Abstract
The distribution of identical enzymatic activities between different subcellular compartments is a fundamental process of living cells. At present, the Saccharomyces cerevisiae aconitase enzyme has been detected only in mitochondria, where it functions in the tricarboxylic acid (TCA) cycle and is considered a mitochondrial matrix marker. We developed two strategies for physical and functional detection of aconitase in the yeast cytosol: 1) we fused the α peptide of the β-galactosidase enzyme to aconitase and observed α complementation in the cytosol; and 2) we created an ACO1-URA3 hybrid gene, which allowed isolation of strains in which the hybrid protein is exclusively targeted to mitochondria. These strains display a specific phenotype consistent with glyoxylate shunt elimination. Together, our data indicate that yeast aconitase isoenzymes distribute between two distinct subcellular compartments and participate in two separate metabolic pathways; the glyoxylate shunt in the cytosol and the TCA cycle in mitochondria. We maintain that such dual distribution phenomena have a wider occurrence than recorded currently, the reason being that in certain cases there is a small fraction of one of the isoenzymes, in one of the locations, making its detection very difficult. We term this phenomenon of highly uneven isoenzyme distribution “eclipsed distribution.”
INTRODUCTION
The distribution of identical enzymatic activities (isoenzymes) between different subcellular compartments can be achieved by several routes, including existence of two or more genes, alternative transcription and translation initiations, or splicing, all of which lead to synthesis of two translation products. In recent years, there have been discoveries of protein-sorting mechanisms in which a single initial translation product is distributed between membrane-separated subcellular compartments. For example, a subset of the Aky2 molecules in yeast are targeted to the intermembrane space of mitochondria, whereas the rest of the molecules fold before their targeting signal accesses the import receptors and are therefore retained in the cytosol (Strobel et al., 2002). The single translation product of the FUM1 gene, encoding fumarase is distributed between the cytosol and mitochondria of Saccharomyces cerevisiae. Unlike Aky2, all fumarase molecules are targeted to mitochondria and are processed to their mature form before distribution. A subset of fumarase polypeptides are further translocated into the mitochondrial matrix, whereas the rest are released into the cytosol by retrograde movement (Stein et al., 1994; Knox et al., 1998). It is important to point out that the cytosolic and mitochondrial isoenzymes of fumarase are identical according to mass spectrometry; thus, no specific modification of the proteins seems to be involved in their subcellular distribution.
Aconitase, a tricarboxylic acid (TCA) enzyme that converts citrate to isocitrate, belongs to the family of iron–sulfur-containing proteins (Gardner, 1997). Interest in aconitase in higher eukaryotes has been motivated by the discovery that a cytosolic aconitase isoenzyme is an iron-regulated RNA binding protein, (Hentze and Kuhn, 1996; Rouault and Klausner, 1996). In S. cerevisiae, the enzyme aconitase functions in the mitochondrial TCA cycle and is not involved in the regulation of iron metabolism. A single nuclear gene in S. cerevisiae, ACO1, encodes the catalytically active aconitase enzyme. Disruption of the ACO1 gene leads to a respiratory-deficient phenotype and glutamate auxotrophy together with the elimination of cellular aconitase activity (Gangloff et al., 1990). Historically, it was suggested that a cytosolic aconitase might be required for the function of the glyoxylate pathway. This pathway enables plant and fungal cells to make four-carbon organic compounds out of two-carbon compounds, thereby allowing them to grow on acetate, ethanol, or oleate as their carbon source. In plants, the glyoxylate cycle occurs in peroxisomes (Charlton et al., 2004), whereas the enzymes of the glyoxylate pathway in yeast (citrate synthase, isocitrate lyase, and malate synthase) are not localized to peroxisomes (McCammon et al., 1990; McAlister-Henn and Small, 1997). Since cloning of the ACO1 gene in yeast (Gangloff et al., 1990), its natural translation product has been detected only in mitochondria and is actually referred to as an exclusive mitochondrial protein.
We decided to reexamine the question of aconitase localization in yeast with the notion that this may be an example of a highly uneven distribution of a single translation product. We developed two strategies for the physical and the functional detection of aconitase in the yeast cytosol. Our results suggest that aconitase in S. cerevisiae is distributed between two distinct subcellular compartments and participates in two different metabolic pathways: the glyoxylate shunt in the cytosol and the TCA cycle in mitochondria.
MATERIALS AND METHODS
Strains, Plasmids, and Growth Conditions
S. cerevisiae strains used were BY4741 (Mat a; his3Δ1; leu2Δ0; met15Δ0; ura3Δ0) and YPH499 (ura3,lys2,ade2,trp1,his3,leu2), BY4742 (Mat α; his3Δ1; leu2Δ0; lys2Δ0; ura3Δ0; YLR304c::kanMX4), and BY4743 (Mat a/α; his3Δ1/his3Δ1; leu2Δ0/leu2Δ0; lys2Δ0/LYS2; MET15/met15Δ0; ura3Δ0/ura3Δ0; YLR304c:: kanMX4/YLR304c). We used BY4743 strain for tetrad analysis after transformation with pAco1-WT plasmid to get chromosomal aconitase haploid knockouts (Δaco1) expressing external wild-type (WT) protein under the GAL1 promoter. To obtain chromosomal knockout strains expressing variant derivatives of aconitase, we transformed the Δaco1 harboring pAco1-WT with the relevant plasmids and subsequently cured the pAco1-WT plasmid.
Growth Conditions. Strains harboring the appropriate plasmids were grown overnight at 30°C in synthetic depleted (SD) medium containing 0.67% (wt/vol) yeast nitrogen base (Difco, Detroit, MI) and 2% glucose, 2% galactose, 3% glycerol, or 3% ethanol + 2% acetate (wt/vol) supplemented with the appropriate amino acids (50 μg/ml). For agar plates, 2% agar was added and when indicated 0.08% X-gal was added dissolved in 100% N,N-dimethylformamide and 1× BU salts (25 mM sodium phosphate buffer titrated to pH 7.0).
