Abstract
MARK/Par-1 is a kinase involved in development of embryonic polarity. In neurons, MARK phosphorylates tau protein and causes its detachment from microtubules, the tracks of axonal transport. Because the target sites of MARK on tau occur at an early stage of Alzheimer neurodegeneration, we searched for interaction partners of MARK. Here we report that MARK2 is negatively regulated by PAK5, a neuronal member of the p21-activated kinase family. PAK5 suppresses the activity of MARK2 toward its target, tau protein. The inhibition requires the binding between the PAK5 and MARK2 catalytic domains, but does not require phosphorylation. In transfected Chinese hamster ovary (CHO) cells both kinases show a vesicular distribution with partial colocalization on endosomes containing AP-1/2. Although MARK2 transfected alone destabilizes microtubules and stabilizes actin stress fibers, PAK5 keeps microtubules stable through the down-regulation of MARK2 but destabilizes the F-actin network so that stress fibers and focal adhesions disappear and cells develop filopodia. The results point to an inverse relationship between actin- and microtubule-related signaling by the PAK5 and MARK2 pathways that affect both cytoskeletal networks.
INTRODUCTION
The observations reported here originated from a study of the neuronal microtubule-associated protein tau and its abnormal changes in Alzheimer's disease. The function of tau in healthy neurons is to stabilize microtubules and to ensure axonal transport along microtubules. In degenerating neurons, tau is hyperphosphorylated, detaches from microtubules, and aggregates into pathological “paired helical filaments.” The detachment from microtubules is achieved most efficiently by phosphorylating the KXGS-motifs in the microtubule-binding domain of tau, and elevated phosphorylation at these sites occurs early in Alzheimer's disease (Augustinack et al., 2002). A search for the responsible kinase lead to the identification of the MARK family of protein kinases (Drewes et al., 1997). They are related to the Par-1 kinases in Caenorhabditis elegans and Drosophila melanogaster, which are involved in the determination of embryonic polarity (reviews, Pellettieri and Seydoux, 2002; Fortini, 2004), and indeed the activity of MARK is important for neuronal polarity as well (Biernat et al., 2002). MARK kinases consist of an N-terminal catalytic domain, followed by UBA, spacer, and tail domains. One requirement for activity is the phosphorylation of a threonine in the activation loop (T208 in MARK2), which keeps the active site accessible to the substrate. In the case of MARKs from mammalian brain this is achieved by the kinase MARKK (Timm et al., 2003). Its activation, like that of MARK, leads to detachment of tau and neuronal degeneration, and similar principles appear to hold for other organisms (Nishimura et al., 2004). On the other hand, as with other multidomain kinases, regulation is likely to take place on several levels (Huse and Kuriyan, 2002). Examples are the binding of a pseudosubstrate peptide into the active site, forming a complex with an inhibitor or activator protein, dimerization, or anchoring on scaffolds or in compartments. We therefore set out to identify regulatory partners of MARK and describe here the interaction of MARK2 with PAK5, its inhibition by PAK5, and the effects of these kinases on the cytoskeleton.
PAKs (p21-activated kinases) are Ser/Thr kinases that are activated by small G-proteins such as Rac or Cdc42 in their GTP-bound state (Manser et al., 1994; reviewed by Etienne-Manneville and Hall, 2002; Jaffer and Chernoff, 2002). They are best known for regulating the dynamics of the actin cytoskeleton, e.g., formation of lamellipodia or filopodia, stability of stress fibers, and focal contacts. They have an N-terminal regulatory domain that binds Rac or Cdc42 (PBD = p21-binding domain containing the CRIB motif) and is responsible for the inhibition of kinase activity, a C-terminal catalytic domain, and an intervening domain with proline-rich motifs that can dock onto SH3-domains of other molecules such as PIX, Nck, and others. The x-ray structure of PAK1 catalytic and regulatory domains shows that, in its inactive state, this kinase is a dimer coupled by the two regulatory domains, where the kinase-inhibitory motif (KI) reaches into the catalytic cleft and blocks the activation loop of the kinase (Lei et al., 2000, 2005; Parrini et al., 2002). Binding of the G-protein at the CRIB motif of the regulatory domain causes the dissociation of the dimer and opening of the active site. The kinase then acquires activity by autophosphorylation in the activation loop, and the reattachment of the inhibitory motif is prevented by further autophoshorylation of the regulatory domain (Chong et al., 2001; Buchwald et al., 2001). The PAKs can be subdivided into group I (PAK1, 2, 3) and group II (PAK4, 5, 6). PAK5, the most recently characterized member, contains 719 residues, binds Cdc42, and occurs mainly in the brain (Dan et al., 2002; Pandey et al., 2002). It contains a CRIB motif around residues 9-30 and an inhibitory KI motif around residues 120-133 (Ching et al., 2003). The kinase domain extends from about ∼453-700. Ser602 in the activation loop must be phosphorylated for activity. By analogy with PAK6, additional activating phosphorylation likely occurs at Y608 in the activation loop (by MKK6, an activator of the p38 MAP kinase; Kaur et al., 2005).
The roles of PAK5 are only partly understood. Transgenic PAK5 knockout mice are viable, presumably because of functional redundancy with other PAKs (Li and Minden, 2003). In neuroblastoma cells, PAK5 induces filopodia, neurite outgrowth, and dendritic spines (Dan et al., 2002; Bryan et al., 2004). It has a mainly cytosolic distribution where it can activate the JNK kinase pathway (Dan et al., 2002; Pandey et al., 2002). In conjunction with mitochondria, PAK5 can phosphorylate BAD and inhibit apoptosis (Cotteret et al., 2003). Several members of the PAK family are involved in mediating the development of filopodia at the leading front (reviews Small et al., 2002; Pollard and Borisy, 2003). In addition, PAK1 provides an example of a chain linking the growth of microtubules or actin filaments around focal contacts and the leading edge of migrating cells, mediated by microtubule-bound GTP exchange factors and small G-proteins acting on PAK1 (Krendel et al., 2002; Wittmann et al., 2004). In the present study we find a direct interaction between PAK5 and MARK2, which leads to inhibition of MARK2 and opens a novel link between F-actin and microtubule regulation.
MATERIALS AND METHODS
Plasmids
pGBKT7-MARK2 wild-type (wt) and MARK2 S208A/T212A were generated by inserting a NdeI/SmaI fragment of rat MARK2 wt or S208A/T212A-pEU-HA (Drewes et al., 1997) into the corresponding sites of pGBKT7-vector. pGBKT7-MARK2 deletion-mutants were generated by PCR using primers containing appropriate mutations and restriction sites. The coding region of PAK5 was amplified by PCR from the human fetal brain library (Clontech, Palo Alto, CA) using oligonucleotides that introduce an NdeI-restriction site at the start codon and an NheI-restriction site behind the stop codon. The fragment was cloned into pEU-myc and pGADT7 and examined by sequencing. pGADT7-PAK5 deletion-mutants were generated by PCR using primers containing appropriate mutations and restriction sites. Site directed mutagenesis was performed by QuickChange site directed mutagenesis kit (Stratagene, La Jolla, CA). For recombinant expression the coding region of the kinase and its mutants were subcloned into a modified pVL1393-Vector (PharMingen, San Diego, CA; Timm et al., 2003). All PAK5-ECFP and PAK5-EYFP constructs were generated by inserting the NdeI/NheI-fragment of PAK5-pEU-myc after Klenow-treatment into the SmaI-site of the appropriate fluorescence expression vector. PAK1-ECFP was generated by inserting the BamHI/EcoRI-fragment of PAK-1-pGEX2Ti (a kind gift from A. Wittinghofer). The cloning of MARK2-pEYFP was reported previously (Timm et al., 2003). All plasmids were verified by restriction analysis and DNA sequencing. The sequence of all primers used for PCR will be made available upon request.
Yeast Two-Hybrid Analysis
Yeast two-hybrid screening and assays were performed according to the manufacturer′s instructions (Clontech, Yeast Protocols Handbook, details in Supplementary Materials).
Cell Culture and Transfection
Cell culture and transfection were performed with HEK293, CHO, LAN5, and Sf9 cells following standard protocols (see Supplementary Materials).
Antibodies and Markers
Immunofluorescence, antibodies, markers, and coupling methods are described in the Supplementary Materials.
Immunoprecipitation and Immunoblot Analysis
For the pulldown assay equal amounts (1.5 μM) of different recombinant GST-MARK2 constructs were mixed with lysates of Sf9 cells expressing PAK5 wt, active or inactive mutant (75, 30, and 10 nM, respectively) and incubated overnight. The proteins were precipitated for 2 h using glutathione-Sepharose 4B beads (Amersham, Piscataway, NJ). After centrifugation the beads were washed twice with phosphate-buffered saline, resuspended in Laemmli sample buffer, and analyzed by SDS-PAGE followed by Coomassie staining or Western blotting with PAK5 antibody. PAK1, PAK5, and MARK2 were expressed in HEK293 cells and coimmunoprecipitated and analyzed by standard procedures (see Supplementary Materials). methods of subcellular fractionation are described in the Supplementary Materials.
