Abstract
In families with nonsyndromic X-linked mental retardation (NS-XLMR), >30% of mutations seem to cluster on proximal Xp and in the pericentric region. In a systematic screen of brain-expressed genes from this region in 210 families with XLMR, we identified seven different mutations in JARID1C, including one frameshift mutation and two nonsense mutations that introduce premature stop codons, as well as four missense mutations that alter evolutionarily conserved amino acids. In two of these families, expression studies revealed the almost complete absence of the mutated JARID1C transcript, suggesting that the phenotype in these families results from functional loss of the JARID1C protein. JARID1C (Jumonji AT-rich interactive domain 1C), formerly known as “SMCX,” is highly similar to the Y-chromosomal gene JARID1D/SMCY, which encodes the H-Y antigen. The JARID1C protein belongs to the highly conserved ARID protein family. It contains several DNA-binding motifs that link it to transcriptional regulation and chromatin remodeling, processes that are defective in various other forms of mental retardation. Our results suggest that JARID1C mutations are a relatively common cause of XLMR and that this gene might play an important role in human brain function.
Introduction
Mental retardation (MR), usually defined as cognitive impairment with an IQ <70, is estimated to affect 2%–3% of the population in industrialized countries (Roeleveld et al. 1997). Etiologically, MR is a very heterogeneous condition that involves environmental, stochastic, and/or genetic factors. Numerous studies of large families containing males with MR have highlighted the importance of the X chromosome. Prevalence estimates for X-linked mental retardation (XLMR) range from 0.56/1,000 males for severe forms to 1.83/1,000 for mild and severe forms (Herbst and Miller 1980; Fishburn et al. 1983). Approximately two-thirds of male patients with XLMR exhibit a nonsyndromic form of XLMR (NS-XLMR) in which MR is the only clinical manifestation (Fishburn et al. 1983). NS-XLMR is a very heterogeneous condition. With one exception, all of the relevant genes identified until 2002 (FMR2 [MIM 309548], OPHN1 [MIM 300127], PAK3 [MIM 300142], GDI1 [MIM 300104], IL1RAPL1 [MIM 300206], TM4SF2 [MIM 300096], ARHGEF6 [MIM 300267], MECP2 [MIM 300005], FACL4 [MIM 300147], and ARX [MIM 300382]; see review by Chelly and Hamel [2003]) showed mutations only in a very small proportion of families with XLMR, suggesting that up to 100 different genes might be involved in NS-XLMR. In an analysis of linkage intervals found in families examined in published and unpublished studies, we have recently shown that mutations involved in NS-XLMR seem to cluster in three different regions of the human X chromosome (Ropers et al. 2003). Two clusters, one at Xq28 and the other at Xp22.1-p21.3, coincide with known MR genes. However, the presence of a broad peak on proximal Xp argued for the existence of unknown genes for NS-XLMR in this region. Therefore, we chose a 7.4-Mb interval in Xp11, flanked by the genes ELK1 (MIM 311040) and ALAS2 (MIM 301300), to systematically screen 47 candidate genes for mutations in families with overlapping linkage intervals. This led to the identification of PQBP1 (MIM 300463), which encodes the polyglutamine binding protein 1, and FTSJ1 (MIM 300499), a human homolog of the Escherichia coli 2′-O-rRNA methyltransferase FtsJ/RrmJ gene, the first two XLMR genes found in Xp11 (Kalscheuer et al. 2003; Freude et al. 2004). In the present study, we report the identification of seven different mutations, in a novel brain-expressed gene, that are associated with familial XLMR.
Subjects and Methods
Patients and Controls
Mutation screening was performed in 31 families with XLMR and overlapping linkage intervals and in 179 unlinked families from the European XLMR Consortium. All patients had a normal karyotype, and fragile X syndrome was ruled out. The control panel included 312 unrelated healthy male controls. Samples from patients and their relatives as well as from controls were obtained after receiving informed consent. A summary of clinical data from all patients with mutations in JARID1C is given in table A1 (online only).
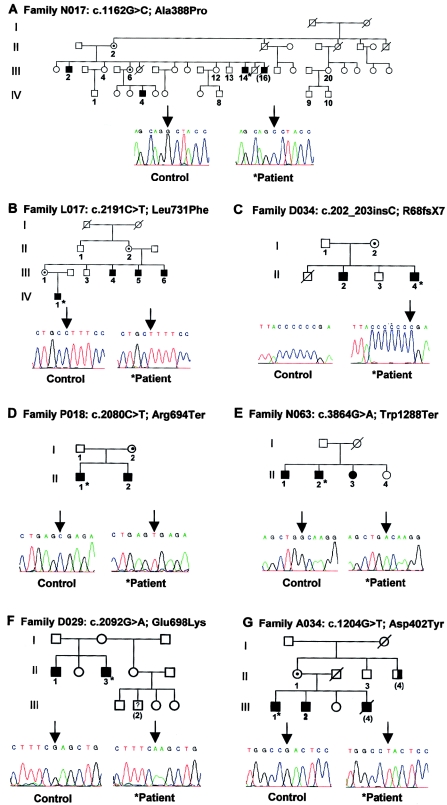
In family N017, with four affected males (fig. 1A), linkage studies had mapped the gene defect to the DXS1003-PGK1P1 interval. The patients in this family present with MR and microcephaly. The index patient, III:14, was born on time with a weight of 1,750 g. He was able to walk at the age of 14 mo and spoke his first words at the age of 3 years. At the age of 41 years, at his last clinical examination, his height was 165 cm (<3rd percentile), his weight was 58.8 kg, and his head circumference was 53.8 cm (∼3rd percentile). He showed mild dysmorphisms, including small forehead, small eyelashes, prognathism, and small testes. His brother, patient III:2, was examined at the age of 53 years: his height was 154 cm (<3rd percentile), and his head circumference was 53.5 cm (∼3rd percentile). He has mild dysmorphic features, including flat philtrum, thin upper lip, prognathism, and a small penis. The third patient, IV:4, was born on time with a weight of 2,500 g. He was able to walk at the age of 2 years and spoke his first words at the age of 3.5 years. His examination at the age of 25 years (height 171 cm [∼25th percentile], weight 65 kg, head circumference 54 cm [∼3rd percentile]) also showed mild dysmorphic features: small, deep-set eyes; upslanting palpebral fissures; and micrognathia. However, his genitals were normal. At that time, his total IQ was measured to be 70 (verbal 60, performance 86). Clinical data were not available for patient III:16.
