Abstract
Although somatic mutations in a number of genes have been associated with development of human tumors, such as lipomas, relatively few examples exist of germline mutations in these genes. Here we describe an 8-year-old boy who has a de novo pericentric inversion of chromosome 12, with breakpoints at p11.22 and q14.3, and a phenotype including extreme somatic overgrowth, advanced endochondral bone and dental ages, a cerebellar tumor, and multiple lipomas. His chromosomal inversion was found to truncate HMGA2, a gene that encodes an architectural factor involved in the etiology of many benign mesenchymal tumors and that maps to the 12q14.3 breakpoint. Similar truncations of murine Hmga2 in transgenic mice result in somatic overgrowth and, in particular, increased abundance of fat and lipomas, features strikingly similar to those observed in the child. This represents the first report of a constitutional rearrangement affecting HMGA2 and demonstrates the role of this gene in human growth and development. Systematic genetic analysis and clinical studies of this child may offer unique insights into the role of HMGA2 in adipogenesis, osteogenesis, and general growth control.
HMGA2 (MIM 600698) encodes an architectural factor belonging to the high-mobility group (HMG) of proteins involved in the pathogenesis of a variety of benign mesenchymal tumors, including uterine leiomyomas, pulmonary chondroid hamartomas, fibroadenomas, pleomorphic salivary gland adenomas, and lipomas (Sandros et al. 1990; Sreekantaiah et al. 1991; Bullerdiek et al. 1993; Schoenberg Fejzo et al. 1995; Kazmierczak et al. 1996). HMG proteins alter chromatin architecture and thus participate in the regulation of gene expression. Exons 1–3 encode AT hook domains that bind to the minor groove of AT-rich DNA. The remaining two exons encode an acidic carboxy terminus, which is of unknown function but has been hypothesized to confer specificity. DNA binding by HMGA2 induces conformational changes in the double helix, allowing formation of transcriptional complexes that, in turn, can either promote or inhibit transcription (Reeves and Beckerbauer 2001).
In vivo data have provided additional biological evidence for the function of HMGA2. Mice with homozygous Hmga2 disruption are born with diminished body size and extremely decreased fat levels (Zhou et al. 1995), whereas mice expressing a truncated Hmga2, including only the first three exons under the regulatory control of the cytomegalovirus (CMV) promoter, develop gigantism and lipomatosis (Battista et al. 1999). Expression of a similarly truncated Hmga2 construct under the control of the murine major histocompatibility complex class I H-2Kb promoter gives rise to mice with increased adipose levels and an unusually high frequency of lipomas (Arlotta et al. 2000). Together, these in vivo studies suggest a role for HMGA2 in adipose metabolism and especially in lipomagenesis.
DGAP103 is an 8-year-old boy with a pericentric inversion of chromosome 12, inv(12)(p11.22q14.3), whose phenotype includes postnatal onset of extreme somatic overgrowth, advanced endochondral bone and dental ages with megaepiphyseal dysplasia, and multiple subcutaneous lipomas. Other features include persistent thrombocytopenia, arthritis, brachydactyly, a stable cerebellar tumor, and facial dysmorphism.
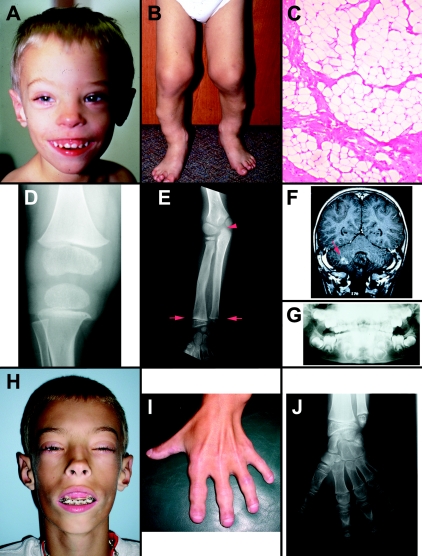
The boy was born at full term, by vaginal delivery, to a 26-year-old gravida 2, para 1–2 woman and an unrelated 25-year-old man. The pregnancy was complicated by premature labor at 30 wk gestation, requiring terbutaline administration through 34 wk of gestation. Birth parameters included weight of 3,267 g (25th–50th percentiles) and length of 49.5 cm (25th–50th percentiles). No anomalies were noted at birth, and the boy had an uneventful neonatal course with mild jaundice that did not require treatment. However, he was referred for genetic evaluation at 9 mo of age for steadily increasing growth parameters, with height, weight, and occipital-frontal circumference all at or above the 95th percentile. In addition to developing mildly dysmorphic facial features and brachydactyly (fig. 1A), he was noted to have bilateral lower extremity subcutaneous nodules (fig. 1B). Biopsy of one of these nodules showed mature adipocytes and fibrovascular tissue (fig. 1C). Findings suggestive of a skeletal dysplasia included bowing of the lower extremities, rhizomelic shortening, and progressive joint enlargement with some reduced range of motion. Radiologic studies at 1, 4, and 8 years of age revealed flared, enlarged epiphyses with shortened long and tubular bones and advanced endochondral bone age (fig. 1D, 1E, and 1J). Additional skeletal abnormalities included megaepiphyseal dysplasia with platyspondyly (not shown); shortened, broad metacarpals; and rhizomelic shortening, as confirmed by the International Skeletal Dysplasia Registry (Cedars-Sinai Medical Center, Los Angeles). He developed premature dentition, with upper incisors erupting at age 3 mo and eight teeth present by 5 mo; a panoramic dental X-ray at 4 years of age showed advanced dental age, with enlarged and possibly supernumerary teeth (fig. 1G). Because of concerns of macrocephaly, he underwent cranial imaging at 27 mo of age. Magnetic resonance imaging revealed a right cerebellar lesion measuring 2 cm, located ventral to the cerebellar nuclei (fig. 1F). On the basis of the imaging studies, the differential diagnosis included hamartoma and low-grade cerebellar glioma. Motor and sensory exams showed normal results. The brain lesion has been followed by use of serial imaging and has not progressed by age 8 years; because the mass appeared stable, biopsy was not performed. Furthermore, mild thrombocytopenia and leukopenia (with platelet counts as low as 60,000) were noted at age 5 years, at which time a bone marrow biopsy was performed that showed trilineage hematopoiesis with mildly reduced cellularity but no evidence of malignancy. Cytogenetic analysis of the bone marrow biopsy was reported as 46,XY,inv(12)(p12.2q15) (fig. 2A). The same cytogenetic result was observed independently when peripheral blood and lipoma specimens were analyzed, confirming this rearrangement to be constitutional in nature.