Preparation of MARK2 and PAK5
MARK2 was prepared and assayed for kinase activity as described (Drewes et al., 1997). The preparation of PAK5 and the in vitro kinase assay is described in the Supplementary Materials.
RESULTS
Identification of PAK5 as a MARK2-interacting Protein
MARK2 has been shown to induce microtubule disruption by phosphorylating microtubule-associated proteins (Drewes et al., 1997). Recently we showed that MARK2 is activated by a Ste20-like kinase purified from brain, termed MARKK (Timm et al., 2003). However, the signaling mechanisms involved in MARK2 regulation remain to be elucidated. To identify regulators or alternative substrates of MARK2, a human fetal brain cDNA library was screened using the MATCHMAKER yeast two-hybrid system. The bait was obtained by cloning the complete coding sequence of a kinase-dead MARK2 cDNA mutant into the two-hybrid vector, thus creating a fusion with the GAL4 DNA-binding domain. This mutation of MARK2 was chosen because it eliminates the phosphorylation sites in the activation loop of MARK2 (T208A/S212A = MARK2AA) and thus stabilizes the interactions with MARK2 partners, as shown for the example of the activating kinase MARKK (Timm et al., 2003). Out of 105 initial transformants, 69 clones were selected because of their growth on medium lacking histidine and positive α-galactosidase assay. Nucleotide sequence analysis revealed 10 positive clones encoding the cytosolic scaffold proteins 14-3-3ζ, 14-3-3β and a fragment of the catalytic domain of the protein kinase PAK5 (residues 502-719). This kinase is a member of the mammalian p21-activated kinase-II subfamily and is predominantly expressed in brain (Dan et al., 2002; Pandey et al., 2002;). For further experiments the full-length PAK5-cDNA was cloned from the human fetal brain library by PCR.
The Catalytic Domain of PAK5 Is Associated with the Catalytic Domain of MARK2
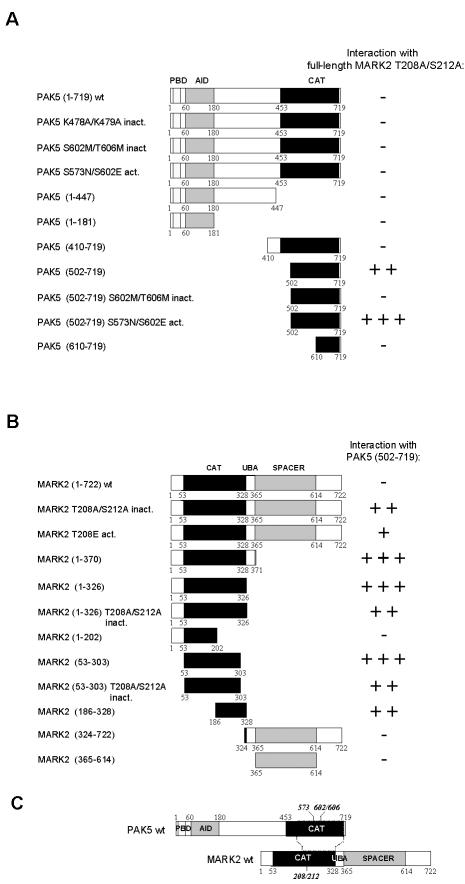
We next determined the region of PAK5 that interacts with MARK2 (Figure 1A). Full-length kinase-dead MARK2AA was used as a bait, and various deletion constructs of PAK5 as prey in a yeast two-hybrid interaction assay. This included constructs with successive truncations from the N- or C-terminus of PAK5, as well as kinase-dead (mutations in the catalytic pocket, K478A/K479A = PAK5AA or in the activation loop, S602M/T606M = PAK5MM), or constitutively active mutants of PAK5 (S573N/S602E = PAK5NE; Dan et al., 2002). Neither full-length PAK5 nor any of the mutants showed any interaction with MARK2 in the yeast two-hybrid assay (Figure 1A). Likewise, the C-terminally truncated forms containing only the PDB and parts of the middle domain were negative. The only successful clone in the interaction assay was the wt and the constitutively active fragment 502-719 comprising most of the catalytic domain of PAK5, whereas the kinase-dead mutant of this fragment, or smaller and larger fragments failed to show interactions with MARK2. Finally, none of the PAK5 constructs interacted with full-length wild-type or constitutively active MARK2 (T208E = MARK2E).
Figure 1.
Interactions of MARK2 and PAK5 mapped by yeast two-hybrid screening. (A) Schematic diagrams of PAK5-constructs used to map the MARK2-binding site. Amino acid residue numbers at boundaries of deletion constructs are indicated below the schematics. Yellow, blue, and red boxes indicate the p21-binding domain (PBD), the autoinhibitory domain (AID), and the kinase domain (CAT). Results of two-hybrid analysis are shown on the right (-, no interaction; +, weak; ++, strong; +++, very strong interaction). (B) Schematic diagrams of MARK2-constructs used to map the PAK5 binding site. Red and blue boxes indicate the kinase domain (CAT) and the SPACER domain, respectively. (C) Model of interaction of MARK2 and PAK5. The catalytic domain of PAK5 binds to the kinase domain (activation loop) of MARK2.
In a reverse set of experiments the catalytic fragment 502-719 of PAK5 was used as a bait and probed against various constructs of MARK2 (Figure 1B). As expected there was an interaction with full-length kinase-dead MARK2AA and its active mutant MARK2E, but not with wild-type MARK2. In addition most derivatives of the catalytic domain of MARK2 showed interactions with the catalytic domain of PAK5 (residues 502-719), independently of whether the phosphorylation sites in the activation loop of MARK2 were mutated or not. By contrast, constructs containing only the UBA, spacer, or tail domains of MARK2 did not interact with PAK5 (Figure 1B). Thus, the two kinases MARK2 and PAK5 interact via their catalytic domains, but this interaction appears to depend on conformational states and/or domain compositions (Figure 1C). In the yeast two-hybrid assay PAK5 interacts with MARK2 only with its catalytic domain (502-719), but the interaction site becomes inaccessible in the full-length PAK5 protein. This result holds independently of whether full-length PAK5 has wild-type activity, is constitutively active or inactive. On the other hand, MARK2 can interact as a full-length protein, provided that it is inactivated, but otherwise the catalytic domain of MARK2 interacts with PAK5 independently of the state of activation (Figure 1B).
Interactions of PAK5 and MARK2 in Cells
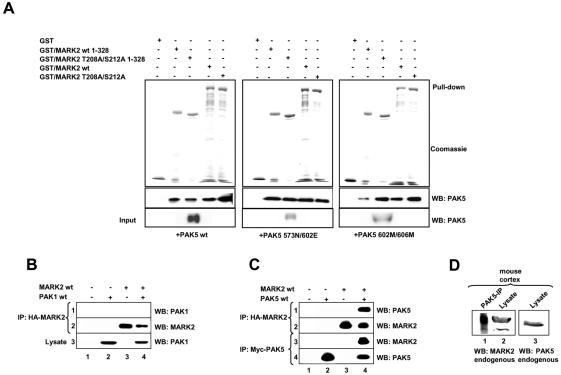
To confirm the results of the direct two-hybrid tests a pulldown assay with bacterial purified GST-tagged MARK2 and his-tagged PAK5 expressed in Sf9 cells was carried out. Both wild-type PAK5 as well as the active and inactive mutants interacted with MARK2 (Figure 2A). This indicates that the kinase activity of PAK5 or MARK2 is not required for the interaction with the partner kinase, although the binding to active PAK5 was most pronounced. Thus, in contrast to the yeast two-hybrid assay, full-length PAK5 can interact in cells with full-length MARK2 (with decreasing strength: active > wild-type > inactive PAK5).
Figure 2.
