Abstract
We have analyzed genetic data for 326 microsatellite markers that were typed uniformly in a large multiethnic population-based sample of individuals as part of a study of the genetics of hypertension (Family Blood Pressure Program). Subjects identified themselves as belonging to one of four major racial/ethnic groups (white, African American, East Asian, and Hispanic) and were recruited from 15 different geographic locales within the United States and Taiwan. Genetic cluster analysis of the microsatellite markers produced four major clusters, which showed near-perfect correspondence with the four self-reported race/ethnicity categories. Of 3,636 subjects of varying race/ethnicity, only 5 (0.14%) showed genetic cluster membership different from their self-identified race/ethnicity. On the other hand, we detected only modest genetic differentiation between different current geographic locales within each race/ethnicity group. Thus, ancient geographic ancestry, which is highly correlated with self-identified race/ethnicity—as opposed to current residence—is the major determinant of genetic structure in the U.S. population. Implications of this genetic structure for case-control association studies are discussed.
Introduction
From an evolutionary point of view, population stratification (genetically distinct subgrouping) and admixture (intermating between genetically distinct groups) are created by human mating patterns. Geographical, social, and cultural barriers have given rise to reproductively isolated human populations, within which random drift has produced genetic differentiation. Numerous recent studies using a variety of genetic markers have shown that, for example, individuals sampled worldwide fall into clusters that roughly correspond to continental lines, as well as to the commonly used self-identifying racial groups: Africans, European/West Asians, East Asians, Pacific Islanders, and Native Americans (Bowcock et al. 1994; Calafell et al. 1998; Rosenberg et al. 2002). One significant consequence of population genetic structure is confounding in case-control association studies. Because of the unique political and social history of the United States, genetic structure in the contemporary U.S. population is extremely complicated. Most prominently, the level of white admixture among African Americans has been estimated at 10%–20% (Parra et al. 1998); more complicated are Hispanic groups, which may have European, Native American, and African ancestries that vary regionally (Hanis et al. 1991). In addition, stratification and admixture occur at finer levels. Such subtle heterogeneity is not readily detected with a limited number of genetic markers, yet their implications in biomedical research may be important.
Epidemiologic designs that aim to detect associations between alleles and disease by use of unrelated cases and controls are popular because of their efficiency and the ease of recruiting subjects. However, spurious associations between a trait and random genetic loci may arise as a result of subtle genetic structure (Lander and Schork 1994). The impact of confounding due to population genetic structure in case-control studies has been debated (Thomas and Witte 2002; Wacholder et al. 2002).
In light of the number of case-control studies that are being performed and planned, the above considerations warrant a careful examination of genetic structure within and between major population groups in the United States. One major goal is to quantify the correspondence between self-identified race/ethnicity (SIRE) and the major genetic structure that exists in the U.S. population. In addition, out of convenience or out of necessity, case and control subjects are sometimes recruited from different geographic regions, matching only at the level of major racial group. An underlying assumption is the relative homogeneity within a single SIRE group. The validity of this assumption must be evaluated. Furthermore, association studies among ethnically admixed populations are particularly vulnerable to spurious association. Although admixed groups have had relatively low representation in the U.S. population in the past, their representation is increasing. Whereas, historically, geneticists have avoided studying such individuals and groups because of the difficulties involved, it is no longer reasonable or fair to exclude such groups from genetic research.
In this study, we examined the genetic structure between and within major racial/ethnic groups by use of data from a large, ethnically diverse sample, the Family Blood Pressure Program (FBPP), which includes self-identified white, African American, Hispanic (Mexican), and East Asian (Chinese and Japanese) subjects (FBPP Investigators 2002). Participants were enrolled, typically as sibships or nuclear families, at 15 field centers (recruitment sites), of which 11 are within the continental United States, 1 is in Hawaii, and 3 are in Taiwan. Details are provided in table A1 (online only). This sample provides a unique opportunity to answer several questions related to population structure. The degree of genetic differentiation can be assessed for this sample with respect to multiple levels of stratification.