Figure 1.
Clinical features of DGAP103. A, Facial features at age 27 mo, showing macrocephaly, flat supraorbital ridges, widely spaced eyes, and prominent alveolar ridges. B, Bilateral bowing and multiple lipomas on lower extremities at age 27 mo. C, Hematoxylin- and eosin-stained tissue sections of right lateral leg subcutaneous soft-tissue mass (biopsied at age 14 mo) confirms a benign lipoma comprised of mature adipocytes and paucicellular collagen. D, Megaepiphyseal flaring of the femur and tibia at the knee joint at 1 year of age. E, Broad distal ulna and radius with enlarged epiphyses (arrows) at 4 years of age. Advanced bone age is indicated by initial ossification of the trochlea of the ulna (arrowhead) which occurs, on average, at ∼7 years of age). F, Cerebellar mass in the right cerebellar hemisphere (arrow). G, Disorganization and advanced dental age evident both for erupted and unerupted teeth on panoramic dental X-ray taken at 4 years of age. H, Facial features with more severe flattening of supraorbital ridges, narrow eye openings, prominent ears with protruding antihelix, and significant gum hypertrophy with recessed mandible at 8 years of age. I, Brachydactyly and enlarged interphalangeal joints with joint restriction at 8 years of age. J, Advanced bone age of 13.5 years, without evidence of epiphyseal closure, on wrist radiograph taken at 8 years of age.
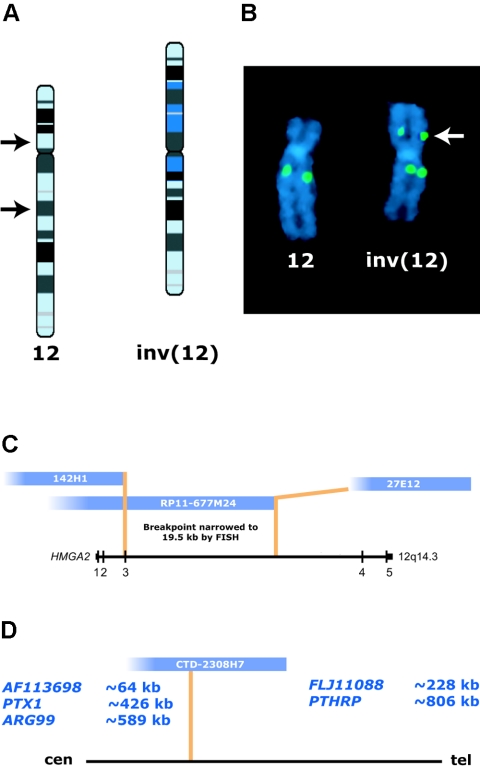
Figure 2.
FISH mapping of chromosome 12 breakpoints. A, Idiograms for normal chromosome 12 (left) and inverted homolog (right) showing inv(12)(p11.22q14.3). B, Representative hybridization pattern following FISH with BAC clone RP11-677M24, which maps to band q14.3 on chromosome 12 and spans the long (q) arm breakpoint. One green signal is present on the normal chromosome 12, whereas two green signals can be seen on the inverted homolog (arrow indicates signal translocated to the short arm by the inversion). C, 12q14.3 breakpoint mapping. Initial FISH mapping of the 12q14.3 breakpoint showed that cosmids 142H1 and 27E12 flank the breakpoint, placing it between exons 2 and 4 of HMGA2. BAC RP11-677M24 was split in the inverted chromosome 12, narrowing the breakpoint region to an ∼19.5-kb interval. D, 12p11.22 breakpoint mapping. BAC CTD-2308H7 is split by the inversion. No genes are known to be disrupted by this breakpoint. Potential candidate genes and their respective distances from the 12p11.22 breakpoint are indicated.
