Abstract
We identified a novel interaction between myosin VI and the GLUT1 transporter binding protein GLUT1CBP(GIPC1) and first proposed that as an adapter molecule it might function to couple vesicle-bound proteins to myosin VI movement. This study refines the model by identifying two myosin VI binding domains in the GIPC1 C terminus, assigning respective oligomerization and myosin VI binding functions to separate N- and C-terminal domains, and defining a central region in the myosin VI tail that binds GIPC1. Data further supporting the model demonstrate that 1) myosin VI and GIPC1 interactions do not require a mediating protein; 2) the myosin VI binding domain in GIPC1 is necessary for intracellular interactions of GIPC1 with myosin VI and recruitment of overexpressed myosin VI to membrane structures, but not for the association of GIPC1 with such structures; 3) GIPC1/myosin VI complexes coordinately move within cellular extensions of the cell in an actin-dependent and microtubule-independent manner; and 4) blocking either GIPC1 interactions with myosin VI or GLUT1 interactions with GIPC1 disrupts normal GLUT1 trafficking in polarized epithelial cells, leading to a reduction in the level of GLUT1 in the plasma membrane and concomitant accumulation in internal membrane structures.
INTRODUCTION
Some years ago, we demonstrated that mutations in the C termini of GLUT1 and GLUT4 altered their cellular distribution (a known effect) but also changed their intrinsic transporter activity (turnover number) (Dauterive et al., 1996). The unique sequence, functional importance of the C terminus of GLUT1, and indirect evidence that it served as a regulatory target, prompted a search for proteins capable of interacting with this unique domain. We subsequently identified a protein termed GLUT1CBP that binds to the GLUT1 C terminus (Bunn et al., 1999). Its binding to GLUT1 is absolutely dependent upon the presence of the four C-terminal amino acids comprising the PDZ domain recognition motif in GLUT1. The protein is 333 amino acids in length, possesses a central PDZ domain required for the recognition of GLUT1, and also contains an N-terminal proline-rich region that may serve as a linkage target recognized by other proteins containing SH3 or WW domains (Figure 1). We also demonstrated that GLUT1CBP uses the PDZ domain to bind not only to GLUT1 but also to the actin cross-linking protein α-actinin and the microtubule linked motor protein KIF-1B (Bunn et al., 1999). The presence of the PDZ domain and the ability of GLUT1CBP to interact with cytoskeletal elements places it among the important class of proteins containing PDZ domains that assemble membrane proteins and signaling cascades into areas of membrane specialization.
Figure 1.
Model of the proposed adapter function for GLUT1CBP(GIPC1), linking potential cargo proteins to myosin VI-catalyzed movement along F-actin filaments. (A)The C termini of GLUT1 or other potential vesicle bound cargo proteins (yellow) bind to the N-terminal region of the PDZ domain (red) of GLUT1CBP(GIPC1). This complex interacts with myosin VI through a C-terminal domain of GLUT1CBP(GIPC1) and a central domain in the tail of myosin VI (blue). Both interacting domains are marked by white boxes. A potential secondary interaction domain in GLUT1CBP(GIPC1) is marked with a dotted rectangle. The motor domain (green) of myosin VI catalyzes movement of the complex toward the negative end of the F-actin filament. A gray box marks the region containing the proline repeats in the N-terminus of GLUT1CBP(GIPC1). Numbers represent the amino acid residues within GLUT1CBP(GIPC1) and in myosin VI that are representative of the corresponding residues of the full-length GLUT1CBP(GIPC1) and pig myosin VI molecules. The two C-terminal inserts present in variant forms of myosin VI (Buss et al., 2001) are absent. (B) Three-dimensional representation of the PDZ domain of neuronal nitric-oxide synthase (nNOS) with bound peptide illustrating that the N-terminal half of the PDZ domain (red), containing the binding pocket for the C-terminal peptide (yellow), and the C-terminal portion of the PDZ domain (green) lie on opposite faces of the molecule. A similar configuration in the homologous GIPC1 molecule would provide a topology allowing cargo proteins (yellow) to bind to the PDZ domain without interference from myosin VI interactions that might involve the myosin VI binding domain D#2 (green) on the opposite face of the molecule. The structure of nNOS was derived from structural data (Tochio et al., 1999) provided by PDB ID# 1BQ8.
Surprisingly, there is a rapidly growing list of other proteins whose C-terminal amino acids interact with the PDZ domain of GLUT1CBP. Accordingly GLUT1CBP also is referred to as TIP-2 (Rousset et al., 1998), GIPC (DeVries et al., 1998), SEMCAP-1 (Wang et al., 1999), NIP-1 (Cai and Reed, 1999), and synectin (Gao et al., 2000), to describe its interactions with the HTLV-1 virus-encoded oncoprotein TAX, the trimeric G protein-directed RGS-GAIP, neuronal semaphorins, neuropilins, and syndecans, respectively. See Table 1 for a more comprehensive list of such interacting proteins.
Table 1.
Three classes of C-terminal PDZ motifs recognized by the PDZ domain of GIPC1 in mammalian systems
| Interacting protein | C-Terminal sequence | Reference |
|---|---|---|
| Class I | ||
| GLUT1 | ...Asp-Ser-Gln-Val | Bunn et al. (1999) |
| KIF-1B | ...Glu-Thr-Thr-Val | Bunn et al. (1999) |
| α-Actinin | ...Glu-Ser-Asp-Leu | Bunn et al. (1999) |
| RGS-GAIP | ...Ser-Ser-Glu-Ala | DeVries et al. (1998) |
| Neuropilin | ...Tyr-Ser-Glu-Ala | Cai and Reed (1999) |
| SemF | ...Glu-Ser-Ser-Val | Wang et al. (1999) |
| SemC | ...Asp-Ser-Val-Val | Wang et al. (1999) |
| Tax | ...Glu-Thr-Glu-Val | Rousset et al. (1998) |
| GP75(Trp-1) | ...Gln-Ser-Val-Val | Liu et al. (2001) |
| Megalin(LDLR) | ...Asp-Ser-Glu-Val | Gotthardt et al. (2000); Lou et al. (2002) |
| 5T4 | ...Xxx-Ser-Asp-Val | Awan et al. (2002) |
| Integrin α6A | ...Thr-Ser-Asp-Ala | Tani and Mercurio (2001) |
| Integrin α6B | ...Glu-Ser-Tyr-Ser | Tani and Mercurio (2001) |
| TGF-βIII receptor | ...Ser-Ser-Thr-Ala | Blobe et al. (2001) |
| β-Adrenergic receptor | ...Glu-Ser-Lys-Val | Hu et al. (2003) |
| Class II | ||
| Syndecan-4 | ...Glu-Phe-Tyr-Ala | Gao et al. (2000) |
| Syndecan-2 | ...Glu-Phe-Tyr-Ala | Gao et al. (2000) |
| Class III | ||
| Insulin-like growth factor-1 receptor | ...Ser-Ser-Thr-Cys | |
| Human LH receptor | ...Tyr-Thr-Glu-Cys | Hirakawa et al. (2003) |
| Dopamine D2 receptor | ...Ile-Leu-His-Cys | Jeanneteau et al. (2003) |
| Dopamine D3 receptor | ...Ile-Leu-Ser-Cys | Jeanneteau et al. (2003) |
| Rat TOM-L1 | ...His-Ser-Glu-Cys | Reed et al., unpublished observations |
| CD93 | ...Gly-Thr-Asp-Cys | Bohlson et al. (2004) |
Historically, this protein was identified first as TIP-2, GIPC, and then GLUT1CBP. As GIPC use predominates the current literature and two homologues to GIPC have been identified recently (Kirikoshi and Katoh, 2002a, b; Saitoh et al., 2002), the term GIPC1 is synonymous to GLUT1CBP and is used interchangeably when referring to the GLUT1CBP protein described in our earlier work (Bunn et al., 1999). That study was the first to identify an interaction between the amino acid sequences flanking the PDZ domain in GLUT1CBP and the actin linked motor protein myosin VI, as well as to identify the ability of GLUT1CBP to dimerize (or form higher order oligomers) with other molecules of GLUT1CBP (Bunn et al., 1999). In that article, two of the three models proposed for the function of GLUT1CBP were to 1) cluster or anchor GLUT1 or any of the additional proteins recognized by the PDZ domain to actin filaments via α-actinin and 2) link them to microtubule directed transport via the kinesin KIF-1B. These models were depicted (Bunn et al., 1999) for GIPC1 interactions with GLUT1, but potentially include any other protein that interacts with the PDZ domain of GIPC1. The data presented in this article address the third and most intriguing of the proposed models (Bunn et al., 1999) in which GIPC1 links PDZ-bound proteins (soluble, or integrated into vesicle membranes) to F-actin-directed transport via myosin VI (Figure 1).
GLUT1CBP(GIPC1) was the first potential adapter protein identified to interact with the unique myosin VI tail. This interaction is significant because mounting evidence now strongly implicates myosin VI as a motor protein involved in the trafficking of vesicle-bound proteins (for recent reviews, see Buss et al., 2002; Hasson, 2003). The postulated role of this motor in protein trafficking is unusual for myosin motors because myosin Vl moves toward the negative end of the actin filament (Wells et al., 1999). Thus, interactions of myosin VI with GIPC1 would provide cargo proteins bound to the PDZ domain with the capacity to move in a direction along actin fibers that opposes the positive end directed movement characteristic of other myosin motor proteins. Understanding when, where, and with which potentially numerous and important cargo proteins (Table 1) GIPC1 is associated when it is bound to or released from myosin VI could enhance our current understanding of the selectivity of the process of protein sorting within the cell.
The studies in this article address the proposed myosin VI-linked adapter function for GIPC1 by 1) further characterizing the domains required for interactions between GIPC1 and myosin VI; 2) demonstrating that GIPC1 can bind to the C-terminal domain of myosin VI and move as a complex coordinately with myosin VI along F-actin filaments within cellular extensions; and 3) illustrating that faulty trafficking of one of the many potential cargo proteins (GLUT1) occurs when interactions are disrupted between GLUT1 and the PDZ domain of GIPC1, or between GIPC1 and myosin VI.
MATERIALS AND METHODS
Cell Culture
Cell lines were maintained in 10-cm Falcon dishes (Lincoln Park, NJ) and 5% CO2 using Dulbecco's modified Eagle's media (DMEM) buffered with KHCO3 (high glucose) for Chinese hamster ovary (CHO) (CHO-K1-HIR) cells, or standard DMEM (high glucose) for 293 cells and clone 5 Madin-Darby canine kidney (MDCK) cells (a generous gift of Dr. Mike Roth, Southwestern Medical School, Dallas, TX).
Reagents
Purified His6-GLUT1CBP(GIPC1) was prepared as described previously (Bunn et al., 1999) after expressing PET30-GLUT1CBP in bacteria. The S-Tag, protease cleavage site, and cloning site encoded amino acids that join the N-terminal His6 tag sequences to the N terminus of GIPC1 are encoded by the PET30 vector and generate a His6-GLUT1CBP that migrates as a larger protein than native GIPC1 in SDS-PAGE. Rabbit anti-GLUT1 was raised against a KLH-peptide conjugate identical to the C-terminal 14 amino acids of GLUT1 and affinity purified on a peptide affinity column. Rabbit anti-GIPC1 was raised against a KLH-peptide conjugate identical to the first 12 amino acids of rat GIPC1 and purified on a peptide affinity column.
Interactions of the Purified Myosin VI C Terminus with Native and Purified GIPC1
For pull-down assays of endogenous native MDCK GIPC1, cell extracts were prepared by scraping confluent 10-cm dishes of MDCK cells into 0.5 ml/plate of ice-cold phosphate-buffered saline (PBS) containing 5% NP-40, 0.1 mM phenylmethylsulfonyl fluoride, and 40 μl/ml stock protease inhibitor cocktail (Roche Diagnostics, Indianapolis, IN). Lysates were treated eight times with 10 strokes each in a glass-teflon homogenizer with a total incubation time of 1 h on ice and then were centrifuged at 100,000 × g for 1 h at 4°C. One milliliter of clear supernatant was added to 40 μl of a 50% suspension of glutathione beads containing bound glutathione S-transfersae (GST) or GST-myosin VI(955-1254) and incubated for 3 h at 4°C. The beads were washed three times quickly and 1 × 10 min with 1 ml of PBS/5% NP-40, three times quickly and 1 × 10 min with 1 ml of PBS/0.1% NP-40, and once with Laemmli buffer lacking SDS and urea.
For pull-down assays of purified His6-GLUT1CBP(GIPC1), glutathione beads containing bound GST and GST-myosin VI(955-1254) were incubated for 1 h at 4°C with or without 20 μg of purified His6-GLUT1CBP(GIPC1) in buffer containing 10 mM HEPES, pH 7.5, 1 M NaCl, 1 mM EDTA, and 1 mM β-mercaptoethanol. The beads were washed two times quickly and 2 × 10 min with 1 ml of PBS/0.1% NP-40, and then once with Laemmli buffer lacking SDS and urea.
Bound proteins were eluted from the washed beads in 70 μl of gel-loading buffer (Laemmli) containing 4% SDS/6 M urea by heating 8 min at 80°C. Twenty-five microliters of the supernatant was resolved by SDS-PAGE in 7.5% gels, and the separated proteins transferred to a polyvinylidene fluoride membrane in Tris-glycine buffer containing 20% (vol/vol) methanol. Membranes were blocked overnight in buffer containing 5% dry milk and 0.1% Tween 20 and then incubated with a 1:1000 dilution of either purified rabbit anti-N-terminal-GIPC1 or rabbit anti-C-terminal GLUT1 as indicated. Membranes were washed, and bound anti-GIPC1 or anti-GLUT1 was detected with horseradish peroxidase-conjugated monoclonal anti-rabbit IgG (Jackson ImmunoResearch Laboratories) and enhanced chemiluminescence. Images were captured using a Molecular Dynamics PhosphorImager and Image-Quant software.
