Abstract
Cytokeratins 8 and 19 concentrate at costameres of striated muscle and copurify with the dystrophin-glycoprotein complex, perhaps through the interaction of the cytokeratins with the actin-binding domain of dystrophin. We overexpressed dystrophin's actin-binding domain (Dys-ABD), K8 and K19, as well as closely related proteins, in COS-7 cells to assess the basis and specificity of their interaction. Dys-ABD alone associated with actin microfilaments. Expressed with K8 and K19, which form filaments, Dys-ABD associated preferentially with the cytokeratins. This interaction was specific, as the homologous ABD of βI-spectrin failed to interact with K8/K19 filaments, and Dys-ABD did not associate with desmin or K8/K18 filaments. Studies in COS-7 cells and in vitro showed that Dys-ABD binds directly and specifically to K19. Expressed in muscle fibers in vivo, K19 accumulated in the myoplasm in structures that contained dystrophin and spectrin and disrupted the organization of the sarcolemma. K8 incorporated into sarcomeres, with no effect on the sarcolemma. Our results show that dystrophin interacts through its ABD with K19 specifically and are consistent with the idea that cytokeratins associate with dystrophin at the sarcolemma of striated muscle.
INTRODUCTION
Mutations in the gene for dystrophin cause Duchenne or Becker Muscular Dystrophy, but we still know little about dystrophin's functions in normal muscle or how its absence leads to the loss of myofibers. Dystrophin is the major cytoskeletal component of a large, transmembrane complex (Ahn and Kunkel, 1993; Matsumura and Campbell, 1994; Ozawa et al., 1998) that plays a role in signaling from the plasma membrane of skeletal myofibers (sarcolemma) to the cytoplasm (Rando, 2001; Lapidos et al., 2004) and in transmitting the force of contraction across the sarcolemma to extracellular structures (Campbell, 1995; Bloch and Gonzalez-Serratos, 2003). Here, we focus on the latter role, and particularly, on the nature of the connections made between the contractile apparatus and the sarcolemma, at sites termed “costameres” (Bloch and Gonzalez-Serratos, 2003; Ervasti, 2003).
Costameres are riblike structures that surround each myofiber at the sarcolemma and that are enriched in a number of integral and peripheral membrane proteins (Bloch and Gonzalez-Serratos, 2003; Ervasti, 2003), including dystrophin (Masuda et al., 1992; Minetti et al., 1992; Porter et al., 1992; Straub et al., 1992; Williams and Bloch, 1999a; Rybakova et al., 2000). Costameres and the connections between the contractile apparatus and the sarcolemma that they anchor are present at the plasma membrane overlying both the Z and M lines of superficial myofibrils of fast-twitch muscles (Porter et al., 1992; Williams et al., 2000). They also can be oriented at the plasma membrane parallel to the longitudinal axis of the myofibers, creating a rectilinear, structural lattice at the sarcolemma. Dystrophin and its associated proteins are enriched at all these sites (Porter et al., 1992; Williams and Bloch, 1999a; Williams et al., 2000). When it is present, the dystrophin complex helps to link the contractile apparatus to the sarcolemma and through the membrane to the extracellular matrix (Ibraghimov-Beskrovnaya et al., 1992; Ervasti and Campbell, 1993; Rybakova et al., 2000; Rybakova et al., 2002; Michele et al., 2003). When dystrophin is absent, links between the contractile apparatus and the sarcolemma can weaken and eventually break (Porter et al., 1992; Ehmer et al., 1997; Williams and Bloch, 1999b; Rybakova et al., 2000), damaging the sarcolemma (Mokri and Engel, 1975), and, ultimately, killing the myofiber (reviewed in Emery, 1993).
Several filamentous proteins, including actin, link the contractile apparatus to the dystrophin complex at costameres. Dystrophin binds actin (Senter et al., 1993; Renley et al., 1998; Sutherland-Smith et al., 2003), and this activity is thought to be physiologically important for the health of the muscle fiber (Corrado et al., 1996; Eraslan et al., 1999; Warner et al., 2002). In healthy muscle, fragments of sarcolemma can be isolated that are linked to filamentous actin at costameres, but this actin is lost from sarcolemmal fragments isolated from muscle lacking dystrophin (Rybakova et al., 2000). The near ubiquitousness of actin in skeletal muscle cells makes its interaction with dystrophin difficult to study in situ. The fact that actin is linked primarily to the Z-disks of striated muscle suggests that it is unlikely to mediate all the connections between the contractile apparatus and costameres, however. Other filament-forming proteins, and especially intermediate filaments, are likely to be involved.
Desmin is the major intermediate filament protein of skeletal muscle (Lazarides, 1980; Thornell, 1992; Capetanaki and Milner, 1998). Ultrastructural evidence, as well as studies of mice lacking desmin due to homologous recombination, suggest that desmin-based intermediate filaments are important in linking the Z-disks of superficial myofibrils to costameres at the sarcolemma (Granger and Lazarides, 1979; Pierobon-Bormioli, 1981; Street, 1983; Shear and Bloch, 1985; O'Neill et al., 2002), perhaps through a plectin cross-linker (Hijikata et al., 2003). Consistent with this, two desmin-associated proteins, syncoilin and desmuslin or synemin, have been shown to bind dystrobrevin, a ligand of dystrophin (Mizuno et al., 2001; Newey et al., 2001). In addition, synemin, which copolymerizes with desmin (Granger et al., 1982; Bellin et al., 1999; Hirako et al., 2003), binds vinculin (Bellin et al., 2001), the quintessential costameric protein, and may concentrate at costameres (Mizuno et al., 2004). Like actin filaments, however, desmin-based intermediate filaments are concentrated near the Z-disks of each sarcomere and between the Z-disks and the sarcolemma (Granger and Lazarides, 1978; Lazarides, 1978; Granger and Lazarides, 1979; Richardson et al., 1981; Capetanaki and Milner, 1998; O'Neill et al., 2002), making them unlikely candidates to mediate connections between the contractile apparatus and the domains of costameres overlying M-lines or oriented longitudinally along the sarcolemma.
We recently reported that some fast twitch muscle fibers in the desmin-null mouse retained normal costameres, suggesting that desmin is not necessary to align costameres with the underlying contractile apparatus (O'Neill et al., 2002). Because cytokeratins are expressed in developing muscle and in muscle tumors (Langbein et al., 1989; Miettinen and Rapola, 1989; Kosmehl et al., 1990), and reverse transcription-PCR procedures suggested that mRNAs encoding cytokeratins were present in muscle extracts (Ursitti et al., 2004), we looked for cytokeratins in adult skeletal muscle and found them concentrated near the sarcolemma at costameres (O'Neill et al., 2002). Cytokeratins are present at all costameric domains and not simply those overlying Z-disks (O'Neill et al., 2002), suggesting that they may play a more widespread role at the sarcolemma than desmin or actin. We cloned and sequenced K8 and K19 from striated muscle and localized these cytokeratins at the costameres of cardiac muscle (Ursitti et al., 2004). We showed further that K8 and K19 copurify with the dystrophin-glycoprotein complex. K8, a type II cytokeratin, and K19, a type I cytokeratin, coassemble to form intermediate filaments in other cells (Fuchs and Weber, 1994; Coulombe and Omary, 2002) and are likely to do so in muscle as well. Indeed, in addition to their presence at costameres, K8 and K19 also associated with the Z-disks of sarcomeres deep in the myoplasm (Ursitti et al., 2004). Our results suggested that cytokeratin-based intermediate filaments composed of K8 and K19 interact with the dystrophin complex at costameres, linking it to underlying structures in the myoplasm.
Here, we address the possibility that cytokeratins associate with costameres by binding directly to dystrophin. We chose to focus on the actin-binding domain (ABD) of dystrophin, because the homologous structures of βII-spectrin, and fimbrin and plectin, bind to neurofilaments and vimentin, respectively (Correia et al., 1999; Macioce et al., 1999; Sevcik et al., 2004). Expanding an earlier series of experiments (Ursitti et al., 2004), we use cotransfection of COS-7 cells to show that the ABD of dystrophin (Dys-ABD) indeed associates preferentially and specifically with filaments formed by K8 and K19. We characterize this interaction in transfected cells and in vitro to show that it is mediated preferentially by K19. Finally, we demonstrate that overexpression of K19, but not K8, in skeletal muscle fibers can disrupt the organization of proteins normally found in costameres at the sarcolemma. Our results support the hypothesis that cytokeratin filaments composed of K8 and K19 help to link the contractile apparatus to dystrophin at the costameres of striated muscle.
MATERIALS AND METHODS
Tissue Culture and Transfection
COS-7 cells were purchased from the American Type Culture Collection (Manassas, VA). Cells were grown to confluence in DMEM supplemented with 10% fetal bovine serum and 1% streptomycin. They were subcultured into 10-cm culture dishes or onto 25-mm coverslips in 35-mm dishes (Falcon, BD Biosciences, San Jose, CA) at ∼20% confluence.
Creation of Plasmids
We used PCR to amplify cDNAs encoding keratin 8 and the actin-binding domains of dystrophin, utrophin, and βI-spectrin. Plasmid P8.1.1, containing the human isoform of keratin 8 (American Type Culture Collection), was used as template for PCR of K8, with the following primers (accession no. NM002273/M34225): A, 5 GCCCGAATTCGGATGTCCATCAGG-GTGACC 3′ (sense) and B, 5′ CCGCGGTACCCTAGGGTTTGGCAGAGCTAG 3′ (antisense). The sense primer contained an EcoRI restriction site and the antisense primer a KpnI site, which were used for insertion into the pCMV-myc vector (BD Biosciences Clontech, Palo Alto, CA), resulting in plasmid pCMV-mycK8.
Plasmid pK18 was kindly provided by Dr. M. Bishr Omary (Stanford University, Palo Alto, CA). Plasmid GW1-CMV, containing the cDNA encoding keratin 19, was the gift of Dr. P. Coulombe (Johns Hopkins University, Baltimore, MD). Plasmid pRcCMV1023, containing the DNA encoding desmin, was generously provided by Dr. Y. Capetanaki (Baylor University College of Medicine, Houston, TX).