Material and Methods
Subjects
The FBPP is a collaborative effort of four research networks (GenNet, GENOA, HyperGEN, and SAPPHIRe) that aims to investigate high blood pressure and related conditions in multiple racial/ethnic groups (FBPP Investigators 2002). Each network has been funded by the National Heart, Lung, and Blood Institute (NHLBI) since 1995. In total, DNA samples from 10,527 participants were genotyped at 326 autosomal genome screen microsatellite markers by the NHLBI-sponsored Mammalian Genotyping Service (Marshfield, WI) (screening set 8) and had sufficient marker data for analysis (i.e., at most 40 missing genotypes).
Race/ethnicity information was obtained by self-description. HyperGEN focused their recruitment on whites and African Americans. Subjects were given a response card and were allowed to endorse any of the following categories: “non-Hispanic white,” “non-Hispanic black,” “Hispanic,” “Asian,” “Pacific Islander,” “American Indian/Alaska Native,” or “other.”
GENOA concentrated their sampling on three groups: whites, African Americans, and Hispanics. They also employed a response card and allowed subjects to endorse any of the following categories: “non-Hispanic white,” “African American,” “Hispanic/Mexican,” or “other.”
GenNet focused their recruitment on white and African American subjects. Participants were asked for a self-description of their race/ethnicity without a list of choices. Responses other than “Caucasian/white” or “African American”—including “Hispanic”—were recorded, but, in the pooled data set, they were listed as “other.”
For all three of these networks, there were neither questions nor requirements regarding the race/ethnicity or ancestry of the participants' parents or grandparents for inclusion in the study. SAPPHIRe focused their study on Asian populations. Specifically, they required subjects to report being Chinese and having four Chinese grandparents or being Japanese and having four Japanese grandparents to be included in the study.
Thus, in summary, each study participant identified him/herself as belonging to one of five categories: white non-Hispanic (CAU), black non-Hispanic (AFR), Hispanic (HIS), Chinese (CHI), and Japanese (JAP). Therefore, in our analysis, SIRE corresponds to four major distinctions: CAU, AFR, HIS, and EAS, the latter referring to East Asians (Chinese and Japanese combined), and one minor distinction, that between Chinese and Japanese. In the first analyses, which involved computing genetic distances and comparing SIRE with genetic structure obtained from genetic cluster analysis, we randomly selected one participant with STR genotype information from each nuclear family and treated these participants as unrelated individuals; the resulting set consisted of 3,648 individuals. Table A1 (online only) summarizes the collection site and SIRE information of these individuals. In total, this analysis included 1,349 self-identified CAU, 1,308 AFR, 412 HIS, 407 CHI, 160 JAP, and 12 OTH. Three of the “others” came from HyperGEN (one each from Salt Lake City, Minneapolis, and Framingham, MA), eight came from GenNet (from Tecumseh, MI), and one came from SAPPHIRe (from Honolulu). The rate of missing genotypes was <2%.
Because of its focus on linkage analysis of hypertension, the FBPP recruited sibships or nuclear families that typically had at least one hypertensive index subject, although precise ascertainment criteria varied among networks (FBPP Investigators 2002). For analyses focusing on genetic stratification bias with respect to blood pressure, we selected the hypertensive individual (“case”) from those families with a single hypertensive subject and no other relatives and a single, randomly selected hypertensive individual from families with multiple hypertensive subjects and at most one normotensive subject. To obtain “controls,” we selected the normotensive subject from those families with a single normotensive subject and no relatives and a single, randomly selected normotensive individual from families with multiple normotensive subjects and at most one hypertensive individual. For the networks and field centers that included only hypertensive subjects, this analysis was not possible. If a family contained exactly one hypertensive subject and one normotensive subject or more than one hypertensive subject and more than one normotensive subject, the family was not included in this analysis.
Genetic Distance Analysis
We created 18 subpopulations on the basis of the participants’ SIRE and the recruitment site (the few individuals who identified their race/ethnicity as “other” were excluded from this analysis). As a measure of genetic distance, we computed the “coancestry coefficient” among groups (Reynolds et al. 1983). The coancestry coefficient is a measure of distance that is closely related to an average value of FST across genes. To visualize these genetic distances, we performed multidimensional scaling (MDS) analysis (Mardia et al. 1980). In simple terms, this analysis provides a configuration of 18 points on a two-dimensional plane, such that the Euclidean distances among these points match the genetic distance matrix as closely as possible.
