Abstract
Myosin II filament assembly in Dictyostelium discoideum is regulated via phosphorylation of residues located in the carboxyl-terminal portion of the myosin II heavy chain (MHC) tail. A series of novel protein kinases in this system are capable of phosphorylating these residues in vitro, driving filament disassembly. Previous studies have demonstrated that at least three of these kinases (MHCK A, MHCK B, and MHCK C) display differential localization patterns in living cells. We have created a collection of single, double, and triple gene knockout cell lines for this family of kinases. Analysis of these lines reveals that three MHC kinases appear to represent the majority of cellular activity capable of driving myosin II filament disassembly, and reveals that cytokinesis defects increase with the number of kinases disrupted. Using biochemical fractionation of cytoskeletons and in vivo measurements via fluorescence recovery after photobleaching (FRAP), we find that myosin II overassembly increases incrementally in the mutants, with the MHCK A-/B-/C- triple mutant showing severe myosin II overassembly. These studies suggest that the full complement of MHC kinases that significantly contribute to growth phase and cytokinesis myosin II disassembly in this organism has now been identified.
INTRODUCTION
Myosin II plays fundamental roles in a variety of cellular contractile processes, ranging from cytokinesis to cell migration and developmental morphogenesis (Ridley et al., 2003; Baumann, 2004;Van Haastert and Devreotes, 2004). A critical feature of nonmuscle myosin II isoforms is that cellular function requires the assembly of this motor protein into bipolar filament structures that can interact with cortical actin arrays. Filament assembly in nonmuscle cells is dynamic and subject to both spatial and temporal regulation. In mammalian nonmuscle cells, phosphorylation of the myosin II regulatory light chain (RLC) is a widely cited model for assembly regulation, with RLC phosphorylation favoring filament assembly (Scholey et al., 1980). However, there is also strong experimental support for other models of mammalian nonmuscle myosin II assembly control, including evidence for monomer sequestration by the S100 protein metastasin 1 (Li et al., 2003), and biochemical evidence that MHC phosphorylation may modulate filament assembly (Murakami et al., 1998; Murakami et al., 2000). Definitive in vivo studies to distinguish relative contributions of each mechanism have as yet not been performed.
The simple amoeba Dictyostelium discoideum contains a single myosin II heavy chain gene, which serves cellular roles that are conserved with those of myosin II in other organisms. Biochemical and cellular studies in this system have established that myosin II filament assembly in vivo involves a dynamic equilibrium between a large pool of disassembled molecules and a pool of assembled filaments that associate with the cortical cytoskeleton. This equilibrium appears regulated by a variety of events ranging from chemoattractant receptor stimulation, to osmotic stress, to mechanical compression (for reviews see de la Roche et al., 2002a; Yumura and Uyeda, 2003). Filament assembly is regulated via phosphorylation of sites at the tip of the MHC tail. Three threonine residues were mapped biochemically (Vaillancourt et al., 1988; Luck-Vielmetter et al., 1990), and mutagenesis of these MHC residues to nonphosphorylatable alanines (3XALA MHC) produces a myosin II molecule that displays severe overassembly into the cytoskeleton, as determined by cytoskeletal fractionation and immunofluorescence (Egelhoff et al., 1993) and as measured in vivo using fluorescence recovery after photobleaching (FRAP) with GFP-myosin II fusion proteins (Yumura, 2001). Cells expressing 3XALA myosin II also display defects in chemotactic migration. Although these 3XALA myosin cells are capable of detecting and responding to chemoattractant gradients, during migration they display poor polarization, frequent formation of inappropriately oriented pseudopodia, and a high frequency of turns in the wrong direction (Stites et al., 1998). They take longer time to initiate new extensions when locally stimulated with a chemoattractant (Yumura and Uyeda, 1997). The 3XALA myosin cells furthermore display defects in chemoattractant-stimulated F-actin assembly dynamics (Heid et al., 2004), indicating that actin and myosin II dynamics during chemotaxis are directly integrated by unknown mechanisms.
Chemotaxis of Dictyostelium amoebae in natural waves of cAMP during developing involves a reiterative series of behaviors, with cells displaying different behaviors in each zone of the natural chemotactic wave (for review see Soll et al., 2002). Although these complex behaviors are poorly understood, a number of upstream signaling pathways have been implicated in this cyclical regulation of polarization and migration, including protein kinase A and internal cAMP signaling (Wessels et al., 2000), Rac GTPase signaling (Park et al., 2004), Ras signaling (Tuxworth et al., 1997; Lee et al., 1999; Wessels et al., 2004), cGMP signaling (Bosgraaf et al., 2002; Goldberg et al., 2002), and phosphatidylinositol 3,4,5-trisphosphate signaling (Iijima et al., 2004). Although myosin II is established as having critical roles in chemotactic behavior, the pathways that link receptor activation to myosin II assembly and subcellular localization are not understood.
Myosin II also plays fundamental conserved roles in cytokinesis. Gene targeting studies in Dictyostelium have revealed complex roles for myosin II in cleavage furrow dynamics, with this motor protein being essential for contractile ring function and furrowing in suspension (De Lozanne and Spudich, 1987; Manstein et al., 1989). When cells divide while attached to an adherent surface, however, myosin II has been found to be less critical, with MHC null mutants displaying an ability to complete mitotically linked cell furrowing, albeit with frequent delays during furrowing, reduced efficiency, and frequent asymmetrical partitioning into daughter cells (Neujahr et al., 1997; Zang et al., 1997; Weber et al., 2000; Uyeda and Nagasaki, 2004). Several signaling pathways, particularly involving low-molecular-weight GTPases, have been implicated in regulating cytokinesis in this system, but as with chemotaxis, the pathways that regulate myosin II assembly and localization to the contractile ring are not understood (For review see Uyeda et al., 2004).
A critical step in understanding the integration of upstream signaling in both these settings is to establish the full set of enzymes that directly regulate myosin II assembly. Earlier mutagenesis studies with myosin II have established that the three threonine residues in the tail region mediate filament assembly, with phosphorylation driving disassembly and dephosphorylation driving assembly. Little attention has been directed to understanding MHC phosphatases in this system, but studies to date suggest that the major identified MHC phosphatase (a PP2A complex) is cytosolic in distribution (Murphy et al., 1999) and is not directly regulated by several common second messengers such as cyclical nucleotides or calcium/calmodulin (Murphy and Egelhoff, 1999).
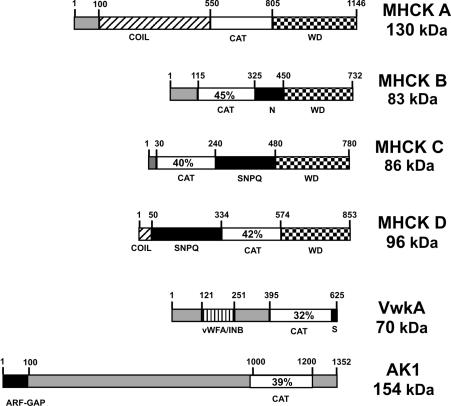
In contrast, Dictyostelium cells appear to have a series of MHC kinases (MHCKs), which drive myosin II filament disassembly both in vitro and in vivo, and the enzymes characterized to date display differing cellular localization patterns and distinct regulation in vitro (Steimle et al., 2001b; Liang et al., 2002; Nagasaki et al., 2002; Rico and Egelhoff, 2003). Single gene disruptions of three of these enzymes have been published, including MHCK A (Kolman et al., 1996), MHCK B (Rico and Egelhoff, 2003), and MHCK C (Nagasaki et al., 2002), and in each case disruption phenotypes supported the model for a role in the in vivo regulation of myosin II assembly. Molecular analysis revealed these enzymes to be founding members of a novel family of eukaryotic protein kinases (Côté et al., 1997; de la Roche et al., 2002a), now recognized throughout the metazoan phyla and commonly referred to as “alpha kinases” (Drennan and Ryazanov, 2004). Intriguingly, the largely completed Dictyostelium genome project contains three other protein kinase genes that encode alpha kinases in addition to the three validated MHCKs. The fourth alpha kinase displays flanking domain organization highly similar to MHCK A, B, and C, with possible coiled-coil segments and a carboxyl-terminal WD repeat domain (Figure 1), and we provisionally named this gene mhkD and its encoded protein MHCK D, based upon this similarity; for further information on MHCK domain organization see (de la Roche et al., 2002a). The fifth member of the Dictyostelium alpha kinase family displays flanking domain organization completely distinct from MHCK A, B, C, and D, with no WD repeat domains or coiled-coil character, and bearing an amino-terminal segment displaying motif similar to the conserved beta-integrin (IntB) and von Willibrand factor A (vWFA) fold (Figure 1). We have named this fifth alpha kinase VwkA. Biochemical analysis and cellular studies indicate that VwkA does not function as a direct MHC kinase (Betapudi et al., 2005). The sixth alpha kinase gene has been provisionally named AK1 (alpha kinase 1) during genome annotation. The deduced protein sequence contains an Arf-GAP motif at the N-terminus and an alpha kinase domain at its C-terminus, but to our knowledge no functional studies have been performed with the gene or protein.