Plasmids. pG1αwt (αc) and pYES/M15 (ωc) were kindly provided by D. Picard (Abbas-Terki and Picard, 1999). pCyb2-α and pFumΔSP-α are described previously (Karniely, Reizner, Sass, and Pines, unpublished data). pSP65Aco was kindly provided by G. Schatz (Biezentrum, University of Basel, Reinach, Switzerland).
pAco1-WT was created by cutting pSP65Aco with BamHI and HindIII and cloning into p425Gal1 vector (Mumberg et al., 1994). pAco1-α was created by cutting pAco1-WT with Eco72I and ApaI and ligating at the C terminus to α fragment cut by SmaI and ApaI from the pFumα plasmid (the α-peptide coding sequence corresponds to the first 81 amino acids of wild-type Escherichia coli β-galactosidase). For pAco1-ΔSP, aconitase sequence was amplified from valine 18 to the end of the open reading frame and subcloned into p425Gal1 vector (Mumberg et al., 1994). For pAco1-Ura3, URA3 was amplified using primers 5′-CCCCCGGGGGATCCCAAGATCCAAGC-3′ and 5′-CCCTCGAGTTAGTTTTGCTGGCCGCATC-3′. The product URA3 was cut with SmaI and XhoI and cloned into pAco1-WT cut with Eco72I and XhoI. The mitochondrial processing peptidase (MPP) cleavage mutant (pAco1-CSP) was created from pAco1-WT as a template and by QuikChange II site-directed mutagenesis kit (Stratagene, La Jolla, CA). We used primer 5′-CCATCAAG AGACCCATTGTTGGTGGTGAAGCGACAGTCTCCAACTTGAC-3′.
Metabolic Labeling
Induced cultures (in galactose) were harvested and labeled with 10 μCi/ml [35S]methionine and further incubated for 30 min at 30°C. When required, 20 μM carbonyl cyanide m-chlorophenylhydrazone (CCCP) was added before labeling. Labeling was stopped by addition of 10 mM sodium azide. Labeled cells were collected by centrifugation, resuspended in Tris/EDTA buffer, pH 8.0, containing 1 mM phenylmethylsulfonyl fluoride, broken with glass beads for 2 min, and centrifuged to obtain the supernatant fraction. Supernatants were denatured by boiling in 1% SDS, immunoprecipitated with anti-aconitase rabbit antiserum and protein A-Sepharose (Amersham Biosciences, Piscataway, NJ), and then analyzed by SDS-PAGE.
Subcellular Fractionation
Induced yeast cultures were grown to 1.5 A at 600 nm. Mitochondria were isolated as described previously (Knox et al., 1998). Spheroplasts were prepared in the presence of Zymolyase-20T (MP Biomedicals, Irvine, CA). Each of our subcellular fractionation experiments were assayed for cross-contaminations using αHsp60 or αmtHsp70 as mitochondrial markers and αBmh1 as a cytosolic marker.
α-Complementation β-Galactosidase
Yeast cells were transformed with α peptide plasmids (pAco1-α, pCyb2-α, and pFumΔSP-α separately) and with pωc. Colonies were streaked on X-gal plates and incubated at 30°C for 72 h.
Enzymatic Assays: β-Galactosidase Activity
Yeast cultures were grown on medium containing 2% galactose as the sole carbon source to 0.5 A at 600 nm. Cells were collected, disrupted, and assayed as described previously (Ausubel et al., 1991). The data represent averages of two independent experiments (each performed in duplicate).
Aconitase Activity
Cellular aconitase activity was measured by monitoring the decrease in the absorbance of the substrate cis-aconitate at 240 nm as described previously (Fansler and Lowenstein, 1969).
URA3–5-Fluoroorotic acid (5-FOA) Selection
S. cerevisiae strains harboring pAco1-URA3 plasmid (contains Leu marker) were streaked on 2% galactose plate supplemented with 5-fluoroorotic acid (0.1%) to select for loss of the pAco1-URA3 in the cytosol. Colonies were picked and streaked on galactose media lacking either leucine or uracil to confirm loss of the Ura3 cytosolic activity.
RESULTS
Subcellular Localization of Aconitase in S. cerevisiae
Yeast aconitase had been proposed to have two distinct subcellular located enzymatic functions as part of the TCA cycle in mitochondria and glyoxylate shunt in the cytosol/peroxisome. Nevertheless, from the time the yeast Aco1 gene was cloned (Gangloff et al., 1990), the protein has been detected only in mitochondria, so much so, that aconitase has been used as a mitochondrial marker protein (e.g., Gerber et al., 2003; Waizenegger et al., 2004).
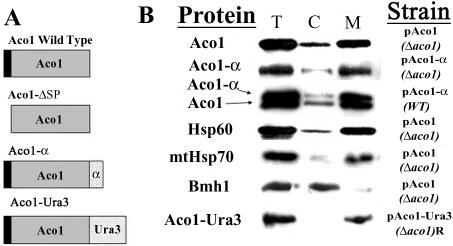
Our first approach was to reexamine aconitase subcellular distribution by standard subcellular fractionation. We have generated a new, highly specific polyclonal antiserum against Aco1 for this purpose. As shown in Figure 1B, the pattern of aconitase distribution either expressed from a plasmid (top) or from the chromosome (third, lower bands) resembles that of Hsp60 (fourth) and mtHsp70 (fifth), which are well-characterized mitochondrial markers. This distribution is the direct opposite of the distribution of Bmh1, which is a cytosolic marker (sixth). The light bands of Hsp60 and mtHsp70 that occur in the cytosolic fraction can be attributed to damage and leakage of mitochondria upon the procedure of subcellular fractionation. Yet in our experiments, it always seems that the “mitochondrial leakage” band of Aco1 is relatively stronger than that of Hsp60 and mtHsp70. This may be attributed to an alleged minor amount of cytosolic aconitase. We have tried to increase this presumed targeting of aconitase to the cytosol by growth of the cells on oleic acid, ethanol, and acetic acid that require the function of the glyoxylate shunt. So far, we have observed essentially insignificant differences in distribution of aconitase under the various conditions (our unpublished data).
Figure 1.
(A) Schematic representation of aconitase variants; wild-type Aco1, Aco1-ΔSP lacking the mitochondrial targeting sequence (16 a.a. indicated in black), Aco1-α containing 85 a.a. of the β-galactosidase α fragment (indicated with dots) inserted after the last amino acid of aconitase, and Aco1-Ura3 containing 358 a.a. of a Ura3 derivative (indicated with diagonal lines) inserted after the last amino acid of aconitase. (B) Subcellular fractionation of yeast cells expressing aconitase variants. Equivalent portions from fractions of the total (T) cytosol (C) and mitochondria (M) were analyzed by Western blotting using antibodies against the indicated proteins.