On most recent evaluation, at 8 years of age, the patient's growth parameters were remarkable for extreme overgrowth, with stature of 169 cm (50th percentile for a 15-year-old boy), weight of 50.9 kg (50th percentile for a 14-year-old boy), and head circumference of 56 cm (50th percentile for an adult male). Limited range of motion was noted for the knees, ankles, wrists, and fingers, most likely secondary to markedly enlarged epiphyses. He had brachydactyly of hands and feet, as well as shortened distal phalanges and curved, redundant nails (fig. 1I). Partial flexion contractures were present in the fingers, and the lower legs were bowed bilaterally. He reported arthritic symptoms that became especially severe during the winter. Bone age was advanced and was equivalent to that of a 13.5-year-old male, with no evidence of epiphyseal closure (fig. 1J). Progressive craniofacial dysmorphism also was observed and included flat supraorbital ridges, ocular hypertelorism, upslanting narrow palpebral fissures, large ears with prominent antihelices, prominent nasal bridge with anteverted nares, hypertrophy of alveolar ridges with inability to close the mouth, and retrognathia (fig. 1H). Several gooseflesh-like, hypopigmented lesions were present on the lower segments of the upper extremities and on the face, as well as prominent papillae on the tongue; these lesions appeared to be transient over several examinations. The visible lipomas became less prominent, and their number appeared to diminish over the course of progressive evaluations. At this same examination, peripheral blood was analyzed for levels of total and ionized calcium, vitamin D, phosphate, alkaline phosphate, parathyroid hormone (PTH), and associated receptor (PTHrP; data not shown). Levels of each of these analytes were considered to be within normal physiological limits.
Breakpoint mapping studies were undertaken to fine map the chromosomal rearrangement, using FISH and other molecular methods. For this purpose, a peripheral blood specimen was collected after informed consent was obtained, in accordance with institutional policies and procedures for human research at Partners Health Care System. Cell transformation was performed at the Massachusetts General Hospital Cell Transformation Core, using standard protocols. Transformed lymphoblastoid cells were then cultured to generate sufficient material for GTG banding, FISH mapping, and nucleic acid extraction using standard protocols (see below).
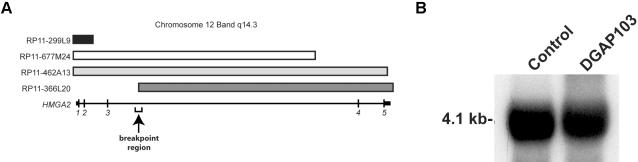
Metaphase chromosome spreads were prepared using standard cytogenetic protocols (Ney et al. 1993). FISH was performed with direct-labeled cosmid or BAC probes to map each inversion breakpoint. BAC clones were selected using the University of California–Santa Cruz (UCSC) Genome Browser (May 2004 freeze) and were hybridized in differentially labeled pairs (Spectrum Green and Spectrum Red or Orange [Amersham]). At least 10 metaphases were scored per hybridization. FISH was performed initially with cosmid clones 142H1 and 27E12 at the HMGA2 locus (Schoenmakers et al. 1995). In a structurally normal chromosome 12, HMGA2 is oriented at 12q14.3 from centromere (5′) to telomere (3′). Cosmid 27E12, which includes the last two exons of HMGA2, remained on the chromosome 12 long arms of both the normal and the inverted homologs (fig. 2C), indicating that the breakpoint was distal to this clone. In contrast, signals corresponding to cosmid 142H1, which includes exons 1 and 2 of HMGA2, were transposed to the short arm of the inverted chromosome 12, indicating an intragenic rearrangement of HMGA2. Successive FISH experiments were then performed using BAC clones to refine this breakpoint further. The hybridization of two BAC clones (RP11-677M24 and RP11-462A13) was split on the inverted homolog (fig. 2B). As a result, the 12q breakpoint was narrowed to an interval flanked by BAC clones RP11-366L20 (defining the distal boundary) and RP11-677M24, for which the signal was split between the short and long arms (defining the proximal boundary) (fig. 2B and 2C). This refined interval flanking the long arm breakpoint was ∼19.5 kb. On the basis of our FISH mapping data, one of the bands involved in this break was reassigned to q14.3, rather than to q15, as defined by initial GTG banding.
Mapping of the short arm breakpoint was accomplished by similar, successive BAC hybridizations. As diagrammed in figure 2D, this breakpoint interval was narrowed to a region of ∼19.5 kb flanked by clones CTD-2308H7, which was split by the inversion, and RP11-954D8 (not shown), which was present on the long arm. On the basis of this set of FISH mapping experiments, the short arm breakpoint was reassigned to p11.22, rather than to p12.2. Interestingly, the UCSC Genome Browser indicates relatively few known or putative genes in the 12p11.22 region. PTX1, which encodes a nuclear protein down-regulated in prostate carcinoma (fig. 2D), and ARG99, which encodes an UDP-N-acetylglucosamine-peptide N-acetylglucosaminyl-transferase 110-kDa subunit, map within the same cytogenetic band but centromeric to the breakpoint (∼426 kb and ∼589 kb away, respectively) (fig. 2D), as do sequences for the hypothetical protein AF113698 (∼64 kb away); hypothetical protein FLJ11088 maps ∼228 kb telomeric to the breakpoint. The only known gene in 12p11.22 with a corresponding entry in the Protein Data Bank is PTHRP (parathyroid hormone–related peptide, also known as “PTHLH” or “parathyroid hormone–like hormone” [MIM 168470]). At a distance of ∼806 kb telomeric to the 12p11.22 breakpoint, it is possible that a fusion product would form between exons 1–3 of HMGA2 and PTHRP (both transcribed centromere to telomere); however, no such fusion product was detected using specific HMGA2-PTHRP primers and 3′ rapid amplification of cDNA ends (RACE) (data not shown). Furthermore, no known genes map directly within BAC CTD-2308H7, suggesting that disruption of a specific gene at 12p11.22 is unlikely to contribute to the phenotype of our patient.
