Abstract
Asthma affects nearly 14 million people worldwide and has been steadily increasing in frequency for the past 50 years. Although environmental factors clearly influence the onset, progression, and severity of this disease, family and twin studies indicate that genetic variation also influences susceptibility. Linkage of asthma and related phenotypes to chromosome 6p21 has been reported in seven genome screens, making it the most replicated region of the genome. However, because many genes with individually small effects are likely to contribute to risk, identification of asthma susceptibility loci has been challenging. In this study, we present evidence from four independent samples in support of HLA-G as a novel asthma and bronchial hyperresponsiveness susceptibility gene in the human leukocyte antigen region on chromosome 6p21, and we speculate that this gene might contribute to risk for other inflammatory diseases that show linkage to this region.
We conducted a genomewide screen of families who participated in the Collaborative Study on the Genetics of Asthma (CSGA) (Collaborative Study on the Genetics of Asthma 1997; Xu et al. 2001). The strongest linkage signal in 129 white families was on chromosome 6p21 at marker D6S1281 (LOD=1.91; P=.003), which is 2.5 cM telomeric to the human leukocyte antigen (HLA) complex. All the evidence of linkage to asthma was found in the 35 white families ascertained in Chicago (LOD=3.6) (fig. 1A). We focused our subsequent studies on these families, as well as on 46 white child-parent trios with asthma (MIM 600807) also ascertained in Chicago and two other populations that had previously shown evidence of linkage of asthma-associated phenotypes to markers in this region (table 1) (Ober et al. 2000; Koppelman et al. 2002). Details of the statistical methods are described in appendix A (online only).
Figure 1.
Linkage to 6p21 in the Chicago families. A, The dashed line shows the results of the initial genome screen with framework markers. The solid line shows the results of five additional STRPs between framework markers D6S1281 and D6S1019. B, 1-Mb region from D6S258 to D6S265 with positional candidate genes. (All known genes [blue]—but not all pseudogenes [brown], STRPs [green], and intragenic SNPs [purple]—are included.) Circles = SNPs; triangles = in/dels; squares = STRPs; rectangle = HLA-A genotype, which is comprised of multiple SNPs. See appendix A (online only) for allele frequencies and results of association studies.
Table 1.
Clinical Characteristics of Patient Samples[Note]
| Characteristic | Chicago Familiesa,b(n=138) | Chicago Triosa,c(n=46) | Hutterite Familiesd(n=156) | Dutch Familiese(n=200) | Descendents ofDutch Familiese(n=327) |
| Sex (% male) | 59.4 | 60.6 | 53.2 | 62.0 | 42.3 |
| Asthma (%) | 100 | 100 | 48.0 | 100 | 40.7 |
| BHR (%) | 100 | 100 | 100 | 100 | 100 |
| Mean % predicted FEV1 (SD) | 88.64 (17.12) | 80.13 (16.83) | 95.58 (15.71) | 69.62 (24.68) | 89.49 (12.10) |
| Mean FEV1/FVC (SD) | .82 (.09) | .77 (.10) | .79 (.10) | .597f (.149) | .80f (.086) |
| Mean log IgE (SD) IU | 2.11 (.73) | 2.19 (.71) | 1.60 (.77) | 1.95 (.69) | 1.95 (.76) |
| Atopyg (%) | 67.2 | 79.8 | 51.0 | 81.9 | 59.1 |
Note.— These studies were approved by the institutional review boards at each institution. Not all individuals were genotyped at all marker loci.
CSGA families were ascertained through two siblings with asthma and were extended to include other affected relatives, never skipping more than one unaffected relative (Lester et al. 2001). Asthma was diagnosed as follows: (1) either a fall in baseline FEV1 by ⩾20% at ⩽25 mg/ml methacholine (BHR) or an increase of ⩾15% in baseline FEV1 after bronchodilator use; (2) two of the following symptoms: cough, wheeze, or shortness of breath (dyspnea); and (3) current medication use or doctor’s diagnosis of asthma. All participating relatives of the siblings were studied.
Family size ranged from 4 to 18 members (mean family size, 7 members).
Trios were individuals meeting the same criteria as the CSGA patients with asthma and their parents, who were not evaluated (n=46).
Hutterites (n=693), who are related to each other in a 13-generation, 1,623-member pedigree, were evaluated using a modified CSGA protocol (Ober et al. 2000). BHR in this group was defined as a fall in baseline FEV1 by ⩾20% at ⩽25 mg/ml methacholine.
The 200 Dutch families were recruited ∼30 years ago through a proband who was diagnosed as having asthma; they were reevaluated in the 1990s, along with their spouses, children, and grandchildren (Panhuysen et al. 1995). The families included 1,183 individuals. BHR in this group was defined as a fall in baseline FEV1 by ⩾20% at ⩽32 mg/ml histamine (30′ protocol).
FEV1/VC.
Defined as a positive skin-prick test to airborne allergens.
To further narrow the linked region, we genotyped the Chicago families for five additional STRPs—two that reside within the HLA region (DQ.CAR and TNFa) and three flanking markers (D6S258, MOGc, and D6S1680) (fig. 1A). The LOD score increased to 3.8, peaking at MOGc. The information content (Nicolae and Kong 2004) in this region was 95%, indicating that we could not increase the LOD score or improve the resolution by adding more markers. Furthermore, the MOGc 136-bp allele was overtransmitted to asthmatic children in the families (42 transmissions [TR]:23 nontransmissions [NT]; P [corrected for relatedness and number of alleles] = .06). In the trios, the MOGc 134-bp allele was overtransmitted to asthmatic children (17 TR:6 NT). Because of the extensive linkage disequilibrium (LD) in the HLA region (Begovich et al. 1992), the disease locus could have been located at a far distance, despite the evidence of association between MOGc alleles and asthma in two independent samples. To determine whether the haplotype shared among affected individuals extended into the HLA region, we genotyped the families at the HLA-A gene and at six STRPs spanning the HLA class I region between TNFa and HLA-A. There was no evidence of association with HLA-A or with markers proximal to it (data not shown).
Chromosome 6p21 is one of the best characterized regions in the human genome, and the nucleotide sequence between D6S258 and D6S265 is known (Mungall et al. 2003; Stewart et al. 2004). This region is gene rich, with 20 known or predicted genes and at least 30 pseudogenes in the 1-Mb region from HLA-A to OR2B3 (fig. 1B). To localize the susceptibility-associated variation, we genotyped the Chicago families and trios for an additional 59 polymorphisms in 19 genes, two pseudogenes, and the intergenic regions flanking the HLA-A and HLA-G loci (fig. 1B; tables A1 and A4 [online only]). Our strategy was to genotype one common SNP every 10–20 kb across each gene between D6S258 and D6S265, as well as all nonsynonomous SNPs, when possible. SNPs were selected from dbSNP or were discovered in our laboratory. We previously extensively characterized the haplotype structure of the HLA-G gene (MIM 142871) in the Hutterites (Ober et al. 1996, 2003). In this gene, we selected SNPs that identify clusters of variants either that are in perfect or near-perfect LD or that uniquely define all of the common HLA-G alleles, as in our previous study (Ober et al. 2003). SNPs in intergenic regions were selected to characterize the LD pattern in the proximal end of the region.
To investigate whether any of these variants explained some or all of the original evidence of linkage in the families, we conditioned on the genotype at each polymorphic site and reexamined the evidence of linkage. Multipoint analysis conditional on the genotypes of one variant in the HLA-G gene (1489C/T, His93His) yielded a LOD score of 0.9, which is substantially lower than the maximum LOD score (3.8) in this region. Analyses conditional on the other variants led to LOD scores >0.9. Thus, variation in the HLA-G gene accounted for most of the linkage in the families; the remaining evidence of linkage could be due to chance sharing among affected family members (Sun et al. 2002) or to the presence of a second susceptibility locus in the linked region. Nonetheless, it is notable that conditioning on a single SNP could result in a reduction in the LOD score of this magnitude (Sun et al. 2002).
We next examined the pattern of LD across this region in unrelated individuals from the Chicago families and trios and identified five LD blocks (Zhang et al. 2002a) (fig. 2A). To determine which LD block contains variation that contributes to asthma susceptibility, we examined pairwise combinations of SNPs within each block by the transmission/disequilibrium test (TDT), conditional on the evidence of linkage at each position. Only pairwise combinations of SNPs in block 2 showed significant nonrandom transmission of haplotypes (P<.001) in both the families and the trios (fig. 2B; table A2 [online only]).