The Aco1-α Fusion Can Fully Complement an Aco1 Knockout Strain
The suspected relatively small amount of aconitase in the cytosol makes the research of this isoenzyme problematic. What we needed was a method designed to detect only the cytosolic protein in vivo, if in fact it exists. For this, we have exploited an approach for proteomic research developed by Karniely, Reizner, Sass, and Pines (unpublished data). This approach uses the α-complementation of the E. coli β-galactosidase enzyme. The α complementation is based on the ability of two artificially expressed peptide fragments of β-galactosidase (designated α and ω) to assemble in vivo into an enzymatically active complex, as has been demonstrated in bacteria, yeast, and mammalian cells (Ullmann et al., 1967; Moosmann and Rusconi, 1996; Abbas-Terki and Picard, 1999). The rationale of our screen is the requirement for coexpression of both α and the ω polypeptides in the same subcellular compartment to reconstitute the β-galactosidase activity. We have fused the α fragment of β-galactosidase to the C terminus of aconitase (Aco1-α) and expressed this in yeast, whereas the ω polypeptide was expressed in the cytosol (ωc). The two plasmids that allow expression of Aco1-α and ωc under the GAL1 promoter were designated pAco1-α and pωc, respectively. The first step in using the Aco1-α fusion for analysis of aconitase distribution was to show that the apparent characteristics of the fusion protein are identical to those of the wild-type protein.
We have examined the hybrid protein's subcellular distribution (Figure 1B), enzymatic activity in vivo (Figure 2A) and in vitro (Figure 2B) and processing of the protein in mitochondria (Figure 2C). Subcellular fractionation experiments reveal that the majority of the Aco1-α molecules occur in the mitochondrial fraction (Figure 1B, 2). The rest are found in the cytosolic fraction, a pattern that is reminiscent of the Aco1 wild-type protein (Figure 1B, top). This also can be seen when both the wild-type and Aco1-α are coexpressed in yeast cells (Figure 1B, third).
Figure 2.
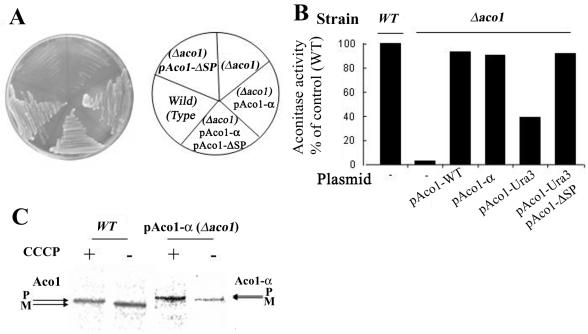
(A) Aco1-α can complement a Δaco1 strain. Wild-type and Δaco1 strains harboring the indicated plasmids encoding aconitase-α hybrid protein (pAco1-α) or aconitase lacking the mitochondrial targeting sequence (pAco1-ΔSP) were grown on galactose medium plates. (B) Aconitase-specific activity. Wild-type and Δaco1 harboring the indicated plasmids were grown in galactose medium, and cell extracts were assayed for aconitase activity. (C) Processing of Aco1-α fusions. Aco1-α processing is blocked by inhibiting import into mitochondria with the proton ionophore CCCP. Wild-type and Δaco1 yeast strains induced for expression of Aco1-α were labeled with [35S]methionine for 30 min, either in the absence (-) or presence (+) of 20 μM CCCP. Total cell extracts were prepared, immunoprecipitated with aconitase antiserum, and analyzed by SDS-PAGE. Dual arrows show positions of the precursor (P) and mature (M) proteins.
The Aco1-α fusion protein can complement an aconitase chromosomal knockout strain; a yeast Δaco1 strain does not grow on galactose plates, whereas the same strain expressing the fusion protein (Aco1-α) readily grows on the same medium and grows similarly to knockout strains expressing the native Aco1 (Figure 2A). Worth pointing out is that in many yeast strains, efficient catabolism of galactose in contrast to glucose requires respiration of the cells. If respiration is blocked by a mutation of a TCA cycle enzyme such as aconitase, growth is blocked or severely inhibited. This has been termed the Kluyver effect, which has recently been suggested to be caused by low-level transporter activities (e.g., gal2p transporter for galactose) that are not sufficient to sustain a high sugar flow necessary for fermentative growth (Fukuhara, 2003). Another explanation has been that galactose in contrast to glucose does not cause the Crabtree effect (repression of the TCA cycle and respiration), thus making growth on galactose extremely inefficient when the TCA cycle is blocked. Obviously, the growth of cultures on glycerol also is defective under such conditions (our unpublished data) but because in many experiments galactose is used for induction of the GAL1 promoter, we preferred to show results with this carbon source.
In vitro enzymatic activity assays performed on yeast cell extracts showed the same trend as found in vivo. Yeast extracts of Aco1 knockout strains exhibited insignificant aconitase activity, whereas the same strains expressing the Aco1-α fusion protein exhibited 91% of the wild-type strain's specific activity (Figure 2B). The aconitase chromosomal knockout strain expressing the native protein from a plasmid exhibited 93% activity of the wild-type strain (Figure 2B).
Aco1-α is processed efficiently, as detected by labeling experiments in the presence (+) and absence (-) of CCCP, which dissipates membrane potential and blocks import. As shown in Figure 2C, in the presence of CCCP both Aco1 and Aco1-α occur as higher molecular weight bands corresponding to the precursors (P) of these proteins and in the absence of CCCP, the mature proteins (M) are detected. These results indicate that the Aco1-α protein has similar features to the wild-type protein.
Aco1-α Associates with a β-Galactosidase ω-Fragment in the Yeast Cytosol
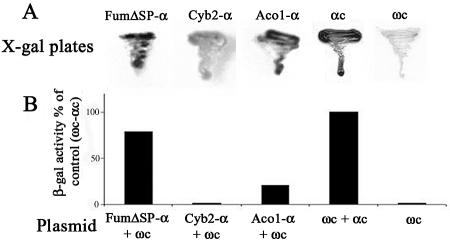
To examine the localization of Aco1-α protein in vivo using the α-complementation system, plasmids encoding Aco1-α and a cytosolically localized ω fragment of β-galactosidase (pωc) were simultaneously expressed in S. cerevisiae. When these cells were grown on agar plates containing the X-gal indicator and galactose as the carbon source, blue colonies were obtained, indicating that at least some of the Aco1-α molecules were located in the cytosol (Figure 3A). As controls, we show that Cyb2-α (a cytochrome b2 mitochondrial targeting signal fused to the β-galactosidase α fragment), which is exclusively targeted to mitochondria (negative control), produces noncolored colonies similar to a strain lacking α expression (Figure 3A). Cells expressing FumΔSP-α (fumarase lacking a mitochondrial targeting sequence and exclusively localized in the cytosol), a positive control (Karniely, Reizner, Sass, and Pines, unpublished data), produce blue colonies, similarly to cells expressing the cytosolic ωc and cytosolic αc fragments (Figure 3A). We have performed additional controls for those fusions in which α seems to be exclusively targeted to mitochondria, thereby producing noncolored colonies. By coexpressing them with ωm (ω fused to a mitochondrial targeting sequence), which is partially targeted to mitochondria, these strains produced blue colonies on X-gal plates, confirming that their α-complementation is only dependent on colocalization of the β-galactosidase fragments (our unpublished data).