Plasmids pDys246 (Amann et al., 1998) and FLAG-Utrophin (pet23Utr261; Galkin et al., 2002) were kindly provided by Dr. J. Ervasti (University of Wisconsin, Madison, WI) and served as templates for generating the actin-binding domains of dystrophin and utrophin. Primers A, 5′ GCGAATTCTATGTTGTTGTGGGAAGAAGTA 3′ (sense), and B, 5′ GCAGGTACCCTATTCAATGCTCACTTGTTG 3′ (antisense), were used to obtain the first 738 bases of the dystrophin gene (accession no. M18533). The sense primer contained an EcoRI site and the antisense primer a KpnI site for cloning into the DsRed2-c1 vector (BD Biosciences Clontech). An identical set of primers carrying EcoRI and SalI recognition sites were used for insertion into the pET-16b vector, generating a polyhistidine, or His, tag at the N terminus of the actin-binding domain of dystrophin. The first 783 bases of the open reading frame of utrophin (accession no. Y12229) were cloned with primers A, 5′ TATAAGCTTC-TATGGCCAAGTATGGGAC 3′ (sense) and B, 5′ TGTGGATCCCTAATCTATCGTGACTT-GCTG 3′ (antisense). The sense primer contained a HinDIII site and the antisense primer contained a BamHI restriction site for cloning into the DsRed2-c1 vector (BD Biosciences Clontech).
Plasmid B259 served as the template to amplify the first 942 base pairs of the coding sequence of the ABD of βI-spectrin (NM_000347). Primers A, 5′ CGTAGAAT TCCTATG-ACATCGGCCACAG 3′ (sense), and B, 5′ TCCCCGCGGCTAAGGTGAG CAGGTCCGA 3′ (antisense), were used in PCR. The sense primer contained an EcoRI site, and the antisense primer contained a SacII site for cloning into the DsRed2-c1 plasmid.
Expression of the recombinant 6X-His form of the Dys-ABD by pDys246 (see above) was induced with 0.5 mM isopropyl β-d-thioglucopyranoside for 3 h. The expressed protein was purified by affinity chromatography on TALON resin (BD Biosciences Clontech), following the manufacturer's instructions.
Transfection
COS-7 cells, plated as described above, received fresh medium 24 h after replating (see above) and were transfected with 1-10 μg of plasmid DNA, introduced as calcium phosphate precipitates (Sambrook et al., 1989) 3 h later. After 24-h incubation, cultures were washed and either fixed and processed on coverslips or harvested by scraping into phosphate-buffered saline (PBS).
We used 14-d-old Sprague Dawley rats (Zivic Miller, Zelienople, PA), anesthetized with Metofane, for transfection of myofibers in situ (Wolff et al., 1990). Briefly, plasmid DNA constructs (5 μg/ml, in normal saline and 2% India ink) were injected with a 28-gauge tuberculin syringe into the Tibialis anterior (TA) muscle. The rats were killed under anesthesia 7-10 d later by cardiac perfusion with 2% paraformaldehyde in PBS (10 mM NaP, 145 mM NaCl, pH 7.2), supplemented with protease inhibitors (Complete, EDTA-free; Roche Diagnostics, Indianapolis, IN). Injected muscles were removed and snap frozen in a slush of liquid nitrogen. Frozen longitudinal sections, 20 μm in thickness, were prepared on a cryostat (Reichert-Jung; Cambridge Instruments, Deerfield, IL) and collected on slides pretreated with chrom-alum gelatin. All procedures involving the use of animals and animal tissue were approved by the Institutional Animal Care and Use Committee of the University of Maryland School of Medicine.
Antibodies
Polyclonal rabbit antibodies to desmin were used at a concentration of 1:20 and monoclonal antibodies to K19 (both from Sigma-Aldrich., St. Louis, MO) were used at a concentration of 1:50. Monoclonal antibodies to K18 (Lab Vision, Fremont, CA) were used at a dilution of 1:200. Guinea pig antibodies to K19 (GP67; Progen, Heidelberg, Germany) were used at 1:100. Monoclonal antibodies to the Myc and His epitope tags (Invitrogen, Carlsbad, CA) were used at dilutions of 1:500. Rabbit antibodies to dystrophin (Lab Vision) were used at a dilution of 1:100. Mouse monoclonal antibodies to α-actinin were from Sigma-Aldrich and were used at 1:500. Monoclonal mouse antibodies to β-dystroglycan (Novacastra, Newcastle-upon-Tyne, United Kingdom) were used at dilutions of 1:10. Affinity-purified chicken antibodies to βI-spectrin (Ursitti et al., 2001) were used at 2 μg/ml. Rabbit antibodies to DS-Red (BD Biosciences Clontech) were used at 1:20. Goat anti-mouse IgG and goat anti-rabbit IgG linked to Alexa-488 or Alexa-568 (Molecular Probes, Eugene, OR) were diluted 1:200. Cy5-conjugated goat anti-mouse IgG or goat anti-rabbit IgG antibodies (Molecular Probes) were used at 1:50. Fluoresceinated donkey anti-chicken IgY and rabbit anti-goat IgG were from Jackson ImmunoResearch Laboratories (West Chester, PA) and were each diluted 1:100 before use.
Fluorescent Immunolabeling
Transfected COS-7 were fixed in 2% paraformaldehyde in PBS for 20 min at room temperature, washed in PBS, permeabilized in 0.5% Triton X-100, washed again, incubated in PBS/bovine serum albumin (BSA; PBS containing 1 mg/ml BSA and 10 mM NaN3) for 20 min, and then placed in primary antibody in PBS/bovine serum albumin for 2 h at room temperature or overnight a 4°C. Samples were washed and then incubated with species-specific secondary antibodies (see above) diluted in PBS/bovine serum albumin. Frozen longitudinal sections of transfected TA muscle were labeled similarly, except that the sections were not treated with detergents before labeling.
After additional washing, samples were mounted in Vectashield (Vector Laboratories, Burlingame, CA). Slides were observed with a Zeiss 410 confocal laser scanning microscope (Carl Zeiss, Tarrytown, NY) equipped with a 63×, numerical aperture 1.4, plan-apochromatic objective. The pinholes for all fluorophores were set to 18. Images were collected and stored with software provided by Carl Zeiss (Jena, Germany).
For quantitation of the effects of overexpressing hK19 or myc-K8 in skeletal muscle fibers, transfected myofibers were identified in cryosections of muscle as those labeled by either anti-myc or anti-hK19 antibodies. All transfected fibers were scored for labeling of intracellular aggregates or vesicles (dystrophin, β-spectrin, small ankyrin 1, β-dystroglycan, and α-actinin), or for the presence of periodic punctate structures at the sarcolemma, typical of costameres (β-dystroglycan). Control, untransfected fibers assessed for the presence of costameres consisted of those fibers visible in the same microscopic fields as transfected myofibers. No fibers were excluded because of the angle of section or other features, although this tended to lower the number of controls scored as having intact costameres. Results were evaluated for statistical significance with Fisher's exact test.
Images were arranged into montages, labeled and given scale bars with Corel Draw (Corel, Ottawa, Ontario, Canada). Inset pictures were prepared with MetaMorph and magnified twofold with Corel Draw. Zeiss 510 imaging software (LSM Image Examiner) was used to quantitate the extent of colocalization of two fluorescent labels. R values obtained with Pearson's coefficient were analyzed by analysis of variance (Origin 4.1; OriginLab, Northampton, MA) software.
Dot Blot Assay
COS-7 cells were harvested and lysed with Cell Lytic M (Sigma-Aldrich) for 20 min at room temperature. Soluble and pellet fractions were prepared by centrifugation at 14,000 × g for 15 min. The soluble fractions of untransfected cells or cells expressing myc-K8 or hK19 were dotted onto nitrocellulose (3 μg of protein) and incubated overnight with bacterially expressed, purified His-tagged Dys-ABD (4 μg/ml) in PBS containing 3% milk, 0.05% Tween 20, and 10 mM NaN3. After rapid washing, blots were probed with monoclonal anti-His antibody (1:5000) and goat anti-mouse IgG conjugated to alkaline phosphatase (1:10,000), and visualized by chemiluminescence (Western Light Detection; Tropix Laboratories, Bedford, MA). MetaMorph imaging software (Universal Imaging, West Chester, PA) was used for quantitation.
Surface Plasmon Resonance
A BIAcore 3000 instrument (BIAcore, Uppsala, Sweden) was used at room temperature with a CM5 chip, coupled to a rabbit antibody to mouse IgG following the manufacturer's recommendations.
To assay binding of the Dys-ABD to cytokeratins enriched in cell extracts, prepared as described above for dot blots, rabbit anti-mouse IgG was captured onto the surface of flow cells 3 and 4 at 6000 resonance units (RUs). Flow rates for these and all other steps with the BIAcore 3000 were 20 μl/min. Nonimmune MopC 21 antibody was immobilized at 50 μg/ml as a control in flow cell 3. Monoclonal antibody (mAb) to human K19 antibody at 200 μg/ml was introduced into flow cell 4, to give a signal of 400 RUs. Homogenates of cells expressing human K19 were passed over flow cells 3 and 4 at 200 μg/ml (∼5 μM). The His-tagged Dys-ABD, diluted in HBS-EP buffer (BIAcore) to 25 μM, was flowed through both cells. The control signals were subtracted from the experimental samples to give specific binding.
To assay binding of His-tagged Dys-ABD to pure cytokeratin subunits, we coupled rabbit anti-mouse IgG to the surfaces of all flow cells, to give signals of 12,600-16,000 RUs. Aliquots (30 μl) containing 25 μg/ml anti-K8 antibodies were immobilized in flow cells 1 and 3, yielding signals of 615 and 400 RUs, respectively. Aliquots of antibodies to K19 (60 μl, 15 μg/ml) were immobilized in flow cell 4, yielding 310 RUs. Aliquots (40 μl, 25 μg/ml) of antibodies to K18 were immobilized in flow cell 2, yielding a signal of 780 RUs. Aliquots (40 μl, 25 μg/ml) of pure K18 (Research Diagnostics, Flanders, NJ) were introduced into cells 1 and 2 over a period of 3 min, yielding 4 RUs of binding in flow cell 1 and 140 RUs in flow cell 2. Aliquots (60 μl, 20 μg/ml) of pure K19 (Research Diagnostics) were introduced into flow cells 3 and 4 over a 3-min period, yielding 5 RUs in flow cell 3 and 100 RUs in flow cell 4. His-Dys-ABD (5 μM) was passed over flow cells 1-4, and binding was measured.
Materials
Unless otherwise noted, all materials were purchased from Sigma-Aldrich and were the highest grade available.
RESULTS
Our experiments were designed to learn whether the ABD of dystrophin, Dys-ABD, interacts with cytokeratins, specifically with K8 and K19, which are expressed in striated muscles and concentrated at costameres (Ursitti et al., 2004). We used transfection of COS-7 cells in vitro and skeletal muscle fibers in vivo to study the association of the Dys-ABD with structures containing K8 and K19. We also examined the ability of the Dys-ABD to bind to K8 and K19 in dot blots and by surface plasmon resonance. The results of our experiments suggest that the Dys-ABD associates preferentially with K19 and that the binding is direct and physiologically significant.
