Abstract
Human genetic linkage maps are based on rates of recombination across the genome. These rates in humans vary by the sex of the parent from whom alleles are inherited, by chromosomal position, and by genomic features, such as GC content and repeat density. We have examined—for the first time, to our knowledge—racial/ethnic differences in genetic maps of humans. We constructed genetic maps based on 353 microsatellite markers in four racial/ethnic groups: whites, African Americans, Mexican Americans, and East Asians (Chinese and Japanese). These maps were generated using 9,291 subjects from 2,900 nuclear families who participated in the National Heart, Lung, and Blood Institute–funded Family Blood Pressure Program, the largest sample used for map construction to date. Although the maps for the different groups are generally similar, we did find regional and genomewide differences across ethnic groups, including a longer genomewide map for African Americans than for other populations. Some of this variation was explained by genotyping artifacts—namely, null alleles (i.e., alleles with null phenotypes) at a number of loci—and by ethnic differences in null-allele frequencies. In particular, null alleles appear to be the likely explanation for the excess map length in African Americans. We also found that nonrandom missing data biases map results. However, we found regions on chromosome 8p and telomeric segments with significant ethnic differences and a suggestive interval on chromosome 12q that were not due to genotype artifacts. The difference on chromosome 8p is likely due to a polymorphic inversion in the region. The results of our investigation have implications for inferences of possible genetic influences on human recombination as well as for future linkage studies, especially those involving populations of nonwhite ethnicity.
Introduction
Genetic linkage maps describe the relative locations of genetic markers on chromosomes. Distances between genetic markers are determined by measuring the frequency of meiotic recombination between markers. Genetic linkage maps can be used to identify the location of genes responsible for traits and diseases. Human genetic linkage maps are important for two reasons. First, genetic linkage maps can be used as a tool in linkage analysis, association studies, and the building of physical maps. The first constructed maps of the human genome were genetic linkage maps, built by measuring the recombination rates between genetic markers, which usually were blood groups and serum proteins. Second, genetic linkage maps can be used to study rates and patterns of recombination across the genome.
Variation exists in human genetic map length. Rates of recombination vary by chromosome position, GC content, and the density of selected repeat units (Yu et al. 2001). Genetic linkage maps based on maternal inheritance are, on average, >50% longer than maps based on paternal inheritance. This difference is likely due to the different underlying biological processes of male and female meiotic recombination.
Some differences in recombination rates have been shown to be under genetic control. In humans, variation in maternal recombination rates has been shown to be specific to individuals and is not explained by maternal age (Broman et al. 1998). Significant variation between individuals has been noted for human spermatocytes (Cullen et al. 2002; Lynn et al. 2002) and oocytes (Tease et al. 2002). Rates of recombination have also been shown to be subject to genetic control in other organisms (Page and Hawley 2003). Specifically, the existence of both genomewide control (Catcheside 1977) and chromosome-wide control (Hillers and Villeneuve 2003) has been demonstrated in other species. Variation in recombination rates across different strains of mice has been noted elsewhere (Koehler et al. 2002).
Variation in recombination rates (and, therefore, in genetic map length) across human ethnic groups has not been studied. The most recent and extensive genomewide human genetic linkage maps (Broman et al. 1998; Kong et al. 2002) have been constructed using either entirely white samples or combined information from individuals of various ethnicities (Matise et al. 2003), making ethnic comparisons impossible. Here, we made such comparisons on the basis of large samples from four major racial/ethnic groups: whites, African Americans, Mexican Americans (Hispanics), and East Asians (Chinese and Japanese).
Material and Methods
Family Blood Pressure Program (FBPP) Data Set
The FBPP consists of four component networks: GenNet, GENOA, HyperGEN, and SAPPHIRe. Recruitment strategies for each network have been described elsewhere (FBPP Investigators 2002). The genetic maps were constructed using the pooled data version 3.15 from the FBPP. A total of 353 microsatellite markers were selected that had been genotyped in all four racial/ethnic groups. A total of 9,291 subjects from families with two or more children were included in the construction of the maps (table A1 [online only]). The total number of children examined for the combined sample of all four ethnicities was 8,428—including 3,301 in the white sample, 1,564 in the African American sample, 1,610 in the Hispanic sample, and 1,953 in the Asian sample (table A2 [online only]. A total of 863 parents were available, including 220 in the African American sample, 408 in the white sample, and 235 in the Asian sample. There were no parents available for the Hispanic sample. The FBPP data set contains nuclear families with primarily full sibships, as well as some half siblings. The families with half sibs were split into independent full sibships. Whereas a parent can be a member of more than one nuclear family subdivision, children appear only once in the final data set, and thus all families provide independent information.
Genetic Markers
Maps were constructed using 353 autosomal microsatellite markers from Marshfield screening set 8. All genotyping was performed by the Mammalian Genotyping Service of the Marshfield Center for Medical Genetics (see Marshfield Web site).
Statistical Methods
Genetic maps were constructed using the ASPEX package program sib_map (see ASPEX Web site). The sib_map program generates two-point and multipoint maximum-likelihood estimates of map distances between markers, on the basis of data from nuclear families. The multipoint maximization algorithm determines the complete set of distances that gives the maximum likelihood globally for the marker data across all markers on a given chromosome. The two-point algorithm considers each pair of adjacent markers separately, ignoring information from more-distant markers. In a second mode of operation, do_shuffle, the sib_map program calculates three-point distances for one marker against all other pairs of adjacent markers along a map. This method can be used to verify map orders or to position new markers on an already determined map.
The order of the microsatellite markers has been established elsewhere and is available at the Marshfield Web site. The first step in the construction of the FBPP genetic maps was to verify the previously established order for whites by use of the do_shuffle function of sib_map. A typographical mistake in the Marshfield map was identified, and its correction placed marker GATA7G07 on chromosome 6 rather than chromosome 8. No other inconsistencies were noted. Sex-averaged and sex-specific genetic intermarker distances were estimated using the Kosambi map function of sib_map. Maps were constructed for each of the four racial/ethnic groups.
Genotyping Errors
An earlier report describing the Marshfield map (Broman et al. 1998) determined that errors in genotyping that lead to misinheritance can dramatically inflate genetic map distances, by as much as 25% in that reported case. Misinheritances were eliminated during several rounds of data cleaning of the FBPP microsatellite data. Genotype errors can persist even after the elimination of inheritance errors. Because of observed racial/ethnic differences in genetic maps in our preliminary analyses, we decided to examine more carefully the genotype data for systematic problems. We tested for deviation from Hardy-Weinberg expectations that results from the presence of null alleles (i.e., alleles with null phenotypes). Null alleles leave two marks on genotype data: apparent excess homozygosity and an increased proportion of null phenotypes (subjects having no value for a particular marker genotype). We estimated the frequency of null alleles for each marker and each race/ethnicity by use of maximum-likelihood analysis. We tested all markers for each racial/ethnic group by use of the Null Allele Test (NAT), which tests whether the frequency of null alleles is different from 0 at a given marker, by use of a likelihood-ratio test. The test is one sided because we constrained the frequency of null alleles at >0; the statistic is then distributed as a 50:50 mixture of a χ2 distribution with 1 df and a point mass at 0.
Allele frequencies were calculated using (independent) subjects from each ethnic group. Frequencies were calculated separately for the Japanese and Chinese groups, and the resulting null-allele frequencies for the two groups were found to be quite similar. The total number of individuals in each group was 1,818 African Americans, 1,657 whites, 416 Hispanics, 162 Japanese, and 409 Chinese.
Nonrandom Missing Data
In the construction of the genetic maps, it was also determined that nonrandom missing genotype data inflated intermarker distances. In particular, subjects at one site—Jackson, MS (1,725 subjects)—had a greater proportion of missing genotypes than the other groups. For this reason, we dropped this site from the map construction, and subjects from this site are not included in the sample size counts, although they are included in the genotype error calculations. Although the maximum-likelihood algorithm that was used to determine map lengths provides unbiased results in the presence of random missing data, results are not necessarily unbiased in the presence of nonrandom missing data (Little and Rubin 2002). We add nonrandom missing data to the list of caveats to be considered in constructing genetic maps.