Genetic Cluster Analysis
In this analysis, we studied genetic similarity at an individual level by use of the program structure (Pritchard et al. 2000). This approach is similar to that of a previous analysis (Rosenberg et al. 2002), except that the FBPP population primarily represents a United States–based sample. Because our goal is classification, we used the “NOADMIX” option in structure, so that the entire genome of each individual was assumed to have been derived from a single homogeneous population. We examined the correspondence rate between SIRE and genetic cluster classification by crossclassifying subjects on the basis of these two criteria.
Tests of Stratification
To examine allele-frequency differentiation between pairs of groups defined either by geography or by disease status, we computed χ2 tests of independence on the basis of the 2×2 table of allele frequencies by group. Levels of significance were determined empirically by permutation analysis, with 10,000 permutations. For the microsatellite markers, each distinct allele was tested, provided that there were at least 50 occurrences of that allele in the two tested groups combined. We used this threshold to ensure adequate power to detect modest differences, given the sample sizes employed. Because of the small number of Chinese families recruited in Hawaii (n=25) and the small number of Japanese families recruited in Stanford, CA (n=16), these two field centers were excluded from this analysis. Since all Japanese individuals in this analysis are from Hawaii and all Hispanic individuals are from Starr County, TX, comparison between sites was not performed within these two SIRE categories.
Results
Genetic Distance Analysis
In table 1, the diagonal elements represent the mean (SD) of genetic distances between recruitment sites within a SIRE group; the corresponding figures across SIRE groups are indicated by the off-diagonal elements. The greatest genetic distances occur between populations with ancestries from different continents and little mixing (i.e., between East Asians and African Americans, followed by East Asians and whites). The second largest genetic distances are between the groups with some shared ancestry—namely, East Asians and Hispanics (whose Native American ancestry resembles that of Asians) and whites and African Americans (who have white admixture). Most similar are whites and Hispanics (who have substantial white admixture) and Chinese and Japanese. As can be seen by comparing the genetic distances on and off the diagonals in table 1, continental ancestry and separation time play more-important roles than current geographic distance. Thus, for example, Hawaiian Chinese bear much more genetic resemblance to Chinese from Stanford, CA, and from Taiwan than they do to Hawaiian Japanese. In fact, the genetic distances between recruitment sites within SIRE categories are uniformly very small.
Table 1.
Average Genetic Distances (×10−2) between SIRE/Site Pairs [Note]
|
Average Genetic Distance (SD) between Pair |
|||||
| CAU | AFR | HIS | CHI | JAP | |
| CAU | .07 (.05) | 2.90 (.13) | 1.05 (.05) | 4.20 (.12) | 4.26 (.16) |
| AFR | .01 (.006) | 2.88 (.09) | 4.62 (.10) | 4.67 (.16) | |
| HIS | … | 3.09 (.01) | 3.03 (.16) | ||
| CHI | .02 (.02) | .60 (.06) | |||
| JAP | .00 | ||||
Note.— Genetic distances were calculated by use of the coancestry coefficient of Reynolds et al. (1983).
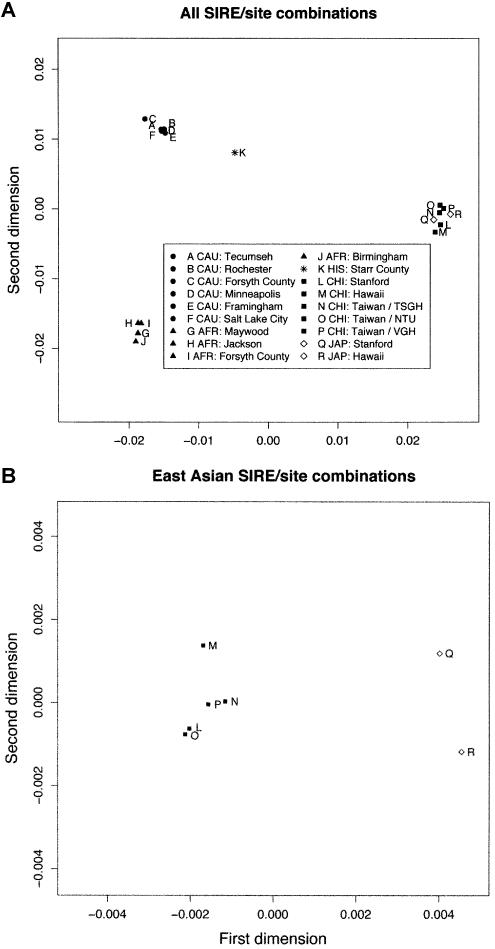
The MDS analysis for all 18 SIRE/site combinations is shown in figure 1A. As we expect, subpopulations of the same SIRE tend to cluster closely. Essentially, the X-axis separates the East Asians from the other groups, whereas the Y-axis separates the African Americans from the other groups. The MDS places the Hispanic group between the white cluster and the East Asian cluster, which is consistent with this being an admixed group with European and Native American ancestries and with Native Americans being closer, genetically, to the East Asians (Calafell et al. 1998). Although the Chinese and the Japanese groups appear clustered together in this plot, they are separable on another dimension. In other words, MDS with only the Asians produces excellent separation between the Chinese and the Japanese groups (fig. 1B).