Figure 1.
Domain organization of Dictyostelium complement of alpha kinases. Amino acid residue positions indicated above each sequence. COIL refers to segments with strong predicted or experimentally demonstrated coiled-coil character; CAT, the conserved alpha kinase catalytic domain of each enzyme, with percent identity to MHCK A indicated for each family member; WD, the WD-repeat domain, implicated in targeting MHCKs to the myosin II substrate; N, a polyasparagine-rich segment; SNPQ, a low complexity segment of MHCK C that is rich in serine, asparagines, proline, and glutamine; S, a polyserine segment; vWFA/INB, the vWFA motif/Integrin B motif domain of VwkA.
Another structurally unrelated enzyme, named “MHC-protein kinase C” in early studies, was proposed to affect myosin II assembly via direct MHC phosphorylation (Abu-Elneel et al., 1996; Dembinsky et al., 1997). More recent studies (de la Roche et al., 2002b), as well as the completed Dictyostelium genome project, have confirmed that this gene product is in fact a highly conserved diacylglycerol (DG) kinase and is unlikely to function as a protein kinase in vivo. This protein has been renamed DgkA based upon that analysis (de la Roche et al., 2002b).
Analysis to date thus suggests that MHCK A, MHCK B, MHCK C, and possibly MHCK D represent the likely complement of MHC kinases among this family. Target sites in the MHC tail region were biochemically mapped for MHCK A to residues 1823, 1833, and 2029 of MHC in earlier studies (Vaillancourt et al., 1988; Luck-Vielmetter et al., 1990), and it was subsequently shown that mutation of these three sites to alanines (the 3XALA mutation) not only caused myosin II overassembly in vivo (Egelhoff et al., 1993), but also rendered the 3XALA myosin II resistant to disassembly in vivo when MHCK A was overexpressed (Kolman and Egelhoff, 1997). This resistance to disassembly confirmed the biochemically mapped target sites to be the relevant target sites in vivo as well. Although target sites for MHCK B and MHCK C have never been mapped via in vitro phosphopeptide mapping, MHCK B and MHCK C robustly phosphorylate MHC in vitro to drive filament disassembly, and 3XALA myosin II is resistant to disassembly in phosphorylation tests with MHCK B or MHCK C (Liang et al., 2002;Rico and Egelhoff, 2003; and unpublished data), supporting the idea that MHCK B and MHCK C probably phosphorylate the same three target sites as MHCK A. Biochemical studies addressing site preference or target site order of phosphorylation by the individual kinases have not been performed to date, but the simplest hypothesis consistent with available evidence suggest that the three target sites likely contribute to assembly control in a roughly additive manner, although it is possible that some sites contribute to a greater degree than others. For example, one published study analyzing 1X, 2X, and 3X aspartic acid mutations in Dictyostelium myosin II (created to mimic phosphorylation events) reported a possibly greater role for the 1823 site in controlling myosin II assembly levels in vivo (Nock et al., 2000).
To test the relative importance of each of these candidate MHC kinases in the in vivo regulation of myosin II assembly and to assess whether other as yet unidentified kinases might exist, we have generated and evaluated an array of single, double, and triple gene disruption cell lines of MHCK A, MHCK B, MHCK C, and MHCK D. Biochemical fractionation of cytoskeletons and analysis of the in vivo dynamics of a GFP-myosin II reporter have been used to assess the effects of these multiple gene disruptions. The mhck A-/B-/C- triple gene disruption line displays levels of myosin II overassembly that closely mimic the severe overassembly of 3XALA myosin II, suggesting that these three enzymes may represent the full complement of MHC kinases that have critical roles in assembly control during interphase and cytokinesis. Analysis of these cell lines further reveals previously unrecognized defects in cytokinesis in suspension culture in the mutants that display myosin II overassembly, indicating that dynamic control of myosin II filament disassembly is critical for efficient contractile ring function.
MATERIALS AND METHODS
Cell Culture and Gene Disruption and GFP Constructs
Cell lines were maintained in plastic Petri dishes in HL5 medium (Sussman, 1987) supplemented with penicillin/streptomycin (Life Technologies, Rockville, MD) for the parental line Ax2. Media for blasticidin-based gene disruption cell lines was also supplemented with 2 μg/ml blasticidin (Calbiochem, La Jolla, CA). The mhkA- cell line (CW0371) was generated via disruption of the mhkA locus in the cell line JH10 using a Thy1 marker, as described previously (Kolman et al., 1996). Single disruptions of mhkB (line CW0166) and mhkC (line CW0403) were generated with a blasticidin-resistance disruption cassette. The double mhkA-/mhkB- line (CW0394) was generated via mhkB-blasticidin cassette targeting into the mhkA- line listed above. The double mhkA-/mhkC- line (CW0403) was generated via mhkC-blasticidin cassette targeting in the mhkA- cell line above. The triple MHCK gene disruption line mhkA-/mhkB-/mhkC- (CW0431) was generated by cotransfecting a nonselected mhkB gene disruption cassette together with a selected mhkC disruption cassette, with resultant blasticidin-resistant colonies screen via PCR for the simultaneous disruption of both genes. The full methodology and technical details of this coselection method were described recently in detail (Betapudi et al., 2004).
Disruption of the mhkD gene was performed as follows. A short DNA sequence of 466 base pairs length was PCR amplified using MHCKE934 (5′-GCAGGTACCGGTAGAATGCCATCAACTGGC-3′) and MKLA1386 (5′-GCATCTAGACAATTGATATGCATTTCTAAGTGCACC-3′) from the N-terminus of mhkD gene. The purified DNA fragment was cloned as KpnI and XbaI fragment in pBsr-Nsi plasmid vector (Betapudi et al., 2005) to generate pBsr-Nsi-5′-MHCKD vector. Similarly, another DNA fragment of 526 base pairs length was PCR amplified using MHCKE2236 (5′-CAGATCCAAGCTTCAGCATCTGCTGATGGTTACG-3′) and MHCKE2762 (5′-ACTAGAGGCGCCCCAAACATACGTAATGATGTAATTGG-3′) primers from the C terminus of mhkD gene. The purified fragment was digested with HindIII and SfoI and cloned in pBsr-Nsi-5′-MHCKD vector to create pBsr-Nsi-MHCKD-KO plasmid vector. From this vector DNA, the entire knock-out cassette carrying blasticidin resistance cartridge in the middle was PCR amplified using MHCKD-5′ (5′-GGTAGAATGCCATCAACTGGC-3′) and MHCKD-3′ (5′-CCAAACATACGTAATGATGTAATTGG-3′) primers lacking cloning sites.
The amplified PCR DNA was purified and used for electroporation of Ax2 cells as described earlier (Betapudi et al., 2004). Transformants were selected in the presence of 3 μg/ml blasticidin in the growth medium. Individual clones were isolated, amplified, and subjected to further analyses. Genomic DNA isolated from each clone was analyzed by PCR to confirm presence of the knockout cassette and absence of a wild-type copy of the gene, using a general strategy described recently (Betapudi et al., 2004).
Previously reported cell lines expressing the 3XALA mutant MHC gene were generated via either G418 selection or hygromycin selection (Egelhoff et al., 1993). For the work reported here, we have generated a new 3XALA myosin cell line that does not carry an antibiotic resistance marker. This new cell line, designated CW0401, was generated as follows. A plasmid was constructed that carries the previously described 3XALA MHC gene fused to a Dictyostelium actin 15 promoter (Egelhoff et al., 1993), in an Escherichia coli plasmid carrying an ampicillin selection marker. This plasmid, pMHC3XALA, does not carry a Dictyostelium selectable marker. The MHC null cell line HS1 (Manstein et al., 1989) was transfected with this plasmid, and direct functional selection was used to identify clonal cell lines expressing the 3XALA MHC construct. Selection involved initial growth in suspension culture, followed by plating on live bacterial lawns to identify clonal cell lines capable of multicellular development, as has been described previously with the wild-type MHC gene (Egelhoff et al., 1990).