Southern blot analysis was initiated to refine the data generated by FISH. Total genomic DNA was isolated from DGAP103 and control lymphoblastoid cells by use of the PureGene DNA isolation kit (Gentra Systems). Restriction enzymes BclI and XbaI were used independently to digest 8 μg of DNA. Digested DNA was electrophoresed in a 1% agarose gel, transferred onto Nylon-N+ membrane (Amersham), and crosslinked using a Stratalinker (Stratagene). For probe preparation, primers were designed to amplify DNA within the overlapping region between these two fragments. These included primers 103-4Lb (5′-TGAACATGGCCATCTCTGTG-3′) and 103-4Rb (5′-CAGAGCCTACCATTGTCTTGG-3′). The resulting 707-bp amplicon was labeled with 32α-dCTP using the MegaPrime DNA labeling mix (Amersham). Digestion with the restriction enzyme XbaI identified a novel fragment (fig. 3A) and refined the breakpoint interval to a 2,115-bp segment within the third intron of HMGA2.
Figure 3.
Molecular analyses. A, Southern blot analysis refines the 12q14.3 breakpoint within HMGA2 to a 2,115-bp interval that maps within intron 3. This intron is a frequent target of disruption in mesenchymal tumors. B, Northern blot analysis performed using a cDNA probe against the entire HMGA2 coding sequence identifies a wild-type 4.1-kb transcript in total RNA from lymphoblastoid cells established from DGAP103 (right) and control (left) peripheral blood specimens.
By use of standard protocols (Sambrook and Russell 2001), northern blot analysis was performed to test for expression of aberrant transcripts. A full-length HMGA2 cDNA clone (kindly provided by B. Kazmierczak) was used as a probe against total RNA isolated from lymphoblastoid cultures established from DGAP103 and from a control lymphoblastoid sample.
From this nonquantitative analysis, a 4.1-kb band representing the wild-type transcript (fig. 3B) was observed in both samples, as was a faint and slightly smeared band (∼10 kb) in the RNA from DGAP103 (not shown). Although it is possible that this larger product represents a fusion transcript, detection of a run-on transcription product cannot be excluded. However, repeated attempts at 3′ RACE performed by use of a poly-T primer for first-strand synthesis were unsuccessful at amplifying any product (data not shown). To assess quantitative differences in HMGA2 expression, real-time PCR was performed by use of an iCycler Optical System (Bio-Rad), with total RNA derived from DGAP103 lymphoblasts and control lymphoblasts without rearrangement affecting chromosome 12 (data not shown). Primers selected included 5′-AGTCCCTCTAAAGCAGCTCAAAAG-3′, within exon 2 of HMGA2, and 5′-GCCATTTCCTAGGTCTGCCTC-3′, within exon 3. The probe, 5′-AGAAGCCACTGGAGAAAAACGGCCA-3′, was labeled with 5′-FAM/3′-BHQ-1 (Biosearch Technologies) and is within the amplified sequence. Expression levels were determined from an average of triplicate readings, by use of the same sample preparation. This analysis indicated slightly increased levels of the 5′ RNA transcript, including exon 3, in DGAP103, relative to the levels in the control. Specifically, lymphoblasts from DGAP103 had a relative HMGA2 expression level of 1.4 times that in the control.
Of individuals with balanced de novo chromosomal rearrangements, ∼6% are born with anatomical, biochemical, developmental, or behavioral abnormalities, some of which will be attributable to disruption of genes critical to growth and development. In analysis of DGAP103, the unusual phenotype and constitutional chromosomal inversion implicated a gene, HMGA2, that is known to be involved in both normal and pathologic development of mesenchymal tissues. On the basis of the hypothesis that HMGA2 involvement was likely, FISH mapping and Southern blot analysis of the 12q14.3 region were used to confirm and narrow the suspected HMGA2 breakpoint to an ∼2-kb interval within the ∼140-kb third intron. Involvement of a gene at the 12p11.22 breakpoint region, either directly or by position effect, was not detected and seems unlikely.
Because DGAP103 developed numerous lipomas on his lower extremities and because of the known role of HMGA2 in lipomagenesis, discovery of a constitutional disruption of HMGA2 became especially relevant. Lipomas are one of the most common neoplasms in humans, and rearrangement of HMGA2 in these and other mesenchymal tumors is well established (Sreekantaiah et al. 1991; Fedele et al. 2001). Approximately 50% of lipomas are karyotypically abnormal, and, of those, the most frequently rearranged region involves chromosome 12q14-q15 (Ashar et al. 1995); both translocations and inversions have been described (Mitelman Database of Chromosome Aberrations in Cancer).
Somatic rearrangements of HMGA2 may result in several different forms of gene disruption and, subsequently, different forms of transcripts. Intragenic disruption of HMGA2 in mesenchymal tumors has been shown to involve formation of chimeric transcripts. For lipomas in particular, these partner genes include LPP, a gene with highly conserved LIM motifs that are active in a variety of developmentally regulated processes (Freyd et al. 1990). Chimeric HMGA2 gene products are uncommon among uterine leiomyomata and other generally nonlipomatous mesenchymal tumors (Quade et al. 2003). Interestingly, in vitro experiments demonstrated that either truncated HMGA2 (exons 1–3) or an HMGA2 fusion (HMGA2-LPP) can transform NIH 3T3 murine fibroblasts (Fedele et al. 1998). It is possible, therefore, that HMGA2 contributes to tumorigenesis following either type of structural alteration. Although some data support a role in regulation of cellular proliferation, the specific contribution of either truncated or chimeric HMGA2 transcripts to tumorigenesis is unknown. For DGAP103, it is likely that the lipomatous lesions noted are due to disruption of HMGA2.