Figure 3.
α Complementation. (A) Yeast cultures harboring ωc and the indicated plasmids were streaked on galactose medium plates containing X-gal. Blue colonies detect α fragments that are in the cytosol and associate with cytosolic ω (ωc); Aco1-α fusion protein, cytochrome b2-α (fully imported into mitochondria), FumΔSP-α (lacking a mitochondrial targeting signal and localized only in the cytosol), αc (cytosolic α fragment). (B) β-Galactosidase activity. Yeast cultures harboring the indicated plasmids were grown in galactose medium, and culture extracts were assayed for β galactosidase activity.
β-Galactosidase activity was also determined in extracts of yeast cultures expressing either Aco1-α, FumΔSP-α (cytosolic), or Cyb2-α (mitochondrial) together with ωc. As shown in Figure 3B, the strain expressing the plasmids encoding Aco1-α and ωc displayed significant β-galactosidase enzymatic activity compared with the FumΔSPα/ωc combination. The control coexpressing cytosolic ωc and cytosolic αc fragments displayed the highest β-galactosidase activity (the 100% reference). Neither the Cyb2-α/ωc combination nor the Aco1-α nor the ωc when expressed on their own exhibited enzymatic activity. In addition, cells encoding ωc and a sole mitochondrial derivative of fumarase fused to α produced noncolored colonies (our unpublished data). These results allow us to conclude that a fraction of the Aco1-α molecules is localized in the cytosol.
An Aco1-Ura3 Hybrid Protein, Exclusively Targeted to Mitochondria, Does Not Allow Growth on Ethanol-Acetate
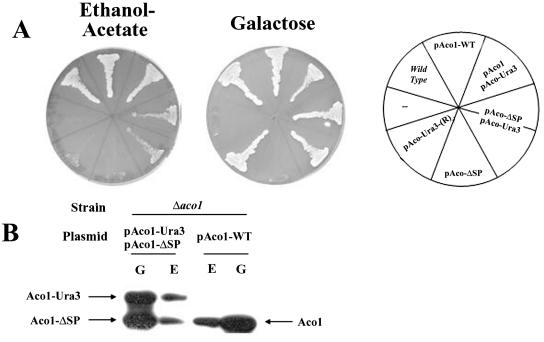
A second approach to the study of aconitase distribution was to demonstrate that it has a specific function in the cytosol. To identify a phenotype associated with aconitase depletion from the cytosol, we used a powerful selection that is based on the creation of ACO1-URA3 hybrid genes (Alani and Kleckner, 1987). The C terminus of aconitase was fused to a functional URA3 gene product (orotidine-5′-phosphate decarboxylase). If an Aco1-Ura3 product is even partially localized to the cytoplasm, such yeast strains are sensitive to the analog 5-FOA. However, if mutations in the aconitase gene on the plasmid or mutations elsewhere in the yeast chromosome cause the Ura3 fusion protein to be targeted exclusively to mitochondria, the strain should be resistant to 5-FOA. Yeast harboring an aconitase chromosomal knockout (Δaco1) and expressing Aco1-Ura3 under the GAL1 promoter on a plasmid (pAco1-Ura3) were grown on galactose agar plates containing 5-FOA. We isolated 5-FOA-resistant strains presumably selected for misdistribution of Aco1-Ura3. Two facts are worth mentioning; the first fact is that such 5-FOA-resistant strains occurred at very high frequency, indicating that all these mutations did not occur in a single loci. The second fact is that plasmids rescued from such resistant strains did not provide 5-FOA resistance to naïve strains transformed with the rescued plasmids, suggesting that the 5-FOA resistance trait was not plasmid borne.
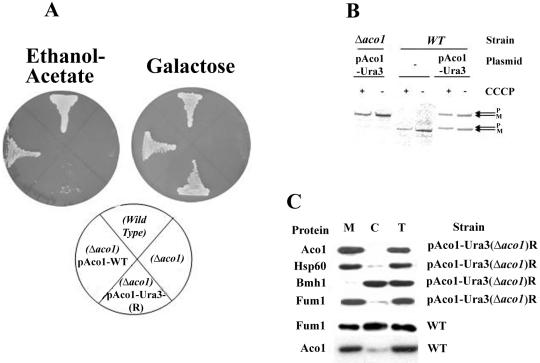
These 5-FOA-resistant strains, which presumably have exclusive mitochondrial targeting of the Aco1-Ura3, were examined for a specific phenotype. Loss of cytosolic aconitase was found to result in a glyoxylate shunt defect as indicated by the yeast mutant's inability to grow on plates with ethanol-acetate (Figure 4A, left) or oleate (our unpublished data) as the sole carbon source. As shown in Figure 4A, the Aco1-Ura3-(R) strain did not grow on ethanol-acetate in contrast to the wild-type strain or a Δaco1 strain expressing the wild-type protein. The Aco1-Ura3-(R) mitochondrial function was not abolished because the same mutant strain maintained growth (although somewhat slower) on galactose (Figure 4A, right) and glycerol plates (our unpublished data). Accordingly, the Aco1-Ura3 hybrid is targeted and processed properly in the mitochondria as evident from a labeling experiments in the presence or absence of CCCP (Figure 4B, compare + and - CCCP, see the P and M bands). In summary, the results mentioned above imply that aconitase in the wild-type is localized to, and functions in, the yeast cytosol. One fact worth mentioning is that the distribution of chromosomal encoded fumarase also is compromised in an Aco1-Ura3-(R) strain (Figure 4C fourth) and that a cured strain expressing Aco1-α and ωc develops white colonies on X-gal plates (our unpublished data).
Figure 4.
(A) Isolation of yeast strains expressing Aco1-Ura3 exclusively in mitochondria. Yeast strains expressing Aco1-Ura3 in a Δaco1 background were selected for 5-FOA resistance. The 5-FOA-resistant yeast strain (R) harboring a plasmid encoding aconitase-Ura3 hybrid protein (pAco1-Ura3) together with Wild-type, Δaco1, and Δaco1 harboring pAco1-WT were grown on galactose or ethanol-acetate agar plates. (B) Processing of Aco1-Ura3. Cells expressing Aco1-Ura3 in a wild-type or a Δaco1 background were labeled with [35S]methionine for 30 min, either in the absence (-) or presence (+) of CCCP (as described in legend Figure 2C). Total cell extracts were prepared, immunoprecipitated with aconitase antiserum, and analyzed by SDS-PAGE. Dual arrows show positions of the precursor (P) and the mature (M) proteins. (C) Subcellular fractionation of yeast cells expressing aconitase variants. Equivalent portions from fractions of the total (T), cytosol (C), and mitochondria (M) were analyzed by Western blotting using antibodies against the indicated proteins.