Dys-ABD Associates Specifically with Filaments Containing K8 and K19
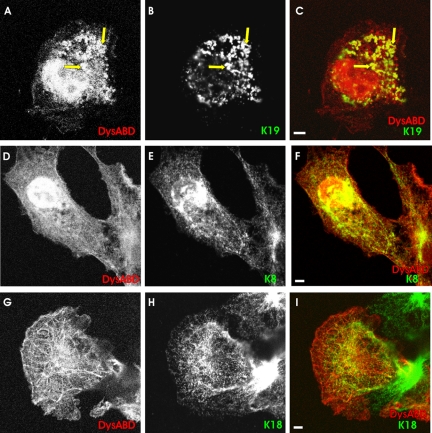
We first transfected COS-7 cells with a plasmid encoding a myc-tagged form of K8 (myc-K8) and a plasmid encoding the human sequence of K19 (hK19), to learn whether they were able to form filaments when expressed alone or together. COS-7 cells contain vimentin and endogenous cytokeratins (Niessen et al., 1997) that, although not fully characterized, include K8 and K18 (our unpublished data). Overexpression of myc-K8 in COS-7 cells resulted in its incorporation into filamentous structures that labeled with antibodies to the myc epitope tag (Figure 1A), perhaps due to the ability of K8 to associate with endogenous K18 (Fuchs and Weber, 1994; Coulombe and Omary, 2002). The overexpression of hK19 in COS-7 cells produced two types of structures. In ∼25% of the cells, that seemed to express relatively low levels of hK19, the protein incorporated into filaments, perhaps through its ability to associate with endogenous K8 (Figure 1B′). In most cells, K19 was expressed at higher levels and, rather than incorporating into filaments, it instead accumulated in punctate structures in the cytoplasm that labeled with antibodies specific for hK19 (Figure 1B). Expressed together, myc-K8 and hK19 coassembled into thickly bundled, filamentlike arrays (Figure 1, D-F), as expected (Fuchs and Weber, 1994; Coulombe and Omary, 2002).
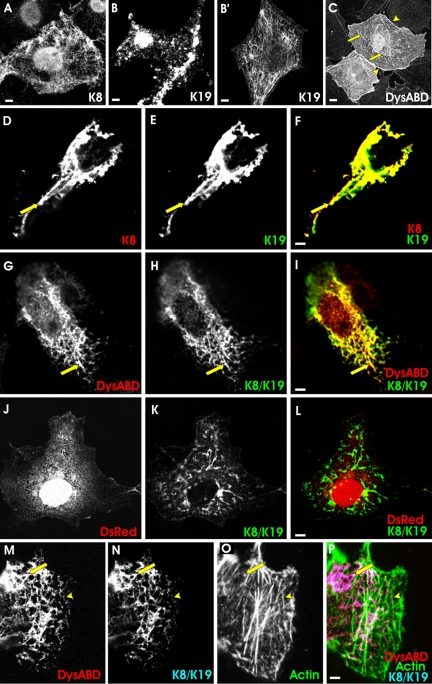
Figure 1.
Distribution of the actin-binding domain of dystrophin, cytokeratins, and actin in COS-7 cells. COS-7 cells were transfected with plasmids encoding myc-K8, hK19, and a DS-Red fusion protein of the Dys-ABD, in different combinations. One day later, cells were fixed and labeled, to localize the exogenous proteins. Fluorescence of DS-Red served to localize the Dys-ABD, whereas K8 and K19 were immunolocalized with antibodies to the myc epitope tag and hK19, respectively. In some cases, actin was localized with Alexa 488-phalloidin. (A) myc-K8 alone incorporated into endogenous filaments. (B and B′) hK19 alone incorporated into large aggregates (B) or, less frequently, endogenous filaments (B′). (C) DS-Red-Dys-ABD, expressed alone, associated preferentially with stress fibers (arrows) and the plasma membrane (arrowheads), which also labeled with Alexa 488-phalloidin (not shown). (D-F) myc-K8 (D) and hK19 (E) together incorporated into filaments (arrow). In F, myc-K8 is shown in red, hK19 in green, and structures that label for both proteins in yellow. (G-I) DS-Red-Dys-ABD (G; red in I) was localized along filaments formed by myc-K8 (H; green in I) and hK19 (not labeled). Structures labeled for myc-K8 and DS-Red-Dys-ABD are shown in yellow in I. (J-L) DS-Red without the Dys-ABD fusion partner (J; red in L) failed to localize to filaments formed by myc-K8 (K; green in L) and hK19 (not labeled). (M-P) As in G-I, but labeled with Alexa 488-phalloidin to reveal the location of actin filaments (O; green in P). In this case, structures containing both DS-Red-Dys-ABD (M; red in P) and myc-K8 (N; blue in P) and hK19 (not labeled) are shown in pink in P, and structures containing these proteins that overlap with labeling for actin are shown in white. DS-Red-ABD associated preferentially with filaments formed by K8 and K19, not with actin. Bars, 5 μm.
We next expressed the Dys-ABD as a DS-Red fusion protein in COS-7 cells, either alone or with the cytokeratins. When expressed alone, Dys-ABD occasionally associated with endogenous filaments that, we have reported previously (Ursitti et al., 2004), can be labeled with antibodies to cytokeratins (not shown), usually in cells that lacked actin in well organized stress fibers. More typically, however, the DS-Red-tagged Dys-ABD associated preferentially with bundles of microfilaments, labeled with phalloidin (Figure 1C, arrows), and with the periphery of the cells (Figure 1C, arrowheads). Its ability to associate with these structures did not depend on the DS-Red fusion partner, because DS-Red alone failed to associate with filaments or with the cell periphery (Figure 1J). Although DS-Red accumulated in the nucleus (Figure 1J), it reached significant levels in the cytoplasm and so could be detected if it were concentrated at actin-rich structures. Thus, under otherwise normal conditions, the DS-Red fusion protein of the Dys-ABD preferentially associated with actin-rich structures in the cytoplasm of COS-7 cells, consistent with the ability of the Dys-ABD to associate with actin filaments in vitro (Senter et al., 1993; Renley et al., 1998; Orlova et al., 2001).
When we coexpressed the Dys-ABD with myc-K8 and hK19, its distribution in COS-7 cells changed dramatically. Instead of concentrating along bundles of actin filaments and at the cell periphery, the Dys-ABD associated with the now prevalent cytokeratin filaments, recognized with antibodies to the myc epitope tag associated with K8 (Figure 1, G-I), or with antibodies to hK19 (our unpublished data). This result, too, was independent of the DS-Red fusion partner, because DS-Red alone did not associate with the cytokeratins (Figure 1, J-L), and similar results have been obtained with a green fluorescent protein (GFP) tag (Ursitti et al., 2004). Thus, the Dys-ABD associated preferentially with cytokeratin filaments composed of K8 and K19 when these are present at high levels in COS-7 cells. This was true even when bundles of actin filaments were readily visible in transfected cells, after labeling with fluorescent derivatives of phalloidin (Figure 1, M-P). Notably, although the Dys-ABD can associate with actin and cytokeratin filaments, it did not colocalize with both simultaneously (Figure 1, M-P), suggesting that it is incapable of cross-linking these two types of filaments in the cytoplasm of COS-7 cells.
The association of the Dys-ABD with cytokeratin filaments was specific both for the Dys-ABD and for K8 and K19. When we expressed desmin together with the Dys-ABD in COS-7 cells, the desmin formed arrays of intermediate filaments, but in most cells the Dys-ABD failed to associate with these filaments (Figure 2, A-C). We also examined the ability of the Dys-ABD to associate with filaments composed of K8 and K18. These two cytokeratin subunits together form filaments (Fuchs and Weber, 1994; Coulombe and Omary, 2002), but K18 is not expressed at significant levels in mature striated muscle (Kosmehl et al., 1990; Ursitti et al., 2004). When coexpressed in COS-7 cells, K8 and K18 assembled into filamentous arrays (Figure 2E), but these did not associate with the Dys-ABD (Figure 2, D and F). When we cotransfected COS-7 cells with K8, hK19, and the ABD of βI-spectrin, the spectrin ABD failed to concentrate along K8/K19 filaments (Figure 2, G-I), suggesting that the ability to associate with these cytokeratins is not a general property of actin-binding domains. Finally, we used 10 μM colchicine to break up microtubules, or 1 μM cytochalasin D to disrupt microfilaments, in cells that coexpressed the Dys-ABD with K8 and hK19. (We confirmed the effectiveness of these concentrations of the drugs in independent experiments in which we used fluorescent phalloidin or antibodies to tubulin; our unpublished data). The Dys-ABD remained associated with cytokeratin filaments in the presence of either colchicine (Figure 2, M-O) or cytochalasin D (Figure 2, J-L). Our results suggest that the Dys-ABD associates with cytokeratins K8 and K19 but not with other intermediate filament proteins, that this association is not mediated by microfilaments or microtubules, and that it is specific for the Dys-ABD.
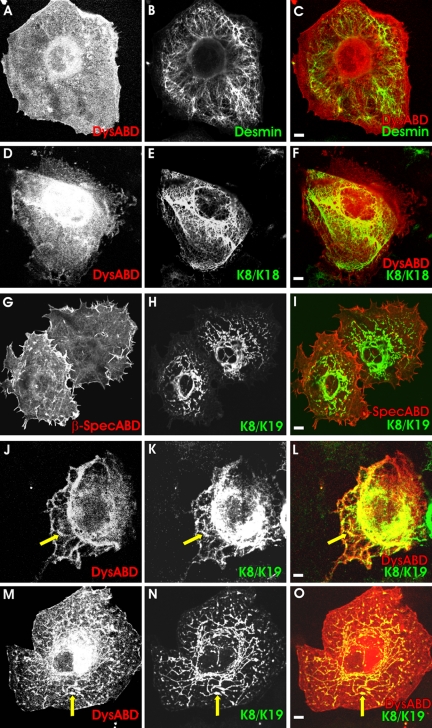
Figure 2.