Race/Ethnicity Comparisons
ASPEX provides marker-allele frequency estimates from the individuals in the sample analyzed. Allele frequencies of microsatellite markers vary by race. For the purpose of comparison of ethnic-specific maps, a combined map for a pair of ethnicities was constructed to allow for different allele frequencies for each racial/ethnic group in the analysis, by use of a modification of the sib_map program. ASPEX provides a LOD score for each marker interval, comparing the likelihood of the estimated interval length with the likelihood for the case in which the two markers are assumed to be unlinked. By taking the LOD score for the same marker interval of two racial/ethnic group maps and the LOD score for the combined map, we were able to perform a likelihood-ratio test on each marker interval. Since the result of each likelihood-ratio test is distributed as a χ2 with 1 df, we converted the result to a normally distributed Z score by taking the square root and assigning either a positive or negative sign (+ or −) on the basis of which of the two map intervals was larger. The order of ethnicities used for this calculation was African American, white, Hispanic, and Asian, so that all intervals for which the African American map was longer were given a “+” sign, and all intervals for which the Asian map was longer were given a “−” sign. We used the derived Z scores to determine the significance of the differences in map length between racial/ethnic groups.
Z scores for the difference in total genetic map length between groups were calculated by summing the Z scores of each of the 331 individual map intervals and dividing the sum by the square root of 331. The same procedure was employed to calculate Z scores for individual chromosome arms, as well as centromeric and telomeric Z scores. Centromeric intervals were defined as intervals that span the known location of the centromere. Telomeric intervals were defined as the intervals that are covered by the two most telomeric markers on a chromosome arm. Microsatellite markers do not exist in the telomere tandem repeats, and so our markers cover regions that are telomeric but are not at the physical end points of the chromosomes. The physical distance from the midpoint of the two telomeric markers to the physical end of the chromosome ranged from 1.8 Mb to 17.9 Mb, with an average distance of 6.2 Mb (table A3 [online only]).
Results
Total Map Lengths
The total genetic map lengths for each chromosome and for the entire genome were calculated for sex-averaged, maternal, and paternal maps (table 1). The total African American map is 1%–2.5% longer than the maps of the other groups. The difference between the African American and Hispanic sex-averaged maps was nominally significant, although not when the results of multiple testing (which allowed for six comparisons) were considered.
Table 1.
Paternal, Maternal, and Sex-Averaged Map Lengths
|
Map Length (cM) for Group |
||||||||||
| African American |
White |
Asian |
||||||||
| Chromosome | Paternal | Maternal | Averaged | Paternal | Maternal | Averaged | Paternal | Maternal | Averaged | HispanicAveraged |
| 1 | 209 | 359 | 277 | 209 | 356 | 276 | 213 | 344 | 273 | 272 |
| 2 | 216 | 345 | 274 | 213 | 354 | 276 | 213 | 334 | 267 | 277 |
| 3 | 165 | 281 | 218 | 158 | 281 | 212 | 160 | 271 | 210 | 217 |
| 4 | 146 | 271 | 201 | 154 | 263 | 203 | 157 | 277 | 207 | 202 |
| 5 | 158 | 263 | 203 | 154 | 270 | 205 | 153 | 259 | 199 | 198 |
| 6 | 135 | 245 | 185 | 126 | 235 | 176 | 135 | 259 | 191 | 179 |
| 7 | 141 | 237 | 186 | 135 | 237 | 182 | 140 | 227 | 180 | 190 |
| 8 | 133 | 219 | 170 | 123 | 220 | 167 | 125 | 213 | 164 | 170 |
| 9 | 118 | 230 | 161 | 118 | 190 | 150 | 121 | 207 | 158 | 158 |
| 10 | 135 | 234 | 178 | 141 | 233 | 180 | 135 | 220 | 172 | 173 |
| 11 | 137 | 209 | 160 | 131 | 201 | 157 | 129 | 209 | 155 | 159 |
| 12 | 133 | 217 | 171 | 127 | 209 | 162 | 123 | 205 | 160 | 165 |
| 13 | 101 | 138 | 117 | 105 | 154 | 127 | 108 | 148 | 126 | 123 |
| 14 | 104 | 126 | 114 | 94 | 123 | 107 | 90 | 137 | 111 | 111 |
| 15 | 107 | 154 | 123 | 110 | 128 | 115 | 106 | 140 | 119 | 117 |
| 16 | 100 | 159 | 125 | 84 | 163 | 120 | 87 | 154 | 117 | 123 |
| 17 | 129 | 182 | 149 | 125 | 169 | 141 | 125 | 174 | 142 | 144 |
| 18 | 89 | 144 | 113 | 82 | 149 | 112 | 88 | 146 | 114 | 113 |
| 19 | 94 | 133 | 108 | 90 | 130 | 103 | 93 | 136 | 107 | 103 |
| 20 | 79 | 131 | 99 | 85 | 125 | 101 | 84 | 126 | 101 | 101 |
| 21 | 53 | 85 | 67 | 59 | 74 | 65 | 62 | 67 | 62 | 63 |
| 22 | 31 | 53 | 41 | 34 | 57 | 44 | 37 | 53 | 45 | 45 |
| Total | 2,711 | 4,415 | 3,440 | 2,654 | 4,320 | 3,379 | 2,685 | 4,306 | 3,378 | 3,398 |
Individual Map Intervals
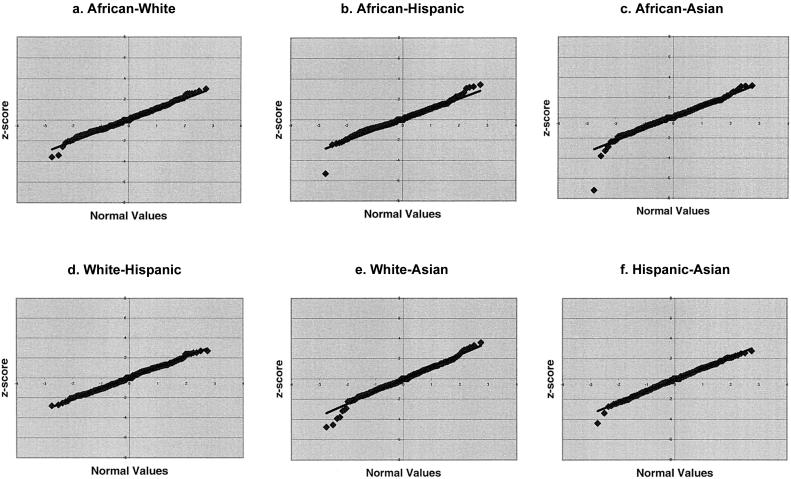
To determine whether the Z scores for individual marker-interval-length differences fit the normal distribution as expected, we created quantile-quantile (Q-Q) plots of the Z scores for individual marker-interval comparisons for each pair of racial/ethnic groups (fig. 1a–1f). Q-Q plots compare an observed distribution with an expected distribution. The expected distribution of Z scores is the normal distribution, and so deviations from the y=x line indicate deviation of the observed score from the expected distribution. Plots involving the African American sex-averaged map (fig. 1a–1c) fell largely above the y=x line, consistent with the overall greater average interval length of the African American maps. The white versus Hispanic plot (fig. 1d) shows data points very close to the y=x line, indicating little difference in map interval lengths for these two maps. We expect, a priori, to find the smallest difference between the white and Hispanic groups on the basis of genetic distances, since Hispanics have ∼60% white ancestry (Tang et al. 2005 [in this issue]). Plots involving the Asian map (fig. 1c, 1e, and 1f) show the largest number of outliers.
Figure 1.
Q-Q plots of Z scores for individual interval-length differences between racial/ethnic groups. a, African Americans versus whites. b, African Americans versus Hispanics. c, African Americans versus Asians. d, Whites versus Hispanics. e, Whites versus Asians. f, Hispanics versus Asians.
Outliers
To examine the distribution of outliers in our sample, we calculated the number of Z scores, for each comparison group, that exceeded two cutoffs (table 2). The cutoffs chosen were 1.96 (i.e., nominal significance) and 3.48 (i.e., 1 expected false positive in 1,986 tests). The total (+ and −) number of Z scores for each comparison group exceeded the expected number for all groups, except for the white-Hispanic comparison.
Table 2.