Figure 2.
LD block structure in the extended class I region. A, Graph of LD map, showing LDUs on the Y-axis and distance on the X-axis (Zhang et al. 2002a). Shaded boxes show the five blocks in this region. B, Pairwise TDT of variants within each block. Results for Chicago families are shown in the lower half, and results for Chicago trios are in the upper half. P values were derived by simulations that were conditioned on the evidence of linkage but were not corrected for multiple comparisons.
Analyses of the individual variants revealed that polymorphisms in HLA-G (block 2) were associated with asthma in both the families and the trios; SNPs in two genes in block 4 (OR12D2 and OR10C1) and one gene each in block 3 (GABBR1) and block 5 (OR5V) were associated with asthma in the families only (table A3 [online only]). Thus, the HLA-G gene in block 2 was the only gene that showed evidence of association with asthma in both the families and the trios. However, in the families, the association was with a haplotype carrying the −964G allele (43 TR:25 NT), whereas the association in the trios was with a haplotype carrying the −964A allele (23 TR:12 NT).
To further localize and characterize the susceptibility locus, we genotyped selected markers in two populations that previously showed linkage of asthma-related phenotypes to 6p21: (1) the Hutterites, a founder population of European descent, and (2) Dutch families (Ober et al. 2000; Koppelman et al. 2002). Because of the different ascertainment schemes, we used different approaches in each sample. Ascertainment in the Hutterites was population based; individuals in this single, large pedigree were not selected on the basis of any particular phenotype. As a result, we used a test of association that was designed for large, multigenerational pedigrees (Bourgain et al. 2003). Here, we defined “cases” as individuals with bronchial hyperresponsiveness (BHR [MIM 600807]) (n=156) and “controls” as individuals with a negative history of asthma symptoms and without BHR (n=434). We note that all of the affected individuals in the Chicago families and trios also had BHR (table 1). Only 3 SNPs in HLA-G (of 21 SNPs genotyped) showed evidence of association with BHR in the Hutterites (P<.05) in whom the −964G allele was associated, similar to the results in the Chicago families.
The 200 Dutch families were ascertained through a parent in whom asthma was diagnosed ∼30 years ago (Panhuysen et al. 1995). Therefore, we first examined the prevalence of BHR and atopy (MIM 147050) in the descendents of each proband. The prevalence of atopy differed significantly by genotype at HLA-G −964 (60% of AA children, 49% of AG children, and 41% of GG children were atopic; P=.006); there was no association with BHR (table 2). However, because of the method of ascertainment used in the Dutch families, because HLA-G is an important immunomodulatory molecule during pregnancy (Hunt et al. 2000; Le Bouteiller et al. 2003), and because maternal asthma is a significant risk factor for asthma (Martinez 1997), we next analyzed these data stratified by mothers’ and fathers’ affection status (BHR+ or BHR−). The −964A allele was overtransmitted to children with BHR if the mother was unaffected (57 TR:27 NT; P=.004), whereas the −964G allele was overtransmitted to children with BHR if the mother was affected (61 TR:45 NT; P=.15). The differences in transmission patterns of alleles to children with BHR from mothers with and without BHR was highly significant (P=.0008). Similar analyses stratified by father’s asthma status did not show as significant a trend. Furthermore, the prevalences of BHR are not different if maternal status is ignored, but the prevalence of BHR among children with the GG genotype is significantly influenced by maternal status (table 2). Among children whose mothers have BHR, 56% of GG children have BHR; among children of mothers without BHR, 26% of GG children have BHR (P=.001). No such relationship was observed for AA or AG children. Thus, in the Dutch families, GG children are less likely to be atopic but are more likely to have BHR if their mother also has BHR. None of the other five markers typed in the Dutch families showed associations with either BHR or atopy (table A3 [online only]).
Table 2.
Number of Dutch Children with BHR and Atopy, by Child’s HLA-G −964 Genotype and Mother’s Affection Status[Note]
|
No. of Childrenwith Affected Status of |
||||
| Child’s Genotypeand Mother’sAffection Status | BHR+ | BHR− | Atopy+ | Atopy− |
| AA: | ||||
| BHR+ | 22 | 28 | 33 | 18 |
| BHR− | 27 | 30 | 32 | 25 |
| AG: | ||||
| BHR+ | 57 | 63 | 62 | 59 |
| BHR− | 56 | 68 | 60 | 67 |
| GG: | ||||
| BHR+ | 57 | 45 | 44 | 61 |
| BHR− | 18 | 50 | 28 | 44 |
Note.— The prevalence of BHR in children is influenced by both the child’s genotype and the mother’s affection status. Among GG children, the prevalence of BHR is 56% if the mother has BHR and 26% if the mother does not have BHR (P=.001). The prevalence of atopy in children is influenced by the child’s genotype but not the mother’s affection status (P=.006 for differences between genotypes).
To further examine interactions between mother’s affection status and child’s genotype, we reexamined the Chicago families, stratifying the sample by maternal BHR status (table 3). Genotype distributions in the children with asthma differed by mother’s BHR status for the nine polymorphisms in the HLA-G gene (data not shown), with three being significant at P<.01. Similar to the Dutch families, the −964GG genotype was associated with asthma among children of mothers with a positive BHR affection status, whereas the −964AA genotype was associated with asthma among children of mothers with a negative affection status. Other variants in HLA-G that were not genotyped in the Dutch families showed more striking differences, suggesting that variation in the gene in addition to the promoter region may contribute to risk. A similar trend was observed in the Hutterites: the frequency of the −964G allele among children with BHR whose mothers also had BHR was higher (0.69) than among children with BHR whose mothers did not have BHR (0.60), although this difference was not significant (P=.16).
Table 3.
Number of Children with Asthma in the Chicago Families, by Child’s Genotype and Mother’s Affection Status[Note]
|
No. of Childrenwith Genotype of |
||||||
|
HLA-G −964 |
HLA-G 1489 |
|||||
| Mother’sAffection Status | GG | AG | AA | CC | CT | TT |
| BHR+ | 12 | 17 | 4 | 18 | 15 | 0 |
| BHR− | 9 | 19 | 18 | 9 | 30 | 12 |
Note.— The genotype distribution in the children differs by mother’s affection status. The −964GG genotype is more common among children with asthma whose mothers have BHR, whereas the −964AA genotype is more common among children with asthma whose mothers do not have BHR, although these differences do not reach statistical significance. Genotype differences are more striking for HLA-G 1489 (difference between children with asthma of BHR+ and BHR− mothers, P=.009), the same SNP that explained the evidence of linkage.
Thus, in the Chicago families and trios and in the Hutterite and Dutch families, variants in the HLA-G gene were associated with asthma or BHR. Although no other variation that we examined in this region showed associations in all four populations, we cannot exclude the possibility that unidentified variation in HLA-G or in other genes in block 2 also contributes to susceptibility. Furthermore, because we observed different alleles and haplotypes associated in the different populations and also when stratified by maternal affection status, susceptibility at this locus is complex, influenced by maternal factors, and associated with multiple related phenotypes.
HLA-G is a novel HLA gene that has limited polymorphisms in the coding region and a restricted tissue distribution (Ober and Aldrich 1997). This gene is the most highly expressed HLA gene in placental cells at the maternal-fetal interface, where it plays important immunoregulatory roles, including the inhibition of maternal NK and T cells and the promotion maternal tolerance of the allogeneic fetus (Hunt et al. 2000; Le Bouteiller et al. 2003). Recently, it has been demonstrated that HLA-G is expressed in adult macrophages, dendritic cells, and myoblasts in response to inflammation (Yang et al. 1996; Khosrotehrani et al. 2001; Wiendl et al. 2003), in intestinal biopsies in patients with Crohn disease (Torres et al. 2004), and in malignant and nonmalignant lung diseases (Pangault et al. 2002). Furthermore, in biopsied myocardial cells from transplanted hearts, expression of HLA-G is correlated with prolonged graft survival and transplantation success (Rouas-Freiss et al. 2003). In this context, HLA-G is thought to inhibit Th1-mediated inflammation, perhaps in a concentration-dependent manner (Kapasi et al. 2000). The association of HLA-G variants with asthma is particularly intriguing, because asthma, like pregnancy, is characterized by a predominance of Th2 cytokines (Lin et al. 1993). Furthermore, the interaction between maternal phenotype and the HLA-G genotype in children in the Dutch and Chicago families is notable, given the important role that this gene plays in pregnancy and the fact that maternal asthma is a well-established risk factor for asthma (Martinez 1997).