Interaction of PAK5 with MARK2 determined by GST pulldown assay and coprecipitation. (A) His-tagged PAK5 expressed in Sf9 cells (wt 75 nM, active 30 nM, or inactive mutant 10 nM) was pulled down with equal amounts (1.5 μM) of different bacterially purified GST-MARK2 constructs. The pulldown fractions were analyzed by SDS-PAGE followed by Coomassie staining (GST-constructs) or immunoblotting with anti-PAK5 antibody. Quantification shows that typically 2-3% of PAK5 was pulled down. (B and C) HEK293 cells were transfected with plasmids encoding CFP-PAK1 (B), myc-tagged PAK5 (C) and HA-tagged MARK2 either singly (lanes 2 and 3) or in combination (lanes 4). Empty pEU vector (lane 1) was used to make the total amount of transfected DNA equivalent. Cell lysates were immunoprecipitated with anti-HA antibody (B, row 2, and C, row 2 and 3) and immunoblotted either with anti-PAK1 antibody (B, row 1) or anti-myc antibody (C, row 1). Expression of PAK1 and myc-tagged PAK5 was analyzed by immunoblotting with anti-PAK1 antibody (B, row 3) or anti-myc-antibody (C, row 3). (D) The cortex of an adult mouse was lysed and endogenous PAK5 was immunoprecipitated with anti-PAK5 antibody. The same probe was immunoblotted with anti-MARK2 antibody (lane 1). Endogenous expression of both proteins in the lysate was analyzed with the corresponding antibodies (lanes 2 and 3). IP, immunoprecipitation; WB, Western blotting.
The kinase domains of the PAK-family members are closely related. To examine the specificity of the PAK5-MARK2 interaction, we tested whether PAK-1 can also interact with MARK2. We coexpressed wild-type PAK1 and HA-tagged wild-type MARK2 in HEK293 cells. When lysates of these cells were immunoprecipitated with anti-HA antibody, PAK1 was not detected in the HA-MARK2 immune complex (Figure 2B, lane 4, row 1), even though both proteins were expressed (rows 2 and 3). By comparison, myc-tagged wild-type PAK5 coimmunoprecipitated well under the same conditions with HA-MARK2 (Figure 2C, lane 4, row 1), demonstrating the specificity of this interaction. The same holds for the reciprocal experiment where HA-MARK2 was coimmunoprecipitated with myc-tagged PAK5 (Figure 2C, lane 4, row 3).
To investigate the biological relevance of the PAK5-MARK2 complex, we observed whether the interaction occurs under normal in vivo conditions in which neither protein is overexpressed. The cortex from an adult mouse was lysed and endogenous PAK5 was immunoprecipitated from this extract. Endogenous MARK2 was clearly detected in this immune complex (Figure 2D, lane 1). Lanes 2 and 3 showed the expression of both proteins in the brain lysate, implying that native MARK2 and PAK5 are associated in neurons.
PAK5 Binds to MARK2 and Inhibits Its Kinase Activity
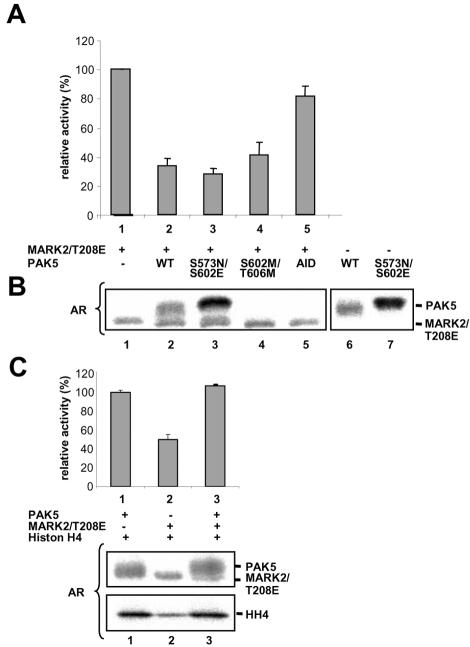
Because the catalytic domains of the two kinases interact with one another, the question arises whether this influences the kinase activities. For example, the activity of MARK2 can be measured by its phosphorylation of the peptide TR1 derived from the first repeat sequence of tau protein. Constitutively active MARK2E (T208E; Timm et al., 2003) and PAK5 or its variants were mixed together with the TR1 peptide, incubated with [32P]ATP, and the phosphorylation of TR1 measured by the incorporation of radioactive phosphate. MARK2 alone (without PAK5) had the highest activity (=100% in Figure 3A, lane 1). By contrast, wild-type PAK5 suppressed the activity of MARK2 significantly (about threefold at equimolar concentrations of PAK5 and MARK2, lane 2, p < 0.05). Similar observations were made for the active mutant PAK5NE and inactive PAK5MM (lanes 3 and 4, p < 0.05). The magnitude of the effect varied depending on the state of activity of PAK5, i.e., the inhibitory effect was strongest with constitutively active PAK5. This correlates well with the interaction data (Figure 2A). The data suggested that the inhibition of MARK2 was due to the mutual binding of both catalytic domains. To corroborate this, the experiment was also done with the noncatalytic N-terminal domain of PAK5 (residues 1-181), which does not bind to MARK2 and consequently has little effect on the activity of MARK2 (lane 5, p > 0.05, no significance). Assaying the activity of MARK2E in a range of different concentrations of PAK5NE yielded Ki values around 1 μM, with some variations between protein preparations (unpublished data).
Figure 3.
PAK5 inhibits the kinase activity of MARK2 but not vice versa. (A) The inhibition of constitutively active MARK2T208E by recombinant PAK5 was measured via the phosphorylation of the tau peptide TR1 by MARK2. The kinase activity of MARK2T208E alone was normalized to 100% (lane 1). PAK5 wild-type and different mutants reduce the kinase activity of MARK2 about threefold (lanes 2-4). The N-terminal domain of PAK5 (1-181) is only marginally inhibitory, consistent with the lack of interaction with MARK2 (lane 5). Triplicate experiments showing mean ± SE. (B) Autoradiograms of combinations of PAK5 and MARK2 in the presence of TR-1 peptide. Lane 1, MARK2 alone; lanes 2-5, combinations of PAK5 and MARK2 mutants; and lanes 6-7, PAK5wt or PAK5NE alone. Each kinase shows some autophosphorylation, but the phosphorylation of MARK2 does not depend on the activity of PAK5 (see lanes 3 and 4). (C) Autoradiograms of PAK5wt and active MARK2T208E in the presence of histone H4. Lane 1, PAK5 shows some autophosphorylation (top) and strongly phosphorylates histone H4 (bottom). Lane 2, MARK2T208E only weakly phosphorylates histone; lane 3, the presence of MARK2T208E does not alter the phosphorylation of histone by PAK5 and shows that PAK5 is not inhibited. AR, autoradiogram.
In principle, it is conceivable that the inhibition of MARK2 kinase activity is due to phosphorylation by PAK5. We therefore checked the state of phosphorylation by autoradiography (Figure 3B). Both kinases show some autophosphorylation, visible best when only one kinase is present (lanes 6 and 7). In the case of PAK5, the autophosphorylation increases when comparing wild-type with the active mutant (top bands in lanes 2 and 3), and it disappears for the kinase-dead mutant (lane 4). In the case of MARK2 there was no change in autophosphorylation, regardless of whether PAK5 was present, active, or inactive (bottom bands in lanes 3 and 4). We conclude that PAK5 does not phosphorylate MARK2, and therefore any change in activity of MARK2 must be due to the binding of PAK5. However, although PAK5 inhibits MARK2 by binding, the reverse does not hold: MARK2 has no influence on PAK5 activity, as seen in Figure 3C (lanes 1 and 3), which shows that the addition of MARK2 causes no change in the phosphorylation of the substrate histone.
PAK5 and MARK2 Colocalize in Vesicle-like Structures
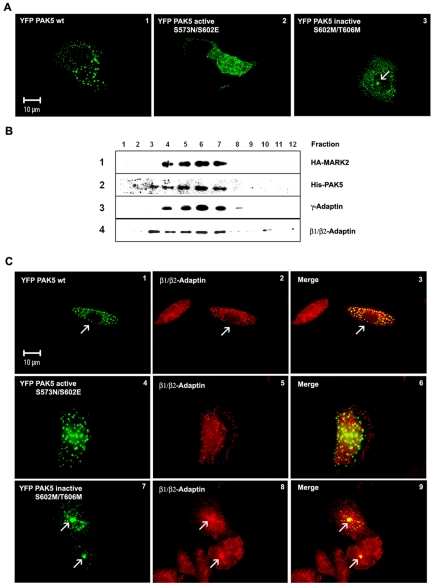
To examine the subcellular localization of MARK2 and PAK5, CHO cells were cotransfected with plasmids of both kinases, singly or jointly, and analyzed by fluorescence microscopy (CFP, YFP) or immunofluorescence. Figure 4A, 1-3, shows cells transfected with YFP-tagged wild-type PAK5, active PAK5NE, or constitutively inactive PAK5MM. The wild-type kinase has a dotted appearance, suggesting a distribution on vesicles (Figure 4A1). The inactive mutant PAK5MM has a similar vesicular distribution, but in addition the centrosome shows enhanced labeling (Figure 4A3; more apparent later, see below). The constitutively active mutant PAK5NE is also mainly vesicular, with an elevated diffuse background throughout the cytosol, suggesting partial detachment from vesicles (Figure 4A2). To check whether the clustering of PAK5MM around the centrosome was due to aggresome formation the cells were immunostained for vimentin; however, this did not reveal the cagelike structures typical of aggresomes (Kopito, 2000; Supplementary Figure S2). To identify the nature of the vesicles carrying PAK5, we labeled the cells with different markers (e.g., for peroxisomes, lysosomes, mitochondria; see Supplementary Figure S1). Partial coincidence of transfected PAK5 was observed with the AP adaptor complex (Figure 4C, 1-9), but not with other cell organelles. More specifically, in a subcellular fractionation experiment the vesicles carrying MARK2 and PAK5 were identified as part of the trans-Golgi network. PAK5 and MARK2 occurred in the same fractions as β1/β2- and γ-adaptin, the subunits of the AP-1 adaptor complex (Figure 4B).