The plasmid constructs for the GFP fusions used in the current work have been described previously for MHCK A (Steimle et al., 2001b), for MHCK B and MHCK C (Liang et al., 2002), and for myosin II (Moores et al., 1996).
Suspension Growth Analysis
Cells were also cultured in suspension with reciprocal shaker at 150 rpm. Cell counting was performed by a hemacytometer. To examine the number of nuclei, an aliquot of cell suspension was centrifuged, and the centrifuged cells were fixed with 2.5% formaldehyde in 15 mM phosphate buffer (pH 6.2). Two microliters of fixed cells were mixed with 1 μg/ml DAPI solution containing 50% glycerol and phosphate buffer on a slide glass. A coverglass was overlaid on the cells and mildly squashed with a forcep. Fluorescence microscopy of DAPI staining was performed by Nikon TM300 with UV fluorescent filter sets (Melville, NY).
Cytoskeletal Fractionation
Assembled myosin II filaments cofractionate with Triton X-100-resistant cytoskeletal ghosts when Dictyostelium cells are lysed under mild conditions (Spudich, 1987). Triton-insoluble cytoskeletons were isolated as described previously (Kolman et al., 1996). Briefly, cells (typically 2 × 106) were washed in starvation buffer (20 mM MES, pH 6.8, 0.2 mM CaCl2, 2 mM MgCl2), then resuspended in 150 μl of ice cold buffer A (0.1 M MES, pH 6.8, 2.5 mM EGTA, 5 mM MgCl2, 0.5 mM ATP), and chilled on ice. Samples were then mixed with 150 μl of buffer B (same as buffer A, but also contains 1% Triton X-100, and protease cocktail mix PIC I and PIC II described in Steimle et al., 2001a). Samples were vortexed at top speed for 5 s and then subjected to centrifugation in a microfuge at top speed (∼10,000 × g) at 4°C for 2 min. Pellets (cytoskeletal fractions were directly suspend in 2× SDS-PAGE sample buffer and heated at 95°C for 60 s. Supernatants (cytosolic fractions) were precipitated with 700 μl acetone with incubation on ice followed by centrifugation (10 min in microfuge) to collect protein. Dried supernatant protein pellets were then suspended in SDS-PAGE sample buffer and heated in same manner. Samples were subjected to SDS-PAGE, and resultant gels were stained with Coomassie Blue, and myosin heavy chain band intensity in each fraction was quantified via densitometry. The percent myosin II in cytoskeleton was calculated for each sample by dividing the value for MHC in the cytoskeletal pellet by the sum of the pellet and supernatant MHC values for that sample.
Photobleaching Analysis
Photobleaching was performed by a confocal laser system (LSM 510, Carl Zeiss, Thornwood, NY) as described previously (Yumura, 2001). Full power of Argon laser (488 nm lines, 25 mW) was applied. Usually five times-iterated illumination was required for >95% photobleaching. Images were acquired at 12 bits depth gray scale. The change of fluorescence intensity at the bleached area was monitored in respect to time after background subtraction. These data were applied to curve-fitting in Sigma Plot software (Jandel, Corte Madera, CA) as described (Yumura, 2001). The time course of recovery was fitted to the equation for a single exponential rise to maximum for a simple enzymatic reaction as shown below.
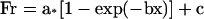 |
In the above equation, a is amplitude of exponential, b is rate constant, x is time (sec), and c is constant. The half time recovery (t1/2) was calculated using the following equation.
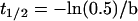 |
RESULTS
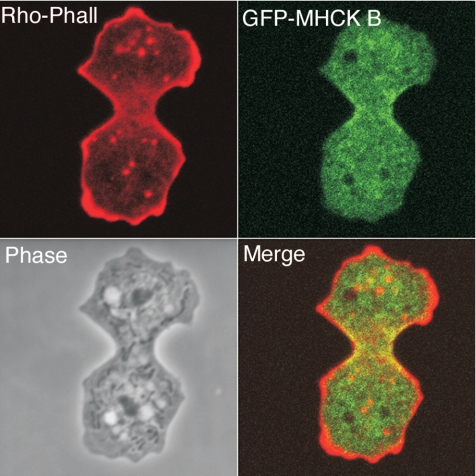
Previous studies have reported MHCK A as displaying anterior localization during migration of Dictyostelium cells and weak polar enrichment during cytokinesis (Steimle et al., 2001b;Liang et al., 2002). GFP-MHCK C fusions, in contrast, have been localized to the posterior of migrating cells and to the contractile ring of dividing cells (Liang et al., 2002; Nagasaki et al., 2002). We have now pursued additional studies addressing possible subcellular localization by MHCK B. Attempts to generate rabbit polyclonal antibodies against MHCK B did not produce a reagent of sufficient sensitivity for immunolocalization (unpublished data). However, GFP-MHCK B was clearly enriched in contractile rings of fixed Dictyostelium cells (Figure 2). With further examination of live cells expressing GFP-MHCK B, contractile ring enrichment could be detected in live cells as well (Figure 3A). This localization contrasts to earlier analysis indicating cytosolic localization (Liang et al., 2002), possibly for two reasons. First, the current work utilized higher magnification imaging, and perhaps most importantly the current live cell imaging was performed with a laser confocal system, in contrast to the earlier analysis that utilized wide field fluorescence. Use of confocal typically reduces the background signal from any cytosolic phase GFP-reporter pool, allowing weak localizations to be more readily observed.
Figure 2.
GFP-MHCK B displays enrichment in the contractile ring. Ax2 cells expressing GFP-MHCK B were fixed and permeabilized and then stained with rhodamine-phalloidin.
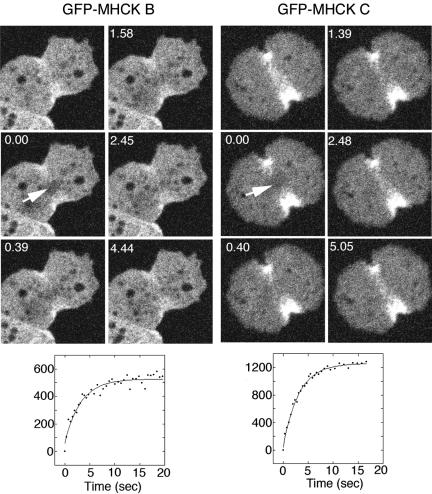
Figure 3.
FRAP analysis reveals a high turnover rate for GFP-MHCK B and C associated with the contractile ring. Photobleaching was performed on the contractile ring of an actively dividing cell (0-s time point). Recovery was monitored in this zone >15 s, revealing a half-life of recovery of 1.72 ± 0.30 s (SE; n = 15), and 2.32 ± 0.25 s (SE; n = 18), respectively
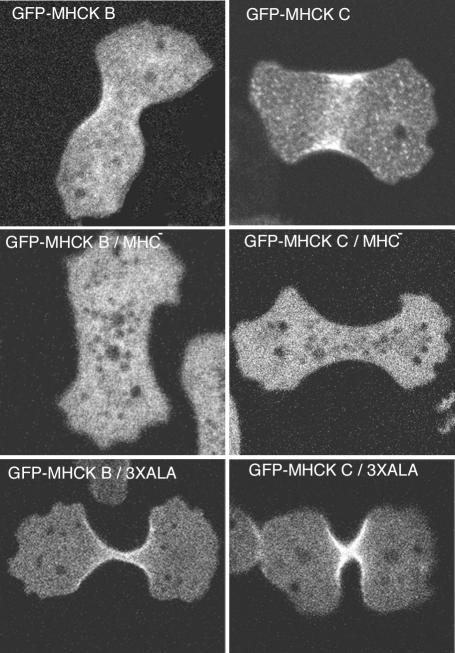
Previous studies using FRAP with cells expressing GFP-myosin II indicated that myosin II rapidly turns over in the contractile ring. However, 3XALA myosin II showed defects in turnover in similar experiments, suggesting that MHC phosphorylation is crucial for rapid turnover of myosin II (Yumura, 2001). To examine possible turnover or dynamics of MHCKs in the contractile ring, FRAP was used to further evaluate the contractile ring localization. Subsequent to photobleaching of the contractile ring zone in live cells, GFP-MHCK B and C signal within the bleached zone recovered rapidly, with a half time of recovery of 1.72 ± 0.30 s (SE; n = 15), and 2.32 ± 0.25 (SE; n = 18), respectively (Figure 3). This rapid recovery indicates that during cytokinesis MHCK B and C are dynamically assembling and disassembling in the contractile ring, apparently associating with a low-affinity binding partner within the ring. The half time of recovery of the MHCKs was shorter than that of myosin II (7.01 ± 0.65 s; Yumura, 2001). We previously demonstrated that GFP-MHCKC expressed in myosin II-null cells does not localize to the cleavage furrows (Nagasaki et al., 2002). Likewise, GFP-MHCKB expressed in myosin II null cells did not localize to the contractile ring, suggesting that the localization of MHCK B and C in the contractile ring depends on myosin II (Figure 4). To investigate further whether MHCK B and C directly bind to phosphorylation sites on myosin II molecules, the GFP constructs were expressed in cells carrying 3XALA myosin II instead of wild-type myosin II. GFP-MHCK B and C still localized at the cleavage furrow in the background of 3XALA myosin II (Figure 4). Therefore, MHCK B and C associate with myosin II molecules in vivo, likely recognizing a target zone on the myosin II tail that is larger than just the phosphorylation sites alone.