For pull-down assays of labeled native GIPC1(1-333) and mutant GIPC1(1-300), [35S]methionine-labeled GIPC1(1-333) and GIPC1(1-300) proteins were produced by in vitro transcription and translation from the DNA templates pcDNA3.1(+)-GLUT1CBP(1-333) and pcDNA3.1(+)-GLUT1CBP(1-300) by using the TNT-coupled reticulocyte system (Promega, Madison, WI). Labeled proteins were incubated with beads containing bound GST, GST-myosin VI(955-1254), GST-myosin VI(1065-1160), or GST-myosin VI(1065-1254) as indicated for 1 h at room temperature in PBS containing 0.1% NP-40 (PBS-NP40) and then washed extensively in PBS-NP40. Bound labeled proteins were separated by SDS-PAGE and detected using a Molecular Dynamics PhosphorImager after exposure of the dried gel to a PhosphorImager screen.
Construction and Expression of Cyan Fluorescent Protein (CFP), Yellow Fluorescent Protein (YFP), and Green Fluorescent Protein (GFP) Fusion Proteins
YFP-GIPC1(1-333), YFP-GIPC1(1-249), CFP-myosin VI(955-1254), GFP-GLUT1, GFP-GLUT1Δ4, GFP-GLUT1Δ25, CFP-myosin VI(1-1254), and GFP-GLUT1synd4ctrm were constructed by inserting the cDNA fragments generated from restriction digests or PCR amplification of cDNA's encoding the appropriate amino acid sequences into pECFP, pEYFP, or pEGFP vectors (BD Biosciences Clonetech, Palo Alto, CA). The sequences derived from PCR-generated fragments were verified by sequencing. GIPC1 sequences were derived from pBSKII-GLUT1CBP (Bunn et al., 1999). GLUT1 sequences were from isolated mouse GLUT1 cDNA (Reed et al., 1990). Myosin VI(955-1254) sequences were derived from previously isolated rat myosin VI(955-1254) (Bunn et al., 1999) or from full-length pig myosin VI(1-1254), a generous gift of Dr. Tama Hasson (University of California, San Diego, CA). cDNAs were introduced into the appropriate cell lines using Lipofectamine 2000 according to manufacturer's protocols and analyzed after allowing 18 h or 2-4 d for protein expression as indicated.
Fluorescence Microscopy
For Figure 7A alone, a Bio-Rad MRC1000 confocal scope and Nikon plan apo 60× (numerical aperture [N.A.] 1.4) lens was used. Cells expressing GFP-GIPC1 were fixed for 1 h in 2% paraformaldehyde/phosphate-buffered saline and then permeabilized by incubation with 0.2% Triton X-100/phosphate-buffered saline for 0.5 h before incubation with rhodamine-phalloidin and mounting in Vectashield mounting media (Vector Laboratories, Burlingame, CA). A 488-nm excitation laser and 522/34 emission filter were used to collect the GFP-GIPC1 signal, and a 568-nm excitation laser and 605/32 emission filter were used to collect the rhodamine-phalloidin signal. All other microscopy was performed using a Bio-Rad Radiance 2000/AGR-3 (Q) confocal microscope equipped with a three-channel, three-detector system mounted on a Nikon TE 300 inverted microscope and three available lasers: 1) a 25-mW argon ion laser emitting at 457, 476, 488, and 514 nm; 2) a 1-mw green HeNe laser emitting at 543 nm; and 3) a red diode laser emitting at 638 nm. Images were acquired using plan apo 100× oil (N.A. 1.40) lens and Lasersharp I software with 32-bit acquisition, display, and three-dimensional volume rendering. Images of cells expressing native or mutant GFP-GIPC1 or GFP-GLUT1 were obtained by excitation at 488 nm with the argon laser and collecting emitted light through a HQ500LP emission filter. Live cell normal and time-lapse images were obtained using cells grown on temperature regulated ΔT3-plates (Bioptechs, Butler, PA) that were washed and incubated in HEPES-glucose-saline solution (140 mM NaCl, 5.4 mM KCl, 1.3 mM CaCl2, 1.0 mM MgCl2, 25 mM HEPES, and 33 mM glucose, pH 7.4). The temperature was maintained at 25°C using a ΔT3 temperature regulator (Bioptics). Simultaneous imaging of normal and mutant CFP-myosin VI and YFP-GIPC1 fusion proteins were collected using the strobe mode to block cross-channel signal bleed by sequentially exciting with either the 457- or 514-nm single laser excitation line and collecting with emission filters HQ485/30 for CFP emissions and HQ545/40 for YFP emissions. Three-dimensional reconstructions of z-sections or generation of videos from time-lapse images were obtained using MetaMorph imaging software (Universal Imaging, Downigntown, PA). GFP signals with rhodamine isothiocyanate (RITC)-dextran, LysoTracker red, or Cy3-EEA1 were collected by exciting with the 488 laser line and the HQ515/30 emission filter or with the 543 laser line and HQ590/70 emission filter, respectively, in strobe mode. GFP signals with LysoTracker Red and Cy5-EEA1 were collected with the 488 laser line and the HQ515/30 emission filter, the 543 laser line and the HQ590/70 emission filter, and the 637 laser line and the HQ660LP filter setting, respectively, in strobe mode.
Figure 7.
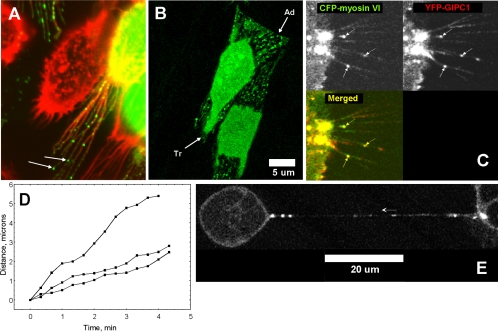
Movement of GIPC1 and GIPC1-myosin VI complexes in CHO and 293 cells. A and B demonstrate that particles containing GFP-GIPC1 colocalize with F-actin filaments in cellular extensions of CHO cells and move toward the cell body. In A, transfected CHO cells expressing full-length GFP-GIPC1(1-333) were fixed with 2% paraformaldehyde, permeabilized with 0.2% Triton X-100, and the F-actin stained with rhodamine-phalloidin. Green particles of GFP-GIPC1 (arrows) can be observed along with F-actin (red) within the cellular extensions. In B, CHO cells were transfected with GFP-GIPC1(1-333) and images were collected from live cells at 30-s intervals using a confocal microscope. GFP-GIPC1(1-333) particles are evident in the cellular extensions shown in the still image presented in B. As illustrated in the supplemental video Fig 7B.mov constructed from the individual time-lapse images, GFP-GIPC1(1-333) particles move toward the cell body within cellular extensions protruding from both the advancing (Ad) and trailing (Tr) edge of the cell. C and D demonstrate that CFP-myosin VI and YFP-GlPC1 form complexes in 293 cells that move coordinately toward the cell body within cellular extensions. Time-lapse images of live 293 cells expressing CFP-myosin VI and YFP-GIPC1 were collected at 20-s intervals using settings for the confocal microscope like those for Figure 6 to separate CFP and YFP emissions. The beginning frame of the series is presented in C, where the CFP-myosin VI distribution is presented in the top left quadrant, YFP-GIPC1 in the top right quadrant, and the merged image in the bottom left quadrant. Three complexes (solid arrows) observable in the CFP, YFP, and merged channels indicate that expressed YFP-GIPC1 and CFP-myosin VI colocalize in the cellular extensions and move coordinately within the extensions toward the cell body. Plots of their movement indicate that the rate of movement of each of the complexes differ with the slowest moving at 0.5 μm/min and the fastest at 2.0 μm/min. Although the rates differ slightly for each of the three marked complexes (arrows), the movement of CFP-myosin VI and YFP-GIPC1 in each instance is as a single complex. The direction and coordinate nature of movement of each complex is best observed in the supplemental movie (Fig 7C.mov). E illustrates a long bridge between two 293 cells on the left and right extremes of the image that are expressing both CFP-myosin VI and YFP-GIPC1. This structure is identical in appearance to the actin-containing nanotubes reported to transport particles between adjoining cells (Rustom et al., 2004). The direction of movement of a myosin VI/YFP-GIPC1 complex is marked by an adjacent arrow. The measured velocity of this complex was 1.2 μm/min. In A-C, the images were captured within the plane of the retraction fibers near the base of the cell, and near the midpoint for the cells in E.
Quantitation of total cellular and plasma membrane fluorescence from either CFP-GLUT1 or CFP-GLUT1Δ4 in Figure 10 was obtained by collecting z-sections in 0.35-μm steps throughout the entire height of each cell. The total cellular fluorescence was obtained by integration of the fluorescent intensity of each section within a software defined boundary paralleling the outer contour of the cell using MetaMorph software. Likewise the total plasma membrane-associated fluorescence was determined by integrating and summing the fluorescence intensity within a 0.4-μm-thick boundary region paralleling the contour of the outer and inner surfaces of the plasma membrane for each z-section. The contribution of the basal plasma membrane fluorescence was obtained by integrating the z-section image centered within the plane of the membrane lying parallel to the coverslip. The calculated vertical resolution is 0.70 μm and therefore collects fluorescence within a 0.35-μm boundary either side of the basal membrane. Both total and plasma membrane fluorescence were corrected for endogenous fluorescence by integrating identical volumes in adjacent fields containing cells not expressing CFP-GLUT1 or CFP-GLUT1Δ4. The laser settings were held constant throughout the integration of all cells in Figure 10 and set initially to maintain all pixel intensities within the linear response range of the system. Statistical differences in the means were evaluated by a Student's t test.
Figure 10.
Removal of the C-terminal four amino acids of GLUT1 or expression of the myosin VI tail in MDCK cells disrupts normal trafficking of GLUT1. An ECFP-C1 vector driving the expression of CFP fused to the N terminus of GLUT1 was introduced into MDCK cells with either an EYFP vector or an EYFP vector driving the expression of YFP fused to the N terminus of myosin VI(955-1254). Eighteen hours after transfection, the distributions of CFP-GLUT1, YFP, and YFP-myosin VI(955-1254) were determined by collecting X-Y plane images near the midpoint of the cell with a confocal microscope. (A) Image of a representative cell coexpressing CFP-GLUT1 (green) and YFP (red) illustrating that YFP expression does not alter the normal distribution of CFP-GLUT1. CFP-GLUT1 (left) is concentrated within the plasma membrane with only low levels of internal CFP-GLUT1 visible. This distribution is identical to that observed for GFP-GLUT1 expressed in the absence of YFP (Figure 9G, left, day 2). (B) Image of cells coexpressing CFP-GLUT1 (green) and YFP-myosin VI(955-1254) (red) illustrating that YFP-myosin VI(955-1254) expression disrupts the normal distribution of CFP-GLUT1. Cells expressing either no, intermediate, or high levels of YFP-myosin VI expression (middle) exhibit respectively either no, intermediate, or high levels of CFP-GLUT1 accumulation in internal membranes (respectively, marked 1, 2, or 3 in the left panel). (C) Enlarged view of the area marked by a bracket in the merged view of B. YFP-myosin VI(955-1254) (red) is associated with membranes and individual vesicles (arrows) containing inappropriately targeted CFP-GLUT1 (green). (D) Eighteen hours after transfection, z-section images were collected though the entire cell volume from five random cells expressing CFP-GLUT1 and five random cells expressing CFP-GLUT1Δ4. The total plasma membrane associated fluorescence and total cell fluorescence were integrated and summed for all z-sections and the level of plasma membrane or intracellular transporter is expressed as a percentage of the total cellular transporter. For cells transfected with CFP-GLUT1Δ4 the amount of fluorescence transporter in the plasma membrane (22.3 ± 3.2%) is significantly smaller than the amount (51.3 ± 2.4%) for cells transfected with CFP-GLUT1 (p < 0.001). Accordingly cells expressing CFP-GLUT1Δ4 display a correspondingly 1.6-fold higher internal distribution of transporter (77.7%) than those expressing CFP-GLUT1 (48.7%). (E) An experimental approach identical that of D was used to determine the average CFP-GLUT1 distribution (mean ± SE) in five cells coexpressing CFP-GLUT1 and YFP and five cells coexpressing CFP-GLUT1 and YFP-myosin VI(955-1254). The cells selected were expressing matched levels of YFP and YFP-myosin VI(955-1254). In cells expressing YFP, the level of CFP-GLUT1 in the plasma membrane was 53.2 ± 3.8% of the total cellular CFP-GLUT1. This value is nearly identical to the distribution of CFP-GLUT1 expressed in the absence of YFP (Figure 10D), demonstrating that YFP expression itself does not alter CFP-GLUT1 trafficking. For cells expressing CFP-GLUT1 and YFP-myosin VI(955-1254) the amount of fluorescence transporter in the plasma membrane (27.6 ± 2.5%) is significantly smaller than the amount (53.2 ± 3.8%) for cells expressing CFP-GLUT1 and YFP (p < 0.001). Accordingly cells expressing CFP-GLUT1 and YFP-myosin VI(955-1254) display a correspondingly 1.5-fold higher internal distribution of transporter (72.4%) than those expressing CFP-GLUT1 and YFP (46.8%). These results demonstrate that blocking the interactions of GLUT1 (GLUT1Δ4) with GIPC1 or blocking the interactions of GIPC1 with native myosin VI by overexpressing myosin VI(955-1254) both disrupt GLUT1 trafficking and raise the internal concentration of transporter to the same extent. Percentages are expressed as the mean ± SE of the mean. Error bars represent the SE of the mean.
Labeling MDCK Vesicle Compartments
Primary mouse anti-EEA1 antibody was purchased from BD Transduction Laboratories (Lexington, KY) and diluted to a final concentration of 1/100 in bovine albumin/saponin/phosphate-buffered saline, pH 7.4 (BSP) and incubated with monolayers for 1 h at room temperature. Secondary antibodies of donkey anti-mouse IgG conjugated with either Cy3 (550/570) or Cy5 (650/670) (Jackson ImmunoResearch Laboratories) were similarly diluted 1/100 in BSP and incubated for 1 h at ambient temperature. LysoTracker Red (577/590) was incubated with cultures for 10 min before fixation with 4% paraformaldehyde to label intracellular acidic vesicles. Monolayers were incubated in DMEM/10%fetal calf serum at 37°C for 20 min with RITC-dextran, and then after washing were either fixed or incubated in fresh media and serum for an additional 60-min chase before fixing for 10 min with 4% paraformaldehyde to label, respectively, early endosomes or late endosomes and lysosomes. LysoTracker Red (DND-99) and antifade mounting solutions Slow-fade and Prolong were purchased from Molecular Probes (Eugene, OR) and RITC-dextran was from Sigma (St. Louis, MO). Images were collected as described above using the strobe mode with the appropriate laser and emission filter settings.