Figure 4.
Subcellular distribution of PAK5. (A) Cytoplasmic distribution of different PAK5 constructs in CHO cells. The cells transfected with different YFP-PAK5 plasmids (wild-type, active PAK5NE, and inactive PAK5MM mutants) were cultured for 16 h and fixed, and the intracellular distribution of the proteins was visualized by fluorescence microscopy. PAK5 wild-type and kinase dead mutant are distributed in vesiclelike dots (A1 and A3), whereas the constitutively active mutant shows vesicular and cytosolic distribution throughout the cell (A2; PAK5 shown in green). (B) Total vesicle proteins from Sf9 cells expressing MARK2 wt and PAK5 wt were fractionated on 2.5-30% discontinuous iodixanol gradients. Twenty microliters of each fraction was separated by SDS-PAGE and immunoblotted with anti-HA (row 1), anti-PAK5 (row 2), anti-γ-adaptin (Golgi/TGN marker) (row 3), and anti-β1/β2-adaptin antibodies (row 4). Fractions are numbered from top of gradient (left) to bottom (right). (C) (1-3) Colocalization of transfected YFP-PAK5 wt (green, 1) with endogenous β-adaptin visualized by fixing and staining with an antibody against β1/β2-adaptin (red, 2) which shows a vesicular distribution with a cytosolic background. PAK5 and β1/β2-adaptin colocalize on the vesicles (3, yellow merge). (4-6) Similar experiment with constitutively active YFP-PAK5NE showing a vesicular and cytosolic distribution throughout the cell and is no longer colocalized with β1/β2-adaptin. (7-9) Similar experiment with inactive YFP-PAK5MM. The colocalization is concentrated on the pericentriolar region (merge, 9, arrows), reminiscent of MARK4 (Trinczek et al., 2004). The highest coincidence is observed with wild-type PAK5.
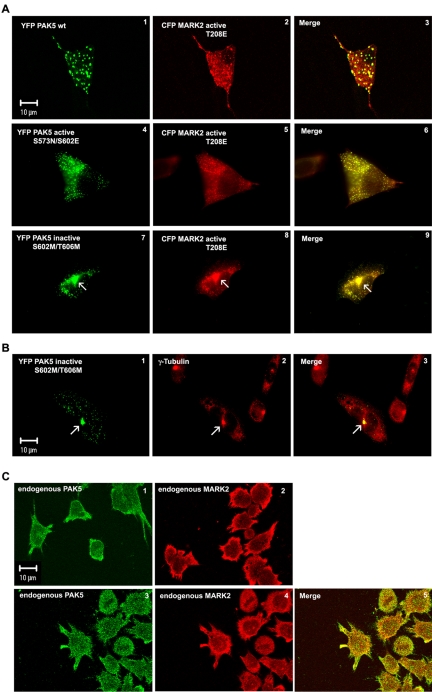
Next we asked how the localization of MARK2 was related to that of PAK5. We had shown earlier that MARK2 has a punctate distribution with a diffuse background, suggesting that it was partially associated with vesicles (Drewes et al., 1997). To study the localization of the two kinases, CHO cells were cotransfected with wild-type YFP-labeled PAK5 and CFP-labeled active MARK2E. As can be seen in Figure 5A, 1-3, both kinases colocalize on vesicles. The diffuse background of MARK2 did not colocalize with PAK5.
Figure 5.
Subcellular localization of MARK2 and PAK5. (A) Colocalization of different YFP-PAK5 constructs with transfected CFP-MARK2 (red). (A1-3) Cotransfection of PAK5wt and active MARK2E shows colocalization of both kinases on vesicles and a diffuse background of MARK2E. (A4-6) Cotransfection of active PAK5NE and active MARK2E shows colocalization on vesicles and a diffuse background of both kinases. (A7-9) Cotransfection of inactive PAK5MM and active MARK2E shows colocalization on vesicles and accumulation of both kinases around the centrosome (arrow). (B) Colocalization of transfected inactive YFP-PAK5MM (green) with centrosomes, visualized by fixation and labeling with an antibody against γ-tubulin (red). The merge (yellow, B3) confirms that inactive PAK5 preferentially localizes on the centrosome (arrow). (C) Localization of endogenous PAK5 (C1) or endogenous MARK2 (C2) stained with specific PAK5 or MARK2 antibodies, and TRITC secondary antibody in differentiated LAN5 cells. Colocalization of endogenous PAK5 (C3, green) visualized by first staining with a PAK5-specific antibody plus Cy5 secondary antibody and then staining endogenous MARK2 in differentiated LAN5 cells (C4, red) with a MARK2 antibody (SA 2117) directly coupled to Alexa 488.
When the experiment was done with active PAK5NE and active MARK2E we observed colocalization of the vesicular components as well as the cytosolic background that is typical of active PAK5NE and MARK2E (Figure 5A, 4-6). When inactive PAK5MM was used together with active MARK2E (Figure 5A, 7-9), we find fewer but larger vesicles with a high degree of colocalization, notably at the centrosome where the vesicles are concentrated. In summary, PAK5 and MARK2 largely colocalize with each other, independently of the state of activation of PAK5. However, the state of activation influences the distribution: wild-type PAK5 is on larger vesicles, active PAK5NE is on smaller vesicles with a high diffuse cytosolic background, kinase-dead PAK5MM is concentrated around the centrosomes. In each case, the distribution of MARK2 follows that of PAK5, suggesting that the two kinases are associated throughout. To confirm the centrosomal staining of inactive PAK5, we immunostained the cells with an antibody against γ-tubulin, a marker of centrosomes. Inactive PAK5 is vesicular but strongly concentrated on the centrosome where it overlaps with γ-tubulin (Figure 5B, 1-3).
To examine colocalization of endogenous PAK5 and MARK2, we differentiated human neuroblastoma cells (LAN5) and costained for both proteins with specific antibodies (Figure 5C, 1-5). Both proteins show the same distribution at the cell membrane and in neurites (Figure 5C, 1 and 2, single staining) and colocalize partly (Figure 5C, 3 and 4, double staining with Alexa 488- and Cy5-labeled antibodies).
PAK5 Inhibits MARK2 and Regulates the Stability of Microtubule and Actin Networks
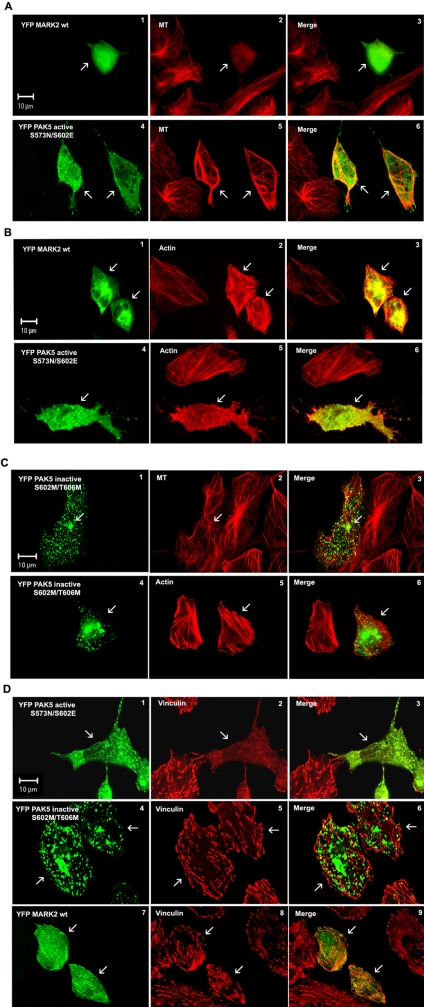
The next question was: Does the colocalization and interaction of PAK5 and MARK2 have a functional significance in cells? We had shown in vitro that PAK5 inhibits MARK2 by binding to it, rather than by phosphorylation. In addition our earlier work had shown that MARK2 has a strong influence on the dynamics of the microtubule network by phosphorylating microtubule-associated proteins, detaching them from microtubules, and thus destabilizing them (Ebneth et al., 1998; Illenberger et al., 1998). On the other hand, PAK family members tend to make the actin cytoskeleton more dynamic (Manser, 2002; Bokoch, 2003). Thus we asked how the interaction of the two kinases would affect the cytoskeleton in cells. We first consider the microtubule network. When we transfect wild-type YFP-MARK2 alone into CHO cells, microtubules disappear and the cell eventually dies (Figure 6A2), in agreement with our earlier studies on MARK (Drewes et al., 1997; Timm et al., 2003; note that constitutively active MARK2E is toxic so that no transfected cells would be found). By contrast, the overexpression of active PAK5NE leads to the stabilization of microtubules, which frequently form bundles in the cytoplasm (Figure 6A, 4-6, arrow). Opposite effects are found with the actin network. Overexpression of wild-type MARK2 leads to the stabilization of actin stress fibers (Figure 6B, 1-3, arrows), whereas active PAK5NE causes the disappearance of actin stress fibers, actin becomes dynamic, and the cell develops filopodia (Figure 6B, 4-6, arrow).