Distribution of Nominally Significant Z Scores, by Comparison Group
|
No. of Significant Z Scores |
|||||||||
| For Comparison Group |
|||||||||
| Z ScoreCutoff | Expectedper Group | AfricanAmerican–White | AfricanAmerican–Hispanic | AfricanAmerican–Asian | White-Hispanic | White-Asian | Hispanic-Asian | Total | TotalExpected |
| +1.96 | 8.28 | 13 | 13 | 13 | 9 | 11 | 12 | 71 | 49.65 |
| +3.48 | .08 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | .50 |
| −1.96 | 8.28 | 6 | 6 | 8 | 8 | 14 | 17 | 59 | 49.65 |
| −3.48 | .08 | 1 | 1 | 2 | 0 | 4 | 1 | 9 | .50 |
| Total: | |||||||||
| 1.96 | 16.55 | 19 | 19 | 21 | 17 | 25 | 29 | 130 | 99.30 |
| 3.48 | .17 | 1 | 1 | 2 | 0 | 5 | 1 | 10 | 1.00 |
In an effort to identify regions of the genome in which significant Z scores clustered, we identified all marker intervals that had one or more Z scores >3.48 (table 3). We found three regions, on chromosomes 6p, 8p, and 12q.
Table 3.
Regions with Significant Differences in Interval Length for Comparison Groups
|
Z Score for Difference in Interval Length for Comparison Group |
||||||
| Chromosomeand Marker Interval | African American–White | African American–Hispanic | African American–Asian | White-Hispanic | White-Asian | Hispanic-Asian |
| 6: | ||||||
| ATA50C05–GATA29A01 | 2.56 | 1.55 | −.68 | −1.01 | −3.78 | −2.49 |
| GATA29A01–GATA163B10 | 1.99 | .30 | −.71 | −1.94 | −3.06 | −1.17 |
| GATA163B10–GGAA15B08 | 3.02 | .43 | −.64 | −2.80 | −3.91 | −1.15 |
| GGAA15B08–GGAT3H10 | .21 | .21 | −3.78 | .00 | −4.53 | −4.38 |
| 8: | ||||||
| AFM143xd8–AFM198wd2 | 2.56 | .91 | 3.19 | −1.78 | .83 | 2.51 |
| AFM198wd2–GATA25C10 | −3.57 | −5.29 | −7.18 | −2.40 | −4.75 | −2.06 |
| GATA25C10–GATA23D06 | 2.47 | 3.18 | 3.12 | 1.30 | .96 | .43 |
| 12: | ||||||
| GATA4H01–GATA32F05 | −.37 | −.21 | 2.62 | .64 | 3.60 | 2.56 |
In three of four adjoining intervals on 6p, the Asian map is significantly longer than the white map, and, in one of those intervals, the Asian map is significantly longer than all other maps. This difference is apparent in the sex-specific maps as well. The Asian maternal map is longer than both the African American and white maternal maps, whereas the white paternal map appears to be smaller than both the African American and Asian paternal maps (table 4).
Table 4.
Interval Length for Markers on Chromosomes 6p, 8p, and 12q
|
Interval Length (in cM)for Group |
||||
| Chromosome, Map Type,and Marker Interval | AfricanAmerican | White | Hispanic | Asian |
| 6p: | ||||
| Sex-averaged: | ||||
| ATA50C05–GATA29A01 | 12.2 | 8.9 | 10.2 | 12.3 |
| GATA29A01–GATA163B10 | 9.7 | 8.3 | 9.6 | 10.9 |
| GATA163B10–GGAA15B08 | 13.4 | 10.5 | 12.3 | 13.4 |
| GGAA15B08–GGAT3H10 | 4.6 | 4.5 | 3.8 | 7.7 |
| Total | 39.9 | 32.2 | 35.9 | 44.3 |
| Maternal: | ||||
| ATA50C05–GATA29A01 | 17.6 | 13.6 | … | 17.9 |
| GATA29A01–GATA163B10 | 11.2 | 14.6 | … | 15.7 |
| GATA163B10–GGAA15B08 | 17.9 | 15.3 | … | 21.2 |
| GGAA15B08–GGAT3H10 | 6.5 | 5.9 | … | 9.2 |
| Total | 53.2 | 49.4 | … | 64 |
| Paternal: | ||||
| ATA50C05–GATA29A01 | 7.8 | 4.9 | … | 8.1 |
| GATA29A01–GATA163B10 | 8.2 | 3.1 | … | 6.9 |
| GATA163B10–GGAA15B08 | 9.8 | 6.6 | … | 7.6 |
| GGAA15B08–GGAT3H10 | 2.8 | 3.2 | … | 6.3 |
| Total | 28.6 | 17.8 | … | 28.9 |
| 8p: | ||||
| Sex-averaged: | ||||
| AFM143xd8–AFM198wd2 | 17.1 | 12.2 | 14.7 | 11.2 |
| AFM198wd2–GATA25C10 | 3.1 | 5.5 | 8.4 | 10.4 |
| GATA25C10–GATA23D06 | 7.6 | 4 | 3.3 | 3.2 |
| Total | 27.8 | 21.7 | 26.4 | 24.8 |
| Maternal: | ||||
| AFM143xd8–AFM198wd2 | 9.3 | 7.6 | … | 7.4 |
| AFM198wd2–GATA25C10 | 5.3 | 7.9 | … | 12.1 |
| GATA25C10–GATA23D06 | 10.3 | 4.8 | … | 4.3 |
| Total | 24.9 | 20.3 | … | 23.8 |
| Paternal: | ||||
| AFM143xd8–AFM198wd2 | 27.9 | 17 | … | 15.6 |
| AFM198wd2–GATA25C10 | .4 | 3.3 | … | 8.5 |
| GATA25C10–GATA23D06 | 5 | 3.3 | … | 2.1 |
| Total | 33.3 | 23.6 | … | 26.2 |
| 12q: | ||||
| Sex-averaged: | ||||
| GATA4H01–GATA32F05 | 12.9 | 14.1 | 13.5 | 9.8 |
| Maternal: | ||||
| GATA4H01–GATA32F05 | 13.8 | 12.8 | … | 8.1 |
| Paternal: | ||||
| GATA4H01–GATA32F05 | 12 | 15.2 | … | 12.1 |
One interval on chromosome 8p has the largest Z scores of any interval comparison. Interestingly, the neighboring intervals on each side of the significant interval have high Z scores in the opposite direction. Examining the interval lengths, we see that the African American map, which has the shortest interval length for the middle markers, has the longest interval lengths for the neighboring markers (table 4). The opposite is true for the Asian map.
The last significant interval-length difference occurs on chromosome 12q. The white map has the longest map length, whereas the Asian map has the shortest (table 4). The adjoining intervals are not significantly different. It is clear that the statistical significance is a result of the sex-averaged map in Asians being shorter than that in the other three groups; this observation is also reproduced in the maternal but not the paternal maps.
Having identified marker intervals with significant differences in map length, we sought to examine chromosomal regions to see if any regional differences in map length exist. We calculated Z scores for the arms of each chromosome (39 arms in all) and identified several highly significant differences in genetic map length. The p arm of chromosome 6 gave the most significant results, including a Z score of −6.40 in the white-Asian comparison group and significant Z scores in other comparison groups. As described above, chromosome 6p contains several marker intervals with high Z scores, particularly those in the white-Asian comparison group. When considered together, the marker-interval-length difference is highly significant.