Mapping of the Myosin VI and GIPC1 Interaction Domains Using the Yeast Two-Hybrid System
Plasmid vectors pGBT9 (BD Biosciences Clontech) or pGBDC3 were engineered to contain GLUT1CBP constructs fused to the Gal4 DNA binding domain (DBD) and are marked with TRP1 for nutritional selection in yeast. The plasmid vector pGADT7 containing rat myosin VI constructs fused to the Gal4 activation domain (ACT) were marked with LEU2 for nutritional selection in yeast. The procedures for selection and elimination of false positives are described in detail previously (BD Biosciences Clontech two-hybrid manuals). For Figure 3, plasmids encoding Gal4 DBD and ACT fusion proteins were cotransfected into yeast strains HF7c and the transformants were plated onto selection media lacking tryptophan and leucine. For each interaction tested, three or four viable colonies containing both plasmids were patched onto an identical plate and after 2-3 d were replicate plated onto selection plates lacking tryptophan and leucine or plates containing 30 mM 3-aminotriazole that lacked tryptophan, leucine, and histidine. Significant growth relative to the control plates indicated a positive interaction. For Figure 4, plasmids were introduced into yeast strain PJ69-4A (James et al., 1996) and selected in a similar manner on plates lacking tryptophan, leucine, and adenine. Appropriate controls were included for each fusion protein with the appropriate corresponding empty vector and a positive control to eliminate any self-activating sequences that would yield false positives.
Figure 3.
Sequences in GIPC1 critical for dimerization (or oligomer formation) and binding to myosin VI are separate and reside in respective N-terminal and C-terminal domains. Using the yeast two-hybrid system, GBD fusion proteins to full-length GIPC1(1-333), GIPC1(107-333) missing the N terminus, GIPC1(1-249) missing the C terminus, and GIPC1(107-249) retaining the PDZ domain but missing both the N and C termini, were tested for their interactions with Gal4 activation domain (ACT) fusion proteins to full-length GIPC1(1-333) and myosin VI(955-1254). The sequences comprising the PDZ domain are represented by the gray rectangle. The microtubule motor protein KIF-1B and α-actinin are known to bind to the PDZ domain of GIPC1 and serve as positive controls. Strong interactions indicated by rapid growth on -Trp, -Leu, -His plates containing 30 mM aminotriazole are designated by a “+” and no interactions (no growth) are indicated by a “-.” Loss of N-terminal GIPC1 sequences blocks only dimerization with GIPC1 (row 2), whereas loss of C-terminal sequences destroys interactions only with myosin VI (row 3). Both interactions are lost when both the N-terminal and C-terminal sequences are removed, whereas binding of the PDZ domain to KIF-1B and α-actinin is retained (row 4).
Figure 4.
Mapping of the interaction domains in GIPC1 and myosin VI using a yeast two-hybrid system. DBD fusions to full-length GIPC1(1-333) or to smaller fragments tested in this study are depicted on the left side of the diagram. The sequences comprising the PDZ domain are represented by the dark gray rectangle. A plasmid expressing each of these fusion proteins was cotransfected into yeast with a selected plasmid expressing either ACT fusion protein to amino acids 955-1254 of myosin VI containing a portion of the coiled-coil domain (jagged line) and the entire tail domain (white box) or smaller regions of myosin VI as depicted across the top of the diagram. Strong interactions (indicated by rapid growth on -Trp, -Leu, -Ad plates) are designated by a “+,” weaker interactions (moderate growth) by “±,” and no interactions (no growth) are indicated by a “-.” Results defining the region to which GIPC1 binds in the myosin VI tail are presented in row 1. A single minimal interacting domain identified in the myosin VI tail is represented by the myosin VI(1065-1160) fragment and is bounded by the light gray horizontal rectangle. Two interacting domains were identified in GIPC1. The results from column 1, rows 1-6 define domain #1, which is represented by the GIPC1(261-333) fragment and is marked by the light gray vertical rectangle labeled #1. This region interacts with all Gal4 ACT fusion proteins to myosin VI containing the myosin VI(1065-1160) region, including those that also contain a portion of the coiled-coil domain (rows 4 and 5). Smaller regions (i.e., 279-333) are self-activating (SA) and because growth is no longer dependent upon interaction with a GAL4 ACT fusion protein, their interaction with myosin VI fragments cannot be evaluated in this system. The data in rows 7-16 define domain #2 and demonstrate its inability to interact with fragments containing the coiled-coil domain. Domain #2 is represented by the GIPC1(173-231) fragment and is marked by the light gray vertical rectangle labeled #2. As shown by the data in row 15, it interacts with myosin VI fragments containing a more C-terminal extension of the myosin VI (1065-1160) region, and lacking the coiled-coil domain.
RESULTS
Figure 1 presents the basic model proposed for GIPC1 acting as an adapter molecule. Potential cargo proteins bound to the PDZ domain of GIPC1 are coupled to myosin VI-catalyzed movement via interactions between the C-terminal domains of myosin VI and GIPC1. The C-terminal sequence of the rat myosin VI protein isolated in our original two-hybrid screen is identical to that of amino acids 955-1254 of pig myosin VI. The studies presented below use the pig myosin VI(1-1254) amino acid numbering scheme (Hasson and Mooseker, 1994) to define myosin VI C-terminal protein fragments and use the numbering for the full-length rat GIPC1 (amino acids 1-333) (Bunn et al., 1999).
A Characterization of the Interactions between Purified GIPC1 and Myosin VI Proteins and Their Endogenous Cellular Counterparts
A His6 fusion protein containing full-length GIPC1 when expressed in bacteria and purified by nickel affinity chromatography retains its ability to bind to the C terminus of GLUT1 expressed as a GST-fusion protein (Bunn et al., 1999). In addition, purified His6-GIPC1 covalently coupled to agarose beads can bind to and pull-down native GLUT1 present in detergent extracts of MDCK cells (Bunn et al., 1999). These previous studies establish the ability of GLUT1 to interact with purified GIPC1. Before defining the interacting domains between myosin VI and GIPC1, it was necessary to establish that both native and purified forms of GIPC1 and myosin VI were capable of forming a complex and that other unidentified proteins were not required for their interaction.
Purified His6-GIPC1 coupled to agarose beads is fully capable of binding tightly to native full-length myosin VI present in detergent extracts of MDCK cells (Bunn et al., 1999). Conversely, to demonstrate the ability of endogenous native GIPC1 to interact with the purified C terminus of myosin VI, MDCK cell extracts were incubated with GST or the purified GST-myosin VI(955-1254) fusion protein bound to agarose beads, and the associated proteins were separated by SDS-gel electrophoresis. Western blots of the gels probed with rabbit anti-GIPC1 antibody demonstrated that only GST-myosin VI beads interact with endogenous GIPC1 (Figure 2A, lane 4) present in MDCK cell extracts. This confirms the ability of the C-terminal amino acids 955-1254 of myosin VI to bind to GIPC1 protein synthesized and folded in its native environment. The purified GST-myosin VI(955-1254) fusion protein bound to agarose beads also interacts with purified His6-GIPC1(Figure 2B, lane 8). GST bound to agarose beads does not interact with either native GIPC1 (Figure 2A, lane 3) or purified His6-GIPC1 (Figure 2B, lane 7). These data establish the ability of the myosin VI C terminus to interact directly with His6-GIPC1, and they eliminate a concern that the interactions studied in the yeast two-hybrid or other cell systems might be mediated solely by an unknown protein or proteins capable of binding simultaneously to both GIPC1 and myosin VI.
Figure 2.
Western blots illustrating the interactions of cellular and purified GLUT1CBP(GIPC1) with the C-terminal tail of myosin VI and the formation of a trimeric complex between the myosin VI tail, GIPC1, and GLUT1. In A and B, purified GST (lanes 3 and 7) or GST-myosin VI(955-1254) (lanes 4 and 8) fusion proteins bound to glutathione-agarose beads were incubated with or without MDCK cell extracts (A) or purified His6-GLUT1CBP(GIPC1) (B) as indicated. The Western blots of bound proteins released from the washed beads were probed with rabbit anti-GIPC1 to detect associated GIPC1. Only the myosin VI(955-1254) fusion protein interacts with endogenous GIPC1 from MDCK cells (lane 4) or with purified His6-GLUT1CBP(GIPC1) (lane 8). GST beads do not interact (lanes 3 and 7) with native or purified GLUT1CBP(GIPC1), respectively. Arrows point to endogenous GLUT1CBP(GIPC1) from MDCK extracts (A) or to purified His6-GLUT1CBP(GIPC1) (B) applied directly to the gel. His6-GIPC1 (lane 8) migrates with a higher apparent molecular weight than native GIPC1 (lane 4) due to fusion protein sequences linking GIPC1 and the His6 residues (see Materials and Methods). In C, beads containing bound purified GST (lanes 11 and 13) or GST-myosin VI(955-1254) (lanes 12 and 14) were incubated with cell extracts without (lanes 11 and 12) or with (lanes 13 and 14) 40 μg of added purified His6-GIPC1 as indicated. The Western blots of bound proteins released from the washed beads were probed with rabbit anti-GLUT1 to detect associated GLUT1 transporter. Only the beads containing myosin VI(955-1254) react with endogenous GIPC1 and pull down associated GLUT1 (lane 12). This GLUT1 signal is dramatically enhanced by the addition of purified His6-GIPC1 to GST-myosin VI(955-1254) containing beads (lane 14). No GLUT1 is associated with beads containing GST (lane 11) or GST with added His6-GIPC1 (lane 13). Analysis of proteins released directly from beads containing purified GST or GST-myosin VI without incubation with cell extracts or His6-GIPC1 demonstrate no contaminants are present that cross-react with either anti-GIPC1 (lanes 1, 2, 5, and 6) or anti-GLUT1 (lanes 9 and 10) antibodies.
GIPC1-dependent Formation of Complexes Containing Myosin VI(955-1254) and GLUT1
Agarose beads containing bound GST-myosin VI(955-1254) that pull-down GIPC1 (Figure 2A, lane 4) also pull-down cellular GLUT1 bound to endogenous GIPC1 (Figure 2C, lane 12) from MDCK cell extracts. Addition of purified His6-GIPC1 to the extract dramatically increases the amount of GLUT1 associating with GST-myosin VI(955-1254) (Figure 2C, lane 14), emphasizing the role of GIPC1 to mediate the interaction of GLUT1 with myosin VI. Agarose beads containing bound GST fail to interact with cellular GLUT1 in the absence (Figure 2C, lane 11) or presence (Figure 2C, lane 13) of added purified His6-GIPC1. These data establish the ability of GLUT1, GIPC1, and the myosin VI tail to form a trimeric complex.
Mapping of the Myosin VI and GIPC1 Interaction Domains
With the knowledge that mediating proteins were not required for binding, the mapping of the regions required for direct binding of GIPC1 to myosin VI was accomplished using a yeast two-hybrid system.
Our initial studies documented that removal of both N-terminal and C-terminal sequences flanking the PDZ domain blocked dimerization (or oligomerization) of GIPC1 with another molecule of GIPC1 in addition to destroying the ability to interact with myosin VI (Bunn et al., 1999). Previously, it was demonstrated (Gao et al., 2000) that removal of N-terminal and not C-terminal flanking sequences of synectin (GIPC1) destroyed the ability of GIPC1 to form dimers or higher oligomers. However, the data did not determine whether the dimerization and myosin VI interaction domains resided together in the N-or C-terminal regions of GIPC1 or whether they were located in separate regions.
To resolve this issue, the experiment presented in Figure 3 was performed. A diagram of each of the Gal4 binding domain (GBD)-GIPC1 fusion proteins tested for their interaction with the activation domain fusion proteins to myosin VI(955-1254) and GIPC1(1-333) are presented in the left column. Strong interactions (+) are indicated by rapid growth on -Trp, -Leu, -His plates containing 30 mM aminotriazole. The results demonstrate that constructs lacking the N-terminal flanking sequences (row 2) lose the capacity to interact with GIPC1(1-333) but maintain the ability to interact with myosin VI(955-1254). The converse is true for GIPC1 constructs lacking the C-terminal flanking sequence (row 3). GIPC1 constructs missing both N-terminal and C-terminal flanking sequences lose both the ability to dimerize and interact with myosin VI (row 4). The microtubule based motor protein KIF-1B and α-actinin were included as positive controls, and consistent with their known ability to bind to the PDZ domain of GIPC1 (Bunn et al., 1999), interacted with all forms of GIPC1 tested. Thus, the stretches of amino acid sequences required for either dimerization or binding to myosin VI are separate and reside in the N-terminal and C-terminal regions of GIPC1, respectively.
With the knowledge that the myosin VI binding domain resides in the C-terminal half of the GIPC1 protein and is separate from the dimerization domain, a more thorough characterization of the GIPC1 and myosin VI interaction domains was undertaken. A diagram of each of the Gal4 DBD-GIPC1 fusion proteins and ACT-myosin VI fusion proteins tested is presented in Figure 4. Strong interactions tested in a more stringent system (yeast strain PJ69-4A) are indicated by rapid growth on -Trp, -Leu, -Ad plates and are designated by a “+,” whereas no interactions (no growth) are indicated by a “-”. The rat myosin VI fragment we isolated during the initial two-hybrid screen for proteins interacting with GLUT1CBP(GIPC1) extends from amino acid 955 to the C-terminal amino acid 1254 (using numbering for pig myosin VI (Hasson and Mooseker, 1994). This includes a portion of the conserved coiled-coil domain and the entire unique C-terminal domain of myosin VI (Figure 1). This myosin VI isoform is identical in this region to pig myosin VI and lacks both the short and long inserts that have been reported to be present in less common isoforms of myosin VI (Buss et al., 2001).