Aco1 Lacking a Mitochondrial Targeting Sequence Complements Aco1-Ura3-(R) for Growth on Ethanol-Acetate
To validate the function of aconitase in the cytosol, we have constructed a plasmid designed to express Aco1 exclusively in the cytosol. The sequence encoding the first 17 amino acids was replaced by a single methionine codon to initiate translation without the mitochondrial targeting signal. As with the other constructs, transcription was under the regulation of the GAL1 promoter. We examined Aco1-ΔSP localization using a combination of subcellular fractionation and protease sensitivity experiments and found that it is absent from the mitochondrial matrix (our unpublished data). On transformation and expression of Aco1-ΔSP into an Aco1-Ura3-(R) strain (5-FOA resistant), the resultant strain succeeded in growing on ethanol-acetate plates (Figure 5A, left). Although growth of the strain was less efficient than the Δaco1 strain expressing native Aco1, it was in clear contrast to Aco1-Ura3-(R) strains, which failed to grow on ethanol-acetate altogether. Δaco1 strains expressing only Aco1-ΔSP are unable to use either ethanol-acetate or galactose as the sole carbon source because they lack mitochondrial aconitase activity required for growth on both media (Figure 5A). Aco1-ΔSP can complement the cytosolic glyoxylate shunt function but not the TCA cycle function of aconitase (Figure 5A). For clarity, it is important to point out that growth of these strains on ethanol-acetate results in lower expression of the plasmid encoded proteins that are under the GAL1 promoter (Figure 5B, compare E with G). Nevertheless, Aco1-ΔSP, Aco1-Ura3 and Aco1 are detected and apparently functional at this lower level. The data mentioned above indicate that aconitase is required for a specific metabolic function in the cytosol.
Figure 5.
(A) Aco1-ΔSP can complement ΔAco1-pAco1-Ura3-(R) cells for growth on ethanol-acetate. Yeast wild-type and Δaco1 strains harboring the indicated plasmids and the Δaco1-5FOA-resistant strain (R) harboring pAco1-URA3 encoding the Aco1-Ura3 hybrid protein were grown on galactose or ethanol-acetate medium agar plates. (B) Aco1-Ura3 and Aco1-ΔSP coexpression. Yeast Δaco1 strains expressing the indicated aconitase derivatives were grown in media containing galactose (G) or ethanol-acetate (E) as the sole carbon source. Total cell extracts were prepared, blotted, and assayed with aconitase antiserum and analyzed by SDS-PAGE.
Aconitase's MPP Cleavage Site and the Generation of a Cleavage Site Mutant
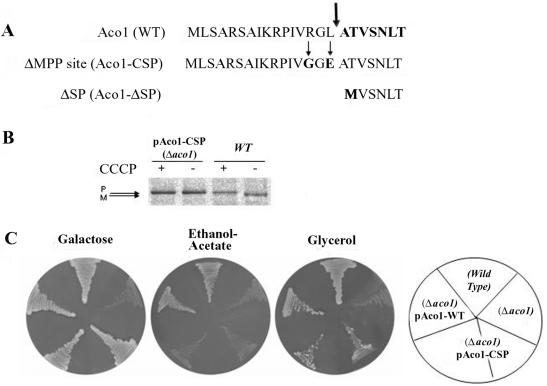
To characterize aconitase translation products, one of our first efforts was to determine the precise position of aconitase processing by MPP in mitochondria. Aconitase was purified from yeast by immunoprecipitation using our specific aconitase antibodies and was subjected to sequential Edman degradation. The amino terminal sequence detected was ATVSNLT and as shown in Figure 6A (top, sequence in bold), this means that yeast aconitase is cleaved by MPP between leucine 16 and alanine 17 (Figure 6A, thick arrow).
Figure 6.
(A) Amino terminus of aconitase. Sequences of the amino termini of wild-type aconitase, aconitase with a mutated MPP cleavage site (Aco1-CSP), and aconitase lacking the mitochondrial targeting sequence (Aco1-ΔSP). The thick arrow indicates the position of the MPP cleavage site determined by Edman degradation. The thin arrows indicate amino acids changes in the MPP cleavage site mutant. (B) Aconitase's MPP cleavage site mutant is not processed. Aconitase processing was blocked by inhibiting import into mitochondria with CCCP. Wild-type or Δaco1 yeast cells induced in galactose medium for the expression of the Aco1 MPP cleavage site mutants (Aco1-CSP) were labeled with [35S]methionine for 30 min, either in the absence (-) or presence (+) of CCCP. Total cell extracts were prepared, immunoprecipitated with aconitase antiserum and analyzed by SDS-PAGE. Dual arrows show positions of the precursor (P) and mature (M) proteins. (C) pAco1-CSP encoding an unprocessable Aco1 does not complement cells for growth on ethanol-acetate. Cells expressing an unprocessed aconitase were examined for growth on ethanol-acetate as the carbon source. Wild-type and Δaco1 harboring the indicated plasmids were grown on galactose, ethanol-acetate, or glycerol medium agar plates.
Worth mentioning is the fact that in contrast to our experimental data mentioned above, aconitase has been previously predicted to be cleaved twice (Branda and Isaya, 1995), once by MPP between phenylalaine 11 and isoleucine 12 (see below) and a second time by mitochondrial intermediate peptidase between valine 19 and serine 20 (see below): [aconitase amino terminus: MLSARSAIKRP(*)IVRGLATV(*)SNLTR].
In addition, according to mitochondrial targeting sequence prediction programs: MitoProt II (http://ihg.gsp.de/cgi-bin/paolo/mitofilter) and PSORT II (http://psort.nibb.ac.jp/cgi-bin/rupsort.pl), the predicted cleavage site is between tryptopan 23 and arginine 24, which also is different from what we have found.
