Abstract
We examined the spatio-temporal activity of RhoA in migrating cells and growth factor-stimulated cells by using probes based on the principle of fluorescence resonance energy transfer. In HeLa cells migrating at a low cell density, RhoA was activated both at the contractile tail and at the leading edge. However, RhoA was activated only at the leading edge in MDCK cells migrating as a monolayer sheet. In growth factor-stimulated Cos1 and NIH3T3 cells, the activity of RhoA was greatly decreased at the plasma membrane, but remained high at the membrane ruffles in nascent lamellipodia. These observations are in agreement with the proposed role played by RhoA in stress fiber formation, but they also implicated RhoA in the regulation of membrane ruffling, the induction of which is a typical phenotype of activated Rac. In agreement with this view, dominant negative RhoA was found to inhibit membrane ruffling induced by active Rac. Furthermore, we found that Cdc42 activity was also required for high RhoA activity in membrane ruffles. Finally, we found that mDia1, but not ROCK, was stably associated with membrane ruffles. In conclusion, these results suggested that RhoA cooperates with Rac1 and Cdc42 to induce membrane ruffles via the recruitment of mDia.
INTRODUCTION
Cell migration is an essential process in embryonic development, wound repair, immune surveillance, and tumor cell metastasis, and such migration is known to be regulated by growth factors, the extracellular matrix, and other extracellular components. Such extracellular cues are known to elicit various intracellular responses in the organization of both the actin and the microtubule cytoskeletons, as well as that of vesicular transport pathway and gene transcription (Mitchison and Cramer, 1996; Lauffenburger and Horwitz, 1996; Michaelson et al., 2001). It is generally accepted that the driving force for short-term cell movement is provided by the dynamic reorganization of the actin cytoskeleton, which directs the protrusion of the cell membrane at the front of the cell, as well as retraction at the rear. On the other hand, efficient and long-term migration requires the stabilization of cell polarity, which is achieved through the reorganization of the microtubule cytoskeleton (Lauffenburger and Horwitz, 1996; Gundersen and Cook, 1999; Etienne-Manneville and Hall, 2002; Raftopoulou and Hall, 2004). Therefore, the spatial regulation of the actin as well as the microtubule cytoskeleton is a critical component in the regulation of cell migration.
Rho-family GTPases function as critical molecular switches in transducing extracellular signals to both the actin and microtubule cytoskeleton (Ridley, 2001; Etienne-Manneville and Hall, 2002). For example, RhoA triggers actin stress fiber formation; Rac1 induces lamellipodia and membrane ruffles; and Cdc42 elicits the formation of filopodia, i.e., protrusive actin (Mitchison and Cramer, 1996; Hall, 1998). At the leading edge of motile cells, filopodia, lamellipodia, and membrane ruffles are often recognized, suggesting the local activation of Rac and Cdc42. Previous studies using in vivo probes based on the principle of fluorescent resonance energy transfer (FRET) have demonstrated such localized activation of Rac1 and Cdc42 in migratory cell (Kraynov et al., 2000; Itoh et al., 2002).
On the other hand, Rho is thought to function at the tail of migratory cells by promoting actomyosin contraction and thereby causing retraction forces to retract the cytoplasmic tail. Active Rho (RhoA or RhoC) recruits and activates ROCK, which phosphorylates several cytoskeletal proteins and thereby promotes the contraction of actin stress fibers to generate contractile forces (Kimura et al., 1996; Amano et al., 1997). Because the activity of Rho is suppressed by Rac (Sander et al., 1999; Zondag et al., 2000; Nimnual et al., 2003), it is reasonable to speculate that Rho activity is low at the leading edge of migratory cells. However, Rho may also be active at the front of motile cells: it has been shown that Rho stabilizes microtubules oriented toward the leading edge of cells via its effector, mDia (Nagasaki and Gundersen, 1996; Cook et al., 1998; Palazzo et al., 2001; Wen et al., 2004).
To gain a better understanding of the role played by RhoA in cell migration, we examined the spatio-temporal control of RhoA activity in motile cells. We found that RhoA is activated not only at the rear of motile cells, but also at the front. It appears that RhoA, in concert with Rac and Cdc42, evokes membrane ruffles at the leading edge of the migrating cells.
MATERIALS AND METHODS
FRET Probes and Plasmids
FRET probes for the Rho-family GTPases, namely, Raichu-RhoA/1237X, Raichu-Rac1/1011X, and Raichu-Cdc42/1054X, have been described previously (Itoh et al., 2002; Yoshizaki et al., 2003). The cDNA of a GTPase-deficient Rac1 mutant, Rac1-V12, was subcloned into pCAGGS-ECFP to generate pCAGGS-ECFP-Rac1-V12. Similarly, cDNAs of mutants of Rho-family GTPases, i.e., RhoA-V14, RhoA-N19, Cdc42-V12, Cdc42-N17, and Rac1-N17, as well as that of wild-type Ras, were subcloned into pERedNLS, which, at the 3′-side of the cloning site, contained an internal ribosomal entry site followed by the cDNA of express Red (BD Biosciences, Franklin Lakes, NJ) fused to a nuclear localization signal. The cDNA of Rac1-V12 was also subcloned into pERedMit, which contained an internal ribosomal entry site followed by the cDNA of express Red fused to a mitochondria-targeting signal. Use of these two red fluorescent proteins with different targeting signals allowed us to identify cells that express both of the two expression plasmids. cDNAs of wild-type and mutant mDia1 proteins (Tsuji et al., 2002) were obtained from S. Narumiya (Kyoto University, Japan). KAEDE (Ando et al., 2002) and Dronpa (Ando et al., 2004) were obtained from A. Miyawaki (the Brain Science Institute, RIKEN, Japan). The coding region of mDia1 was subcloned into pCAGGS-Flag-KAEDE, which was an expression vector derived from pCAGGS and contained a Flag tag and the cDNA of the KAEDE in front of the cloning site. pIRM-21-N-WaspCRIB encoding the CRIB domain of N-Wasp (aa 124–274) was reported previously (Kurokawa et al., 2004a).
Cells, Antibodies, and Reagent
Cos1 cells, HeLa cells, and MDCK cells were purchased from the Human Science Research Resources Bank (Sennan-shi, Osaka, Japan). The cells were maintained in DMEM (Sigma, St. Louis, MO) supplemented with 10% fetal bovine serum. Before cell imaging, the medium was exchanged for phenol red-free MEM (Nissui, Tokyo, Japan). Anti-RhoA antibody was purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Epidermal growth factor (EGF) and platelet-derived growth factor (PDGF) were purchased from Sigma. Y27632 was purchased from Calbiochem (La Jolla, CA).
Recombinant C3 Protein
C3 exoenzyme was produced by using the pGEX-6P expression vector (Amersham Biosciences, Piscataway, NJ). The GST-fused C3 proteins were purified by use of glutathione-Sepharose and they were cleaved with PreScission protease (Amersham Biosciences). The purity of the eluted C3 proteins was verified by SDS-PAGE.
