Abstract
Instability of the fragile X CGG repeat involves both maternally derived expansions and deletions in the gametes of full-mutation males. It has also been suggested that the absence of aberrant CpG methylation may enhance repeat deletions through an unknown process. The effect of CGG tract length, DNA replication direction, location of replication initiation, and CpG methylation upon CGG stability were investigated using an SV40 primate replication system. Replication-dependant deletions with 53 CGG repeats were observed when replication was initiated proximal to the repeat, with CGG as the lagging-strand template. When we initiated replication further from the repeat, while maintaining CGG as the lagging-strand template or using CCG as the lagging-strand template, significant instability was not observed. CpG methylation of the unstable template stabilized the repeat, decreasing both the frequency and the magnitude of deletion events. Furthermore, CpG methylation slowed the efficiency of replication for all templates. Interestingly, replication forks displayed no evidence of a block at the CGG repeat tract, regardless of replication direction or CpG methylation status. Templates with 20 CGG repeats were stable under all circumstances. These results reveal that CGG deletions occur during replication and are sensitive to replication-fork dynamics, tract length, and CpG methylation.
Introduction
Fragile X syndrome (FRAXA [MIM 309550]), the leading cause of inherited mental retardation, is caused by an expansion of the CGG trinucleotide repeat (TNR) located within the 5′ UTR of the FMR1 gene (Verkerk et al. 1991). TNR diseases, such as FRAXA, bring to the forefront important issues regarding DNA replication and its effect on the dynamic nature of repeat stability. Experiments on multiple transgenic mice have been unable to reproduce the parent-of-origin effects observed in humans, with the most instability observed for knock-in and human-YAC mice (Lavedan et al. 1998; Bontekoe et al. 2001; Peier and Nelson 2002; Pearson 2003). Furthermore, the majority of investigations into the underlying mechanism(s) of TNR instability are based on CAG/CTG model systems. However, there are striking differences between CGG- and CAG/CTG-associated diseases, including parent of origin, tissue specificity, somatic instability, aberrant CpG methylation, and the formation of unusual DNA structures. Given the numerous differences between CGG and CAG/CTG genetics, conclusions based on CAG/CTG experiments may not be applicable to the mechanisms of CGG instability; therefore, parallel studies of CGG instability should be performed.
Somatic and gametic instability play a crucial role in the etiology of most TNR diseases. Unlike the extraordinarily high level of somatic CTG instability—both within and between tissues—displayed by patients with myotonic dystrophy (DM1 [MIM 160900]) (Martorell et al. 1995; Wohrle et al. 1995), patients with FRAXA do not exhibit postnatal somatic CGG instability (Reyniers et al. 1999). Distinct patterns of CGG repeat–length heterogeneity formed during fetal development persist postnatally in patients with FRAXA (Wohrle et al. 1995), which contrasts DM1, in which the CTG length may vary over the patient’s life (Martorell et al. 1995; Wohrle et al. 1995). This lack of postnatal somatic instability, along with data from studies published elsewhere (Wohrle et al. 1993; Malter et al. 1997; Moutou et al. 1997), supports the suggestion that FRAXA expansions are the result of either meiotic events in oocytes or mitotic processes restricted to the earliest postzygotic cell divisions (i.e., before days 7–12). It is important to note, however, that only unmethylated premutation repeat lengths (50–200 repeats) exist in the sperm of males with full-mutation FRAXA, even though all other tissues display disease-length alleles (>200 repeats) (Reyniers et al. 1993; Tassone et al. 1999). The absence of full-mutation alleles in the sperm of males with FRAXA suggests that deletions in the male germline arise later in fetal development, after the initial expansion event (Wohrle et al. 1993; Malter et al. 1997; Moutou et al. 1997; reviewed in Pearson 2003).
Unlike other TNR diseases, the FRAXA phenotype is modulated not only by repeat tract length (Nolin et al. 2003; Rife et al. 2004) but also by aberrant CpG methylation. Patients with FRAXA exhibit complete methylation of their expanded CGG tract, whereas rare individuals classified as “high-functioning” males or “methylation mosaics” possess unmethylated CGG repeats within the disease-length range and display considerable levels of postnatal somatic CGG instability (mostly deletions), highlighting the increased instability of the unmethylated repeats (Wohrle et al. 1998; Taylor et al. 1999). Interestingly, not only does CpG methylation modulate phenotype and influence CGG instability (Wohrle et al. 1998; Nichol and Pearson 2002), but it also has also been documented to delay the already slowed replication of the FRAXA chromosomal locus (Hansen et al. 1993; Subramanian et al. 1996).
Previous work of our lab, performed using an SV40 system of primate replication, revealed that the location of replication initiation and fork direction, relative to the repeat, can lead to different types and frequencies of repeat instability in CTG/CAG repeat tracts (Cleary et al. 2002). By taking advantage of this SV40 system, it was possible to examine not only the influence of replication-fork dynamics on CGG stability but also how CpG methylation may modulate this stability. Our results support a role for replication-mediated events in CGG instability, with the frequency of deletions depending on length, sequence type (i.e., CGG or CCG), and location of replication initiation. Furthermore, the occurrence and magnitude of deletions, as well as the efficiency of replication, could be modulated by CpG methylation. Interestingly, replication-induced deletions arose independently of a replication-fork block at the CGG repeat.