Centromere and Telomere Effects
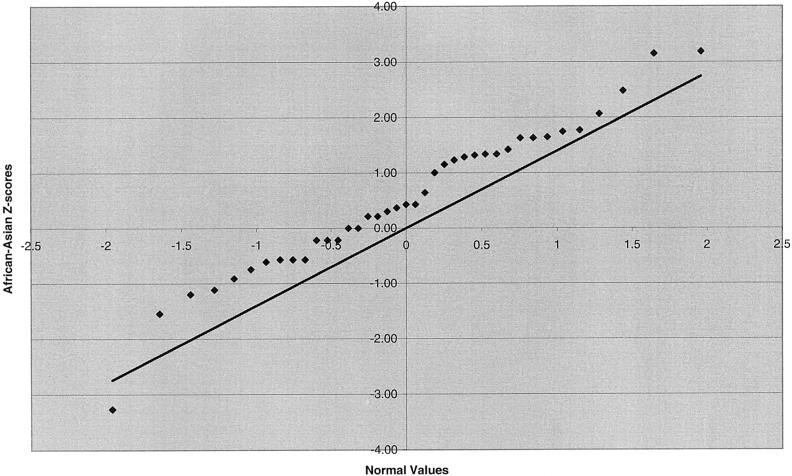
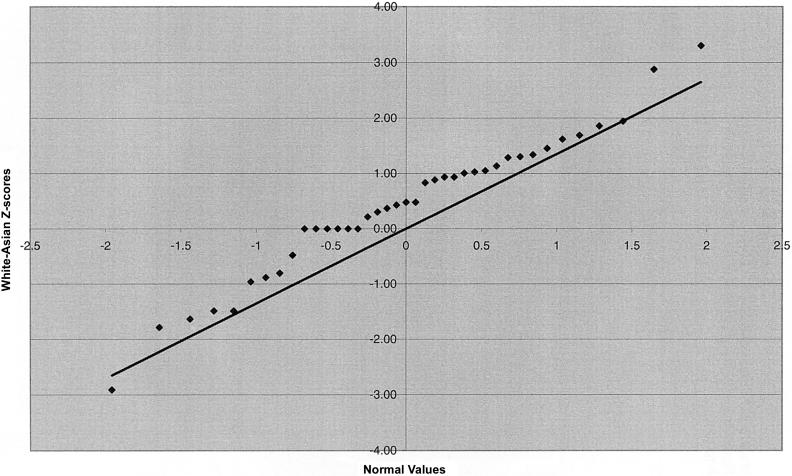
We also examined marker intervals that straddle the centromere and intervals that are most telomeric (table 5). No significant Z scores were identified in the centromeric interval comparisons, nor were the centromeric interval lengths especially disparate. A significant difference in telomeric length was observed for the African American–Asian and white-Asian comparison groups as a result of shorter interval lengths in the Asian group, with length differences between the Asian and the other groups in the range of 4%–11%. These differences are not due to outliers; instead, they appear to be due to a universal phenomenon involving all telomeric intervals (fig. 2a and 2b). A regression of telomeric interval length on the distance of the midpoint of the two telomeric markers from the physical end of the chromosome was significant for each of the four ethnic groups, with interval length increasing with distance from the telomere. A regression of the differences in telomeric length between ethnic groups on distance from the physical end of the chromosome was not significant.
Table 5.
Centromere and Telomere Comparisons
|
Z Scores for Comparison Group |
||||||
| Region | African American–White | African American–Hispanic | African American–Asian | White-Hispanic | White-Asian | Hispanic-Asian |
| Centromere | .97 | .92 | .39 | .22 | −.54 | −.55 |
| Telomere | 1.17 | .98 | 3.25 | −.12 | 2.62 | 1.59 |
Figure 2.
Q-Q plots of Z scores for telomeric interval-length differences. a, African Americans versus Asians. b, Whites versus Asians.
Marker Heterozygosity
Heterozygosity of microsatellite markers varies by race/ethnicity (Calafell et al. 1998). We calculated the average heterozygosity of all microsatellite markers for each racial/ethnic group. The African American group had the highest heterozygosity (79.1%), followed by whites (76.4%), Hispanics (75.5%), and Asians (73.1%). Because average heterozygosity differs between these racial/ethnic groups, we examined whether the average heterozygosity difference between two groups for a given pair of markers affected the estimated interval-length difference for that pair of markers. The correlation of average heterozygosity difference and interval-length difference was very low in our sample (median absolute value, 0.03), and none of the differences were statistically significant.
Null Alleles
We next sought to determine the extent to which genotyping artifacts could explain the differences we observed. Several causes for the appearance of genotype errors have been described, including the simple misreading of allele sizes, gene-conversion events, and mutations in lymphoblastoid cell lines. In fact, tetranucleotide repeat microsatellite markers have a particularly high mutation rate in lymphoblastoid cell lines—as high as 1% (Zahn and Kwiatkowski 1995). Tetranucleotide repeat markers are preferable to dinucleotide repeat markers, because their allele sizes are typically easier to score. The majority of microsatellite markers in Marshfield screening set 8 are tetranucleotide repeat markers.
Genotype error has been reported to inflate genetic distance estimates by as much as 25% (Broman et al. 1998). Most genotype errors due to misread alleles, gene conversion, and mutation in cell lines can be eliminated through data cleaning, which removes suspect genotypes due to misinheritance (nonpaternity or nonmaternity, >4 alleles at a given locus in a group of full siblings, etc.) and unlikely tight double recombinants. Because the distribution of microsatellite markers in our data set averages 10 cM between markers, we have no tight double recombinants. The FBPP microsatellite genotype data had undergone several cleaning steps prior to the construction of the genetic maps. The cleaning process also identified individuals whose familial relationship was misassigned (half sibs identified as full sibs, MZ twins as DZ twins, etc.). These errors are designated “Mendelian errors.”
In addition to cleaning the data of Mendelian errors, it is also possible to identify potential problems, such as the presence of null alleles at microsatellite markers, by testing for deviations from Hardy-Weinberg expectations. The FBPP data were not previously examined for these types of errors. Deviations from Hardy-Weinberg expectations can be examined by comparing the observed versus expected genotype frequencies, calculated from the allele frequencies for a given marker. When null alleles occur at a high rate, the data present two features. First, a subject with two null alleles will appear to have a missing genotype. Therefore, the first indication of null alleles is an excess of missing genotypes for a given marker. Second, if a subject has one copy of a null allele and one copy of a normal allele, the subject will appear to be homozygous for the visible allele. Thus, the second indication of null alleles is deviation from Hardy-Weinberg expectations, as manifest by an excess of homozygotes.
We examined the markers on chromosome 6 in the region of the significant deviations in map length for null alleles. By comparing the number of subjects missing genotypes at each marker with the number missing them at marker GGAT3H10 (chosen because, of the five markers in the region, it had the least number of subjects with missing genotypes in three of the four groups), we detected an excess of missing data for at least two markers, GATA29A01 and GGAA15B08, for all four groups. Marker GATA163B10 was also suggestive of excess missing data in Asians. We tested for and estimated the frequency of null alleles for each marker within each racial/ethnic group. Chinese and Japanese subjects were considered separately. We tested whether the estimate of the null-allele frequency was significantly different from zero, using the NAT (see the “Methods” section).
In the three regions with significant deviations in map length for racial/ethnic groups, we identified three markers with significant null-allele frequency estimates (table 6). On chromosome 6, the two markers that had an excess of individuals with missing data, GATA29A01 and GGAA15B08, also had null-allele frequency estimates for which the difference from zero was highly significant. The null-allele frequency estimates for both markers were highest in the Chinese and Japanese groups. The null-allele frequency estimates for the white group were the lowest of the groups, for both markers. On chromosome 8, the null-allele frequency estimate for marker AFM143xd8 was highly significant in the African American group but not in the other groups. No group had significant estimates for the other three markers. On chromosome 12, neither marker showed any evidence of the presence of null alleles.
Table 6.