Figure 4.
Distribution of MHCK B and C in the contractile ring depends on myosin II. Cells were fixed and observed by confocal fluorescence microscopy. Neither GFP-MHCK B nor C displayed its localization to the cleavage furrow in myosin heavy chain null cells (MHC-). MHCK B and C still localized at the cleavage furrow in the background of 3XALA myosin II, indicating that these MHCKs recognize a target zone of the myosin II tail that is larger than just the phosphorylation site alone.
The localization of GFP-MHCK B, together with earlier studies, supports a model that MHCKs in this system may have both overlapping and unique roles in the spatial control of myosin II filament assembly. To understand the relative contributions of theses MHCKs in the in vivo control of myosin II filament assembly, we have created a series of double and triple MHCK gene knockout mutants.
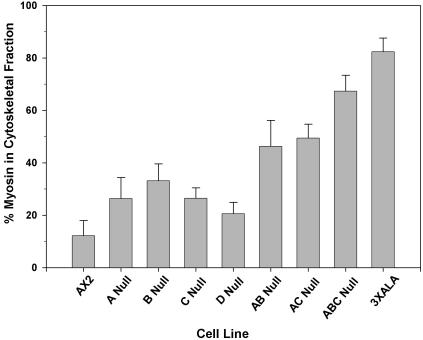
As an initial test of myosin II assembly levels in MHCK null lines, bulk myosin II assembly levels were assessed by isolating Triton X-100-resistant cytoskeletal ghosts from populations of cells growing in Petri dish culture. As has been reported earlier (Kolman et al., 1996), the parental line Ax2 displays ∼10% of total myosin II assembled into the cytoskeletal fraction with the extraction conditions used here (Figure 5). Also in keeping with earlier reports, mhkA null cells and mhkB null cells display partial overassembly of ∼25–35% myosin II into the cytoskeletal fraction. This contrasts to the 3XALA myosin II cell line, which displayed 80–90% myosin II association with the cytoskeleton (Figure 5), as reported for earlier 3XALA cell lines (Egelhoff et al., 1993). Evaluation of mhkC null cells reveals partial overassembly, similar to the single mhkA and mhkB null lines, confirming that MHCK C contributes to myosin II assembly control in vivo. Evaluation of mhkD null cells revealed a slight but more modest increase in filament assembly. Effects of MHCK gene disruption become more dramatic in this assay when multiple knockout lines are evaluated. The mhkA-/mhkB- double knockout, and the mhkA-/mhkC- double knockout both display a significant increase in filament overassembly to ∼50% association with the cytoskeleton. The mhkA-/mhkB-/mhkC- triple knockout displays even more severe overassembly, with ∼70% of the myosin II associating with the cytoskeletal fraction. This assembly level approaches the level of overassembly in the 3XALA myosin cell line, but is still statistically slightly lower, suggesting that another kinase that phosphorylates the target sites (threonines 1829, 1833, and 2029 of MHC) may contribute to a small degree to phosphorylation and subsequent filament disassembly in vivo. However, this result suggests that MHCK A, MHCK B, and MHCK C together represent the majority of MHC kinase activity affecting filament assembly in growth phase cells.
Figure 5.
Assembly of Myosin II into Triton-resistant cytoskeletal fractions. Cells were grown in Petri dishes to near-confluence and then harvested and lysed with Triton X-100. Cytoskeletal ghosts were isolated via centrifugation and SDS-PAGE, and densitometry was used to determine percent of myosin II associated with the cytoskeletal fractions. Bars, SEM; n = 7–9 samples for each cell line.
Although biochemical fractionation provides a useful gauge of myosin II assembly, it is a static measure that does not reveal dynamic or spatially resolved features of assembly control. We therefore performed FRAP analysis, evaluating GFP-myosin II dynamics in parental versus MHCK gene knockout cell lines during both interphase and during cytokinesis. As reported previously, assembled GFP-myosin II displays rapid turnover both in the cortex of interphase Dictyostelium cells and within the contractile ring of dividing cells (t1/2 ∼ 7 s), whereas in both settings GFP-3XALA-myosin II displays dramatically greater stability, with <10% fluorescence recovery occurring within a 70-s image collection period (Yumura, 2001). For the current analysis we transfected a wild-type myosin II GFP fusion (GFP-myosin II) into the single mhkA-, mhkB-, mhkC-, and mhkD- knockout cell lines. In the background of mhkA-, mhkB-, or mhkC- lines, GFP-myosin II displayed slower recovery at the contractile ring than wild-type cells, with half-time of fluorescence recovery values increasing from 7.89 ± 0.86 s for the parental Ax2 to 11.60 ± 0.99, 12.20 ± 0.64, and 21.40 ± 1.17 s, for the three mutants, respectively (see Figure 6 and Table 1). In the mhkD null background, GFP-myosin II in the contractile ring showed a recovery rate indistinguishable from the parental (Table 1).
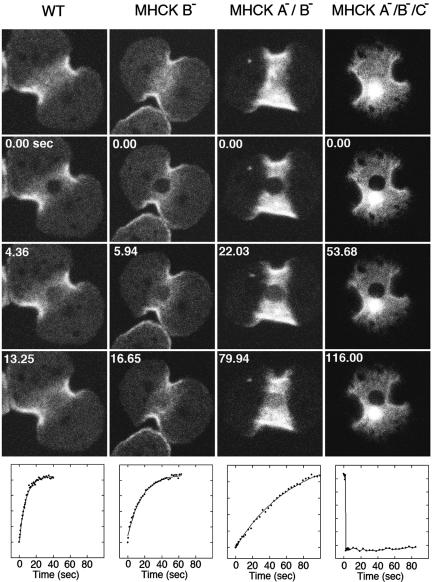
Figure 6.
Photobleaching analysis of GFP-myosin II turnover in the contractile ring in single, double, and triple MHCK gene disruption lines. One example of each class of gene disruption is presented. For each series the top image was collected just before photobleaching, and the next three images beneath that present examples of the fluorescence recover time course (time in sec). Below each series is a graph reflecting fluorescence recovery rate for the bleached spot. The X axis is presented in seconds, and the Y axis is arbitrary units reflecting rate of fluorescence recovery. The solid lines were generated by curve-fitting as described in Materials and Methods. These examples illustrate the decrease in GFP-myosin II turnover, which occurs to an increasing degree as more MHCK genes are disrupted. A large spherical fluorescence in the cleavage furrow in the MHCK A-/B-/C- cell was an aggregation of myosin II due to its overassembly. Aggregates of transfected MHC gene constructs have been observed in several previous reports, particularly in the triple MHCK knockout cells and with transfection of 3XALA MHC constructs (Egelhoff et al., 1993; Nagasaki et al., 2002).
Table 1.
Half-time of fluorescence recovery of cortical GFP myosin II in the contractile rings during cytokinesis, determined via FRAP measurement
| Parental A×2 | 7.01 ± 0.65 (16) |
| MHCK A- | 22.60 ± 1.63 (36) |
| MHCK B- | 11.00 ± 0.77 (31) |
| MHCK C- | 33.10 ± 2.27 (34) |
| MHCK D- | 7.78 ± 0.44 (20) |
| MHCK A-/MHCK B- | 46.30 ± 5.11 (13) |
| MHCK A-/MHCK C- | 38.17 ± 3.04 (41) |
| MHCK A-/MHCK B-/MHCK C- | ND, estimate >>> 2 min |
| 3XALA myosin cells | ND, estimate >>> 2 min |
ND: The half-time of recovery of triple mutant cells and 3XALA myosin cells could not measured because it took very long time or recovery was not observed. We estimate that the half-time of recovery for these cell lines is likely much greater than 2 min, but it is not possible to collect data for a long enough period to quantify this value, because of photodamage of cells or cell movement with longer imaging times. Parentheses show the number of examined cells. There are no significant differences between parental A×2 and mhkD-. Error values represent SE of the mean, defined as the SD divided by the square root of n for each data set.