Material and Methods
Replication Templates
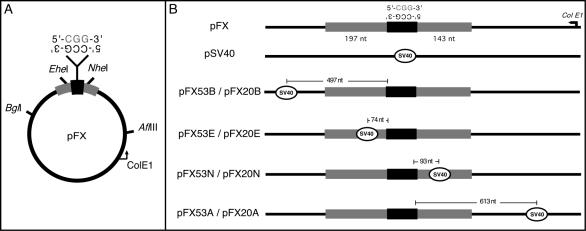
Genomic FRAXA sequence (nucleotides 2523–2911 of pE5.1 [GenBank accession number X61378]) was cloned into pPCR-Script Amp (Stratagene), with the repeat in the stable orientation, relative to the unidirectional bacterial ColE1 origin of replication (with CCG as the lagging-strand template) (Pearson et al. 1998). SV40 viral sequence (nucleotides 5171–128 [GenBank accession number NC_001669]), containing the origin of bidirectional replication (nucleotide 5210/5211), was inserted into the FRAXA vector at various locations (fig. 1A).
Figure 1.
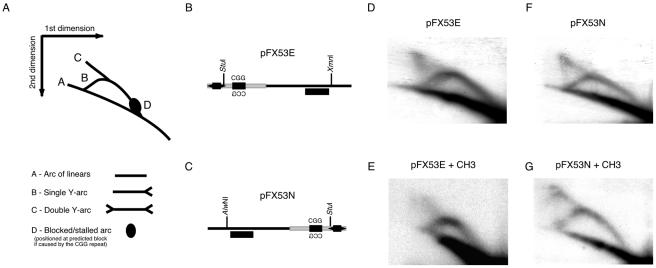
Replication templates. A, Templates containing human FRAXA CGG repeats (20 or 53 repeats) (blackened box), with genomic nonrepeating sequences (gray boxes) flanking the repeat. The SV40-ori fragment encompassed the unique origin of bidirectional replication (OBR) (Hay and DePamphilis 1982) and was inserted at the BglI, EheI, NheI, or AflIII site. The location of the SV40-ori, relative to the repeat, defined the direction of replication, as well as which strand (CGG or CCG) would serve as the leading- or lagging-strand template. B, Diagram of templates. The distance to the repeat tract was measured from the SV40 OBR (circled) to the nucleotide preceding either the 5′ or the 3′ end of the CGG repeat.
CpG Methylation
pFX/SV40 clones were methylated with SssI methylase (New England Biolabs), in accordance with the manufacturer’s protocol, by use of a buffer without MgCl2, to avoid topoisomerase activity (Matsuo et al. 1994). A small amount of each plasmid was digested with AciI to confirm the presence of CpG methylation (data not shown).
Cell Transfections
COS-1 cells (American Type Culture Collection) (Gluzman 1981) were grown in Dulbecco modified Eagle medium (Invitrogen) with 10% fetal bovine serum (HyClone). Cells were incubated with 5 μg of each pFX/SV40 template and 50 μl of LipoTAXI (Stratagene), in accordance with the manufacturer’s specifications. Episomal DNAs were extracted 48 h posttransfection with 100 mM Tris-HCl (pH 7.5), 1 mM EDTA, and 0.6% SDS (Hirt 1967).
Mutation Analysis
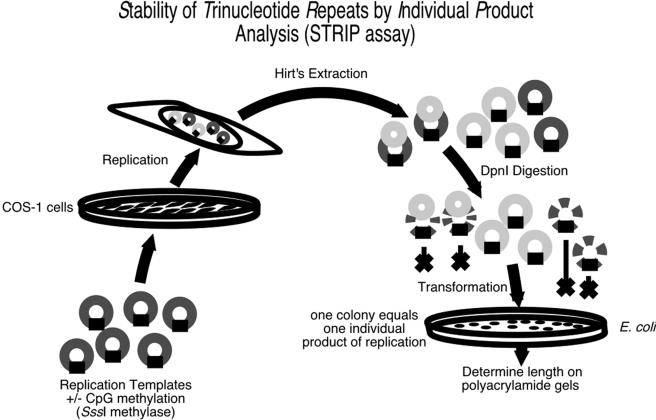
Mutations were assessed using the stability of trinucleotide repeats by individual product (STRIP) assay (Cleary et al. 2002). Episomal DNAs were digested with DpnI (New England Biolabs) to eliminate unreplicated parental templates. DpnI-resistant material was transformed, and plasmid DNA from individual bacterial colonies, each representing an individual product of primate replication, was prepared. To minimize repeat instability due to propagation in Escherichia coli, the XL1-Blue MR cell strain (Stratagene) was selected for analysis of all pFX/SV40 replication products except pFX53A/pFX20A, for which the XLmutS cell strain (Stratagene) was used.