Using this approach, one binding domain for GIPC1 was identified in the myosin VI tail. The results of experiments defining the minimal myosin VI domain capable of interacting with full-length GIPC1 are illustrated by row 1 in Figure 4. A region as small as myosin VI(1055-1169) interacts well with full-length GIPC1(1-333), and a smaller myosin VI(1065-1160) segment also binds, but less effectively as evidenced by marginal growth (±) of yeast expressing these constructs. Thus, these data define a minimal interaction domain within the myosin VI tail that is marked by the light-gray horizontal rectangle (Figure 4) and the solid white box (Figure 1). In agreement with this assignment, GIPC1(1-333) fails to interact with a myosin VI fragment containing a portion of the coiled-coil region (955-1066) or with more C-terminal regions such as myosin VI(1127-1254), (1162-1254), and (1182-1254) shown in Figure 4, row 1.
Rows 2-6 in Figure 4 depict the N-terminal truncations of GIPC1 used to determine the N-terminal boundary of the GIPC1 domain (D#1) that recognizes myosin VI. Fusion protein fragments as small as GIPC1(261-333) retain binding activity to all myosin VI constructs containing amino acids 1065-1160 and indicate amino acids 1-260 of GIPC1 are not required for binding of the C-terminal portion of GIPC1 to myosin VI. DBD fusion proteins containing further N-terminal truncations equal to or smaller in length than GIPC1(279-333) inexplicably become self-activating (SA; Figure 4, row 6) and could not be evaluated in this system.
Experiments depicted by rows 7-10, column 1, in Figure 4 represent the experiments to define the C-terminal boundary of the myosin VI binding domain D#1 in GIPC1. No interactions were detected when the binding to the myosin VI(955-1254) fragment containing the coiled-coil domain was tested using a truncated GIPC1(1-300) or any other constructs missing more of the GIPC1 C terminus. Thus, when evaluated in the yeast system, the region defined by amino acids 261-333 of GIPC1 contains amino acid sequences critical for interactions of GIPC1 with the myosin VI(955-1254) tail. These data demonstrated that the first of two myosin VI binding domains in GIPC1 resides within the region marked by the light gray vertical rectangle and designated D#1 in Figure 4.
An unexpected finding in these studies was the existence of a potential second myosin VI binding domain (D#2, Figure 4) located within the C-terminal half of the PDZ domain in GIPC1. This domain is revealed (Figure 4, row 7) only when the binding of GIPC1(1-300) is tested with myosin VI fusion proteins that lack the coiled-coil domain, but retain amino acids 1065-1160; for example, the myosin VI(1065-1204) fragment (Figure 4, column 5, row 7). Apparently, the conformation of the myosin VI(955-1254) fragment containing a portion of the coiled-coil domain blocks domain D#2 interaction with myosin VI.
To locate the C-terminal boundary of the second binding domain D#2, additional C-terminal amino acids were removed from GIPC1(1-300) to form the GIPC1(1-277), (1-249), and (1-231) fusion proteins. Binding of each of these proteins was tested with myosin VI constructs lacking the coiled-coil domain. All of the GIPC1 truncations (Figure 4, rows 8-10) produced a marginal interaction (marginal growth) with both the myosin VI(1065-1160) and (1055-1169) fusion proteins, but they maintained a strong interaction with each of the myosin VI(1065-1204) and (1065-1254) fusion proteins.
Experiments to define the N-terminal boundary of domain D#2 are presented in rows 11-13. Results using successive N-terminal truncation mutants of GIPC1(1-300) demonstrate that both GIPC1(173-300) and GIPC1(189-300) are unable to bind to the shorter myosin VI(1065-1160) and myosin VI(1065-1169) fragments, but they retain their ability to bind to myosin VI(1065-1254) and myosin VI(1065-1204) constructs that contain additional C-terminal amino acid residues within the myosin VI tail (rows 11 and 12). Further truncation to form the GIPC1(214-300) fragment (row 13) abolishes binding to any of the shorter or longer myosin VI fragments tested.
The data from rows 7-13 indicate that the smallest region comprising this second binding domain should reside within amino acids 189-231 of GIPC1. This short fragment, however, must not contain sufficient structural information as it fails to bind to any of the myosin VI fragments tested (row 16). The shortest interacting sequence capable of binding to myosin VI was GIPC1(173-231) (Figure 4, row 15) and defines the potential second interaction domain in GIPC1 marked by the light gray vertical rectangle and labeled D#2. The interaction pattern exhibited by domain D#2 in GIPC1 suggests that it binds to a myosin VI region that either overlaps with the myosin VI region recognized by GIPC1 domain D#1, or is located slightly more toward the C terminus of myosin VI. This is illustrated in Figure 4, row 15, by the ability of fusion proteins containing only the D#2 domain to interact with myosin VI(1065-1204) but not myosin VI(1065-1160) or myosin VI(1055-1169).
Anomalous Results Observed for In Vitro Binding of GIPC1(1-333) and GIPC1(1-300) to GST-myosin VI(955-1254)
To further explore the requirement of the C-terminal 33 amino acids of GIPC1 for binding to forms of the myosin VI tail containing or missing the coiled-coil domain, three GST fusion proteins containing either the minimal myosin VI binding domain (amino acids 1065-1160), the full C terminus and a portion of the coiled-coil domain (amino acids 955-1254), or the full C terminus lacking the coiled-coil domain (amino acids 1065-1254) were constructed, expressed in bacteria and purified. Each purified protein was bound to glutathione beads, incubated with in vitro translated 35S-labeled GIPC1(1-333) or GIPC1(1-300), and after extensive washing any bound 35S-labeled full-length or truncated GIPC1 was eluted from the beads, separated by SDS-PAGE using a 10% gel, and detected by autoradiography. The full-length GIPC1(1-333) (Figure 5A) is recognized and adsorbed to beads containing either myosin VI(955-1254) (lane 3) or the minimal recognition domain, myosin VI(1065-1160) (lane 2). These results are consistent with the two-hybrid studies indicating that regardless of the presence (lane 3) or absence (lane 2) of the coiled-coil region, full-length GIPC1(1-333) interacts with the C terminus of myosin VI. Furthermore, these data indicate that the minimal myosin VI domain (1065-1160) can interact with GIPC1 in both an in vitro or intracellular environment. However, when the C-terminal truncated form of GIPC1(1-300) was tested (Figure 5B), it was found to interact with the myosin VI(1065-1254) fusion protein lacking the coiled-coil domain (lane 7) as expected, but to our surprise it also interacted with the myosin VI(955-1254) fusion protein containing the coiled-coil region (lane 6). Neither GIPC1(1-333) nor GIPC1(1-300) interacted with beads containing only bound GST (lanes 4 and 8, respectively), which demonstrates the specificity of the binding to myosin VI-derived sequences. The in vitro experiments presented in Figure 5 indicate that interactions of GIPC1(1-300) with the myosin VI C terminus seem possible when using GST fusions to myosin VI(955-1254) containing a portion of the coiled-coil domain, whereas identical interactions are blocked when using DBD-fusions to GIPC1(1-300) and ACT-fusions to myosin VI(955-1254) in the yeast system. Removal of the coiled-coil domain of myosin VI apparently is required in yeast to allow D#2 of GIPC1 to interact with myosin VI(955-1254), but it is not required in the in vitro system where protein folding patterns might differ.
Figure 5.
In vitro interactions between the myosin VI C terminus and the native and truncated forms of GIPC1. (A) Autoradiogram of full-length in vitro-translated and 35S-labeled GIPC1(1-333) protein applied to the gel either directly (lane 1) or after incubation and elution from glutathione beads containing bound GST-myosin VI(1065-1160) (lane 2), GST-myosin VI(955-1254) (lane 3), or GST alone (lane 4). (B) Autoradiogram of truncated in vitro translated and 35S-labeled GIPC1(1-300) applied either directly (lane 5) or after incubation and elution from beads containing bound GST-myosin VI(955-1254) (lane 6), GST-myosin VI(1065-1254) (lane 7), or GST alone (lane 8). Labeled proteins eluted from the beads were separated by SDS-PAGE using a 10% gel. Both forms of the myosin VI C terminus that either contain (lanes 3 and 6) or are missing (lanes 2 and 7) a portion of the coiled-coil domain are able to bind both the full-length GLUT1CBP(1-333) (A) and truncated GLUT1CBP(1-300) (B) forms.
Analysis of Intracellular Interactions between Full-Length GIPC1(1-333) or GIPC1(1-249), and Myosin VI Using a Mammalian Cell System
To analyze the binding interactions in a normal environment in which these proteins exist, CHO cells were used to process and fold native and mutant proteins in the most natural environment (an intact mammalian cell) to test for myosin VI and GIPC1 domain interactions. The cells are convenient to use for binding assays because the exogenous YFP-tagged native GIPC1 and GIPC1 mutants form punctuate intracellular distributions when expressed alone in CHO cells. Under the conditions of this assay, exogenous native and mutant myosin VI proteins conveniently exhibit a diffuse distribution unless coexpressed with an interacting form of GIPC1, in which case they will redistribute and colocalize with the punctuate GIPC1 proteins. To illustrate the behavior of the assay with full-length GIPC1 and myosin VI proteins, a CFP fusion protein to the N terminus of full-length pig myosin VI and a YFP fusion protein to the N terminus of full-length GIPC1(1-333) were constructed and expressed in CHO cells. When cells expressing only CFP-myosin VI(1-1254) were analyzed (Figure 6A, solid arrows), CFP-myosin VI was excluded from the nucleus, but it was otherwise distributed diffusely throughout the cell. Although YFP-GIPC1 is a soluble protein, the native and mutant fusion proteins nevertheless exhibit a punctate distribution (A-E, middle), presumably by binding to the termini of integral membrane proteins associated with membranous and vesicular structures. When YFP-GIPC1 and CFP-myosin VI were expressed in the same cell (A, dotted arrows), the normal diffuse distribution of CFP-myosin VI shifted to the plasma membrane and to internal vesicular-like structures that colocalize with YFP-GIPC1 (Figure 6A merged). The redistribution and colocalization of CFP-myosin VI with YFP-GIPC1 is more apparent in a separate cell viewed at a higher magnification (Figure 6B).
Figure 6.
In the absence of domain D#1, domain D#2 in YFP-GIPC1 is insufficient for effective recruitment and colocalization with full-length or truncated forms of CFP-myosin VI in a mammalian cell system. Vectors expressing CFP-fusion proteins to either full-length (1-1254) or truncated (955-1254) myosin VI were cotransfected into CHO cells with vectors expressing either full-length (1-333) or truncated (1-249) YFP-fusion proteins to GIPC1 as indicated. Two days after transfection, images of the distribution of CFP-myosin VI fusion proteins (left column) and YFP-GIPC1 fusion proteins (middle column) and their merged images (right column) were collected using a confocal microscope focused near the midpoint of each cell. Excitation lasers and emission filters were set as described in Materials and Methods such that no significant CFP emissions were detected in the YFP channel (middle column) and no significant YFP emissions in the CFP channel (left column). Row A, distribution of full-length CFP-myosin VI(1-1254) is diffuse in cells not expressing YFP-GIPC1 (solid arrows), but in cells expressing full-length YFP-GIPC1(1-333) (dotted arrows) CFP-myosin VI redistributes into membrane bound and punctate cytosolic structures that colocalize with CFP-GIPC1. Row B, higher magnification of a CHO cell illustrating the effective redistribution and colocalization of full-length CFP-myosin VI(1-1254) with YFP-GIPC1(1-333). Full-length YFP-GIPC1(1-333) also induces the redistribution and colocalization of truncated CFP-myosin VI(955-1254) containing the C terminus and a portion of the coiled-coil domain (row D), whereas truncated CFP-GIPC1(1-249) containing domain D#2, but lacking domain D#1, fails to induce redistribution and colocalization with either full-length myosin VI(1-1254) or truncated myosin VI(955-1254) as shown in rows C and E, respectively.
To analyze the requirement of domain D#1 for the interaction of GIPC1 with myosin VI in a mammalian cell system, a YFP-GIPC1(1-249) variant that contains the D#2 domain and is missing the D#1 domain was coexpressed with full-length CFP-myosin VI(1-1254). YFP-GIPC1(1-249) still displayed a punctate and membrane associated distribution like that of full-length GIPC1, but CFP-myosin VI(1-1254) retained a diffuse distribution and no longer redistributed or colocalized with YFP-GIPC1(1-249) (Figure 6C).
In experiments that are directly analogous to the yeast two-hybrid tests, full-length YFP-GIPC1(1-333) was coexpressed with the truncated CFP-myosin VI(955-1254) containing the C-terminal domain of myosin VI and a portion of the coiled-coil domain. As expected, the redistribution of the myosin VI fusion protein to a punctate pattern that colocalizes with YFP-GIPC1(1-333) was observed (Figure 6D, merged). However, as noted in the yeast studies, when truncated YFP-GIPC1(1-249) containing the D#2 domain and missing the D#1 domain was coexpressed with truncated CFP-myosin VI(955-1254), the redistribution and colocalization of CFP-myosin VI(955-1254) with YFP-GIPC1(1-249) did not occur (Figure 6E). These experiments establish the importance of D#1 for proper interactions of GIPC1 with myosin VI in the intact mammalian cell. When it is missing, GIPC1 does not bind to myosin VI(955-1254) and more importantly does not interact with full-length myosin VI. Domain D#2 alone simply is not sufficient.
Demonstration of Actin-dependent and Microtubule-independent Movement of GIPC1
If GIPC1 functions as an adapter molecule that links cargo proteins to myosin VI movement as proposed, then it should be possible to demonstrate motion of GIPC1 that is actin dependent and characteristic of motion catalyzed by a myosin VI motor protein.
While analyzing the characteristic distributions of GFP-GIPC1(1-333) in a number of cell lines, it was noted that GFP-GIPC1 is often concentrated in small structures or aggregates within tubular extensions from the cell that seemed similar to retraction fibers and in many cases, filopodia. Small GFP-GIPC1-containing structures were evident in tubular extensions protruding from both the leading and trailing edges of a migrating cell. Because retraction fibers and filopodia often are difficult to discern from shape alone and can interconvert (Litman et al., 2000; Svitkina et al., 2003), these processes are referred to collectively as cellular extensions (Svitkina et al., 2003) with the realization that we are analyzing potentially both types of structures.