NAT and Null-Allele Frequency Estimates for Chromosomes 6p, 8p, and 12q
|
Results of NAT (χ2) for Group |
Null-Allele Frequency Estimates for Group |
|||||||||
| Chromosomeand Marker | African American | White | Hispanic | Chinese | Japanese | African American | White | Hispanic | Chinese | Japanese |
| 6p: | ||||||||||
| ATA50C05 | 0 | .01 | 1.24 | .40 | .32 | 0 | .0025 | .0134 | .0062 | .0089 |
| GATA29A01 | 42.60 | 3.03 | 22.20 | 130.72 | 50.64 | .1278 | .0173 | .0845 | .2729 | .2871 |
| GATA163B10 | 0 | .29 | .08 | 0 | 0 | 0 | .0053 | .0021 | 0 | 0 |
| GGAA15B08 | 38.14 | 5.23 | 36.87 | 89.92 | 29.87 | .0649 | .0308 | .0778 | .1663 | .1299 |
| GGAT3H10 | 0 | 0 | 0 | 0 | .40 | 0 | 0 | 0 | 0 | .0137 |
| 8p: | ||||||||||
| AFM143xd8 | 68.10 | 1.93 | 0 | 0 | 0 | .0994 | .0149 | 0 | 0 | 0 |
| AFM198wd2 | 0 | 0 | 0 | .17 | 0 | 0 | 0 | 0 | .0045 | 0 |
| GATA25C10 | 0 | 0 | .85 | 0 | 0 | 0 | 0 | .0105 | 0 | 0 |
| GATA23D06 | .01 | 1.51 | 0 | 0 | .35 | .0018 | .0134 | 0 | 0 | .0153 |
| 12q: | ||||||||||
| GATA4H01 | .55 | 0 | 0 | 0 | .31 | .0089 | .0007 | 0 | 0 | .0162 |
| GATA32F05 | 0 | 0 | 1.40 | 1.42 | .01 | 0 | 0 | .0153 | .0093 | .0030 |
In an effort to determine whether the markers with significant levels of null alleles affected the estimates of genetic map length, we constructed maps without the markers GATA29A01 and GGAA15B08 on chromosome 6 (table 7). The map-length estimates for the region decreased across all groups but decreased the most in the Asian group. Map lengths appeared to be quite similar across groups, once the markers with null alleles were removed.
Table 7.
Chromosome 6p Length With and Without Two Markers with Null Alleles
|
Chromosome 6p Length (cM) for Group |
||||
| Map | African American | White | Hispanic | Asian |
| With markers | 39.9 | 32.2 | 35.9 | 44.3 |
| Without markers | 34.5 | 29.6 | 31.9 | 32.6 |
The significance of the map-length differences was based on two-point analysis. Because the two markers on chromosome 8p, AFM198wd2 and GATA25C10, and the two markers on chromosome 12q, GATA4H01 and GATA32F05, that showed significant map-length differences between racial/ethnic groups were unaffected by null alleles, the significant differences seen for these intervals remain unexplained.
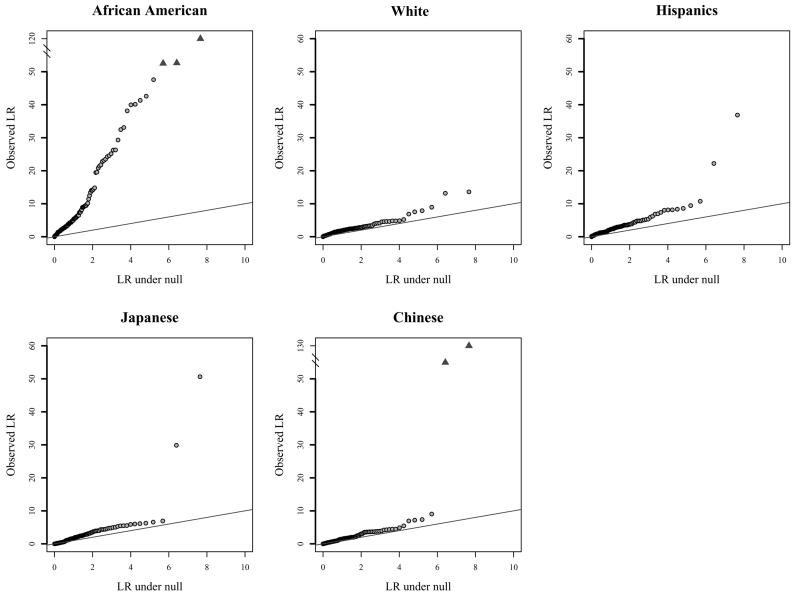
We also examined the number of significant NAT scores across racial/ethnic groups for all markers. Because sample sizes varied by racial/ethnic group, we calculated NAT results by selecting 400 independent subjects (if available) from each group, to compare the frequency of null alleles in these groups. The Japanese group did not have 400 independent subjects from which to sample for any marker, so we chose the maximum number of independent subjects available for each marker in that group. The African American group had, by far, the greatest number of significant NAT scores of any group (fig. 3). This is consistent with the greater genetic diversity in African Americans. The large excess of markers with null alleles in African Americans also provides an explanation for the greater overall map length observed in this population. The presence of null alleles in the other racial/ethnic groups was largely confined to a few markers.
Figure 3.
Distribution of observed versus expected results of null-allele frequency tests in African Americans (a), whites (b), Hispanics (c), Japanese (d), and Chinese (e)
With regard to the results for telomeric intervals, it is difficult to evaluate the true degree of difference between the African American and Asian interval lengths because of the high frequency of null alleles in the African American group. However, for whites, there is only one telomeric interval containing a marker with a significant NAT result and, for Asians, there are none. The difference between white and Asian intervals is significant and uninfluenced by null alleles, and the effect appears to be greater in the maternal map (mean difference, 0.82 cM) than in the paternal map (mean difference, 0.43 cM), despite the longer telomeric map lengths in males.
Discussion
Although the two most recent human genetic maps—the Marshfield map (Broman et al. 1998) and the deCODE map (Kong et al. 2002)—have included a large number of microsatellite markers (8,325 for Marshfield and 5,136 for deCODE), the accuracy of their estimates of genetic distance has been limited by the number of meioses available for mapping (188 for Marshfield and 1,257 for deCODE). The FBPP human genetic maps were constructed with the largest number of subjects to date. In our case, however, map construction was not completed without some challenges. Because we had only limited genotype data on parents, certain types of genotyping error were difficult to identify. Our original maps were constructed with a subset of data that was missing a substantial amount of genotype data. We found that the resulting maps were biased (i.e., had inflated map lengths) compared with the results obtained when the subjects with missing data were excluded. Thus, we discovered that nonrandom missing data is another pitfall to be avoided in genetic map construction. In addition, because our analysis was based on a genome scan (albeit a large one), only a sparse map was generated, compared with the high-density maps reported elsewhere. Our maps should nonetheless be useful for analysis of other genome-scan data sets by use of the same microsatellite marker set, particularly for various ethnic groups.
In the process of generating microsatellite genotype data, many laboratories “multiplex” their samples—that is, multiple markers of different sizes are run on the same gel. The FBPP microsatellite genotype data were generated by the Marshfield Clinic Center for Medical Genetics. To multiplex on a large scale, each marker is read in a window determined through optimization. This optimization occurs by running large (mainly white) CEPH families for each marker and by determining the appropriate marker window in which alleles should be read. Usually, three to five markers are included in each multiplex (see PCR Protocol Web site). Although multiplexing cuts the cost of generating genotypes, problems can arise as a result of this method, specifically through the creation of null phenotypes for some alleles (“null alleles”). Null alleles can occur in two ways for microsatellite markers. First, a null allele can be generated as a result of narrow multiplexing windows or allele size ranges. When a window is established, alleles that fall outside the window are not read. These null alleles will be read as missing values. In the case of our microsatellite marker data, it is possible that multiplexing windows established on mainly white samples are too narrow to encompass all the allelic diversity present in nonwhite ethnic groups. Second, variation in the primer-binding sequence can lead to a failure of the allele to amplify. No allele can be read in this case, which leads to a missing value. This problem arises even when loci are not multiplexed. It is also consistent with the higher rate of null alleles we found in African Americans, since Africans are known to have greater levels of sequence variation and hence to have potential variation in the primer sequences. We do not know whether one or both of these phenomena are occurring in our samples. The failure of amplification because of variation in the primer-binding sequence, if common, will continue to be a problem in maps—of linkage or association—based on SNPs. We suggest that SNP markers should also be vetted for null alleles in multiple ethnically diverse populations as part of the optimization of SNP-based maps.
A third possibility is that some fragments are only weakly amplified and detected, leading to an excess of apparent homozygotes. This might lead to the preferential loss of larger alleles. We examined this possibility by determining whether there was a size bias toward excess homozygosity of shorter alleles among our markers. We detected no such size bias, so this explanation seems less likely.
Null alleles inflate map distance by increasing the perceived rate of recombination between markers. The presence of null alleles creates false homozygotes. The false homozygotes may lead to an incorrect inference with regard to allelic inheritance and may indicate that a recombination event has taken place between the marker with null alleles and neighboring markers when, in fact, no recombination event has occurred. The presence of null alleles will therefore result in increased map distance.