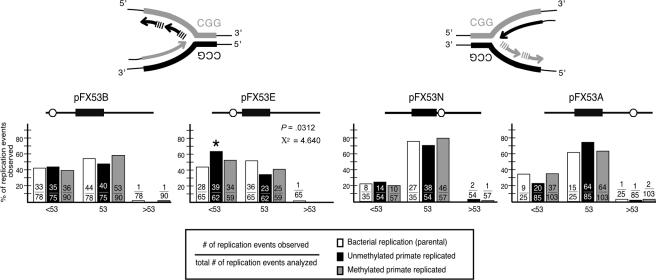
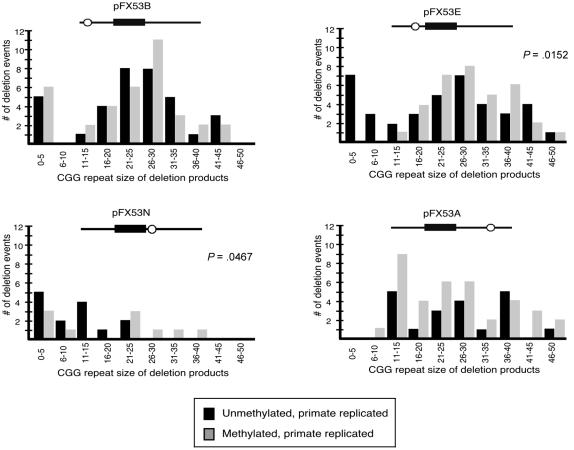
Repeat-containing restriction fragments were resolved on 4% polyacrylamide gels, with tract lengths scored relative to starting material and a 123-bp ladder (Invitrogen). As described in detail elsewhere (Cleary et al. 2002), corrected mutation frequencies were calculated only for repeat-length distributions that were significantly different from the parental template (by use of χ2 analysis) (fig. 2), by subtracting the background-length heterogeneity within the parental plasmid from the frequencies generated by primate replication (table 1). Repeat-length distributions that were not significantly different from the parental background (P>.05) were classified as either stable or having no primate cell–induced events (“0”) (table 1). Statistical analysis of the magnitude data was performed using t-test analysis (fig. 3). To confirm that the deletions or expansions observed were repeat-length changes, samples were electrotransferred and hybridized with a 32P-(CGG)10 oligonucleotide. Selected expansion and deletion products were also sequenced.
Figure 2.
Mutation analysis. As shown elsewhere (Cleary et al. 2002), bacterial preparations of the starting parental templates contained a distribution of repeat lengths that were classified into one of three categories: <53 repeats, 53 repeats, and >53 repeats (unblackened bars). After primate replication, the repeat-length distribution of the primate-replicated material for each template (unmethylated template [blackened bars] and CpG premethylated template [gray bars]) was similarly determined. The bar height (Y-axis) reflects the percentage of replication events within each length category. For each length category, the number of replication events observed within that length category, over the total number of replication events analyzed, is indicated inside the bar. After primate replication, templates with a statistically significant (P<.05) increase in the number of molecules with lengths <53 repeats were marked with an asterisk (*). All constructs were transfected a minimum of three times.
Table 1.
Summary of pFX Data[Note]
|
CorrectedFrequencya (%) of |
||||
| Template | Expansions | Deletions | Expansion Length(s)b | ReplicationEfficiencycNormalizedto pFX53E(%) |
| pFX53B +CH3 | 0 | 0 | 68 (67) | 43 |
| pFX53B | 0 | 0 | NA | 80 |
| pFX53E +CH3 | 0 | 0 | NA | 71 |
| pFX53E | 0 | 20 | NA (58) | 100 |
| pFX53N +CH3 | 0 | 0 | 69 | 54 |
| pFX53N | 0 | 0 | 54, 55 (NA) | 57 |
| pFX53A +CH3 | 0 | 0 | 60, 74 | 27 |
| pFX53A | 0 | 0 | 57 (62) | 45 |
| pFX20E/pFX20N +/−CH3 | 0 | 0 | NA | ND |
| pFX53E in CV-1d | 0 | 0 | NA | ND |
| pFX53E +CH3 CV-1d | 0 | 0 | ND | ND |
Note.— As described elsewhere (Cleary et al. 2002) (see the “Material and Methods” section), corrected mutation frequencies were calculated only for repeat-length distributions that were significantly different from the parental template background (fig. 2). NA = not applicable; ND = not determined.
Expansion and deletion frequencies mediated by primate-cell replication were corrected for background by subtracting the frequencies of >53 or <53 events, respectively, for the parental template molecules from the frequencies of the same category for the replicated products. Repeat-length distributions that were not significantly different from the parental background (P>.05) were classified as stable or as having no primate-replication–induced events (“0”).
In cases in which expansions were observed, the length of the primate-replicated expanded CGG repeat was noted. The length of the bacterially replicated expanded CGG repeat is noted in parentheses.
Replication efficiencies were determined as in figure 4 (see “Material and Methods” section).
Replication not permitted.
Figure 3.
Length distributions of observed deletion events. All deletion events were sized and graphed as the number of events within each CGG size category. Unmethylated primate-replicated deletions (blackened bars) and CpG premethylated primate-replicated deletions (gray bars) are noted. Statistical analysis comparing the difference in distribution between the two groups was performed using t-test analysis, with P values noted when significant (P<.05).
Replication Efficiency
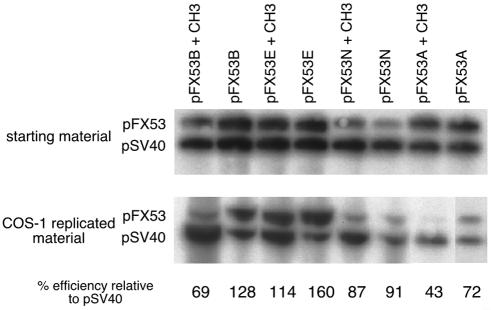
Replication efficiency (fig. 4) was calculated by cotransfecting the pFX53 plasmids, with and without CpG methylation, with unmethylated, repeat-free pSV40 (fig. 1). Posttransfection, episomal DNA was digested with AlwNI and DpnI, resolved on a 1% agarose gel, transferred to Biodyne B membrane (Pall Corporation), and probed with the 32P-SV40 origin of replication (SV40-ori). An aliquot of the starting mixture used for the cotransfection was linearized with AlwNI and was analyzed on the same gel. ImageQuant version 1.2 (Molecular Dynamics) was used to quantitate the ratio of starting materials between pFX53 and pSV40 and the ratio of completely replicated materials. These ratios were used to determine the replication efficiency of each template in relation to unmethylated pSV40.
Figure 4.