To evaluate whether the HLA-G gene could contribute toward the immunologic milieu in the asthmatic lung, we studied the expression pattern of HLA-G in lung tissues from two individuals with asthma, one individual without asthma, and one individual without asthma but with a history of cigarette smoking. We demonstrated by immunohistochemistry the expression of HLA-G in bronchial epithelial cells (fig. 3). Expression in the lung was limited to the soluble isoform, HLA-G5 (also called “soluble G1”) (Fujii et al. 1994). Neither the transmembrane G1 and G2 isoforms nor the soluble G6 isoform were identified in these tissues. Thus, expression of HLA-G in the lung might contribute toward the aberrant immunologic response to inhaled allergens in genetically susceptible individuals.
Figure 3.
A, Control using an irrelevant IgG antibody. B–F, Sections labeled with the anti-HLA-G5 antibody. Epithelial cell labeling may be increased in basal epithelial cells immediately above the basement membrane (arrows in panels B, C, and E) but also may label columnar cells (arrowheads in panel B). In areas of damage and focal denudation (D), labeling may be found in remaining epithelial cells. Labeling is also seen in mucosal and submucosal gland cells (F) in the airway, which are of epithelial origin. Panels A and E are from individuals without asthma; panel C is from an individual without asthma but with a 30-year smoking history; panels B, D, and F are from individuals with asthma. mAB 1-2C3 detects soluble HLA-G5 in human bronchial epithelial cells. Human donor lungs that could not be used for transplantation were obtained under an institutional review board–approved protocol from the Regional Organ Bank of Illinois. Diagnoses were extracted from medical records. Bronchi were dissected and frozen in OCT. Five-micron sections were stained for antibodies against HLA-G1, HLA-G2, HLA-G5, and HLA-G6 isoforms, as described elsewhere (Morales et al. 2003). Original magnification was ×400 for all images. Antibodies specific for HLA-G1, HLA-G2, and HLA-G6 were negative in lung sections from patients with asthma as well as those without asthma (not shown).
Although the LD pattern in this region makes it impossible to rule out the possibility that variation in other genes in block 2 contributes to susceptibility, several lines of evidence suggest that HLA-G is an asthma susceptibility gene. First, a SNP in HLA-G accounted for nearly all the linkage in this region. For other variation to be causal, it would have to be in nearly perfect LD with and at a similar frequency to the HLA-G 1489C/T SNP (Sun et al. 2002) and not in LD with the other SNPs tested. Moreover, if the effect size of our initial linkage signal was biased upwards because of chance sharing in the families (Sun et al. 2002), then HLA-G may be the sole asthma susceptibility gene in this region. Even if the signal was not biased, additional variation in HLA-G that we did not sample could also contribute to risk. Second, among the variants that were surveyed across the linked region, only SNPs in HLA-G were associated with asthma or BHR in four different populations, which were ascertained using different strategies. Third, we demonstrated by immunohistochemistry that the soluble HLA-G isoform, G5, was highly expressed in bronchial epithelial cells in asthmatic lungs. The presence of soluble HLA-G protein in bronchial epithelial cells indicates that it could participate in a local inflammatory response to airborne allergens or other agents. On the basis of these combined data, we propose that HLA-G is an asthma susceptibility gene on chromosome 6p21. The differential association of alleles (or haplotypes) with childhood disease on the basis of maternal affection status is intriguing. Although the contribution of prenatal events to metabolic diseases such as diabetes and obesity is well established (Osmond and Barker 2000), the fetal origins of immune-mediated diseases have received less attention (Jones et al. 2000; Adams and Nelson 2004). Nonetheless, the programming of the fetal immune system likely begins in utero, and our data suggest that the maternal immunologic milieu (captured in this study by the mother's disease status) may influence the child’s subsequent response to inhaled allergens in a genotype-specific manner. One potential mechanism is through differential methylation, which has been previously implicated in regulating the expression levels of HLA-G (Onno et al. 1997; Moreau et al. 2003; Ober et al. 2003), a hypothesis that we are currently investigating.
Finally, many other inflammatory diseases—such as multiple sclerosis (GAMES and Transatlantic Multiple Sclerosis Genetics Cooperative 2003), psoriasis (Zhang et al. 2002b), atopic dermatitis (Soderhall et al. 2001), inflammatory bowel disease (Mathew and Lewis 2004), and schizophrenia (Wright et al. 2001)—have been linked to the HLA region. To date, all of the variation underlying these linkages has not been identified. It is tempting to speculate that variation in the HLA-G gene may contribute to susceptibility to a wide range of inflammatory diseases, including asthma. Thus, this gene that likely evolved to promote tolerance in pregnancy may contribute to risk for many common diseases, suggesting that novel therapeutic strategies could have broad relevance to these immune-mediated diseases.
Acknowledgments
We acknowledge the following individuals and organizations: our CSGA collaborators (Malcolm Blumenthal, David Marsh, Terri Beaty, and Susan Banks-Schlegel); Rhonda Peterson, Jennifer Anderson, Heidi Gidley, Stephanie Willadsen, Patrick Klimczyk, Rebecca Brown, and Natasha Phillips, for coordinating the study and enrolling families in Chicago; Rhonda Morse and Vera Braun, for assistance on Hutterite field trips; Sue Platt, April Chan, Raluca Nicolae, and Megan Burkart, for technical assistance; Harvey Dytch, for computer programming; Henry Ehrlich (Roche Molecular Systems), for providing HLA-A genotyping reagents; The University of Chicago General Clinical Research Center (National Institutes of Health [NIH] grant M01 RR00055); and the many families that participated in these studies. The studies of Chicago families and Hutterites were supported by the NIH (grants HL72414, HL66533, and HL56399) and the Mammalian Genotyping Service (Marshfield, WI). The Dutch family studies were supported by the Netherlands Asthma Foundation (grants AF 95.09 and AF 3.2.00.38) and the NIH (grant HL66393).
Appendix A: Statistical Analyses
Linkage analyses were performed with the program ALLEGRO (Gudbjartsson et al. 2000) with the use of affecteds-only allele-sharing methods. We used the exponential allele-sharing model (Kong and Cox 1997) on the scoring function Spairs. The identity-by-descent (IBD) status was determined using multipoint calculations; P values were calculated on the basis of large-sample approximations. To search for variants that explain the evidence of linkage, we calculated conditional LOD scores as described elsewhere (Sun et al. 2002). Given a marker, the distribution of the IBD sharing, conditional on the marker genotypes, was calculated and was used in deriving the conditional multipoint LOD scores for the markers in the region. The maximum conditional LOD score was reported. This approach assumes no recombination between the markers in the tested region.
The TDT was performed on the affected individuals in the families from whom both parents were genotyped (Spielman et al. 1993). We used the χ2 statistic—and, in markers with more than two alleles, the sum of the individual χ2 functions—as the test statistic. For the markers with more than two alleles and in the data sets in which there were more than two affected individuals per family, the significance of the test was evaluated using simulations. The simulations were performed conditional on the IBD process at that location. The missing genotype patterns were identical in the simulated and observed data sets.
Two-marker TDT was performed for selected pairs of markers within each of five blocks (Zhang et al. 2002a), which were defined so that adjacent blocks were separated by an increase in LD units (LDU) of ∼0.30 or greater and so that markers in each block had small estimated LD measures. This procedure leads to a smaller number of tests and, therefore, to a less stringent threshold of significance. Only trios without missing genotypes for the pair of markers were used. The transmission counts were estimated on the basis of the two-marker haplotype frequencies that were calculated from unrelated individuals. The test statistic for one pair of markers is the maximum over all the haplotypes of 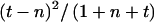 , where t is the estimated number of transmissions and n is the estimated number of nontransmissions of the haplotype from heterozygous parents. The statistical significance was evaluated by simulation. Founder haplotypes were simulated within each block of markers, and Mendelian transmissions, under the assumption of no recombination, were simulated conditional on the IBD process. The missing genotype patterns were identical in the simulated and observed data sets. Two-marker analyses, identical to those described above, were performed in each simulated data set. The P values were calculated as the number of simulated data sets in which the test statistic was larger than the observed statistic. The difference in prevalences was calculated using χ2 statistics. P values were obtained by simulation, conditional on the pedigree structure and the IBD process.