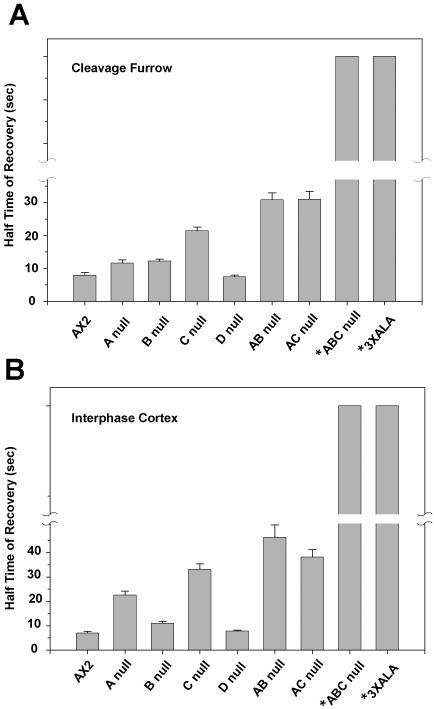
We further assessed GFP-myosin II FRAP behavior in the contractile rings of the double and triple MHCK knockout cell lines. In double knockout mutants (mhkA-/mhkB-and mhkA-/mhkC-), recovery became much slower than single mutants (Figure 6 and Table 1), with recovery values of 30.83 ± 2.09 and 31.00 ± 2.42 s, respectively. In the triple knockout mutants (mhkA-/mhkB-/mhkC-), <10% recovery was observed during imaging periods of 80–100 s (Figure 6 and Table 1), comparable to the extremely slow recovery cited above for GFP-3XALA myosin constructs tested in cell lines wild-type for MHCKs (Yumura, 2001). The extremely slow recovery of fluorescence in this mutant prevented accurate determination of half-life of recovery values. Similar slow recovery was also observed when FRAP was performed on the cortex of mhkA-/mhkB-/mhkC- cells during interphase cells (summarized graphically in Figure 7, and in Table 2). Fluorescence recovery values determined in the cortex of interphase cells for the single and double mutant cell lines followed the same general pattern as the contractile ring values, with single mutants showing partial increases in time of recovery, and double mutants showing greater increases (Figure 7 and Table 2).
Figure 7.
Graphical comparison of fluorescence recovery times for GFP-myosin II assembled into (A) the contractile ring of dividing cells and (B) the cortex of interphase cells. Error bars, SE of mean. See Tables 1 and 2 for n values. Asterisks indicate that recovery values for these samples could not be determined due to very slow turnover. Bars for these samples indicate only that values are large relative to the other samples.
Table 2.
Half-time of fluorescence recovery of cortical GFP myosin II in the cortex during interphase, determined via FRAP measurement
| Parental A×2 | 7.89 ± 0.86 (12) |
| MHCK A- | 11.60 ± 0.99 (28) |
| MHCK B- | 12.20 ± 0.64 (41) |
| MHCK C- | 21.40 ± 1.17 (51) |
| MHCK D- | 7.41 ± 0.53 (25) |
| MHCK A-/MHCK B- | 30.83 ± 2.09 (39) |
| MHCK A-/MHCK C- | 31.00 ± 2.42 (33) |
| MHCK A-/MHCK B-/MHCK C- | ND, estimate ⋙ 2 min |
| 3XALA myosin cells | ND, estimate ⋙ 2 min |
ND: The half-time of recovery of triple mutant cells and 3XALA myosin cells could not measured because it took very long time or recovery was not observed.
This analysis reveals defects in assembly turnover both in the contractile rings of dividing cells and in the cortices of interphase cells, with the degree of impairment roughly proportional to the number of MHCK genes disrupted. These results suggest that MHCK A, MHCK B, and MHCK C constitute the major complement of MHCK activities responsible for controlling myosin II filament disassembly kinetics in Dictyostelium cells in these settings. MHCK D does not seem to contribute to turnover of myosin II in either of these two settings, although the Triton-resistant cytoskeleton data (Figure 5) suggest that it could perhaps play a role in some other structure within interphase cells.
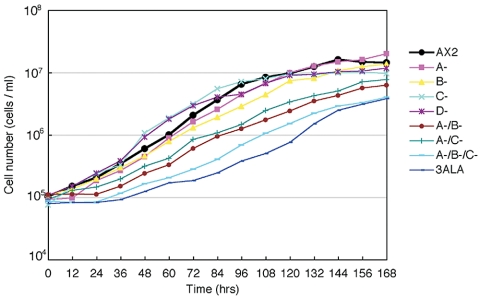
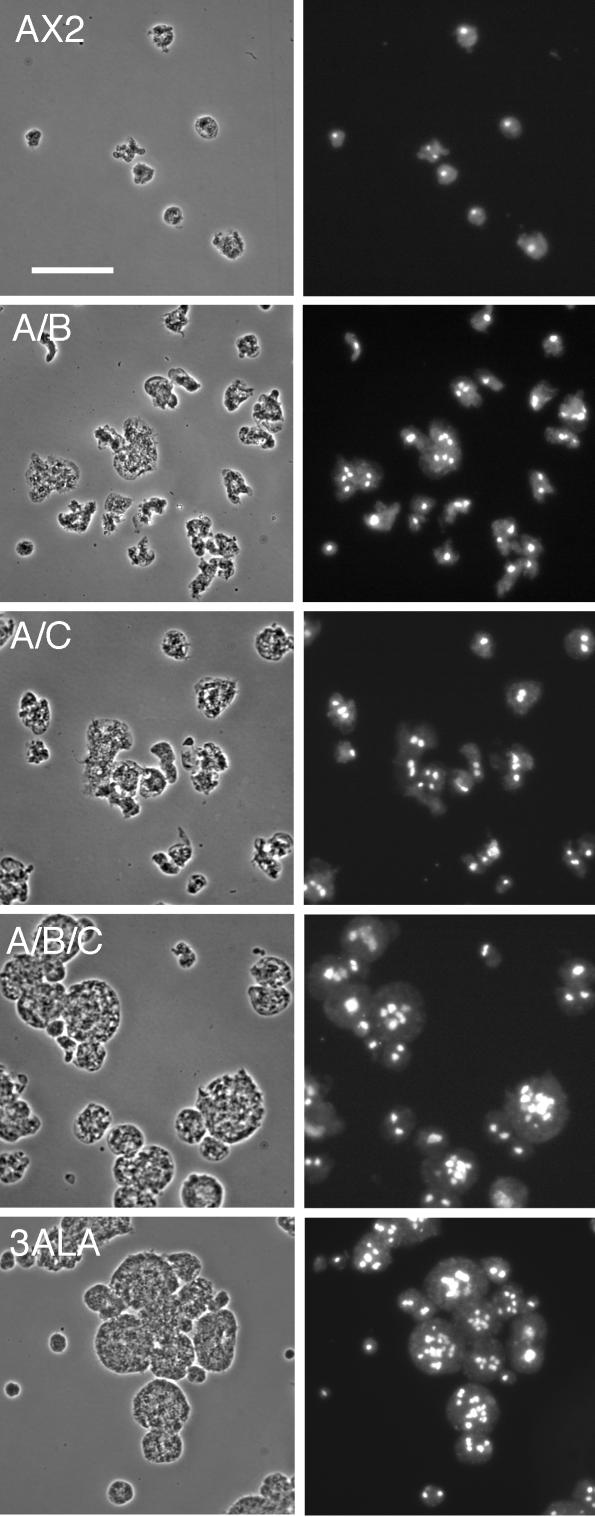
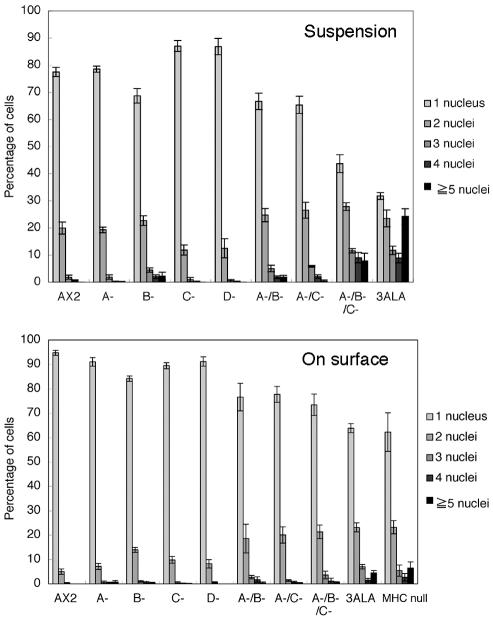
Earlier immunofluorescence studies (Yumura and Uyeda, 1997), GFP imaging studies (Sabry et al., 1997), and FRAP analysis (Yumura, 2001) have demonstrated that 3XALA-myosin II over assembles into the contractile ring. To assess the effects of myosin II overassembly on myosin function during cytokinesis induced either by the 3XALA MHC mutation, or by MHCK gene disruption, we evaluated growth rates and multinucleation in suspension culture. When placed in suspension culture, 3XALA cells displayed a markedly slower growth rate than the parental control Ax2, as did the mhkA-/mhkB-/mhkC- triple knockout line (Figure 8). The two double MHCK knockout cell lines display a similar but more modest reduction in growth in suspension. To evaluate whether this growth defect correlated with incomplete cytokinesis, cells were collected from suspension cultures, fixed, and stained with DAPI. This analysis revealed substantial multinucleation in both 3XALA myosin cells and in the mhkA-/mhkB-/mhkC- triple knockout line, and more modest multinucleation in the two double MHCK knockout cell lines (Figure 9). We next compared degree of multinucleation for all cell lines, with cells collected either from suspension culture or when harvested from attached Petri dish cultures (Figure 10). These analyses revealed that the multinucleation for the double and triple MHCK knockout lines, and for the 3XALA myosin II line, is most severe in suspension culture, similar in severity to the HS1 cell line (MHC null). The four single MHCK gene disruption lines displayed no increase in multinucleation relative to Ax2 in either growth setting. These data demonstrate that regulated myosin II filament disassembly is critical for efficient completion of cytokinesis and further suggest that multiple MHCKs contribute to this process.