Figure 6.
Effects of PAK5 and MARK2 on the stability of microtubules and actin filament networks. CHO cells transfected with different YFP-PAK5 and YFP-MARK2 (green) constructs were cultured for 16 h, fixed, and costained with YL1/2- and TRITC-secondary antibody (MT-staining, red) or with anti-vinculin antibody and Cy5-secondary antibody, respectively (red). Actin was stained using rhodamine-conjugated phalloidin (red). Transfected cells are indicated by arrows. (A and B) In cells expressing wild-type YFP-MARK2 (A1 and B1), microtubules disappear (A2, arrow) and actin stress fibers are stabilized (B2). In contrast, constitutively active PAK5 (A4 and B4) stabilizes MT (A5), but the actin stress fibers are dissolved (B5). (C) Inactive PAK5 has no effect on stability of microtubules and actin networks (C1-6). (D) Active PAK5 causes a dissolution of focal adhesions (D1 and D2), whereas cells expressing inactive PAK5 or MARK2 show normal vinculin staining (D4, D5, D7, and D8).
In both types of experiments, the transfection of cells with the inactive kinases has no major effect on the relative stability of the microtubule or actin networks. This is demonstrated in Figure 6C, where CHO cells were transfected with inactive PAK5MM. Microtubules and actin filaments appear normal (Figure 6C, 1-6, arrows), and PAK5MM is enriched near the centrosomes (Figure 6C, 1-3, arrows). The effect of active PAK5NE can also be demonstrated by visualizing focal adhesions (stained by vinculin). Active PAK5NE causes their dissolution while inactive PAK5MM preserves focal adhesions in parallel with actin stress fibers (Figure 6D, 1-6, arrows).
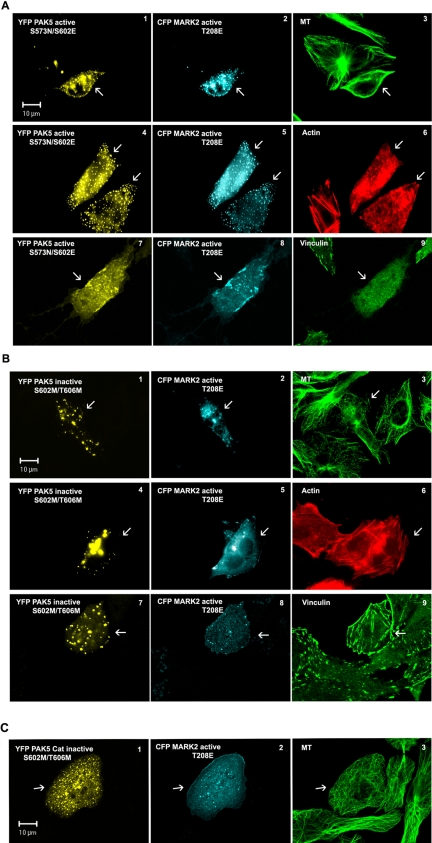
How can these results be interpreted in the light of the PAK5-MARK2 interactions described above? One key finding is that PAK5 and its mutants inhibit MARK2 activity in vitro by binding to the catalytic domain, and the two proteins are closely colocalized in cells. One would therefore expect that active or inactive PAK5 eliminates the effect of MARK2 on the cytoskeleton (i.e., the destabilization of microtubules). This is indeed the case. Figure 7 shows that if active PAK5NE is coexpressed with active MARK2E the microtubule network is protected (Figure 7A3, arrow), whereas actin stress fibers and focal adhesions are dissolved, which correlates with the emergence of filopodia (Figure 7A, 6 and 9, arrows). A similar effect of MARK2 inhibition and microtubule preservation is obtained by coexpressing inactive PAK5MM with active MARK2E. However, in this case the actin stress fibers are not rendered dynamic, and consequently focal adhesions are preserved and filopodia do not evolve (Figure 7B, 3, 6, and 9). This experiment shows that active PAK5NE has two independent effects on the cytoskeleton: First, it stabilizes microtubules by binding and inhibiting MARK2, and second, it makes actin dynamic by dissolving stress fibers and focal adhesions and inducing the formation of filopodia. Inactive PAK5MM only shows the first effect (on microtubules, Figure 7B3), but the second effect is absent because it would require PAK5 activity (Figure 7B, 6 and 9). The experiment was also done with the noncatalytic N-terminal domain of PAK5 (residue 1-181, PAK-AID), which does not bind to MARK2. In agreement with the results of the kinase assay, this PAK5 domain has no effect on the activity of MARK2, which causes microtubules to disappear and the cell eventually to die (Drewes et al., 1997, and Supplementary Figure S4, 1-6). Furthermore, the inactive catalytic domain of PAK5 colocalizes and inhibits active MARK2 (Figure 7C), demonstrating that the binding of the two catalytic domains causes the inhibition (and not an indirect signaling via the N-terminal domain of PAK5). To measure the protective effect on microtubules provided by PAK5, an antibody against Glu-tubulin was used in cells singly transfected with PAK, revealing a strong increase in stable microtubules (Supplementary Figure S3A). To check the inhibition of MARK2 in cells, the ability of PAK5 to suppress the phosphorylation of tau by MARK2 was tested using htau40 stably transfected CHO cells (Supplementary Figure S5, 1-9). The data clearly confirm that the PAK5 acts as a MARK2 inhibitor.
Figure 7.
PAK5 inhibits the MARK2 effect on the microtubule and actin networks. (A) CHO cells coexpressing the constitutively active form of PAK5 (A1, A4, and A7, yellow) and MARK2 (A2, A5, and A8, cyan) show a stabilized microtubule network (A3, green) and a dynamic actin cytoskeleton discernible by loss of actin stress fibers (A6, red) and focal adhesions (A9, green). (B) Coexpression of inactive PAK5 (B1, B4, and B7, yellow) and active MARK2 (B2, B5, and B8, cyan) results in an inhibition of MARK2 and stabilization of microtubules (B3). Actin stress fibers (B6, red) and focal adhesions (vinculin, B9, green) also remain stable because only active PAK5 makes the actin organization dynamic. Transfected cells labeled by arrows. (C) Coexpression of inactive catalytic domain of PAK5MM (residues 502-719) and active MARK2E shows partial colocalization (C1 and C2) and inhibition of MARK2 activity, as seen by the intact microtubule network (C3).
DISCUSSION
This article describes the interaction of two types of protein kinases, both of which have a strong and complementary effect on the cytoskeleton. Kinases of the PAK family are known for their influence on remodeling the actin filaments, whereas MARK kinases regulate microtubule dynamics. These polymer networks have different compositions, distributions, and tasks in the cell. Each of them is regulated by their distinct set of signaling pathways and molecules whose mechanisms of cross-talk are gradually emerging. Our study opens up a new connection between microtubule- and actin-based signaling. The initial observation was that kinases of the MARK family can make microtubules dynamic by phosphorylating and detaching their stabilizing MAPs. MARK homologues (e.g., Par-1) may have different roles in various cell types and organisms (see below), but in our context the main feature is that in neuronal cells the local destabilization of microtubules can be achieved by the phosphorylation of tau (or related MAPs) and is important for neurite outgrowth and growth cone advance (Biernat and Mandelkow, 1999; Biernat et al., 2002). Another noteworthy aspect is that this type of phosphorylation is an early event in the neuronal degeneration of Alzheimer's disease, suggesting that the overactivation of MARK might play a role in the breakdown of microtubules and the interruption of axonal transport (Augustinack et al., 2002). We were therefore interested in the regulation of MARK. One pathway is the phosphorylation of the catalytic domain by the recently identified upstream kinase MARKK, a member of the Ste20-like kinases (Timm et al., 2003; also known as TAO-1 in the context of JNK signaling, Hutchison et al., 1998). On the other hand, MARK is an unusually large kinase, it contains several domains and could therefore interact with other potential regulatory partners. We searched for them by a yeast two-hybrid screen using a fetal brain library. The most conspicuous hits were 14-3-3 proteins (β and ζ isoforms) and the PAK5 isoform of p21-activated kinases. This family of kinases is best known for its role in signaling to the actin cytoskeleton after activation by the small GTPases Rac or Cdc42 (Etienne-Manneville and Hall, 2002; Bokoch, 2003).