, where t is the estimated number of transmissions and n is the estimated number of nontransmissions of the haplotype from heterozygous parents. The statistical significance was evaluated by simulation. Founder haplotypes were simulated within each block of markers, and Mendelian transmissions, under the assumption of no recombination, were simulated conditional on the IBD process. The missing genotype patterns were identical in the simulated and observed data sets. Two-marker analyses, identical to those described above, were performed in each simulated data set. The P values were calculated as the number of simulated data sets in which the test statistic was larger than the observed statistic. The difference in prevalences was calculated using χ2 statistics. P values were obtained by simulation, conditional on the pedigree structure and the IBD process.
The difference in the prevalence of atopy and BHR in the Dutch children was tested using χ2 statistics. The significance was assessed using simulations. For each simulation, the family structure and phenotypes were the same as in the original data. Genotypes were simulated in the founders of the families, by use of the observed genotype frequencies, and in the nonfounders, conditional on the pedigree structure and the IBD process. The difference in genotype frequencies in the Chicago children with asthma whose mothers had positive BHR status compared with those whose mothers had negative BHR status was also tested using χ2 statistics on 2×3 tables. The significance was assessed using simulated data sets obtained by permuting the mother’s BHR status.
Table A1.
Allele Frequencies in Study Samples[Note]
|
Allele Frequency in |
|||||||
| Locus | Distance from pter(in Mb) | Allele | Patients with Asthma in Chicago Families(n=124) | Patients with Asthma in Chicago Trios(n=46) | Hutterite Patients with BHR(n=156) | Dutch Subjects with BHR(n=598) | Controlsa(n=123) |
| RFP*2 | 28.98 | T | .679 | .725 | |||
| RFP*1 | 29.03 | A | .564 | .537 | |||
| HS6M1-4P*1 | 29.17 | A | .566 | .575 | |||
| OR2J3*1 | 29.25 | T | .828 | .890 | .880 | ||
| OR2J2*1 | 29.25 | A | .568 | .534 | |||
| OR5V1*1 | 29.43 | T | .851 | .792 | .823 | .915 | |
| OR5V1*2 | 29.43 | C | .881 | .902 | .694 | ||
| OR12D3*3 | 29.44 | T | .639 | .655 | .509 | ||
| OR12D3*2 | 29.44 | C | .595 | .525 | |||
| OR12D3*1 | 29.46 | A | .592 | .526 | |||
| OR12D2*3 | 29.47 | G (47V) | .598 | .625 | .401 | ||
| OR12D2*2 | 29.47 | T (56P) | .578 | .557 | .448 | .537 | |
| OR12D2*1 | 29.47 | G (159I) | .571 | .554 | .410 | .611 | |
| OR11A1*1 | 29.50 | G | .686 | .652 | |||
| OR10C1*1 | 29.52 | C | .835 | .878 | .775 | .817 | |
| OR10C1*2 | 29.52 | C | .572 | .488 | .500 | .477 | |
| OR2H1*1 | 29.54 | C | .768 | .767 | .752 | ||
| OR2H1*2 | 29.54 | C | .605 | .511 | .524 | ||
| MRG*1 | 29.56 | A | .768 | .795 | |||
| UBD*1 | 29.63 | G | .538 | .585 | |||
| UBD*2 | 29.64 | C | .516 | .598 | |||
| OR2H3*1 | 29.66 | T (587L) | .758 | .729 | |||
| OR2H3*2 | 29.66 | C (642A) | .581 | .559 | |||
| GABBR1*8 | 29.68 | T | .942 | .893 | |||
| GABBR1*7 | 29.68 | T | .966 | .952 | |||
| GABBR1*1 | 29.68 | G | .930 | .900 | |||
| GABBR1*19 | 29.68 | G | .914 | .988 | |||
| GABBR1*6 | 29.68 | C | .925 | .881 | |||
| GABBR1*5 | 29.68 | T/C | .986 | .988 | |||
| GABBR1*2 | 29.68 | T (658F) | .835 | .875 | .860 | .870 | |
| GABBR1*21 | 29.69 | T | .844 | .867 | |||
| GABBR1*22 | 29.69 | T | .918 | .900 | |||
| GABBR1*4 | 29.69 | ΔA | .956 | ||||
| GABBR1*3 | 29.70 | G | .858 | .814 | |||
| GABBR1*24 | 29.70 | C | .917 | .977 | .938 | ||
| GABBR1*23 | 29.71 | C | .997 | .989 | .980 | ||
| MOG*3 | 29.74 | G | .906 | .859 | |||
| MOG*2 | 29.74 | G | .749 | .756 | .665 | .745 | |
| MOG*4 | 29.74 | T | .762 | .854 | .842 | ||
| LOC222567*1 | 29.75 | G | .780 | .782 | |||
| HLA-F*2 | 29.80 | G | .521 | .633 | .470 | .624 | |
| HLA-F*1 | 29.80 | A | .880 | .897 | |||
| HCGIV.9*1 | 29.87 | C | .553 | .560 | |||
| HLA-G*-1306 | 29.90 | A | .510 | .529 | .367 | ||
| HLA-G*-964 | 29.90 | G | .500 | .423 | .633 | .432 | |
| HLA-G*-725b | 29.90 | C | .886 | .837 | .800 | .850 | .844 |
| HLA-G*-689 | 29.90 | A | .524 | .443 | .671 | .508 | |
| HLA-G*-666 | 29.90 | A | .520 | .556 | .329 | ||
| HLA-G*-201 | 29.90 | G | .511 | .434 | .633 | ||
| HLA-G*1489 | 29.90 | C (93H) | .611 | .618 | .747 | ||
| HLA-G*1538 | 29.90 | C (110L) | .940 | .936 | .926 | ||
| HLA-G*3741ins | 29.90 | 14-bp deletion | .549 | .511 | .647 | .590 | .561 |
| IG.6-1 | 29.93 | A | .563 | .500 | |||
| IG.6-2b,c | 29.96 | C | .324 | .211 | |||
| HCGIV.9*2b,c | 29.99 | A | .422 | .276 | |||
| HCGIV.9*1b,c | 30.00 | A | .435 | 0.324 | |||
| IG.6-3b | 30.00 | C | .615 | .538 | |||
| IG.6-4 | 30.05 | G | .568 | .550 | |||
Note.— Genotypes at all loci were in Hardy-Weinberg proportions in all samples (nominal P>.01), except for three markers (IG.6-2, IG.6-3, and IG.6-4) that showed deviations from Hardy-Weinberg equilibrium (nominal P<.01) in the Chicago trios only. These were genotyped at the same time as the Chicago families and by the same method, suggesting that the deviations are not due to genotyping errors. The Chicago trios, Hutterite families, and Dutch families were genotyped for a subset of the markers typed in the Chicago families.
Controls were white adults (⩾18 years old), who were recruited in Chicago and who reported a negative personal and family history (among first-degree relatives) of asthma.
Triallelic markers.
Markers residing within a polymorphic deletion (Geraghty et al. 1992); the deletion was scored as a third allele.