Replication efficiency. An aliquot of the starting mixture, composed of each pFX53 construct with unmethylated pSV40, was linearized with AlwNI. Primate-replicated DNA resulting from the cotransfection of this same starting mixture was also linearized with AlwNI, was further digested with DpnI to remove the unreplicated parental template, and was then probed with the SV40-ori fragment. Replication efficiency was calculated for each template in relation to pSV40 (as described in the “Material and Methods” section).
Two-Dimensional (2D) Gel Electrophoresis
Replication intermediates of pFX53E and pFX53N were digested with StuI/XmnI and StuI/AlwNI, respectively (fig. 5B and 5C). Samples were run in the first dimension for 24 h at 1 V/cm on a 0.4% agarose gel. Lanes were then cut out, embedded in 1% agarose gel with 0.5 μg/ml ethidium bromide, and run in the second dimension for 8 h at 5 V/cm at 4°C, with buffer recirculation (Cordeiro-Stone et al. 1997; Krasilnikova and Mirkin 2004). Gels were transferred as described above and were probed with the 32P-XmnI-AlwNI fragment of pPCR-Script Amp (Stratagene).
Figure 5.
Replication-fork progression, 2D gel electrophoresis. A, Schematic representation of predicted Y arcs, as well as expected position of a block (if resulting from the CGG repeat tract) with respect to the restriction digests used for analysis. B and C, Maps of CGG repeat (blackened box), FRAXA genomic flanking sequences (dark gray lines), SV40-ori (blackened arrows), and the restriction digests used for 2D gel analysis of pFX53E (B) and pFX53N (C). The black rectangle below the line denotes the Xmn I/AlwNI fragment used as a probe. D and F, Replication forks of pFX53E and pFX53N. E and G, Replication forks of CpG premethylated pFX53E and pFX53N.
Results
Primate Replication Assay for CGG Stability
The role of primate DNA replication in FRAXA CGG repeat stability was investigated using an established SV40 replication system (Cleary et al. 2002). The SV40-ori was inserted once within a construct containing both the CGG tract and FRAXA human genomic flanking sequence (pFX). By positioning the SV40-ori on either side of the repeat, it was possible to change which strand (CGG or CCG) was used as the lagging-strand template in a series of replication constructs comprising either 20 or 53 pure CGG repeats (fig. 1).
pFX/SV40 replication templates were transfected into SV40 T-antigen (T-Ag)–expressing COS-1 cells and were replicated by primate proteins. The episomally replicated DNAs were isolated and assayed for repeat stability by use of STRIP analysis (Cleary et al. 2002) (fig. A1 [online only]). The STRIP assay allowed for length analysis of individual products of primate replication, with the unreplicated starting material removed by DpnI digestion prior to transformation into E. coli.
Primate-Replication–Induced Deletions of a (CGG)53 Template
In pFX53E, the SV40-ori location permitted replication to initiate 74 nt 5′ of the CGG tract, with CGG as the template for lagging-strand synthesis (fig. 1). Replication of pFX53E in primate cells yielded significantly more deletions than the background (fig. 2) (P=.031; χ2=4.640), with a corrected frequency of deletion events (<53) calculated as 20% ( 100 × [39/62 (primate replicated) − 28/65 (background)] ≃ 20) (see table 1 and the “Material and Methods” section).
To confirm that the deletions were products of primate replication, pFX53E was transfected into CV-1 cells, the precursor of COS-1 cells (Gluzman 1981), which do not express T-Ag and therefore cannot replicate the template. DpnI digestion of episomal DNA recovered from the transfection of pFX53E into CV-1 cells did not yield bacterial colonies after transformation, confirming a lack of replication. Furthermore, CGG length distribution of non–DpnI-treated episomal DNA extracted from CV-1 cells did not differ significantly from background (table 1), indicating a causative role for DNA replication in deletions observed when CGG was used as the template for lagging-strand synthesis.
Replication Direction and the Location of Replication Initiation
Data from bacterial and yeast models suggest a role for direction of replication in CGG instability (Shimizu et al. 1996; Hirst and White 1998; White et al. 1999). To determine whether this phenomenon extends to primate replication, pFX53N was constructed such that CCG was lagging-strand template (fig. 1). In contrast to pFX53E, the frequency of deletions did not differ significantly from background (fig. 2 and table 1) (P=.669; χ2=0.183), supporting a role for replication direction in CGG instability. Thus, primate-replicated deletions were observed when CGG, but not CCG, was used as the lagging-strand template.
For CTG/CAG repeats, changing the location of the SV40-ori, relative to the repeat tract, affected repeat instability (Cleary et al. 2002). To examine the influence of the location of replication initiation on CGG stability, the SV40-ori was placed at varying distances from the repeat (fig. 1). For pFX53B (fig. 1B), replication proceeded through the repeat in the same direction as pFX53E (CGG as lagging-strand template), but initiation occurred further from the repeat tract (497 nt vs. 74 nt). The stable replication of pFX53B (fig. 2 and table 1) (P=.637; χ2=0.223) contrasted the significant increase in deletion events observed for pFX53E, which was replicated in the same orientation. This shift—from high levels of deletions to levels that do not differ from background—results from a change in the distance between replication initiation and the repeat without changing the direction of replication, highlighting the contribution of cis-elements to CGG instability.
To further address the influence of the location of replication initiation on instability, pFX53A was analyzed. In pFX53A (fig. 1), replication proceeded in the same direction as pFX53N (with CCG as the lagging-strand template), but it was initiated further from the repeat tract (613 nt vs. 93 nt). Both pFX53N and pFX53A (fig. 2 and table 1) (P=.182; χ2=1.781) did not show significant levels of deletions. Therefore, the effect of initiation location on instability was evident only when CGG, not CCG, served as the lagging-strand template.
