Abstract
Fragile X-related 1 protein (FXR1P) is a member of a small family of RNA-binding proteins that includes the Fragile X mental retardation 1 protein (FMR1P) and the Fragile X-related 2 protein (FXR2P). These proteins are thought to transport mRNA and to control their translation. While FMR1P is highly expressed in neurons, substantial levels of FXR1P are found in striated muscles and heart, which are devoid of FMRP and FXR2P. However, little is known about the functions of FXR1P. We have isolated cDNAs for Xenopus Fxr1 and found that two specific splice variants are conserved in evolution. Knockdown of xFxr1p in Xenopus had highly muscle-specific effects, normal MyoD expression being disrupted, somitic myotomal cell rotation and segmentation being inhibited, and dermatome formation being abnormal. Consistent with the absence of the long muscle-specific xFxr1p isoform during early somite formation, these effects could be rescued by both the long and short mRNA variants. Microarray analyses showed that xFxr1p depletion affected the expression of 129 known genes of which 50% were implicated in muscle and nervous system formation. These studies shed significant new light on Fxr1p function(s).
INTRODUCTION
Fragile X-related 1 (FXR1) belongs to a small gene family that includes Fragile X mental retardation 1 (FMR1) and Fragile X-related 2 (FXR2) (reviewed in Khandjian, 1999; Bardoni and Mandel, 2002). Human FMR1 is located on chromosome X at q27.3 (Sutherland, 1977), whereas FXR1 and FXR2 are autosomal genes mapping at 3q28 and 17p13.1, respectively (Coy et al., 1995; Zhang et al., 1995). Inactivation of FMR1 expression is the cause of the Fragile X syndrome in human (Imbert et al., 1998; O'Donnell and Warren, 2002), whereas FXR1 and FXR2 have not been associated so far with any known pathology or defect. The three genes code for proteins that are highly conserved in vertebrate evolution. They are structurally very similar and share high degree of sequence homologies in clustered regions. They contain two KH domains and a RGG box that are characteristic motifs in RNA-binding proteins as well as nuclear localization and export signals (Siomi et al., 1995; Zhang et al., 1995; Eberhart et al., 1996). Similarly to its other homologues, FXR1P is predominantly associated with polyribosomal messenger ribonucleoparticles (RNPs) and is thought to play a yet unknown role in translation control (Siomi et al., 1996; Khandjian, 1999; Dubé et al., 2000).
The expression pattern of FXR1P is rather complex in mammals. Six distinct protein isoforms have been detected, and their respective levels were shown to be cell-type specific. Four isoforms of 70, 74, 78, and 80 kDa are widely expressed in all cell lines and mouse tissues so far tested, whereas these isoforms are absent in heart and skeletal muscles and are replaced by longer isoforms of 82-84 kDa (Khandjian et al., 1998; Dubé et al., 2000). The shifts in apparent molecular weights are clearly observed during in vitro differentiation of myocytes and transition of myoblasts to myotubes as the 70-80 kDa isoforms are progressively replaced by the 82-84 kDa species generated by the insertion of a small peptide pocket of 27 aa (Dubé et al., 2000). This short peptide stretch is coded by exon15 corresponding to the 81-base pair insertion present in mRNA variants that have been detected in striated muscles and in adult testis (Khandjian et al., 1998; Kirkpatrick et al., 1999; Huot et al., 2001) and seems to be conserved in evolution having been detected in human, mouse, and more recently in zebrafish (Engels et al., 2004; Tucker et al., 2004).
Although viable mouse knockout (KO) models for both Fmr1 and Fxr2 have been generated (Bakker and The Dutch-Belgian Fragile X Consortium, 1994; Bontekoe et al., 2002), homozygous Fxr1-/- neonates die shortly after birth most likely due to cardiac and/or respiratory failure (Mientjes et al., 2004), making it difficult to study these phenotypes. A second mouse model that expresses reduced levels of Fxr1p also has been generated and displays reduced limb musculature and a reduced life span of ∼18 wk. This latter animal model has suggested the implication of Fxr1p in skeletal muscle development (Mientjes et al., 2004).
In our approach to study the function of Fxr1p in vivo, we chose the Xenopus laevis model because a large number of eggs is easily obtained and it offers rapid embryonic development external to the mother. It is a highly flexible model because gene expression can be perturbed by microinjection of antisense Morpholino-oligonucleotides (MO; Summerton and Weller, 1997; Heasman, 2002) to directly test the role and/or functions of the protein of interest. Most importantly, Xenopus offers the possibility to inject different amounts of MO to study the dose-dependent inactivation of the target sequences, allowing a wide range of induced phenotypes to be studied. Here, we show that xFxr1 plays a vital role during Xenopus embryogenesis because its complete or partial inactivation has dramatic impact on myotome formation. The inactivation of xFxr1 also has an impact on the expression of many genes implicated in embryogenesis and more particularly in myogenesis.
MATERIALS AND METHODS
DNA Manipulations
Isolation of xFxr1 cDNAs. In total, 105 plaques from a X. laevis stage 30 tail cDNA library in modified Blue Script pRN3 (established by N. Garrett, E. Bellefroid, and A. M. Zorn, Wellcome/CRC Institute, Cambridge, United Kingdom) were screened using PCR fragments described below. The entire sequence of the open reading frame was determined at the DNA core facilities at Laval University. Sequence data from this article have been deposited with the GenBank Data Library under accession nos. DQ083374-DQ083375.
Reverse Transcription (RT) and PCR. RT was performed on 0.2 μg of total RNA according to the manufacturer's protocol (Invitrogen, Carlsbad, CA). For xFxr1 mRNA variants detection, one-fifth of the resulting reaction was used to perform a PCR by using the following oligonucleotides: XL-F: 5′-GTGCTTAAGGATCCAGACAGCA-3′ (X. laevis Fxr1 forward primer); XL-R: 5′-GACACCCCATTCATTATGGCT-3′ (X. laevis Fxr1 reverse primer); MM-F: 5′-CGTCGTAGGCGTCTCGTAG-3′ (Mus musculus Fxr1 forward primer); and MM-R: 5′-ACCATTCAGGACTGCTGCTT-3′ (M. musculus Fxr1 reverse primer).
PCR were performed initially by denaturation at 95°C for 5 min, followed by 30 cycles of denaturation at 95°C for 30 s, annealing at 64°C for 30 s, extension at 72°C for 1 min, and a final extension step at 72°C for 5 min. Amplified DNA fragments were fractionated on 1.5% agarose gels, stained with ethidium bromide, eluted, and sequenced to validate the RNA variants.
xFxr1 3′ -Untranslated Region (UTR) Cloning. 3′-UTR were reverse transcribed from total RNA prepared from ovary and muscle and amplified using the SMART-RACE cDNA amplification kit (BD Biosciences Clontech, Palo Alto, CA). Fragments were cloned into pBlue Script SK and sequenced.
Embryo Manipulations
Morpholino. The antisense (match) xFxr1.Morpholino (xFxr1.MO) (5′-GCACTTCCACCGTCATGTCCTCCAT-3′) and mismatch Morpholino (xFxr1mm-MO) (5′-GCAgTTgCACCGTgATGTCgTCgAT-3′) (Gene Tools, Philomath, OR) corresponded to nucleotides +1 to +25 of xFXR1 mRNA relative to the ATG. The Morpholino against the human globin also was used as a second negative control: hGlob.MO (5′-CCTCTTACCTCAGTTACAATTTATA-3′). The Morpholino was diluted in water, and 0.1-1.0 pmol was injected into one or two blastomeres of two cell stage embryos. These were allowed to develop to the appropriate stage before analyses. Rescue of the Morpholino effects was achieved by coinjecting mRNA encoding the full-length hemagglutinin (HA)-tagged xFxr1. The xFxr1 cDNA encoding amino acids 2-677 was subcloned into pT7TS (obtained from P. Krieg, University of Arizona, Tucson, AZ) in-frame with an N-terminal HA1-epitope as described previously (Bisson et al., 2003). Capped RNA was transcribed from linearized plasmid by using T7 (sense transcripts) (mMESSAGE-mMACHINE; Ambion, Austin, TX), and RNA dissolved in water was injected into one or two blastomeres of two cell Xenopus embryos. Eggs were recovered in MMR (0.1 M NaCl, 2 mM KCl, 1 mM MgSO4, 2 mM CaCl2, 5 mM HEPES, and 0.1 mM EDTA, pH 7.8), in vitro fertilized, injected with the MO and/or mRNA in 0.2× MMR, 5% Ficoll, and then cultured in 0.1× MMR at 18°C. Embryo stages were determined according to Nieuwkoop and Faber (1967).