Specificity of the association of the actin-binding domain of dystrophin with intermediate filaments formed by K8 and K19. COS-7 cells were cotransfected with plasmids expressing DS-Red-Dys-ABD (A-F, J-O) or the ABD of βI-spectrin, also as a DS-Red fusion protein (G-I), together with myc-K8 and hK19 (G-O), myc-K8 and K18 (D-F), or desmin (A-C). In some samples expressing DS-Red-Dys-ABD, myc-K8 and hK19, cells were treated with 1 μM cytochalasin D (J-L) or 10 μM colchicine (M-O), to break up microfilaments or microtubules, respectively, before further processing (see Figure 1). (A-C, D-F) Dys-ABD (A and D; red in C and F) failed to accumulate along desmin filaments (B; green in C) or cytokeratin filaments formed by myc-K8 (E; green in F) and K18 (not labeled). (G-I) The ABD of βI-spectrin (G; red in I) failed to accumulate along cytokeratin filaments formed by myc-K8 (H; green in I) and hK19 (not labeled). (J-L, M-O) The Dys-ABD (J and M; red in L and O) accumulates along filaments formed by myc-K8 (K and N; green in L and O) and hK19 (not labeled) when microfilaments (J-L) or microtubules (M-O) are disrupted. Yellow in the color overlays shows structures containing both the DS-Red-ABD and cytokeratins. Bars, 5 μm.
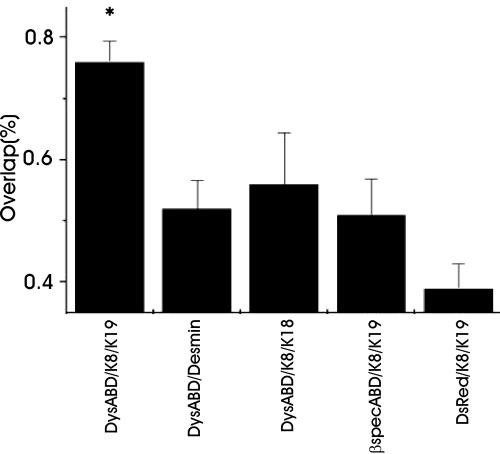
We quantitated the differences in the distributions of the various intermediate filament proteins and ABDs with image analysis software (see Materials and Methods). Our measurements (Figure 3; n ≥15 cells for each condition) indicate that the differences in the interactions between K8/K19 filaments and the Dys-ABD and other intermediate filament proteins and ABDs are highly significant (p < 0.001).
Figure 3.
Quantitation of immunofluorescence labeling. Images such as those shown in Figures 1 and 2 were subjected to quantitative analysis, as described in Materials and Methods. The results show that the extent of codistribution of the DS-Red-ABD with cytokeratin filaments formed by K8 and K19 is significantly greater (p < 0.001) than the extent of codistribution with filaments formed by desmin or K8 and K18. It is also significantly greater (p < 0.001) than the codistribution of the ABD of βI-spectrin, or DS-Red alone, with filaments formed by K8 and K19.
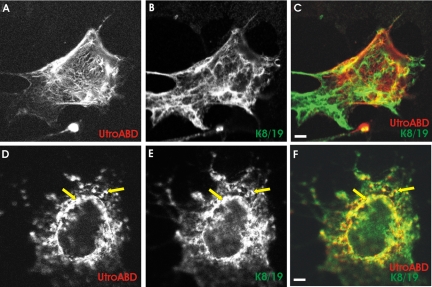
Utrophin is a homologue of dystrophin that is widely expressed in mammalian tissues (Love et al., 1989; Khurana et al., 1990, 1991). When up-regulated in dystrophic muscle, utrophin mitigates the severest effects of dystrophinopathy (Tinsley et al., 1996), suggesting that it may be an effective replacement for dystrophin in Duchenne Muscular Dystrophy (Tinsley and Davies, 1993). We tested the ability of the ABD of utrophin to interact with K8/K19 filaments in COS-7 cells, to learn whether it was as active as the Dys-ABD in this assay. We found that the ABD of utrophin, expressed as a DS-Red fusion protein, did not selectively target K8/K19 filaments (Figure 4, A-C) but instead associated preferentially with longer, straighter filaments as well as with the cell periphery—structures that are enriched in actin (Figure 1, C and O). When actin filaments were destabilized with cytochalasin D, however, the utrophin ABD associated with filaments formed by K8 and K19 (Figure 4, D-F, arrows). The colocalization of the ABD of utrophin with K8 and K19 filaments in COS-7 cells treated with cytochalasin D was statistically significant (p < 0.005). Our results suggest that, although utrophin's ABD may normally prefer actin filaments, consistent with its higher affinity for actin (Winder et al., 1995), it shares with the Dys-ABD the ability to interact with K8/K19 filaments.
Figure 4.
The actin-binding domain of utrophin associates with K8/K19 filaments when actin filaments are disrupted. COS-7 cells were transfected with plasmids expressing a DS-Red fusion protein of the ABD of utrophin, together with myc-K8 and hK19, and processed, with (D-F) and without (A-C) exposure to 1 μM cytochalasin D, as in Figures 1 and 2. The organization of the ABD of utrophin (A and D; red in C and F) did not correspond to that of the filaments formed by myc-K8 (B and E; green in C and F) unless actin filaments were first disrupted by cytochalasin D. Bars, 5 μm.
The Dys-ABD Associates Preferentially with K19
The association of the Dys-ABD with filaments composed of K8 and K19, but not K8 and K18, suggests that K19 harbors a sequence that mediates specific binding. We used cellular transfections, blot overlay and surface plasmon resonance experiments to test this. We first cotransfected cells with the plasmid encoding the DS-Red-Dys-ABD fusion protein with the plasmids encoding hK19, K18, or K8, alone. When overexpressed, hK19 typically forms large punctate structures (Figure 1B). The Dys-ABD accumulated in these same structures when it was coexpressed with hK19 (Figure 5, A-C, arrows). By contrast, the Dys-ABD incorporated poorly into the filamentous structures that contain overexpressed myc-K8 (Figure 5, D-F) or K18 (Figure 5, G-I). These results suggest that the Dys-ABD interacts directly or indirectly with K19.
Figure 5.
Preferential association of the actin-binding domain of dystrophin with K19 in COS-7 cells. COS-7 cells were transfected with DS-Red-Dys-ABD together with hK19 (A-C), myc-K8 (D-F) or K18 (G-I), as in Figure 1. (A-C) The Dys-ABD (A; red in C) accumulated in large punctate structures formed by hK19 (B; green in C). (D-F) The Dys-ABD (D; red in F) failed to accumulate to a significant extent in the structures containing myc-K8 (E; green in F). (G-I) The Dys-ABD (G; red in I) failed to accumulate to a significant extent in the filaments containing K18 (H; green in I). Yellow structures in C and F and I contained both markers indicated in each color overlay. Bars, 5 μm.
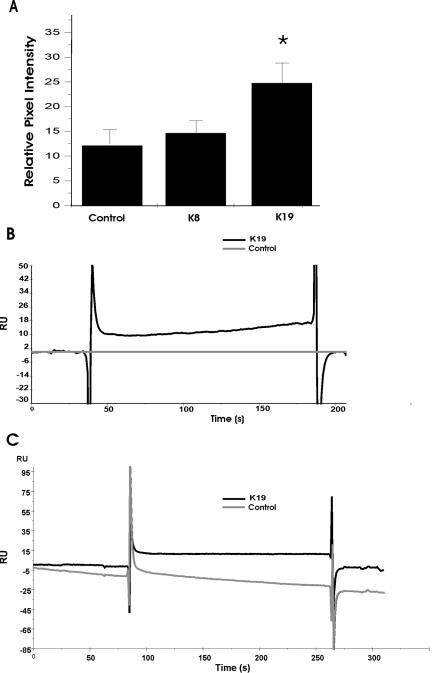
We confirmed the association of the Dys-ABD for K19 in dot blot assays. We expressed either K8 or K19 alone in COS-7 cells and then applied the soluble fractions, obtained from the transfected cultures, to dot blots. The blots were incubated with a His-tagged form of the Dys-ABD (His-Dys-ABD; see Materials and Methods), which was then detected with antibodies specific for the His epitope. The results (Figure 6A) show that the extract of COS-7 cells containing hK19 retained significantly more (p < 0.05) His-Dys-ABD in this experiment than extracts of control cells or extracts enriched in myc-K8. These results, which we obtained in five separate experiments (p < 0.01, t test), supports the conclusion that K19, but not K8, associates directly or indirectly with the Dys-ABD.
Figure 6.
Association of the actin-binding domain of dystrophin with K19 in vitro. Soluble extracts, prepared from untransfected COS-7 cells, or cells that overexpressed myc-K8 or hK19, were applied to dot blots (A) or to the surface of a sensor chip, for surface plasmon resonance measurements (B). Alternatively, purified keratin subunits, K18 and K19, were applied to a sensor chip for additional surface plasmon resonance experiments (C). Binding of the Dys-ABD to dot blots or to the sensor chip was subsequently assayed. For details, see Materials and Methods. (A) Dot blots prepared with extracts of cells that overexpressed hK19 bound significantly larger amounts of His-tagged Dys-ABD, detected with antibodies to the His epitope tag, than extracts of control cells or of cells that overexpressed myc-K8. (B) Chip surfaces charged with extracts of cells that overexpressed hK19 (dark line) bound His-tagged Dys-ABD specifically. (C) Chip surfaces charged with purified K19 (dark line) bound more His-tagged Dys-ABD (12 RU) than surfaces charged with purified K18 (≤0 RU). The results suggest that K19 binds the Dys-ABD specifically and directly.
We also used surface plasmon resonance, measured with a BIAcore 3000 Biosensor, to examine real time binding of the His-Dys-ABD to extracts of COS-7 cells enriched in hK19. In this experiment, we first coated the surfaces of the two flow cells of the biosensor chip with anti-mouse IgG antibodies. We then introduced a mouse mAb specific for hK19 into both cells, to create surfaces that would selectively retain hK19. The soluble fractions of control cells or cells expressing hK19 were then each applied to one flow cell, followed by recombinant His-Dys-ABD, introduced into both flow cells. The Dys-ABD showed no binding to the control surface, exposed to extracts of untransfected cells (≤1 RU). By contrast, the His-Dys-ABD bound at significant levels (17 RU) to the surface charged with anti-hK19 and exposed to soluble extracts of cells expressing hK19 (Figure 6B, top curve). After the minimal correction for the signal seen in the control (Figure 6B, bottom curve), our results clearly demonstrated binding of the Dys-ABD to the surface charged with K19. We also tried to assay binding of His-Dys-ABD to cell extracts enriched in K8 with this technology, but soluble extracts of COS-7 cells expressing K8 showed irreversible binding to the chip surface, making interpretation and comparison to K19 difficult.