Effect of CpG Premethylation on Stability
To examine the possible role of CpG methylation in CGG stability, the pFX/SV40 constructs were premethylated with SssI methylase prior to transfection. CpG premethylation of pFX53E decreased not only the frequency of deletion events incurred during primate replication (from 20% to 0%) (see table 1 and the “Material and Methods” section) but also significantly reduced the magnitude of CGG length changes within the observed deletions (fig. 3) (P=.0152). CpG premethylation did not modulate the frequency of deletions for pFX53B, pFX53N, or pFX53A (fig. 2). However, CpG premethylation of pFX53N significantly decreased the magnitude of CGG length change of deletions observed when compared with unmethylated pFX53N (fig. 3) (P=.0467). Only CpG methylation was changed in this experiment, providing strong support for the hypothesis that CpG methylation stabilizes the CGG repeat during replication.
Length Dependence on Instability
To assess whether the stability of shorter CGG repeat tracts was affected by replication, an identical series of pFX/SV40 templates with 20 CGG repeats (fig. 1B) was replicated in COS-1 cells. None of the pFX20 templates displayed significant levels of instability, regardless of replication direction and location or CpG methylation status (tables 1 and 2), suggesting an association between repeat length and instability; longer pure repeat tracts were more prone to mutation.
Table 2.
Analysis of Templates Containing 20 CGG Repeats
| Template | No. of Expansions | No. of Deletions | Level of Stablility (%) |
| pFX20E +CH3 | 0 | 1 | 32 (97) |
| pFX20E | 0 | 0 | 38 (100) |
| pFX20N +CH3 | 0 | 1 | 36 (97) |
| pFX20N | 0 | 0 | 38 (100) |
Rare Expansions of the (CGG)53 Templates
One hallmark of FRAXA is the ability of premutation alleles containing 50–200 CGG repeats to expand upon transmission (Nolin et al. 2003; Rife et al. 2004). Episomal replication of templates with 53 CGG repeats resulted in rare expansion events, which were not statistically significant, as their numbers were not greater in magnitude or frequency than those observed in the bacterial background (table 1). However, it was interesting to note that most of the expansions occurred when CCG was used as the lagging-strand template (six of the seven expansion events; pFX53N and pFX53A) (table 1), perhaps suggesting a role for sequence specificity not only in deletion events but in expansion events as well.
Replication Efficiency Is Correlated with CGG Stability
The timing of replication through the FRAXA chromosomal locus occurs late in the cell cycle (Howell and McDermott 1982) and is further slowed by CpG methylation of the expanded CGG repeat (Hansen et al. 1993). To address whether the replication rate is correlated to the observed deletion frequency, the replication efficiency of each template (with and without CpG premethylation) was measured relative to an unmethylated internal control SV40-ori template containing no CCG repeats (fig. 1B). pFX53E, which displayed the highest frequency of deletions, was also the most efficiently replicated (fig. 4 and table 1). Interestingly, CpG premethylation of pFX53E decreased not only its levels of instability but also its replication efficiency. The effect of CpG premethylation on replication efficiency was observed for all templates when compared with their unmethylated replication efficiencies (fig. 4 and table 1).
Replication Efficiency Is Not Associated with a Block in the Replication Fork at the CGG Tract
A decrease in replication efficiency may be due to a block in replication-fork progression in or around the CGG repeat tract (Samadashwily et al. 1997). By use of 2D gel electrophoresis (fig. 5), replication forks derived from replication intermediates of pFX53E and pFX53N were assessed for the presence of a replication block. Neither pFX53E nor pFX53N, regardless of CpG methylation, displayed a block in replication that was localized to the CGG tract within their respective replication forks, by analysis of either y arcs (fig. 5D–5G) or bubble arcs (data not shown). The lack of a replication block may indicate that, during primate-mediated replication, instead of forming complete replication stalls, the CGG tract may actually incur pauses that are rapidly processed and overcome.
Discussion
Our data suggest that the location of replication initiation, in terms of proximity to the CGG tract, as well as the sequence of the lagging-strand template, is a major determinant of CGG deletions. Furthermore, CpG methylation stabilizes the repeat by decreasing the frequency and magnitude of replication-mediated CGG deletions. We also demonstrate, using a sensitive assay, that a block in replication-fork progression does not occur at the repeat tract and, therefore, is not related to CGG deletions. The lack of a distinct block in replication-fork progression (regardless of CpG methylation status) is consistent with previous observations in cells from patients with FRAXA (Hansen et al. 1993). Our observation that the context of the CGG tract, relative to its location within the replication fork, influences repeat stability may reflect the increased instability observed in knock-in and YAC transgenic mice—supporting the contribution of cis-elements to CGG instability (Bontekoe et al. 2001; Peier and Nelson 2002).
Replication Initiation
Studies focusing on stability of the FRAXA CGG repeat within fetal tissues (Devys et al. 1992; Wohrle et al. 1993; Malter et al. 1997) have provided evidence suggesting that maternally inherited CGG expansions either are meiotic in origin or occur early in postzygotic development and are then stably maintained. The ability to incur deletions within a construct containing 53 pure CGG repeats, simply by moving the position of replication initiation, highlights the potential impact of the location of replication initiation, with respect to the location of the repeat within the replication fork (Cleary et al. 2002; J. D. Cleary and C. E. Pearson, unpublished data), on early postzygotic CGG instability. This importance is further demonstrated by the rare occurrence of expansions, in our system, mainly when CCG is used as the lagging-strand template.