Whole Mount In Situ Hybridization, Cryosections, and Hoechst Staining.Whole mount in situ hybridization was carried out on pigmented X. laevis (Nasco, Fort Atkinson, WI) as described previously (Hemmati-Brivanlou et al., 1990; Jowett and Lettice, 1994). Embryos used for whole mount in situ hybridizations were rehydrated in phosphate-buffered saline (PBS) and transferred to 1.6 M sucrose in PBS for >12 h before being placed in a drop of Tissue-Tek embedding solution (Bayer Corp., Emeryville, CA) and frozen on a metal block on dry ice. Sections of 16 μm were cut with a Microm HM 500 cryostat microtome and picked up on gelatin-coated slides and allowed to air dry for 30-60 min. Slides were incubated in water for 2 min, transferred to a Hoechst solution (Sigma, St. Louis, MO; 0.2 mM in water) for 10 min, and washed 5 min in water. Sections were analyzed under fluorescence with a Leica DMIRE2 microscope and Openlab software (Improvision, Lexington, MA).
RNA Studies
Northern Blot Analysis. Total RNA was extracted from Xenopus and mice tissues by using the TRIzol reagents (Invitrogen) and quantified by UV densitometry. Northern blot analyses were performed as described previously (Khandjian, 1986) with the exception that RNA was transferred to nylon membranes by vacuum blotting. 32P-labeled probes were prepared by random-priming and used at ∼106 cpm/ml hybridization solution.
Microarrays. RNA from whole embryos was extracted using the TRIzol reagents. RNA was then cleaned of any residual DNA and protein contaminants using the RNAeasy column (QIAGEN, Valencia, CA). Purified RNA was quantified using a Biochrom GeneQuant spectrometer. Microarray analyses were performed using Affymetrix chips (no. 900492) containing 14,400 transcripts. Analyses were done in triplicate with RNA extracted from three individual series of injections. Microarray and statistics analyses were performed at the Genome Quebec Innovation Centre/McGill University core facility.
Protein Studies
Immunoblot Analyses. Organs were removed from animals, processed according to standard protocols, and protein extracts were prepared for SDS-PAGE as described previously (Khandjian et al., 1995). Immunoblot analyses were performed using mAb3FX and antisera #27-17 directed against FXR1P (dilution 1:2000 and 1:5000, respectively). Other antibodies and sera used were anti-β-tubulin mAbE7 and anti-actin mAbJLA20 (obtained from the Developmental Studies Hybridoma Bank, University of Iowa, Iowa City, IA), anti-L7 ribosomal protein (obtained from A. Ziemiecki, University of Bern, Bern, Switzerland) and anti-FMRP mAb1C3 (Devys et al., 1993). Detection of bound antibodies was performed with horseradish peroxidase-coupled secondary antibodies followed by enhanced chemiluminescence reaction as described previously (Corbin et al., 1997).
Immunohistochemistry. Xenopus and mice embryos were fixed in a freshly prepared 4% paraformaldehyde solution in PBS for 18 h at 4°C. After dehydration in increasing ethanol series, tissues were embedded in paraffin. Five-micrometer sections were processed for deparaffinization and for microwave oven treatments. Endogenous peroxidase was inhibited by a 15-min treatment with 0.5% H2O2 solution in PBS, and the sections were next incubated for 18 h at 4°C with antiserum #27-17 (dilution 1:500) or mAb3FX (dilution 1:500). Bound immunoglobulin was reacted with biotinylated secondary antibodies followed by a streptavidin-peroxidase conjugate and stained with the AEC chromogen (Histomouse-SP kit; Zymed Laboratories, South San Francisco, CA). The slides were then mounted in GVA Mount (Zymed Laboratories). Light microscopy was performed using a Nikon TE300 microscope connected to a CoolSnap camera (RS Photometrics, Trenton, NJ) by using 4×, 10×, 40×, and 60× objectives. Images were transferred to the Adobe Photoshop program.
RESULTS
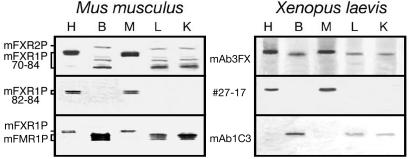
Previous studies have shown that FXR1P 82-84-kDa isoforms are expressed in skeletal and cardiac muscle in mammals, whereas the other isoforms are absent from these tissues (Khandjian et al., 1998). These specific long-isoforms containing 27 additional aa accumulate during myoblastic differentiation of mammalian cultured cells (Dubé et al., 2000). Of the several available antibodies raised against different mouse and human FXR1P isoforms, two, namely, mAb3FX and rabbit serum #27-17, also recognized the frog protein. Whereas mAb3FX reacted with all Fxr1p isoforms in mouse tissues, only two species of 84 and 88 kDa were observed in Xenopus (Figure 1). It also should be noted that this monoclonal antibody reacts with mammalian FXR2P (Khandjian et al., 1998), but it does not detect a putative frog ortholog. In contrast, #27-17 revealed the presence of both mouse 82- to 84- and frog 88-kDa-long isoforms in heart and muscle (Figure 1). On the other hand, FMRP was predominantly detected in frog brain extract, as is the case for mouse. Due to the fact that the mAb1C3 reacts slightly with mouse Fxr1p, additional faints bands also were detected in heart and muscle extracts that contain high amounts of the long Fxr1p isoforms (Figure 1). These results indicate that the tissue specificities of FXR1P and FMR1P are conserved from mammals to amphibians.
Figure 1.
Expression of Fxr1p in mouse and frog. Immunoblot analyses of Fxr1p in selected tissues of adult mouse (left) and adult Xenopus (right). Antibody mAb3FX is directed against an epitope common to all Fxr1p isoforms from both mouse and frog, whereas #27-17 is directed against the 27-amino acid pocket coded by the exon 15 that also is present in Xenopus. Fmr1p is detected predominantly in brain extracts from both mouse and frog. Note the cross-reactivity between mAb1C3 and Fxr1p in mouse heart and muscle as described previously (Khandjian et al., 1998; Chemicon International information sheet, MAB2160). H, heart; B, brain; M, muscle; L, liver; and K, kidney.
Fxr1 mRNA Variants in Xenopus
Although extensive alternative splicing of FXR1 primary transcript has been observed in mouse and in human (Kirkpatrick et al., 1999), only a single mRNA species has been described in Xenopus (Siomi et al., 1995). Because the FXR1 gene is highly conserved in evolution and in the mouse model Fxr1 mRNA variants are tissue specific, we hypothesized that different mRNA variants also should be present in Xenopus.