These results are consistent with the idea that the Dys-ABD associates with hK19, but, as they were obtained with cell extracts, we could not conclude if the binding was direct or indirect. We performed a similar experiment with purified K19 to address this question. We bound antibodies to K8 to two flow cells, and antibodies to K18 and K19 to one flow cell each. We introduced purified K18 over the pair of surfaces charged with antibodies to K8 and K18, and purified K19 over the pair charged with antibodies to K8 and K19. This allowed us to compare specific binding of His-Dys-ABD to two type I cytokeratin filaments: K18, which our cotransfection experiments suggested to be a poor ligand, and K19, which our experiments suggested to be the preferred ligand. When we introduced a solution of the His-Dys-ABD into all four flow cells and measured changes in surface plasmon resonance, we found that His-Dys-ABD bound specifically and reversibly to the surface charged with K19 (12 RUs of specific binding; Figure 6C, top curve), compared with K18 (≤0 RUs of specific binding; Figure 6C, bottom curve). We repeated these experiments eight times, with similar results each time (chips binding 73.4 ± 13.2 RUs of K19 retained 9.9 ± 5.5 RUs of the Dys-ABD; both figures are mean ± SE; p < 0.02, one sample t test, for Dys-ABD binding).
The fact that the His-Dys-ABD binds to purified K19 on the surface of the chip as well as to hK19 in soluble extracts of COS-7 cells, suggests that the Dys-ABD interacts directly with this cytokeratin subunit. The rapid time course of binding and dissociation of the his-Dys-ABD to the surface of the biosensor chip charged with K19, as well as the high concentrations of the fusion protein required to observe binding, are consistent with affinities in the micromolar range or higher, the same range documented for the binding of the Dys-ABD to actin (Senter et al., 1993; Winder et al., 1995; Renley et al., 1998; Sutherland-Smith et al., 2003).
Overexpression of K8 and K19 in Muscle Fibers
We overexpressed K8 and K19 in skeletal myofibers to learn whether either of these proteins can influence the organization of the myoplasm, and especially of dystrophin and other proteins at costameres. For these experiments, we injected naked plasmid cDNAs encoding myc-K8 or hK19 into the hindlimbs of young rats (Wolff et al., 1990). After allowing 7-10 d for expression of the exogenous proteins, we fixed the hindlimb muscles in situ and prepared longitudinal frozen sections through the injected regions. We used antibodies to hK19, and to the myc tag carried by our K8 construct, to identify transfected myofibers and to localize the exogenous proteins at the subcellular level.
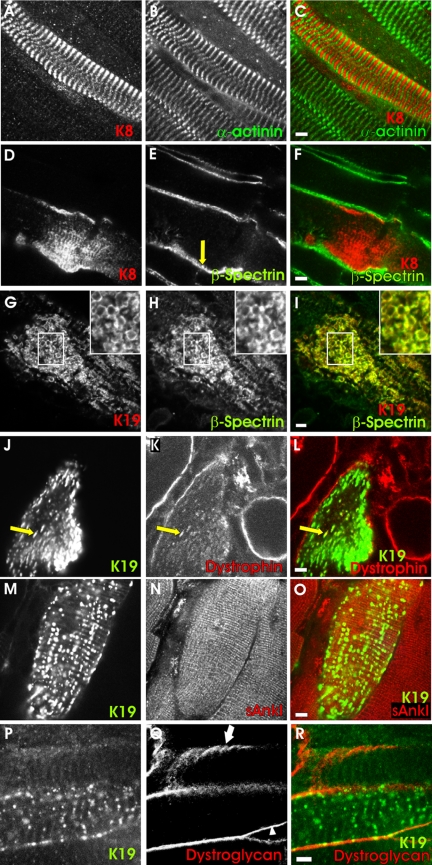
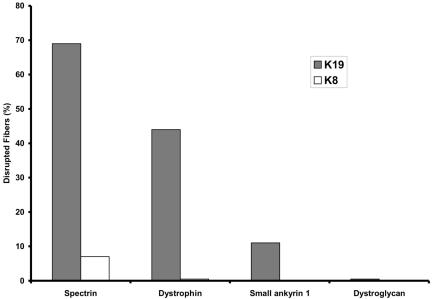
As in COS-7 cells, overexpression of K8 and K19 in skeletal myofibers yielded distinct results. K8 had no significant effect on myoplasmic organization but instead associated with myofibrillar structures in a regular, periodic manner (Figure 7). Colabeling with antibodies to α-actinin showed these structures to be located in the middle of sarcomeres (Figure 7, A-C), possibly at M-lines. Overexpression of K8 had no effect on the distributions of either β-spectrin (Figure 7, D-F) or dystrophin (our unpublished data) at the sarcolemma, which remained in punctate structures associated with costameres, typical of control myofibers (Figure 7E, arrow; Porter et al., 1992; Williams and Bloch, 1999a,b). By contrast, overexpression of hK19 produced irregular aggregates of this protein in the myoplasm and altered the distribution of dystrophin and β-spectrin. Many of the fibers expressing exogenous K19 showed neither β-spectrin nor dystrophin in punctate sarcolemmal structures typical of costameres (Figure 7, G-I, J-L). Instead, these proteins accumulated in intracellular aggregates (Figure 7, J-L, arrow). Some fibers also concentrated K19 intracellularly at the periphery of round structures that resembled vesicles (Figure 7G, inset). These membranelike profiles always contained β-spectrin (Figure 7, H and I, inset) and sometimes contained dystrophin (our unpublished data). Quantitation of the differences between the distributions of dystrophin and β-spectrin in muscle fibers overexpressing K8 and K19 (Figure 8) showed them to be highly significant (p < 0.0002, Fisher's exact test).
Figure 7.
Overexpression of K8 or K19 in skeletal muscle fibers. T. anterior muscles of young rats were injected with plasmid DNA encoding myc-K8 (A-F) or hK19 (G-R). After 7-10 d, muscles were fixed, collected, sectioned and immunolabeled for the exogenous cytokeratin (A, D, G, J, M, and P) and for several endogenous proteins: α-actinin (B), normally at the Z-disks of the contractile apparatus; dystrophin (K) and βI-spectrin (E and H) or β-dystroglycan (Q), normally at costameres at the sarcolemma; and small ankyrin 1 (N), normally in the network sarcoplasmic reticulum. Transfected fibers were identified under fluorescence optics by the presence of the exogenous cytokeratin. (A-C, D-F) Overexpression of myc-K8 (A and D; red in C and F) has no effect on the organization of α-actinin, at Z-disks (B; green in C) or of βI-spectrin, in punctate, costameric structures at the sarcolemma (E; green in F). myc-K8 itself incorporates into striations (A and D). (G-I, J-L, M-O) Overexpression of hK19 (G, J, and M; red in I; green in L and O) disrupted the sarcolemmal pattern of labeling for βI-spectrin (H; green in I) and dystrophin (K; red in L), but not small ankyrin 1 (N, red in O). (P-R) Overexpression of hK19 (P; green in R) did not displace β-dystroglycan (Q; red in R) from the sarcolemma, but it disrupted the normally costameric distribution of β-dystroglycan (arrowhead). Untransfected fibers showed β-dystroglycan in costameres (arrow). Yellow in the color overlays indicates structures labeled for both of the markers indicated. The results show that overexpression of K19, but not K8, can displace βI-spectrin and dystrophin, but not β-dystroglycan, from the sarcolemma into the myoplasm, simultaneously altering the organization of the sarcolemma. Bars, 5 μm.
Figure 8.
Quantitation of the effects of overexpressing K8 and K19 in skeletal myofibers. Transfected myofibers, such as those shown in Figure 7, were counted and scored for the presence or absence of normal structures in the myoplasm (small ankyrin 1, K19 only, n = 21) or at the sarcolemma (dystrophin, n = 36 for K19, n = 22 for K8; βI-spectrin, n = 26 for K19, n = 28 for K8; β-dystroglycan, K19 only, n = 20). Differences in the effects of K19 and K8 on the organization of dystrophin and βI-spectrin were highly significant (p < 0.0002).
Not all membrane markers were affected by the overexpression of K19, however. For example, the organization of small ankyrin 1 in the sarcoplasmic reticulum around Z-disks and M-lines (Zhou et al., 1997) remained largely unchanged in myofibers expressing exogenous K19 (Figures 7, M-O, and 8). Perhaps more remarkably, the distribution of β-dystroglycan between the sarcolemma and the myoplasm was also unaffected by overexpression of K19 (Figures 7R and 8). β-Dystroglycan remained concentrated at the sarcolemma in all 20 transfected myofibers we examined that contained K19 in aggregates or vesicle-like structures in the myoplasm; it never accumulated intracellularly. The absence of small ankyrin 1 and β-dystroglycan from intracellular aggregates was significantly different from the results obtained for dystrophin and β-spectrin (p < 0.01, Fisher's exact test). The distribution of β-dystroglyan within the sarcolemma was altered by overexpression of K19, however. In particular, its typical distribution in costameres tended to be lost. We found costameric patterns of labeling for β-dystroglycan in only six of the 20 transfected myofibers we examined, whereas 19 of the 26 nearby, untransfected fibers in the same microscopic fields showed β-dystroglycan in costameres (Figure 7P, arrow and arrowhead). These differences were highly significant (p < 0.01, Fisher's exact test). These results, as well as our results for small ankyrin 1, suggest that the overexpression of K19 has limited and specific, not pleiotropic, effects on the organization of myofibers.
DISCUSSION
The sarcolemma of fast-twitch skeletal muscle fibers is organized into costameres that are linked to the Z-disks and M-lines of the underlying contractile apparatus, and, in some fibers, are also present in longitudinally oriented structures. Because the disruption of costameres may contribute to the instability of the sarcolemma observed in various myopathies and muscular dystrophies, we have been studying their organization and the biochemical basis for their formation. Previous results suggest that both actin and desmin are involved in linking the contractile apparatus to the sarcolemma (Rybakova et al., 2000; O'Neill et al., 2002), but because these proteins are concentrated only at the level of Z-disks, they are unlikely to be able to link the M-line and longitudinally oriented domains of costameres to the contractile apparatus. We have cloned K8 and K19 from striated muscle and shown them to be present at all three domains of costameres (Ursitti et al., 2004). We postulated that the filaments formed by K8 and K19 could interact directly with the N-terminal ABD of dystrophin, much as neurofilaments, and vimentin and plectin, associate with the ABDs of βII-spectrin and fimbrin, respectively (Correia et al., 1999; Macioce et al., 1999; Sevcik et al., 2004). Here, we confirm this prediction, characterize the specificity of the association, and demonstrate its potential importance in situ.