We addressed the question whether a precursor form of aconitase can be functional in the cytosolic glyoxylate shunt. Based on the determination of the aconitase MPP cleavage site, we have constructed a mutant aconitase gene, which encodes an unprocessed protein. This was accomplished by creating two point mutations at critical points within the MPP cleavage site of aconitase as indicated in Figure 6A, middle (amino acid changes are indicated by thin arrows and by G and E in bold). Arginine 14 at position 3 to our deduced processing site was replaced by a glycine and a negatively charged glutamic acid was introduced instead of leucine 16 (R14G, L16E). Cultures of S. cerevisiae harboring a plasmid encoding the aconitase MPP cleavage site mutant (pAco1-CSP) were grown on galactose as the carbon source to induce the expression under the GAL1 promoter. Metabolic labeling of these cultures was performed in the absence (-) or presence (+) of CCCP. As shown in Figure 6B, these two point mutations abolished the processing of aconitase in vivo, which occurred as an identical precursor size band in the presence and absence of the uncoupler (compare differences in size for Aco1 and no change of Aco1-CSP mutant).
This mutant pAco1-CSP could not complement Δaco1 strains on ethanol-acetate plates (Figure 6C, middle), indicating a defect in the cytosolic glyoxylate shunt. The same strain exhibits no difficulty growing on both galactose (Figure 6C, left) and glycerol (Figure 6C, right) plates, indicating that their mitochondrial TCA is functional. In contrast, strains expressing wild-type Aco1 were able to restore the ability of the Δaco1 strain to grow on ethanol-acetate medium. These results indicate that the mutations in Aco1-CSP affect the protein's activity only in the cytosol. In other words, the full unprocessed precursor of aconitase is essentially nonfunctional in the cytosolic glyoxylate pathway.
Aco1 Can Be Imported Posttranslationally
There are several possible mechanisms that can explain aconitase distribution in yeast. One of these possibilities is a mechanism similar to that of yeast fumarase. Two unique features of fumarase targeting and distribution in yeast are 1) all fumarase translation products are targeted and processed in mitochondria before distribution (Sass et al., 2001, 2003) and 2) translocation of fumarase into mitochondria must occur during translation or immediately after its completion (Knox et al., 1998). To approach these questions, wild-type yeast cultures were pulse labeled for 10 min in the absence or presence of CCCP, and cell extracts were immunoprecipitated with Aco1 antiserum followed by SDS-PAGE analysis. A single mature form of aconitase was detected in the absence of CCCP (Figure 7, lane 1), whereas all molecules occurred as precursors in the presence of CCCP (Figure 7, lane 2). Importantly, note that no precursor was detected in the absence of CCCP, even when the experiment described above was performed with higher specific radioactivity or when the gel was overexposed to sensitive film.
Figure 7.
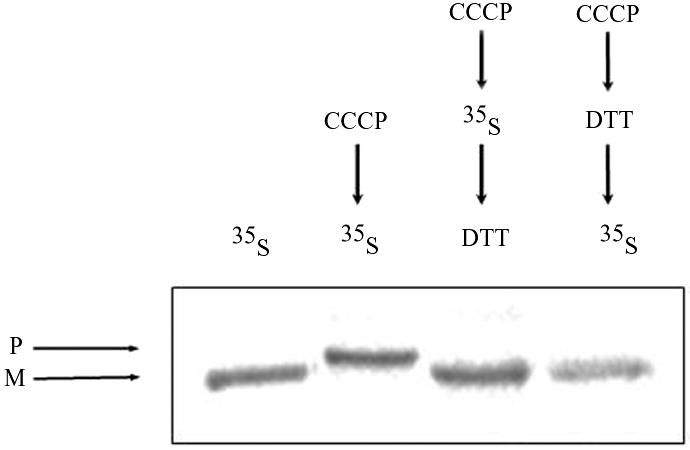
Reversal of an import block allows posttranslational translocation of aconitase. Pulse-labeled (35S Met) processed translation product (M) of aconitase was detected in lane 1, whereas there was no trace of the precursor (P) (as described in the legend of Figure 2). Pulse-labeled (35S Met) aconitase precursors (P) that accumulated in the presence of CCCP (lane 2 and legend of Figure 2C) were efficiently chased into mitochondria and processed after treatment with DTT (lane 3). As a control, cells were first treated with CCCP followed by incubation with DTT and then labeled (lane 4).
In the same experiment described above, cells were labeled in the presence of CCCP to accumulate precursors in the cytosol, but then they were treated with the reducing agent dithiothreitol (DTT) (for 10 min) to restore the inner membrane potential. The addition of DTT resulted in the import of aconitase molecules as detected by the processing of all detectable aconitase molecules (Figure 7, lane 3). As a control for this experiment, cells were treated with CCCP for 10 min followed by a 10-min treatment with DTT, and only then were the cells labeled with [35S]methionine (Figure 7, lane 4), showing that the treatments of CCCP and DTT do not affect the import of nascent polypeptides. Together, these results show that the aconitase precursor can be posttranslationally imported into mitochondria in contrast to fumarase (Knox et al., 1998).
DISCUSSION
In contrast to the celebrated examples in which there are significant amounts of a specific enzyme in more than one subcellular compartment (see Introduction), we maintain that the isoenzyme distribution phenomenon has a wider occurrence than recorded currently. The reason being that in many cases there may be a small, overlooked amount of one of the isoenzymes in one of the cell compartments. We term this phenomenon “eclipsed distribution,” in which the large amount of one of the isoenzymes obscures detection of the small amount of the other. This study indicates that aconitase may be an excellent example of such an eclipsed distribution because it seems to be localized to and function in both cytosol and mitochondria of S. cerevisiae. Dual localization of aconitase is supported by the following observations: 1) An Aco1-α fusion (which seems to have identical behavior to wild-type Aco1) associates with an ω fragment in the cytosol. 2) An Aco1-Ura3 hybrid protein targeted exclusively to mitochondria displays a cytosolic glyoxylate shunt defect (manifested in its inability to grow on ethanol-acetate or oleic acid as the carbon source). 3) The glyoxylate shunt defect in strain Aco1-Ura3-(R) can be complemented with an Aco1-ΔSP, which is exclusively located in the cytosol. 4) According to standard subcellular fractionation a small amount of Aco1 probably exists in the cytosol. 5) In experiments that will be published elsewhere, a “degron” was added to the C terminus of aconitase, thus targeting the cytosolic isoenzyme for degradation by the proteosome, whereas the mitochondrial isoenzyme is stable. Strains expressing the aconitase-degron show a specific glyoxylate shunt defect that can be complemented with Aco1-ΔSP. In fact, an unstable Aco1-degron expressed in the cytosol can be stabilized by proteosome inhibitors (Shlevin, Regev-Rudzki, Karniely, and Pines, unpublished data).