Green fluorescent GIPC1 particles can be observed throughout the length of such cellular extensions (Figure 7A, arrows), when CHO cells expressing GFP-GIPC1(1-333) are fixed and the F-actin is counterstained with rhodamine phalloidin. Phalloidin staining (red) of the F-actin filaments is evident throughout the entire length of these cellular extensions. Furthermore, when analyzing time-lapse images of live cell preparations, GFP-GIPC1(1-333) particles were observed to stream continuously toward the cell body. This movement toward the cell body was observed in cellular membrane extensions from both the advancing (Ad) and trailing (Tr) edge of motile cells (Figure 7B). This is more readily observed in time-lapse images of live cells presented in the supplemental movie (Fig 7B.mov) from which the representative single frame image shown in Figure 7B was taken. In this movie, membrane protrusions are being extended on the advancing edge (Ad) and are being retracted from the trailing edge (Tr) of the cell, whereas the direction of GFP-GIPC1 particle movement from either the trailing or leading edge is always toward the cell body and therefore, importantly, toward the negative end of the F-actin filament. This is consistent with its movement being catalyzed by a myosin VI motor that has been shown to move toward the negative end of the actin filament (Wells et al., 1999).
Because GIPC1 is known to associate with the microtubule-associated kinesin motor protein KIF-1B (Bunn et al., 1999) (Figure 3), and we cannot rule out the possibility that microtubule filaments also might be present within many of the cellular extensions being analyzed, it was necessary to test whether the observed GIPC1 movement was dependent upon the integrity of actin or microtubule structures within these cellular extensions. Time-lapse images were collected to establish the rate of GFP-GIPC1 movement along the extensions of living CHO cells. Then, colchicine or nocodazole was added at concentrations known to disrupt microtubules, or cytochalasin D or latrunculin B was added at concentrations known to disrupt F-actin filaments. The acquisition of images was continued after the addition of each agent to assess its effect on GFP-GIPC1 particle movement. The right panels in Figure 8 show single-frame images of a representative cell used for each experiment. The associated videos are provided as Fig 8a, b, c, and d.mov in the supplement. Individual particles were tracked using MetaMorph software to determine the distance each traveled along the cellular extensions during the interval between successive image acquisitions. For each particle tracked, the data for the distance traveled versus time were plotted (Figure 8, left). The slope of this plot provides an estimate of the average speed of each particle before and after the addition of each disruptive agent.
Figure 8.
Movement of GIPC1(1-333) within cellular extensions of CHO cells is actin dependent and microtubule independent. Time-lapse images of live transfected CHO cells expressing GFP-GIPC1 were collected at 30-s intervals using a confocal microscope focused within the plane of the extended fibers near the base of each cell. GFP-GIPC1 particles were tracked using MetaMorph software to measure the distance moved during the collection period before and after the addition of the drugs colchicine (100 μM) and nocodazole (33 μM) to disrupt microtubule structure (A and B, respectively) or cytochalasin D (4 μM) and latrunculin (10 μM) to disrupt F-actin (C and D, respectively). Representative subsets of plots of the distance of individual particle movement versus time are presented in the left column. Positive slopes indicate movement toward, and negative slopes movement away, from the cell body. The images of representative cells used to collect the data and for which videos are provided are presented in the right column. The data indicate that microtubule disrupting agents (A and B) do not inhibit particle movement, and that both the F-actin-disrupting agents (C and D) block particle movement. The videos offer an enhanced view of this phenomenon and demonstrate that particle movement is toward the cell body and consistent with the direction predicted for myosin VI catalyzed movement. They also dramatically illustrate the continued movement toward the cell body of a large sampling of particles after the addition of either colchicine or nocodazole, and their block in progressive movement toward the cell body after the addition of cytochalasin D or latrunculin. Videos are presented in the supplement as Fig 8a, b, c, and d.mov. E demonstrates that coexpression of CFP-myosin VI(955-1254) lacking the motor domain with YFP-GIPC1 blocks movement of GIPC1 toward the cell body. Consistent with the model presented in Figure 1, mutant CFP-myosin VI(955-1254) by interacting with YFP-GIPC1 (see Figure 6D) blocks movement by preventing the interaction of a fully functional myosin VI with YFP-GIPC1. A video demonstrating this effect is presented in the supplement as Fig 8e.mov.
The addition of colchicine or nocodazole to disrupt microtubules does not stop or retard the movement of the GFP-GIPC1 particles (Figure 8, A and B). Colchicine in fact seemed to accelerate their movement by at least twofold. In contrast, cytochalasin D, after a small delay, and latrunculin, immediately, stopped particle movement after disrupting F-actin structure (Figure 8, C and D). This behavior is consistent with an actin-dependent and myosin VI-catalyzed movement of GIPC1 as proposed. The videos provided for each of the experimental drug treatments more emphatically demonstrate the direction of movement and the dependence of GFP-GIPC1 movement on intact F-actin (Supplemental Figures 8A-D.mov). The rate of movement of GFP-GIPC1 particles before drug treatment was estimated from measurements taken for 24 particles from three different cells. The mean velocity ± SE was 0.94 ± 0.024 μm/min. The maximum velocity over any given 30-s interval was 5.7 μm/min.
Although the direction of movement of the GFP-GIPC1 complexes is consistent with myosin VI-catalyzed motion, it is also consistent with the movement that could arise from actin tread-milling. To further substantiate that the observed motion arises from association with a functional myosin VI molecule, cells expressing both YFP-GIPC and a CFP-myosin VI(955-1254) mutant lacking the motor domain were also analyzed. As presented in the left panel of Figure 8E, after 10 min the majority of the particles have shown little to no movement toward the cell body. One particle has moved at most 1 μm closer after 10 min, compared with the 9-10 μm traveled in 10 min by those particles tracked before drug treatment in Figure 8B. The average velocity and SE obtained from measurements of a total of 53 YFP-GIPC1 particles from 12 different cells was -0.009 ± 0.001 μm/min (-signifies movement away from the cell body) in the presence of CFP-myosin VI(955-1254), whereas in its absence the average rate is nearly 1 μm/min. Thus, for all practical purposes motion toward the cell is stopped by disrupting potential interactions of GIPC1 with native myosin VI. This is also illustrated in the supplemental video Fig 8E.mov illustrating particle movement in the representative cell presented in the right panel of Figure 8E.
Occasionally, a particle can be found that is moving more rapidly away from the cell, for example the track with the negative slope in Figure 8E (velocity -0.25 μm/min). This suggests that other forces might function to move GIPC1 toward the tips of the cell extensions when myosin VI interactions are disrupted. This possibility is also supported by the observation that some particles reverse their direction and begin moving outward when F-actin structure is disrupted by cytochalasin D or latrunculin (see the left panels of Figure 8, C and D, and movies Fig 8C.mov and Fig 8D.mov).
Assays for Coordinated Movement of GIPC1-Myosin VI Complexes
This experiment was designed to demonstrate that GIPC1 movement within cellular extensions is not restricted to CHO cells and to test the model for myosin VI-assisted GIPC1 movement by determining whether GIPC1-myosin VI complexes could be observed to move together along an actin network within cellular extensions. This was accomplished by cotransfecting 293 cells with CFP-myosin VI(1-1254) and YFP-GIPC1(1-333). Then, 2 d after transfection, time-lapse images were collected with the confocal microscope using the same settings as before (Figure 6) to separate CFP and YFP emission signals. Particles of CFP-myosin VI in the CFP channel (Figure 7C, top left) and YFP-GIPC1 in the YFP channel (top right) were located in identical positions along the cellular extensions (marked by arrows). The merged image (Figure 7C, bottom left) of the CFP-myosin VI (green) and YFP-GIPC1 (red) signals supports their colocalization (yellow) and coordinated movement as a complex toward the cell body. An analysis of their rate of movement using MetaMorph software indicates that each of the three GIPC1-myosin VI complexes was moving at a different rate (Figure 7D), whereas the CFP-myosin VI and YFP-GIPC1 components of each complex moved together and did not separate. The movie (Figure 7C.mov) provided in the supplement more readily illustrates this point. Measurement of the movement of 22 separate particles from five different cells gave a mean velocity ± SE of 0.76 ± 0.037 μm/min for the CFP-myosin VI/GIPC1 particle complexes. The maximum rate observed within the 20-s sampling interval was 5.0 μm/min.
In a recent report (Rustom et al., 2004), structures arising from extending filopodia that establish an actin containing bridge between two well separated cells were reported to serve as conduits for unidirectional transport of small membrane organelles between the two cells. These structures were named nanotubes. A bridge between 293 cells identical in appearance to the nanotubes described in that report was noted in our study of myosin VI/GIPC1 complex movement (Figure 7E). Several CFP-myosin VI/YFP-GIPC1 complexes were noted within this bridging structure. The complex marked by the arrow was moving in the direction indicated at a rate of 1.2 μm/min.
Disruption of GLUT1 Interactions with GIPC1 Induces Faulty Trafficking of the Cargo Protein GLUT1
The studies described above support the proposed adapter function for GIPC1 to link PDZ bound proteins to actin-dependent movement catalyzed by myosin VI. As further evidence of the potential trafficking function for GIPC1, we have analyzed the processing of one of its potential cargo proteins, GLUT1, after disrupting interactions between the two proteins.
MDCK cells were used in these experiments because GLUT1 is efficiently targeted to the basolateral membrane (Pascoe et al., 1996), and in addition to GLUT1, they normally express both GIPC1 and myosin VI. If the interaction of GLUT1 with the GIPC1-myosin VI complex is important for the delivery of GLUT1 to this region of the plasma membrane, then disrupting the interactions should alter proper GLUT1 movement within the MDCK cell.
To test this hypothesis, a GFP-fusion protein to GLUT1 was expressed in polarized MDCK cells to confirm that the N-terminal fluorescent fusion protein would not interfere with normal GLUT1 targeting to the basolateral membrane. After 2 d, reconstructed X-Z sections from confocal scans of MDCK cells expressing GFP-GLUT1 (Figure 9C) demonstrate the predicted basolateral distribution for GFP-GLUT1. This is also evident in the three-dimensional reconstruction of the GFP-GLUT1 protein distribution provided as a supplemental video (Fig 9A.mov) that is represented by the single-frame image (Figure 9A). The majority of the protein is confined to the basolateral membrane and very little transporter is concentrated in either the apical membrane or in any large vesicle populations within the cell (Figure 9C).
Figure 9.
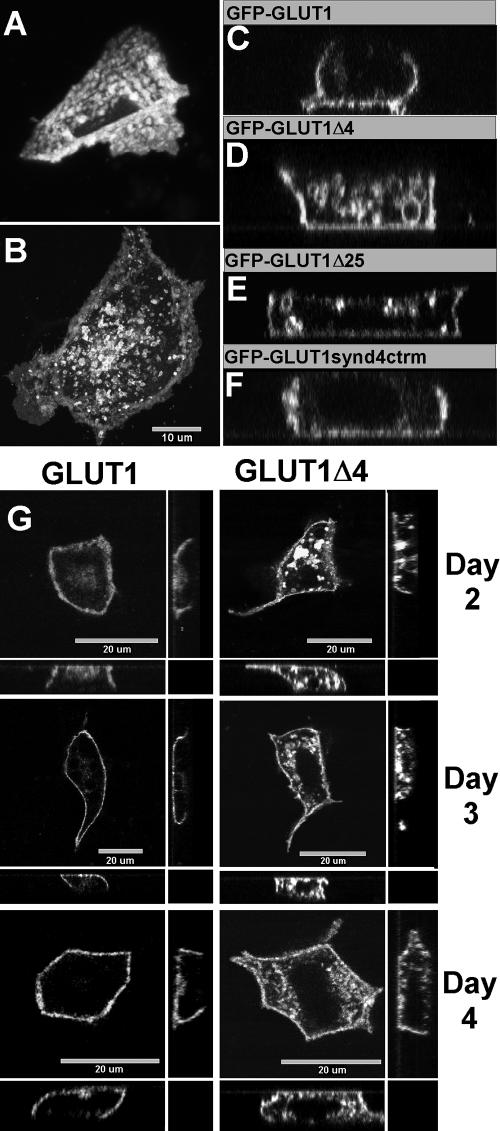
The C-terminal four-amino acid sequence recognized by the PDZ domain of GIPC1 is required for normal distribution of GLUT1 between the plasma membrane and intracellular membranous structures in MDCK cells. EGFP-C1 vectors driving the expression of GFP fused to the N terminus of GLUT1, GLUT1Δ4, GLUT1Δ25, and GLUT1synd4ctrm were introduced separately into monolayers of MDCK cells using Lipofectamine 2000. The expressed proteins represent, respectively, GLUT1 with the native C terminus, GLUT1 missing the last four amino acids or the last 25 amino acids, and GLUT1 with the last four amino acids replaced by the C-terminal four amino acid sequence from syndecan 4. Two days after transfection, the distribution of each GFP fusion protein was determined with a confocal microscope by collecting a series of z-section images. A and C, three-dimensional projection and X-Z plane reconstruction, respectively, demonstrating the normal basolateral distribution for GFP-GLUT1 and the normal low concentration of transporter in intracellular membranes relative to that present in the basolateral membrane. B and D, three-dimensional projection and X-Z plane reconstruction, respectively, demonstrating high concentrations of mutant GFP-GLUT1Δ4 in intracellular membranes. A comparison of the single frame images presented in A and B is best observed by viewing the three-dimensional reconstructions in the supplemental videos Fig 9A.mov and Fig 9B.mov. They effectively demonstrate the low internal GLUT1 distribution in A and the high internal GLUT1Δ4 distribution in B. (C, D, E, and F) X-Z plane reconstructions of cells expressing GFP-GLUT1, GFP-GLUT1Δ4, GFP-GLUT1Δ25, and GFP-GLUT1synd4ctrm, respectively. These results illustrate that the observed abnormal intracellular distribution of GFP-GLUT1Δ4 (D) and GFP-GLUT1Δ25 (E) relative to GFP-GLUT1 (C) reverts to a normal distribution when the missing amino acids in GFP-GLUT1Δ4 are replaced by those of syndecan 4 to form GFP-GLUT1synd4ctrm (F). The apical domain is orientated toward the top of the panel in the X-Z reconstructions presented in C, D, E, and F. Native and mutant transporters present in the plasma membrane remain in the basolateral domain with little to no targeting to the apical domain (C-F). The experiments presented in G demonstrate that extended intervals of expression do not correct the abnormal elevated intracellular distribution observed for mutant GFP-GLUT1Δ4 relative to native GFP-GLUT1. Vectors driving the expression of GFP-GLUT1 and GFP-GLUT1Δ4 were transfected into MDCK cells and analyzed as described above, at 2-, 3-, and 4-d intervals after transfection. The X-Y plane images collected near the midpoint of the cell are presented in the large panel with the associated reconstructed X-Z plane (bottom) and Y-Z plane (right) presented for each mutant and interval of expression. The apical domain is oriented toward the bottom of the X-Z image and toward the right of the Y-Z image. The contrast and brightness settings have been adjusted to enhance the visibility of the basolateral distribution of native and mutant transporters in the plasma membrane and to permit a visual comparison of the distribution of native and mutant transporters between internal and plasma membrane compartments. A quantitative comparison obtained from unprocessed images is presented in Figure 10.