Figure 8.
Growth rates in suspension culture for MHCK gene disruption cell lines. Cells collected from Petri dishes were inoculated into shaker flasks at a density of 105 cells/ml and counted daily.
Figure 9.
Multinucleation in suspension culture of MHCK knockout cell lines. Cells were grown in suspension culture for 3 d, fixed, and stained with DAPI to assess degree of multinucleation. Bar, 50 μm.
Figure 10.
Multinucleation in MHCK-deficient cells and 3XALA myosin cells is most severe during suspension culture. Cells were collected either from suspension culture as in previous figure or from Petri dishes and processed as in previous figure and then scored for number of nuclei per cell.
DISCUSSION
The results presented here demonstrate that in growth phase cells, MHCK A, MHCK B, and MHCK C account for the majority of MHC kinase activity responsible for maintaining myosin II in its normal equilibrium between cytoskeletal assembled filaments and the unassembled cytosolic pool. Triton-resistant cytoskeleton analysis suggests that MHCK D may have a small role in assembly control, but a difference is not apparent with the FRAP method of analysis. If MHCK D has a role during growth phase, it is likely in a setting other than basal control of cortical myosin II levels. Further analysis with cells at various stages of development, and focused on specific structures such as phagocytic cups (Muller-Taubenberger et al., 2002) will be of interest to determine whether MHCK D may have roles in other settings. FRAP analysis also demonstrated that myosin II is severely over assembled in the mhkA-/mhkB-/mhkC- triple kinase knockout cell line. However, because of technical limitations on long-term imaging of cells, the FRAP approach cannot resolve whether there is a difference in myosin II assembly behavior in the triple kinase knockout line and the behavior of 3XALA myosin II, bearing mutations in the mapped MHC phosphorylation target sites. It is noteworthy that the Triton method consistently reported slightly milder overassembly in the triple kinase knockout line relative to the 3XALA line. This could reflect a minor assembly control contribution from either MHCK D or perhaps another yet to be identified MHCK.
Another important aspect of assembly control revealed by these studies is that MHCK A, MHCK B, and MHCK C all appear to contribute to assembly control. This is a consistent feature of the Triton-cytoskeleton data, the interphase FRAP analysis, and the FRAP analysis performed on contractile rings. Focusing on cytokinesis, previous studies and the current work establish that MHCK B and MHCK C tend to be enriched in the cleavage furrow, whereas MHCK A displays no furrow enrichment, and in fact displays slight enrichment at the cell poles during cytokinesis (Liang et al., 2002). MHCK B and MHCK C likely enhance contractile ring myosin II filament turnover because they are enriched in the furrow, where they phosphorylate and disassemble filament. But how can MHCK A contribute to contractile ring myosin filament disassembly when it is enriched at cell poles, not in the furrow? We suggest two possible explanations. One possibility is that perhaps elevated myosin II assembly at polar pseudopods in the cell cortex occurs in the mhkA- cells, resulting in greater delivery of filaments via cortical flow (Yumura, 2001), which in some indirect manner alters turnover rates within the ring. A second possible explanation is that the wild-type parental line, cytoplasmic phase amounts of MHCK A, some of which exist in the vicinity of the contractile ring, may be sufficient to enhance disassembly in the furrow. More detailed analysis of MHCK localization and dynamics are needed to resolve these questions, perhaps utilizing further FRAP analysis of kinase behavior and TIRF microscopy approaches.
The analysis presented here reveals previously unrecognized defects in cytokinesis that occur when myosin II over assembles, which occur either when myosin II overassembly is caused directly via mutagenesis of MHC target sites (the 3XALA myosin II mutant), or when myosin II overassembly is induced indirectly (i.e., elimination of the MHCKs that phosphorylate these target sites). Earlier analysis revealed partial myosin II overassembly into the furrow and slightly reduced furrowing rates in 3XALA cells and mhkC null cells (Sabry et al., 1997; Yumura and Uyeda, 1997), but the current analysis demonstrates that persistent myosin II overassembly causes a significant rate of complete cytokinesis failure, leading to multinucleation in suspension culture. One possible explanation for the defect is that over assembled myosin II at the furrow site may physically block terminal membrane fusion events necessary for abscission. Further studies are needed to test this and other possible causes of cytokinesis failure in these cell lines.
The analysis presented here further appears to define the primary complement of MHC kinases that participate in growth-phase cells myosin II assembly control. A critical next step is to begin clarifying the upstream regulation of these enzymes. At the biochemical level, little is known about regulation of MHCK B and MHCK C, except that MHCK C can be activated in vitro by autophosphorylation (Liang et al., 2002). A bit more information is available for MHCK A. This kinase is activated in vitro by autophosphorylation, which in turn can be stimulated by acidic phospholipids and by high concentrations of myosin II (Medley et al., 1990, 1992). Very recent studies have demonstrated that MHCK A is also activated via binding to F-actin (Egelhoff et al., 2005), suggesting that translocation to the cytoskeleton may be a trigger for activation of this enzyme. Critically lacking at present are clear connections between these biochemical observations and signaling pathways further upstream.
The identification and in vivo validation of MHC phosphatases in this system is another important topic needing further investigation. The work presented in this report identifies the critical complement of MHC kinases that act during interphase and cytokinesis. Earlier biochemical studies identified from Dictyostelium cell lysates a trimeric PP2A complex as the major class of protein phosphatase capable of dephosphorylating the heavy chain of MHCK A-phosphorylated myosin II in vitro (Murphy et al., 1999; Murphy and Egelhoff, 1999), but further studies are needed to address in vivo relevance and regulation of this candidate MHC phosphatase. The approaches applied to study MHC kinases in this report may prove useful in this regard.
Recent studies on cGMP signaling pathways have implicated a family of cGMP binding proteins as upstream regulators of myosin II assembly in Dictyostelium (Bosgraaf et al., 2002; Goldberg et al., 2002), but whether these cGMP mediators directly affect MHCKs is currently unknown. A protein kinase in the p21-activated kinase (PAK) family known as PAKa or DPAKa was implicated as a possible upstream modulator of myosin II function (Chung and Firtel, 1999; Chung et al., 2001), although more recent studies have reported conflicting results regarding localization and presence of cytokinesis defects in cell lines deficient in this PAK (Muller-Taubenberger et al., 2002). Further studies are clearly needed to establish the relevance of cGMP signaling, p21 kinase signaling, and other pathways in regulating myosin II assembly in this system. The identification here of MHCK A, MHCK B, and MHCK C as the critical modulators of myosin II assembly in growth phase cells will facilitate these studies. On the basis of the precedence of the work presented here, we suggest that FRAP analysis, in conjunction with gene disruption approaches, has the potential to provide valuable new insights and resolution regarding importance of upstream regulatory pathways in control of myosin II assembly dynamics in this system.