The genomic structure of HMGA2 includes an extremely large third intron (∼140 kb) that has been described as the most frequent target of cytogenetic rearrangements in human neoplasms (Kazmierczak et al. 1998). Several HMGA2 rearrangements mapping within intron 3 have been described in detail (Ashar et al. 1995; Schoenmakers et al. 1995); thus, we speculated initially that the breakpoint would map there. If so, the chromosomal inversion in DGAP103 would result in translocation of the first three DNA-binding domain exons to the short arm of chromosome 12. Indeed, both FISH and Southern blotting confirmed that the breakpoint mapped within intron 3 and that it separated the three DNA-binding AT hook domains from the acidic carboxy terminus of the protein encoded by exons 4 and 5.
Constitutional disruption of HMGA2 could contribute to the phenotype in DGAP103 through a number of different mechanisms. First, formation of a fusion transcript between the 5′ end of HMGA2 and the 3′ portion of another gene, similar to what has been observed in lipomas, might favor a gain-of-function mechanism with a novel function or altered specificity of the chimeric protein. This scenario, however, is not supported by our data. Notably, a prospective fusion partner within a plausible distance and in the correct orientation relative to the breakpoint on the short arm of chromosome 12 was not identified. Furthermore, 3′ RACE experiments did not identify any chimeric transcripts (data not shown). In a second model, expression of a truncated HMGA2 allele could create a dysregulated protein with novel function or altered specificity related to the developmental abnormalities in DGAP103. Although analysis by northern blotting and real-time PCR did not detect such a truncated transcript in lymphoblast RNA (i.e., derived from B cells), we cannot exclude the possibility that a truncated HMGA2 transcript might not be expressed stably in lymphoblasts, the only cell type available for testing. A third potential mechanism is up-regulation of the wild-type allele following disruption of the translocated allele (Tkachenko et al. 1997; Ashar et al. 2003). HMGA2 transcript abundance, measured by northern blotting and real-time PCR in RNA from the DGAP103 lymphoblastoid cell line, was slightly elevated in DGAP103 compared with a control. The full-length transcript detected by northern blot analysis presumably is attributed to the normal allele. In this scenario, it is possible that a compensatory response might up-regulate HMGA2 expression in other tissues.
Battista and coauthors (1999) reported that expression of a truncated murine Hmga2 induced gigantism associated with lipomatosis. In these transgenic animals, expression of only the first three exons of Hmga2 was driven by the CMV promoter. By 12 mo of age, these mice showed an average 15% increase in body length and a 36% weight gain compared with wild-type littermates. An expansion of retroperitoneal adipose tissue was observed, consistent with a role for Hmga2 in murine lipoma development. The authors proposed that disruption of Hmga2 led to up-regulation of the gene and subsequent stimulation of adipogenesis. No abnormalities of the skeletal system or dentition were described.
Subsequently, Arlotta and coauthors (2000) described transgenic mice expressing a truncated Hmga2 (also lacking the carboxy terminus) driven by the H-2Kb promoter. Although lipomas are exceedingly common in humans, they rarely arise spontaneously in mice (Arlotta et al. 2000). Therefore, it was significant when a high frequency of lipomas was seen, lending further support to a role for truncated Hmga2 in adipocyte growth. As in the model reported above, no abnormalities of the skeletal system or dentition were described.
In contrast to murine models with overexpression of the three AT hook domains, Hmga2−/− mice demonstrate the “pygmy” phenotype, in which dramatic reductions in body fat and small stature are hallmark features (Zhou et al. 1995). Hmga2−/− mice are resistant to diet-induced obesity, and reduction of Hmga2 expression protects mice from leptin-deficiency–induced obesity (Anand and Chada 2000). This transgenic model differs from the genotype in our patient in that haploinsufficiency fails to eliminate HMGA2 expression, suggesting a different mechanism of altered expression underlying the DGAP103 phenotype. To date, HMGA2 function has not been associated with regulation of any of the known somatic overgrowth genes, underscoring the need for further study of its role in growth.
These murine models support a role for HMGA2 in adipogenesis and implicate expression of the truncated HMGA2 in the appearance of numerous lipomas and somatic overgrowth in our patient. However, some of the other clinical features may also be explained simply by HMGA2 disruption. For example, the stable right-sided cerebellar mass and the prominent tongue papillomas may represent benign hamartomatous lesions, analogous to those described in other overgrowth conditions such as Cowden/Bannayan-Riley-Ruvalcaba syndrome (MIM 158350 and MIM 153480) and Proteus syndrome (MIM 176920) (Eng 2003).
Low-level expression of HMGA2 has been demonstrated in a few human adult tissues (e.g., lung and kidney), but more-abundant levels of transcript exist in certain tissues, primarily during embryonic and fetal development (Gattas et al. 1999). HMGA2 has been shown to be expressed in hematopoietic stem cells and in certain leukemias (Rommel et al. 1997; Santulli et al. 2000). Andrieux and coauthors (2004) observed rearrangements of 12q13-15 in the hematopoietic cells of patients with a chronic myeloproliferative disorder (myelofibrosis with myeloid metaplasia) and proposed that dysregulation and increased expression of HMGA2 in myeloid progenitor cells may contribute to disease. Furthermore, extragenic rearrangement of HMGA2 also has been reported in acute lymphocytic leukemia (Pierantoni et al. 2003). Both studies raise the question of the role HMGA2 might play in normal hematopoiesis and, by extension, the role of HMGA2 in the chronic thrombocytopenia and neutropenia in DGAP103.