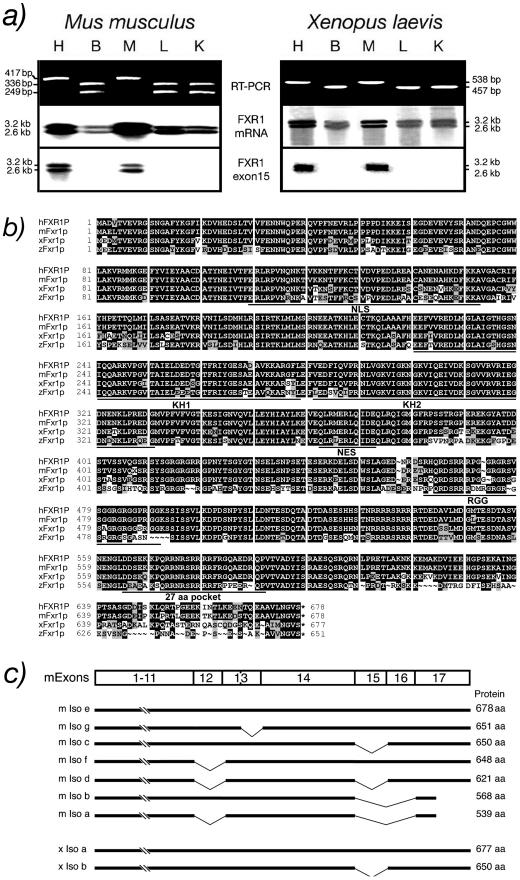
Using primers targeting sequences found in mouse Fxr1 exon 14 and 17, we performed RT-PCR on RNA prepared from different mouse tissues. Consistent with previous reports (Kirkpatrick et al., 1999; Huot et al., 2001), we observed two amplicons of 336 and 249 base pairs in brain, liver, and kidney, whereas in heart and muscle a single amplicon of 417 base pairs harboring an additional muscle specific 81-base pair stretch (Khandjian et al., 1998) was observed. In Xenopus, only a single 457-base pair amplicon was detected in brain, liver, and kidney, but heart and muscle displayed a 538-base pair amplicon containing an additional 81 base pairs (Figure 2a). These analyses showed that similarly to human and mouse, an additional exon corresponding to mammalian exon 15 is very likely to also be maintained in the heart and muscle mRNA variant of Xenopus. Northern blot analyses of RNA isolated from various murine tissues revealed the presence of two Fxr1 transcript species estimated at 2.4 and 3.2 kb (Kirkpatrick et al., 1999; Huot et al., 2001; Garnon et al., 2005). The same distribution also was observed for Xenopus tissues (Figure 2a). We determined that the only difference between the two RNA transcripts in both mouse and frog resides in the presence of either a short or a long 3′-UTR. The short xenopus 3′-UTR (Figure 3a) corresponds to 244 nt and retains 84% homology with human (254 nt) and 82% with mouse (247 nt). On the other hand, the conserved frog long 3′-UTR counterpart (Figure 3b) is made of 595 nt and shares 71 and 82% homologies with human (617 nt; Kirkpatrick et al., 1999) and mouse (615 nt; Huot et al., 2001), respectively. These results highlight the importance of maintaining two 3′-UTR in evolution.
Figure 2.
Fxr1p is conserved in vertebrates. (a) Detection of xFxr1 mRNA variants by RT-PCR in different tissues from mouse and frog by using primers flanking the muscle-specific exon. Northern blots containing RNA isolated from murine and frog tissues (H, heart; B, brain; M, muscle; L, liver; and K, kidney) were hybridized with probes specific for mouse and frog Fxr1. The muscle sequence (exon 15) is found in both mouse and Xenopus. (b) Amino acid sequence of Xenopus Fxr1p (xFxr1p) aligned with human (hFXR1P), mouse (mFxr1p), and zebrafish (zFxr1p) sequences. Conserved amino acids are boxed in black, similar amino acids are in light gray and divergent amino acids are in white. (c) Representation of the mouse (m Iso a-g) and Xenopus (x Iso a and b) Fxr1 mRNA-spliced variants.
Figure 3.
Nucleotide alignment of human, mouse, and frog 3′-UTR. (a) Short 3′-UTR. (b) Long 3′-UTR. Nucleotides are numbered according to Kirkpatrick et al. (1999) and GenBank accession no. U25165.
To further study Xenopus Fxr1 RNA, both 457- and 538-base pair amplicons were cloned and used as probes to screen a cDNA library prepared from X. laevis tail at stage 30. Ten positive clones were selected and sequence analyses revealed the presence of two cDNA species, one corresponding to the previously characterized xFxr1 cDNA (Siomi et al., 1995), whereas the second contained an additional 81-nucleotide insert. This short additional sequence was shown to be highly homologous to that of human FXR1 exon 15 (Khandjian et al., 1998; Kirkpatrick et al., 1999). Amino acid alignment of human, mouse, and Xenopus revealed that the corresponding proteins are highly conserved during evolution, because 94% similarity and 88% identity in amino acids were observed (Figure 2b). Of high interest, the 27-aa pocket is maintained in Xenopus and differs from mammalian exon 15 by a single change in amino acid K-568-Q.
In spite of our efforts to detect additional amplicons, no other mRNA-spliced variants were identified by RT-PCR analyses, using different Xenopus primers. This allows us to propose that in contrast to human and mouse, only two Fxr1 mRNA variants are detected in Xenopus (Figure 2c).
Expression of FXR1P in Xenopus
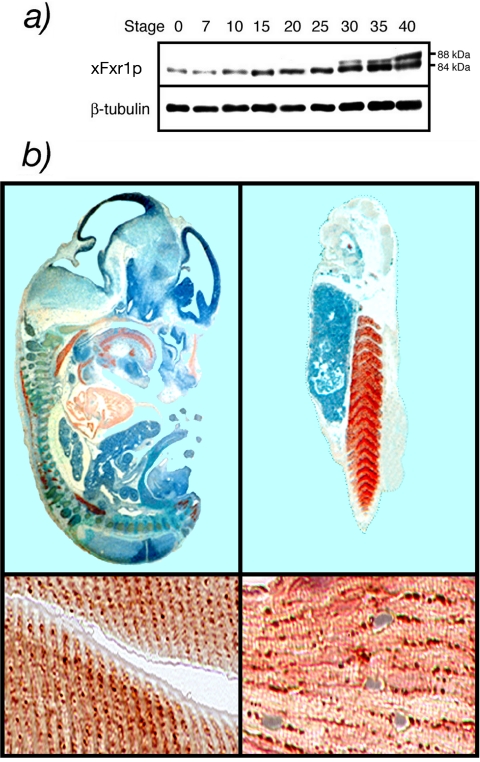
Using mAb3FX, the distribution of the different xFxr1p isoforms could be followed throughout the first stages of embryogenesis of Xenopus. Although only the shorter 84 kDa was present from stage 0 (fertilization), the long 88 kDa was detectable after stage 30 (Figure 4a). Immunohistostaining on sections of both mice and frog embryos were performed using antibody #27-17, specific to the 27-amino acid pocket. At stage 12.5 days postcoitum (dpc), mouse embryos showed a positive staining in heart, tongue, and all other striated muscle (Figure 4b). Similar results were observed in Xenopus embryos at stage 36 as the somites forming the caudal muscle showed a strong positive staining. A clear but small signal also was detected in the eye. As reported previously (Dubé et al., 2000), mFxr1p is detected in granular structures reminiscent of the costameres that contain RNA and proteins (Cripe et al., 1993; Morris and Fulton, 1994; Fulton and Alftine, 1997). This association with these RNP structures is consistent with the RNA-binding properties of Fxr1p. In Xenopus muscles, the same granular distribution was observed (Figure 4b), indicating again the conserved role of the protein in evolution.
Figure 4.
(a) Developmental profile of xFxr1p accumulation during the first stages of embryogenesis. β-tubulin was used as an internal marker. (b) Immunostained sections of a 12.5 dpc mouse embryo and stage 36 Xenopus embryos reacted with antibody #27-17-specific to the muscle-specific isoforms. Positive signals for Fxr1p were revealed with chromophore AEC (red staining). Counterstaining with hematoxylin (blue).