Imaging of RhoA Activity in Living Cells Stimulated with Growth Factors
RhoA activity was imaged with Raichu-RhoA probe essentially as described previously (Mochizuki et al., 2001). Cos1 and NIH3T3 cells were plated on a collagen-coated 35-mm-diameter glass-base dish (Asahi Techno Glass Co., Tokyo, Japan), and the cells were transfected with expression plasmids with Polyfect (Qiagen, Valencia, CA). After 24 h, the cells were serum-starved for 6 h and stimulated with 10 ng/ml EGF or 50 ng/ml PDGF. The cells were imaged on an Olympus IX71 inverted microscope (Olympus Optical Co., Tokyo, Japan) equipped with a cooled CCD camera, CoolSNAP HQ (Roper Scientific, Tucson, AZ), and this imaging system was controlled by MetaMorph software (Universal Imaging, West Chester, PA). For dual-emission ratio imaging, we used a 440AF21 excitation filter, a 455DRLP dichroic mirror, and two emission filters, 480AF30 for CFP and 535AF26 for YFP (Omega Optical, Brattleboro, VT), which were alternated by a filter changer. Cells were illuminated with a 75-W xenon lamp through a 12% ND filter (Olympus Optical) and an x60× Plan Apo oil immersion objective lens (Olympus Optical). The exposure time was 0.5 s when binning was set to 4 × 4. In most experiments, the cells were imaged every 30 s for 30 min. After background subtraction, the ratio image of YFP/CFP was created with MetaMorph software and the ratio was used to represent FRET efficiency. Kymographs were generated along 10-pixel-wide box regions oriented in the direction of individual protrusions using MetaMorph software, as described previously (Bear et al., 2002). Each cell imaging session was repeated at least five times in order to confirm the reproducibility of the results.
Imaging of Migrating HeLa Cells
HeLa cells were transfected with plasmids by using Polyfect (Qiagen). Twenty-four hours after transfection, the cells were replated onto a 35-mm-diameter collagen-coated glass-base dish. Starting at 1 h after replating, the cells were imaged as described above.
Wound Healing Assay
MDCK cells were seeded at a high density on a 35-mm-diameter collagen-coated glass-base dish. Expression plasmids of Raichu probes were transfected into the cells with Lipofectamine 2000 (Invitrogen, San Diego, CA). When the cells formed a confluent monolayer, they were wounded by being scraped with a needle, and then the cells were rinsed with phosphate-buffered saline (PBS) and fed with fresh medium containing 10% serum. Beginning at 1 h after wounding, the cells were imaged every 4 min, as described above. In some of the experiments, the cells at the edge of the wound were microinjected with GST (0.2 mg/ml), C3 protein (0.175 mg/ml), or GST-Rhotekin RBD (0.1 mg/ml) and FITC-dextran 30 min before the time-lapse analysis. The cells were then imaged for FITC and phase contrast.
Microinjection
GST (0.2 mg/ml), C3 protein (0.175 mg/ml), or GST-Rhotekin RBD (0.1 mg/ml) was microinjected into Cos1 or MDCK cells using a manipulator set (Micromanipulator 5171 and FemtoJet; Eppendorf, Hamburg, Germany). As a marker for the microinjected cells, 5 mg/ml fluorescein isothiocyanate (FITC)-coupled dextran (Sigma) was coinjected with the fusion proteins.
Imaging of Proteins Tagged with KAEDE and Dronpa
Cos1 cells were plated on collagen-coated 35-mm-diameter glass-base dishes and transfected with plasmids by the use of Polyfect. After 24 h, the cells were serum-starved for 6 h and stimulated with 10 ng/ml EGF. The cells were observed with an Olympus confocal microscope Fluoview FV500 (Olympus Optical Co., Tokyo, Japan) equipped with an Argon laser, an He:Ne laser, a Laser Diode 405 nm laser, and a 100× Plan Apo oil immersion objective lens (Olympus Optical). For the photoconversion of the KAEDE protein, a region of interest was illuminated with an LD laser at 30% of the laser power. For dual-emission ratio imaging of KAEDE protein with the Argon laser for the Green-KAEDE assay, or with the He:Ne laser for the Red-KAEDE assay, we used a dichroic mirror, DM405/488/543, a beam splitter SDM560, and two emission filters, BA505–525 for Green-KAEDE and BA560IF for Red-KAEDE (Olympus Optical Co.). Before imaging of Dronpa fluorescence protein, fluorescence was erased to background levels with a strong 488-nm laser. Dronpa protein was then photoactivated with an illumination of LD laser at 1% of the laser power in a region of interest. For the series of imaging of Dronpa protein, we used a dichroic mirror, DM405/488/543, and an emission filters, BA505–525 (Olympus Optical Co.).
Phalloidin Staining
HeLa cells were fixed with 3.7% formaldehyde in the medium, permeabilized with 0.1% Triton-X 100 in PBS, and stained with Alexa488-conjugated phalloidin (Molecular Probes, Eugene, OR).
Pulldown Analysis of RhoA
GTP-RhoA was quantitated according to the pulldown method modified by Yamaguchi et al. (Yamaguchi et al., 2001; Ren et al., 1999). Briefly, the cells were harvested in lysis buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 30 mM MgCl2, 0.1% Triton X-100, 10% glycerol, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, 1 μg/ml aprotinin, and 1 μg/ml leupeptin) containing GST-Rhotekin-RBD. Cleared lysates were incubated with glutathione-Sepharose beads (Amersham Biosciences) for 1 h at 4°C. Proteins bound to the beads were separated by SDS-PAGE and analyzed by immunoblotting with anti-RhoA monoclonal antibody. Bound antibodies were detected by an ECL chemiluminescence system (Amersham Pharmacia) and analyzed with an LAS-1000 image analyzer (Fuji Film, Toyo, Japan).
RESULTS
RhoA Is Activated at the Leading Edge of Migrating Cells
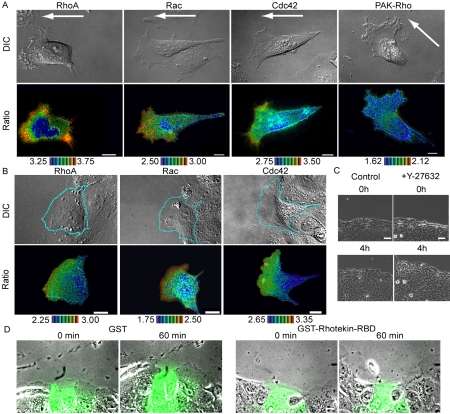
To clarify the role played by RhoA in cell migration, the subcellular distribution of RhoA activity was examined in stochastically migrating HeLa cells by the use of the Raichu-RhoA probe (Figure 1A and Supplementary Video Fig 1HeLa.mov). We found that RhoA was activated not only at the rear, but also at the front of migrating cells, where lamellipodia, membrane ruffles, and filopodia were prominent. Of note, the two terms, lamellipodia and membrane ruffles, are often used interchangeably and both are regarded as signs of Rac activation. Here, we define the sheetlike plasma membrane protrusions as lamellipodia, and plasma membrane folding back and being transported rearward as membrane ruffles, because these two morphological changes are regulated by different mechanisms, as will be described later. Rac1 activity increased gradually toward the tip of the leading edge, whereas Cdc42 activity increased suddenly at the tip of the leading edge, as has been observed in HT1080 cells (Itoh et al., 2002). In the cells expressing Raichu-Pak-Rho, which was used as a negative control, the efficiency of the FRET probe was uniform throughout the entire cell. We obtained very similar results with Raichu probes fused to either the carboxy-terminus of the authentic GTPases or the carboxy-terminus of K-Ras. For the brevity, here we show results obtained with Raichu probes fused to the carboxy-terminus of K-Ras. Data obtained with Raichu-RhoA fused to the carboxy-terminus of RhoA was available as supplementary data (Supplementary Figure 1 and Supplementary Video SupFig1HeLa.mov). The detailed comparison of the probes has been described elsewhere (Kurokawa et al., 2004b).