Moreover, Broberg and coauthors (1999, 2001) showed that a rearranged HMGA2 is expressed in the synovia of patients with osteoarthritis; these rearrangements seem to occur without any known propensity for mesenchymal tumor formation at the growth plate. Therefore, it is possible that disruption of HMGA2 may also play a role in mesenchymal hyperplasia rather than being restricted uniquely to the neoplastic process. HMGA2 rearrangements have recently been reported in bone and soft-tissue tumors, a subset of which express only a truncated HMGA2 transcript (exons 1–3) (Dahlen et al. 2003). Abnormal HMGA2 expression may be the cause of epiphyseal overgrowth in DGAP103, and, although we cannot exclude the possibility that his arthritis arose as a secondary effect of enlarged epiphyses, this documented expression pattern of HMGA2 in synovial tissues also raises the possibility that aberrant HMGA2 expression is a contributing factor to the joint pain.
The complete phenotype of DGAP103 need not be explained entirely by disruption of HMGA2, and analysis of the second inversion breakpoint could confer insight into potential genetic causes for the remainder of the clinical features. However, the split BAC clone (CTD-2308H7) identified at the p11.22 breakpoint included no known genes, a finding consistent with our inability to identify a rearrangement partner for HMGA2 by 3′ RACE. This observation also supports the hypothesis that truncation of HMGA2, rather than fusion with a novel sequence, could be responsible for dysregulating this gene.
In an effort to consider a role for position effect, we analyzed the annotated genomic sequence for a distance of 1 Mb on either side of the p11.22 inversion breakpoint. Centromeric to the p11.22 breakpoint is PTX1, which has a postulated role in prostate cancer is but unlikely to be involved in the current phenotype (Kwok et al. 2001). In the 1-Mb interval telomeric to the p11.22 breakpoint, only a single gene corresponded to a known protein product: PTHLH (i.e., PTHRP). The protein encoded by PTHRP, parathyroid hormone–related peptide (MIM 168470), is involved in regulation of endochondral bone development and epithelial-mesenchymal interactions, and it signals specifically through the parathyroid hormone receptor (PTHR1). Mice homozygous for a null mutation in PTHRP show abnormalities of endochondral bone development, including premature maturation of chondrocytes and accelerated bone formation similar to that observed in some forms of a dwarfing chondrodysplasia (Karaplis et al. 1994). Unfortunately, there is no existing animal model for overexpression of PTHRP to allow comparison of phenotypes.
PTHRP maps at a great distance from the p11.22 inversion breakpoint, and its currently recognized genomic structure is not disrupted by the rearrangement. However, there is precedence for position effect acting over comparable distances (∼806 kb between the p11.22 breakpoint and PTHRP). SOX9, a gene involved in campomelic dysplasia and, coincidentally, a target of PTHRP signaling in bone development, has been reported to exert an effect at a distance of 950 kb away from a chromosomal breakpoint (Pfeifer et al. 1999; Huang et al. 2001). Similarly, expression of POU3F4, involved in X-linked deafness, can be effected over a distance of ∼900 kb (de Kok et al. 1996). However, despite the intriguing possibility that PTHRP might somehow contribute to the DGAP103 phenotype, particularly with regard to the advanced bone and dental ages, normal peripheral blood levels of PTH and PTHrP do not support the possibility that position effect exerted through PTHRP is a contributing factor in the bone abnormalities.
Certainly, position effects of unknown genes mapping within a critical interval surrounding the p11.22 breakpoint—or even the q14.3 breakpoint—remain possibilities in this novel phenotype. Recently, a novel category of mammalian genomic sequences more conserved than protein-encoding sequences has been identified. These conserved nongenic sequences (CNGs) could be candidates for disruption in chromosomal rearrangements such as the pericentric inversion in our patient. CNGs appear to be located in random intergenic regions and do not share any distance requirements with any previously characterized regulatory elements (Dermitzakis et al. 2003). It has been suggested that CNGs could be involved in either cis or trans chromosomal interactions affecting genome function and regulation (Dermitzakis et al. 2004). To date, CNGs have only been studied for human chromosome 21. It is worth noting, however, that the region surrounding the p11.22 breakpoint in DGAP103 also shares significant homology with the murine genome and could harbor undiscovered coding or noncoding elements such as CNGs.
In summary, we report an 8-year-old male with a de novo chromosomal inversion resulting in aberrant adiposity and somatic overgrowth. These phenotypic features are attributed to a constitutional disruption of HMGA2—to our knowledge, an event not previously observed in humans. This phenotype, particularly with respect to overgrowth and lipomagenesis, is strikingly similar to that of murine models expressing truncated Hmga2 transcripts. The possibility that HMGA2 participates in human bone or tooth development must also be considered, on the basis of the phenotype of DGAP103. A pleiotropic effect of HMGA2 in this human developmental disorder and the potential contribution of any position effect of nearby genetic elements warrant further study.
Acknowledgments
The authors wish to acknowledge the patient and his family, for their participation in the Developmental Genome Anatomy Project (DGAP); Drs. Roberta Pagon and Louanne Hudgins, for additional clinical evaluation; Dr. Stephen Done, for radiological expertise; Drs. Henry Kronenberg and Richard Maas, for insightful discussions; Dr. Weining Lu, for his guidance with real-time PCR analysis; Dr. Natalia Leach, for her assistance with figure 2; the International Skeletal Dysplasia Registry, Cedars-Sinai Hospital, Los Angeles; and Ms. Amy Bosco of DGAP. This work was supported by National Institutes of Health grant GM61354 (to C.C.M.).