To identify the binding regions, several truncated forms of MARK2 and PAK5 were generated and probed by a two-hybrid interaction assay. The results show that the interaction is based on the two catalytic domains, which are located on opposite ends of the respective molecules (N-terminal for MARK2, C-terminal for PAK5, Figure 1). A more detailed analysis of the interaction assay shows that the catalytic domain of one kinase binds the catalytic domain of the other, preferentially when the noncatalytic domains are absent, suggesting that some conformation or steric hindrance of the whole kinase molecules weakens the interaction in the yeast assay (Figure 1, A and B). By contrast, the full-length proteins can interact in cells, as shown by coimmunoprecipitation (Figure 2). Furthermore, the interaction is strengthened by the inactive form MARK2AA (Figure 1A), possibly because the conformation of the regulatory loop contributes to the binding.
In the case of the MARK2cat-PAK5cat complex it is surprising that the interaction has distinct consequences for the kinase activities: MARK2 is inhibited by binding of PAK5, but PAK5 remains active when bound to MARK2 (Figure 3). The inhibition of MARK2 is based on the binding of PAK5, not on phosphorylation (trans- or autophosphorylation; Figure 3A). Consistent with the binding and activity data, the two proteins largely colocalize in cells, as judged by confocal imaging of fluorescent derivatives transfected into CHO cells (Figure 5). The staining is punctate, reminiscent of the staining of vesicles, and similar to the distribution of MARK1 or MARK2 reported earlier (Drewes et al., 1997). There is partial overlap of PAK5/MARK2 with endosomal vesicles carrying the adaptor complexes AP-1/2 (Figure 4). In addition there is a noticeable shift in distribution of PAK5, depending on its activity: the most active variant PAK5NE has a higher cytosolic (and lower vesicular) fraction than the inactive PAKMM. This suggests that sequestration of PAK5 on vesicles is another mechanism of regulation.
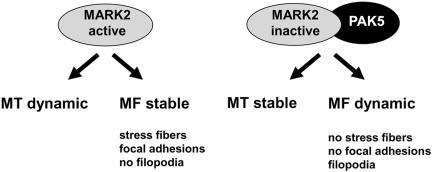
When transfecting the kinases into CHO cells, the cellular effects can be described roughly in terms of their effects on the cytoskeleton: To a first approximation, the initial effect of MARK2 alone is to render microtubules dynamic, whereas actin stress fibers remain stabilized, and in fact appear to be overstable (Figure 6; note that cell degeneration sets in at longer exposures or higher concentrations of MARK2; see Drewes et al., 1997). Conversely, the effect of PAK5 alone is to dissolve the actin stress fibers and focal adhesions and to promote filopodia, whereas microtubules are stabilized and form bundles (Figure 6A5). These effects on the actin and microtubule cytoskeleton depend on the activity of the kinases and are therefore not observed with kinase-deficient mutants (Figure 6). However, when both kinases are coexpressed in the cells, MARK2 can no longer dissolve microtubules, whereas PAK5 still causes the dissolution of stress fibers and focal adhesions and induces filopodia. This illustrates that on a cellular level MARK2 is inhibited by PAK5, in agreement with the inhibition seen with the activity assays in vitro. Moreover, this effect is also observed with inactive PAK5, confirming the in vitro result that MARK2 is inhibited by binding of PAK5 and not by phosphorylation (Figure 7). It therefore appears that the interplay between actin stress fibers and microtubules can be controlled by the switches PAK5 and MARK2. The unexpected feature is that the actin switch PAK5 can down-regulate the microtubule switch MARK2, but not vice versa. From the viewpoint of MARK/microtubule signaling this result is remarkable because of the curious asymmetry of switching: MARK is switched on by an upstream kinase, MARKK, and therefore the activation of MARKK has the same effect as that of MARK, namely microtubule destabilization (Timm et al., 2003). However, switching MARK off can be achieved by a kinase of the PAK family that is embedded in a different cellular signaling network controlling the “competing” actin cytoskeleton. These conclusions are summarized in the diagram of Figure 8.
Figure 8.
Summary of effects of PAK5 and MARK2 on cytoskeleton. For details see text.
From a structural perspective, how could PAK5 be inhibitory for MARK2? This result was unexpected, considering that known kinase-regulatory proteins (e.g., cyclins, p35 for the CDK family) are usually not kinases themselves (Huse and Kuriyan, 2002). Some kinases contain inhibitory sequences that can become pseudosubstrates after phosphorylation and block their own active site (e.g., Ser9 in GSK-3β phosphorylated by PKB, Dajani et al., 2001) or sequences that can block the active site of a partner molecule in a dimer (e.g., the autoinhibitory domain of PAK-1; Parrini et al., 2002). MAP kinase/ERK provides an example where dimerization of the catalytic domain regulates signaling (Cobb and Goldsmith, 2000). A further variation is that of Par-1b (homologous to MARK2) whose phosphorylation at Thr595 by aPKCζ creates a 14-3-3 binding site and subsequent inhibition (Hurov et al., 2004; Suzuki et al., 2004). Such control sequences tend to lie outside the core of the catalytic domain, unlike our present case where the catalytic core domain of PAK5 can bind and inhibit the core domain of MARK2. This is reminiscent of kinases that can activate each other by trans- or autophosphorylation of regulatory loops. It requires the regulatory loop of one kinase molecule to fit transiently into the catalytic cleft of the other. The analogy suggests that the regulatory loop of PAK5 might occupy the catalytic site of MARK2. At any rate, it appears that the mechanism of MARK2 inhibition requires binding of the catalytic cores but not phosphorylation; although both kinases display some autophosphorylation, the levels are independent of each other (Figure 3).
The overall picture of the MARK-PAK interplay outlined above must be filled in with details of molecular interactions by further experimental work. But we can ask whether connections between the MARK and PAK kinase families can be deduced from the available literature. Several suggestions can be made that are based on the complementary effects of PAKs (inducing actin dynamics) and MARKs (inducing microtubule dynamics):
1. In the case of cell migration or growth cone advancement, the actin network is responsible for the initial decisions at the leading edge (formation of lamellipodia, extension of filopodia), but dynamic microtubules must subsequently back up and reinforce the initial gains in territory (Wittmann and Waterman-Storer, 2001; Pollard and Borisy, 2003). The most characteristic effect of PAKs is the promotion of F-actin bundles that generate filopodia, which occurs under the control of activated Cdc42GTP (triggered by extracellular cues and involving receptors, G-proteins, and PAK-associated proteins such as PIX; Buchwald et al., 2001; Chong et al., 2001). On the other hand, activated Cdc42GTP triggers the membrane recruitment of polarity-inducing proteins of the Par family (Etienne-Manneville and Hall, 2002; Macara, 2004). This includes Par-3, Par-6, and atypical protein kinase C (PKC), whose shuttling is in turn mediated by Par-1 alias MARK. It involves, among others, the scaffold protein 14-3-3 (alias Par-5), which also appeared as a MARK2 interaction partner in our screen, and it is noteworthy that 14-3-3 plays a role in the phosphorylation of tau (Hashiguchi et al., 2000).
2. Members of the PAK family (e.g., PAK1) exert a direct effect on microtubule dynamics through the phosphorylation of stathmin/Op18 at Ser16 (Daub et al., 2001; Wittmann et al., 2004). This prevents stathmin from scavenging free tubulin subunits and thus contributes to microtubule stabilization. In this sense PAK is antagonistic to MARK, which causes the destabilization of microtubules through the withdrawal of MAPs.