Figure 1.
Imaging of RhoA, Rac, and Cdc42 activities in migrating cells. (A) HeLa cells expressing Raichu-RhoA, Raichu-Rac, Raichu-Cdc42, or Raichu-PAK-Rho were replated onto glass-bottom dishes coated with collagen to observe their stochastic migration. YFP, CFP, and differential interference contrast (DIC) images were recorded every 2 min for 60 min (Fig 1HeLa.mov is available as a Supplementary Video). A ratio image of YPF/CFP was created to represent FRET efficiency, which correlated with the activities of the G proteins. Shown here are representative pseudocolor images of the Ratio (FRET efficiency) and DIC images. Ratio ranges are shown at the bottom of each panel. The arrows point in the direction of cell migration. White scale bars, 10 μm. (B) MDCK monolayers expressing Raichu-RhoA, Raichu-Rac, or Raichu-Cdc42 were wounded and, from 1 h later, were imaged as in A. A video file, Fig 1MDCK.mov is available as supplementary information. (C) MDCK monolayers were wounded and, 1 h later, were imaged for DIC in the presence or absence of 30 μM Y-27632. Scale bars, 50 μm. (D) MDCK monolayers were wounded and, 1 h later, the cells at the wound edge were microinjected with FITC-dextran and GST or GST-Rhotekin-RBD. Shown here are the overlay FITC and phase contrast images at 0 and 60 min after microinjection. Scale bars, 20 μm.
Because the observation of high levels of RhoA activity at the front of migrating cell was unexpected, we confirmed this finding in MDCK cells showing directional movement. As shown in Figure 1B and Supplementary Video Fig 1MD-CK.mov, an increasing gradient of RhoA activity toward the tip of leading edge was observed, as was the case in the HeLa cells, but, in the MDCK cells, no activation of RhoA was observed at the rear end of the cells. It should be noted, however, that high RhoA activity at the tail was observed when the MDCK cells were cultured at a low cell density and were allowed to move stochastically (unpublished data). Therefore, high levels of RhoA activity may not be required when cells are migrating as a monolayer sheet. The activity maps of Rac1 and Cdc42 in the migrating MDCK cells were very similar to those of Rac1 and Cdc42 in migrating HeLa cells.
It has been demonstrated that RhoA activation, but not ROCK activation, is required for the migration of monolayer cells; wound closure of REF cells is inhibited by a Rho inhibitor, C3 transferase, but is conversely promoted by a ROCK inhibitor, Y-27632 (Nobes and Hall, 1999). We reproduced this observation in MDCK monolayer cells. First, the addition of Y27632 was found to accelerate the process of wound healing (Figure 1C). Second, perturbation of RhoA signaling by the microinjection of the Rho-binding domain of Rhotekin inhibited wound healing and membrane protrusion in the presence of Y27632 (Figure 1D). Thus, RhoA effectors other than ROCK appear to play an important role at the leading edge of migrating MDCK cells.
High RhoA Activity Persists at EGF- and PDGF-induced Membrane Ruffles
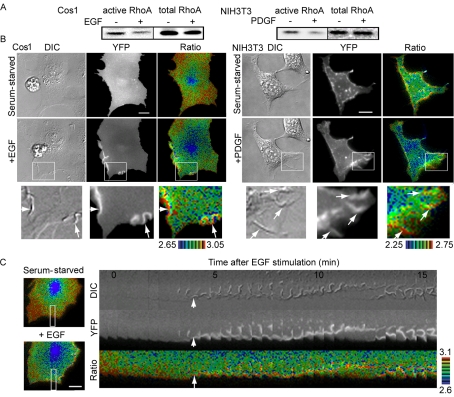
Membrane ruffles are also induced in growth factor-stimulated cells. Thus, we next examined the distribution of RhoA activity in Cos1 and NIH3T3 cells stimulated with EGF and PDGF, respectively. We first examined activity changes in RhoA by Bos' pulldown assay using GST-Rhotekin-RBD. We found that both EGF and PDGF led to a reduction in the amount of GTP-RhoA (Figure 2A), a finding that agreed with those of previous reports (Chang et al., 1995; Sander et al., 1999; Haskell et al., 2001). By using the Raichu-RhoA probe, it was confirmed that RhoA activity decreased diffusely and rapidly upon stimulation with these growth factors (Figure 2B and Supplementary Video SupFig2-RhoA.mov). Surprisingly, however, high RhoA activity persisted at the membrane ruffles (white arrows, Figure 2B). This finding was more clearly demonstrated by kymograph analysis (Figure 2C). Similar results were obtained by using Raichu-RhoA probes fused to the carboxyl-terminus of the authentic RhoA (Supplementary Figure 2 and Supplementary Video SupFig2RhoA.mov). The high level of FRET efficiency observed here was not considered to be the result of an accumulation of the probe, because such high levels of FRET efficiency were not detected at the membrane ruffles in the cells expressing the control probe (unpublished data).
Figure 2.
Spatiotemporal activity of RhoA after stimulation with growth factor. (A) Serum-starved Cos1 cells or NIH3T3 cells were stimulated with 10 ng/ml EGF or 50 ng/ml PDGF for 5 min and were analyzed according to the pulldown method using GST-Rhotekin-RBD. Proteins bound to GST-Rhotekin-RBD and total lysates were subjected to SDS-PAGE and were probed with anti-RhoA antibody. (B) Cells expressing Raichu-RhoA were stimulated with 10 ng/ml EGF or 50 ng/ml PDGF and were imaged for YFP, CFP, and DIC every 20 s for 30 min. Representative pseudocolor images of the Ratio (FRET efficiency) and DIC images before and after stimulation are shown (Fig2Rho.mov is available as supplementary information). The panels in the third column show magnified images of the membrane ruffles that are boxed in the middle panels. The white arrows point to the membrane ruffles. Ratio ranges are shown on the bottom. The experiments were performed at least five times. Scale bars, 10 μm. (C) Kymographic analysis of Cos1 cells expressing Raichu-RhoA. The right panel shows the RhoA activity at the membrane ruffles in the boxed regions of the left panels. The white arrows point to the initiation of membrane ruffles. Ratio ranges are shown on the right.