Electronic-Database Information
URLs for data presented herein are as follows:
- Mitelman Database of Chromosome Aberrations in Cancer, http://cgap.nci.nih.gov/Chromosomes/Mitelman
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/
- UCSC Genome Browser, May 2004 assembly, http://genome.ucsc.edu/
References
- Anand A, Chada K (2000) In vivo modulation of Hmgic reduces obesity. Nat Genet 24:377–380 [DOI] [PubMed] [Google Scholar]
- Andrieux J, Demory JL, Dupriez B, Quief S, Plantier I, Roumier C, Bauters F, Lai JL, Kerckaert JP (2004) Dysregulation and overexpression of HMGA2 in myelofibrosis with myeloid metaplasia. Genes Chromosomes Cancer 39:82–87 [DOI] [PubMed] [Google Scholar]
- Arlotta P, Tai AK-F, Manfioletti G, Clifford C, Jay G, Ono SJ (2000) Transgenic mice expressing a truncated form of the high mobility group I-C protein develop adiposity and an abnormally high prevalence of lipomas. J Biol Chem 275:14394–14400 [DOI] [PubMed] [Google Scholar]
- Ashar HR, Schoenberg Fejzo M, Tkachenko A, Zhou X, Fletcher JA, Weremowicz S, Morton CC, Chada K (1995) Disruption of the architectural factor HMGIC: DNA-binding AT hook motifs fused in lipomas to distinct transcriptional regulatory domains. Cell 82:57–65 [DOI] [PubMed] [Google Scholar]
- Ashar HR, Tkachenko A, Shah P, Chada K (2003) HMGA2 is expressed in an allele-specific manner in human lipomas. Cancer Genet Cytogenet 143:160–168 [DOI] [PubMed] [Google Scholar]
- Battista S, Fidanza V, Fedele M, Klein-Szanto JP, Outwater E, Brunner H, Santoro M, Croce CM, Fusco A (1999) The expression of a truncated HMGIC gene induces gigantism associated with lipomatosis. Cancer Res 59:4793–4797 [PubMed] [Google Scholar]
- Broberg K, Hoglund M, Limon J, Lindstrand A, Toksvik-Larsen S, Mandahl N, Mertens F (1999) Rearrangement of the neoplasia-associated gene HMGIC in synovia from patients with osteoarthritis. Genes Chromosomes Cancer 24:278–282 [PubMed] [Google Scholar]
- Broberg K, Tallini G, Hoglund M, Lindstrand A, Toksvig-Larsen S, Mertens F (2001) The tumor-associated gene HMGIC is expressed in normal and osteoarthritis-affected synovia. Mod Pathol 14:311–317 [DOI] [PubMed] [Google Scholar]
- Bullerdiek J, Wobst G, Meyer-Bolte K, Chilla R, Haubrich J, Thode B, Bartnetizke S (1993) Cytogenetic subtyping of 220 salivary gland pleomorphic adenomas: correlation to occurrence, histological subtype, and in vitro cellular behavior. Cancer Genet Cytogenet 65:27–31 [DOI] [PubMed] [Google Scholar]
- Dahlen A, Mertens F, Rydholm A, Brosjo O, Wejde J, Mandahl N, Panagopoulos I (2003) Fusion, disruption, and expression of HMGA2 in bone and soft tissue chondromas. Mod Pathol 16:1132–1140 [DOI] [PubMed] [Google Scholar]
- de Kok YJ, Vossenaar ER, Cremers CW, Dahl N, Laporte J, Hu LJ, Lacombe D, Fischel-Ghodsian N, Friedman RA, Parnes LS, Thorpe P, Bitner-Glindzicz M, Pander HJ, Heilbronner H, Graveline J, den Dunnen JT, Brunner HG, Ropers HH, Cremers FP (1996) Identification of a hot spot for microdeletions in patients with X-linked deafness type 3 (DFN3) 900 kb proximal to the DFN3 gene POU3F4. Hum Mol Genet 5:1229–1235 [DOI] [PubMed] [Google Scholar]
- Dermitzakis ET, Kirkness E, Schwarz S, Birney E, Reymond A, Antonarakis SE (2004) Comparison of human chromosome 21 conserved nongenic sequences (CNGs) with the mouse and dog genomes shows that their selective constraint is independent of their genic environment. Genome Res 14:852–859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dermitzakis ET, Reymond A, Scamuffa N, Ucla C, Kirkness E, Rossier C, Antonarakis SE (2003) Evolutionary discrimination of mammalian conserved nongenic sequences (CNGs). Science 302:1033–1035 [DOI] [PubMed] [Google Scholar]
- Eng C (2003) PTEN: one gene, many syndromes. Hum Mutat 22:183–198 [DOI] [PubMed] [Google Scholar]
- Fedele M, Battista S, Manfioletti G, Croce CM, Giancotti V, Fusco A (2001) Role of the high mobility group A proteins in human lipomas. Carcinogenesis 22:1583–1591 [DOI] [PubMed] [Google Scholar]
- Fedele M, Berlingiere MT, Scala S, Chiariotti L, Viglietto G, Rippel V, Bullerdiek J, Santoro M, Fusco A (1998) Truncated and chimeric HMGIC genes induce neoplastic transformation of NIH3T3 murine fibroblasts. Oncogene 17:413–418 [DOI] [PubMed] [Google Scholar]
- Freyd G, Kim SK, Horvitz HR (1990) Novel cysteine-rich motif and homeodomain in the product of the Caenorhabditis elegans cell lineage gene lin-11. Nature 344:876–879 [DOI] [PubMed] [Google Scholar]
- Gattas GJF, Quade BJ, Nowak RA, Morton CC (1999) HMGIC expression in human adult and fetal tissues and in uterine leiomyomata. Genes Chromosomes Cancer 25:316–322 [PubMed] [Google Scholar]