The last four amino acids comprising the class I PDZ recognition motif can be removed from GFP-GLUT1 to form a GFP-GLUT1Δ4 mutant that no longer binds to the PDZ domain of GIPC1 (Bunn et al., 1999). When cells expressing this mutant form of GLUT1 are analyzed after the same interval of expression used for GFP-GLUT1 (2 d), a significantly higher percentage of the total cellular transporter mass seems to accumulate in an abundant vesicle population within the cell (Figure 9, B and D). This is evident when comparing both the three-dimensional projections as well as the X-Z section images of cells expressing “native” GFP-GLUT1 (Figure 9A, 9A.mov and C) or mutant GFP-GLUT1Δ4 (Figure 9B, 9B.mov and D).
To determine whether the abnormal concentration of GLUT1Δ4 protein in internal vesicle pools observed 2 d after transfection was a transient phenomenon, the protein distribution of native GFP-GLUT1 and mutant GFP-GLUT1Δ4 proteins were analyzed at 2, 3, and 4 d after transfection. The level of GFP-GLUT1 present in internal structures (Figure 9G) remains very low relative to the amount present in the plasma membrane. In addition, GFP-GLUT1 remains effectively excluded from the apical domain throughout the 4-d period of expression. In contrast, the level of GFP-GLUT1Δ4 present in internal membranous structures remains high relative to that present in the plasma membrane throughout the same interval. The majority of the GFP-GLUT1Δ4 in the plasma membrane, however, was still restricted to the basolateral domain of the cell (Figure 9G, day 4). Small amounts of GFP-GLUT1Δ4 protein are evident near the apical region of the cell throughout the interval of expression, but it is unclear whether this small signal resides in the apical membrane or may actually represent a small portion of the enlarged vesicle pool of mutant transporter residing below the apical membrane surface.
These data strongly suggest that relative to cells expressing native GLUT1, cells expressing the mutant GLUT1Δ4 exhibit some defect in processing of GLUT1Δ4 that persists throughout the entire 4-d period of expression. It is also clear that the C-terminal four amino acids are not required for newly synthesized GLUT1Δ4 to reach the plasma membrane. Furthermore, the presence of the C-terminal four amino acids of GLUT1 does not seem necessary to target GLUT1 to the basolateral membrane as their loss does not result in faulty targeting of the transporter to the apical membrane.
An alternative interpretation for the inefficient trafficking of GLUT1Δ4 could be that it arises as an artifact in processing of newly synthesized transporter that is related to the structure or amino acid sequence of the newly created C terminus, rather than the removal of the motif recognized by the PDZ domain of GIPC1. Replacement of the single C-terminal valine with an alanine, for example, significantly interferes with Golgi processing of CD8α (Iodice et al., 2001). To address this possibility two experiments were performed.
First, 25 rather than four amino acids were removed to create a mutant GFP-GLUT1Δ25 with an entirely new C terminus. This transporter mutant is fully capable of inserting into the plasma membrane and effectively transporting glucose when expressed in Xenopus oocytes (Bunn, 1999); thus, the structural features important to these functions are not disrupted by the mutation. Nevertheless, a significant percentage of the total cellular GFP-GLUT1Δ25 transporter was restricted to large intracellular vesicles (Figure 9E), similar to GFP-GLUT1Δ4.
In a second experiment, the Asp-Ser-Gln-Val sequence removed from native GLUT1 to form the GLUT1Δ4 mutant, was replaced by the syndecan-4 C-terminal sequence Glu-Phe-Tyr-Ala to form a GFP-GLUT1synd4ctrm mutant. This substitution not only provides an entirely different C-terminal four-amino acid sequence but also converts the normal class I PDZ recognition motif present in the native protein to a class II motif. The substitution is useful because it takes advantage of the surprising and diverse property of the GIPC1 PDZ domain to recognize protein sequences representing all three classes of recognition motifs (see discussion, Table 1). When the GFP-GLUT1synd4ctrm is introduced into MDCK cells, the mutant protein (Figure 9F) regains the targeting behavior of native GLUT1 (Figure 9C). In direct contrast to GFP-GLUT1Δ4 and GFP-GLUT1Δ25 (Figure 9, D and E, respectively), only a very low percentage of GFP-GLUT1synd4ctrm transporter is evident in internal vesicles (Figure 9F). Thus, although this sequence is entirely different from the native four terminal amino acids of GLUT1, an alternative and known target sequence for the PDZ domain of GIPC1 is sufficient to restore normal distribution of the transporter primarily to the plasma membrane and restrict the accumulation of transporter in large internal vesicles.
These data provide reasonable evidence that the interaction of GLUT1 with GIPC1 functions at one or more points in the sorting pathway in MDCK cells to ensure efficient protein distribution to the plasma membrane such that only low amounts of transporter reside in internal vesicle membrane compartments compared with that present in the plasma membrane.
Blocking the Interaction of the GLUT1/GIPC1 Complex with Native Myosin VI Also Disrupts Normal GLUT1 Trafficking
If loss of the C-terminal four amino acids of GLUT1 leads to faulty accumulation of mutant GLUT1Δ4 in internal membrane structures through its inability to form a GLUT1/GIPC1/myosin VI complex, then blocking the interaction of a GLUT1/GIPC1 complex with myosin VI also should lead to internal accumulation of native GLUT1. To test this hypothesis, MDCK cells were cotransfected with plasmids encoding the expression of CFP-GLUT1 and either YFP or YFP-myosin VI(955-1254). YFP-myosin VI(955-1254) should compete with native myosin VI for binding to a GLUT1/GIPC1 complex and block interactions of the complex with endogenous native myosin VI, whereas YFP expression alone should have no effect. Because the expression of YFP-myosin VI(955-1254) drops significantly 2 d after transfection, the incubation period was shortened from 2 d to 18 h to maintain a higher uniform level of expression. This shorter period of incubation does not significantly alter the efficient processing of CFP-GLUT1 because its distribution between the plasma membrane and internal pools at 18 h and 2 d after transfection is indistinguishable.
Cells coexpressing CFP-GLUT1 and YFP (Figure 10A) show a high level of CFP-GLUT1(green) in the plasma membrane and a low internal concentration of CFP-GLUT1, like that observed for GFP-GLUT1 expressed in the absence of YFP (Figure 9G). However, when cells coexpressing CFP-GLUT1 and YFP-myosin VI(955-1254) were examined, an entirely different result was obtained. In cells expressing no YFP-myosin VI(955-1254) (Figure 10B, cell 1), normal low levels of intracellular CFP-GLUT1 were observed, but as the level of YFP-myosin VI(955-1254)(red) expression is elevated (Figure 10B, cells 2 and 3) the amount of internal CFP-GLUT1(green) increases dramatically. These data demonstrate that increased levels of YFP-myosin VI(955-1254) expression lead to faulty internal accumulation of CFP-GLUT1 identical to that observed for the mutant CFP-GLUT1Δ4. Furthermore, in an enlarged view YFP-myosin VI(955-1254) fluorescence (Figure 10C, red) in all cases colocalizes with internal membranous structures containing CFP-GLUT1 (Figure 10C, green). In several regions (marked by arrows in Figure 10C), YFP-myosin VI(955-1254) seems to be bound to vesicles containing CFP-GLUT1.
Quantitative Estimates of the Changes in Plasma Membrane and Internal Membrane Transporter Distribution Induced by Removal of the C-Terminal Four Amino Acids of GLUT1, or by Overexpression of Myosin VI(955-1254)
MDCK cells were transfected with CFP-GLUT1 or CFP-GLUT1Δ4 (Figure 10D) or cotransfected with CFP-GLUT1 and either YFP or YFP-myosin VI(955-1254) (Figure 10E), and then they were analyzed after 18 h by collecting z-sections through the entire height of the cell. Both total cellular and plasma membrane-associated CFP-GLUT1 or CFP-GLUT1Δ4 fluorescence were integrated throughout the entire cell volume, and the total plasma membrane fluorescence was expressed as a percentage of the total cellular fluorescence (Figure 10, D and E).
Data obtained from the integration of five random cells from the population provide a mean ± SE value for CFP-GLUT1 located within or intimately associated with the plasma membrane equal to 51.3 ± 2.4% of the total cellular CFP-GLUT1 (Figure 10D), whereas the remaining 48.7% is internal. In cells expressing CFP-GLUT1Δ4 (Figure 10D), the plasma membrane content of the mutant protein drops to 22.3 ± 3.2% (p < 0.001) of the total content, whereas the internal content rises to 77.7% of the total expressed CFP-GLUT1Δ4. This 1.6-fold increase in internal CFP-GLUT1Δ4 would not be that noticeable if it remained dispersed evenly throughout the cell. However, there is an apparent redistribution and concentration of the transporter into a unique vesicle population that does not occur when CFP-GLUT1 is expressed in MDCK cells.
Figure 10E presents the results obtained when CFP-GLUT1 is coexpressed with either YFP or YFP-myosin VI(955-1254). Integrations of total and plasma membrane CFP-GLUT1 fluorescence were obtained from 10 total cells, half coexpressing YFP and half coexpressing equivalent levels of YFP-myosin VI(955-1254). When expressed with YFP, ∼53.2 ± 3.8% of the CFP-GLUT1 is localized to the plasma membrane, whereas the remaining 46.8% is internal. This distribution is nearly identical to that observed when CFP-GLUT1 is expressed in the absence of YFP (Figure 10D). This demonstrates that the expression of YFP itself has no effect on the distribution of CFP-GLUT1. However, coexpression of YFP-myosin VI(955-1254) with CFP-GLUT1 reduces the plasma membrane content of CFP-GLUT1 to 27.6 ± 2.5% (p < 0.001) of the total with a concomitant rise of internal CFP-GLUT1 to 72.4% of total. As with GLUT1Δ4, this internal accumulation (1.5-fold) of CFP-GLUT1 due to the disruptive effects of myosin VI(955-1254) expression also is accompanied by a shift in distribution of internal CFP-GLUT1 to larger more visible internal membranous components not observed when only YFP is expressed.
CFP-GLUT1Δ4 Is Inappropriately Targeted to Membrane Vesicles of Endosomal Origin
To determine the nature of the intracellular vesicles that accumulated large amounts of the GLUT1Δ4 protein, MDCK cells were transfected with pECFP-GLUT1Δ4, and 18 h later various vesicle compartments were labeled and examined by fluorescence and immunofluorescence microscopy. Results from immunofluorescence microscopy revealed that an appreciable portion of CFP-GLUT1Δ4 vesicles (green) were positive for the early endosome marker EEA1 (red) (Figure 11, C and D). In contrast, incubating transfected cells with the acidic vesicle marker LysoTracker Red showed that the majority of the CFP-GLUT1Δ4-positive vesicles (green) were positive for the dye (red) (Figure 11, A, B, and E). This result indicates that the lumen of the CFP-GLUT1Δ4 vesicles were acidic pH <5.5, a property associated with late endosomes and lysosomes. Typical maturation of vesicles from early endosomes into late endosomes is marked by the loss of EEA1 and accumulation of late endosomal components that coincides with vesicle acidification. Because CFP-GLUT1Δ4 vesicles were found to associate with both early endosomal (EEA1) and late endosomal (acidic pH) components, it was necessary to further characterize the distribution of this protein within the endosomal pathway. MDCK cells transfected with pECFP-GLUT1Δ4 were stained for acidic vesicles, fixed, and then stained with anti-EEA1 antibodies to reveal whether any aberrant accumulation of EEA1 occurred on acidic CFP-GLUT1Δ4-positive vesicles. It was observed that those CFP-GLUT1Δ4 vesicles (green) positive for EEA1 (blue) were not acidic (red) (Figure 11E), suggesting that CFP-GLUT1Δ4 protein associates with distinct populations of both early and late endosomes.
Figure 11.
GFP-GLUT1Δ4 accumulates in endosome derived compartments, primarily late endosomes and lysosomes. In all panels, MDCK cells were transfected with a vector expressing GFP-GLUT1Δ4 and after 18 h were treated to label various vesicle compartments. Images were collected at the midpoint of each cell using a confocal microscope. (A) Monolayers were exposed for 10 min to LysoTracker red to label acidic compartments, washed, and then fixed with paraformaldehyde. A majority of the large GFP-GLUT1Δ4-containing vesicles (green) contained an acidic lumen (red, pH < 5.5) marked by the LysoTracker dye. (B) An enlarged view of the region bounded by the dotted rectangle shown in A illustrating the acidic vesicles (red) bounded by membranes containing high concentrations of GFP-GLUT1Δ4. (C) Monolayers were fixed, permeabilized, and stained with a mouse anti-EEA1 antibody followed by a Cy3-conjugated goat anti-mouse IgG to label the early endosomal compartment. The image demonstrates that an appreciable number of GFP-GLUT1Δ4 containing vesicles (green) are associated with the early endosome marker EEA1 (red). (D) An enlarged view of the region bounded by the large rectangle in C that more clearly demonstrates GFP-GLUT1Δ4-containing vesicles (green) decorated by EEA1 (red). (E) To confirm that EEA1-positive vesicles were not inappropriately associated with the large number of acidic GFP-GLUT1Δ4-enriched vesicles, monolayers were pulsed for 10 min with LyosTracker red, washed, fixed, permeabilized, and incubated with mouse anti-EEA1 and Cy5-conjugated goat anti-IgG to label respective acidic and early endosomal compartments. EEA1-positive compartments (blue) do not coincide with acidic vesicles (red) containingaccumulatedGFP-GLUT1Δ4.(F)Monolayers were pulsed with RITC-dextran for 20 min then fixed to label early endosomal compartments. In this image a small number of the GFP-GLUT1Δ4 containing vesicles (green) contain RITC-dextran (red). (G) Monolayers were pulsed with RITC-dextran as in F, washed, chased for an interval of 60 min, and then fixed to label late endosomal and lysosomal structures. Much like the labeling pattern in A, a majority of vesicles enriched in GFP-GLUT1Δ4 (green) contain RITC-dextran (red).