Figure 1.
MDS of the genetic distance matrix for 18 SIRE/site combinations (A) and 7 East Asian SIRE/site combinations (B).
Genetic Clusters versus SIRE
Genetic cluster analysis using structure was performed, allowing, sequentially, for k=2, 3, 4, or more clusters (Pritchard et al. 2000). The results can be summarized as follows. When k=2 clusters was specified, the Chinese and Japanese emerged as a combined cluster; when k=3 clusters was specified, the African Americans separated from the whites and Hispanics; when k=4 clusters was specified, an additional cluster was formed that was nearly exclusively Hispanic (99.8%). All but one of the Hispanic individuals analyzed were included in this new cluster. The four-cluster results are given in table 2, with crossclassification by SIRE. Our sequential cluster results are completely consistent with what we observed from the genetic distance measures and from figure 1—namely, that the East Asians are the most distant from the other groups, followed by the African Americans, and then the Hispanics. Allowing for more than four clusters did not yield stable results: multiple runs of structure produced varying cluster configurations; in many runs, one cluster was nearly empty. However, when we repeated the cluster analysis with only the East Asian subjects, two clusters did emerge that almost perfectly distinguished between the two ethnicities, with a total of 6 (2 Chinese and 4 Japanese) (1.1%) of 567 subjects being differentially classified. No such consistent subclusters emerged from separate analyses of the African American, white, or Hispanic groups. Thus, the structure we observed at the population level using MDS is recaptured here at an individual level. For the group reporting a major SIRE category, the correspondence between genetic cluster and SIRE is remarkably high, with only 5 (0.14%) of 3,636 individuals being differentially classified (table 2). Accordingly, in this case, major SIRE category and genetic cluster are effectively synonymous. Overall, our cluster analysis results are completely consistent with previous theoretical predictions regarding the ease of separating these groups on the basis of the number of markers tested (Risch et al. 2002). Nearly all individuals had a cluster assignment probability of ∼1. Only two subjects had a probability <.95: one of these subjects self-reported as Hispanic but fell into the white genetic cluster, and the other subject self-reported as African American but fell into the white genetic cluster. We note that this analysis was not based on determination of individuals’ “racial” ancestry (e.g., estimating individual European, African, and Native American ancestry for the African American and Hispanic subjects). To do so would require inclusion of the nonadmixed ancestral groups (such as Africans and Native Americans) and the use of the “ADMIX” option of structure. What our results do show is that the (admixed) groups included have approximated within-group random mating sufficiently long enough to give rise to distinct genetic clusters.
Table 2.
Results of Genetic Cluster Analysis versus SIRE for Entire Sample
|
No. of Subjects in Genetic Cluster |
||||
| SIRE | A | B | C | D |
| CAU | 1,348 | 0 | 0 | 1 |
| AFR | 3 | 0 | 1,305 | 0 |
| HIS | 1 | 0 | 0 | 411 |
| CHI | 0 | 407 | 0 | 0 |
| JAP | 0 | 160 | 0 | 0 |
| OTH | 1 | 2 | 0 | 9 |
There were 12 individuals who reported “other” in response to the race/ethnicity question. Of these individuals, nine were classified genetically in the Hispanic cluster, two in the East Asian cluster, and one in the white cluster. Eight of the nine subjects who fell into the Hispanic cluster were from GenNet (Tecumseh, MI), a site where the recruitment focused on whites. Tracing back to the original interview records we found that, in fact, all eight subjects self-reported as “Hispanic” but were categorized as “other” when included in the pooled data set.