Figure 1.
Mutations identified in the JARID1C gene in families N017 (A), L017 (B), D034 (C), P018 (D), N063 (E), D029 (F), and A034 (G). The sequence chromatograms of the index patients (asterisks) are shown beside the wild-type sequences. The corresponding pedigrees are presented above the chromatograms. Family members who have been analyzed for sequence changes are numbered. For patients with numbers in parentheses, only clinical data were available (no DNA was available for mutation analysis).
Linkage data and clinical findings for family L017 (fig. 1B) have been described elsewhere (Claes et al. 2000). In this family, which has a syndromic form of XLMR, four males in two generations present with severe MR, slowly progressive spastic paraplegia, facial hypotonia, and maxillary hypoplasia. Additional features are aggressive behavior and strabismus.
In family D034 (fig. 1C), the two affected brothers exhibit a severe form of MR. The index patient, II:4, was born after an uneventful pregnancy. Developmental delay was noticed at the age of 1 year. The patient shows strabismus divergens. He understands spoken language well, but the only word he speaks is “yes.” At the age of 5 years, electroencephalogram examination revealed abnormalities (groups of sharp theta/delta waves), but epileptic seizures have never been observed. His behavior is characterized by frequent unmotivated aggressive outbursts along with indolence. He has good memory skills and can perform simple tasks such as shopping at the local bakery. His older brother shows an almost identical phenotype. In addition, he exhibits abundant body hair and diastema of the teeth. Because of gallstones, the gallbladders of both brothers had been removed at the age of 20 years and 31 years, respectively. No other medical abnormalities have been noticed.
The two boys in family P018 (fig. 1D) have MR and short stature (patient II:1, body height 105 cm at the age of 6.5 years [<3rd percentile]; patient II:2, body height 91 cm at the age of 4.5 years [<3rd percentile]). Apart from strabismus and myopia in the younger brother (II:2), no other clinical abnormalities have been reported. The nonconsanguineous parents are clinically inconspicuous.
In family N063 (fig. 1E), two brothers and their sister have MR. They share a conspicuous phenotype of severe MR, spasticity, epileptic seizures, short stature, microcephaly, hypermetropia, and small feet. Both males have cryptorchism. Additional symptoms include alopecia areata in patient II:1. The sister (II:3) was described to have an unstable mood disorder. The parents of these children are distantly related.
In family D029, with two affected brothers (fig. 1F), the index patient (II:3) has severe MR. His height (169 cm [∼3rd percentile]) and head circumference (59 cm [∼97th percentile]) at the age of 22 years were in the normal range. The testis volume was 25 ml (>97th percentile), but there was no expansion of the CGG repeat in the FMR1 gene. His older brother (II:1) is also affected with a severe form of MR. Height (169 cm [∼3rd percentile]), head circumference (58 cm [∼97th percentile]), and testis volume at the age of 25 years are normal, and he has no additional abnormalities. The son of a half sister (III:2) is said to suffer from learning impairment, but no clinical details are available.
In family A034 (fig. 1G), four affected males have been reported. Little is known about the early lives of the three affected brothers (III:1, III:2, and III:4) except that development was slow and all were institutionalized in early childhood. Patient III:4 died as a result of accidental drowning at the age of 12 years. The two other brothers (III:1 and III:2), now aged 61 years and 56 years, respectively, reside in a group home. Both had short stature (154 cm [<3rd percentile] and 156 cm [<3rd percentile], respectively), with normal head circumferences (57 cm [∼75th percentile] and 55 cm [25th–50th percentile], respectively). Both were severely handicapped, and neither spoke more than a few single words. Occasional short outbursts of aggression were reported. In his 30s, patient III:2 started to have generalized seizures about once a year, which sometimes led to hospitalization. Their uncle (II:4) is mildly handicapped and lives at home. Recent reexamination of this patient revealed a very different phenotype, strongly suggesting a different disorder. He was previously known as the “sunny boy” of the family and had a “big head at birth, which had to be carried on a pillow.” He did not walk until the age of 6 years, and, later on, his walk was unsteady and awkward. He had “plenty of speech, which was unclear.” Now 74 years of age, he sits and smiles and is very unsteady on his feet. He is of normal height, but he has a large head circumference of 62 cm (>97th percentile).
Mutation Analysis by Denaturing High-Performance Liquid Chromatography (DHPLC)
For each patient, the complete JARID1C (Jumonji AT-rich interactive domain 1C [MIM 314690]) coding sequence and ∼40 nucleotides of the neighboring intronic sequences were amplified in 34 independent PCRs from genomic DNA. All primer sequences and PCR product sizes are given in table A2 (online only). In general, PCR amplifications were performed in 20-μl reaction volumes that contained 100 ng genomic DNA, 1× buffer (50 mM KCl, 15 mM Tris-HCl [pH 9.0], 2.5 mM MgCl2, 0.01% Tween 20), 0.2 μM of each primer, 100 μM dNTPs, 0.5 M betaine (Sigma), 1 U Taq polymerase, and 0.025 U Pfu polymerase. The PCR profile consists of a touchdown PCR with an initial denaturing step at 96°C for 3 min and then 20 cycles at 95°C for 30 s, 65°C for 30 s (lowered 0.5°C at each cycle), and 72°C for 30 s, followed by 30 cycles at 95°C for 30 s, 55°C for 30 s, and 72°C for 30 s. The PCR was finalized by a 5-min extension at 72°C. PCR products were checked by 1.5% agarose gel electrophoresis before they were subjected to DHPLC by the WAVE nucleic-acid–fragment analysis system (Transgenomic). For DHPLC, we pooled PCR products from two patients. Melting profiles and resolution temperatures were predicted by the Transgenomic WAVEMAKER software, version 4.1. Conditions are available on request. PCR products that gave rise to abnormal elution profiles either were gel purified by use of the QIAquick gel extraction kit (Qiagen) and then subjected to sequencing or were directly sequenced in both directions by use of BigDye Terminator chemistry (PE Biosystems). Separation was performed on an Applied Biosystems 3730xl DNA Analyzer.