Specific inactivation of xFxr1 mRNA
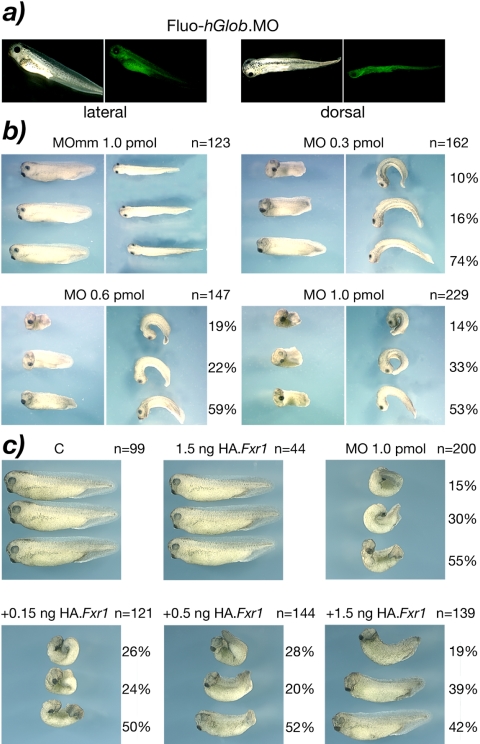
X. laevis is a model of choice to study gene functions during early development. The highly specific and efficient MOs (Summerton and Weller, 1997) have been shown to be very efficient gene silencing reagents in this system. Morpholinos were designed to specifically target sequences surrounding the initiation codon of xFxr1 mRNA. The strategy we adopted was to inject one blastomere of a two-cell embryo and to follow development. We first tested the diffusion potential of the MO by injecting a fluorescein tagged control MO directed against the human globin (Summerton et al., 1997) and known to have no effect on Xenopus embryogenesis and larval development (Figure 5a). The Fluo-hGlobin.MO was distributed throughout the length of the embryo, and as expected, was restricted to the side of the injection. Neither this hGlobin.MO nor a MO containing five mismatched bases (MOmm) had detectable effects on the development (Figure 5b). In contrast, response to the match xFxr1.MO was clearly dose dependent. Although injection of 0.1 pmol of match xFxr1.MO induced no visible phenotype (our unpublished data), at 0.3 pmol all embryos showed a curved appearance (Figure 5b). Injection of 0.6 pmol of MO produced a dramatic phenotype from stages 22 to 25 and onward, with all embryos being tightly curled. In addition, these embryos were ∼30% shorter than their uninjected siblings. Finally, injection of 1.0 pmol of xFxr1.MO resulted in a very severe phenotype, all embryos surviving beyond stage 25 having a tightly curled morphology. To confirm that this phenotype was solely due to silencing of the xFxr1 mRNA, a rescue was attempted by coinjecting an HA-tagged xFxr1 mRNA. Because the HA-tag coding sequence disrupted the MO-targeted sequence of the wild-type xFxr1 mRNA, the tagged mRNA could not interact with the Morpholino and hence could not be silenced by it. The modified mRNAs coding the long and short forms of xFxr1p were shown to support translation of the full-length HA-tagged proteins in a rabbit reticulocyte lysate (our unpublished data), and their injection into two cell embryos gave no observable phenotype, even up to 1.5 ng per embryo (Figure 5c). Coinjection of 1 pmol of xFxr1.MO with increasing amounts of long-form HA-xFrx1 mRNA gave a dose-dependent rescue, and at 1.5 ng ∼80% of embryos showed no curled phenotype and >40% recovered their near normal length (Figure 5c). The short form HA-xFxr1 mRNA was only slightly less effective in rescuing the xFxr1.MO phenotype. For example, at 1.5 ng/embryo, 48% of embryos showed no curling (our unpublished data), the phenotype being closely comparable with 0.5 ng of the long mRNA variant (Figure 5c).
Figure 5.
Morphological alterations induced in Xenopus embryos after injection of Fxr1.Morpholino (Fxr1.MO). (a) Lateral and dorsal views of control embryos for the diffusion of the fluorescein tagged Morpholino directed against the human globin (hGlob.MO) injected in one blastomere of two-cell stage embryos. (b) Injected embryos with xFxr1.MO mismatch (mm) as control, and with increasing concentrations of match MO. Number of injected embryos (n =) is on the right of each panel, as well as percentage of penetrance of the depicted phenotypes. The severity of morphological alterations is proportional to the concentration of injected Fxr1.MO (0.3-1.0 pmol). (c) Rescue of the phenotypes is dose-dependent of the injected HA.Fxr1 mRNA.
Reduction of xFxr1p Steady-State Levels Prevents Normal Somitogenesis
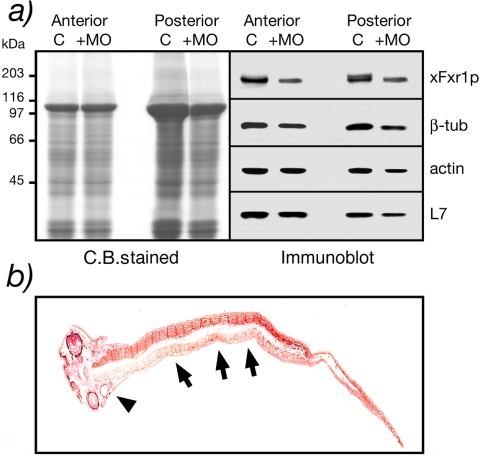
Because high levels of Fxr1p are present in the somites of the swimming tadpole, we investigated the effects of Fxr1.MO injection on the steady-states levels of the protein. Two cell embryos were injected with 0.3 pmol of match or mismatch xFxr1.MO in one blastomere and total protein extracted from the dissected head and trunk at stage 36. Using antibody mAb3FX, a single xFxr1p species was detected in the head (anterior) extracts, and this isoform also was present in the trunk (posterior) that contained an additional band corresponding to the larger 88-kDa muscle isoform (Figure 6a). In xFxr1.MO injected embryos, an ∼40% reduction in the level of the shorter 84-kDa xFxr1p was observed in both head and trunk. Given that only half the embryo was injected with xFxr1.MO, this represented a very significant level of silencing. Interestingly, the longer 88-kDa muscle-specific isoform was no longer detected. The decrease in xFxr1p levels particularly in the somites of the injected side was further illustrated after immunostaining of a longitudinal section with mAb3FX (Figure 6b).
Figure 6.
Biochemical evidence that xFxr1p levels decrease after xFxr1.MO treatment. (a) Coomassie-stained proteins after SDS-PAGE analysis of anterior and posterior extracts obtained from one embryo at stage 36 injected with 0.3 pmol of either xFxr1.MOmm (C, control) or xFxr1.MO (+MO). Western blot analysis using antibody mAb3FX for detection of xFxr1p, and antibodies to tubulin, actin, and ribosomal L7 proteins. (b) Partial inactivation of xFxr1correlates with decreased xFxr1p levels in the forming tail. Immunostaining view of a longitudinal section reacted with mAb3FX to detect Fxr1p. Note the reduced levels of xFxr1p in myotomes (arrows) as well as in the head (arrow head) in the injected side of the embryo.
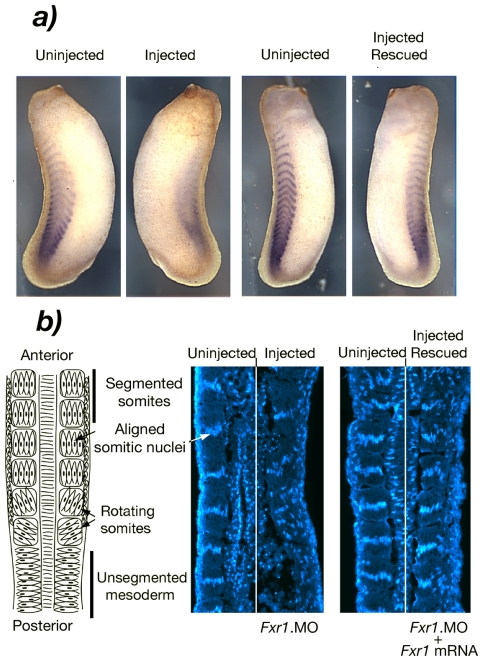
To determine more precisely the stage at which xFxr1p was required for somitic muscle formation, we studied the effects of low doses of xFxr1.MO on the expression of an early muscle marker and somite formation. When two cell embryos were injected with 0.3 pmol of xFxr1.MO in one blastomere, no morphological changes were observed during early embryogenesis, whereas a mildly curled phenotype was observed from stage 22-25 (Figure 5b). At stage 25, embryos showed a diffused pattern of expression of the early mesodermal marker MyoD on the injected side of the embryo (Figure 7a). More importantly, the normally striped somite pattern of MyoD expression was clearly disrupted, suggesting that segmentation of the trunk mesoderm to form the somites had been affected. This effect was highly specific because the normal striped MyoD expression pattern was fully restored by coinjection of xFxr1 mRNA. In Xenopus, the trunk mesoderm segments in an anterior-posterior sequence, each somite forming by the rotation of a small group of myotomal cells as depicted in Figure 7b (Hamilton, 1969; Youn and Malacinski, 1981). By stage 25, 16 somites have already segmented (Nieuwkoop and Faber, 1967). One overt characteristic of completed somite rotation is the transverse alignment of the nuclei of the myotomal cells in each somite (Figure 7b). When the same embryos depicted in Figure 7a were longitudinally sectioned and nuclei stained with Hoechst, near normal numbers of myotomal cells were evident. However, somite segmentation and nuclear alignment in myotomal cells was clearly disrupted on the injected side of the embryo. The lack of visible somite boundaries and the random positioning of the myotomal cell nuclei demonstrated that the process of myotomal rotation had been inhibited or at least, significantly delayed. Again, this effect was highly specific because normal somite segmentation and the characteristic alignment of myotomal cell nuclei were rescued by the coinjection of xFxr1 mRNA (Figure 7b, right). In Xenopus, the dermatome initially separates from the myotome as a single layer of mesodermal cells. After somite segmentation, these cells remain as an unsegmented primary muscle cell layer underlying the prospective epiderm (Youn and Malacinski, 1981). Hoechst staining revealed that the dermatome in xFxr1.MO injected embryos also was disorganized and the dermatomal cell layer seemed thickened (Figure 7b) as might be expected if somite segmentation and/or the preceding separation of myotome and dermatome had been prevented or delayed. These data suggest that xFxr1p may not be required for definition of trunk mesoderm nor for the onset of myotomal cell differentiation, but it becomes essential during the formation of functional muscle.