RhoA Is Required for the Induction of Membrane Ruffles
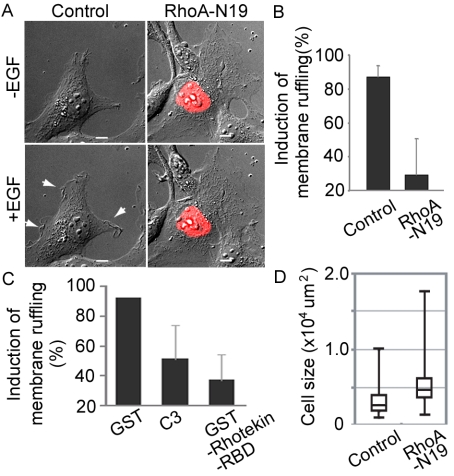
The global decrease in the activity of RhoA was found to correlate with a decrease in the amount of stress fibers in the cell body (unpublished data); nevertheless, the persistence of high RhoA activity at membrane ruffles also suggested that localized high RhoA activity may also play important roles upon stimulation of cells with growth factors. To examine this possibility, we expressed dominant negative RhoA and stimulated cells with EGF (Figure 3). Cells expressing the dominant negative RhoA mutants showed a characteristic flattened morphology with extensive lamellipodial plasma membrane extension and increased cell size (Figure 3, A and D). In these cells, EGF could not induce membrane ruffling (Figure 3A). The requirement of RhoA in EGF-induced membrane ruffling was further confirmed by microinjecting C3 transferase or GST-Rhotekin-RBD into Cos1 cells (Figure 3C). Thus, even though RhoA activity dropped markedly on average, persistent RhoA activity at the peripheral lamellipodia appeared to contribute to the induction of membrane ruffles in growth factor-stimulated cells.
Figure 3.
Requirement of RhoA for EGF-induced membrane ruffling. (A) DIC images of Cos1 cells expressing RhoA-N19 before and 5 min after EGF stimulation. The white arrows point to the membrane ruffles. Scale bars, 10 μm. (B and C) Cos1 cells transfected with RhoA-N19 (B) or microinjected with GST, C3, or GST-Rhotekin-RBD (C) were video-imaged and the induction of membrane ruffling was scored 5 min after stimulation. The averaged data from at least four independent experiments are shown with the SD. (D) Cell area was calculated by MetaMorph software. At least 120 cells were assessed in each experiment. Box-and-Whisker plots show (top down) maximum, third percentile, median, first percentile, and minimum.
RhoA Cooperates with Rac to Induce Membrane Ruffles
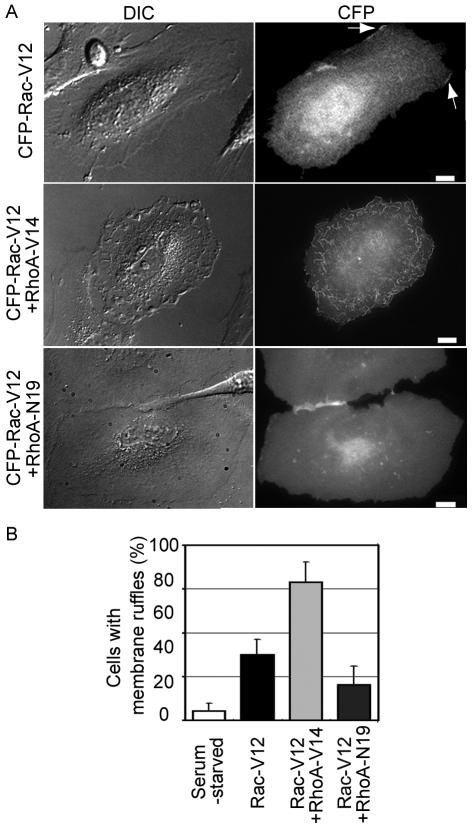
In agreement with previous findings (Ridley and Hall, 1992), we found that the expression of constitutively active RhoA alone did not induce membrane ruffling in HeLa and MDCK cells. Meanwhile, membrane ruffling, as well as lamellipodial protrusion, have been known to be canonical phenotypes of the activation of Rac (Ridley et al., 1992). Thus, we investigated whether or not RhoA cooperates with Rac in the induction of membrane ruffling (Figure 4). The expression of constitutively active Rac1 in HeLa cells induced membrane ruffling, and lamellipodial protrusion at the cell periphery. Coexpression of constitutively active RhoA with constitutively active Rac1 markedly enhanced membrane ruffling, and this phenotype was most clearly evidenced by the appearance of dorsal ruffling across the entire cell surface. In contrast, cells expressing both the constitutively active Rac1 mutant and the dominant negative RhoA mutant displayed an extremely flattened phenotype with lamellipodial protrusion (Figure 4). These cells entirely lacked membrane ruffles because their lamellipodia could not be lifted up from the substrate to fold back on themselves. Phalloidin staining revealed that the expression of the dominant negative RhoA mutant diminished the number of stress fiber without affecting the accumulation of F-actin at the tip of lamellipodia (Supplementary Figure 3). Therefore, these data suggested that RhoA cooperated with Rac1 in the membrane ruffling but not in the lamellipodia protrusion.
Figure 4.
Cooperation of Rac and RhoA in the induction of membrane ruffles. (A) DIC and CFP images of HeLa cells expressing CFP-Rac-V12 alone, CFP-Rac-V12 with RhoA-V14, or CFP-Rac-V12 with RhoA-N19 are shown. The white arrows indicate the membrane ruffles in HeLa cells expressing only CFP-Rac-V12. Scale bars, 10 μm. (B) Cells exhibiting a ruffling phenotype were scored. Values are presented as percentages of the total number of transfected cells. Averaged data from five independent experiments, with an average of 10 cells scored per experiment, are shown with the SD.
mDia1 Associates with Membrane Ruffles
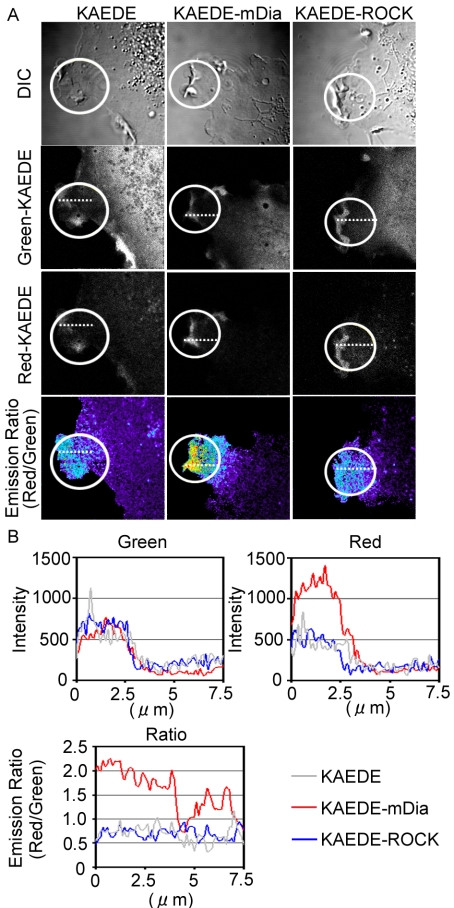
We next attempted to determine whether ROCK and mDia, two canonical effectors of RhoA, are recruited at membrane ruffles. The accumulation of cytoplasmic proteins at membrane ruffles is often used as a sign of the plasma membrane recruitment of signaling molecules. However, because of the perpendicular positioning of membranes at the ruffles, the accumulation of cytoplasmic proteins caused by a thickening of cytoplasm is often misinterpreted as the recruitment of cytoplasmic proteins to membrane ruffles. To avoid this artifact, we developed a novel assay to reliably demonstrate the stable association of Rho effectors with membrane ruffles. We expressed ROCK and mDia1 as fusion proteins with KAEDE, the fluorescence of which changes from green to red upon UV illumination (Ando et al., 2002). On the photoconversion of KAEDE in a small area, cytoplasmic Red-converted KAEDE fusion proteins are expected to diffuse rapidly, whereas those associated tightly with the cell membrane are expected to remain at the site of photoconversion. By measuring the ratio of Red-KAEDE proteins versus Green-KAEDE proteins, it is possible to efficiently compare the level of slow motility and rapidly diffusing KAEDE fusion proteins.