RNA Isolation and Northern Blot Analysis
Total RNA was isolated from patient and control lymphoblastoid cell lines by use of TRIzol (Invitrogen), in accordance with the manufacturer's protocol. Poly-A+ RNAs, which were each obtained from 100 μg of total RNA by use of Dynabeads (Dynal), were separated in a formaldehyde-containing gel in 1 × MOPS buffer, transferred to a Hybond N+ membrane, and crosslinked by UV. A human multiple-tissue northern blot was obtained from BioChain. The JARID1C (GenBank accession number L25270) and JARID1D (GenBank accession number NM_004653) probes corresponding to nucleotides c.4563 to c.5292 (730 bp) and c.4400 to c.5128 (729 bp), respectively, were synthesized using the following primer pairs: JARID1C (forward): 5′-GAACCAGAATGGCTTGGAAC-3′ and JAIRD1C (reverse): 5′-GGACTGAGGTGGTTGTGAGG-3′; JARID1D (forward): 5′-CTCGGAGCTCAGGGATTATG-3′ and JARID1D (reverse): 5′-GAGTCATGAAATTCTTGTTTTTCC-3′. The probes were purified, and their sequences were verified. Subsequently, probes were labeled with 32[P]dCTP and were hybridized to the blots in ULTRAhyb buffer (Ambion). The blots were washed in accordance with the manufacturer’s protocol and were exposed to Fuji medical X-ray films at −80°C for 6 h for up to 8 d by use of intensifying screens. Autoradiograms were scanned using an Epson Expression 1680 Pro scanner. To control for RNA loading, blots were reprobed with a β-actin probe that was provided by BioChain.
Results
Identification of JARID1C Mutations in Families with XLMR
In a recent analysis of linkage data from published and unpublished studies of families with NS-XLMR, we showed that mutations involved in XLMR seem to cluster in three distinct regions on the X chromosome (Ropers et al. 2003). Therefore, we chose a 7.4-Mb interval on Xp11.23-p11.21, flanked by the genes ELK1 and ALAS2, to systematically screen 47 brain-expressed candidate genes for mutations in families with overlapping linkage intervals. One of these genes, JARID1C, was found to be mutated in 2 of 31 families investigated. The JARID1C gene contains 26 exons and encodes a member of a conserved protein family, which carries several DNA-binding motifs potentially involved in transcriptional regulation and chromatin remodeling. In family N017, we found a missense mutation, c.1162G→C, that resulted in an alanine-to-proline exchange at amino acid 388 (fig. 1A). Another single-nucleotide substitution (c.2191C→T) was identified in family L017; this substitution changed the amino acid leucine into phenylalanine at position 731 (fig. 1B). Sequencing of PCR-amplified JARID1C transcripts from available family members showed that, in both families, the respective mutations cosegregate with the disease (data not shown). None of these changes was found in 312 male controls.
These findings prompted us to analyze 179 additional unrelated patients from smaller families with established or probable XLMR. In five of these families, we found additional mutations, including one frameshift mutation, two nonsense mutations, and two missense mutations. A single-nucleotide insertion of a cytidine (c.202_203insC) introduces a frameshift mutation in patients from family D034 (fig. 1C), eventually resulting in a premature stop codon at position 75 of JARID1C. In family P018, the single-nucleotide substitution c.2080C→T replaces the normal arginine by a termination codon at position 694 of the protein (fig. 1D). Another nonsense mutation (Trp1288Ter) was identified in family N063 and is caused by the sequence change c.3864G→A (fig. 1E). In family D029, a guanine-to-adenine mutation (c.2092G→A) results in a glutamic acid–to-lysine exchange at amino acid 698 (Glu698Lys) (fig. 1F). The second missense mutation found in the patient cohort that is present in family A034 (c.1204G→T) changes an aspartic acid residue into tyrosine at position 402 (fig. 1G).
Reinvestigation of the respective sequences in all available affected and unaffected family members confirmed the cosegregation of the mutations with the phenotype (data not shown). All the mutations that were identified in these families were absent in the remaining 205 analyzed patients with XLMR and also in 312 male controls. In addition to the seven most likely disease-causing mutations, we have found several other sequence variants that were not known before. However, these variants either were silent mutations or were found in intronic sequences and, therefore, for the time being, were not considered as disease causing. All sequence changes found in patients with MR are listed in table 1.
Table 1.
Summary of Sequence Changes in JARID1C Found in Patients with MR
| Nucleotide Change | Amino Acid Variation | Position | Type of Sequence Change | Family Code | No. of Linked Families with Sequence Change(n=31) | No. of UnlinkedFamilies withSequence Change(n=179) | Frequency in ControlX Chromosomes |
| c.202_203insC | R68fsX7 | Exon 2 | Frameshift mutation | D034 | 0 | 1 | 0/312 |
| c.1162G→C | Ala388Pro | Exon 9 | Missense mutation | N017 | 1 | 0 | 0/312 |
| c.1204G→T | Asp402Tyr | Exon 9 | Missense mutation | A034 | 0 | 1 | 0/312 |
| c.2080C→T | Arg694Ter | Exon 15 | Nonsense mutation | P018 | 0 | 1 | 0/312 |
| c.2092G→A | Glu698Lys | Exon 15 | Missense mutation | D029 | 0 | 1 | 0/312 |
| c.2191C→T | Leu731Phe | Exon 15 | Missense mutation | L017 | 1 | 0 | 0/312 |
| c.3864G→A | Trp1288Ter | Exon 23 | Nonsense mutation | N063 | 0 | 1 | 0/312 |
| c.−239G→A | 5′ UTR | Intronic alteration | 0 | 1 | 0/312 | ||
| c.−90G→A | 5′ UTR | Intronic alteration | 1 | 0 | 0/312 | ||
| c.522+19G→A | Intron 4 | Intronic alteration | 3 | 3 | 9/96 | ||
| c.564G→A | Lys188Lys | Exon 5 | Silent mutation | 0 | 3 | 3/94 | |
| c.1764G→A | Gln588Gln | Exon 13 | Silent mutation | 0 | 1 | 0/312 | |
| c.1794C→T | Pro598Pro | Exon 13 | Silent mutation | 0 | 2 | 0/312 | |
| c.2243+11G→T | Intron 15 | Intronic alteration | 0 | 5 | 2/96 | ||
| c.2517-7_8insTAC | Intron 17 | Intronic insertion | 0 | 1 | 0/312 |
Expression Analysis of JARID1C in Patients
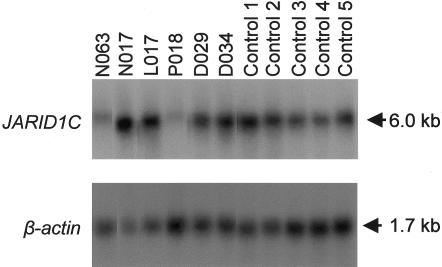
To check the stability of all mutated transcripts, we performed northern blot hybridizations of poly-A+ cell line RNA from one affected male in each family (except family A034) and five male controls. To avoid crosshybridization with the highly similar Y-chromosomal gene JARID1D, probes were employed that did not show significant sequence similarity to each other upon BLAST2 alignment. JARID1C transcripts were almost undetectable in the patients from families P018 and N063, who had premature terminations at codon 694 and codon 1288 (fig. 2), respectively. Thus, the phenotype in these patients most likely results from functional loss of JARID1C protein. In contrast, no reduction was observed in the patient from family D034, in whom a frameshift mutation introduces a premature termination at codon 75 of JARID1C (fig. 2). In patients with missense mutations, JARID1C RNA levels were not conspicuously changed.