Figure 7.
Depletion of Fxr1p alters somite formation. (a) In situ hybridization showing the presence of the lateral xMyoD mRNA in the control side, whereas a reduction is observed in the injected side. xMyoD mRNA levels are restored after coinjection with HA.xFr1 mRNA. (b) Left, schematic overview of somite formation in Xenopus. Middle, disruption of nuclear alignment in the injected side of the same embryo depicted in a, as detected by Hoechst staining (right side). Right, somitic nuclei are realigned after rescue with xFxr1 mRNA (same embryo as in a).
Inactivation of xFxr1 mRNA Affects the Expression of a Range of Genes Associated with Muscle Formation
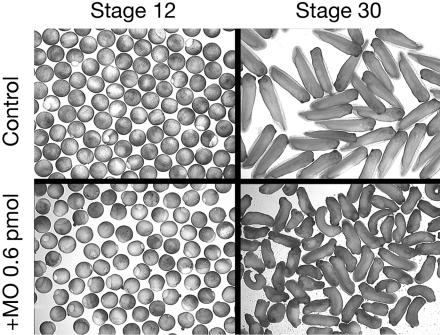
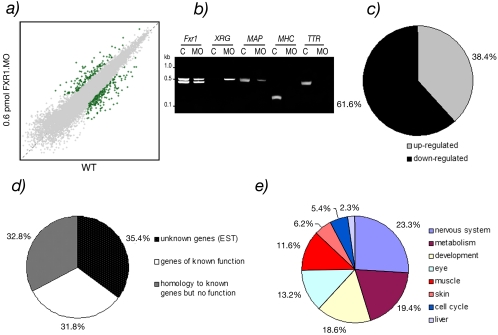
To determine the impact of the inactivation of xFxr1p on gene expression, 0.6 pmol of xFxr1.MO was injected in both blastomeres of two-cell stage embryos that were then allowed to develop until stage 30. Macroscopically, embryos inactivated for xFxr1 seemed to be shorter by ∼40% compared with the controls (Figure 8). Total RNA was extracted from mock- and MO-injected embryos and were simultaneously analyzed with high-density cDNA microarrays covering 14,400 genes expressed in X. laevis. Among the genes showing measurable differential levels of expression, a significant change (p ≤ 0.05) of >2.5-fold was observed in 409 transcripts, of which 38.4 and 61.6% were, respectively, up- or down-regulated (Figure 9, a and c). Of these 409 genes, 129 (31.8%) have a known function (Figure 9, d and e; also see Supplemental Table 1), whereas 35.4% of the differentially expressed genes have no currently assigned function, nor presented any sequence homology with known genes, whereas the remaining 32.8% showed variable homologies with characterized genes in other organisms with no yet assigned function in Xenopus (Figure 9d). The known genes (31.8% mRNAs) were subdivided into several functional subcategories (Figure 9e). 70% of these mRNAs corresponded to tissue-specific messengers, which reflected the phenotype observed. The others were essentially metabolism-related (18.6%), general developmental factors (15.5%), or cell cycle-related (2.2%). Among the tissue-specific mRNAs, approximately one-fifth (19.2%) represented cardiac or striated skeletal muscle-specific messengers, whereas the remaining mRNAs were linked to the nervous system, eye specific, skin and liver specific (26.2, 13.1, 5.4, and 2.3%, respectively). The affected muscle specific mRNAs are presented in Table 1, whereas the list of the 129 affected mRNAs is available in Supplemental Table 1. To validate the microarray data, we performed semiquantitative RT-PCR analyses on mRNAs that were either stable (Fxr1), up-regulated (IRG), or down-regulated (MAP2, MHC, and TTR). The results of these analyses (Figure 9b) are in agreement with the data presented in Supplemental Table 1.
Figure 8.
Injection of 0.6 pmol of xFxr1.MO in both blastomeres at the two-cell stage induces alteration at stage 30. RNA extracted from the control and from the injected embryos were subjected to microarray analyses presented in Figure 7.
Figure 9.
Microarray analyses of injected embryos with 0.6 pmol of xFxr1.MO versus wild-type embryos. (a) The data set in the squatter plot shows the expression levels of genes on a logarithmic scale as the median value of uninjected (x-axis) and xFxr1.MO injected (y-axis) embryos. The threshold for significancy (p < 0.05) was set at 2.5-fold of change. (b) Examples of validation of RT-PCR analyses of stable (Fxr1), increased (XIRG), and decreased (MAP2, TTR, and MHC) mRNAs in knockdown experiments. (c) Percentage of up- and down-regulated transcripts. (d) Percentage of transcripts of unknown (EST) or known functions, and those presenting homologies to genes not yet described in Xenopus. (e) Tissue distribution of the 129 known transcripts (31.8% in Figure 8d) affected. Note that due to overlapping of these transcripts in different tissues the distribution is only relative and cannot be considered as absolute.
Table 1.
Myospecific mRNAs affected by xFxr1 mRNA knockdown in Xenopus embryos stage 30
| Clone ID on chip | Fold of change | BLAST best hit | Similarity | GenBank ID |
|---|---|---|---|---|
| Striated muscle-skeletal | ||||
| Xl.16736.1.S1_at | 3.02 | Myosin binding subunit 85 [hs] | + | BU906692 |
| Xl.15784.1.A1_at | 2.57 | Voltage-dependent calcium channel γ-1 subunit [hs] | + | BJ078880 |
| Xl.1285.2.S1_x_at | −2.50 | Myosin light chain [xl] | = | BG346399 |
| Xl.8919.3.S1_x_at | −2.50 | Tubulin α6 [xl] | = | CD360597 |
| Xl.21921.1.S1_at | −2.52 | Myozenin 1 [xl] | = | BC042928.1 |
| Xl.1285.2.S2_at | −2.53 | Myosin light chain [xl] | = | BM172707 |
| Xl.11094.1.S1_at | −3.28 | Skeletal muscle. calcium channel α subunit [xl] | = | CD100310 |
| Xl.23777.1.S1_at | −3.63 | Troponin T [hs] | + | BG730694 |
| Xl.1251.1.S1_at | −3.71 | Na.K+-ATPase γ subunit [xl] | = | Y11587.1 |
| Xl.17369.1.S1_at | −5.29 | μ/m-Calpain large subunit [xl] | = | AB061521.1 |
| Striated muscle-cardiac | ||||
| Xl.1407.1.S1_at | 3.43 | K+ channel β2 subunit [xl] | = | BJ086795 |
| Xl.1120.1.S1_at | 3.33 | Acetylcholine receptor γ subunit [xl] | = | BF231684 |
| Xl.15700.1.S1_at | −3.69 | Calsequestrin 2 [xl] | = | BC041283.1 |
| Xl.5188.1.A2_at | −4.64 | Cardiac myosin heavy chain [xl] | = | BG730874 |
An additional exhaustive list of other affected tissue-specific transcripts is given as Supplemental Table 1. Clone ID corresponds to the position of the gene on the Affymetrix chip. BLAST best hit indicates the clone identity (xl, X. laevis; hs, Homo sapiens; mm, M. musculus). The fold of change indicates the relative level of expression of each gene in xFxr1.MO double-injected embryos relative to wild-type embryos. Only genes displaying a 2.5-fold, or greater, changes were considered. =, Xenopus gene; +, weak similarity; ++, moderate similarity; and +++, strong similarity.