Acknowledgments
This work was supported by a Grant-in-Aid for scientific research from Ministry of Education, Science, and Culture of Japan to S.Y. and National Institutes of Health Grant GM50009 to T.T.E.
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05-03-0219) on June 29, 2005.
References
- Abu-Elneel, K., Karchi, M., and Ravid, S. (1996). Dictyostelium myosin II is regulated during chemotaxis by a novel protein kinase C (PKC). J. Biol. Chem. 271, 977-984. [DOI] [PubMed] [Google Scholar]
- Baumann, O. (2004). Spatial pattern of nonmuscle myosin-II distribution during the development of the Drosophila compound eye and implications for retinal morphogenesis. Dev. Biol. 269, 519-533. [DOI] [PubMed] [Google Scholar]
- Betapudi, V., Mason, C., Licate, L., and Egelhoff, T. T. (2005). Identification and Characterization of a novel α-kinase with a von Willebrand factor A-like motif localized to the contractile vacuole and Golgi complex in Dictyostelium discoideum. Mol. Biol. Cell 16, 2248-2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betapudi, V., Shoebotham, K., and Egelhoff, T. T. (2004). Generation of double gene disruptions in Dictyostelium discoideum using a single antibiotic marker selection. Biotechniques 36, 106-112. [DOI] [PubMed] [Google Scholar]
- Bosgraaf, L., Russcher, H., Smith, J. L., Wessels, D., Soll, D. R., and Van Haastert, P. J. (2002). A novel cGMP signalling pathway mediating myosin phosphorylation and chemotaxis in Dictyostelium. EMBO J. 21, 4560-4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung, C. Y., and Firtel, R. A. (1999). PAKa, a putative PAK family member, is required for cytokinesis and the regulation of the cytoskeleton in Dictyostelium discoideum cells during chemotaxis. J. Cell Biol. 147, 559-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung, C. Y., Potikyan, G., and Firtel, R. A. (2001). Control of cell polarity and chemotaxis by Akt/PKB and PI3 kinase through the regulation of PAKa. Mol. Cell 7, 937-947. [DOI] [PubMed] [Google Scholar]
- Côté, G. P., Luo, X., Murphy, M. B., and Egelhoff, T. T. (1997). Mapping of the novel protein kinase catalytic domain of Dictyostelium myosin II heavy chain kinase A. J. Biol. Chem. 272, 6846-6849. [DOI] [PubMed] [Google Scholar]
- de la Roche, M. A., Smith, J. L., Betapudi, V., and Egelhoff, T. T. (2002a). Signaling pathways regulating Dictyostelium myosin II. J. Muscle Res. Cell Motil. 23, 703-718. [DOI] [PubMed] [Google Scholar]
- de la Roche, M. A., Smith, J. L., Rico, M., Carrasco, S., Merida, I., Licate, L., Cote, G. P., and Egelhoff, T. T. (2002b). Dictyostelium discoideum has a single diacylglycerol kinase gene with similarity to mammalian theta isoforms. Biochem. J. 368, 809-815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Lozanne, A., and Spudich, J. A. (1987). Disruption of the Dictyostelium myosin heavy chain gene by homologous recombination. Science 236, 1086-1091. [DOI] [PubMed] [Google Scholar]
- Dembinsky, A., Rubin, H., and Ravid, S. (1997). Autophosphorylation of Dictyostelium myosin II heavy chain-specific PKC is required for its activation and membrane dissociation. J. Biol. Chem. 272, 828-834. [DOI] [PubMed] [Google Scholar]
- Drennan, D., and Ryazanov, A. G. (2004). Alpha-kinases: analysis of the family and comparison with conventional protein kinases. Prog. Biophys. Mol. Biol. 85, 1-32. [DOI] [PubMed] [Google Scholar]
- Egelhoff, T. T., Croft, D., and Steimle, P. A. (2005). Actin activation of myosin heavy chain kinase A in Dictyostelium: a biochemical mechanism for the spatial regulation of myosin II filament disassembly. J. Biol. Chem. 280, 2879-2887. [DOI] [PubMed] [Google Scholar]
- Egelhoff, T. T., Lee, R. J., and Spudich, J. A. (1993). Dictyostelium myosin heavy chain phosphorylation sites regulate myosin filament assembly and localization in vivo. Cell 75, 363-371. [DOI] [PubMed] [Google Scholar]
- Egelhoff, T. T., Manstein, D. J., and Spudich, J. A. (1990). Complementation of myosin null mutants in Dictyostelium discoideum by direct functional selection. Dev. Biol. 137, 359-367. [DOI] [PubMed] [Google Scholar]
- Goldberg, J. M., Bosgraaf, L., Van Haastert, P. J., and Smith, J. L. (2002). Identification of four candidate cGMP targets in Dictyostelium. Proc. Natl. Acad. Sci. USA 99, 6749-6754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heid, P. J., Wessels, D., Daniels, K. J., Gibson, D. P., Zhang, H., Voss, E., and Soll, D. R. (2004). The role of myosin heavy chain phosphorylation in Dictyostelium motility, chemotaxis and F-actin localization. J. Cell Sci. 117, 4819-4835. [DOI] [PubMed] [Google Scholar]
- Iijima, M., Huang, Y. E., Luo, H. R., Vazquez, F., and Devreotes, P. N. (2004). Novel mechanism of PTEN regulation by its phosphatidylinositol 4,5-bisphosphate binding motif is critical for chemotaxis. J. Biol. Chem. 279, 16606-16613. [DOI] [PubMed] [Google Scholar]
- Kolman, M. F., and Egelhoff, T. T. (1997). Dictyostelium myosin heavy chain kinase A subdomains. Coiled-coil and WD repeat roles in oligomerization and substrate targeting. J. Biol. Chem. 272, 16904-16910. [DOI] [PubMed] [Google Scholar]
- Kolman, M. F., Futey, L. M., and Egelhoff, T. T. (1996). Dictyostelium myosin heavy chain kinase A regulates myosin localization during growth and development. J. Cell Biol. 132, 101-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, S., Parent, C.A., Insall, R., and Firtel, R. A. (1999). A novel Ras-interacting protein required for chemotaxis and cyclic adenosine monophosphate signal relay in Dictyostelium. Mol. Biol. Cell 10, 2829-2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Z. H., Spektor, A., Varlamova, O., and Bresnick, A. R. (2003). Mts1 regulates the assembly of nonmuscle myosin-IIA. Biochemistry 42, 14258-14266. [DOI] [PubMed] [Google Scholar]
- Liang, W., Licate, L., Warrick, H., Spudich, J., and Egelhoff, T. (2002). Differential localization in cells of myosin II heavy chain kinases during cytokinesis and polarized migration. BMC Cell Biol. 3, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luck-Vielmetter, D., Schleicher, M., Grabatin, B., Wippler, J., and Gerisch, G. (1990). Replacement of threonine residues by serine and alanine in a phosphorylatable heavy chain fragment of Dictyostelium myosin II. FEBS Lett. 269, 239-243. [DOI] [PubMed] [Google Scholar]
- Manstein, D. J., Titus, M. A., De Lozanne, A., and Spudich, J. A. (1989). Gene replacement in Dictyostelium: generation of myosin null mutants. EMBO J. 8, 923-932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medley, Q. G., Bagshaw, W. L., Truong, T., and Côté, G. P. (1992). Dictyostelium myosin II heavy-chain kinase A is activated by heparin, DNA and acidic phospholipids and inhibited by polylysine, polyarginine and histones. Biochim. Biophys. Acta 1175, 7-12. [DOI] [PubMed] [Google Scholar]
- Medley, Q. G., Gariepy, J., and Côté, G. P. (1990). Dictyostelium myosin II heavy-chain kinase A is activated by autophosphorylation: studies with Dictyostelium myosin II and synthetic peptides. Biochemistry 29, 8992-8997. [DOI] [PubMed] [Google Scholar]
- Moores, S. L., Sabry, J. H., and Spudich, J. A. (1996). Myosin dynamics in live Dictyostelium cells. Proc. Natl. Acad. Sci. USA 93, 443-446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller-Taubenberger, A., Bretschneider, T., Faix, J., Konzok, A., Simmeth, E., and Weber, I. (2002). Differential localization of the Dictyostelium kinase DPAKa during cytokinesis and cell migration. J. Muscle Res. Cell Motil. 23, 751-763. [DOI] [PubMed] [Google Scholar]