Many of our experiments use transfection of COS-7 cells to examine interactions between cytokeratins and ABDs under conditions prevalent in mammalian cytoplasm. Although COS cells fail to show a clear association of full-length dystrophin with actin filaments (Lee et al., 1991), they have proved useful for our studies because the distribution of ABDs, and especially the Dys-ABD (expressed without the rod and C-terminal regions), can be easily localized to actin or other filamentous structures. Untransfected COS-7 cells have intermediate filaments, of course, but unlike K8/K19 filaments, these do not associate avidly with the Dys-ABD, although they do so occasionally (Ursitti et al., 2004), perhaps due to the presence of low levels of K19 or other subunits to which it can bind (see below). More frequently, the Dys-ABD codistributes with actin in stress fibers and at the cell periphery. Both the endogenous intermediate filaments and actin-rich structures are easily distinguished from the structures formed after overexpression of cytokeratins. This facilitated the discovery of how efficiently the Dys-ABD colocalized with filaments formed by the overexpression of K8 and K19.
The association of the Dys-ABD with actin in endogenous stress fibers and at the plasma membrane of most COS-7 cells is consistent with in vitro studies of its ability to bind to actin and to cross-link microfilaments (Senter et al., 1993; Renley et al., 1998; Sutherland-Smith et al., 2003). Despite its cross-linking activity, linked to its ability to dimerize (Sutherland-Smith et al., 2003), the Dys-ABD does not significantly alter the organization of K8/K19 filaments, suggesting that the Dys-ABD does not cross-link these filaments to each other. It also seems unable to cross-link K8/K19 filaments to actin (Figure 1P), suggesting that it is incapable of binding to both types of filamentous proteins simultaneously. Instead, it associates preferentially with K19, without changing the distribution of the aggregates or filaments formed by this cytokeratin.
The specificity of the Dys-ABD for K19 and the filaments it forms with K8 is remarkable, indeed. The Dys-ABD does not associate to a significant extent with filaments composed of desmin or of K8 paired with K18 (like K19, a type I cytokeratin), nor does the homologous ABD of βI-spectrin associate with K8/K19 filaments. The ability of the Dys-ABD to bind purified K19 in vitro indicates that binding is direct as well as specific, but we cannot rule out the possibility that other endogenous proteins promote their association in vivo. Although, in addition to K8, K19 also can copolymerize with K5 and K7 (Coulombe and Omary, 2002), we are not aware of any evidence of K5 or K7 in striated muscle and have so far been unsuccessful in detecting transcripts encoding K5 in that tissue (Ursitti, Lee, McNally, and Bloch, unpublished data). More extensive studies of the cytokeratins and their roles in muscle are clearly needed.
The basis for the specificity of binding of the Dys-ABD to K19 remains unclear. One possibility is that the Dys-ABD shares some homology with sequences within the cytokeratins that are required for oligomerization. We found significant homology between amino acids 42-65 of the Dys-ABD and two distinct sequences shared by K19 (amino acids 180-209 and 294-320), and by K8 (amino acids 190-219 and 305-332) (Figure 9). Although these sequences share other key features, we suggest the term “I/LE(G)L motif “ for this region, for the amino acid residues that are conserved in four of the five sequences (Figure 9; the G is in parentheses to indicate that it is missing in one sequence). Although not unique to cytokeratins 8 and 19, these amino acids are not conserved in human K18, with which K8 can heterodimerize, or in human K5 or K7, with which K19 can heterodimerize. It is also absent in other classes of intermediate filament proteins, including type III proteins (desmin, vimentin, synemin, or GFAP), neurofilaments (L, M, or H) and nuclear lamins, consistent with the specificity we have described for the interaction of Dys-ABD with K8/K19 filaments. Notably, the I/LE(G)L motif is present only in the first, more N-terminal CH domain of the Dys-ABD. Preliminary experiments confirm that this domain, and not the more COOH-terminal CH domain of the Dys-ABD, has binding activity for K19, in agreement with a recent report that the first, more N-terminal CH domain of plectin harbors the binding activity of the plectin ABD for vimentin (Sevcik et al., 2004).
Figure 9.
Sequence homologies between the Dys-ABD and cytokeratins 8 and 19. The sequence of the C-helix and nearby residues of the Dys-ABD (human, accession number NP031894) was compared with the sequences of K8 (human, accession number P05787) and K19 (human, accession number NP002267). Two regions of homology were identified in each cytokeratin and aligned to show the I/LE(G)L motif (underlined) and other conserved residues. Residues in bold italics are conserved among K8, K19, and the Dys-ABD. Residues in Bold are conserved only between K8 and the Dys-ABD. Residues in italics are conserved only between K19 and the Dys-ABD. The numbers to the right indicate the number of homologies found, out of the total number of residues compared in each pair of sequences.
Structural studies show that the sequences containing the I/LE(G)L motifs of the Dys-ABD, K8, and K19 are all helical, although the flanking N- and C-terminal residues in the Dys-ABD are in nonhelical linkers (Norwood et al., 2000). The I/LE(G)L motif of the Dys-ABD, contained in its third or C-helix, has not been implicated in actin binding (Norwood et al., 2000; Gimona et al., 2002), nor is it present in human actins. It contains two residues, Q45 and D52, that distinguish it from the homologous region in the ABD of βI-spectrin and that may account in part for its preference for the cytokeratins. The Dys-ABD shares D52, but not Q45, with the ABD of utrophin, suggesting that this residue may contribute to cytokeratin binding. Both these side chains are exposed to the solvent in a shallow groove on the surface of the Dys-ABD (Norwood et al., 2000) and so could help form a binding site. Site-directed mutagenesis will be required to test this idea further.
Whatever confers the specificity of the Dys-ABD for K19, our results suggest that the binding is of only moderate affinity. Values in the low micromolar range are consistent with the rapid on- and off-rates for binding observed in surface plasmon resonance experiments at the micromolar concentrations of reagents we used (Figure 6) as well as with the results of our dot blots. This range is comparable to the affinity of the Dys-ABD for actin filaments of ∼10-50 μM (Senter et al., 1993; Winder et al., 1995; Renley et al., 1998; Sutherland-Smith et al., 2003). The affinity of intact, full-length dystrophin for actin is considerably higher, due to the presence of clusters of basic residues in its spectrin repeat region that greatly enhance actin binding (Amann et al., 1998; Warner et al., 2002). We have not yet assayed the other regions of dystrophin for their possible contributions to binding to cytokeratin filaments, nor have we examined any of the proteins that are normally complexed with dystrophin (Ahn and Kunkel, 1993; Matsumura and Campbell, 1994; Ozawa et al., 1998) for similar activity. Nevertheless, the comparable affinities of the Dys-ABD for cytokeratin and actin filaments suggest that the severity of dystrophinopathies linked to mutations in the ABD may be due to changes in binding not only to actin but also to cytokeratins. This may explain the ability of a mutation (L54R) in the C-helix of the first CH domain of the Dys-ABD to cause Duchenne Muscular Dystrophy (Prior et al., 1993).
The similarities and differences between the effects of overexpressing K8 and K19 alone in COS-7 cells and in muscle fibers are significant and revealing. In both types of cells, K8 incorporates into endogenous structures, whereas K19 forms intracellular aggregates. K19 in both cell types can associate with dystrophin (myofibers) or its ABD (COS-7), whereas K8 is targeted to other structures. This suggests that the COS-7 cell is a valid model in which to study some aspects of the behavior of cytokeratins of muscle. Nevertheless, the differences in the behavior of K8 and K19 in myofibers are considerable and potentially relevant to understanding the architecture of striated muscle cells in health and disease.
Unlike its activity in COS-7 cells, K8 overexpressed in skeletal myofibers seems to be unable to incorporate into endogenous cytokeratin filaments, which in the interior of striated muscle fibers are concentrated around Z-disks (Ursitti et al., 2004). The fact that exogenous K8 concentrates midway between Z-disks suggests that A-bands or M-lines may harbor binding sites for this cytokeratin. Although this would be consistent with the presence of K8 at the domains of costameres that are linked to M-lines in underlying myofibrils, it does not explain its absence from the keratin filaments surrounding Z-disks. Access to those sites may be limited, perhaps by high local protein densities or by slow turnover of endogenous structures. More insight into these questions should be obtained through the identification of the proteins, other than the type I cytokeratin subunits, with which K8 interacts in striated muscle.
When expressed at high levels in myofibers, K19 disrupts costameres. Although we cannot rule out other plausible explanations for its activity, the ability of K19 to displace dystrophin from the sarcolemma in vivo is consistent with our studies of the association of the Dys-ABD to K19 in COS-7 cells. Whatever the mechanism, our results suggest that K19 can compete with the normal binding sites for dystrophin, which would otherwise concentrate at the sarcolemma. Because previous studies have shown that the costameric distribution of dystrophin-associated proteins, including β-dystroglycan, is compromised when dystrophin is absent from the sarcolemma in dystrophic muscle (Williams and Bloch, 1999b), it is not surprising that the ability of β-dystroglycan to concentrate at costameres is altered in myofibers that overexpress K19. Dystroglycan also redistributes in the plane of the sarcolemma in other muscular dystrophies and myopathies (O'Neill et al., 2002; Reed et al., 2004).
Our finding that β-dystroglycan is absent from the vesicular structures that occur in many of the fibers that overexpress K19 is unexpected, however. It suggests that these vesicles, present in a subset of fibers that overexpress K19, represent a distinct membrane population that associates with membrane-cytoskeletal proteins, such as β-spectrin and dystrophin, but that does not contain all of the integral membrane proteins that anchor these proteins at the sarcolemma. The presence of β-spectrin at these structures is unlikely to be due to interactions of that molecule or its ABD with K19 directly (Figure 2, G-I), but is instead more likely linked to the ability of β-spectrin to associate with muscle membranes in a manner that is independent of dystrophin (Porter et al., 1992; Ehmer et al., 1997; Williams and Bloch, 1999b). Further research will be needed to establish the nature of the aggregates and vesicles induced by the overexpression of K19, the mechanism by which they form, and their relationship to structures seen in myopathies (De Bleecker et al., 1993).
The ability of excess K19 to displace membrane-cytoskeletal proteins from the sarcolemma, to disrupt costameres, and to induce cytoplasmic aggregates and vesicles, reminiscent of vesicular myopathies (De Bleecker et al., 1993), raises the possibility that some muscular dystrophies or myopathies may be caused not only by changes in the binding of the cytokeratins to the Dys-ABD but also by changes in their levels of expression, or by mutations that affect their ability to bind to each other. Studies of skeletal and cardiac muscle of mice lacking K8 or K19, due to homologous recombination, are now in progress in our laboratory to test this idea.
Acknowledgments
We thank C. Antolik, J. Strong, A. Bowman, and Drs. J. A. Ursitti and A. Kontrogianni-Konstantopoulos for useful discussions; D. W. Pumplin for assistance with statistical analyses; and Drs. M. Bishr Omary and D. Toivola for comments on an earlier version of this article. Our research has been supported by grants to R.J.B. from the National Institutes of Health (NS 17282 and NS43976) and the Muscular Dystrophy Association and by fellowships to M.R.S. provided by training grants T32 NS 07375 (P.I., Dr. B. E. Alger) and T32 HL 072751 (P.I., Dr. C. W. Balke).