DISCUSSION
We have shown that two xFxr1 mRNA variants are detected in X. laevis, as opposed to the seven previously identified in mammals (Kirkpatrick et al., 1999). Although a simpler animal model, Xenopus displays the same tissue-specific expression of a long, 88-kDa Fxr1p isoform as the 82-84 kDa found in mammals (Khandjian et al., 1998; Dubé et al., 2000). Also, in mouse and frog, the same distribution is observed for Fmrp because the highest levels are present in brain extracts. We observed that Fxr1p expressed in both mouse and frog are highly conserved and antibodies directed against mammalian FXR1P also recognize the frog counterpart. The additional 81 nucleotide stretch (Khandjian et al., 1998) corresponding to mammalian exon 15 (Kirkpatrick et al., 1999) also is detected in Xenopus striated muscle and heart mRNAs. Twenty-six of the 27 aa composing the pocket coded by this muscle-specific exon are maintained in frog, mouse, and human (Figure 2b). This pocket also was detected recently in zebrafish zFxr1p (Engels et al., 2004; Tucker et al., 2004). Conservation in such distant species supports the notion that FXR1P serves an essential function. However, the tissue-specific expression of the long Fxr1p isoform is regulated early during embryogenesis, because we observed that it is already present in developing striated muscle of 12.5 dpc mouse embryos (48-52 somite pairs), whereas in Xenopus it is detected only after stage 25-30, when 16-25 somites have already formed (Nieuwkoop and Faber, 1967). This suggests that it is not essential for the formation of functional somatic muscle and is perhaps a special adaptation to support later stages of muscle differentiation, maintenance, or growth.
To inactivate xFxr1 mRNA, we used the Morpholino approach, because these molecules are highly specific, stable, and are known for their low level of toxicity compared with other methods of inactivation (Summerton and Weller, 1997). Moreover, the Morpholinos specifically block translational activity by interacting with the targeted mRNA in an RNAse H-independent manner and thus do not alter its steady-state level (Summerton, 1999). The strategy was to inject one or two blastomeres of the two-cell stage embryos, knowing that the first cleavage furrows delimits the dorso-ventral axis that determines the left and right sides of the embryo (Klein, 1987). Interestingly, phenotypic effects were observed only after stage 22, corresponding with the first stages in somitogenesis (Nieuwkoop and Faber, 1967), but well before the expression of the muscle-specific long xFxr1p isoform. The external curled phenotype corresponded with an inhibition or a significant delay in the formation of the somites and was proportional with the level of xFxr1p silencing. Embryos injected with 0.3 pmol of xFxr1-MO did not show a significant loss of presomitic muscle tissue, but they did display an abnormal formation of the somites, implying that a certain minimal level of xFxr1p is required to complete myogenesis. This phenotype, and even more severe phenotypes in which trunk muscle tissues was severely reduced, could be rescued by complementation with the muscle-specific long form of xFxr1 mRNA. Coinjection of 1.5 ng of this mRNA together with 1.0 pmol of the xFxr1.MO, which silenced both long and short forms, resulted in the rescue of ∼80% of the injected embryos as their morphology could be corrected (Figure 5) and expression of MyoD (Figure 7) and xFr1p (our unpublished data) were restored. However, ∼50% rescue also could be achieved by coinjecting the mRNA encoding the short xFxr1p isoform. This is consistent with the observation that the long form of xFxr1p is detected only after 25 and more trunk somites have already formed. Thus, both the long and short xFxr1p isoforms can support the formation of early skeletal muscle in Xenopus and perhaps in mammals. These data strongly suggest that the conservation of the long protein isoform from fish to human indicates an essential role only after the initial trunk muscle is formed, perhaps for maintenance and growth, or perhaps for more specialized tissues such as cardiac muscle.
When the defect in somite formation was more closely investigated, it was found that although levels of xMyoD mRNA, a very early marker essential for muscle formation, were near normal, their striped expression pattern was disrupted. Sectioning of these embryos showed that though myotomal cells were present in the presomitic mesoderm, their rotation and alignment to form the segmented functional somitic myotome was prevented or severely delayed. The formation of the muscle dermatome also seemed to be affected. Somite segmentation was, however, fully rescued by the xFxr1 mRNA. Consistent with these observations, many transcripts encoding cardiac and striated skeletal muscle proteins were found to be down-regulated in the xFxr1.MO injected embryos (Table 1). mRNA for proteins expressed during late myogenesis, such as the myosin light chain (Latinkic et al., 2004), skeletal muscle Ca2+ channel α-subunit (Lhuillier and Tabti, 1998), troponin T (Warkman and Atkinson, 2004), and cardiac myosin heavy chain (Cox and Neff, 1995) were significantly less abundant.
Neurospecific mRNAs also were strongly affected by silencing of xFxr1: 71.4% of these showing a drastic down-regulation, in particularly mRNAs encoding proteins controlling or implicated in neurites elongation and direction (Supplemental Table 1). These results underline the important role Fxr1p also plays in the control of nervous system development, a role probably interrelated to that in muscle. Finally, 13.1% of the modulated mRNAs were eye specific and all showed a down-regulation. Indeed, our preliminary results show that xFxr1p is expressed in the developing Xenopus eye and its inactivation most probably has significant consequences on the development of the retina and lens since down-regulation of mRNA levels related to the occular apparatus, such as crystallins (Brunekreef et al., 1997; Smolich et al., 1994) or Otx5 (Vignali et al., 2000) is observed. Moreover, down-regulated proteins such as tyrosinase-related 1 (Kumasaka et al., 2003), retinol-binding protein (Quadro et al., 1999) are implicated in retina pigmentation, whereas down-regulation of cone arrestin (Craft and Whitmore, 1995) and retinal G protein-coupled receptor (Maeda et al., 2003) are likely to affect signal transduction in the differentiated eye. However, the phenotypes associated with this down-regulation would not be observed until much later developmental stages than studied here.
In vertebrate muscle, mFxr1 expression is preferentially restricted to granular structures reminiscent to costamere-like structures (Dubé et al., 2000), and this distribution is conserved in lower organisms such as the zebrafish (Engels et al., 2004). These costameric structures have been reported to contain specific mRNAs as well as proteins implicated in muscle contraction and maintenance and are thought to be reservoirs of mRNA required for local de novo protein synthesis (Cripe et al., 1993; Morris and Fulton, 1994; Fulton and Alftine, 1997; Jockusch et al., 2003). It is tempting to draw a parallel between FMRP and FXR1P functions in two different highly polarized and large cells such as neurons and muscle cells. In neurons, FMRP is thought to maintain certain classes of mRNAs in a repressed state during their transport in motile granules through neurites to their destinations, the synapses (De Diego Otero et al., 2002; Mazroui et al., 2002; Antar et al., 2004). These cargoes are then targeted and anchored to dentritic spines or filopodia, which are the sites where FMRP may regulate synthesis of proteins essential for spine development and maintenance (Miyashiro et al., 2003; Antar et al., 2004; Weiler et al., 2004). Similar to FMRP in neuron, FXR1P may play an equivalent role in muscle to maintain silent mRNAs in costameres until protein synthesis is required. We propose that in neuron, FMRP is not essential for its development, because the homologues FXR1P and FXR2P, which colocalize to the same repressed granules (De Diego Otero et al., 2002; Mazroui et al., 2002) could partially compensate for the absence of FMRP in fragile X syndrome. In contrast, because FXR1P is the sole member of the FXR protein family to be found in muscle (Khandjian et al., 1998; Bakker et al., 2000) its presence, as seen in the Xenopus experimental system, is essential for muscle development and maintenance. Given that xFxr1p silencing leads to down-regulation of many muscle-specific mRNAs, our data strongly suggest that as an RNA-binding protein, it might control specific mRNAs that are translated into proteins essential for muscle differentiation. Alternatively, xFxr1p might control a restricted number of mRNAs upstream of a complex cascade event leading to the alteration of the steady state levels of a high number of down-stream transcripts. We suggest that the Xenopus experimental system is a unique model to study whether FMR1P or FXR2P, which both have similar functions as FXR1P, could rescue the phenotype induced by xFr1p depletion, and thus to determine whether FXR1P and FMRP target a different spectrum of mRNAs. Work in this direction is in progress.