When KAEDE alone was expressed and UV-converted within a small region containing membrane ruffles, the diffusion of KAEDE occurred very rapidly, resulting in a small and diffuse increase in the Red-KAEDE/Green-KAEDE ratio. Very similar results were obtained with KAEDE-ROCK-expressing cells. In contrast, in KAEDE-mDia1 expressing cells, a high Red-KAEDE/Green-KAEDE ratio was observed at the membrane ruffles, indicating a stable association of KAEDE-mDia1 at the membrane ruffles (Figure 5, A and B). To further examine whether the slow diffusion of KAEDE-mDia1 depended on active RhoA, we used two mDia1 mutants, KAEDE-mDia1 ΔN3 and KAEDE-mDia1 ΔN3 (HindIII) (Tsuji et al., 2002; Supplementary Figure 4). KAEDE-mDia1 ΔN3, a constitutively active mutant, lacked the RhoA binding domain and KAEDE-mDia1 ΔN3 (Hin-dIII) lacked the RhoA binding domain and FH2 domain. We found that the diffusion of KAEDE-mDia1 ΔN3 was slow, but that it was not concentrated at the tip of membrane ruffles as was seen in the wild-type KAEDE-mDia1. In contrast, KAEDE-mDia1 ΔN3 (HindIII) showed rapid diffusion as did KAEDE alone.
Figure 5.
Retention of mDia at the membrane ruffles. Cells expressing KAEDE, KAEDE-ROCK, or KAEDE -mDia were used for the analysis. At time point 0, KAEDE proteins in the white-circled regions were photo-converted from green to red by laser irradiation. (A) Cells were imaged for Green-KAEDE, Red-KAEDE, and DIC every 5 s. Pseudocolor Ratio images (Red-KAEDE/Green-KAEDE) were also created to demonstrate the retention of photo-converted KAEDE proteins. Representative images created after photoconversion are shown. (B) The intensities of Green-KAEDE and Red-KAEDE and Ratio values (Red/Green) along the white-dotted line are plotted against the distance from the edge of the cells.
In the period of revision, another fluorescent protein, named Dronpa, became available (Ando et al., 2004). Dronpa's unique photochromic property of light-induced conversion between the bright and dark statuses is ideal for the measurement of rapid diffusion; therefore, we reexamined the association of mDia1 with membrane ruffles by the use of Dronpa-fused mDia1 proteins (Supplementary Figure 5). We set two regions within a cell, a membrane ruffle-containing region and a region without membrane ruffles. By doing this, we could detect the persistence of mDia1 on the membrane ruffles more precisely. As was observed with KAEDE-fusion proteins, Dronpa-mDia1-WT and Dronpa-mDia1-ΔN3 diffused out slower than Dronpa-mDia1 ΔN3 (HindIII), indicating that the FH2 domain significantly decreased the diffusion rate. Importantly, Dronpa-mDia1, but not Dronpa-mDia1-ΔN3, exhibited a slower rate of diffusion at the membrane ruffle-containing region than in the region without membrane ruffles. Although other proteins such as Cdc42 and/or Rac1 may also contribute to the membrane recruitment of mDia1, these data strongly suggested that localized RhoA activity contributes, at least partially, to recruit mDia1 to the membrane ruffles.
Cdc42 Regulate the Distribution of RhoA Activity
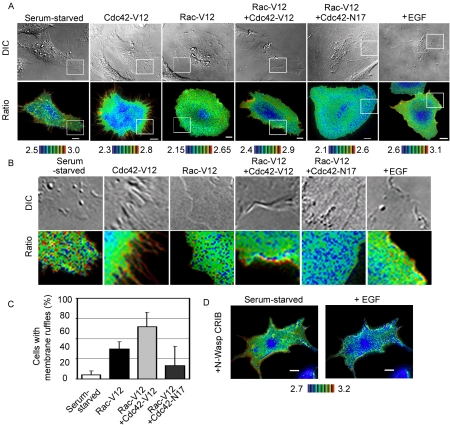
Lastly, we asked why RhoA activity persisted at the tip of membrane ruffles of the EGF-stimulated cells. The localized high activity of RhoA was similar to the localized activation of Cdc42 at membrane ruffles in EGF-stimulated cells (Kurokawa et al., 2004a), which prompted us to investigate whether or not Cdc42 activity might exert an effect on the distribution of RhoA activity (Figure 6, A and B). In HeLa cells, the expression of constitutively active Cdc42 induced a number of filopodia, and, at these filopodia, RhoA activity was found to be very high. In contrast, HeLa cells expressing constitutively active Rac1 displayed prominent lamellipodial protrusions with or without membrane ruffles. Notably, lamellipodia without membrane ruffles did not show any local activation of RhoA. HeLa cells expressing both constitutively active Rac1 and Cdc42 mutants displayed lamellipodia with prominent membrane ruffles at the cell periphery (Figure 6, A–C). In these cells, high RhoA activity was observed at the tip of membrane ruffles, as was also observed in EGF-stimulated HeLa cells. On the other hand, cells expressing the constitutively active Rac1 mutant and the dominant negative Cdc42 mutants exhibited only slight membrane ruffling and an extremely flattened phenotype as did cells expressing the dominant negative RhoA (Figure 4). In these cells, lamellipodial protrusion was observed all around the cell and the activity of RhoA was remarkably low at the cell periphery. Furthermore, in the cells expressing CRIB domain of N-Wasp, which was used to sequester the active Cdc42, EGF-stimulation suppressed RhoA not only at the central region but also a the peripheral plasma membrane (Figure 6D). These results strongly suggested that high Cdc42 activity contributed to maintain high RhoA activity at the membrane ruffles.
Figure 6.
Effect of Cdc42 on the distribution of RhoA activity. (A) HeLa cells expressing Raichu-RhoA with or without expression vectors, indicated at the top of each panel, were imaged for YFP, CFP, and DIC. Representative pseudocolor images of the Ratio (FRET efficiency) and DIC images are shown. The rightmost panels show EGF-stimulated cells as a control. (B) Magnified images of the peripheral regions boxed in white in A. (C) The appearance of membrane ruffling was scored. The values are presented as percentages of total transfected cells. The data were averaged over three independent experiments and are shown with the SD (D) Cos1 cells expressing Raichu-RhoA and CRIB domain of N-Wasp was stimulated with EGF as in Figure 2.