Table A2.
|
Locus |
P Value for |
|||
| 1 | 2 | Block | Families | Trios |
| RFP*2 | RFP*1 | 5 | .7120 | .8550 |
| RFP*2 | HS6M1-4P*1 | 5 | .8330 | .8050 |
| RFP*2 | OR2J3*1 | 5 | .4790 | .3250 |
| RFP*2 | OR2J2*1 | 5 | .7620 | .7750 |
| RFP*2 | OR5V1*1 | 5 | .1390 | .5000 |
| RFP*2 | OR5V1*2 | 5 | .4170 | .7550 |
| RFP*2 | OR12D3*3 | 5 | .2150 | .8550 |
| RFP*2 | OR12D3*2 | 5 | .0470 | .9300 |
| RFP*2 | OR12D3*1 | 5 | .4500 | .4600 |
| RFP*1 | HS6M1-4P*1 | 5 | .7490 | .3500 |
| RFP*1 | OR2J3*1 | 5 | .5720 | .6200 |
| RFP*1 | OR2J2*1 | 5 | .7680 | .9250 |
| RFP*1 | OR5V1*1 | 5 | .1000 | .3750 |
| RFP*1 | OR5V1*2 | 5 | .1770 | .4300 |
| RFP*1 | OR12D3*3 | 5 | .6030 | .5400 |
| RFP*1 | OR12D3*2 | 5 | .4310 | .8600 |
| RFP*1 | OR12D3*1 | 5 | .4250 | .4800 |
| HS6M1-4P*1 | OR2J3*1 | 5 | .4890 | .5800 |
| HS6M1-4P*1 | OR2J2*1 | 5 | .7520 | .5250 |
| HS6M1-4P*1 | OR5V1*1 | 5 | .1550 | .4500 |
| HS6M1-4P*1 | OR5V1*2 | 5 | .3490 | .3650 |
| HS6M1-4P*1 | OR12D3*3 | 5 | .3470 | .2950 |
| HS6M1-4P*1 | OR12D3*2 | 5 | .0900 | .6750 |
| HS6M1-4P*1 | OR12D3*1 | 5 | .2280 | .2900 |
| OR2J3*1 | OR2J2*1 | 5 | .6690 | .3150 |
| OR2J3*1 | OR5V1*1 | 5 | .1690 | .8150 |
| OR2J3*1 | OR5V1*2 | 5 | .3500 | .4300 |
| OR2J3*1 | OR12D3*3 | 5 | .3460 | .4100 |
| OR2J3*1 | OR12D3*2 | 5 | .1780 | .4950 |
| OR2J3*1 | OR12D3*1 | 5 | .3050 | .5000 |
| OR2J2*1 | OR5V1*1 | 5 | .1450 | .6250 |
| OR2J2*1 | OR5V1*2 | 5 | .0620 | .3450 |
| OR2J2*1 | OR12D3*3 | 5 | .5010 | .3950 |
| OR2J2*1 | OR12D3*2 | 5 | .2710 | .8550 |
| OR2J2*1 | OR12D3*1 | 5 | .5140 | .6300 |
| OR5V1*1 | OR5V1*2 | 5 | .2220 | .2950 |
| OR5V1*1 | OR12D3*3 | 5 | .0290 | .5800 |
| OR5V1*1 | OR12D3*2 | 5 | .2120 | .8500 |
| OR5V1*1 | OR12D3*1 | 5 | .2190 | .4550 |
| OR5V1*2 | OR12D3*3 | 5 | .0930 | .3400 |
| OR5V1*2 | OR12D3*2 | 5 | .1460 | .3950 |
| OR5V1*2 | OR12D3*1 | 5 | .0480 | .3450 |
| OR12D3*3 | OR12D3*2 | 5 | .4470 | .5550 |
| OR12D3*3 | OR12D3*1 | 5 | .5370 | .3750 |
| OR12D3*2 | OR12D3*1 | 5 | .0730 | .4900 |
| OR12D2*3 | OR12D2*2 | 4 | .0090 | .9450 |
| OR12D2*3 | OR12D2*1 | 4 | .0020 | .8000 |
| OR12D2*3 | OR11A1*1 | 4 | .0110 | .7750 |
| OR12D2*3 | OR10C1*1 | 4 | .0160 | .8400 |
| OR12D2*3 | OR10C1*2 | 4 | .2030 | .3500 |
| OR12D2*3 | OR2H1*1 | 4 | .1000 | .6800 |
| OR12D2*3 | OR2H1*2 | 4 | .0190 | .4400 |
| OR12D2*3 | MRG*1 | 4 | .0660 | .2200 |
| OR12D2*3 | UBD*1 | 4 | .2300 | .7950 |
| OR12D2*3 | UBD*2 | 4 | .4160 | .5600 |
| OR12D2*2 | OR12D2*1 | 4 | .1290 | .6850 |
| OR12D2*2 | OR11A1*1 | 4 | .4260 | .8000 |
| OR12D2*2 | OR10C1*1 | 4 | .5020 | .7750 |
| OR12D2*2 | OR10C1*2 | 4 | .9190 | .8350 |
| OR12D2*2 | OR2H1*1 | 4 | .4530 | .5600 |
| OR12D2*2 | OR2H1*2 | 4 | .2280 | .5500 |
| OR12D2*2 | MRG*1 | 4 | .3510 | .7200 |
| OR12D2*2 | UBD*1 | 4 | .6850 | .7050 |
| OR12D2*2 | UBD*2 | 4 | .5030 | .8350 |
| OR12D2*1 | OR11A1*1 | 4 | .0820 | .8300 |
| OR12D2*1 | OR10C1*1 | 4 | .4060 | .6500 |
| OR12D2*1 | OR10C1*2 | 4 | .6620 | .8050 |
| OR12D2*1 | OR2H1*1 | 4 | .3120 | .8100 |
| OR12D2*1 | OR2H1*2 | 4 | .1100 | .5500 |
| OR12D2*1 | MRG*1 | 4 | .0980 | .6300 |
| OR12D2*1 | UBD*1 | 4 | .1450 | .5200 |
| OR12D2*1 | UBD*2 | 4 | .1860 | .9950 |
| OR11A1*1 | OR10C1*1 | 4 | .3050 | .7100 |
| OR11A1*1 | OR10C1*2 | 4 | .8130 | .5050 |
| OR11A1*1 | OR2H1*1 | 4 | .2610 | .7050 |
| OR11A1*1 | OR2H1*2 | 4 | .7410 | .2050 |
| OR11A1*1 | MRG*1 | 4 | .1560 | .8100 |
| OR11A1*1 | UBD*1 | 4 | .8140 | .7800 |
| OR11A1*1 | UBD*2 | 4 | .7600 | .9950 |
| OR10C1*1 | OR10C1*2 | 4 | .2230 | .4700 |
| OR10C1*1 | OR2H1*1 | 4 | .4740 | .7650 |
| OR10C1*1 | OR2H1*2 | 4 | .3370 | .5900 |
| OR10C1*1 | MRG*1 | 4 | .5220 | .6000 |
| OR10C1*1 | UBD*1 | 4 | .0730 | .3500 |
| OR10C1*1 | UBD*2 | 4 | .0770 | .6100 |
| OR10C1*2 | OR2H1*1 | 4 | .2560 | .3700 |
| OR10C1*2 | OR2H1*2 | 4 | .7550 | .1050 |
| OR10C1*2 | MRG*1 | 4 | .1970 | .6400 |
| OR10C1*2 | UBD*1 | 4 | .7020 | .2750 |
| OR10C1*2 | UBD*2 | 4 | .9520 | .8050 |
| OR2H1*1 | OR2H1*2 | 4 | .5660 | .5750 |
| OR2H1*1 | MRG*1 | 4 | .0710 | .8100 |
| OR2H1*1 | UBD*1 | 4 | .1050 | .7700 |
| OR2H1*1 | UBD*2 | 4 | .3350 | 1.0000 |
| OR2H1*2 | MRG*1 | 4 | .3610 | .7150 |
| OR2H1*2 | UBD*1 | 4 | .8740 | .2700 |
| OR2H1*2 | UBD*2 | 4 | .5420 | .7650 |
| MRG*1 | UBD*1 | 4 | .1080 | .7900 |
| MRG*1 | UBD*2 | 4 | .1480 | .8850 |
| UBD*1 | UBD*2 | 4 | .8720 | .8400 |
| OR2H3*1 | OR2H3*2 | 3 | .6910 | NA |
| OR2H3*1 | GABBR1*8 | 3 | .1080 | NA |
| OR2H3*1 | GABBR1*7 | 3 | .4670 | NA |
| OR2H3*1 | GABBR1*1 | 3 | .5680 | NA |
| OR2H3*1 | GABBR1*19 | 3 | .5570 | NA |
| OR2H3*1 | GABBR1*6 | 3 | .2860 | NA |