In summary, we show the localization and function of cytosolic aconitase in yeast cells, probably as part of the glyoxylate cycle. For aconitase, not only is its cytosolic location eclipsed by the relatively large amount of mitochondrial aconitase but also its function is eclipsed. In the absence of mitochondrial aconitase, cells are petite and are essentially unable to grow on ethanol, acetic, or oleic acids. Thus, lack of a probable function in the glyoxylate shunt cannot be detected by a simple knockout of the ACO1 gene from the yeast chromosome. Only by creating a strain in which the Aco1 mitochondrial function is intact, while its cytosolic function is defective, could we observe a glyoxylate shunt defect. This defect is the result of the inability of the cells to make four-carbon organic compounds out of two-carbon-containing compounds via the glyoxylate shunt. In this regard, it is interesting to note that neither mitochondrial nor cytosolic aconitase is able to substitute one for the other, as opposed to fumarase for which an exclusively located cytosolic protein (lacking a mitochondrial targeting sequence) can complement its mitochondrial TCA function (Wu and Tzagoloff, 1987; Stein et al., 1994).
As pointed out, we think that aconitase may be one example of a broad group of proteins whose dual targeting was missed due to eclipsed distribution. Another example of eclipsed distribution may be the enzyme Nfs1 that has been reported to have desulfurase activity. The NFS1 gene encodes a translation product that harbors both an amino-terminal mitochondrial targeting sequence and a more recently identified nuclear targeting signal (NLS) in the midst of the protein (Nakai et al., 2001). Mitochondrial Nfs1 mediates the assembly of iron–sulfur clusters in mitochondria, which are destined for both mitochondrial and cytosolic iron–sulfur proteins (Muhlenhoff et al., 2004). Initially, when the nfs1 null mutant was found to be inviable, it was assumed that this was only because its crucial iron–sulfur assembly function was lost. This was supported by the fact that Nfs1 was only detected in the mitochondrial matrix (Nakai et al., 2004). The fact that a nonmitochondrial Nfs1 also is essential for cell viability became apparent only when Nakai et al. (2001) demonstrated that Nfs1 contains an NLS and alteration of this NLS was lethal. Subsequently, Nakai et al. (2004) and Muhlenhoff et al. (2004) have shown that Nfs1 is responsible for an essential thiolation of tRNAs in the nucleus (Muhlenhoff et al., 2004; Nakai et al., 2004). To date, Nfs1 has not been physically identified in the nucleus or for a matter of fact anywhere else besides the mitochondrion. Therefore, this seems to be an extreme case of eclipsed distribution, which was detected fortuitously by the combination of identifying the NLS and the strong phenotype of the loss of function (lethality) in both of its subcellular locations.
A second topic that this study begins to address is the mechanism of Aco1 distribution. As pointed out in Introduction, many proteins are distributed by creation of two translation products: one containing an amino terminal signal peptide and one whose translation starts downstream, after the encoded mitochondrial targeting signal. These mechanisms require a second in frame methionine initiation codon (ATG) in the dual targeted protein's mRNA. ACO1 has no second translation start codon in the vicinity of the first ATG; the closest one being Met101, which is preceded by four ATGs not in frame. Thus, the amino terminal sequence of all aconitase molecules is predicted to harbor a mitochondrial targeting signal, and the situations in which two products are translated from two in-frame initiation codons (encompassing a mitochondrial targeting sequence) are not relevant for the ACO1 gene in yeast.
Because Aco1 has a mitochondrial targeting signal that is removed upon translocation into mitochondria, there are two more likely alternative scenarios for aconitase distribution. 1) The majority of the aconitase precursor molecules are imported into mitochondria and processed, whereas a small fraction are not imported and remain in precursor form in the cytosol. 2) All the Aco1 precursors are translocated across the mitochondrial membranes and are cleaved by the matrix protease. The majority of the molecules are fully imported, whereas the rest (maybe a few percent) of the protein is released from the organelle back into the cytosol. Although we have only limited data concerning this question, our data support the second alternative in which all the aconitase is processed in mitochondria before distribution: 1) No precursor-sized molecules can be detected in yeast cell extracts by Western blotting or immunoprecipitation, even after long exposures or heavy metabolic labeling. 2) A mutant aconitase that is not processed and remains in precursor form does not complement the cytosolic glyoxylate shunt function of aconitase, whereas it readily complements its mitochondrial TCA function. No aconitase activity can be detected in the cytosol of this mutant. 3) In contrast, a mutant aconitase lacking the mitochondrial targeting signal complements the cytosolic glyoxylate shunt function. Similarly, Aco1-ΔSP can complement the mutant aconitase that is not processed, together suggesting that this processed-like molecule is probably the cytosolic active form in vivo.
The mechanism of distribution of aconitase may be similar to the unique mechanism of fumarase subcellular localization in yeast (Stein et al., 1994; Knox et al., 1998); for both, a single nuclear gene encodes both the mitochondrial and cytosolic isoenzymes. The same initial translation products of these genes are distributed between the cytosol and the mitochondria (Stein et al., 1994). When import into mitochondria is blocked, all the aconitase or fumarase molecules accumulate in precursor form, whereas all are processed under normal conditions. This supports the notion that like fumarase all aconitase molecules are processed in mitochondria before distribution. However, there are differences in targeting and distribution of aconitase versus fumarase. First, a very small fraction of aconitase is localized to one of the two subcellular locations, whereas for fumarase large fractions of the enzymes are found in both locations. Second, aconitase import can occur in vivo (this study) and in vitro posttranslationally (Dubaquie et al., 1998), whereas fumarase import seems to be coupled to translation (Stein et al., 1994; Knox et al., 1998). Therefore, although we favor a fumarase-like distribution model, the present data do not rule out the possibility of a different mechanism. For example, it is possible, although not likely, that aconitase is fully imported, and a small fraction of the molecules is exported. Such questions will be the focus of our future studies.
Acknowledgments
We thank Nabieh Ayoub for help with tetrad analysis and Ariel Gaaton (Hebrew University, Jerusalem, Israel) for Edman degradation. Special thanks to Yeudit Karp for assistance and to Doron Rapaport and Israel Goldberg for critical reading of the manuscript. This research was supported by the Israel Science Foundation, German Israeli Foundation, and German Israeli Project Cooperation (to O. P.).
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E04-11-1028) on June 22, 2005.