DISCUSSION
For the directional migration of cells, the activity of Rho-family GTPases must be coordinately regulated both in space and time (Raftopoulou and Hall, 2004). In migrating leukocytes, Rho functions at the tail of the cells by promoting actomyosin contraction, thereby yielding retraction forces to retract the cytoplasmic tail (Xu et al., 2003). Furthermore, it has been shown that the RhoA-induced activation of ROCK is essential for cell detachment at the tail of migrating leukocytes (Alblas et al., 2001). In agreement with these previous studies, we observed high RhoA activity at the rear of migrating HeLa cells (Figure 1) and MDCK cells (unpublished data). However, we did not detect high RhoA activity at the rear of MDCK cells migrating as a monolayer sheet, nor did we observe any inhibitory effect of the ROCK inhibitor on MDCK cell migration (Figure 1). This observation indicated a lack of the involvement of the Rho-ROCK pathway in the migration of monolayer cells and was in agreement with a previous report showing that ROCK activity is dispensable in the migration of monolayer REF cells (Nobes and Hall, 1999) Thus, the requirement of the RhoA-ROCK pathway at the tail of migrating cells appears to be cell context-dependent.
Our findings of high levels of RhoA activity at the leading edge of migrating cells and at the membrane ruffles of growth factor-stimulated cells (Figures 1 and 2) are in agreement with those of several previous reports demonstrating that RhoA plays positive roles in cell migration and in the induction of membrane ruffles. RhoA is required both to maintain cell adhesion during the movement of monolayer REF cells (Nobes and Hall, 1999) and to stabilize microtubules oriented toward the leading edge (Nagasaki and Gundersen, 1996; Cook et al., 1998). Although membrane ruffling is attributed to Rac in many cell types (Hall, 1998), Rho is also known to regulate membrane ruffling in epithelial cells (Nishiyama et al., 1994; Fukata et al., 1998). Furthermore, the translocation of RhoA to the membrane ruffles at the edge of lamellipodia has been demonstrated in integrin-stimulated colon carcinoma cells (O'Connor et al., 2000). Because RhoA translocation to the plasma membrane reflects its release from RhoGDI and, therefore, its conversion from the GDP-bound inactive form to the GTP-bound active form, this result appears to support our findings that RhoA activation was observed only at the edge of membrane ruffles in growth factor-stimulated cells. However, in our investigations of HeLa cells, we have not observed the translocation of RhoA to membrane ruffles, because the thickening of the cytoplasm at membrane ruffles hinders evaluation of the translocation of cytoplasmic proteins to the membrane ruffles, as already mentioned in Results.
In many cell types, activation of Rac is indispensable and sufficient for the induction both of lamellipodia (sheetlike protrusions) and membrane ruffling (plasma membrane folding back and being transported rearward) (Ridley et al., 1992; Nobes and Hall, 1999). However, this observation does not necessarily indicate that these two morphological changes are operated by the same mechanism or are induced sequentially. We propose that, in addition to Rac activation, accumulation of RhoA-mDia complex is required for the induction of membrane ruffling, based on the observation that mDia1, but not ROCK, associated stably with membrane ruffles in a manner dependent on active RhoA (Figure 5 and Supplementary Figures 4 and 5). Because mDia1 regulates actin polymerization through its FH2 domain (Li and Higgs, 2003; Shimada et al., 2004) and the length of actin filaments is proposed to be critical for the induction of membrane ruffles in an “elastic Brownian ratchet model” (Mogilner and Oster, 1996), mDia1 remains a strong candidate as a determinant of membrane ruffling. In fact, a mDia1 ΔFH2 mutant inhibits EGF-induced membrane ruffling in Cos1 cells, supporting the positive role of mDia1 in the induction of membrane ruffling by regulating actin polymerization in this cell type (Supplementary Video SupFig5.mov). Furthermore, the local activation of RhoA at membrane ruffles in the leading edge may mediate mDia activation after the stabilization of microtubules oriented toward the leading edge (Palazzo et al., 2001). Alternatively, mDia may induce membrane ruffling via Rac activation. In Swiss 3T3 cells, Y-27632, a ROCK inhibitor, increases Rac activity and induces membrane ruffles in a manner dependent on mDia1 (Tsuji et al., 2002). However, in growth factor-stimulated Cos1 cells, the area of increased Rac activity was more widespread than that of persistent RhoA activity, rendering mDia-dependent Rac activation unlikely under this condition. Of note, in epithelium-derived cells, ROCK/Rho kinase is involved in Rho-dependent membrane ruffling (Nishiyama et al., 1994; Fukata et al., 1998). Therefore, the mechanism by which Rho regulates membrane ruffling may be different among various cell types. Furthermore, the contribution of each of the mDia isoforms, mDia1 (Watanabe et al., 1997), mDia2 (Alberts et al., 1998), and mDia3/hDia2 (Bione et al., 1998), may also be cell type specific and should be investigated in future studies.
Our results have strongly suggested that, of the two well-known phenotypes of Rac activation, membrane ruffling, but not lamellipodial protrusion, depends on RhoA and mDia1. However, we have also found that inhibition of RhoA signaling by the microinjection of the Rho-binding domain of Rhotekin into MDCK cells inhibited not only the membrane ruffling but also the lamellipodial protrusion of MDCK cells (Figure 1D). We speculate that MDCK cells microinjected with Rhotekin-RBD loose the polarity, which hampers the activation of Rac at the wound-side of MDCK cells, resulting in the loss of lamellipodia. Alternatively, the discrepancy between the effect of the dominant negative RhoA and that of Rhotekin-RBD might be aroused by the difference in their specificity. The dominant negative RhoA mutant is assumed to sequester GEFs for RhoA. Because many GEFs activate not only RhoA but also other Rho-family GTPases, the effect of the dominant negative RhoA mutant may not be specific RhoA. On the other hand, Rhotekin-RBD might also inhibit other Rho-family GTPases such as RhoB. These issues must be resolved in the future studies.
It has been reported that local periodic contractions of lamellipodia generate waves of rearward moving of F-actin in a manner dependent on MLCK (Giannone et al., 2004). We have found that ML7, a MLCK inhibitor, suppresses EGF-induced lamellipodia and membrane ruffling in Cos1 cells (unpublished data). However, it is currently unknown whether there is any cross-talk between the RhoA-mDia pathway and MLCK.
Seemingly against our findings and those of the aforementioned previous reports, it has been proposed that the ubiquitin-mediated degradation of RhoA must occur for the successful formation of lamellipodia and filopodia (Wang et al., 2003). In this model, a Cdc42-PAR6-protein kinase Cζ complex activates Smurf1, an E3 ligase, and Smurf1 promotes the ubiquitination and degradation of RhoA at the lamellipodia- and filopodia-like protrusions of Mv1Lu and NIH3T3 cells. Because it has been proposed that Smurf1 binds to RhoA in a GEF-dependent manner, the high GEF activity detected by the Raichu-RhoA probe may indicate the rapid degradation of RhoA at the edge of membrane ruffles. In this case, the rapid turnover of RhoA-GTP, rather than the accumulation of RhoA-GTP, may play an important role in the induction of lamellipodia and filopodia.