| OR2H3*1 | GABBR1*5 | 3 | .3630 | NA |
| OR2H3*1 | GABBR1*2 | 3 | .3430 | NA |
| OR2H3*1 | GABBR1*21 | 3 | .0430 | NA |
| OR2H3*1 | GABBR1*22 | 3 | .2570 | NA |
| OR2H3*1 | GABBR1*4 | 3 | .1320 | NA |
| OR2H3*1 | GABBR1*3 | 3 | .4040 | NA |
| OR2H3*1 | GABBR1*24 | 3 | .6010 | NA |
| OR2H3*1 | GABBR1*23 | 3 | .2050 | NA |
| OR2H3*2 | GABBR1*8 | 3 | .3770 | NA |
| OR2H3*2 | GABBR1*7 | 3 | .8130 | NA |
| OR2H3*2 | GABBR1*1 | 3 | .3620 | NA |
| OR2H3*2 | GABBR1*19 | 3 | .7830 | NA |
| OR2H3*2 | GABBR1*6 | 3 | .6720 | NA |
| OR2H3*2 | GABBR1*5 | 3 | .7630 | NA |
| OR2H3*2 | GABBR1*2 | 3 | .1630 | NA |
| OR2H3*2 | GABBR1*21 | 3 | .1550 | NA |
| OR2H3*2 | GABBR1*22 | 3 | .2600 | NA |
| OR2H3*2 | GABBR1*4 | 3 | .3790 | NA |
| OR2H3*2 | GABBR1*3 | 3 | .4640 | NA |
| OR2H3*2 | GABBR1*24 | 3 | .3050 | NA |
| OR2H3*2 | GABBR1*23 | 3 | .1970 | NA |
| GABBR1*8 | GABBR1*7 | 3 | .0140 | .6450 |
| GABBR1*8 | GABBR1*1 | 3 | .0040 | .2650 |
| GABBR1*8 | GABBR1*19 | 3 | .0002 | .4950 |
| GABBR1*8 | GABBR1*6 | 3 | .0400 | .7200 |
| GABBR1*8 | GABBR1*5 | 3 | .0015 | .2550 |
| GABBR1*8 | GABBR1*2 | 3 | .0035 | .6450 |
| GABBR1*8 | GABBR1*21 | 3 | .0040 | .6750 |
| GABBR1*8 | GABBR1*22 | 3 | .0070 | .1400 |
| GABBR1*8 | GABBR1*3 | 3 | .3930 | .5250 |
| GABBR1*8 | GABBR1*24 | 3 | .0110 | .7000 |
| GABBR1*8 | GABBR1*23 | 3 | .0080 | .2400 |
| GABBR1*7 | GABBR1*1 | 3 | .0005 | .6950 |
| GABBR1*7 | GABBR1*19 | 3 | .0230 | .3450 |
| GABBR1*7 | GABBR1*6 | 3 | .0280 | .6550 |
| GABBR1*7 | GABBR1*5 | 3 | .0360 | .3850 |
| GABBR1*7 | GABBR1*2 | 3 | .0130 | .6950 |
| GABBR1*7 | GABBR1*21 | 3 | .0090 | .7550 |
| GABBR1*7 | GABBR1*22 | 3 | .0023 | .3650 |
| GABBR1*7 | GABBR1*3 | 3 | .7590 | .6800 |
| GABBR1*7 | GABBR1*24 | 3 | .1610 | .3500 |
| GABBR1*7 | GABBR1*23 | 3 | .1480 | .3600 |
| GABBR1*1 | GABBR1*19 | 3 | .0140 | .4750 |
| GABBR1*1 | GABBR1*6 | 3 | .0030 | .4750 |
| GABBR1*1 | GABBR1*5 | 3 | .0030 | .2750 |
| GABBR1*1 | GABBR1*2 | 3 | .0100 | .4050 |
| GABBR1*1 | GABBR1*21 | 3 | .0230 | .5350 |
| GABBR1*1 | GABBR1*22 | 3 | .0660 | .1250 |
| GABBR1*1 | GABBR1*3 | 3 | .5930 | .5550 |
| GABBR1*1 | GABBR1*24 | 3 | .0360 | .5000 |
| GABBR1*1 | GABBR1*23 | 3 | .0420 | .3750 |
| GABBR1*19 | GABBR1*6 | 3 | .0130 | .4550 |
| GABBR1*19 | GABBR1*5 | 3 | .1170 | .0200 |
| GABBR1*19 | GABBR1*2 | 3 | .0070 | .5400 |
| GABBR1*19 | GABBR1*21 | 3 | .0110 | .6450 |
| GABBR1*19 | GABBR1*22 | 3 | .0010 | .3250 |
| GABBR1*19 | GABBR1*3 | 3 | .8800 | .1050 |
| GABBR1*19 | GABBR1*24 | 3 | .3580 | .0100 |
| GABBR1*19 | GABBR1*23 | 3 | .2130 | .0300 |
| GABBR1*6 | GABBR1*5 | 3 | .0090 | .3900 |
| GABBR1*6 | GABBR1*2 | 3 | .0063 | .4200 |
| GABBR1*6 | GABBR1*21 | 3 | .0090 | .6200 |
| GABBR1*6 | GABBR1*22 | 3 | .0050 | .2300 |
| GABBR1*6 | GABBR1*3 | 3 | .3560 | .7500 |
| GABBR1*6 | GABBR1*24 | 3 | .0280 | .4750 |
| GABBR1*6 | GABBR1*23 | 3 | .0230 | .5250 |
| GABBR1*5 | GABBR1*2 | 3 | .0050 | .2600 |
| GABBR1*5 | GABBR1*21 | 3 | .0130 | .4400 |
| GABBR1*5 | GABBR1*22 | 3 | .0040 | .1700 |
| GABBR1*5 | GABBR1*3 | 3 | .8890 | .4850 |
| GABBR1*5 | GABBR1*24 | 3 | .1780 | .0350 |
| GABBR1*5 | GABBR1*23 | 3 | 1.0000 | 1.0000 |
| GABBR1*2 | GABBR1*21 | 3 | .0060 | .2300 |
| GABBR1*2 | GABBR1*22 | 3 | .0030 | .3200 |
| GABBR1*2 | GABBR1*3 | 3 | .1150 | .6300 |
| GABBR1*2 | GABBR1*24 | 3 | .0170 | .2950 |
| GABBR1*2 | GABBR1*23 | 3 | .0110 | .1450 |
| GABBR1*21 | GABBR1*22 | 3 | .0400 | .1750 |
| GABBR1*21 | GABBR1*3 | 3 | .1900 | .5700 |
| GABBR1*21 | GABBR1*24 | 3 | .0140 | .3000 |
| GABBR1*21 | GABBR1*23 | 3 | .0110 | .4400 |
| GABBR1*22 | GABBR1*3 | 3 | .5530 | .4600 |
| GABBR1*22 | GABBR1*24 | 3 | .0220 | .2750 |
| GABBR1*22 | GABBR1*23 | 3 | .0640 | .1150 |
| GABBR1*3 | GABBR1*24 | 3 | .8150 | .2250 |
| GABBR1*3 | GABBR1*23 | 3 | .7500 | .8350 |
| GABBR1*24 | GABBR1*23 | 3 | .1250 | .0250 |
| MOG*3 | MOG*2 | 2 | .0760 | .6100 |
| MOG*3 | MOG*4 | 2 | .5470 | .1800 |
| MOG*3 | LOC222567*1 | 2 | .4830 | .0300 |
| MOG*3 | HLA-F*2 | 2 | .8780 | .5600 |
| MOG*3 | HLA-F*1 | 2 | .5100 | .6700 |
| MOG*3 | HCGIV.9*1 | 2 | .3930 | .0750 |
| MOG*3 | HLA-G*-1306 | 2 | .1100 | .1000 |
| MOG*3 | HLA-G*-964 | 2 | .3370 | .0300 |
| MOG*3 | HLA-G*-725 | 2 | .6260 | .1550 |
| MOG*3 | HLA-G*-689 | 2 | .3680 | .0900 |
| MOG*3 | HLA-G*-666 | 2 | .2470 | .0600 |
| MOG*3 | HLA-G*-201 | 2 | .1790 | .0500 |
| MOG*3 | HLA-G*1489 | 2 | .3050 | .0800 |
| MOG*3 | HLA-G*1538 | 2 | .2720 | .1000 |
| MOG*3 | HLA-G*3741ins | 2 | .1620 | .6550 |
| MOG*3 | IG.6-1 | 2 | .1080 | .2850 |
| MOG*2 | MOG*4 | 2 | .4390 | .0950 |