During early embryogenesis, replication initiation occurs randomly throughout the genome before becoming localized to specific replication origins (Hyrien et al. 1995; Sasaki et al. 1999). If localized replication initiation becomes set at a very early time point, the window for random replication initiation within human embryogenesis, and, therefore, the window for postzygotic CGG instability, may be limited to a very early stage in development. This limited window for instability can explain fetal-tissue data displaying minimal CGG heterogeneity within and between various tissues, a pattern that does not change throughout life.
CpG Methylation Stabilizes the CGG Repeat and Prevents Deletions
For most TNR diseases, the parent-of-origin effect plays an important role in repeat instability (reviewed in Pearson 2003). What is unique to FRAXA, however, is that expansion to the disease range occurs only upon maternal transmission. If expansions were occurring postzygotically and were modulated only by repeat length, then premutation alleles should behave similarly (i.e., show a tendency toward expansion), regardless of the transmitting parent; yet, they do not. This disparity in CGG instability may be driven by the differing CpG methylation status of maternally and paternally inherited DNA during zygote formation. In murine zygotes, the paternally derived alleles undergo rapid demethylation before the very first round of replication (Tamanini et al. 1997). This rapid demethylation contrasts with the maintenance of CpG methylation found for the maternally inherited alleles. Our data show that replication-mediated deletion events can be stabilized by the presence of CpG methylation, which perhaps hints at why paternally inherited premutation alleles, void of CpG methylation and potentially prone to deletions, do not expand during early fetal development.
The presence of only unmethylated premutation-length alleles in the sperm of full-mutation males diverges from the stable maintenance, within the rest of their tissues, of aberrantly methylated full-mutation–length CGG repeats. Whereas sperm contain only premutation-length alleles, CGG length analysis of the adult testis revealed the presence of both full-mutation and unmethylated premutation lengths (Reyniers et al. 1993; Tassone et al. 1999). Furthermore, expression studies of the fragile X mental retardation protein (FMRP) in fetal primordial germ cells (PGCs) from male fetuses with FRAXA demonstrate that FMRP is being expressed in some PGCs by 17 wk of development (Malter et al. 1997; Tamanini et al. 1997). This would suggest that, within the PGCs expressing FMRP, a CGG deletion event occurred after the original expansion event, resulting in unmethylated premutation-length CGG alleles.
Early in embryogenesis, male PGCs develop, migrate to the extraembryonic mesoderm, proliferate, and then return to the developing fetus to form the gonadal tissue (Sutton 2000). While in the extraembryonic mesoderm, paternal—not maternal—X chromosomes become preferentially inactivated by CpG methylation (Goto et al. 1997). The lack of CpG methylation on the maternally inherited X chromosome, as inherited by a male with FRAXA, may provide an opportunity for CGG deletions to occur before the PGCs return to the fetus and become mitotically arrested. As development continues, the cells that have incurred deletions to premutation lengths are perhaps selectively maintained by their expression of FMRP, which is thought to be necessary for gonadal development (Tamanini et al. 1997).
Absence of a Block in Replication at the CGG Repeat
Similar to the chromosomes of patients with FRAXA, which show delayed and/or slowed replication but no distinct pause (Hansen et al. 1993), primate replication of the pFX/SV40 constructs displayed reduced replication efficiency when they were CpG methylated but did not display a replication block at the CGG tract, regardless of replication direction or CpG methylation. The reduced replication efficiency we observed may reflect the generally slowed replication over the FRAXA locus, reported by Hansen et al. (1993), which may suggest delayed initiation or slowed fork progression through the region. The latter may permit enhanced replication fidelity due to an increase in the time available for replication-fork progression. The absence of a pause contrasts the strong replication-fork blocks observed at the CGG tract in both yeast and bacterial systems (Samadashwily et al. 1997; Pelletier et al. 2003). The difference in the ability to replicate through CGG repeats (primate replication vs. yeast or bacteria replication) may result from the fact that neither bacterial nor yeast genomes contain endogenous CCG/CGG repeats (Astolfi et al. 2003) and therefore may not have the mechanisms in place to properly replicate through the repeat tract without experiencing blockages. However, other explanations for the differences are possible, including the unidirectional replication versus bidirectional replication or the location of the initiation relative to the repeat tract (the yeast study used distances ⩾2.5 kbp, greater than the 74–613 bp distances used herein). Supporting this, replication stalls were recently observed for (CGG)n repeats in a primate SV40-derived episomal system, while positioned further apart from the origin (Sergei Mirkin, personal communication). However, the CGG deletions or their stabilization by CpG methylation reported here do not appear to correlate with a detectable propensity to pause replication-fork progression.
CGG deletions, similar to CAG/CTG instability, are influenced by tract length and replication-fork dynamics (J. D. Cleary and C. E. Pearson, unpublished data). However, CGG deletions are further influenced by CpG methylation, which highlights the need to study dynamic DNA mutations, not only in the proper sequence context, but also with as many epigenetic modifiers as allowed by the experimental system.
Acknowledgments
The authors thank S. M. Mirkin for assistance with 2D gel electrophoresis and J. D. Cleary for many hours of insightful discussions. M.R.L. was supported by an Ontario Graduate Scholarship and by a studentship from the Ontario Student Opportunity Trust Fund—Hospital for Sick Children Foundation Student Scholarship Program. This work was supported by grants from the Canadian Institutes of Health Research (CIHR) (to C.E.P.), the Fragile X Research Foundation Canada (to C.E.P.), and the University of Toronto Dean’s Fund (to C.E.P.). C.E.P. is a CIHR Scholar and a Canadian Genetic Disease Network Scholar.
Appendix A
Figure A1.