Our study deliberately sampled whites, African Americans, Hispanics, and East Asians; therefore, a more general survey would likely have produced a larger representation of individuals with other self-descriptions (e.g., Native Americans, Pacific Islanders, and South Asians). Nonetheless, our results do reflect an unbiased sampling of individuals who self-describe within the major categories we included.
Stratification by Geography
We tested for differences in the frequency of alleles at each of the 326 microsatellite (STR) markers between subpopulations defined by SIRE and recruitment site. Table 3 displays the proportion of tests that were significant at the P=.05 level. Stratification across SIRE groups is uniformly high, with ⩾40% of allele-frequency differences significant. The one exception, as expected, is the Chinese-Japanese comparison, involving two East Asian ethnicities, for which the proportion that are significant is ∼18%. Perhaps of greater interest are the comparisons within a SIRE group, which are indicated by the diagonal elements in table 3. Here, we see only a modest increase of significant tests over expected (5.3% for AFR and 6.3% for CAU). Thus, stratification within SIRE groups on the basis of current geography may lead to confounding, but the lack of significant geographic differences in allele frequencies suggests that the impact is not likely to be large.
Table 3.
Allele-Frequency Difference between SIRE/Site Combinations[Note]
|
Proportion of Tests (±SE) Significant at P = .05 |
|||||
| CAU | AFR | HIS | CHI | JAP | |
| CAU | .063 (±.008) | .576 (±.062) | .414 (±.079) | .493 (±.059) | .566 (±.047) |
| AFR | .053 (±.006) | .640 (±.036) | .554 (±.065) | .642 (±.018) | |
| HIS | … | .482 (±.077) | .557 | ||
| CHI | .047 (±.005) | .182 (±.034) | |||
Note.— On average, 1,660 alleles were tested between each pair of SIRE/site combinations. SEs are estimated on the basis of SIRE/site combinations.
Tests of Stratification in Comparisons of Hypertensive Subjects with Normotensive Subjects
To examine this question in the FBPP data, we selected “cases” (hypertensive subjects) and “controls” (normotensive subjects) in accordance with a scheme described in the “Material and Methods” section. We then tested for differences in the frequency of alleles at each of the 326 microsatellite markers between the “cases” and “controls” and calculated the proportion of tests significant at the P=.05 level. We saw no trend toward an excess of significant tests (table 4). We also examined Q-Q plots of the entire distribution of P values for the alleles at the 326 markers and compared this distribution with the expected uniform distribution. None of these plots revealed any significant deviations from expectation. Thus, it appears that, at least in the context of these analyses of hypertension, sampling hypertensive cases and controls from the same local population does not create a serious confounding problem.
Table 4.
Test of Stratification between Unrelated Normotensive Subjects and Hypertensive Subjects, for Various SIRE/Site Combinations
|
No. of Subjects |
||||
| SIRE andRecruitment Site | Normotensive | Hypertensive | Proportion ofSignificant Allelesa | No. of Alleles Tested |
| AFR: | ||||
| Birmingham, AL | 35 | 368 | .055 | 1,799 |
| Forsyth, NC | 49 | 149 | .058 | 1,055 |
| Jackson, MS | 61 | 389 | .042 | 1,753 |
| Maywood, IL | 164 | 55 | .048 | 1,173 |
| CHI: | ||||
| Taiwan | 72 | 156 | .044 | 1,160 |
| HIS: | ||||
| Starr, TX | 175 | 114 | .057 | 1,375 |
| CAU: | ||||
| Tecumseh, MI | 216 | 27 | .043 | 1,265 |
Proportion of alleles with frequencies that are significantly different at the level of P=.05.
Because the study sample was largely based on the presence of hypertension—and hypertension is age related—age might also be acting as a confounder, if allele frequencies are age dependent. We therefore also undertook an analysis to determine whether there was genetic stratification in the sample on the basis of age, particularly in the admixed groups (African Americans and Mexican Americans). Each race/ethnicity group was divided in half at the median age (which ranged from 50 years to 58 years), and allele frequencies were compared between the two age groupings for each allele. Examination of Q-Q plots of the distribution of P values from this analysis also showed near-perfect conformity with expectation, a result that suggests no age trends in allele frequencies.