To further establish the endosomal origin of vesicles in which CFP-GLUT1Δ4 accumulates, monolayers were incubated with RITC-dextran for a 20-min pulse to label early endosomes (Figure 11F) or pulsed for 20 min followed by a 60-min chase period to label late endosomes (Figure 11G). After the 20-min pulse only a small number of the CFP-GLUT1Δ4-positive vesicles (green) were RITC-dextran positive (red). In contrast, after the 60-min chase period, the majority of the CFP-GLUT1Δ4-containing vesicles (green) also were RITC-dextran positive (red). Together, these results demonstrate that CFP-GLUT1Δ4 vesicles are found predominantly within the endosomal pathway where these vesicles communicate with functional early endosomes and to a much greater extent, late endosomes.
DISCUSSION
The data presented support our earlier proposed adapter function for GLUT1CBP(GIPC1) (Bunn et al., 1999) and provide a basis for a refined model (Figure 1) wherein movement of a cargo protein bound to the PDZ domain of GLUT1CBP(GIPC1) is mediated through interactions of its C-terminal domain with the tail of the myosin VI motor protein.
Several observations support this model: First, the binding of the C-terminal domain of GIPC1 to myosin VI can be demonstrated in both an intracellular and in vitro environment. Second, no additional proteins are required to mediate their binding, because these interactions occur between the purified GIPC1 and myosin VI proteins. Third, the movement of GIPC1 within the cellular extensions of intact cells is consistent with motion catalyzed by myosin VI, because the movement is actin dependent, toward the negative end of the actin filament, opposite that of other actin-dependent motor proteins, and blocked by the expression of a myosin VI mutant lacking the motor domain. Fourth, a functional interaction between these two proteins was substantiated by the ability of CFP-myosin VI and YFP-GIPC1 to form complexes in 293 cells that move coordinately toward the negative end of the actin filament within cellular extensions. Fifth, these complexes can move at maximum rates comparable with those measured using purified proteins in a reconstituted system (Altman et al., 2004). Sixth, the formation of trimeric complexes between the myosin VI tail, GIPC1, and GLUT1 can be demonstrated. Seventh, removing the last four amino acids of GLUT1 to block its ability to interact with GIPC1 disrupts normal GLUT1 trafficking, whereas replacing them with amino acids belonging to a different class PDZ recognition sequence (syndecan class II) known to interact with GIPC1 restores normal trafficking of GLUT1. Finally, disruption of GIPC1 interactions with native myosin VI by overexpression of myosin VI(955-1254) also disrupts normal GLUT1 trafficking.
Two binding domains in GIPC1 capable of interacting with myosin VI were identified in this study. Domain D#1 comprises, or is contained within the C-terminal amino acids 261-333. The second domain D#2 is defined by amino acids 173-231 located in the C-terminal half of the PDZ domain. The typical cargo binding site resides in the N-terminal half of a PDZ domain. This site exhibits sufficient spatial separation from domain D#1 to allow simultaneous binding of a cargo protein to the PDZ domain and myosin VI to domain D#1 as presented by the model (Figure 1A). The situation becomes more problematical for simultaneous binding if myosin VI were to interact with GIPC1 via domain D#2, because the cargo and myosin VI interaction sites are significantly closer within the primary sequence of GIPC1. However, a comparison of the topology of these sites in the PDZ domain of GIPC1 to the structure of the PDZ domains of several homologous proteins suggests both interactions might be accommodated. For example, the solution structure of the PDZ domain for nitric oxide synthase is depicted in Figure 1B. The N-terminal half of the PDZ domain (red) is shown interacting with the target C-terminal peptide sequence (yellow). The location of the C-terminal half of the PDZ domain (green) represents the hypothetical location of domain D#2 if GIPC1 assumed a similar structure. These sites lie on opposite faces of the PDZ domain; thus, if such a spatial structure exists for the PDZ domain of GIPC1, both cargo and D#2-mediated interactions with myosin VI conceivably could still occur.
Whether domain D#2 can ever participate in binding interactions with myosin VI under physiological conditions, nevertheless, remains unresolved. Of most importance, in the normal mammalian environment for GIPC1 and myosin VI, expression experiments presented in this study demonstrated that domain D#2 in the absence of domain D#1 was insufficient to support interactions of GIPC1 with either full-length myosin VI or with the myosin VI tail containing a only a portion of the coiled-coil domain. In the yeast system, removal of the coiled-coil domain in myosin VI was necessary before interactions of the D#2 domain of GIPC1 with myosin VI(955-1254) tail could be demonstrated in the absence of D#1. Thus, the interactions between D#2 with myosin VI do not seem to be physiologically relevant in intact cell systems. Nevertheless, the data to date cannot rule out a cooperative or sequential interaction scheme in which the binding of D#1 to native myosin VI must occur first, facilitating the subsequent interactions of D#2 with sequences residing more toward the C terminus of myosin VI. Alternatively, the mammalian cell system used may contain unknown proteins that bind to domain D#2 and block its interaction with myosin VI until unknown signals remove that block. For example, Farquhar's group has recently demonstrated that a region in the PDZ domain (separate from the C-terminal binding pocket) interacts with an internal amino acid sequence of the TrkA receptor (Lou et al., 2001). Occupancy of this site might prevent any D#2 interactions with myosin VI. Further studies will be needed to ascertain the validity of these alternatives.
Finally, the observed inability of GIPC1(1-300) to interact with myosin VI(955-1254) in yeast (Figure 4), but to interact in the in vitro system (Figure 5) when the domain D#1 has been compromised by the removal of 33 amino acids in domain D#1, is interesting. These data suggest that either differences in protein folding, the respective presence and absence of proteins capable of blocking D#2 interactions in the two systems, or both are likely explanations for the observed binding differences. These or other alternatives also must be resolved in future studies.
The adapter role observed for GIPC1 with GLUT1 could extend to other potential cargo proteins represented by the diverse list (Table 1) of proteins recognized by the PDZ domain of the mammalian GIPC1. As discussed in a recent review (Harris and Lim, 2001), PDZ domains generally recognize ligands that fall into one of three classes. The majority of proteins (Table 1) interacting with the PDZ domain of GIPC1 fall into class I in which the last four amino acids are X-S/T-X-hydrophobic (X can be any amino acid). However, the interacting syndecans contain a class II motif and end with a X-hydrophobic-X-hydrophobic sequence, whereas the C termini of LHR, rat TomL1, and the dopamine D2 and D3 receptors express the class III X-X-C motif. Thus, GIPC1 exhibits a very diverse binding specificity, not only by interacting with a large number of proteins in class I but also by crossing typical specificity boundaries and interacting with members of all three classes of recognition sequences. This diversity, however, could provide a more generalized function for GIPC1 to sort vesicles containing one or more of these proteins by linking them to myosin VI-directed movement within the cell.
This is not meant to imply that the GIPC1 PDZ domain lacks binding specificity, only that it is more diverse than many. Mercurio's laboratory reported that an acidic amino acid in either or both of the -1 or -3 positions is required for integrin binding to GIPC1 (Tani and Mercurio, 2001). The proteins listed in Table 1 obey this binding requirement with the two exceptions of GP75(Trp-1) and the transforming growth factor (TGF)-βIII receptor. In agreement with this sequence requirement, we have shown that human GLUT3 fails to bind to GIPC1 (Bunn et al., 1999). Although it contains a typical class I binding motif (Thr-Thr-Asn-Val), it lacks acidic amino acids in both the -1 and -3 positions.
What trafficking functions might such a GIPC1-myosin VI complex perform for the potential cargo proteins listed in Table 1? The orientation of actin filaments underlying the plasma membrane and those near the Golgi suggest several functions. Myosin VI that is located near the trans-Golgi network might function in conjunction with GIPC1 to sort PDZ bound cargo proteins departing the trans-Golgi network and direct them toward the plasma membrane. Several earlier studies implicate GIPC1 functioning to move the newly synthesized TGFβIII receptor (Blobe et al., 2001) and GP75 protein (Liu et al., 2001) toward the plasma membrane. Conversely, at the plasma membrane, myosin VI in conjunction with GIPC1 could function in the clustering of receptors destined for endocytosis in coated pits or in the movement of newly formed endosomes to as yet unidentified locations within the cell. Electron microscopy studies by Farquhar's group demonstrate that GIPC1, although normally a soluble protein, can associate with endosomes (DeVries et al., 1998). Hasson's laboratory has recently demonstrated that myosin VI associates with newly uncoated endosomes and that GIPC1 can reside in a portion of this vesicle population (Aschenbrenner et al., 2003). In addition, myosin VI seems necessary for some endosomes to penetrate the actin cortex beneath the plasma membrane (Aschenbrenner et al., 2004). Additional reports indicate that GIPC1 participates in the recycling of recognized endosomal proteins back to the plasma membrane and/or away from degradative pathways (Blobe et al., 2001; Hirakawa et al., 2003; Jeanneteau et al., 2003).
The studies presented in this article demonstrate the direct association of GIPC1 with myosin VI and the coordinate movement of such a GIPC1-myosin VI complex along actin filaments for extended distances. When CFP-myosin VI is expressed in the absence of exogenous YFP-GIPC1, a substantial portion of the myosin VI seems diffusely distributed. YFP-GIPC1(1-249) missing the D#1 binding domain for myosin VI can easily associate with plasma membrane and internal vesicle structures, presumably by binding to the PDZ recognition motifs of potential membrane bound cargo proteins listed in Table 1, but it fails to recruit significant amounts of myosin VI to these structures. Full-length GIPC1 also binds to these structures, but it induces an efficient redistribution and colocalization of myosin VI to these same regions. This recruitment via domain D#1 of myosin VI to membrane bound cargo proteins may be an important step in connecting these cargo proteins to myosin VI-catalyzed movement within the cell. A more detailed analysis of the potential order and localization of these interactions was published during the preparation of this article (Dance et al., 2004). These observations are consistent with a trafficking function of the GIPC1-myosin VI complex for membrane-bound proteins.
A recent report (Inukai et al., 2004) that was published during this study demonstrated that C-terminal sequences of GLUT1 were not required for its proper targeting to the basolateral domain of MDCK cells. Consistent with that report, our studies confirm that the ability of GLUT1Δ4 to be delivered primarily to the basolateral rather than the apical membrane of MDCK cells is unaffected by the removal of the four C-terminal amino acids. However, the results of this study do strongly support the need for a functional GLUT1-GIPC1-myosin VI complex to maintain normal internal and basolateral membrane distributions of GLUT1 in MDCK cells. Abnormal accumulation of GLUT1Δ4 in intracellular vesicles and reduced levels of transporter in the basolateral domain occur when GLUT1Δ4 lacking a C-terminal PDZ recognition motif is unable to interact with GIPC1 (Bunn et al., 1999) in the GIPC1-myosin VI complex. Proper myosin VI interactions with the GLUT1-GIPC1 complex also are important in this process, because overexpression of myosin VI(955-1254) also disrupts normal trafficking of GLUT1 and results in a similar accumulation and concentration of GLUT1 in larger intracellular vesicles. The results of the EEA1 labeling, LysoTracker dye staining, and RITC-dextran pulse-chase experiments suggest that vesicles in which GLUT1Δ4 accumulates are of endosomal origin and ultimately comprise a population of acidic late endosomes and lysosomes in which GLUT1 normally does not reside at such high concentrations. These data suggest that interaction of GLUT1 with the GIPC1-myosin VI complex is important for proper routing of endocytosed vesicles containing GLUT1 away from late endosomes and lysosomes and toward recycling pathways to the plasma membrane. More complex kinetic studies of GLUT1 processing will be needed to determine whether reduced rates of movement of newly synthesized GLUT1Δ4 to the plasma membrane or direct diversion to lysosomal structures also might contribute to the observed trafficking defects. In summary, these studies strongly support an adapter role for GIPC1 in which the ability to form a trimeric GLUT1-GIPC1-myosin VI complex is required for normal trafficking and distribution of GLUT1 within the cell.
The adapter function of GIPC1 was further supported by the ability of GIPC1 and myosin VI to form a complex and move coordinately within the cellular extensions. The average rate of movement for the GIPC1-myosin VI complex was 0.76 μm/min with a maximum rate of 5.0 μm/min. Rates as low as 4.8 μm/min for movement have been measured at 25°C for an unloaded myosin VI molecule using an in vitro sliding actin filament assay (Altman et al., 2004). At the same temperature myosin VI-catalyzed movement of intracellular vesicles in live cells was reported to be 2.3 μm/min with a maximum rate of 4.8 μm/min measured within a single 20-s collection interval (Aschenbrenner et al., 2004). In our study, the maximum rate observed in a given 20-s sampling interval was comparable with values observed in the sliding filament assay and to the maximum rate of vesicle movement; however, the average rate was slower than both. Our observed slower movement of the myosin VI-GIPC1 complex might arise from the ability of GIPC1 to interact with less mobile membrane bound cargoes or to dimerize and incorporate two or more myosin VI molecules into the complex. Either could then interfere with normal rapid movement of a single unloaded myosin VI molecule. Alternatively, it should be noted when GIPC1 particle movement toward the cell is disrupted either by the addition of drugs that affect F-actin integrity (latrunculin or cytochalasin D) or by expressing a myosin VI mutant to block GIPC1 interactions with a functional mysosin VI, some particles reverse their direction of motion and move toward the tips of the cellular extensions. The forces driving these particles outward are unknown, but they also might contribute to the slower than normal average rate of myosin VI-catalyzed movement toward the cell observed in the absence of these agents.
It is unclear whether the observed movement of GIPC1-myosin VI complexes along cellular extensions toward the cell body serves to regulate GLUT1 distribution in these structures. This movement, however, also could serve to retrieve additional soluble or integral membrane proteins from the distal tips of these processes and return them to central cellular locations for signaling, processing, or degradative purposes. This process would be similar to the retrograde retrieval of vesicles from the distal portions of axonal processes in neurons. In this regard, Farquhar's group has noted the presence of GIPC1 and TrkA in retrograde vesicles within the processes of differentiated PC12 cells (Lou et al., 2001).