3. Dynamic microtubules represent a locus for the anchoring or release of guanosine exchange factors (GEFs) that control the activity of Rho, Rac, or Cdc42, especially in the vicinity of focal adhesions. They are attached via end-binding proteins such as EB1, APC, or CLIP-170 and include GEF-H1, Asef, or IQGAP (for review see Small and Kaverina, 2003; Gundersen et al., 2004), which in turn activate small G-proteins with corresponding effects on the stability of actin stress fibers and focal adhesions. Thus, factors that make microtubules dynamic (e.g., MARK) would be expected to make stress fibers stable, and vice versa. The caveat in the above considerations is that PAK5 is the least well characterized member of the PAK family so that the examples must be borrowed from studies on other PAKs. Nevertheless, differences between PAKs occur mainly in the noncatalytic domains, whereas the kinase domains are highly homologous, suggesting that the signaling pathways for PAK activation may be different, whereas the downstream effect of making the actin cytoskeleton dynamic is comparable. Furthermore, PAKs can substitute one another because PAK5-deficient mice appear to be normal (Li and Minden, 2003) and PAK5 cooperates with other PAK family members in neurite outgrowth (Bryan et al., 2004).
It is interesting to compare the interactions obtained in this study using the yeast two-hybrid screen with the proteomic approach involving tandem affinity purification (Brajenovic et al., 2004). The TAP approach revealed a number of proteins expected to interact with MARK/Par-1 kinases during the development of cell polarity, notably Cdc42, ARH-GEF2, atypical PKC, and the scaffold proteins 14-3-3 (Par-5), Par-3, and Par-6. The two data sets overlap for the case of 14-3-3, which is a ubiquitous scaffolding protein mediating protein interactions, and its phosphorylation by MARK/Par-1 creates binding sites for other kinase-related proteins (Benton et al., 2002; Muller et al., 2003). The TAP results also reveal an interaction with the kinase LKB1. This tumor suppressor is able to activate members of the subfamily of AMP-activated kinases, including isoforms of MARK and is responsible for oncogenic transformation in Peutz-Jeghers syndrome (Lizcano et al., 2004), consistent with the earlier description of this protein as a tumor marker (Parsa, 1988). The TAP results do not reveal MARKK/TAO-1, which in our hands efficiently activates MARK2/Par-1 and causes phosphorylation of MAPs and microtubule dynamics (Timm et al., 2003). Finally, the TAP data do not show a connection between MARK2 and PAK5 or other PAK isoforms, even though the two kinases interact strongly in vitro and in cells, as shown above. A possible explanation of the differences is that each method is based on an initial set of potential interactors (in our case a fetal human cDNA library), follows different experimental selection procedures, and the readout of the effects is done in different cell types and conditions.
Kinases are often involved in more than one signaling cascade. For example, MARK isoforms are involved not only in regulating microtubule dynamics and the Par cell polarity determinants but also in Wnt signaling (Sun et al., 2001), and the upstream kinase MARKK/TAO-1 activates the p38 stress pathway by phosphorylating MEK3 and MEK6 (Hutchison et al., 1998). On the other hand, PAK5 is not only involved in remodelling the actin cytoskeleton, but also in inducing the JNK stress pathway (Dan et al., 2002; Pandey et al., 2002) and in the apoptotic pathway (Cotteret et al., 2003). It is interesting to note that the two kinases whose interaction we have studied in the context of the cytoskeleton also have a relationship to the cell's stress response. Consistent with this, the activation of MARK and the phosphorylation of its downstream target tau is elevated by cellular stress (Jenkins and Johnson, 2000; Schneider et al., 2004). This might explain the increased phosphorylation of tau at early stages of neurodegeneration in Alzheimer's disease and frontotemporal dementias (Augustinack et al., 2002). The impact of PAK5 on MARK during neurodegeneration will be an interesting question to pursue.
Supplementary Material
Acknowledgments
We thank Jacek Biernat, Edda Thies, Thomas Timm, Martin von Bergen, and Katrin Eckermann for expert advice on experimental procedures, and Anna Takacs for excellent technical assistance. This project was supported by the Deutsche Forschungsgemeinschaft.
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05-01-0081) on July 12, 2005.
Abbreviations used: CHO, CHO cells; MAP, microtubule-associated protein; MARK, MAP/microtubule affinity regulating kinase; PAK, p21-activated kinase.
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
References
- Augustinack, J. C., Schneider, A., Mandelkow, E.-M., and Hyman, B. (2002). Specific tau phosphorylation sites correlate with severity of neuronal cytopathology in Alzheimer's disease. Acta Neuropathol. 103, 26-35. [DOI] [PubMed] [Google Scholar]
- Benton, R., Palacios, I. M., St. Johnston, D. (2002). Drosophila 14-3-3/PAR-5 is an essential mediator of PAR-1 function in axis formation. Dev. Cell 3, 659-671. [DOI] [PubMed] [Google Scholar]
- Biernat, J., and Mandelkow, E.-M. (1999). The development of cell processes induced by tau protein requires phosphorylation of serine 262 and 356 in the repeat domain and is inhibited by phosphorylation in the proline-rich domains. Mol. Biol. Cell 10, 727-740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biernat, J., Wu, Y. Z., Timm, T., Zheng-Fischhofer, Q., Mandelkow, E., Meijer, L., and Mandelkow, E.-M. (2002). Protein kinase MARK/par-1 is required for neurite outgrowth and establishment of neuronal polarity. Mol. Biol. Cell 13, 4013-4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokoch, G. M. (2003). Biology of the p21-activated kinases. Annu. Rev. Biochem. 72, 743-781. [DOI] [PubMed] [Google Scholar]
- Brajenovic, M., Joberty, G., Kuster, B., Bouwmeester, T., and Drewes, G. (2004). Comprehensive proteomic analysis of human Par protein complexes reveals an interconnected protein network. J. Biol. Chem. 279, 12804-12811. [DOI] [PubMed] [Google Scholar]
- Bryan, B., Kumar, V., Stafford, L. J., Cai, Y., Wu, G., and Liu, M. (2004). GEFT, a rho family guanine nucleotide exchange factor, regulates neurite outgrowth and dendritic spine formation. J. Biol. Chem. 279, 45824-45832. [DOI] [PubMed] [Google Scholar]
- Buchwald, G., Hostinova, E., Rudolph, M. G., Kraemer, A., Sickmann, A., Meyer, H. E., Scheffzek, K., and Wittinghofer, A. (2001). Conformational switch and role of phosphorylation in PAK activation. Mol. Cell. Biol. 21, 5179-5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ching, Y. P., Leong, V. Y., Wong, C. M., and Kung, H. F. (2003). Identification of an autoinhibitory domain of p21-activated protein kinase 5. J. Biol. Chem. 278, 33621-33624. [DOI] [PubMed] [Google Scholar]
- Chong, C., Tan, L., Lim, L., and Manser, E. (2001). The mechanism of PAK activation. Autophosphorylation events in both regulatory and kinase domains control activity. J. Biol. Chem. 276, 17347-17353. [DOI] [PubMed] [Google Scholar]
- Cobb, M. H., and Goldsmith, E. J. (2000). Dimerization in MAP-kinase signaling. Trends Biochem. Sci. 25, 7-9. [DOI] [PubMed] [Google Scholar]
- Cotteret, S., Jaffer, Z. M., Beeser, A., and Chernoff, J. (2003). p21-Activated kinase 5 (Pak5) localizes to mitochondria and inhibits apoptosis by phosphorylating BAD. Mol. Cell. Biol. 23, 5526-5539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dajani, R., Fraser, E., Roe, S. M., Young, N., Good, V., Dale, T. C., and Pearl, L. H. (2001). Crystal structure of glycogen synthase kinase 3 beta: structural basis for phosphate-primed substrate specificity and autoinhibition. Cell 105, 721-732. [DOI] [PubMed] [Google Scholar]