STRIP assay and replication of the pFX/SV40 templates (dark gray rings) by primate proteins within COS-1 cells. At 48 h posttransfection, the episomally replicated DNA was extracted and digested by DpnI to remove the unreplicated (dark gray rings) and partially replicated (dark gray rings with small light gray rings on top) templates. The DpnI-resistant primate-replicated templates (light gray rings) were transformed into E. coli, and individual colonies, each an individual product of primate replication, were cultured. The resulting DNA was restriction digested and analyzed on 4% polyacrylamide gels for CGG length changes.
Electronic-Database Information
Accession numbers and URLs for data presented herein are as follows:
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for SV40 [accession number NC_001669] and pE5.1 [accession number X61378])
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for FRAXA and DM1)
References
- Astolfi P, Bellizzi D, Sgaramella V (2003) Frequency and coverage of trinucleotide repeats in eukaryotes. Gene 317:117–125 [DOI] [PubMed] [Google Scholar]
- Bontekoe CJ, Bakker CE, Nieuwenhuizen IM, van der Linde H, Lans H, de Lange D, Hirst MC, Oostra BA (2001) Instability of a (CGG)98 repeat in the Fmr1 promoter. Hum Mol Genet 10:1693–1699 [DOI] [PubMed] [Google Scholar]
- Cleary JD, Nichol K, Wang YH, Pearson CE (2002) Evidence of cis-acting factors in replication-mediated trinucleotide repeat instability in primate cells. Nat Genet 31:37–46 [DOI] [PubMed] [Google Scholar]
- Cordeiro-Stone M, Zaritskaya LS, Price LK, Kaufmann WK (1997) Replication fork bypass of a pyrimidine dimer blocking leading strand DNA synthesis. J Biol Chem 272:13945–13954 [DOI] [PubMed] [Google Scholar]
- Devys D, Biancalana V, Rousseau F, Boue J, Mandel JL, Oberle I (1992) Analysis of full fragile X mutations in fetal tissues and monozygotic twins indicate that abnormal methylation and somatic heterogeneity are established early in development. Am J Med Genet 43:208–216 [DOI] [PubMed] [Google Scholar]
- Gluzman Y (1981) SV40-transformed simian cells support the replication of early SV40 mutants. Cell 23:175–182 [DOI] [PubMed] [Google Scholar]
- Goto T, Wright E, Monk M (1997) Paternal X-chromosome inactivation in human trophoblastic cells. Mol Hum Reprod 3:77–80 [DOI] [PubMed] [Google Scholar]
- Hansen RS, Canfield TK, Lamb MM, Gartler SM, Laird CD (1993) Association of fragile X syndrome with delayed replication of the FMR1 gene. Cell 73:1403–1409 [DOI] [PubMed] [Google Scholar]
- Hay RT, DePamphilis ML (1982) Initiation of SV40 DNA replication in vivo: location and structure of 5′ ends of DNA synthesized in the ori region. Cell 28:767–779 [DOI] [PubMed] [Google Scholar]
- Hirst MC, White PJ (1998) Cloned human FMR1 trinucleotide repeats exhibit a length- and orientation-dependent instability suggestive of in vivo lagging strand secondary structure. Nucleic Acids Res 26:2353–2358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirt B (1967) Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol 26:365–369 [DOI] [PubMed] [Google Scholar]
- Howell RT, McDermott A (1982) Replication status of the fragile X chromosome, fra(X)(q27), in three heterozygous females. Hum Genet 62:282–284 [DOI] [PubMed] [Google Scholar]
- Hyrien O, Maric C, Mechali M (1995) Transition in specification of embryonic metazoan DNA replication origins. Science 270:994–997 [DOI] [PubMed] [Google Scholar]
- Krasilnikova MM, Mirkin SM (2004) Analysis of triplet repeat replication by two-dimensional gel electrophoresis. Methods Mol Biol 277:19–28 [DOI] [PubMed] [Google Scholar]
- Lavedan C, Grabczyk E, Usdin K, Nussbaum RL (1998) Long uninterrupted CGG repeats within the first exon of the human FMR1 gene are not intrinsically unstable in transgenic mice. Genomics 50:229–240 [DOI] [PubMed] [Google Scholar]
- Malter HE, Iber JC, Willemsen R, de Graaff E, Tarleton JC, Leisti J, Warren ST, Oostra BA (1997) Characterization of the full fragile X syndrome mutation in fetal gametes. Nat Genet 15:165–169 [DOI] [PubMed] [Google Scholar]
- Martorell L, Martinez JM, Carey N, Johnson K, Baiget M (1995) Comparison of CTG repeat length expansion and clinical progression of myotonic dystrophy over a five year period. J Med Genet 32:593–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo K, Silke J, Gramatikoff K, Schaffner W (1994) The CpG-specific methylase SssI has topoisomerase activity in the presence of Mg2+. Nucleic Acids Res 22:5354–5359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moutou C, Vincent MC, Biancalana V, Mandel JL (1997) Transition from premutation to full mutation in fragile X syndrome is likely to be prezygotic. Hum Mol Genet 6:971–979 [DOI] [PubMed] [Google Scholar]