| MOG*2 | LOC222567*1 | 2 | .0490 | .0002 |
| MOG*2 | HLA-F*2 | 2 | .0560 | .0600 |
| MOG*2 | HLA-F*1 | 2 | .0910 | .0900 |
| MOG*2 | HCGIV.9*1 | 2 | .0800 | .1950 |
| MOG*2 | HLA-G*-1306 | 2 | .0030 | .8200 |
| MOG*2 | HLA-G*-964 | 2 | .0190 | .0040 |
| MOG*2 | HLA-G*-725 | 2 | .0090 | .0090 |
| MOG*2 | HLA-G*-689 | 2 | .0750 | .5450 |
| MOG*2 | HLA-G*-666 | 2 | .0090 | .1250 |
| MOG*2 | HLA-G*-201 | 2 | .0210 | .0250 |
| MOG*2 | HLA-G*1489 | 2 | .0260 | .0060 |
| MOG*2 | HLA-G*1538 | 2 | .0800 | .7550 |
| MOG*2 | HLA-G*3741ins | 2 | .0320 | .3750 |
| MOG*2 | IG.6-1 | 2 | .0530 | .0150 |
| MOG*4 | LOC222567*1 | 2 | .0140 | .0500 |
| MOG*4 | HLA-F*2 | 2 | .0830 | .6950 |
| MOG*4 | HLA-F*1 | 2 | .0550 | .1550 |
| MOG*4 | HCGIV.9*1 | 2 | .1850 | .0050 |
| MOG*4 | HLA-G*-1306 | 2 | .0350 | .2150 |
| MOG*4 | HLA-G*-964 | 2 | .0040 | .2250 |
| MOG*4 | HLA-G*-725 | 2 | .0033 | .3850 |
| MOG*4 | HLA-G*-689 | 2 | .0001 | .0650 |
| MOG*4 | HLA-G*-666 | 2 | .2920 | .0070 |
| MOG*4 | HLA-G*-201 | 2 | .0060 | .0800 |
| MOG*4 | HLA-G*1489 | 2 | .0700 | .0260 |
| MOG*4 | HLA-G*1538 | 2 | .0050 | .0460 |
| MOG*4 | HLA-G*3741ins | 2 | .0160 | .4300 |
| MOG*4 | IG.6-1 | 2 | .0160 | .5750 |
| LOC222567*1 | HLA-F*2 | 2 | .3690 | .0100 |
| LOC222567*1 | HLA-F*1 | 2 | .0007 | .1750 |
| LOC222567*1 | HCGIV.9*1 | 2 | .0730 | .3800 |
| LOC222567*1 | HLA-G*-1306 | 2 | .6890 | .0050 |
| LOC222567*1 | HLA-G*-964 | 2 | .1180 | .2050 |
| LOC222567*1 | HLA-G*-725 | 2 | .1160 | .0150 |
| LOC222567*1 | HLA-G*-689 | 2 | .0500 | .5550 |
| LOC222567*1 | HLA-G*-666 | 2 | .0220 | .0350 |
| LOC222567*1 | HLA-G*-201 | 2 | .3960 | .0110 |
| LOC222567*1 | HLA-G*1489 | 2 | .0480 | .8050 |
| LOC222567*1 | HLA-G*1538 | 2 | .0730 | .1400 |
| LOC222567*1 | HLA-G*3741ins | 2 | .0520 | .0200 |
| LOC222567*1 | IG.6-1 | 2 | .0210 | .9450 |
| HLA-F*2 | HLA-F*1 | 2 | .0910 | .0800 |
| HLA-F*2 | HCGIV.9*1 | 2 | .5390 | .0300 |
| HLA-F*2 | HLA-G*-1306 | 2 | .0050 | .4450 |
| HLA-F*2 | HLA-G*-964 | 2 | .1770 | .1000 |
| HLA-F*2 | HLA-G*-725 | 2 | .2160 | .0850 |
| HLA-F*2 | HLA-G*-689 | 2 | .2110 | .1650 |
| HLA-F*2 | HLA-G*-666 | 2 | .0830 | .3350 |
| HLA-F*2 | HLA-G*-201 | 2 | .1750 | .1650 |
| HLA-F*2 | HLA-G*1489 | 2 | .1900 | .1000 |
| HLA-F*2 | HLA-G*1538 | 2 | .1620 | .3350 |
| HLA-F*2 | HLA-G*3741ins | 2 | .2160 | .1650 |
| HLA-F*2 | IG.6-1 | 2 | .0710 | .1250 |
| HLA-F*1 | HCGIV.9*1 | 2 | .1070 | .1250 |
| HLA-F*1 | HLA-G*-1306 | 2 | .3670 | .3500 |
| HLA-F*1 | HLA-G*-964 | 2 | .4130 | .1000 |
| HLA-F*1 | HLA-G*-725 | 2 | .1680 | .6500 |
| HLA-F*1 | HLA-G*-689 | 2 | .4470 | .0250 |
| HLA-F*1 | HLA-G*-666 | 2 | .0510 | .5600 |
| HLA-F*1 | HLA-G*-201 | 2 | .1140 | .2400 |
| HLA-F*1 | HLA-G*1489 | 2 | .1610 | .1750 |
| HLA-F*1 | HLA-G*1538 | 2 | .1880 | .0200 |
| HLA-F*1 | HLA-G*3741ins | 2 | .0680 | .1300 |
| HLA-F*1 | IG.6-1 | 2 | .0560 | .7500 |
| HCGIV.9*1 | HLA-G*-1306 | 2 | .2250 | .3650 |
| HCGIV.9*1 | HLA-G*-964 | 2 | .4330 | .0700 |
| HCGIV.9*1 | HLA-G*-725 | 2 | .0190 | .9100 |
| HCGIV.9*1 | HLA-G*-689 | 2 | .1170 | .9100 |
| HCGIV.9*1 | HLA-G*-666 | 2 | .0030 | .9600 |
| HCGIV.9*1 | HLA-G*-201 | 2 | .3850 | .5450 |
| HCGIV.9*1 | HLA-G*1489 | 2 | .0220 | .8800 |
| HCGIV.9*1 | HLA-G*1538 | 2 | .0400 | .4700 |
| HCGIV.9*1 | HLA-G*3741ins | 2 | .2490 | .0030 |
| HCGIV.9*1 | IG.6-1 | 2 | .0370 | .5350 |
| HLA-G*-1306 | HLA-G*-964 | 2 | .0470 | .7450 |
| HLA-G*-1306 | HLA-G*-725 | 2 | .0680 | .7400 |
| HLA-G*-1306 | HLA-G*-689 | 2 | .0360 | .4300 |
| HLA-G*-1306 | HLA-G*-666 | 2 | .0440 | .8350 |
| HLA-G*-1306 | HLA-G*-201 | 2 | .1180 | .8850 |
| HLA-G*-1306 | HLA-G*1489 | 2 | .0070 | .9650 |
| HLA-G*-1306 | HLA-G*1538 | 2 | .2930 | .9100 |
| HLA-G*-1306 | HLA-G*3741ins | 2 | .0180 | .0450 |
| HLA-G*-1306 | IG.6-1 | 2 | .0250 | .3900 |
| HLA-G*-964 | HLA-G*-725 | 2 | .1380 | .2800 |
| HLA-G*-964 | HLA-G*-689 | 2 | .0060 | .0250 |
| HLA-G*-964 | HLA-G*-666 | 2 | .0100 | .0450 |
| HLA-G*-964 | HLA-G*-201 | 2 | .0170 | .0150 |
| HLA-G*-964 | HLA-G*1489 | 2 | .0230 | .4950 |
| HLA-G*-964 | HLA-G*1538 | 2 | .0190 | .8850 |
| HLA-G*-964 | HLA-G*3741ins | 2 | .0310 | .2500 |
| HLA-G*-964 | IG.6-1 | 2 | .0330 | .1700 |
| HLA-G*-725 | HLA-G*-689 | 2 | .0540 | .4900 |
| HLA-G*-725 | HLA-G*-666 | 2 | .2720 | .0850 |
| HLA-G*-725 | HLA-G*-201 | 2 | .0290 | .0140 |
| HLA-G*-725 | HLA-G*1489 | 2 | .0180 | .0250 |
| HLA-G*-725 | HLA-G*1538 | 2 | .0220 | .0150 |
| HLA-G*-725 | HLA-G*3741ins | 2 | .0040 | .7550 |
| HLA-G*-725 | IG.6-1 | 2 | .2170 | .5450 |
| HLA-G*-689 | HLA-G*-666 | 2 | .0080 | .6250 |