Interestingly, the tips of several cellular extensions in Figure 8, B and C, seem to be in direct contact with adjacent cells. More specialized filopodia-like projections have recently been reported to be involved in the formation of cell-cell communication “tunnels” termed “tunneling nanotubes” in which actin-dependent particle transport between cells has been demonstrated (Rustom et al., 2004). If the direction of transport were toward the minus end of the actin filament contained within the nanotube connection, GIPC1-myosin VI complexes conceivably could participate in such transport processes between cells joined by nanotube structures. This seems possible, because we have noted in this study the movement of GIPC1-myosin VI complexes within such a structure joining two 293 cells.
A final point to be noted about the potential cellular function of GIPC1 is that the altered expression of GIPC1 has been shown to alter a variety of signal transduction events (Blobe et al., 2001; Booth et al., 2002; Hu et al., 2003; Garzon et al., 2004). The ability of GIPC1 to form homodimers (Bunn et al., 1999; Gao et al., 2000) and heterodimers with syntenin (Gao et al., 2000) or with other yet unidentified binding partners reinforce the fact that GIPC1 also may function to cluster proteins or otherwise participate in protein complex assembly required for structural or signal transduction purposes. These roles may or may not require interactions with myosin VI for proper localization or for anchoring cargo proteins to cytoskeletal structures. It will be interesting to ascertain during the ensuing years which signaling and trafficking roles are fulfilled by GIPC1 and its homologues.
Supplementary Material
Acknowledgments
We express our appreciation to Dr. Tama Hasson (University of California, San Diego, CA) for helpful discussions provided during the early stages of this study and to Jeanne LeBlanc for technical assistance. This work was supported by grant-in-aids from the American Diabetes Association and the Feist-Weiller Cancer Center, Louisiana State University Health Sciences Center, and also in part by the American Heart Association Southern Affiliate Grant GS-14, National Institutes of Health Grant DK-42647, and Department of Defense award PC030680.
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E04-11-0978) on June 22, 2005.
Abbreviations used: ACT, Gal4-activation domain; CHO, Chinese hamster ovary; DBD, Gal4-DNA binding domain; EEA1, early endosomes antigen 1; GLUT1CBP, GLUT1 C-terminal binding protein; GST, glutathione S-transferase; KIF-1B, kinesin superfamily protein 1B; MDCK, Madin-Darby canine kidney; RITC, rhodamine isothiocyanate.
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
References
- Altman, D., Sweeney, H. L., and Spudich, J. A. (2004). The mechanism of myosin VI translocation and its load-induced anchoring. Cell 116, 737-749. [DOI] [PubMed] [Google Scholar]
- Aschenbrenner, L., Lee, T., and Hasson, T. (2003). Myo6 facilitates the translocation of endocytic vesicles from cell peripheries. Mol. Biol. Cell 14, 2728-2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aschenbrenner, L., Naccache, S. N., and Hasson, T. (2004). Uncoated endocytic vesicles require the unconventional myosin, Myo6, for rapid transport through actin barriers. Mol. Biol. Cell 15, 2253-2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awan, A., Lucic, M. R., Shaw, D. M., Sheppard, F., Westwater, C., Lyons, S. A., and Stern, P. L. (2002). 5T4 interacts with TIP-2/GIPC, a PDZ protein, with implications for metastasis. Biochem. Biophys. Res. Commun. 290, 1030-1036. [DOI] [PubMed] [Google Scholar]
- Blobe, G. C., Liu, X., Fang, S. J., How, T., and Lodish, H. F. (2001). A novel mechanism for regulating transforming growth factor β (TGF-β) signaling. functional modulation of type III TGF-β receptor expression through interaction with the PDZ domain protein, GIPC. J. Biol. Chem. 276, 39608-39617. [DOI] [PubMed] [Google Scholar]
- Bohlson, S. S., Zhang, M., Ortiz, C. E., and Tenner, A. J. (2004). CD93 interacts with the PDZ domain-containing adaptor protein GIPC: implications in the modulation of phagocytosis. J. Leukoc. Biol. 77, 80-89. [DOI] [PubMed] [Google Scholar]
- Booth, R. A., Cummings, C., Tiberi, M., and Liu, X. J. (2002). GIPC participates in G protein signaling downstream of insulin-like growth factor 1 receptor. J. Biol. Chem. 277, 6719-6725. [DOI] [PubMed] [Google Scholar]
- Bunn, R., Jensen, M., and Reed, B. (1999). Protein interactions with the glucose transporter binding protein GLUT1CBP that provide a link between GLUT1 and the cytoskeleton. Mol. Biol. Cell 10, 819-832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunn, R. C. (1999). Identification of Domains within Facilitative Glucose Transporter GLUT1 Which Are Critical for Activation and Cytoskeletal Association of the Transporter. Ph.D. Thesis. Shreveport, LA: Louisiana State University.
- Buss, F., Arden, S. D., Lindsay, M., Luzio, J. P., and Kendrick-Jones, J. (2001). Myosin VI isoform localized to clathrin-coated vesicles with a role in clathrin-mediated endocytosis. EMBO J. 20, 3676-3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buss, F., Luzio, J. P., and Kendrick-Jones, J. (2002). Myosin VI, an actin motor for membrane traffic and cell migration. Traffic 3, 851-858. [DOI] [PubMed] [Google Scholar]
- Cai, H., and Reed, R. R. (1999). Cloning and characterization of neuropilin-1-interacting protein: a PSD-95/Dlg/ZO-1 domain-containing protein that interacts with the cytoplasmic domain of neuropilin-1. J. Neurosci. 19, 6519-6527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dance, A. L., Miller, M., Seragaki, S., Aryal, P., White, B., Aschenbrenner, L., and Hasson, T. (2004). Regulation of Myosin-VI Targeting to Endocytic Compartments. Traffic 5, 798-813. [DOI] [PubMed] [Google Scholar]
- Dauterive, R., Laroux, S., Bunn, R. C., Chaisson, A., Sanson, T., and Reed, B. C. (1996). C-Terminal mutations that alter the turnover number for 3-O-methylglucose transport by GLUT1 and GLUT4. J. Biol. Chem. 271, 11414-11421. [DOI] [PubMed] [Google Scholar]
- DeVries, L., Lou, X. J., Zhao, G., Zheng, B., and Farquhar, M. G. (1998). GIPC, a PDZ domain containing protein, interacts specifically with the C terminus of RGS-GAIP. Proc. Natl. Acad. Sci. USA 95, 12340-12345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, Y., Li, M., Chen, W., and Simons, M. (2000). Synectin, syndecan-4 cytoplasmic domain binding PDZ protein, inhibits cell migration. J. Cell. Physiol. 184, 373-379. [DOI] [PubMed] [Google Scholar]
- Garzon, J., Rodriguez-Munoz, M., Lopez-Fando, A., Garcia-Espana, A., and Sanchez-Blazquez, P. (2004). RGSZ1 and GAIP regulate micro- but not delta-opioid receptors in mouse CNS: role in tachyphylaxis and acute tolerance. Neuropsychopharmacology 3, 3. [DOI] [PubMed] [Google Scholar]
- Gotthardt, M., Trommsdorff, M., Nevitt, M. F., Shelton, J., Richardson, J. A., Stockinger, W., Nimpf, J., and Herz, J. (2000). Interactions of the low density lipoprotein receptor gene family with cytosolic adaptor and scaffold proteins suggest diverse biological functions in cellular communication and signal transduction. J. Biol. Chem. 275, 25616-25624. [DOI] [PubMed] [Google Scholar]
- Harris, B. Z., and Lim, W. A. (2001). Mechanism and role of PDZ domains in signaling complex assembly. J. Cell Sci. 114, 3219-3231. [DOI] [PubMed] [Google Scholar]
- Hasson, T. (2003). Myosin VI: two distinct roles in endocytosis. J. Cell Sci. 116, 3453-3461. [DOI] [PubMed] [Google Scholar]
- Hasson, T., and Mooseker, M. S. (1994). Porcine myosin-VI: characterization of a new mammalian unconventional myosin. J. Cell Biol. 127, 425-440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirakawa, T., Galet, C., Kishi, M., and Ascoli, M. (2003). GIPC binds to the human lutropin receptor (LHR) through an unusual PDZ domain binding motif and it regulates the sorting of the internalized human choriogonadotropin (hCG) and the density of cell surface LHR. J. Biol. Chem. 278, 49348-49357. [DOI] [PubMed] [Google Scholar]
- Hu, L. A., Chen, W., Martin, N. P., Whalen, E. J., Premont, R., and Lefkowitz, R. J. (2003). GIPC interacts with the β 1-adrenergic receptor and regulates β 1-adrenergic receptor mediated ERK activation. J. Biol. Chem. 278, 26295-26301. [DOI] [PubMed] [Google Scholar]
- Inukai, K., Shewan, A. M., Pascoe, W. S., Katayama, S., James, D. E., and Oka, Y. (2004). Carboxy terminus of glucose transporter 3 contains an apical membrane targeting domain. Mol. Endocrinol. 18, 339-349. [DOI] [PubMed] [Google Scholar]
- Iodice, L., Sarnataro, S., and Bonatti, S. (2001). The carboxyl-terminal valine is required for transport of glycoprotein CD8α from the endoplasmic reticulum to the intermediate compartment. J. Biol. Chem. 276, 28920-28926. [DOI] [PubMed] [Google Scholar]
- James, P., Halladay, J., and Craig, E. A. (1996). Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144, 1425-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeanneteau, F., Diaz, J., Sokoloff, P., and Griffon, N. (2003). Interactions of GIPC with dopamine D2, D3 but not D4 receptors define a novel mode of regulation of G protein-coupled receptors. Mol. Biol. Cell. 15, 696-705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirikoshi, H., and Katoh, M. (2002a). Expression of human GIPC1 in normal tissues, cancer cell lines, and primary tumors. Int. J. Mol. Med. 9, 509-513. [PubMed] [Google Scholar]
- Kirikoshi, H., and Katoh, M. (2002b). Up-regulation of GIPC2 in human gastric cancer. Int. J. Oncol. 20, 1183-1187. [PubMed] [Google Scholar]
- Litman, P., Amieva, M. R., and Furthmayr, H. (2000). Imaging of dynamic changes of the actin cytoskeleton in microextensions of live NIH3T3 cells with a GFP fusion of the F-actin binding domain of moesin. BMC Cell Biol. 1, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, T. F., Kandala, G., and Setaluri, V. (2001). PDZ-domain protein GIPC interacts with the cytoplasmic tail of melanosomal membrane protein gp75 (tyrosinase related protein-1). J. Biol. Chem. 276, 35768-35777. [DOI] [PubMed] [Google Scholar]
- Lou, X., McQuistan, T., Orlando, R. A., and Farquhar, M. G. (2002). GAIP, GIPC and G{α}i3 are concentrated in endocytic compartments of proximal tubule cells: putative role in regulating Megalin's function. J. Am. Soc. Nephrol. 13, 918-927. [DOI] [PubMed] [Google Scholar]
- Lou, X., Yano, H., Lee, F., Chao, M. V., and Farquhar, M. G. (2001). GIPC and GAIP form a complex with TrkA: a putative link between G protein and receptor tyrosine kinase pathways. Mol. Biol. Cell 12, 615-627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascoe, W. S., Inukai, K., Oka, Y., Slot, J. W., and James, D. E. (1996). Differential targeting of facilitative glucose transporters in polarized epithelial cells. Am. J. Physiol. 40, C547-C554. [DOI] [PubMed] [Google Scholar]
- Reed, B. C., Shade, D., Alperovich, F., and Vang, M. (1990). 3T3-L1 adipocyte glucose transporter (HepG2 Class): sequence and regulation of protein and mRNA expression by insulin, differentiation, and glucose starvation. Arch. Biochem. Biophys. 279, 261-274. [DOI] [PubMed] [Google Scholar]
- Rousset, R., Fabre, S., Desbois, C., Bantignies, F., and Jalinot, P. (1998). The C-terminus of the HTLV-1 Tax oncoprotein mediates interaction with the PDZ domain of cellular proteins. Oncogene 16, 643-654. [DOI] [PubMed] [Google Scholar]
- Rustom, A., Saffrich, R., Markovic, I., Walther, P., and Gerdes, H.-H. (2004). Nanotubular highways for intercellular organelle transport. Science 303, 1007-1010. [DOI] [PubMed] [Google Scholar]
- Saitoh, T., Mine, T., and Katoh, M. (2002). Molecular cloning and characterization of human GIPC3, a novel gene homologous to human GIPC1 and GIPC2. Int. J. Oncol. 20, 577-582. [PubMed] [Google Scholar]
- Svitkina, T. M., Bulanova, E. A., Chaga, O. Y., Vignjevic, D. M., Kojima, S.-I., Vasiliev, J. M., and Borisy, G. G. (2003). Mechanism of filopodia initiation by reorganization of a dendritic network. J. Cell Biol. 160, 409-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tani, T. T., and Mercurio, A. M. (2001). PDZ interaction sites in integrin α subunits. TIP-2/GIPC binds to a type I recognition sequence in α6A/α5 and a novel sequence in α6B. J. Biol. Chem. 276, 36535-36542. [DOI] [PubMed] [Google Scholar]
- Tochio, H., Zhang, Q., Mandal, P., Li, M., and Zhang, M. (1999). Solution structure of the extended neuronal nitric oxide synthase PDZ domain complexed with an associated peptide. Nat. Struct. Biol. 6, 417-421. [DOI] [PubMed] [Google Scholar]
- Wang, L. H., Kalb, R. G., and Strittmatter, S. M. (1999). A PDZ protein regulates the distribution of the transmembrane semaphorin, M-SemF. J. Biol. Chem. 274, 14137-14146. [DOI] [PubMed] [Google Scholar]
- Wells, A. L., Lin, A. W., Chen, L.-Q., Safer, D., Cain, S. M., Hasson, T., Carragher, B. O., Milligan, R. A., and Sweeney, H. L. (1999). Myosin VI is an actin-based motor that moves backwards. Nature 401, 505-508. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.