One advantage of the use of FRET probes was best manifested by the detection of persistent high RhoA activity at the membrane ruffles of EGF-stimulated cells; under these conditions, the levels of RhoA-GTP decreased diffusely at the plasma membrane (Figure 2). It has been reported that this EGF-induced RhoA suppression is caused by the Rac-dependent activation of p190RhoGAP (Nimnual et al., 2003). Thus, there seems to be a mechanism that maintains high RhoA activity at membrane ruffles under the presence of activated p190RhoGAP. Several lines of evidence have suggested that Cdc42 is involved in this persistence of high RhoA activity at the membrane ruffles. First, we have shown that EGF activated Cdc42 at the membrane ruffles (Kurokawa et al., 2004a). Second, high levels of RhoA activity were observed at the filopodia, an effect which was induced by active Cdc42 (Figure 6). Third, the expression of active Cdc42 with active Rac1 induced the formation of a number of membrane ruffles, where high RhoA activity was also observed (Figure 6). Lastly, the expression of the Cdc42-specific GAP, CdGAP, or CRIB domain of N-Wasp not only inhibited EGF-induced membrane ruffling (Kurokawa et al., 2004a), but also suppressed RhoA activity at the peripheral plasma membrane (Figure 6D). Currently, it is unknown how Cdc42 counteracts Rac-mediated RhoA suppression in EGF-stimulated cells. Some GEFs for the Rho-family GTPases have been shown to associate with specific actin and membrane structures; therefore, such GEFs may contribute to the persistence of RhoA activity in the membrane ruffles.
In conclusion, we have demonstrated the spatiotemporal regulation of RhoA during cell migration and growth factor stimulation. The present findings suggested that the local activation of RhoA activated distinct signaling pathways such as ROCK and mDia pathways; furthermore, such recruitment of different subsets of effectors may account for the different roles played by RhoA.
Supplementary Material
Acknowledgments
We thank A. Miyawaki, J. Miyazaki, and S. Narumiya for the plasmids and N. Yoshida, N. Fujimoto, and K. Fukuhara for their technical assistance. We are grateful to S. Narumiya and the members of the Matsuda laboratory for helpful discussions. This study was supported by grants from the Ministry of Education, Culture, Sports, Science, and Technology of Japan, New Energy and Industrial Development Organization, and the Health Science Foundation of Japan.
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E04-12-1076) on June 29, 2005.
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
References
- Alberts, A. S., Bouquin, N., Johnston, L. H., and Treisman, R. (1998). Analysis of RhoA-binding proteins reveals an interaction domain conserved in heterotrimeric G protein beta subunits and the yeast response regulator protein Skn7. J. Biol. Chem. 273, 8616-8622. [DOI] [PubMed] [Google Scholar]
- Alblas, J., Ulfman, L., Hordijk, P., and Koenderman, L. (2001). Activation of Rhoa and ROCK are essential for detachment of migrating leukocytes. Mol. Biol. Cell 12, 2137-2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amano, M., Chihara, K., Kimura, K., Fukata, Y., Nakamura, N., Matsuura, Y., and Kaibuchi, K. (1997). Formation of actin stress fibers and focal adhesions enhanced by Rho-kinase. Science 275, 1308-1311. [DOI] [PubMed] [Google Scholar]
- Ando, R., Hama, H., Yamamoto-Hino, M., Mizuno, H., and Miyawaki, A. (2002). An optical marker based on the UV-induced green-to-red photoconversion of a fluorescent protein. Proc. Natl. Acad. Sci. USA 99, 12651-12656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ando, R., Mizuno, H., and Miyawaki, A. (2004). Regulated fast nucleocytoplasmic shuttling observed by reversible protein highlighting. Science 306, 1370-1373. [DOI] [PubMed] [Google Scholar]
- Bear, J. E. et al. (2002). Antagonism between Ena/VASP proteins and actin filament capping regulates fibroblast motility. Cell 109, 509-521. [DOI] [PubMed] [Google Scholar]
- Bione, S. et al. (1998). A human homologue of the Drosophila melanogaster diaphanous gene is disrupted in a patient with premature ovarian failure: evidence for conserved function in oogenesis and implications for human sterility. Am. J. Hum. Genet. 62, 533-541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, J. H., Gill, S., Settleman, J., and Parsons, S. J. (1995). c-Src regulates the simultaneous rearrangement of actin cytoskeleton, p190RhoGAP, and p120RasGAP following EGF stimulation. J. Cell Biol. 130, 355-368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook, T. A., Nagasaki, T., and Gundersen, G. G. (1998). Rho guanosine triphosphatase mediates the selective stabilization of microtubules induced by lysophosphatidic acid. J. Cell Biol. 141, 175-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etienne-Manneville, S., and Hall, A. (2002). Rho GTPases in cell biology. Nature 420, 629-635. [DOI] [PubMed] [Google Scholar]
- Fukata, Y., Kimura, K., Oshiro, N., Saya, H., Matsuura, Y., and Kaibuchi, K. (1998). Association of the myosin-binding subunit of myosin phosphatase and moesin: dual regulation of moesin phosphorylation by Rho-associated kinase and myosin phosphatase. J. Cell Biol. 141, 409-418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannone, G., Dubin-Thaler, B. J., Dobereiner, H. G., Kieffer, N., Bresnick, A. R., and Sheetz, M. P. (2004). Periodic lamellipodial contractions correlate with rearward actin waves. Cell 116, 431-443. [DOI] [PubMed] [Google Scholar]
- Gundersen, G. G., and Cook, T. A. (1999). Microtubules and signal transduction. Curr. Opin. Cell Biol. 11, 81-94. [DOI] [PubMed] [Google Scholar]
- Hall, A. (1998). Rho GTPases and the actin cytoskeleton. Science 279, 509-514. [DOI] [PubMed] [Google Scholar]
- Haskell, M. D., Nickles, A. L., Agati, J. M., Su, L., Dukes, B. D., and Parsons, S. J. (2001). Phosphorylation of p190 on Tyr1105 by c-Src is necessary but not sufficient for EGF-induced actin disassembly in C3H10T1/2 fibroblasts. J. Cell Sci. 114, 1699-1708. [DOI] [PubMed] [Google Scholar]
- Itoh, R. E., Kurokawa, K., Ohba, Y., Yoshizaki, H., Mochizuki, N., and Matsuda, M. (2002). Activation of rac and cdc42 video imaged by fluorescent resonance energy transfer-based single-molecule probes in the membrane of living cells. Mol. Cell Biol. 22, 6582-6591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura, K. et al. (1996). Regulation of myosin phosphatase by Rho and Rho-associated kinase (Rho-kinase). Science 273, 245-248. [DOI] [PubMed] [Google Scholar]
- Kraynov, V. S., Chamberlain, C., Bokoch, G. M., Schwartz, M. A., Slabaugh, S., and Hahn, K. M. (2000). Localized rac activation dynamics visualized in living cells. Science 290, 333-337. [DOI] [PubMed] [Google Scholar]