- Dan, C., Nath, N., Liberto, M., and Minden, A. (2002). PAK5, a new brain-specific kinase, promotes neurite outgrowth in N1E-115 cells. Mol. Cell. Biol. 22, 567-577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daub, H., Gevaert, K., Vandekerckhove, J., Sobel, A., and Hall, A. (2001). Rac/Cdc42 and p65PAK regulate the microtubule-destabilizing protein stathmin through phosphorylation at serine 16. J. Biol. Chem. 276, 1677-1680. [DOI] [PubMed] [Google Scholar]
- Drewes, G., Ebneth, A., Preuss, U., Mandelkow, E.-M., and Mandelkow, E. (1997). MARK, a novel family of protein kinases that phosphorylate microtubule-associated proteins and trigger microtubule disruption. Cell 89, 297-308. [DOI] [PubMed] [Google Scholar]
- Ebneth, A., Godemann, R., Stamer, K., Illenberger, S., Trinczek, B., Mandelkow, E.-M., Mandelkow, E. (1998). Overexpression of tau protein alters kinesin-dependent trafficking of vesicles, mitochondria, and endoplasmic reticulum: implications for Alzheimer's disease. J. Cell Biol. 143, 777-794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etienne-Manneville, S., and Hall, A. (2002). Rho GTPases in cell biology. Nature 420, 629-635. [DOI] [PubMed] [Google Scholar]
- Fortini, M. E. (2004). PAR-1 for the course of neurodegeneration. Cell 116, 631-632. [DOI] [PubMed] [Google Scholar]
- Gundersen, G. G., Gomes, E. R., and Wen, Y. (2004). Cortical control of microtubule stability and polarization. Curr. Opin. Cell Biol. 16, 106-112. [DOI] [PubMed] [Google Scholar]
- Hashiguchi, M., Sobue, K., and Paudel, H. K. (2000). 14-3-3zeta is an effector of tau protein phosphorylation. J. Biol. Chem. 275, 25247-25254. [DOI] [PubMed] [Google Scholar]
- Hurov, J., Watkins, J., and Piwnica-Worms, H. (2004). Atypical PKC phosphorylates PAR-1 kinases to regulate localization and activity. Curr. Biol. 14, 736-741. [DOI] [PubMed] [Google Scholar]
- Huse, M., and Kuriyan, J. (2002)., The conformational plasticity of protein kinases. Cell 109, 275-282. [DOI] [PubMed] [Google Scholar]
- Hutchison, M., Berman, K. S., and Cobb, M. H. (1998)., Isolation of TAO1, a protein kinase that activates MEKs in stress-activated protein kinase cascades. J. Biol. Chem. 273, 28625-28632. [DOI] [PubMed] [Google Scholar]
- Illenberger, S., Zheng-Fischhöfer, Q., Preuss, U., Stamer, K., Baumann, K., Trinczek, B., Biernat, J., Godemann, R., Mandelkow, E.-M., Mandelkow, E. (1998). The endogenous and cell-cycle dependent phosphorylation of tau protein in living cells: implications for Alzheimer's disease. Mol. Biol. Cell 9, 1495-1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffer, Z. M., and Chernoff, J. (2002). p21-activated kinases: three more join the Pak. Int. J. Biochem. Cell Biol. 34, 713-717. [DOI] [PubMed] [Google Scholar]
- Jenkins, S. M., and Johnson, G. V. (2000). Microtubule/MAP-affinity regulating kinase (MARK) is activated by phenylarsine oxide in situ and phosphorylates tau within its microtubule-binding domain. J. Neurochem. 74, 1463-1468. [DOI] [PubMed] [Google Scholar]
- Kaur, R., Liu, X., Gjoerup, O., Zhang, A., Yuan, X., Balk, S., Schneider, S., and Lu, M. (2005). Activation of p21-activated kinase 6 by MAP kinase kinase 6 and p38 MAP kinase. J. Biol. Chem. 280, 3323-3330. [DOI] [PubMed] [Google Scholar]
- Kopito, R. R. (2000). Aggresomes, inclusion bodies and protein aggregation. Trends Cell Biol. 10, 524-530. [DOI] [PubMed] [Google Scholar]
- Krendel, M., Zenke, F. T., and Bokoch, G. M. (2002). Nucleotide exchange factor GEF-H1 mediates cross-talk between microtubules and the actin cytoskeleton. Nat. Cell Biol. 4, 294-301. [DOI] [PubMed] [Google Scholar]
- Lei, M., Lu, W., Meng, W., Parrini, M. C., Eck, M. J., Mayer, B. J., and Harrison, S. C. (2000). Structure of PAK1 in an autoinhibited conformation reveals a multistage activation switch. Cell 102, 387-397. [DOI] [PubMed] [Google Scholar]
- Lei, M., Robinson, M. A., and Harrison, S. C. (2005). The active conformation of the PAK1 kinase domain. Structure 13, 769-778. [DOI] [PubMed] [Google Scholar]
- Li, X., and Minden, A. (2003). Targeted disruption of the gene for the PAK5 kinase in mice. Mol. Cell. Biol. 23, 7134-7142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lizcano, J. M. et al. (2004). LKB1 is a master kinase that activates 13 kinases of the AMPK subfamily, including MARK/PAR-1. EMBO J. 23, 833-843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macara, I. G. (2004). Par proteins: partners in polarization. Curr. Biol. 14, R160-R162. [PubMed] [Google Scholar]
- Manser, E. (2002). Small GTPases take the stage. Dev. Cell 3, 323-328. [DOI] [PubMed] [Google Scholar]
- Manser, E., Leung, T., Salihuddin, H., Zhao, Z. S., Lim, L. (1994). A brain serine/threonine kinase activated by Cdc42 and Rac1. Nature 367, 40-46. [DOI] [PubMed] [Google Scholar]
- Muller, J., Ritt, D. A., Copeland, T. D., and Morrison, D. K. (2003). Functional analysis of C-TAK1 substrate binding and identification of PKP2 as a new C-TAK1 substrate. EMBO J. 22, 4431-4442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura, I., Yang, Y., and Lu, B. (2004). PAR-1 kinase plays an initiator role in a temporally ordered phosphorylation process that confers tau toxicity in Drosophila. Cell 116, 671-682. [DOI] [PubMed] [Google Scholar]
- Pandey, A., Dan, I., Kristiansen, T. Z., Watanabe, N. M., Voldby, J., Kajikawa, E., Khosravi-Far, R., Blagoev, B., and Mann, M. (2002). Cloning and characterization of PAK5, a novel member of mammalian p21-activated kinase-II subfamily that is predominantly expressed in brain. Oncogene 30 21, 3939-3948. [DOI] [PubMed] [Google Scholar]
- Parrini, M. C., Lei, M., Harrison, S. C., and Mayer, B. J. (2002). PAK1 kinase homodimers are autoinhibited in trans and dissociated upon activation by Cdc42 and Rac1. Mol. Cell 9, 73-83. [DOI] [PubMed] [Google Scholar]
- Parsa, I. (1988). Loss of Mr 78000 marker in chemically induced transplantable carcinomas and primary carcinoma of human pancreas. Cancer Res. 48, 2265-2272. [PubMed] [Google Scholar]
- Pellettieri, J., and Seydoux, G. (2002). Anterior-posterior polarity in C. elegans and Drosophila—PARallels and differences. Science 298, 1946-1950. [DOI] [PubMed] [Google Scholar]
- Pollard, T. D., and Borisy, G. G. (2003). Cellular motility driven by assembly and disassembly of actin filaments. Cell 112, 453-465. [DOI] [PubMed] [Google Scholar]
- Schneider, A. et al. (2004). Identification of regulated genes during permanent focal cerebral ischaemia: characterization of the protein kinase 9b5/MARKL1/MARK4. J. Neurochem. 88, 1114-1126. [DOI] [PubMed] [Google Scholar]
- Small, J. V., Geiger, B., Kaverina, I., and Bershadsky, A. (2002). How do microtubules guide migrating cells? Nat. Rev. Mol. Cell. Biol. 3, 957-964. [DOI] [PubMed] [Google Scholar]
- Small, J. V., and Kaverina, I. (2003). Microtubules meet substrate adhesions to arrange cell polarity. Curr. Opin. Cell Biol. 15, 40-47. [DOI] [PubMed] [Google Scholar]
- Sun, T. Q., Lu, B., Feng, J. J., Reinhard, C., Jan, Y. N., Fantl, W. J., and Williams, L. T. (2001). PAR-1 is a Dishevelled-associated kinase and a positive regulator of Wnt signalling. Nat. Cell Biol. 3, 628-636. [DOI] [PubMed] [Google Scholar]
- Suzuki, A., et al. (2004). aPKC acts upstream of PAR-16 in both the establishment and maintenance of epithelial polarity. Curr. Biol. 14, 1425-1435. [DOI] [PubMed] [Google Scholar]
- Timm, T., Li, X.-Y., Biernat, J., Jiao, J., Mandelkow, E., Vandekerckhove, J., and Mandelkow, E.-M. (2003). MARKK, a Ste20-like kinase, activates the polarity-inducing kinase MARK/PAR-1. EMBO J. 22, 5090-5101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinczek, B., Brajenovic, M., Ebneth, A., and Drewes, G. (2004). MARK4 is a novel microtubule-associated proteins/microtubule affinity-regulating kinase that binds to the cellular microtubule network and to centrosomes. J. Biol. Chem. 279, 5915-5923. [DOI] [PubMed] [Google Scholar]
- Wittmann, T., and Waterman-Storer, C. M. (2001). Cell motility: can Rho GTPases and microtubules point the way? J. Cell Sci. 114, 3795-3803. [DOI] [PubMed] [Google Scholar]
- Wittmann, T., Bokoch, G. M., and Waterman-Storer, C. M. (2004). Regulation of microtubule destabilizing activity of Op18/Stathmin downstream of Rac1. J. Biol. Chem. 279, 6196-6203. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.