Figure 2.
Northern blot analysis of poly-A+ RNA isolated from lymphoblastoid cell lines obtained from six patients and five controls. The blot was sequentially hybridized with a cDNA probe corresponding to nucleotides 4563–5292 of JARID1C and with a β-actin cDNA probe. Note the significant reduction of JARID1C transcript levels in patients from families N063 and P018.
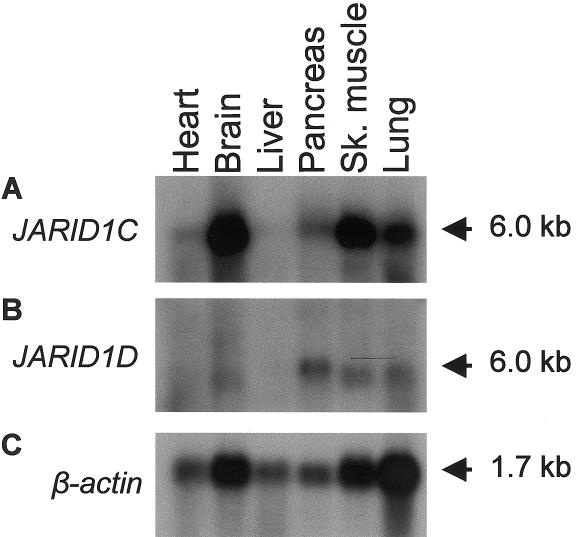
JARID1C Is Mainly Expressed in Brain and Skeletal Muscle
A predominant transcript of ∼6.0 kb was detected in almost all of the human adult tissues investigated, although at very different levels. The strongest expression was observed in brain and skeletal muscle, whereas the lowest expression was seen in heart and liver (fig. 3A). Hybridization of a JARID1D-specific probe to the same blot showed expression that was much lower than that of JARID1C in general, and expression was seen only in pancreas, lung, brain, and skeletal muscle (fig. 3B).
Figure 3.
Sequential hybridization of JARID1C (A), JARID1D (B), and β-actin (C) to a northern blot containing human poly-A+ RNAs from adult heart, brain, liver, pancreas, skeletal muscle, and lung. In panel A, probes correspond to nucleotides 4563–5292 of JARID1C; in panel B, probes correspond to nucleotides 4400–5128 of JARID1D; and, in panel C, probes correspond to a cDNA fragment of β-actin.
The JARID1C Protein Contains Several Conserved DNA-Binding Domains
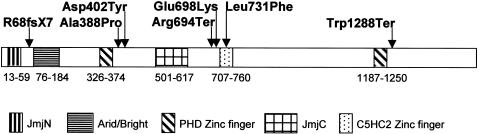
JARID1C (GenBank accession number P41229) encodes a 1,560-aa protein that is homologous to three other proteins of the human JARID1 family. On the basis of a BLAST2 alignment, the amino acid identity of JARID1C to JARID1D (GenBank accession number NP_004644), JARID1A (GenBank accession number NP_005047), and JARID1B (GenBank accession number NP_006609) is 85%, 51%, and 47%, respectively. Human JARID1C is evolutionarily conserved, with homologs in the mouse (Jarid1c [GenBank accession number NP_038696]), fruit fly (Lid [GenBank accession number NP_723140]), and nematode (ZK593.4 [GenBank accession number CAA93426]) that show 94%, 42%, and 34% amino acid identity, respectively. A Conserved Domain Database alignment (Marchler-Bauer et al. 2003) revealed several conserved domains between the JARID proteins, including a JmjN domain (smart00545), an Arid/Bright domain (smart00501), a JmjC domain (smart00558), a C5HC2 zinc-finger domain (pfam02928), and PHD zinc-finger domains (pfam00628) (fig. 4 and data not shown).
Figure 4.
Schematic representation of the human JARID1C protein, showing the positions and types of mutations found. On the basis of the NCBI Conserved Domain Database, the positions of known domains and motifs are marked by different boxes, which are explained at the bottom of the figure.
Discussion
We have shown that patients affected with XLMR carry mutations in a novel XLMR gene, JARID1C. Mutation screening in 210 unrelated families with XLMR revealed at least seven different sequence alterations that are most likely disease causing—including two nonsense mutations, one frameshift mutation, and four missense mutations. The three mutations that introduce premature termination codons are expected to remove >95%, >55%, and >17% of the normal protein, respectively. Moreover, in the two families with the longest residual transcripts, P018 and N063, JARID1C expression was almost undetectable, probably because of the mechanism of nonsense-mediated mRNA decay (NMD) (Holbrook et al. 2004), suggesting a complete loss of JARID1C function in these two families. However, the frameshift mutation in family D034, which truncates the JARID1C protein at position 75, seems to be insensitive to NMD. Similar findings have been reported elsewhere (Asselta et al. 2001), including intertissue (Bateman et al. 2003) and interindividual (Kerr et al. 2001) variations in NMD efficiency. In family D034, the aberrant phenotype may be a result of either extreme shortening of the protein and deletion of nearly all relevant domains or an abnormal protein produced by alternative usage of a methionine downstream of exon 2.
