Denis, C. L. (1998). DBF2 protein kinase binds to and acts through the cell cycle-regulated MOB1 protein. Mol. Cell. Biol. 18, 2100-2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai, Z.-C., Wei, X., Shimizu, T., Ramos, E., Rohrbaugh, M., Nikolaidis, N., Ho, L.-L., and Li, Y. (2005). Control of cell proliferation and apoptosis by Mob as tumor suppressor, Mats. Cell 120, 675-685. [DOI] [PubMed] [Google Scholar]
- Leiserson, W. M., Harkins, E. W., and Keshishian, H. (2000). Fray, a Drosophila serine/threonine kinase homologous to mammalian PASK, is required for axonal ensheathment. Neuron 28, 793-806. [DOI] [PubMed] [Google Scholar]
- Luca, F. C., and Winey, M. (1998). MOB1, an essential yeast gene required for completion of mitosis and maintenance of ploidy. Mol. Biol. Cell 9, 29-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mah, A. S., Jang, J., and Deshaies, R. J. (2001). Protein kinase Cdc15 activates the Dbf2-Mob1 kinase complex. Proc. Natl. Acad. Sci. USA 98, 7325-7330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millward, T. A., Cron, P., and Hemmings, B. A. (1995). Molecular cloning and characterization of a conserved nuclear serine (threonine) protein kinase. Proc. Natl. Acad. Sci. USA 92, 5022-5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millward, T. A., Hess, D., and Hemmings, B. A. (1999). Ndr1 protein kinase is regulated by phosphorylation on two conserved sequence motifs. J. Biol. Chem. 274, 33847-33850. [DOI] [PubMed] [Google Scholar]
- Nelson, B. et al. (2003). RAM: a conserved signaling network that regulates Ace2p transcriptional activity and polarized morphogenesis. Mol. Biol. Cell 14, 3782-3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shraiman, B. I. (2005). Mechanical feedback as a positive regulator of tissue growth. Proc. Natl. Acad. Sci. USA 102, 3318-3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegert, M. R., Tamaskovic, R., Bichsel, S. J., Hergovich, A., and Hemmings, B. A. (2004). Regulation of NDR2 protein kinase by multi-site phosphorylation and the S100B calcium-binding protein. J. Biol. Chem. 279, 23806-23812. [DOI] [PubMed] [Google Scholar]
- Tamaskovic, R., Bichsel, S. J., Rogniaux, H., Stegert, M. R., and Hemmings, B. A. (2003). Mechanism of Ca2+-mediated regulation of NDR1 protein kinase through autophosphorylation and phosphorylation by an upstream kinase. J. Biol. Chem. 278, 6710-6718. [DOI] [PubMed] [Google Scholar]
- Thibault, S. T. et al. 2004. A complementary transposon tool kit for Drosophila melanogaster using P and piggyBac. Nat. Genet. 36, 283-287. [DOI] [PubMed] [Google Scholar]
- Tilney, L. G., Connelly, P. S., Vranich, K. A., Shaw, M. K., and Guild, G. M. (2000). Actin filaments and microtubules play different roles during bristle elongation in Drosophila. J. Cell Sci. 113, 1255-1265. [DOI] [PubMed] [Google Scholar]
- Turner, C. M., and Adler, P. N. (1998). Distinct roles for the actin and microtubule cytoskeletons in the morphogenesis of epidermal hairs during wing development in Drosophila. Mech. Dev. 70, 181-192. [DOI] [PubMed] [Google Scholar]
- Verde, F., Wiley, D. J., and Nurse, P. (1998). Fission yeast orb6, a ser/thr protein kinase related to mammalian rho kinase and myotonic dystrophy kinase is required for maintenance of cell polarity and coordinates cell morphogenesis with the cell cycle. Proc. Natl. Acad. Sci. USA 95, 7526-7531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verheyen, E. M., and Cooley, L. (1994). Profilin mutations disrupt multiple actin-dependent processes during Drosophila development. Development 120, 717-728. [DOI] [PubMed] [Google Scholar]
- Weiss, E. L., Kurischko, C., Zhang, C., Shokat, K., Drubin, D. G., and Luca, F. C. (2002). The Saccharomyces cerevisiae Mob2p-Cbk1p kinase complex promotes polarized growth and acts with the mitotic exit network to facilitate daughter cell-specific localization of Ace2p transcription factor. J. Cell Biol. 158, 885-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong, L. L., and Adler, P. N. (1993). Tissue polarity genes of Drosophila regulate the subcellular location for prehair initiation in pupal wing cells. J. Cell Biol. 123, 209-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, T., and Rubin, G. M. (1993). Analysis of genetic mosaics in developing and adult Drosophila tissues. 117, 1223-1237. [DOI] [PubMed] [Google Scholar]
- Xu, T., Wang, W., Zhang, S., Stewart, R. A., and Yu, W. (1995). Identifying tumor suppressors in genetic mosaics: the Drosophila lats gene encodes a putative protein kinase. Development 121, 1053-1063. [DOI] [PubMed] [Google Scholar]
- Yarden, O., Plamann, M., Ebbole, D. J., and Yanofsky, C. (1992). cot-1, a gene required for hyphal elongation in Neurospora crassa, encodes a protein kinase. EMBO J. 11, 2159-2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zallen, J. A., Peckol, E. L., Tobin, D. M., and Bargmann, C. I. (2000). Neuronal cell shape and neurite initiation are regulated by the Ndr1 kinase SAX-1, a member of the Orb6/COT-1/warts serine/threonine kinase family. Mol. Biol. Cell 11, 3177-3190. [DOI] [PMC free article] [PubMed] [Google Scholar]