- Kurokawa, K., Itoh, R. E., Yoshizaki, H., Nakamura, Y. O., and Matsuda, M. (2004a). Coactivation of Rac1 and Cdc42 at lamellipodia and membrane ruffles induced by EGF. Mol. Biol. Cell 15, 1003-1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurokawa, K., Takaya, A., Terai, K., Fujioka, A., and Matsuda, M. (2004b). Visualizing the signal transduction pathways in living cells with GFP-based FRET probes. Acta Histochem. Cytochem. 37, 347-355. [Google Scholar]
- Lauffenburger, D. A., and Horwitz, A. F. (1996). Cell migration: a physically integrated molecular process. Cell 84, 359-369. [DOI] [PubMed] [Google Scholar]
- Li, F., and Higgs, H. N. (2003). The mouse Formin mDia1 is a potent actin nucleation factor regulated by autoinhibition. Curr. Biol. 13, 1335-1340. [DOI] [PubMed] [Google Scholar]
- Michaelson, D., Silletti, J., Murphy, G., D'Eustachio, P., Rush, M., and Philips, M. R. (2001). Differential localization of Rho GTPases in live cells: regulation by hypervariable regions and RhoGDI binding. J. Cell Biol. 152, 111-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison, T. J., and Cramer, L. P. (1996). Actin-based cell motility and cell locomotion. Cell 84, 371-379. [DOI] [PubMed] [Google Scholar]
- Mochizuki, N., Yamashita, S., Kurokawa, K., Ohba, Y., Nagai, T., Miyawaki, A., and Matsuda, M. (2001). Spatio-temporal images of growth-factor-induced activation of Ras and Rap1. Nature 411, 1065-1068. [DOI] [PubMed] [Google Scholar]
- Mogilner, A., and Oster, G. (1996). Cell motility driven by actin polymerization. Biophys. J. 71, 3030-3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagasaki, T., and Gundersen, G. G. (1996). Depletion of lysophosphatidic acid triggers a loss of oriented detyrosinated microtubules in motile fibroblasts. J. Cell Sci. 109(Pt 10), 2461-2469. [DOI] [PubMed] [Google Scholar]
- Nimnual, A. S., Taylor, L. J., and Bar-Sagi, D. (2003). Redox-dependent down-regulation of Rho by Rac. Nat. Cell Biol. 5, 236-241. [DOI] [PubMed] [Google Scholar]
- Nishiyama, T., Sasaki, T., Takaishi, K., Kato, M., Yaku, H., Araki, K., Matsuura, Y., and Takai, Y. (1994). rac p21 is involved in insulin-induced membrane ruffling and rho p21 is involved in hepatocyte growth factor- and 12-O-tetradecanoylphorbol-13-acetate (TPA)-induced membrane ruffling in KB cells. Mol. Cell Biol. 14, 2447-2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobes, C. D., and Hall, A. (1999). Rho GTPases control polarity, protrusion, and adhesion during cell movement. J. Cell Biol. 144, 1235-1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor, K. L., Nguyen, B. K., and Mercurio, A. M. (2000). RhoA function in lamellae formation and migration is regulated by the alpha6beta4 integrin and cAMP metabolism. J. Cell Biol. 148, 253-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palazzo, A. F., Cook, T. A., Alberts, A. S., and Gundersen, G. G. (2001). mDia mediates Rho-regulated formation and orientation of stable microtubules. Nat. Cell Biol. 3, 723-729. [DOI] [PubMed] [Google Scholar]
- Raftopoulou, M., and Hall, A. (2004). Cell migration: Rho GTPases lead the way. Dev. Biol. 265, 23-32. [DOI] [PubMed] [Google Scholar]
- Ren, X. D., Kiosses, W. B., and Schwartz, M. A. (1999). Regulation of the small GTP-binding protein Rho by cell adhesion and the cytoskeleton. EMBO J. 18, 578-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridley, A. J. (2001). Rho GTPases and cell migration. J. Cell Sci. 114, 2713-2722. [DOI] [PubMed] [Google Scholar]
- Ridley, A. J., and Hall, A. (1992). The small GTP-binding protein rho regulates the assembly of focal adhesions and actin stress fibers in response to growth factors. Cell 70, 389-399. [DOI] [PubMed] [Google Scholar]
- Ridley, A. J., Paterson, H. F., Johnston, C. L., Diekmann, D., and Hall, A. (1992). The small GTP-binding protein rac regulates growth factor-induced membrane ruffling. Cell 70, 401-410. [DOI] [PubMed] [Google Scholar]
- Sander, E. E., ten Klooster, J. P., van Delft, S., van der Kammen, R. A., Collard, J. G. (1999). Rac downregulates Rho activity: reciprocal balance between both GTPases determines cellular morphology and migratory behavior. J. Cell Biol. 147, 1009-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada, A., Nyitrai, M., Vetter, I. R., Kuhlmann, D., Bugyi, B., Narumiya, S., Geeves, M. A., and Wittinghofer, A. (2004). The core FH2 domain of diaphanous-related formins is an elongated actin binding protein that inhibits polymerization. Mol. Cell 13, 511-522. [DOI] [PubMed] [Google Scholar]
- Tsuji, T. et al. (2002). ROCK and mDia1 antagonize in Rho-dependent Rac activation in Swiss 3T3 fibroblasts. J. Cell Biol. 157, 819-830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H. R., Zhang, Y., Ozdamar, B., Ogunjimi, A. A., Alexandrova, E., Thomsen, G. H., and Wrana, J. L. (2003). Regulation of cell polarity and protrusion formation by targeting RhoA for degradation. Science 302, 1775-1779. [DOI] [PubMed] [Google Scholar]
- Watanabe, N., Madaule, P., Reid, T., Ishizaki, T., Watanabe, G., Kakizuka, A., Saito, Y., Nakao, K., Jockusch, B. M., Narumiya, S. (1997). p140mDia, a mammalian homolog of Drosophila diaphanous, is a target protein for Rho small GTPase and is a ligand for profilin. EMBO J. 16, 3044-3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen, Y., Eng, C. H., Schmoranzer, J., Cabrera-Poch, N., Morris, E. J., Chen, M., Wallar, B. J., Alberts, A. S., and Gundersen, G. G. (2004). EB1 and APC bind to mDia to stabilize microtubules downstream of Rho and promote cell migration. Nat. Cell Biol. 6, 820-830. [DOI] [PubMed] [Google Scholar]
- Xu, J., Wang, F., Van Keymeulen, A., Herzmark, P., Straight, A., Kelly, K., Takuwa, Y., Sugimoto, N., Mitchison, T., and Bourne, H. R. (2003). Divergent signals and cytoskeletal assemblies regulate self-organizing polarity in neutrophils. Cell 114, 201-214. [DOI] [PubMed] [Google Scholar]
- Yamaguchi, Y., Katoh, H., Yasui, H., Mori, K., and Negishi, M. (2001). RhoA inhibits the nerve growth factor-induced Rac1 activation through Rho-associated kinase-dependent pathway. J. Biol. Chem. 276, 18977-18983. [DOI] [PubMed] [Google Scholar]
- Yoshizaki, H., Ohba, Y., Kurokawa, K., Itoh, R. E., Nakamura, T., Mochizuki, N., Nagashima, K., and Matsuda, M. (2003). Activity of Rho-family GTPases during cell division as visualized with FRET-based probes. J. Cell Biol. 162, 223-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zondag, G. C., Evers, E. E., ten Klooster, J. P., Janssen, L., van der Kammen, R. A., and Collard, J. G. (2000). Oncogenic Ras downregulates Rac activity, which leads to increased Rho activity and epithelial-mesenchymal transition. J. Cell Biol. 149, 775-782. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.