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05-02-0112) on July 6, 2005.
Abbreviations used: ABD, actin-binding domain; Dys-ABD, actin-binding domain of dystrophin; K8, cytokeratin 8; K19, cytokeratin 19; hK19, human cytokeratin 19; PBS, phosphate-buffered saline; RU, resonance unit.
References
- Ahn, A. H., and Kunkel, L. M. (1993). The structural and functional diversity of dystrophin. Nat. Genet. 3, 283-291. [DOI] [PubMed] [Google Scholar]
- Amann, K. J., Renley, B. A., and Ervasti, J. M. (1998). A cluster of basic repeats in the dystrophin rod domain binds F-actin through an electrostatic interaction. J. Biol. Chem. 273, 28419-28423. [DOI] [PubMed] [Google Scholar]
- Bellin, R. M., Huiatt, T. W., Critchley, D. R., and Robson, R. M. (2001). Synemin may function to directly link muscle cell intermediate filaments to both myofibrillar Z-lines and costameres. J. Biol. Chem. 276, 32330-32337. [DOI] [PubMed] [Google Scholar]
- Bellin, R. M., Sernett, S. W., Becker, B., Ip, W., Huiatt, T. W., and Robson, R. M. (1999). Molecular characteristics and interactions of the intermediate filament protein synemin. Interactions with α-actinin may anchor synemin-containing heterofilaments. J. Biol. Chem. 274, 29493-29499. [DOI] [PubMed] [Google Scholar]
- Bloch, R. J., and Gonzalez-Serratos, H. (2003). Lateral force transmission across costameres in skeletal muscle. Exerc. Sport Sci. Rev. 31, 73-78. [DOI] [PubMed] [Google Scholar]
- Campbell, K. P. (1995). Three muscular dystrophies: loss of cytoskeleton-extracellular matrix linkage. Cell 80, 675-679. [DOI] [PubMed] [Google Scholar]
- Capetanaki, Y., and Milner, D. J. (1998). Desmin cytoskeleton in muscle integrity and function. Subcell. Biochem. 31, 463-495. [PubMed] [Google Scholar]
- Corrado, K., Rafael, J. A., Mills, P. L., Cole, N. M., Faulkner, J. A., Wang, K., and Chamberlain, J. S. (1996). Transgenic mdx mice expressing dystrophin with a deletion in the actin-binding domain display a “mild Becker” phenotype. J. Cell Biol. 134, 873-884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correia, I., Chu, D., Chou, Y. H., Goldman, R. D., and Matsudaira, P. (1999). Integrating the actin and vimentin cytoskeletons. Adhesion-dependent formation of fimbrin-vimentin complexes in macrophages. J. Cell Biol. 146, 831-842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulombe, P. A., and Omary, M. B. (2002). `Hard'and `soft' principles defining the structure, function and regulation of keratin intermediate filaments. Curr. Opin. Cell Biol. 14, 110-122. [DOI] [PubMed] [Google Scholar]
- De Bleecker, J. L., Engel, A. G., and Winkelmann, J. C. (1993). Localization of dystrophin and beta-spectrin in vacuolar myopathies. Am. J. Pathol. 143, 1200-1208. [PMC free article] [PubMed] [Google Scholar]
- Ehmer, S., Herrmann, R., Bittner, R., and Voit, T. (1997). Spatial distribution of β-spectrin in normal and dystrophic human skeletal muscle. Acta Neuropathol. 94, 240-246. [DOI] [PubMed] [Google Scholar]
- Emery, A.E.H. (1993). Duchenne Muscular Dystrophy, Oxford: Oxford University Press.
- Eraslan, S., Kayserili, H., Apak, M. Y., and Kirdar, B. (1999). Identification of point mutations in Turkish DMD/BMD families using multiplex-single stranded conformation analysis (SSCA). Eur. J. Hum. Genet. 7, 765-770. [DOI] [PubMed] [Google Scholar]
- Ervasti, J. M. (2003). Costameres: the Achilles' heel of Herculean muscle. J. Biol. Chem. 278, 13591-13594. [DOI] [PubMed] [Google Scholar]
- Ervasti, J. M., and Campbell, K. P. (1993). A role for the dystrophin-glycoprotein complex as a transmembrane linker between laminin and actin. J. Cell Biol. 122, 809-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs, E., and Weber, K. (1994). Intermediate filaments: structure, dynamics, function, and disease. Annu. Rev. Biochem. 63, 345-382. [DOI] [PubMed] [Google Scholar]
- Galkin, V. E., Orlova, A., VanLoock, M. S., Rybakova, I. N., Ervasti, J. M., and Egelman, E. H. (2002). The utrophin actin-binding domain binds F-actin in two different modes: implications for the spectrin superfamily of proteins. J. Cell Biol. 157, 243-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimona, M., Djinovic-Carugo, K., Kranewitter, W. J., and Winder, S. J. (2002). Functional plasticity of CH domains. FEBS Lett. 513, 98-106. [DOI] [PubMed] [Google Scholar]
- Granger, B. L., and Lazarides, E. (1978). The existence of an insoluble Z disc scaffold in chicken skeletal muscle. Cell 15, 1253-1268. [DOI] [PubMed] [Google Scholar]
- Granger, B. L., and Lazarides, E. (1979). Desmin and vimentin coexist at the periphery of the myofibril Z disc. Cell 18, 1053-1063. [DOI] [PubMed] [Google Scholar]
- Granger, B. L., Repasky, E. A., and Lazarides, E. (1982). Synemin and vimentin are components of intermediate filaments in avian erythrocytes. J. Cell Biol. 92, 299-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hijikata, T., Murakami, T., Ishikawa, H., and Yorifuji, H. (2003). Plectin tethers desmin intermediate filaments onto subsarcolemmal dense plaques containing dystrophin and vinculin. Histochem. Cell Biol. 119, 109-123. [DOI] [PubMed] [Google Scholar]
- Hirako, Y., Yamakawa, H., Tsujimura, Y., Nishizawa, Y., Okumura, M., Usukura, J., Matsumoto, H., Jackson, K. W., Owaribe, K., and Ohara, O. (2003). Characterization of mammalian synemin, an intermediate filament protein present in all four classes of muscle cells and some neuroglial cells: co-localization and interaction with type III intermediate filament proteins and keratins. Cell Tissue Res. 313, 195-207. [DOI] [PubMed] [Google Scholar]
- Ibraghimov-Beskrovnaya, O., Ervasti, J. M., Leveille, C. J., Slaughter, C. A., Sernett, S. W., and Campbell, K. P. (1992). Primary structure of dystrophin-associated glycoproteins linking dystrophin to the extracellular matrix. Nature 355, 696-702. [DOI] [PubMed] [Google Scholar]
- Khurana, T. S., Hoffman, E. P., and Kunkel, L. M. (1990). Identification of a chromosome 6-encoded dystrophin-related protein. J. Biol. Chem. 265, 16717-16720. [PubMed] [Google Scholar]
- Khurana, T. S., Watkins, S. C., Chafey, P., Chelly, J., Tome, F. M., Fardeau, M., Kaplan, J.-C., and Kunkel, L. M. (1991). Immunolocalization and developmental expression of dystrophin related protein in skeletal muscle. Neuromusc. Disord. 1, 185-194. [DOI] [PubMed] [Google Scholar]
- Kosmehl, H., Langbein, L., and Katenkamp, D. (1990). Transient cytokeratin expression in skeletal muscle during murine embryogenesis. Anat. Anz. 171, 39-44. [PubMed] [Google Scholar]
- Langbein, L., Kosmehl, H., Kiss, F., Katenkamp, D., and Neupert, G. (1989). Cytokeratin expression in experimental murine rhabdomyosarcomas. Intermediate filament pattern in original tumors, allotransplants, cell culture and re-established tumors from cell culture. Exp. Pathol. 36, 23-36. [DOI] [PubMed] [Google Scholar]
- Lapidos, K. A., Kakkar, R., and McNally, E. M. (2004). The dystrophin glycoprotein complex: signaling strength and integrity for the sarcolemma. Circ. Res. 94, 1023-1031. [DOI] [PubMed] [Google Scholar]
- Lazarides, E. (1978). The distribution of desmin (100 A) filaments in primary cultures of embryonic chick cardiac cells. Exp. Cell Res. 112, 265-273. [DOI] [PubMed] [Google Scholar]
- Lazarides, E. (1980). Intermediate filaments as mechanical integrators of cellular space. Nature 283, 249-256. [DOI] [PubMed] [Google Scholar]
- Lee, C. C., Pearlman, J. A., Chamberlain, J. S., and Caskey, C. T. (1991). Expression of recombinant dystrophin and its localization to the cell membrane. Nature 349, 334-336. [DOI] [PubMed] [Google Scholar]
- Love, D. R., Hill, D. F., Dickson, G., Spurr, N. K., Byth, B. C., Marsden, R. F., Walsh, F. S., Edwards, Y. H., and Davies, K. E. (1989). An autosomal transcript in skeletal muscle with homology to dystrophin. Nature 339, 55-58. [DOI] [PubMed] [Google Scholar]
- Macioce, P., Gandolfi, N., Leung, C. L., Chin, S. S., Malchiodi-Albedi, F., Ceccarini, M., Petrucci, T. C., and Liem, R. K. (1999). Characterization of NF-L and betaIIsigma1-spectrin interaction in live cells. Exp. Cell Res. 250, 142-154. [DOI] [PubMed] [Google Scholar]
- Masuda, T., Fujimaki, N., Ozawa, E., and Ishikawa, H. (1992). Confocal laser microscopy of dystrophin localization in guinea pig skeletal muscle fibers. J. Cell Biol. 119, 543-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumura, K., and Campbell, K. P. (1994). Dystrophin-glycoprotein complex: its role in the molecular pathogenesis of muscular dystrophies. Muscle & Nerve 17, 2-15. [DOI] [PubMed] [Google Scholar]
- Michele, D. E., et al. (2002). Post-translational disruption of dystroglycan-ligand interactions in congenital muscular dystrophies. Nature 418, 417-422. [DOI] [PubMed] [Google Scholar]
- Miettinen, M., and Rapola, J. (1989). Immunohistochemical spectrum of rhabdomyosarcoma and rhadomyosarcoma-like tumors. Expression of cytokeratin and the 68-kD neurofilament protein. Am. J. Surg. Pathol. 13, 120-132. [DOI] [PubMed] [Google Scholar]
- Minetti, C., Beltrame, F., Marcenaro, G., and Bonilla, E. (1992). Dystrophin at the plasma membrane of human muscle fibers shows a costameric localization. Neuromusc. Disord. 2, 99-109. [DOI] [PubMed] [Google Scholar]