- Huang W, Chung IU, Kronenberg HM, de Crombrugghe B (2001) The chondrogenic transcription factor SOX9 is a target of signaling by the parathyroid hormone-related peptide in the growth plate of endochondral bones. Proc Natl Acad Sci USA 98:160–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karaplis AC, Luz A, Glowacki J, Bronson RT, Tybulewicz VLJ, Kronenberg HM, Mulligan RC (1994) Lethal skeletal dysplasia from targeted disruption of the parathyroid hormone-related peptide gene. Genes Dev 8:277–289 [DOI] [PubMed] [Google Scholar]
- Kazmierczak B, Bullerdiek J, Pham KH, Bartnitzke S, Wiesner H (1998) Intron 3 of HMGIC is the most frequent target of chromosomal aberrations in human tumors and has been conserved basically for at least 30 million years. Cancer Genet Cytogenet 103:175–177 [DOI] [PubMed] [Google Scholar]
- Kazmierczak B, Rosigkeit J, Wanschura S, Meyer-Bolte K, Van de Ven W, Kayser K, Krieghoff B, Kastendick H, Bartnitzke S, Bullerdiek J (1996) HMGIC rearrangements as the molecular basis for the majority of pulmonary chondroid hamartomas: a survey of 30 tumors. Oncogene 12:515–521 [PubMed] [Google Scholar]
- Kwok SC, Liu X, Daskal I (2001) Molecular cloning, expression, localization and gene organization of PTX1, a human nuclear protein that is downregulated in prostate cancer. DNA Cell Biol 20:349–357 [DOI] [PubMed] [Google Scholar]
- Ney PA, Andrews NC, Jane SM, Safer B, Purucker NE, Weremowicz S, Morton CC, Goff SC, Orkin SH, Nienhuis AW (1993) Purification of the human NF-E2 complex: cDNA cloning of the hematopoietic cell-specific subunit and evidence for an associated partner. Mol Cell Biol 13:5604–5612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer D, Kist R, Dewar K, Devon K, Lander ES, Birren B, Korniszewski L, Back E, Scherer G (1999) Campomelic dysplasia translocation breakpoints are scattered over 1 Mb proximal to SOX9: evidence for an extended control region. Am J Hum Genet 65:111–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierantoni GM, Santulli B, Caliendo I, Pentimalli F, Chiappetta G, Zanesi N, Santoro M, Bulrich F, Fusco A (2003) HMGA2 locus rearrangement in a case of acute lymphoblastic leukemia. Int J Oncol 23:363–367 [PubMed] [Google Scholar]
- Quade BJ, Weremowicz S, Neskey DM, Vanni R, Ladd C, Dal Cin P, Morton CC (2003) Fusion transcripts involving HMGA2 are not a common molecular mechanism in uterine leiomyomata with rearrangements in 12q15. Cancer Res 63:1351–1358 [PubMed] [Google Scholar]
- Reeves R, Beckerbauer L (2001) HMGI/Y proteins: flexible regulators of transcription and chromatin structure. Biochim Biophys Acta 1519:13–29 [DOI] [PubMed] [Google Scholar]
- Rommel B, Rogalla P, Jox A, Kalle CV, Kazmierczak B, Wolf J, Bullerdiek J (1997) HMGI-C, a member of the high mobility group family of proteins, is expressed in hematopoietic stem cells and in leukemic cells. Leuk Lymphoma 26:603–607 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell DW (2001) Molecular cloning: a laboratory manual. 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Sandros J, Stenman G, Mark J (1990) Cytogenetic and molecular observations in human and experimental salivary gland tumors. Cancer Genet Cytogenet 44:153–167 [DOI] [PubMed] [Google Scholar]
- Santulli B, Kazmierczak B, Napolitano R, Caliendo I, Chiappetta G, Rippe V, Bullerdiek J, Fusco A (2000) A 12q13 translocation involving the HMGIC gene in Richter transformation of a chronic lymphocytic leukemia. Cancer Genet Cytogenet 119:70–73 [DOI] [PubMed] [Google Scholar]
- Schoenberg Fejzo M, Yoon S-J, Montgomery KT, Rein MS, Weremowicz S, Krauter KS, Dorman TE, Fletcher JA, Mao J, Moir DT, Kucherlapati RS, Morton CC (1995) Identification of a YAC spanning the translocation breakpoints in uterine leiomyomata, pulmonary chondroid hamartoma, and lipoma: physical mapping of the 12q14-q15 breakpoint region in uterine leiomyomata. Genomics 26:265–271 [DOI] [PubMed] [Google Scholar]
- Schoenmakers EFPM, Wanschura S, Mols R, Bullerdiek J, Van den Berghe H, Van de Ven WJM (1995) Recurrent rearrangements in the high mobility group protein gene, HMGIC, in benign mesenchymal tumours. Nat Genet 10:436–443 [DOI] [PubMed] [Google Scholar]
- Sreekantaiah C, Leon SLP, Karakousis CP, McGee DL, Rappaport WD, Villar HV, Neal D, Fleming S, Wankel A, Herrington PN, Carmona R, Sandberg AA (1991) Cytogenetic profile of 109 lipomas. Cancer Res 51:422–433 [PubMed] [Google Scholar]
- Tkachenko A, Ashar HR, Meloni AM, Sandberg AA, Chada KK (1997) Misexpression of disrupted HMGI architectural factors activates alternative pathways of tumorigenesis. Cancer Res 57:2276–2280 [PubMed] [Google Scholar]
- Zhou X, Benson KF, Ashar HR, Chada K (1995) Mutation responsible for the mouse pygmy phenotype in the developmentally regulated factor HMGIC. Nature 376:771–774 [DOI] [PubMed] [Google Scholar]