- Murakami, N., Chauhan, V. P., and Elzinga, M. (1998). Two nonmuscle myosin II heavy chain isoforms expressed in rabbit brains: filament forming properties, the effects of phosphorylation by PKC and casein kinase II, and location of the phosphorylation sites. Biochemistry 37, 1989-2003. [DOI] [PubMed] [Google Scholar]
- Murakami, N., Kotula, L., and Hwang, Y. W. (2000). Two distinct mechanisms for regulation of nonmuscle myosin assembly via the heavy chain: phosphorylation for MIIB and mts 1 binding for MIIA. Biochemistry 39, 11441-11451. [DOI] [PubMed] [Google Scholar]
- Murphy, M. B., and Egelhoff, T. T. (1999). Biochemical characterization of a Dictyostelium myosin II heavy chain phosphatase that promotes bipolar filament assembly. Eur. J. Biochem. 264, 582-590. [DOI] [PubMed] [Google Scholar]
- Murphy, M. B., Levi, S. K., and Egelhoff, T. T. (1999). Molecular characterization and immunolocalization of Dictyostelium discoideum protein phosphatase 2A. FEBS Lett. 456, 7-12. [DOI] [PubMed] [Google Scholar]
- Nagasaki, A., Itoh, G., Yumura, S., and Uyeda, T.Q.P. (2002). Novel myosin heavy chain kinase involved in disassembly of myosin II filaments and efficient cleavage in mitotic Dictyostelium cells. Mol. Biol. Cell 13, 4333-4342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neujahr, R., Heizer, C., and Gerisch, G. (1997). Myosin II-independent processes in mitotic cells of Dictyostelium discoideum: redistribution of the nuclei, re-arrangement of the actin system and formation of the cleavage furrow [In Process Citation]. J. Cell Sci. 110, 123-137. [DOI] [PubMed] [Google Scholar]
- Nock, S., Liang, W., Warrick, H. M., and Spudich, J. A. (2000). Mutational analysis of phosphorylation sites in the Dictyostelium myosin II tail: disruption of myosin function by a single charge change. FEBS Lett. 466, 267-272. [DOI] [PubMed] [Google Scholar]
- Park, K. C., Rivero, F., Meili, R., Lee, S., Apone, F., and Firtel, R. A. (2004). Rac regulation of chemotaxis and morphogenesis in Dictyostelium. EMBO J. 23, 4177-4189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rico, M., and Egelhoff, T. T. (2003). Myosin heavy chain kinase B participates in the regulation of myosin assembly into the cytoskeleton. J. Cell. Biochem. 88, 521-532. [DOI] [PubMed] [Google Scholar]
- Ridley, A. J., Schwartz, M. A., Burridge, K., Firtel, R. A., Ginsberg, M. H., Borisy, G., Parsons, J. T., and Horwitz, A. R. (2003). Cell migration: integrating signals from front to back. Science 302, 1704-1709. [DOI] [PubMed] [Google Scholar]
- Sabry, J. H., Moores, S. L., Ryan, S., Zang, J. H., and Spudich, J. A. (1997). Myosin heavy chain phosphorylation sites regulate myosin localization during cytokinesis in live cells. Mol. Biol. Cell 8, 2605-2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholey, J. M., Taylor, K. A., and Kendrick-Jones, J. (1980). Regulation of non-muscle myosin assembly by calmodulin-dependent light chain kinase. Nature 287, 233-235. [DOI] [PubMed] [Google Scholar]
- Soll, D. R., Wessels, D., Heid, P. J., and Zhang, H. (2002). A contextual framework for characterizing motility and chemotaxis mutants in Dictyostelium discoideum. J. Muscle Res. Cell Motil. 23, 659-672. [DOI] [PubMed] [Google Scholar]
- Spudich, A. (1987). Isolation of the actin cytoskeleton from amoeboid cells of Dictyostelium. Methods Cell Biol. 28, 209-214. [DOI] [PubMed] [Google Scholar]
- Steimle, P. A., Naismith, T., Licate, L., and Egelhoff, T. T. (2001a). WD repeat domains target Dictyostelium myosin heavy chain kinases by binding directly to myosin filaments. J. Biol. Chem. 276, 6853-6860. [DOI] [PubMed] [Google Scholar]
- Steimle, P. A., Yumura, S., Côté, G. P., Medley, Q. G., Polyakov, M. V., Leppert, B., and Egelhoff, T. T. (2001b). Recruitment of a myosin heavy chain kinase to actin-rich protrusions in Dictyostelium. Curr. Biol. 11, 708-713. [DOI] [PubMed] [Google Scholar]
- Stites, J., Wessels, D., Uhl, A., Egelhoff, T., Shutt, D., and Soll, D. R. (1998). Phosphorylation of the Dictyostelium myosin II heavy chain is necessary for maintaining cellular polarity and suppressing turning during chemotaxis. Cell Motil. Cytoskelet. 39, 31-51. [DOI] [PubMed] [Google Scholar]
- Sussman, M. (1987). Cultivation and synchronous morphogenesis of Dictyostelium under controlled experimental conditions. In: Dictyostelium discoideum: Molecular Approaches to Cell Biology, ed. J. A. Spudich, Academic Press, 9-29. [DOI] [PubMed]
- Tuxworth, R. I., Cheetham, J. L., Machesky, L. M., Spiegelmann, G. B., Weeks, G., and Insall, R. H. (1997). Dictyostelium RasG is required for normal motility and cytokinesis, but not growth. J. Cell Biol. 138, 605-614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uyeda, T.Q.P., and Nagasaki, A. (2004). Variations on a theme: the many modes of cytokinesis. Curr. Opin. Cell Biol. 16, 55-60. [DOI] [PubMed] [Google Scholar]
- Uyeda, T.Q.P., Nagasaki, A., and Yumura, S. (2004). Multiple parallelisms in animal cytokinesis. Int. Rev. Cytol. 240, 377-432. [DOI] [PubMed] [Google Scholar]
- Vaillancourt, J. P., Lyons, C., and Côté, G. P. (1988). Identification of two phosphorylated threonines in the tail region of Dictyostelium myosin II. J. Biol. Chem. 263, 10082-10087. [PubMed] [Google Scholar]
- Van Haastert, P. J., and Devreotes, P. N. (2004). Chemotaxis: signalling the way forward. Nat. Rev. Mol. Cell Biol. 5, 626-634. [DOI] [PubMed] [Google Scholar]
- Weber, I., Neujahr, R., Du, A., Kohler, J., Faix, J., and Gerisch, G. (2000). Two-step positioning of a cleavage furrow by cortexillin and myosin II. Curr. Biol. 10, 501-506. [DOI] [PubMed] [Google Scholar]
- Wessels, D. et al. (2004). RasC plays a role in transduction of temporal gradient information in the cyclic-AMP wave of Dictyostelium discoideum. Eukaryot. Cell 3, 646-662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessels, D. J., Zhang, H., Reynolds, J., Daniels, K., Heid, P., Lu, S., Kuspa, A., Shaulsky, G., Loomis, W. F., and Soll, D. R. (2000). The internal phosphodiesterase RegA is essential for the suppression of lateral pseudopods during Dictyostelium chemotaxis. Mol. Biol. Cell 11, 2803-2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yumura, S. (2001). Myosin II dynamics and cortical flow during contractile ring formation in Dictyostelium cells. J. Cell Biol. 154, 137-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yumura, S., and Uyeda, T.Q.P. (1997). Myosin II can be localized to the cleavage furrow and to the posterior region of Dictyostelium amoebae without control by phosphorylation of myosin heavy and light chains. Cell Motil. Cytoskelet. 36, 313-322. [DOI] [PubMed] [Google Scholar]
- Yumura, S., and Uyeda, T.Q.P. (2003). Myosins and cell dynamics in cellular slime molds. Int. Rev. Cytol. 224, 173-225. [DOI] [PubMed] [Google Scholar]
- Zang, J. H., Cavet, G., Sabry, J. H., Wagner, P., Moores, S. L., and Spudich, J. A. (1997). On the role of myosin-II in cytokinesis: division of Dictyostelium cells under adhesive and nonadhesive conditions. Mol. Biol. Cell 8, 2617-2629. [DOI] [PMC free article] [PubMed] [Google Scholar]