- Mizuno, Y., Guyon, J. R., Watkins, S. C., Mizushima, K., Sasaoka, T., Imamura, M., Kunkel, L. M., and Okamoto, K. (2004). Beta-synemin localizes to regions of high stress in human skeletal myofibers. Muscle Nerve 30, 337-346. [DOI] [PubMed] [Google Scholar]
- Mizuno, Y., Thompson, T. G., Guyon, J. R., Lidov, H. G., Brosius, M., Imamura, M., Ozawa, E., Watkins, S. C., and Kunkel, L. M. (2001). Desmuslin, an intermediate filament protein that interacts with alpha-dystrobrevin and desmin. Proc. Natl. Acad. Sci. USA 98, 6156-6161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mokri, B., and Engel, A. G. (1975). Duchenne dystrophy: electron microscopic findings pointing to a basic or early abnormality in the plasma membrane of the muscle fiber. Neurology 25, 1111-1120. [DOI] [PubMed] [Google Scholar]
- Newey, S. E., Howman, E. V., Ponting, C. P., Benson, M. A., Nawrotzki, R., Loh, N. Y., Davies, K. E., and Blake, D. J. (2001). Syncoilin, a novel member of the intermediate filament superfamily that interacts with alpha-dystrobrevin in skeletal muscle. J. Biol. Chem. 276, 6645-6655. [DOI] [PubMed] [Google Scholar]
- Niessen, C. M., Hulsman, E. H., Rots, E. S., Sanchez-Aparicio, P., and Sonnenberg, A. (1997). Integrin alpha 6 beta 4 forms a complex with the cytoskeletal protein HD1 and induces its redistribution in transfected COS-7 cells. Mol. Biol. Cell 8, 555-566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norwood, F. L., Sutherland-Smith, A. J., Keep, N. H., and Kendrick-Jones, J. (2000). The structure of the N-terminal actin-binding domain of human dystrophin and how mutations in this domain may cause Duchenne or Becker muscular dystrophy. Structure 8, 481-491. [DOI] [PubMed] [Google Scholar]
- O'Neill, A., Williams, M. W., Resneck, W. G., Milner, D. J., Capetanaki, Y., and Bloch, R. J. (2002). Sarcolemmal organization in skeletal muscle lacking desmin: evidence for cytokeratins associated with the membrane skeleton at costameres. Mol. Biol. Cell 13, 2347-2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlova, A., Rybakova, I. N., Prochniewicz, E., Thomas, D. D., Ervasti, J. M., and Egelman, E. H. (2001). Binding of dystrophin's tandem calponin homology domain to F-actin is modulated by actin's structure. Biophys. J. 80, 1926-1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozawa, E., Noguchi, S., Mizuno, Y., Hagiwara, Y., and Yoshida, M. (1998). From dystrophinopathy to sarcoglycanopathy: evolution of a concept of muscular dystrophy. Muscle Nerve 21, 421-438. [DOI] [PubMed] [Google Scholar]
- Pierobon-Bormioli, S. (1981). Transverse sarcomere filamentous systems: `Z- and M-cables'. J. Musc. Res. Cell Motil. 2, 401-413. [Google Scholar]
- Porter, G. A., Dmytrenko, G. M., Winkelmann, J. C., and Bloch, R. J. (1992). Dystrophin colocalizes with beta-spectrin in distinct subsarcolemmal domains in mammalian skeletal muscle. J. Cell Biol. 117, 997-1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prior, T. W., Papp, A. C., Snyder, P. J., Burghes, A. H., Bartolo, C., Sedra, M. S., Western, L. M., and Mendell, J. R. (1993). A missense mutation in the dystrophin gene in a Duchenne muscular dystrophy patient. Nat. Genet. 4, 357-360. [DOI] [PubMed] [Google Scholar]
- Rando, T. A. (2001). The dystrophin-glycoprotein complex, cellular signaling, and the regulation of cell survival in the muscular dystrophies. Muscle Nerve 24, 1575-1594. [DOI] [PubMed] [Google Scholar]
- Reed, P., Matthews, K. D., Mills, K. A., and Bloch, R. J. (2004). The sarcolemma in the largemyd mouse. Muscle Nerve 30, 585-595. [DOI] [PubMed] [Google Scholar]
- Renley, B. A., Rybakova, I. N., Amann, K. J., and Ervasti, J. M. (1998). Dystrophin binding to nonmuscle actin. Cell Motil. Cytoskeleton 41, 264-270. [DOI] [PubMed] [Google Scholar]
- Richardson, F. L., Stromer, M. H., Huiatt, T. W., and Robson, R. M. (1981). Immunoelectron and immunofluorescence localization of desmin in mature avian muscles. Eur. J. Cell Biol. 26, 91-101. [PubMed] [Google Scholar]
- Rybakova, I. N., Patel, J. R., Davies, K. E., Yurchenco, P. D., and Ervasti, J. M. (2002). Utrophin binds laterally along actin filaments and can couple costameric actin with sarcolemma when over-expressed in dystrophin-deficient muscle. Mol. Biol. Cell 13, 1512-1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybakova, I. N., Patel, J. R., and Ervasti, J. M. (2000). The dystrophin complex forms a mechanically strong link between the sarcolemma and costameric actin. J. Cell Biol. 150, 1209-1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E. F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual, Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
- Senter, L., Luise, M., Presotto, C., Betto, R., Teresi, A., Ceoldo, S., and Salviati, G. (1993). Interaction of dystrophin with cytoskeletal proteins: binding to talin and actin. Biochem. Biophys. Res. Commun. 192, 899-904. [DOI] [PubMed] [Google Scholar]
- Sevcik, J., Urbanikova, L., Kost'an, J., Janda, L., and Wiche, G. (2004). Actin-binding domain of mouse plectin: crystal structure and binding to vimentin. Eur. J. Biochem. 271, 1873-1884. [DOI] [PubMed] [Google Scholar]
- Shear, C. R., and Bloch, R. J. (1985). Vinculin in subsarcolemmal densities in chicken skeletal muscle: localization and relationship to intracellular and extracellular structures. J. Cell Biol. 101, 240-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small, J. V., Fürst, D. O., and Thornell, L. E. (1992). The cytoskeletal lattice of muscle cells. Eur. J. Biochem. 208, 559-572. [DOI] [PubMed] [Google Scholar]
- Straub, V., Bittner, R. E., Leger, J. J., and Voit, T. (1992). Direct visualization of the dystrophin network on skeletal muscle fiber membrane. J. Cell Biol. 119, 1183-1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Street, S. F. (1983). Lateral transmission of tension in frog myofibers: a myofibrillar network and transverse cytoskeletal connections are possible transmitters. J. Cell Physiol. 114, 346-364. [DOI] [PubMed] [Google Scholar]
- Sutherland-Smith, A. J., Moores, C. A., Norwood, F. L., Hatch, V., Craig, R., Kendrick-Jones, J., and Lehman, W. (2003). An atomic model for actin binding by the CH domains and spectrin-repeat modules of utrophin and dystrophin. J. Mol. Biol. 329, 15-33. [DOI] [PubMed] [Google Scholar]
- Tinsley, J. M., and Davies, K. E. (1993). Utrophin: a potential replacement for dystrophin? Neuromusc. Disord. 3, 537-539. [DOI] [PubMed] [Google Scholar]
- Tinsley, J. M., Potter, A. C., Phelps, S. R., Fisher, R., Trickett, J. I., and Davies, K. E. (1996). Amelioration of the dystrophic phenotype of mdx mice using a truncated utrophin transgene. Nature 384, 349-353. [DOI] [PubMed] [Google Scholar]
- Ursitti, J. A., Lee, P. C., Resneck, W. G., McNally, M. M., Bowman, A. L., O'Neill, A., Stone, M. R., and Bloch, R. J. (2004). Cloning and characterization of cytokeratins 8 and 19 in adult rat striated muscle: interaction with the dystrophin glycoprotein complex. J. Biol. Chem. 279, 41830-41838. [DOI] [PubMed] [Google Scholar]
- Ursitti, J. A., Martin, L., Resneck, W. G., Chaney, T., Zielke, C., Alger, B. E., and Bloch, R. J. (2001). Spectrins in developing rat hippocampal cells. Dev. Brain Res. 129, 81-93. [DOI] [PubMed] [Google Scholar]
- Warner, L. E., DelloRusso, C., Crawford, R. W., Rybakova, I. N., Patel, J. R., Ervasti, J. M., and Chamberlain, J. S. (2002). Expression of Dp260 in muscle tethers the actin cytoskeleton to the dystrophin-glycoprotein complex and partially prevents dystrophy. Hum. Mol. Genet. 11, 1095-1105. [DOI] [PubMed] [Google Scholar]
- Williams, M. W., and Bloch, R. J. (1999a). Differential distribution of dystrophin and β-spectrin at the sarcolemma of fast twitch skeletal muscle fibers. J. Musc. Res. Cell Motil. 20, 383-393. [DOI] [PubMed] [Google Scholar]
- Williams, M. W., and Bloch, R. J. (1999b). Extensive but coordinated reorganization of the membrane skeleton in myofibers of dystrophic (mdx) mice. J. Cell Biol. 144, 1259-1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, M. W., Resneck, W. G., and Bloch, R. J. (2000). Membrane skeleton of innervated and denervated fast- and slow-twitch muscle. Muscle Nerve 23, 590-599. [DOI] [PubMed] [Google Scholar]
- Winder, S. J., Hemmings, L., Maciver, S. K., Bolton, S. J., Tinsley, J. M., Davies, K. E., Critchley, D. R., and Kendrick-Jones, J. (1995). Utrophin actin binding domain: analysis of actin binding and cellular targeting. J. Cell Sci. 108, 63-71. [DOI] [PubMed] [Google Scholar]
- Wolff, J. A., Malone, R. W., Williams, P., Chong, W., Acsadi, G., Jani, A., and Felgner, P. L. (1990). Direct gene transfer into mouse muscle in vivo. Science 247, 1465-1468. [DOI] [PubMed] [Google Scholar]
- Zhou, D., Birkenmeier, C. S., Williams, M. W., Sharp, J. J., Barker, J. E., and Bloch, R. J. (1997). Small, membrane-bound, alternatively spliced forms of ankyrin 1 associated with the sarcoplasmic reticulum of mammalian skeletal muscle. J. Cell Biol. 136, 621-631. [DOI] [PMC free article] [PubMed] [Google Scholar]