Although Laggerbauer et al. (2001) have previously reported that FMRP, but not FXR1P, induces translation repression of reporter constructs in the rabbit reticulocyte lysate system, recent results from Garnon et al. (2005) demonstrate that FXR1P does operate also as a repressor. Our working hypothesis is that the three members of the FXR family, regulate both mRNA stability and translation either individually or in concert, according to their differential levels and expression in various cells, and in this way are able to function in targeting these mRNAs to given subcellular regions. Work is in progress to reevaluate the potential repression activity of FXR1P, because we think that the muscle model will allow us to uniquely characterize the FXR1P RNA targets without interferences from the two other FMRP and FXR2P members.
Supplementary Material
Acknowledgments
We are grateful to Aaron Zorn for the gift of the Xenopus cDNA libraries. We thank Peter Vize for the MyoD cDNA. M.E.H. was a recipient of a scholarship from the Canadian Institutes of Health Research, and L. D. holds a postdoctoral fellowship from the FRAXA Research Foundation (USA). N. B. is a Research Student of the Terry Fox Foundation-National Cancer Institute of Canada. This work was supported by grants from the Natural Sciences and Engineering Research Council of Canada and by the Canadian Institutes of Health Research (to E.W.K.) and by grants from the National Cancer Institute of Canada and from the Canadian Cancer Society (to T. M.).
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05-04-0304) on July 6, 2005.
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
References
- Antar, L. N., Afroz, R., Dictenberg, J. B., Carroll, R. C., and Bassell, G. J. (2004). Metabotropic glutamate receptor activation regulates Fragile X mental retardation protein and Fmr1 mRNA localization differentially in dendrites and at synapses. J. Neurosci. 24, 2648-2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakker, C. E., and The Dutch-Belgian Fragile X Consortium. (1994). Fmr1 knockout mice: a model to study fragile X mental retardation. Cell 78, 23-33. [PubMed] [Google Scholar]
- Bakker, C. E., De Diego Otero, Y., Bontekoe, C., Raghoe, P., Luteijn, T., Hoogeveen, A. T., Oostra, B. A., and Willemsen, R. (2000). Immunocytochemical and biochemical characterization of FMRP, FXR1P, and FXR2P in the mouse. Exp. Cell Res. 258, 162-170. [DOI] [PubMed] [Google Scholar]
- Bardoni, B., and Mandel, J.-L. (2002). Advances in understanding of fragile X pathogenesis and FMRP function, and in identification of X linked mental retardation genes. Curr. Opin. Genet. Dev. 12, 284-293. [DOI] [PubMed] [Google Scholar]
- Bisson, N., Islam, N., Poitras, L., Jean, S., Bresnick, A., and Moss, T. (2003). The catalytic domain of xPAK1 is sufficient to induce myosin II dependent in vivo cell fragmentation independently of other apoptotic events. Dev. Biol., 263, 264-281. [DOI] [PubMed] [Google Scholar]
- Bontekoe, C. J., et al. (2002). Knockout mouse model for Fxr 2, a model for mental retardation. Hum. Mol. Genet. 11, 487-498. [DOI] [PubMed] [Google Scholar]
- Brunekreef, G. A., van Genesen, S. T., Destree, O. H., and Lubsen, N. H. (1997). Extralenticular expression of Xenopus laevis α-, β-, and γ-crystallin genes. Investig. Ophthalmol. Vis. Sci. 38, 2764-2771. [PubMed] [Google Scholar]
- Corbin, F., Bouillon, M., Fortin, A., Morin, S., Rousseau, F., and Khandjian, E. W. (1997). The fragile X mental retardation protein is associated with poly(A)+mRNA in actively translating polyribosomes. Hum. Mol. Genet. 6, 1465-1472. [DOI] [PubMed] [Google Scholar]
- Cox, W. G., and Neff, A. W. (1995). Cardiac myosin heavy chain expression during heart development in Xenopus laevis. Differentiation 58, 269-280. [DOI] [PubMed] [Google Scholar]
- Coy, J. F., Sedlacek, Z., Bachner, D., Hameister, H., Joos, S., Lichter, P., Delius, H., and Poustka, A. (1995). Highly conserved 3′ UTR and expression pattern of FXR1 points to a divergent gene regulation of FXR1 and FMR1. Hum. Mol. Genet. 4, 2209-2218. [DOI] [PubMed] [Google Scholar]
- Craft, C. M., and Whitmore, D. H. (1995). The arrestin superfamily: cone arrestins are a fourth family. FEBS Lett. 362, 247-255. [DOI] [PubMed] [Google Scholar]
- Cripe, L., Morris, E., and Fulton, A. B. (1993). Vimentin mRNA location changes during muscle development. Proc. Natl. Acad. Sci. USA 90, 2724-2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Diego Otero, Y., Severijnen, L.-A., van Cappellen, G., Schrier, M., Oostra, B., and Willemsen, R. (2002). Transport of fragile X mental retardation protein via granules in neurites of PC12 cells. Mol. Cell. Biol. 22, 8332-8341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devys, D., Lutz, Y., Rouyer, N., Bellocq, J. P., and Mandel, J.-L. (1993). The FMR-1 protein is cytoplasmic, most abundant in neurons and appears normal in carriers of a fragile X premutation. Nat. Genet. 4, 335-340. [DOI] [PubMed] [Google Scholar]
- Dubé, M., Huot, M. E., and Khandjian, E. W. (2000). Muscle specific fragile X related protein 1 isoforms are sequestered in the nucleus of undifferentiated myoblast. BMC Genet. 1, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberhart, D. E., Malter, H. E., Feng, Y., and Warren, S. T. (1996). The fragile X mental retardation protein is a ribonucleoprotein containing both nuclear localization and nuclear export signals. Hum. Mol. Genet. 5, 1083-1091. [DOI] [PubMed] [Google Scholar]
- Engels, B., van't Padje, S., Blonden, L., Severijnen, L. A., Oostra, B. A., and Willemsen, R. (2004). Characterization of Fxr1 in Danio rerio; a simple vertebrate model to study costamere development. J. Exp. Biol. 207, 3329-3338. [DOI] [PubMed] [Google Scholar]
- Fulton, A. B., and Alftine, C. (1997). Organization of protein and mRNA for titin and other myofibril components during myofibrillogenesis in cultured chicken skeletal muscle. Cell. Struct. Funct. 22, 51-58. [DOI] [PubMed] [Google Scholar]
- Garnon, J., Lachance, C., Di Marco, S., Hel, Z., Marion, D., Ruiz, M. C., Newkirk, M. M., Khandjian, E. W., and Radzioch, D. (2005). Fragile X-related protein FXR1P regulates proinflammatory cytokine Tumor Necrosis factor expression at the post-transcriptional level. J. Biol. Chem. 280, 5750-5763. [DOI] [PubMed] [Google Scholar]
- Hamilton, L. (1969). The formation of somites in Xenopus. J. Embryol. Exp. Morphol. 22, 253-264. [PubMed] [Google Scholar]
- Heasman, J. (2002). Morpholino oligos: making sense of antisense? Dev. Biol. 243, 209-214. [DOI] [PubMed] [Google Scholar]
- Hemmati-Brivanlou, A., Frank, D., Bolce, M. E., Brown, B. D., Sive, H. L., and Harland, R. M. (1990). Localization of specific mRNAs in Xenopus embryos by whole-mount in situ hybridization. Development 110, 325-330. [DOI] [PubMed] [Google Scholar]
- Huot, M. E., Mazroui, R., Leclerc, P., and Khandjian, E. W. (2001). Developmental expression of the fragile X-related 1 proteins in mouse testis: association with microtubule elements. Hum. Mol. Genet. 10, 2803-2811. [DOI] [PubMed] [Google Scholar]
- Imbert, G., Feng, Y., Nelson, D. L., Warren, S. T., and Mandel, J.-L. (1998). FMR1 and mutations in fragile X syndrome: molecular biology, biochemistry, and genetics. In: Genetic Instabilities and Hereditary Neurological Diseases, ed. S. T. Warren and R. D. Wells, New York: Academic Press, 27-53.