All missense changes affect amino acids, which are identical in the four different members of the human JARID1 family and in homologous proteins of mouse and fruit fly, suggesting an important functional role for these amino acids (fig. 5). The missense change Leu731Phe (family L017), which is located in a predicted, evolutionarily conserved C5HC2 zinc-finger domain, might have a profound effect on that domain as a result of the differences in size and chemical properties of leucine and phenylalanine. Although the three other missense changes—Ala388Pro, Asp402Tyr, and Glu698Lys—are not located in a conserved domain, they can cause considerable changes in the protein structure. The introduction of a proline residue causes the protein backbone to bend. The replacement of aspartic acid by tyrosine, as well as of glutamic acid by lysine, alters the charge of the protein and, presumably, its three-dimensional structure. Together, our data suggest that the seven JARID1C mutations detected affect the function of the protein and are responsible for the cognitive defects found in the patients studied.
Figure 5.
Partial amino acid sequence alignment of the four different members of the human JARID1 family (Hs_JARID1A–Hs_JARID1D), the JARID1C orthologous protein of mouse (Mm_Jarid1c), and the homologous Lid protein of fruit fly (Dm_Lid). Amino acids that differ from the JARID1C sequence are highlighted. The four missense mutations and their positions are indicated by arrows and bold letters. The respective amino acids are identical in the six proteins.
Human JARID1C/SMCX protein—which contains several highly conserved domains, including Arid/Bright and Jumonji—belongs to the JARID1 subfamily of ARID DNA-binding proteins (Kortschak et al. 2000; Wilsker et al. 2002). Apart from JARID1C, three other proteins that are characterized by a core ARID of 81 aa—namely, JARID1D/SMCY, JARID1A/RBBP2, and JARID1B/PLU-1—comprise the JARID1 subfamily. JARID1A/RBBP2 was cloned in a search for retinoblastoma protein (pRB)–binding partners (Fattaey et al. 1993). pRB is a well-known and extensively characterized protein that regulates cell proliferation by inhibiting growth-promoting proteins. It can recruit histone deacetylase to chromatin (Luo et al. 1998), which represses the transcription of genes involved in cell-cycle regulation that contain sites for the E2F transcription factor in their promoters (Dyson 1998). The JARID1B protein, also known as “PLU-1,” is a transcriptional repressor. It binds brain factor 1 (BF1), also known as “FOXG1B,” and paired box 9 (PAX9), both of which are developmental transcription factors (Tan et al. 2003). In mouse embryos and human fetal brain, the expression of BF1/FOXG1B is restricted to the neuronal cells in the telencephalon, with strong expression in the developing dentate gyrus and hippocampus. In Foxg1-null mice, the generation of the earliest born neurons, the Cajal-Retzius cells, is suppressed (Hanashima et al. 2004).
JARID1C/SMCX and JARID1D/SMCY (MIM 426000) constitute an X-Y homologous gene pair, which probably evolved from an identical gene pair on the original mammalian (proto-) sex chromosomes (Delbridge et al. 2004), implicating a functional equivalence of JARID1C and JARID1D. This probable similar function of X and Y copies is strengthened by the fact that the human (as well as the mouse) JARID1C gene was thought to escape X inactivation (Agulnik et al. 1994). However, the mouse Jarid1c gene exhibits partial X inactivation in embryos, in extraembryonic lineages, and in different adult tissues, showing an expression of 20%–70% from the inactive X allele, compared with the active X allele (Sheardown et al. 1996). Furthermore, Jarid1c and Jarid1d are expressed in mouse brain in a sex-specific fashion (i.e., in adulthood, Jarid1c is expressed at a level that is significantly higher in female brains than in male brains, and the expression of Jarid1d in males is not sufficient to compensate for the female bias in X-gene expression [Xu et al. 2002]). Hence, the X-Y gene pair JARID1C/JARID1D might not be functionally equivalent, suggesting an indispensable role of JARID1C for normal brain function. Together with other differentially regulated X-Y gene pairs (e.g., USP9X/Y [MIM 300072 and MIM 400005], RPS4X/Y [MIM 312760 and MIM 470000], and UTX/Y [MIM 300128 and MIM 400009]), JARID1C/JARID1D might also be involved in sex differences in brain development and function and in the prevalence of neuropsychatric disorders (Vawter et al. 2004).
We have found mutations in the gene JARID1C in 7 of 210 families with XLMR, which corresponds to a mutation frequency of 3.3% (CI 0.009–0.057) for XLMR. This number is somewhat biased, since families with XLMR with linkage intervals outside the candidate region in Xp11 have been excluded from our mutation screen. However, the mutation frequency is still 2.8% (CI 0.004–0.052) if only the five changes found in 179 families with proven or probable XLMR are taken into account. Thus, JARID1C seems to be one of the more frequently mutated genes in patients with XLMR, and its mutation frequency is comparable to that described for the creatine transporter SLC6A8 (MIM 300036) (Rosenberg et al. 2004). JARID1C mutations cause not only nonsyndromic but also syndromic XLMR, as seen in families N017, L017, and N063; this is in agreement with findings in other XLMR genes (Merienne et al. 1999; Meloni et al. 2000; Bienvenu et al. 2002; Strømme et al. 2002; Kalscheuer et al. 2003).
We conclude that mutations in JARID1C contribute significantly to the etiology of XLMR. Further studies will include the characterization of mice deficient in Jarid1c and expression profiling of the brain tissues. These investigations should shed more light on the mechanisms by which mutations in JARID1C result in dysfunction of the brain.
Acknowledgments
We thank the members of the families and their clinicians, for their participation in this study, and S. Freier and H. Madle, for help with cell cultures. This project was supported by Deutsches Humangenom-Programm (grant 01KW99087), Nationales Genomforschungsnetzwerk (grant 01GR0105), and the 5th European Union Framework (grant QLG3-CT-2002-01810).
Appendix A: Supplemental Tables
Table A1.