Discussion
Attention has recently focused on genetic structure in the human population. Some have argued that the amount of genetic variation within populations dwarfs the variation between populations, suggesting that discrete genetic categories are not useful (Lewontin 1972; Cooper et al. 2003; Haga and Venter 2003). On the other hand, several studies have shown that individuals tend to cluster genetically with others of the same ancestral geographic origins (Mountain and Cavalli-Sforza 1997; Stephens et al. 2001; Bamshad et al. 2003). Prior studies have generally been performed on a relatively small number of individuals and/or markers. A recent study (Rosenberg et al. 2002) examined 377 autosomal microsatellite markers in 1,056 individuals from a global sample of 52 populations and found significant evidence of genetic clustering, largely along geographic (continental) lines. Consistent with prior studies, the major genetic clusters consisted of Europeans/West Asians (whites), sub-Saharan Africans, East Asians, Pacific Islanders, and Native Americans. It is clear that the ability to define distinct genetic clusters depends on the number and type of markers used (Risch et al. 2002). Reports that document inability to define distinct clusters generally used only a modest number of markers and, hence, had little power to detect clusters (Romualdi et al. 2002). Studies with larger numbers of markers appear to show strong evidence of clustering (Stephens et al. 2001; Rosenberg et al. 2002).
Another major point of discussion has been the correspondence between genetic clusters and commonly used racial/ethnic labels. Some have argued for poor correspondence between these two entities (Lewontin 1972; Wilson et al. 2001), whereas others have suggested a strong correlation (Risch et al. 2002; Burchard et al. 2003). We have shown a nearly perfect correspondence between genetic cluster and SIRE for major ethnic groups living in the United States, with a discrepancy rate of only 0.14%. Perhaps this is not surprising for the major groupings (whites, East Asians, and African Americans), since prior studies would suggest enough genetic differentiation between these groups to produce robust clustering. On the other hand, one prior study of Hispanics did not suggest a distinct cluster for this group, possibly because of the heterogeneous origins of that Hispanic sample (Stephens et al. 2001). From the genetic perspective, Hispanics generally represent a differential mixture of European, Native American, and African ancestry, with the proportionate mix typically depending on country of origin. Our sample was from a single location in Texas and was composed of Mexican Americans. Although the genetic distance analysis suggested relative proximity to the whites in our sample, the distance was still sufficient to allow for creation of a distinct genetic cluster for this group. Again, this is likely because of the large number of markers used in our analysis. On the other hand, in the analysis of the full sample, the two East Asian groups—Chinese and Japanese—did not emerge as distinct subgroups, likely because their distance from one another was too modest to be detectable in the context of the larger sample. However, when the East Asians were analyzed separately, two clusters—corresponding to Chinese and Japanese—did emerge, with only a small amount of discordance (6 [1%] of 567 subjects). In contrast, cluster analysis within the three other major clusters did not produce robust, replicable subgroups, indicating a lack of further subgroups within these entities, at least in the current marker set. This observation does not eliminate the potential for confounding in these populations. First, there may be subgroups within the larger population group that are too small to detect by cluster analysis. Second, there may not be discrete subgrouping but continuous ancestral variation that could lead to stratification bias. For example, African Americans have a continuous range of European ancestry that would not be detected by cluster analysis but could strongly confound genetic case-control studies. Furthermore, our analysis likely underrepresents individuals with recent mixed ancestry (who would require more complex categorization) and other groups typically underrepresented, such as South Asians. Further study is required to evaluate the correlation between genetically determined groupings and SIRE for these individuals.
Our observations also emphasize the importance of SIRE information: although statistical approaches using genetic marker information recapture SIRE with high accuracy, such analyses need to be guided by SIRE information. The outcome of statistical cluster analyses depends on the (relative and absolute) sample size of the subgroups and on the homogeneity within groups relative to distance between groups. Without proper controlling of these nuisance factors, cluster analyses based on genetic markers sometimes overlook important components of population structure, while producing artifact clusters other times.