The potential risk in establishing multiplexing windows largely on the basis of one racial/ethnic group is clear. The generation of null alleles due to narrow multiplexing windows can be avoided by more-careful determination of the windows by use of multiracial samples. In addition, null alleles not due to multiplexing windows, such as those due to failure of PCR and so forth, can be detected by genotyping multiple samples from the same individual and by sequencing the primer-binding sequences for individuals who appear resistant to genotyping.
The implications for previous and future linkage studies are potentially important. The presence of null alleles can lead to false-negative results because of the inflation of distance estimates between the putative disease gene and the genetic marker being tested. Reexamination of previous analyses after the effect of null alleles is taken into account could lead to additional and more-accurate linkage findings. Future studies should test for null alleles, to avoid the loss of statistical power caused by the presence of null alleles.
Although a significant frequency of null alleles exists for markers in all the racial/ethnic groups, including whites, we have shown that the problem is likely to be greatest for Africans and African Americans. The African American group shows the greatest number of markers with null alleles, by use of current genotyping methods. Researchers conducting linkage studies with the use of African and African American samples should be particularly wary of the presence of null alleles.
Our detection of null alleles depended on two observations: an excess of missing genotype data and Hardy-Weinberg deviations of the data that remained. The two sources of information are independent and useful. The NAT is superior to a simple Hardy-Weinberg equilibrium test, in the case of microsatellite markers, because multiple allele sizes require a greater number of degrees of freedom for the Hardy-Weinberg test. The result is that a general Hardy-Weinberg test lacks power, in the case of microsatellite markers, especially relative to the NAT. However, the NAT is specific to detection of a general excess of homozygotes (e.g., as caused by the presence of null alleles) and so would not necessarily be powerful in other scenarios that cause Hardy-Weinberg deviations. We have made the R code for NAT freely available (see NAT Web site).
There remain statistically significant group differences on chromosomes 8 and 12 that cannot be explained by the presence of null alleles. A common large-inversion polymorphism exists on chromosome 8, in the same region as the significant difference we report (Giglio et al. 2001). The frequency of this inversion has been reported for whites (21%) (Broman et al. 2003) and Japanese subjects (27%) (Sugawara et al. 2003) but not for individuals of African or African American descent. These frequencies are based on a small number of subjects, and it is unclear which orientation of the inversion is specified in different studies. There are three ways in which an inversion can affect perceived recombination rates: changing marker order, suppressing recombination, and altering the distance between markers. In our case, it appears that only one marker, GATA25C10, is located within the inversion; therefore, marker order is not affected in our study.
The genetic map impact of a polymorphic inversion generally entails recombination suppression within the inverted interval, in individuals heterozygous for the inversion. Thus, the greatest decrease in recombination, or map length, occurs in populations with the greatest heterozygosity—namely, when the two types of chromosomes are of equal frequency. However, the precise effect for any pair of markers also depends on the locations of the markers involved. For example, if the two markers are within the inversion, only inversion heterozygosity influences the recombination fraction between them, and not their relative locations within the inversion. The same is true for two markers outside and flanking the inversion; in this case, the impact will be a function of how far the markers are from the proximal and distal boundaries, respectively, of the inversion. The situation with one marker inside and the other outside the inversion is more complicated. Here, the observed recombination fraction will also be influenced by the relative location of the internal marker to the boundaries of the inversion. For example, consider three markers, A, B, and C, where B is within the inversion, and A and C flank the inversion on either side. Depending on the relative frequency of the two chromosome types, if interval A–B appears shorter in population 1 than in population 2, then interval B–C will be longer in population 1 than in population 2. This is because, in addition to the overall reduction in the recombination fraction in heterozygotes, there is a difference in the distance from A to B and from B to C in the two types of homozygotes (in whom recombination does take place); these two homozygotes occur at different frequencies in different populations, depending on the population frequencies of the two chromosome types. Specifically, a population that has a higher frequency of homozygotes for the longer A–B interval will also have a higher frequency of homozygotes for the shorter B–C interval.
The data that we showed in table 4 for marker GATA25C10 (inside the inversion) and the two markers AFM198wd2 and GATA23D06 (which flank the inversion) appear consistent with this model. The interval from AFM198wd2 to GATA25C10 appears to be shortest in African Americans, intermediate in whites and Hispanics, and longest in Asians. The opposite pattern applies to the adjacent interval from GATA25C10 to GATA23D06, which appears to be longest in African Americans, intermediate in whites and Hispanics, and shortest in Asians. However, more data are needed to confirm this explanation, because, so far, the inversion frequencies in whites and Asians appear too similar (although they are based on small sample sizes) to cause these results, and there are no data on inversion frequencies in Africans or African Americans. Thus, further work is needed (particularly, the study of this inversion in African Americans) to determine whether it is the cause of the differences we see. We suggest genotyping additional markers in the regions on chromosome 8 and chromosome 12, in particular SNPs, to obtain a more detailed picture of the extent, validity, and potential cause of these differences.
Prior studies have shown individual variation in recombination rates. These observations have been restricted to female meiosis. Hence, one might infer that substantial differences would occur between racial/ethnic groups on this basis and that group differences would be more pronounced for the female maps than for the male maps. However, this was only partially the case. Overall, the group-specific maps were largely similar, with only a few differences, as we have described here. The findings on chromosome 8 were observed in both sexes, whereas those on chromosome 12 appeared specific to female meiosis. On the other hand, the findings regarding the telomeric regions were also more pronounced for female meiosis than for male meiosis. It is noteworthy that we were able to identify such differences even in the presence of potentially considerable individual variation within groups.
Perhaps our most intriguing observation was the racial/ethnic variation in the lengths of telomeric intervals, with Asians having significantly shorter intervals. This difference was uniform across all telomeric intervals and was not due to a small number of outliers; thus, it is unlikely to be a characteristic of the telomeric markers themselves and may be indicative of a biologically based mechanism. We advocate additional studies of the meiotic lengths of telomeric regions across racial/ethnic groups, by use of a variety of markers, to either confirm or refute this interesting observation.
Acknowledgments
This investigation was based on data from the Family Blood Pressure Program (FBPP), which is supported by the National Heart, Lung, and Blood Institute. All authors are members of the FBPP.
Appendix A: Supplemental Data
Table A1.
Subjects Included in the Map Construction (Families with Two or More Children)
| Race/Ethnicity | No. of Families | No. of Children | No. of Parents | No. of Subjects |
| African American | 674 | 1,564 | 220 | 1,784 |
| White | 1,242 | 3,301 | 408 | 3,709 |
| Hispanic | 208 | 1,610 | 0 | 1,610 |
| Asian | 576 | 1,953 | 235 | 2,188 |
| Total | 2,700 | 8,428 | 863 | 9,291 |
Table A2.