Summary of Clinical Data from Patients with Mutations in JARID1C[Note]
|
Family N017 (Ala388Pro) |
Family L017 (Leu731Phe) |
Family D034 (R68fsX7) |
Family P018 (Arg694Ter) |
Family N063 (Trp1288Ter) |
Family D029 (Glu698Lys) |
Family A034 (Asp402Tyr) |
||||||||||||||
| Characteristic | III:14 | III:2 | IV:4 | III:4 | III:5 | III:6 | IV:1 | II:2 | II:4 | II:1 | II:2 | II:1 | II:2 | II:3 | II:1 | II:3 | III:1 | III:2 | III:4 | II:4 |
| Body height (in cm) [percentile] | 165 (<3rd) | 154 [<3rd] | 171 [∼25th] | 177 [25th–50th] | 165 [<3rd] | 166 [<3rd] | 153 [<3rd] | 167 [∼3rd] | 168 [∼3rd] | 105 (at the age of 6.5 years)[<3rd] | 91 (at the age of 4.5 years)[<3rd] | 158 [<3rd] | 154 [<3rd] | 150 [<3rd] | 169 [∼3rd] | 169 [∼3rd] | 154 [<3rd] | 156 [<3rd] | Not known | Normal height |
| Head circumference (in cm) [percentile] | 53.8 [∼ 3rd] | 53.5 [∼3rd] | 54 [∼3rd] | 58.5 [∼97th] | 56.3 [50th] | 56.6 [50th] | 52 [<3rd] | 61 [>97th] | 58 [∼97th] | 52 (at the age of 6.5 years)[25–50th] | 52 (at the age of 4.5 years)[50th] | 53 [3rd] | 54 [3rd] | 51 [<3rd] | 58 [∼97th] | 59 [∼97th] | 57 [∼75th] | 55 [∼25th – 50th] | Not known | 62 [>97th] |
| MR (IQ) | + | + | + (70) | + (25) | + | + | + (32) | + | + | + | + | + | + | + | + | + | + | + | + | + |
| Pronounced verbal deficiency | − | − | + | + | + | + | + | + | + | − | − | − | − | − | + | + | + | + | − | − |
| Spasticity | − | − | − | + | + | + | + | − | − | − | − | + | + | + | − | − | − | − | − | − |
| Epilepsy | − | − | − | − | − | − | − | − | − | − | − | + | + | − | − | − | − | − | − | − |
| Pain insensitivity | − | − | − | − | − | − | − | + | + | − | − | − | − | − | − | − | − | − | − | − |
| Aggressiveness | − | − | − | + | + | + | + | − | + | − | − | − | + | − | − | − | − | − | − | − |
| Strabismus | − | − | − | + | + | + | + | + | + | − | + | + | − | + | − | − | − | − | − | − |
| Hypermetropia | − | − | − | − | − | − | − | − | − | − | − | + | + | + | − | − | − | − | − | − |
| Body hair | Normal | Normal | Normal | Sparse | Abundant | Abundant | Normal | Normal | Sparse | Normal | Normal | Normal | Normal | Normal | Normal | Normal | Normal | Normal | Normal | Normal |
| Additional features | Small forehead, small eye lashes, prognathism, small testes | Flat philtrum, thin upper lip, prognathism, small penis | Small, deep-set eyes, upslanting palpebral fissures, micrognathia | Maxillary hypoplasia | Maxillary hypoplasia | Maxillary hypoplasia | Maxillary hypoplasia | Diastema, gallstones | Gallstones | … | Myopia | Diastema, cryptorchism, alopecia areata | Diastema, cryptorchism | Diastema, unstable mood disorder | … | Macroorchism | … | … | Drowned at the age of 12 years | … |
Note.— Features that have been reported as being present in an individual are indicated with a plus sign (+); features that were absent are shown with a minus sign (−). For those patients for whom the IQ was measured, the value is given in parentheses. All patients listed in the table are male, except for patient II:3 from family N063.
Table A2.
Primer Sequences for the Amplification of the Complete JARID1C Coding Sequence (26 Exons) and ∼40 Nucleotides of the Respective Neighboring Intronic Sequences[Note]
| Amplicon | Forward Primer | Reverse Primer | PCR Product(bp) |
| e1p1 | catcactggcctttaaacgtg | ATCTTGGTTTGTCAGCGTCTC | 295 |
| e1p2 | GAAGGATCCGGTTTGTTGTG | GTTTGAAGCCGAGGCAATG | 288 |
| e1p3 | AGGCGGTAGCAGTAGAGTCAG | GATGTAGCCAAGAGGGTCTCG | 289 |
| e1p4 | ATTGCCTCGGCTTCAAACAG | ctactgcttcattccgtctcg | 289 |
| e2p1 | cactatgctgagataactcaagtcc | ctcaggtatacattctcccaacc | 175 |
| e3p1 | cccaaagatagtggtcagtgg | atcaaggatattggggtttgg | 237 |
| e4p1 | ataaatacacgtggcacatcc | aatctgtgctgaagggtaaagc | 296 |
| e5p1 | ctggagtccatgtcctgacc | actccctccacctcaaagctc | 224 |
| e6p1 | ccccagatagcagtcttgattc | ctgacttttctccccaggtg | 213 |
| e7p1 | gggccctaagtgtcctagacc | ttgaagggacaagaagcagag | 282 |
| e8p1 | tttccaatggagaacttttgc | gaagccctatgaggcagatg | 235 |
| e9p1 | ctttgcctctgttttggtgac | tgatcctagcccaacactacc | 202 |
| e10p1 | gggtgtgatgggccatagtc | taagctcacacagctgacacg | 245 |
| e11p1 | taaccctcatgccctttttac | gagcccacactgacttgattc | 267 |
| e12p1 | tgaccaaggtgtgatttactgtg | aatcactcctgccgcttgtc | 250 |
| e13p1 | caggaaaatctctatctcaacagc | ctacaaccctcctgcttctcc | 215 |
| e14p1 | catcatggcctccagacttg | ccaccagaatagggtgcttg | 284 |
| e15p1 | ttttgctacctggttctggtt | cagctctccatcctccttctt | 286 |
| e16p1 | aagaaggaggatggagagctg | gggaatagaacttgcctgtgg | 229 |
| e17p1 | tctattcaatactgcctactctttgc | ctggatcctcagcaccttatg | 243 |
| e18p1 | aggcactgagtttggacctg | ggtccccttgatccctcatc | 191 |
| e19p1 | cccagctgaggagtttgttc | AACAGTCCTCGCATGACAGC | 279 |
| e19p2new | ATGAGGTGAAACGCACACTG | caactttgatgtttggaataggg | 239 |
| e20p1 | gcagaccacatcagactgagc | cttcccagctggactgaagg | 231 |
| e21p1 | tggcaagttgaactgagctg | aggcgttaagagacgctgtag | 261 |
| e22p1 | gtaagggaagcccagtcattc | tatcatcaccaagcccttctc | 227 |
| e23p1 | gtaggctgctgacccactttg | ATGGAACCAGTCCTGACACAG | 234 |
| e23p2 | CTGTGTGTGTGGGCAGGTG | CCCTCTCTGTGAGGCACTG | 293 |
| e23p3a_new | TCCTGTGTCCACTGTGTATGC | AGGCTCCTCAGGTCTAGGTTC | 248 |
| e23p3b_new | TCTGGCCTCTGAAGATGTGAC | aggggctaggagacaagaatc | 205 |
| e24p1 | cttcacttcagttgcccctac | atgtgcagtcaacagcctacc | 163 |
| e25p1 | accaagggcagcctctaatg | ctactcggcctgacctcctg | 290 |
| e26p1 | actggcccacaacctgtct | CAGTCTCCTCCTCCAGCTCT | 230 |
| e26p2 | CTAGAGCCAAAGAGGGTACGG | AAGAAGCAGGCTTGATGGTC | 338 |
Note.— Large exons (>200 bp) have been amplified in overlapping fragments. Primer sequences given in uppercase letters represent an exonic primer location, and primer sequences given in lowercase letters indicate an intronic primer location. The sizes of the resulting PCR products are listed in the last column.