We note that the genetic cluster results indicate that older geographic ancestry—rather than recent geographic origin—is highly correlated with racial/ethnic categorizations and, thus, is the major determinant of genetic structure in the population. Although our results suggest that genetic stratification may exist within racial/ethnic groups—specifically, whites and African Americans sampled from different geographic locations in the United States—we found the differences based on current geography to be quite modest. On the other hand, geographic matching of Hispanic subjects is likely to be of much greater importance, given the larger genetic differentiation between Hispanic groups on the basis of current geographic origins. In this study, we could not evaluate this question directly, since Hispanics were recruited only from a single site. Also, these geographic analyses do not rule out other potential sources of confounding within geographic regions for these groups (for example, those based on specific ethnic affiliations), which still may require attention.
Our results also suggested little confounding when sampling cases and controls within SIRE and geographic groups for studies of hypertension. We detected little, if any, genetic differentiation at the 326 microsatellite markers between hypertensive and normotensive subjects in any of the ethnic groups we examined. However, this topic merits additional scrutiny—in particular, for the admixed subjects (Hispanics and African Americans)—to determine whether cases and controls have differential levels of admixture, which is likely to be the greatest source of confounding for these populations (H. Tang, personal communication).
In summary, from a very large study of four major racial/ethnic groups within the United States and Taiwan, we found extraordinary correspondence between SIRE and genetic cluster categories but only modest geographic differentiation within each race/ethnicity group. This result indicates that studies using genetic clusters instead of racial/ethnic labels are likely to simply reproduce racial/ethnic differences, which may or may not be genetic. On the other hand, in the absence of racial/ethnic information, it is tempting to attribute any observed difference between derived genetic clusters to a genetic etiology. Therefore, researchers performing studies without racial/ethnic labels should be wary of characterizing difference between genetically defined clusters as genetic in origin, since social, cultural, economic, behavioral, and other environmental factors may result in extreme confounding (Risch et al. 2002).
Appendix A: Supplemental Material
Table A1.
Number of Subjects Included in Genetic Cluster Analysis, by Network, Field Center, and Ethnicity
|
No. of Subjects with SIRE |
|||||||
| Network and Field Center | CAU | AFR | HIS | CHI | JAP | OTH | No. of Subjects Enrolled at Center |
| GenNet: | |||||||
| Tecumseh, MI | 263 | 0 | 0 | 0 | 0 | 8 | 271 |
| Maywood, IL | 0 | 267 | 0 | 0 | 0 | 0 | 267 |
| GENOA: | |||||||
| Rochester, MN | 493 | 1 | 1 | 0 | 0 | 0 | 495 |
| Jackson, MS | 0 | 355 | 0 | 0 | 0 | 0 | 355 |
| Starr County, TX | 0 | 0 | 411 | 0 | 0 | 0 | 411 |
| HyperGEN: | |||||||
| Forsyth County, NC | 69 | 222 | 0 | 0 | 0 | 0 | 291 |
| Minneapolis | 166 | 0 | 0 | 0 | 0 | 1 | 167 |
| Framingham, MA | 178 | 0 | 0 | 0 | 0 | 1 | 179 |
| Salt Lake City | 179 | 0 | 0 | 0 | 0 | 1 | 180 |
| Birmingham, AL | 1 | 463 | 0 | 0 | 0 | 0 | 464 |
| SAPPHIRe: | |||||||
| Stanford, CA | 0 | 0 | 0 | 55 | 16 | 0 | 71 |
| Honolulu | 0 | 0 | 0 | 25 | 144 | 1 | 170 |
| Taiwan/TSGHa | 0 | 0 | 0 | 79 | 0 | 0 | 79 |
| Taiwan/NTUb | 0 | 0 | 0 | 59 | 0 | 0 | 59 |
| Taiwan/VGHc | 0 | 0 | 0 | 189 | 0 | 0 | 189 |
| Total | 1,349 | 1,308 | 412 | 407 | 160 | 12 | 3,648 |
TSGH = Tri-Services General Hospital.
NTU = National Taiwan University.
VGH = Veterans General Hospital.