References
- Abbas-Terki, T., and Picard, D. (1999). α-Complemented β-galactosidase. An in vivo model substrate for the molecular chaperone heat-shock protein 90 in yeast. Eur. J. Biochem. 266, 517-523. [DOI] [PubMed] [Google Scholar]
- Alani, E., and Kleckner, N. (1987). A new type of fusion analysis applicable to many organisms: protein fusions to the URA3 gene of yeast. Genetics 117, 5-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel, F., Brent, R., Kingston, R. E., Moore, D. D., Seidman, J. G., Smith, J. A., and Struhl, K. (eds.) (1991). Current Protocols in Molecular Biology, New York: Greene Publishing Associates.
- Branda, S. S., and Isaya, G. (1995). Prediction and identification of new natural substrates of the yeast mitochondrial intermediate peptidase. J. Biol. Chem. 270, 27366-27373. [DOI] [PubMed] [Google Scholar]
- Charlton, W. L., Johnson, B., Graham, I. A., and Baker, A. (2004). Non-coordinate expression of peroxisome biogenesis, β-oxidation and glyoxylate cycle genes in mature Arabidopsis plants. Plant Cell Rep. 23, 647-653. [DOI] [PubMed] [Google Scholar]
- Dubaquie, Y., Looser, R., Funfschilling, U., Jeno, P., and Rospert, S. (1998). Identification of in vivo substrates of the yeast mitochondrial chaperonins reveals overlapping but non-identical requirement for hsp60 and hsp10. EMBO J. 17, 5868-5876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fansler, B., and Lowenstein, J. M. (1969). Aconitase from pig heart. Methods Enzymol. 13, 26-30. [Google Scholar]
- Fukuhara, H. (2003). The Kluyver effect revisited. FEMS Yeast Res. 3, 327-331. [DOI] [PubMed] [Google Scholar]
- Gangloff, S. P., Marguet, D., and Lauquin, G. J. (1990). Molecular cloning of the yeast mitochondrial aconitase gene (ACO1) and evidence of a synergistic regulation of expression by glucose plus glutamate. Mol. Cell. Biol. 10, 3551-3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner, P. R. (1997). Superoxide-driven aconitase FE-S center cycling. Biosci. Rep. 17, 33-42. [DOI] [PubMed] [Google Scholar]
- Gerber, J., Muhlenhoff, U., and Lill, R. (2003). An interaction between frataxin and Isu1/Nfs1 that is crucial for Fe/S cluster synthesis on Isu1. EMBO Rep. 4, 906-911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentze, M. W., and Kuhn, L. C. (1996). Molecular control of vertebrate iron metabolism: mRNA-based regulatory circuits operated by iron, nitric oxide, and oxidative stress. Proc. Natl. Acad. Sci. USA 93, 8175-8182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knox, C., Sass, E., Neupert, W., and Pines, O. (1998). Import into mitochondria, folding and retrograde movement of fumarase in yeast. J. Biol. Chem. 273, 25587-25593. [DOI] [PubMed] [Google Scholar]
- McAlister-Henn. L., and Small, W. C. (1997). Molecular genetics of yeast TCA cycle isozymes. Prog. Nucleic Acid Res. Mol. Biol. 57, 317-339. [DOI] [PubMed] [Google Scholar]
- McCammon, M. T., Veenhuis, M., Trapp, S. B., and Goodman, J. M. (1990). Association of glyoxylate and β-oxidation enzymes with peroxisomes of Saccharomyces cerevisiae. J. Bacteriol. 172, 5816-5827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moosmann, P., and Rusconi, S. (1996). α Complementation of LacZ in mammalian cells. Nucleic Acids Res. 24, 1171-1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlenhoff, U., Balk, J., Richhardt, N., Kaiser, J. T., Sipos, K., Kispal, G., and Lill, R. (2004). Functional characterization of the eukaryotic cysteine desulfurase Nfs1p from Saccharomyces cerevisiae. J. Biol. Chem. 279, 36906-36915. [DOI] [PubMed] [Google Scholar]
- Mumberg, D., Muller, R., and Funk, M. (1994). Regulatable promoters of Saccharomyces cerevisiae: comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res. 22, 5767-5768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai, Y., Nakai, M., Hayashi, H., and Kagamiyama, H. (2001). Nuclear localization of yeast Nfs1p is required for cell survival. J. Biol. Chem. 276, 8314-8320. [DOI] [PubMed] [Google Scholar]
- Nakai, Y., Umeda, N., Suzuki, T., Nakai, M., Hayashi, H., Watanabe, K., and Kagamiyama, H. (2004). Yeast Nfs1p is involved in thio-modification of both mitochondrial and cytoplasmic tRNAs. J. Biol. Chem. 279, 12363-12368. [DOI] [PubMed] [Google Scholar]
- Rouault, T. A., and Klausner, R. D. (1996). Iron-sulfur clusters as biosensors of oxidants and iron. Trends Biochem. Sci. 21, 174-177. [PubMed] [Google Scholar]
- Sass, E., Blachinsky, E., Karniely, S., and Pines, O. (2001). Mitochondrial and cytosolic isoforms of yeast fumarase are derivatives of a single translation product and have identical amino termini. J. Biol. Chem. 276, 46111-46117. [DOI] [PubMed] [Google Scholar]
- Sass, E., Karniely, S., and Pines, O. (2003). Folding of fumarase during mitochondrial import determines its dual targeting in yeast. J. Biol. Chem. 278, 45109-45116. [DOI] [PubMed] [Google Scholar]
- Stein, I., Peleg, Y., Even-Ram, S., and Pines, O. (1994). The single translation product of the FUM1 gene (fumarase) is processed in mitochondria before being distributed between the cytosol and mitochondria in Saccharomyces cerevisiae. Mol. Cell. Biol. 14, 4770-4778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strobel, G., Zollner, A., Angermayr, M., and Bandlow, W. (2002). Competition of spontaneous protein folding and mitochondrial import causes dual subcellular location of major adenylate kinase. Mol. Biol. Cell 13, 1439-1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullmann, A., Jacob, F., and Monod, J. (1967). Characterization by in vitro complementation of a peptide corresponding to an operator-proximal segment of the β-galactosidase structural gene of Escherichia coli. J. Mol. Biol. 24, 339-343. [DOI] [PubMed] [Google Scholar]
- Waizenegger, T., Habib, S. J., Lech, M., Mokranjac, D., Paschen, S. A., Hell, K., Neupert, W., and Rapaport, D. (2004). Tob38, a novel essential component in the biogenesis of β-barrel proteins of mitochondria. EMBO Rep. 5, 704-709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, M., and Tzagoloff, A. (1987). Mitochondrial and cytoplasmic fumarases in Saccharomyces cerevisiae are encoded by a single nuclear gene FUM1 J. Biol. Chem. 262, 12275-12282. [PubMed] [Google Scholar]