Electronic-Database Information
Accession numbers and URLs for data presented herein are as follows:
- BLAST2, http://www.ncbi.nlm.nih.gov/blast/bl2seq/bl2.html
- Conserved Domain Database, http://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml
- GenBank, http://www.ncbi.nih.gov/Genbank/ (for JARID1C [accession number L25270], JARID1C [accession number P41229], JARID1D [accession number NM_004653], JARID1D [accession number NP_004644], JARID1A [accession number NP_005047], JARID1B [accession number NP_006609], Jarid1c [accession number NP_038696], Lid [accession number NP_723140], and ZK593.4 [accession number CAA93426])
- Human Genome Browser Gateway, http://genome.cse.ucsc.edu/cgi-bin/hgGateway?db=hg10
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/entrez/Omim/ (for FMR2, OPHN1, PAK3,GDI1, IL1RAPL1, TM4SF2, ARHGEF6, MECP2, FACL4, ARX, ELK1, ALAS2, PQBP1, FTSJ1, JARID1C, JARID1D/SMCY, USP9X/Y, RPS4X/Y, UTX/Y, and SLC6A8)
References
- Agulnik AI, Mitchell MJ, Mattei MG, Borsani G, Avner PA, Lerner JL, Bishop CE (1994) A novel X gene with a widely transcribed Y-linked homologue escapes X-inactivation in mouse and human. Hum Mol Genet 3:879–884 [DOI] [PubMed] [Google Scholar]
- Asselta R, Duga S, Spena S, Santagostino E, Peyvandi F, Piseddu G, Targhetta R, Malcovati M, Mannucci PM, Tenchini ML (2001) Congenital afibrinogenemia: mutations leading to premature termination codons in fibrinogen A α-chain gene are not associated with the decay of the mutant mRNAs. Blood 98:3685–3692 10.1182/blood.V98.13.3685 [DOI] [PubMed] [Google Scholar]
- Bateman JF, Freddi S, Nattrass G, Savarirayan R (2003) Tissue-specific RNA surveillance? Nonsense-mediated mRNA decay causes collagen X haploinsufficiency in Schmid metaphyseal chondrodysplasia cartilage. Hum Mol Genet 12:217–225 10.1093/hmg/ddg054 [DOI] [PubMed] [Google Scholar]
- Bienvenu T, Poirer K, Friocourt G, Bahi N, Beaumont D, Fauchereau F, Ben Jeema L, Zemni R, Vinet MC, Francis F, Couvert P, Gomot M, Moraine C, Van Bokhoven H, Kalscheuer V, Frints S, Gécz F, Ohzaki K, Chaabouni H, Fryns JP, Desportes V, Beldjord C, Chelly J (2002) ARX, a novel Prd-class-homeobox gene highly expressed in the telencephalon, is mutated in X-linked mental retardation. Hum Mol Genet 11:981–991 10.1093/hmg/11.8.981 [DOI] [PubMed] [Google Scholar]
- Chelly J, Hamel BCJ (2003) Genetics of X-linked mental retardation. In: Fisch GS (ed) Genetics and genomics of neurobehavioral disorders. Humana Press, Totowa, NJ, pp 263–287 [Google Scholar]
- Claes S, Devriendt K, Van Goethem G, Roelen L, Meireleire J, Raeymaekers P, Cassiman J-J, Fryns J-P (2000) Novel syndromic form of X-linked complicated spastic paraplegia. Am J Med Genet 94:1–4 [DOI] [PubMed] [Google Scholar]
- Delbridge ML, Longepied G, Depetris D, Mattei MG, Disteche CM, Marshall Graves JA, Mitchell MJ (2004) TSPY, the candidate gonadoblastoma gene on the human Y chromosome, has a widely expressed homologue on the X: implications for Y chromosome evolution. Chromosome Res 12:345–356 10.1023/B:CHRO.0000034134.91243.1c [DOI] [PubMed] [Google Scholar]
- Dyson N (1998) The regulation of E2F by pRB-family proteins. Genes Dev 12:2245–2262 [DOI] [PubMed] [Google Scholar]
- Fattaey AR, Helin K, Dembski MS, Dyson N, Harlow E, Vuocolo GA, Hanobik MG, Haskell KM, Oliff A, Defeo-Jones D, Jones RE (1993) Characterization of the retinoblastoma binding proteins RBP1 and RBP2. Oncogene 8:3149–3156 [PubMed] [Google Scholar]
- Fishburn J, Turner G, Daniel A, Brookwell R (1983) The diagnosis and frequency of X-linked conditions in a cohort of moderately retarded males with affected brothers. Am J Med Genet 14:713–724 [DOI] [PubMed] [Google Scholar]
- Freude K, Hoffmann K, Jensen LR, Delatycki MB, Des Portes V, Moser B, Hamel B, Van Bokhoven H, Moraine C, Fryns J-P, Chelly J, Gécz J, Lenzner S, Kalscheuer VM, Ropers H-H (2004) Mutations in the FTSJ1 gene coding for a novel S-adenosyl-methionine–binding protein cause nonsyndromic X-linked mental retardation. Am J Hum Genet 75:305–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanashima C, Li SC, Shen L, Lai E, Fishell G (2004) Foxg1 suppresses early cortical cell fate. Science 303:56–59 10.1126/science.1090674 [DOI] [PubMed] [Google Scholar]
- Herbst DS, Miller JR (1980) Nonspecific X-linked mental retardation II: the frequency in British Columbia. Am J Med Genet 7:461–469 [DOI] [PubMed] [Google Scholar]