- Nichol K, Pearson CE (2002) CpG methylation modifies the genetic stability of cloned repeat sequences. Genome Res 12:1246–1256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolin SL, Brown WT, Glicksman A, Houck GE Jr, Gargano AD, Sullivan A, Biancalana V, Brondum-Nielsen K, Hjalgrim H, Holinski-Feder E, Kooy F, Longshore J, Macpherson J, Mandel JL, Matthijs G, Rousseau F, Steinbach P, Vaisanen ML, von Koskull H, Sherman SL (2003) Expansion of the fragile X CGG repeat in females with premutation or intermediate alleles. Am J Hum Genet 72:454–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson CE (2003) Slipping while sleeping? trinucleotide repeat expansions in germ cells. Trends Mol Med 9:490–495 [DOI] [PubMed] [Google Scholar]
- Pearson CE, Eichler EE, Lorenzetti D, Kramer SF, Zoghbi HY, Nelson DL, Sinden RR (1998) Interruptions in the triplet repeats of SCA1 and FRAXA reduce the propensity and complexity of slipped strand DNA (S-DNA) formation. Biochemistry 37:2701–2708 [DOI] [PubMed] [Google Scholar]
- Peier AM, Nelson DL (2002) Instability of a premutation-sized CGG repeat in FMR1 YAC transgenic mice. Genomics 80:423–432 [DOI] [PubMed] [Google Scholar]
- Pelletier R, Krasilnikova MM, Samadashwily GM, Lahue R, Mirkin SM (2003) Replication and expansion of trinucleotide repeats in yeast. Mol Cell Biol 23:1349–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyniers E, Martin JJ, Cras P, Van Marck E, Handig I, Jorens HZ, Oostra BA, Kooy RF, Willems PJ (1999) Postmortem examination of two fragile X brothers with an FMR1 full mutation. Am J Med Genet 84:245–249 [DOI] [PubMed] [Google Scholar]
- Reyniers E, Vits L, De Boulle K, Van Roy B, Van Velzen D, de Graaff E, Verkerk AJ, Jorens HZ, Darby JK, Oostra B, et al (1993) The full mutation in the FMR-1 gene of male fragile X patients is absent in their sperm. Nat Genet 4:143–146 [DOI] [PubMed] [Google Scholar]
- Rife M, Badenas C, Quinto L, Puigoriol E, Tazon B, Rodriguez-Revenga L, Jimenez L, Sanchez A, Mila M (2004) Analysis of CGG variation through 642 meioses in fragile X families. Mol Hum Reprod 10:773–776 [DOI] [PubMed] [Google Scholar]
- Samadashwily GM, Raca G, Mirkin SM (1997) Trinucleotide repeats affect DNA replication in vivo. Nat Genet 17:298–304 [DOI] [PubMed] [Google Scholar]
- Sasaki T, Sawado T, Yamaguchi M, Shinomiya T (1999) Specification of regions of DNA replication initiation during embryogenesis in the 65-kilobase DNApolα-dE2F locus of Drosophila melanogaster. Mol Cell Biol 19:547–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu M, Gellibolian R, Oostra BA, Wells RD (1996) Cloning, characterization and properties of plasmids containing CGG triplet repeats from the FMR-1 gene. J Mol Biol 258:614–626 [DOI] [PubMed] [Google Scholar]
- Subramanian PS, Nelson DL, Chinault AC (1996) Large domains of apparent delayed replication timing associated with triplet repeat expansion at FRAXA and FRAXE. Am J Hum Genet 59:407–416 [PMC free article] [PubMed] [Google Scholar]
- Sutton KA (2000) Molecular mechanisms involved in the differentiation of spermatogenic stem cells. Rev Reprod 5:93–98 [DOI] [PubMed] [Google Scholar]
- Tamanini F, Willemsen R, van Unen L, Bontekoe C, Galjaard H, Oostra BA, Hoogeveen AT (1997) Differential expression of FMR1, FXR1 and FXR2 proteins in human brain and testis. Hum Mol Genet 6:1315–1322 [DOI] [PubMed] [Google Scholar]
- Tassone F, Hagerman RJ, Gane LW, Taylor AK (1999) Strong similarities of the FMR1 mutation in multiple tissues: postmortem studies of a male with a full mutation and a male carrier of a premutation. Am J Med Genet 84:240–244 [PubMed] [Google Scholar]
- Taylor AK, Tassone F, Dyer PN, Hersch SM, Harris JB, Greenough WT, Hagerman RJ (1999) Tissue heterogeneity of the FMR1 mutation in a high-functioning male with fragile X syndrome. Am J Med Genet 84:233–239 [PubMed] [Google Scholar]
- Verkerk AJ, Pieretti M, Sutcliffe JS, Fu YH, Kuhl DP, Pizzuti A, Reiner O, Richards S, Victoria MF, Zhang FP, et al (1991) Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell 65:905–914 [DOI] [PubMed] [Google Scholar]
- White PJ, Borts RH, Hirst MC (1999) Stability of the human fragile X (CGG)n triplet repeat array in Saccharomyces cerevisiae deficient in aspects of DNA metabolism. Mol Cell Biol 19:5675–5684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohrle D, Hennig I, Vogel W, Steinbach P (1993) Mitotic stability of fragile X mutations in differentiated cells indicates early post-conceptional trinucleotide repeat expansion. Nat Genet 4:140–142 [DOI] [PubMed] [Google Scholar]
- Wohrle D, Kennerknecht I, Wolf M, Enders H, Schwemmle S, Steinbach P (1995) Heterogeneity of DM kinase repeat expansion in different fetal tissues and further expansion during cell proliferation in vitro: evidence for a casual involvement of methyl-directed DNA mismatch repair in triplet repeat stability. Hum Mol Genet 4:1147–1153 [DOI] [PubMed] [Google Scholar]
- Wohrle D, Salat U, Glaser D, Mucke J, Meisel-Stosiek M, Schindler D, Vogel W, Steinbach P (1998) Unusual mutations in high functioning fragile X males: apparent instability of expanded unmethylated CGG repeats. J Med Genet 35:103–111 [DOI] [PMC free article] [PubMed] [Google Scholar]