- Jockusch, B. M., Hüttelmaier, S., and Illenberger, S. (2003). From the nucleus toward the cell periphery: a guided tour for mRNAs. News Physiol. Sci. 18, 7-11. [DOI] [PubMed] [Google Scholar]
- Jowett, T., and Lettice, L. (1994). Whole-mount in situ hybridizations on zebrafish embryos using a mixture of digoxigenin- and fluorescein-labelled probes. Trends Genet. 10, 73-74. [DOI] [PubMed] [Google Scholar]
- Khandjian, E. W. (1986). UV crosslinking of RNA to nylon membrane enhances hybridization signals. Mol. Biol. Rep. 11, 107-115. [DOI] [PubMed] [Google Scholar]
- Khandjian, E. W. (1999). Biology of the fragile X mental retardation protein, an RNA-binding protein. Biochem. Cell Biol. 77, 331-342. [PubMed] [Google Scholar]
- Khandjian, E. W., et al. (1998). Novel isoforms of the fragile X related protein FXR1P are expressed during myogenesis. Hum. Mol. Genet. 7, 2121-2128. [DOI] [PubMed] [Google Scholar]
- Khandjian, E. W., Fortin, A., Thibodeau, A., Tremblay, S., Cote, F., Devys, D., Mandel, J.-L., and Rousseau, F. (1995). A heterogeneous set of FMR1 proteins is widely distributed in mouse tissues and is modulated in cell culture. Hum. Mol. Genet. 4, 783-789. [DOI] [PubMed] [Google Scholar]
- Kirkpatrick, L. L., McIlwain, K. A., and Nelson, D. L. (1999). Alternative splicing in the murine and human FXR1 genes. Genomics 59, 193-202. [DOI] [PubMed] [Google Scholar]
- Klein, S. L. (1987). The first cleavage furrow demarcates the dorsal-ventral axis in Xenopus embryos. Dev. Biol. 120, 299-304. [DOI] [PubMed] [Google Scholar]
- Kumasaka, M., Sato, S., Yajima, I., and Yamamoto, H. (2003). Isolation and developmental expression of tyrosinase family genes in Xenopus laevis. Pigment Cell Res. 16, 455-462. [DOI] [PubMed] [Google Scholar]
- Laggerbauer, B., Ostareck, D., Keidel, E. M., Ostareck-Lederer, A., and Fischer, U. (2001). Evidence that fragile X mental retardation protein is a negative regulator of translation. Hum. Mol. Genet. 10, 329-338. [DOI] [PubMed] [Google Scholar]
- Latinkic, B. V., Cooper, B., Smith, S., Kotecha, S., Towers, N., Sparrow, D., and Mohun, T. J. (2004). Transcriptional regulation of the cardiac-specific MLC2 gene during Xenopus embryonic development. Development 131, 669-679. [DOI] [PubMed] [Google Scholar]
- Lhuillier, L., and Tabti, N. (1998). Influence of muscle cells on the development of calcium currents in Xenopus spinal neurons. Neuroscience 83, 1283-1291. [DOI] [PubMed] [Google Scholar]
- Maeda, T., Van Hooser, J. P., Driessen, C. A., Filipek, S., Janssen, J. J., and Palczewski, K. (2003). Evaluation of the role of the retinal G protein-coupled receptor (RGR) in the vertebrate retina in vivo. J. Neurochem. 85, 944-956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazroui, R., Huot, M. E., Tremblay, S., Filion, C., Labelle, Y., and Khandjian, E. W. (2002). Trapping of messenger RNA by fragile X mental retardation protein into cytoplasmic granules induces translation repression. Hum. Mol. Genet. 11, 3007-3017. [DOI] [PubMed] [Google Scholar]
- Mientjes, E. J., et al. (2004). Fxr1 knockout mice show a striated muscle phenotype: implications for Fxr1p function in vivo. Hum. Mol. Genet. 13, 1291-1302. [DOI] [PubMed] [Google Scholar]
- Miyashiro, K. Y., Beckel-Mitchener, A., Purk, T. P., Becker, K. G., Barret, T., Liu, L., Carbonetto, S., Weiler, I. J., Greenough, W. T., and Eberwine, J. (2003). RNA cargoes associating with FMRP reveal deficits in cellular functioning in Fmr1 null mice. Neuron 37, 417-431. [DOI] [PubMed] [Google Scholar]
- Morris, E. J., and Fulton, A. B. (1994). Rearrangement of mRNAs for costamere proteins during costamere development in cultured skeletal muscle from chicken. J. Cell Sci. 107, 377-386. [DOI] [PubMed] [Google Scholar]
- Nieuwkoop, P. D., and Faber, J. (eds.) (1967). Normal table of Xenopus laevis (Daudin); a systematical and chronological survey of the development from the fertilized egg till the end of metamorphosis, Amsterdam: North-Holland Publishing Company.
- O'Donnell, W. T., and Warren, S. T. (2002). A decade of molecular studies of fragile X syndrome. Annu. Rev. Neurosci. 25, 315-338. [DOI] [PubMed] [Google Scholar]
- Quadro, L., Blaner, W. S., Salchow, D. J., Vogel, S., Piantedosi, R., Gouras, P., Freeman, S., Cosma, M. P., Colantuoni, V., and Gottesman, M. E. (1999). Impaired retinal function and vitamin A availability in mice lacking retinol-binding protein. EMBO J. 18, 4633-4644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi, M. C., Siomi, H., Sauer, W. H., Srinivasan, S., Nussbaum, R. L., and Dreyfuss, G. (1995). FXR1, an autosomal homolog of the fragile X mental retardation gene. EMBO J. 14, 2401-2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi, M. C., Zhang, Y., Siomi, H., and Dreyfuss, G. (1996). Specific sequences in the fragile X syndrome protein FMR1 and the FXR proteins mediate their binding to 60S ribosomal subunits and the interactions among them. Mol. Cell. Biol. 16, 3825-3832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolich, B. D., Tarkington, S. K., Saha, M. S., and Grainger, R. M. (1994). Xenopus γ-crystallin gene expression: evidence that the γ-crystallin gene family is transcribed in lens and nonlens tissues. Mol. Cell. Biol. 14, 1355-1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summerton, J. (1999). Morpholino antisense oligomers: the case for an RNase H-independent structural type. Biochim. Biophys. Acta 1489, 141-158. [DOI] [PubMed] [Google Scholar]
- Summerton, J., and Weller, D. (1997). Morpholino antisense oligomers: design, preparation, and properties. Antisense Nucleic Acid Drug Dev. 7, 187-195. [DOI] [PubMed] [Google Scholar]
- Summerton, J., Stein, D., Huang, S. B., Matthews, P., Weller, D., and Partridge, M. (1997). Morpholino and phosphorothioate antisense oligomers compared in cell-free and in-cell systems. Antisense Nucleic Acid Drug Dev. 7, 63-70. [DOI] [PubMed] [Google Scholar]
- Sutherland, G. R. (1977). Fragile sites on human chromosomes: demonstration of their dependence on the type of tissue culture medium. Science 197, 265-266. [DOI] [PubMed] [Google Scholar]
- Tucker, B., Richards, R., and Lardelli, M. (2004). Expression of three zebrafish orthologs of human FMR1-related genes and their phylogenetic relationships. Dev. Genes Evol. 214, 567-574. [DOI] [PubMed] [Google Scholar]
- Vignali, R., Colombetti, S., Lupo, G., Zhang, W., Stachel, S., Harland, R. M., and Barsacchi, G. (2000). Xotx5b, a new member of the Otx gene family, may be involved in anterior and eye development in Xenopus laevis. Mech. Dev. 96, 3-13. [DOI] [PubMed] [Google Scholar]
- Warkman, A. S., and Atkinson, B. G. (2004). Amphibian cardiac troponin I gene's organization, developmental expression, and regulatory properties are different from its mammalian homologue. Dev. Dyn. 229, 275-288. [DOI] [PubMed] [Google Scholar]
- Weiler, I. J., et al. (2004). Fragile X mental retardation protein is necessary for neurotransmitter-activated protein translation at synapses. Proc. Natl. Acad. Sci. USA 101, 17504-17509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youn, B. W., and Malacinski, G. M. (1981). Somitogenesis in the amphibian Xenopus laevis: scanning electron microscopic analysis of intrasomitic cellular arrangements during somite rotation. J. Embryol. Exp. Morphol. 64, 23-43. [PubMed] [Google Scholar]
- Zhang, Y., O'Connor, J. P., Siomi, M. C., Srinivasan, S., Dutra, A., Nussbaum, R. L., and Dreyfuss, G. (1995). The fragile X mental retardation syndrome protein interacts with novel homologs FXR1 and FXR2. EMBO J. 14, 5358-5366. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.