- Holbrook JA, Neu-Yilik G, Hentze MW, Kulozik AE (2004) Nonsense-mediated decay approaches the clinic. Nat Genet 36:801–808 10.1038/ng1403 [DOI] [PubMed] [Google Scholar]
- Kalscheuer VM, Freude K, Musante L, Jensen LR, Yntema HG, Gécz J, Sefiani A, et al (2003) Mutations in the polyglutamine binding protein 1 gene cause X-linked mental retardation. Nat Genet 35:313–315 10.1038/ng1264 [DOI] [PubMed] [Google Scholar]
- Kerr TP, Sewry CA, Robb SA, Roberts RG (2001) Long mutant dystrophins and variable phenotypes: evasion of nonsense-mediated decay? Hum Genet 109:402–407 10.1007/s004390100598 [DOI] [PubMed] [Google Scholar]
- Kortschak RD, Tucker PW, Saint R (2000) ARID proteins come in from the desert. Trends Biochem Sci 25:294–299 10.1016/S0968-0004(00)01597-8 [DOI] [PubMed] [Google Scholar]
- Luo RX, Postigo AA, Dean DC (1998) Rb interacts with histone deacetylase to repress transcription. Cell 92:463–473 10.1016/S0092-8674(00)80940-X [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A, Anderson JB, DeWeese-Scott C, Fedorova ND, Geer LY, He S, Hurwitz DI, et al (2003) CDD: a curated Entrez database of conserved domain alignments. Nucleic Acids Res 31:383–387 10.1093/nar/gkg087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meloni I, Bruttini M, Longo I, Mari F, Rizzolio F, D’Adamo P, Denvriendt K, Fryns JP, Toniolo D, Renieri A (2000) A mutation in the Rett syndrome gene, MECP2, causes X-linked mental retardation and progressive spasticity in males. Am J Hum Genet 67:982–985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merienne K, Jacquot S, Pannetier S, Zeniou M, Bankier A, Gécz J, Mandel JL, Mulley J, Sassone-Corsi P, Hanauer A (1999) A missense mutation in RPS6KA3 (RSK2) responsible for non-specific mental retardation. Nat Genet 22:13–14 10.1038/8719 [DOI] [PubMed] [Google Scholar]
- Roeleveld N, Zielhuis GA, Gabreels F (1997) The prevalence of mental retardation: a critical review of recent literature. Dev Med Child Neurol 39:125–132 [DOI] [PubMed] [Google Scholar]
- Ropers H-H, Hoeltzenbein M, Kalscheuer V, Yntema H, Hamel B, Fryns J-P, Chelly J, Partington M, Gécz J, Moraine C (2003) Nonsyndromic X-linked mental retardation: where are the missing mutations? Trends Genet 19:316–320 10.1016/S0168-9525(03)00113-6 [DOI] [PubMed] [Google Scholar]
- Rosenberg EH, Almeida LS, Kleefstra T, deGrauw RS, Yntema HG, Bahi N, Moraine C, Ropers H-H, Fryns J-P, deGrauw TJ, Jakobs C, Salomons GS (2004) High prevalence of SLC6A8 deficiency in X-linked mental retardation. Am J Hum Genet 75:97–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheardown S, Norris D, Fisher A, Brockdorff N (1996) The mouse Smcx gene exhibits developmental and tissue specific variation in degree of escape from X inactivation. Hum Mol Genet 5:1355–1360 10.1093/hmg/5.9.1355 [DOI] [PubMed] [Google Scholar]
- Strømme P, Mangelsdorf ME, Shaw MA, Lower KM, Lewis SM, Bruyere H, Lutcherath V, Gedeon AK, Wallace RH, Scheffer IE, Turner G, Partington M, Frints SG, Fryns JP, Sutherland GR, Mulley JC, Gécz J (2002) Mutations in the human ortholog of Aristaless cause X-linked mental retardation and epilepsy. Nat Genet 30:441–445 10.1038/ng862 [DOI] [PubMed] [Google Scholar]
- Tan K, Shaw AL, Madsen B, Jensen K, Taylor-Papadimitriou J, Freemont PS (2003) Human PLU-1 has transcriptional repression properties and interacts with the developmental transcription factors BF-1 and PAX9. J Biol Chem 278:20507–20513 10.1074/jbc.M301994200 [DOI] [PubMed] [Google Scholar]
- Vawter MP, Evans S, Choudary P, Tomita H, Meador-Woodruff J, Molnar M, Li J, Lopez JF, Myers R, Cox D, Watson SJ, Akil H, Jones EG, Bunney WE (2004) Gender-specific gene expression in post-mortem human brain: localization to sex chromosomes. Neuropsychopharmacology 29:373–384 10.1038/sj.npp.1300337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilsker D, Patsialou A, Dallas PB, Moran E (2002) ARID proteins: a diverse family of DNA binding proteins implicated in the control of cell growth, differentiation, and development. Cell Growth Differ 13:95–106 [PubMed] [Google Scholar]
- Xu J, Burgoyne P, Arnold AP (2002) Sex differences in sex chromosomes gene expression in mouse brain. Hum Mol Genet 11:1409–1419 10.1093/hmg/11.12.1409 [DOI] [PubMed] [Google Scholar]