Distribution of Families, by Race/Ethnicity, Number of Children, and Parental Genotypes
|
No. of Families with Available Parental Genotypes for |
||||||
| Race/Ethnicity and No. of Children | No Parents | One Parent | One Father | One Mother | Two Parents | Total No. of Families |
| African American: | ||||||
| 2 | 377 | 129 | 27 | 102 | 14 | 520 |
| 3 | 73 | 21 | 3 | 18 | 10 | 104 |
| 4 | 26 | 10 | 1 | 9 | 4 | 40 |
| 5 | 6 | 2 | 0 | 2 | 0 | 8 |
| 6 | 0 | 2 | 1 | 1 | 0 | 2 |
| Total | 482 | 164 | 32 | 132 | 28 | 674 |
| White: | ||||||
| 2 | 579 | 123 | 39 | 84 | 67 | 769 |
| 3 | 206 | 59 | 25 | 34 | 20 | 285 |
| 4 | 84 | 15 | 4 | 11 | 8 | 107 |
| 5 | 35 | 7 | 4 | 3 | 3 | 45 |
| 6 | 14 | 2 | 1 | 1 | 2 | 18 |
| 7 | 8 | 1 | 0 | 1 | 0 | 9 |
| 8 | 3 | 1 | 0 | 1 | 0 | 4 |
| 9 | 2 | 0 | 0 | 0 | 0 | 2 |
| 10 | 2 | 0 | 0 | 0 | 0 | 2 |
| 11 | 0 | 0 | 0 | 0 | 0 | 0 |
| 12 | 0 | 0 | 0 | 0 | 0 | 0 |
| 13 | 0 | 0 | 0 | 0 | 0 | 0 |
| 14 | 1 | 0 | 0 | 0 | 0 | 1 |
| Total | 934 | 208 | 73 | 135 | 100 | 1,242 |
| Hispanic: | ||||||
| 2 | 100 | 0 | 0 | 0 | 0 | 100 |
| 3 | 100 | 0 | 0 | 0 | 0 | 100 |
| 4 | 81 | 0 | 0 | 0 | 0 | 81 |
| 5 | 48 | 0 | 0 | 0 | 0 | 48 |
| 6 | 36 | 0 | 0 | 0 | 0 | 36 |
| 7 | 25 | 0 | 0 | 0 | 0 | 25 |
| 8 | 9 | 0 | 0 | 0 | 0 | 9 |
| 9 | 7 | 0 | 0 | 0 | 0 | 7 |
| 10 | 2 | 0 | 0 | 0 | 0 | 2 |
| Total | 408 | 0 | 0 | 0 | 0 | 408 |
| Asian: | ||||||
| 2 | 106 | 45 | 16 | 29 | 14 | 165 |
| 3 | 122 | 57 | 10 | 47 | 13 | 192 |
| 4 | 75 | 32 | 7 | 25 | 8 | 115 |
| 5 | 43 | 14 | 1 | 13 | 4 | 61 |
| 6 | 22 | 3 | 2 | 1 | 1 | 26 |
| 7 | 10 | 2 | 0 | 2 | 0 | 12 |
| 8 | 1 | 2 | 0 | 2 | 0 | 3 |
| 9 | 2 | 0 | 0 | 0 | 0 | 2 |
| Total | 381 | 155 | 36 | 119 | 40 | 576 |
Table A3.
Telomeric Interval Lengths
|
Interval Length (cM) for Map |
|||||||||||
| African American |
White |
Asian |
|||||||||
| Chromosome and Marker Interval | Distance to Telomerea | Averaged | Maternal | Paternal | Averaged | Maternal | Paternal | Averaged | Maternal | Paternal | HispanicAveraged |
| 1: | |||||||||||
| AFM280we5–GGAA3A07 | 5.3 | 10.7 | 11.1 | 10.4 | 12.9 | 10.6 | 15.4 | 11.4 | 11.8 | 11 | 12 |
| GATA4A09–GATA50F11 | 10.6 | 4.7 | 4.5 | 4.7 | 4.3 | 2.8 | 5.9 | 4 | 3.4 | 4.5 | 5.1 |
| 2: | |||||||||||
| GATA165C07–GATA116B01 | 4.4 | 18.1 | 12 | 25.4 | 18 | 15 | 21.2 | 13.5 | 9.6 | 18.1 | 20 |
| GATA178G09–AFM112yd4 | 7.2 | 7.2 | 3.7 | 11.1 | 9.9 | 6 | 14.1 | 5.6 | .1 | 12.6 | 8.3 |
| 3: | |||||||||||
| AFM234tf4–GATA164B08 | 4.6 | 5.5 | 5.4 | 5.5 | 4.1 | 3.3 | 4.9 | 4 | 4.8 | 3.2 | 4.9 |
| ATA22E01–AFM254ve1 | 3.2 | 12.7 | 9.1 | 16.9 | 12.5 | 6.6 | 19.6 | 12.4 | 6.1 | 18.9 | 11 |
| 4: | |||||||||||
| GATA22G05–AFM157xg3 | 9.9 | 12.9 | 18.4 | 8 | 16 | 22 | 11 | 14.6 | 17.1 | 12.5 | 13.5 |
| AFM165xc11–GATA5B02 | 4.3 | 14.5 | 8.4 | 21.1 | 15.8 | 10.6 | 22.4 | 15.7 | 7.5 | 29.3 | 15.9 |
| 5: | |||||||||||
| GATA145D10–GATA84E11 | 1.8 | 7.9 | 6 | 9.8 | 8.6 | 7.8 | 9.2 | 6.8 | 7.4 | 6.3 | 7.2 |
| Mfd154–AFM164xb8 | 4.3 | 13.6 | 10.4 | 17.4 | 14.6 | 11 | 19 | 15.2 | 9.9 | 21.4 | 15 |
| 6: | |||||||||||
| SE30–ATA50C05 | 10.0 | 14.6 | 21.7 | 9.1 | 15.8 | 17.4 | 14.2 | 15.7 | 19.7 | 11.9 | 14.8 |
| GATA81B01–ATA22G07 | 4.3 | 11.7 | 8.6 | 15.3 | 10.8 | 9.2 | 12.4 | 11.5 | 9.9 | 13.2 | 10.3 |
| 7: | |||||||||||
| GATA24F03–AFM217yc5 | 7.8 | 16.5 | 18.1 | 14.9 | 14.1 | 17.4 | 11.1 | 12.9 | 17.2 | 9 | 15 |
| GATA30D09–Mfd265 | 5.0 | 6.7 | 6.3 | 7 | 7.1 | 6.1 | 8.1 | 7.1 | 5 | 9.6 | 8.1 |
| 8: | |||||||||||
| AFM143xd8–AFM198wd2 | 4.2 | 17.1 | 9.3 | 27.9 | 12.2 | 7.6 | 17 | 11.2 | 7.4 | 15.6 | 14.7 |
| AFM073yb7–UT721 | 8.5 | 21.9 | 20.6 | 23.2 | 22.2 | 23.4 | 21.2 | 18.8 | 16.5 | 21.3 | 20.9 |
| 9: | |||||||||||
| GATA62F03–GATA27A11 | 7.8 | 25.3 | 36.6 | 18.1 | 22.4 | 22.6 | 21.8 | 27 | 31 | 24.2 | 27.1 |
| ATA59H06–AFM303ZG9 | 17.9 | 15.7 | 11.5 | 20.2 | 15.6 | 10.4 | 21.7 | 15.2 | 10.7 | 20.3 | 17.9 |
| 10: | |||||||||||
| GATA88F09–AFM063xf4 | 4.6 | 13.2 | 12.3 | 14 | 14.3 | 8.9 | 21.1 | 13.1 | 8.1 | 18.9 | 11.8 |
| GGAA23C05–AFM198zb4 | 6.7 | 10.6 | 7.3 | 14.3 | 9 | 4.8 | 13.5 | 7.4 | 3.8 | 11.2 | 10.1 |
| 11: | |||||||||||
| GGAA17G05–ATA33B03 | 3.2 | 10.2 | 8.2 | 12.1 | 8 | 7.6 | 8.6 | 7.7 | 4.6 | 11.1 | 8.9 |
| AFM157xh6–AFM109xc3 | 3.3 | 15.4 | 6.8 | 27.4 | 14.8 | 9.2 | 21.7 | 14.4 | 11.1 | 19 | 14 |
| 12: | |||||||||||
| GATA4H03–GATA49D12 | 5.9 | 13.5 | 11.9 | 15.4 | 12.9 | 9 | 17.5 | 11.2 | 6.3 | 16.7 | 13 |
| ATA29A06–GATA13D05 | 2.5 | 7.5 | 7.8 | 7.2 | 5.5 | 3.7 | 7.4 | 4 | 3.2 | 4.7 | 2.1 |
| 13: | |||||||||||
| GATA51B02–AFM309va9 | 4.0 | 15.5 | 9.3 | 23 | 17.3 | 13.1 | 22.2 | 13.4 | 12.9 | 14.1 | 16.6 |
| 14: | |||||||||||
| GATA168F06–GATA136B01 | 9.5 | 14.4 | 20.1 | 9.6 | 15.3 | 17.3 | 13.6 | 11 | 18.8 | 6.2 | 15.5 |
| 15: | |||||||||||
| GATA22F01–GATA27A03 | 6.5 | 19 | 9.1 | 35.5 | 18 | 7.8 | 32.8 | 16.5 | 7.3 | 29.3 | 16.3 |
| 16: | |||||||||||
| ATA41E04–ATA3A07 | 9.2 | 17.7 | 13.7 | 21.7 | 15.5 | 15.1 | 16 | 13.4 | 11.8 | 14.8 | 14.3 |
| GATA11C06–GATA71F09 | 12.1 | 4.4 | 2.8 | 6.3 | 5.3 | 6.4 | 4.2 | 5.2 | 4 | 6.5 | 5.9 |
| 17: | |||||||||||
| GTAT1A05–GAAT2C03 | 2.1 | 11.4 | 2.9 | 24 | 8.9 | 2.5 | 17.4 | 11.8 | 3.9 | 20.9 | 8.5 |
| AFM044xg3–AFM217yd10 | 3.2 | 5.5 | 2.1 | 9.3 | 6.3 | 3.1 | 9.8 | 6.1 | 1.8 | 11.6 | 5.2 |
| 18: | |||||||||||
| GATA178F11–AFM321xc9 | 2.5 | 4.4 | .6 | 7.9 | 4.5 | 3.5 | 5.6 | 3.3 | 1.8 | 4.9 | 3.8 |
| GATA177C03–ATA1H06 | 5.4 | 5.4 | 5.6 | 5.2 | 3.4 | 3.9 | 3 | 4.3 | 3.2 | 5.1 | 4.1 |
| 19: | |||||||||||
| GATA44F10–GATA21G05 | 4.6 | 10.6 | 9.1 | 12.2 | 10.2 | 8.4 | 11.9 | 11.4 | 7 | 15.1 | 11.1 |
| GATA29B01–Mfd238 | 2.5 | 17.3 | 9.5 | 28.7 | 18.1 | 10.3 | 29.9 | 18.9 | 9.7 | 32.7 | 15.7 |