References
- Bamshad MJ, Wooding S, Watkins WS, Ostler CT, Batzer MA, Jorde LB (2003) Human population genetic structure and inference of group membership. Am J Hum Genet 72:578–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowcock AM, Ruiz-Linares A, Romfohrde J, Minch E, Kidd JR, Cavalli-Sforza LL (1994) High resolution of human evolutionary trees with polymorphic microsatellites. Nature 368:455–457 10.1038/368455a0 [DOI] [PubMed] [Google Scholar]
- Burchard EG, Ziv E, Coyle N, Gomez SL, Tang H, Karter AJ, Mountain JL, Perez-Stable EJ, Sheppard D, Risch N (2003) The importance of race and ethnic background in biomedical research and clinical practice. N Engl J Med 348:1170–1175 10.1056/NEJMsb025007 [DOI] [PubMed] [Google Scholar]
- Calafell F, Shuster A, Speed WC, Kidd JR, Kidd KK (1998) Short tandem repeat polymorphism evolution in humans. Eur J Hum Genet 6:38–49 10.1038/sj.ejhg.5200151 [DOI] [PubMed] [Google Scholar]
- Cooper RS, Kaufman JS, Ward R (2003) Race and genomics. New Engl J Med 348:1166–1170 10.1056/NEJMsb022863 [DOI] [PubMed] [Google Scholar]
- FBPP Investigators (2002) Multi-center genetic study of hypertension: The Family Blood Pressure Program (FBPP). Hypertension 39:3–9 10.1161/hy1201.100415 [DOI] [PubMed] [Google Scholar]
- Haga SB, Venter JC (2003) Genetics: FDA races in wrong direction. Science 301:466 10.1126/science.1087004 [DOI] [PubMed] [Google Scholar]
- Hanis CL, Hewett-Emmett D, Bertin TK, Schull WJ (1991) Origins of U.S. Hispanics: implications for diabetes. Diabetes Care 14:618–627 [DOI] [PubMed] [Google Scholar]
- Lander ES, Schork NJ (1994) Genetic dissection of complex traits. Science 265:2037–2048 [DOI] [PubMed] [Google Scholar]
- Lewontin RC (1972) The apportionment of human diversity. Evol Biol 6:381–398 [Google Scholar]
- Mardia KV, Kent JT, Bibby SM (1980) Multivariate analysis. Academic Press, London [Google Scholar]
- Mountain JL, Cavalli-Sforza LL (1997) Multilocus genotypes, a tree of individuals, and human evolutionary history. Am J Hum Genet 61:705–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parra EJ, Marcini A, Akey J, Martinson J, Batzer MA, Cooper R, Forrester T, Allison DB, Deka R, Ferrell RE, Shriver MD (1998) Estimating African American admixture proportions by use of population-specific alleles. Am J Hum Genet 63:1839–1851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds J, Weir BS, Cockerham CC (1983) Estimation of the coancestry coefficient: basis for a short-term genetic distance. Genetics 105:767–779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risch N, Burchard E, Ziv E, Tang H (2002) Categorization of humans in biomedical research: genes, race and disease. Genome Biol 3:comment2007.1–2007.12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romualdi C, Balding D, Nasidze IS, Risch G, Robichaux M, Sherry ST, Stoneking M, Batzer MA, Barbujani G (2002) Patterns of human diversity, within and among continents, inferred from biallelic DNA polymorphisms. Genome Res 12:602–612 10.1101/gr.214902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg NA, Pritchard JK, Weber JL, Cann HM, Kidd KK, Zhivotovsky LA, Feldman MW (2002) Genetic structure of human populations. Science 298:2381–2385 10.1126/science.1078311 [DOI] [PubMed] [Google Scholar]
- Stephens JC, Schneider JA, Tanguay DA, Choi J, Acharya T, Stanley SE, Jiang R, et al (2001) Haplotype variation and linkage disequilibrium in 313 human genes. Science 293:489–493 10.1126/science.1059431 [DOI] [PubMed] [Google Scholar]
- Thomas DC, Witte JS (2002) Point: population stratification: a problem for case-control studies of candidate-gene associations? Cancer Epidemiol Biomarkers Prev 11:505–512 [PubMed] [Google Scholar]
- Wacholder S, Rothman N, Caporaso N (2002) Counterpoint: bias from population stratification is not a major threat to the validity of conclusions from epidemiological studies of common polymorphisms and cancer. Cancer Epidemiol Biomarkers Prev 11:513–520 [PubMed] [Google Scholar]
- Wilson JF, Weale ME, Smith AC, Gratrix F, Fletcher B, Thomas MG, Bradman N, Goldstein DB (2001) Population genetic structure of variable drug response. Nat Genet 29:265–269 10.1038/ng761 [DOI] [PubMed] [Google Scholar]