| 20: | |||||||||||
| AFM077xd3–GATA51D03 | 2.5 | 10.8 | 2.5 | 21.3 | 11.6 | 2.8 | 23.1 | 13.6 | 5.3 | 25.4 | 9.8 |
| GATA45B10–AFM046xf6 | 10.0 | 17.1 | 21.6 | 13.7 | 16.3 | 19.6 | 13.7 | 16.7 | 25 | 10.4 | 18.8 |
| 21: | |||||||||||
| GATA188F04–GATA70B08 | 4.4 | 20.9 | 17.6 | 24.7 | 18.5 | 15.1 | 22.3 | 18.4 | 12.5 | 26.5 | 17.1 |
| 22: | |||||||||||
| GATA11B12–GGAT3C10 | 14.9 | 4.1 | 7.7 | .5 | 4.6 | 7 | 2.1 | 7.4 | 8.8 | 5.8 | 5 |
| Total length of all telomeric intervals | 6.2 | 486.2 | 410.2 | 600 | 475.2 | 388.9 | 587.6 | 451.8 | 366 | 573.8 | 469.3 |
Physical distance (Mb) from the midpoint of the interval to the physical end of the chromosome.
Electronic-Database Information
The URLs for data presented herein are as follows:
- ASPEX, http://aspex.sourceforge.net/
- Marshfield Center for Medical Genetics, http://research.marshfieldclinic.org/genetics/ (for Mammalian Genotyping Service)
- NAT, http://www.fhcrc.org/labs/tang/NAT/nat.rtf (for R code)
- PCR Protocol, http://research.marshfieldclinic.org/genetics/Lab_Methods/pcr_protocol.htm
References
- Broman KW, Matsumoto N, Giglio S, Martin CL, Roseberry JA, Zuffardi O, Ledbetter DH, Weber JL (2003) Common long human inversion polymorphism on chromosome 8p. In: Goldstein DR (ed) Science and statistics: a festschrift for Terry Speed. IMS Lecture Notes–Monograph Series. Vol 40. Institute of Mathematical Statistics, Bethesda, MD, pp 237–245 [Google Scholar]
- Broman KW, Murray JC, Sheffield VC, White RL, Weber JL (1998) Comprehensive human genetic maps: individual and sex-specific variation in recombination. Am J Hum Genet 63:861–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calafell F, Shuster A, Speed WC, Kidd JR, Kidd KK (1998) Short tandem repeat polymorphism in evolution. Eur J Hum Genet 6:38–49 10.1038/sj.ejhg.5200151 [DOI] [PubMed] [Google Scholar]
- Catcheside DG (1977) The genetics of recombination. University Park Press, London [Google Scholar]
- Cullen M, Perfetto SP, Klitz W, Nelson G, Carrington M (2002) High-resolution patterns of meiotic recombination across the human major histocompatibility complex. Am J Hum Genet 71:759–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- FBPP Investigators (2002) Multi-center genetic study of hypertension: the Family Blood Pressure Program (FBPP). Hypertension 39:3–9 10.1161/hy1201.100415 [DOI] [PubMed] [Google Scholar]
- Giglio S, Broman KW, Matsumoto N, Calvari V, Gimelli G, Neumann T, Ohashi H, Voullaire L, Larizza D, Giorda R, Weber JL, Ledbetter DH, Zuffardi O (2001) Olfactory receptor–gene clusters, genomic-inversion polymorphisms, and common chromosome rearrangements. Am J Hum Genet 68:874–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillers KJ, Villeneuve AM (2003) Chromosome-wide control of meiotic crossing over in C. elegans. Curr Biol 13:1641–1647 10.1016/j.cub.2003.08.026 [DOI] [PubMed] [Google Scholar]
- Koehler KE, Cherry JP, Lynn A, Hunt PA, Hassold TJ (2002) Genetic control of mammalian meiotic recombination. I. Variation in exchange frequencies among males from inbred mouse strains. Genetics 162:297–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong A, Gudbjartsson DF, Sainz J, Jonsdottir GM, Gudjonsson SA, Richardsson B, Sigurdardottir S, Barnard J, Hallbeck B, Masson G, Shlien A, Palsson ST, Frigge ML, Thorgeirsson TE, Gulcher JR, Stefansson K (2002) A high-resolution recombination map of the human genome. Nat Genet 31:241–247 [DOI] [PubMed] [Google Scholar]
- Little R, Rubin D (2002) The problem of missing data. In: Statistical analysis with missing data, 2nd ed. John Wiley and Sons, Hoboken, NJ, pp3–19 [Google Scholar]
- Lynn A, Koehler KE, Judis L, Chan ER, Cherry JP, Schwartz S, Seftel A, Hunt PA, Hassold TJ (2002) Covariation of synaptonemal complex length and mammalian meiotic exchange rates. Science 296:2222–2225 10.1126/science.1071220 [DOI] [PubMed] [Google Scholar]
- Matise TC, Sachidanandam R, Clark AG, Kruglyak L, Wijsman E, Kakol J, Buyske S, et al (2003) A 3.9-centimorgan-resolution human single-nucleotide polymorphism linkage map and screening set. Am J Hum Genet 73:271–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page SL, Hawley RS (2003) Chromosome choreography: the meiotic ballet. Science 301:785–789 10.1126/science.1086605 [DOI] [PubMed] [Google Scholar]
- Sugawara H, Harada N, Ida T, Ishida T, Ledbetter DH, Yoshiura K, Ohta T, Kishino T, Niikawa N, Matsumoto N (2003) Complex low-copy repeats associated with a common polymorphic inversion at human chromosome 8p23. Genomics 82:238–244 10.1016/S0888-7543(03)00108-3 [DOI] [PubMed] [Google Scholar]
- Tang H, Quertermous T, Rodriquez B, Kardia SLR, Zhu X, Brown A, Pankow JS, Province MA, Hunt SC, Boerwinkle E, Schork NJ, Risch NJ (2005) Genetic structure, self-identified race/ethnicity, and confounding in case-control association studies. Am J Hum Genet 76:268–275 (in this issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tease C, Hartshorne GM, Hultén MA (2002) Patterns of meiotic recombination in human fetal oocytes. Am J Hum Genet 70:1469–1479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu A, Zhao C, Fan Y, Jang W, Mungall AJ, Deloukas P, Olsen A, Doggett NA, Ghebranious N, Broman KW, Weber JL (2001) Comparison of human genetic and sequence-based physical maps. Nature 409:140–146 10.1038/35051674 [DOI] [PubMed] [Google Scholar]
- Zahn LM, Kwiatkowski DJ (1995) 37-marker PCR-based genetic linkage map of human chromosome 9: observations on mutations and positive interference. Genomics 28:140–146 10.1006/geno.1995.1124 [DOI] [PubMed] [Google Scholar]