| HLA-G*-689 | HLA-G*-201 | 2 | .0190 | .1500 |
| HLA-G*-689 | HLA-G*1489 | 2 | .0700 | .7700 |
| HLA-G*-689 | HLA-G*1538 | 2 | .0920 | .1250 |
| HLA-G*-689 | HLA-G*3741ins | 2 | .1280 | .3100 |
| HLA-G*-689 | IG.6-1 | 2 | .2620 | .7350 |
| HLA-G*-666 | HLA-G*-201 | 2 | .1180 | .0300 |
| HLA-G*-666 | HLA-G*1489 | 2 | .3110 | .0120 |
| HLA-G*-666 | HLA-G*1538 | 2 | .1970 | .0550 |
| HLA-G*-666 | HLA-G*3741ins | 2 | .1000 | .1300 |
| HLA-G*-666 | IG.6-1 | 2 | .0030 | .6550 |
| HLA-G*-201 | HLA-G*1489 | 2 | .2460 | .0300 |
| HLA-G*-201 | HLA-G*1538 | 2 | .0400 | .0300 |
| HLA-G*-201 | HLA-G*3741ins | 2 | .0140 | .0100 |
| HLA-G*-201 | IG.6-1 | 2 | .1060 | .5500 |
| HLA-G*1489 | HLA-G*1538 | 2 | .0190 | .8000 |
| HLA-G*1489 | HLA-G*3741ins | 2 | .0028 | .7850 |
| HLA-G*1489 | IG.6-1 | 2 | .0030 | .7950 |
| HLA-G*1538 | HLA-G*3741ins | 2 | .0550 | .2800 |
| HLA-G*1538 | IG.6-1 | 2 | .0840 | .4200 |
| HLA-G*3741ins | IG.6-1 | 2 | .0130 | .5800 |
| IG.6-2 | HCGIV.9*2 | 1 | .1440 | .2800 |
| IG.6-2 | HCGIV.9*1 | 1 | .1780 | .5550 |
| IG.6-2 | IG.6-3 | 1 | .1580 | .2150 |
| IG.6-2 | IG.6-4 | 1 | .1030 | .0150 |
| HCGIV.9*2 | HCGIV.9*1 | 1 | .1170 | .3150 |
| HCGIV.9*2 | IG.6-3 | 1 | .1520 | .1850 |
| HCGIV.9*2 | IG.6-4 | 1 | .6240 | .2250 |
| HCGIV.9*1 | IG.6-3 | 1 | .1600 | .0950 |
| HCGIV.9*1 | IG.6-4 | 1 | .5980 | .0650 |
| IG.6-3 | IG.6-4 | 1 | .6390 | .2950 |
Note.— P values < .05 are shown in bold italics. NA = not applicable; SNP not typed.
Table A3.
Associations with Asthma in the Chicago Families and Trios and with BHR in the Hutterite and Dutch Families[Note]
|
P Value for |
||||||||
| Dutch Families with | ||||||||
| Locus | Distance from pter (in Mb) | Polymorphism | dbSNP rs# | Chicago Families | Chicago Trios | Hutterites | BHR− Mother | BHR+ Mother |
| RFP*2 | 28.98 | T/C | 209131 | .778 | .715 | … | … | … |
| RFP*1 | 29.03 | A/G | 1237485 | .641 | .862 | … | … | … |
| HS6M1-4P*1 | 29.17 | A/G | 3131088 | .732 | .398 | … | … | … |
| OR2J3*1 | 29.25 | T/C | 3129157 | .580 | .796 | … | … | … |
| OR2J2*1 | 29.25 | A/G | 3130743 | .948 | .411 | … | … | … |
| OR5V1*1 | 29.43 | T/G | 6930033 | .075 | .414 | .482 | … | … |
| OR5V1*2 | 29.43 | C/T | 4713210 | .0013 | .763 | … | … | … |
| OR12D3*3 | 29.44 | T/C | 4713211 | .873 | .369 | … | … | … |
| OR12D3*2 | 29.44 | C/T | 2354502 | .185 | .612 | … | … | … |
| OR12D3*1 | 29.46 | A/G | 238881 | .273 | .180 | … | … | … |
| OR12D2*3 | 29.47 | G/T (V47F) | 9257834 | .0090 | .670 | .054 | … | … |
| OR12D2*2 | 29.47 | C/T (L56P) | 4987411 | .183 | .835 | .108 | … | … |
| OR12D2*1 | 29.47 | A/G (V159I) | 2073151 | .0620 | .117 | .056 | .80 | .530 |
| OR11A1*1 | 29.50 | G/T | 7770592 | .781 | .384 | … | … | … |
| OR10C1*1 | 29.52 | C/T | 2074469 | .0180 | .564 | .896 | … | … |
| OR10C1*2 | 29.52 | C/T | 2074464 | .771 | .336 | .182 | … | … |
| OR2H1*1 | 29.54 | C/T | 2021729 | .290 | .827 | … | … | … |
| OR2H1*2 | 29.54 | C/T | 3128854 | .913 | .433 | … | … | … |
| MRG*1 | 29.56 | A/G | 1233492 | .132 | .683 | … | … | … |
| UBD*1 | 29.63 | G/C | 444013 | .943 | .493 | … | … | … |
| UBD*2 | 29.64 | C/T | 362513 | .706 | 1.00 | … | … | … |
| OR2H3*1 | 29.66 | T/C (L587F) | 3129034 | .424 | … | .250 | … | … |
| OR2H3*2 | 29.66 | C/T (A642V) | 1233387 | .536 | … | .332 | … | … |
| GABBR1*8 | 29.68 | T/C | .042 | .480 | … | … | … | |
| GABBR1*7 | 29.68 | T/C | .271 | .655 | … | … | … | |
| GABBR1*1 | 29.68 | G/A | 881284 | .130 | .527 | … | … | … |
| GABBR1*19 | 29.68 | A/G | .270 | .317 | … | … | … | |
| GABBR1*6 | 29.68 | T/C | 29261 | .067 | .705 | … | … | … |
| GABBR1*5 | 29.68 | C/T | .842 | .317 | … | … | … | |
| GABBR1*2 | 29.68 | T/C (F658F) | 29230 | .018 | .617 | .352 | .30 | .470 |
| GABBR1*21 | 29.69 | T/C | .037 | 1 | … | … | … | |
| GABBR1*22 | 29.69 | T/C | 29257 | .168 | .206 | … | … | … |
| GABBR1*4 | 29.69 | ΔA | .462 | … | … | … | … | |
| GABBR1*3 | 29.70 | G/C | 29221 | .611 | .670 | … | … | … |
| GABBR1*24 | 29.70 | C/T | 29242 | .296 | .257 | … | … | … |
| GABBR1*23 | 29.71 | C/T | .019 | 1 | … | … | … | |
| MOG*3 | 29.74 | G/A | 3130250 | .466 | .655 | … | … | … |
| MOG*2 | 29.74 | G/A | 1318631 | .083 | .18 | .159 | .027 | .527 |
| MOG*4 | 29.74 | T/G | 2535246 | .154 | .028 | .833 | … | … |
| LOC222567*1 | 29.75 | G/T | 2535238 | .263 | .071 | … | … | … |
| HLA-F*2 | 29.80 | G/A | 136126 | .450 | .086 | .145 | … | … |
| HLA-F*1 | 29.80 | A/C | 176925 | .635 | .617 | … | … | … |
| HCGIV.9*1 | 29.87 | C/T | 1610718 | .176 | .827 | … | … | … |
| HLA-G*-1306 | 29.90 | A/G | 1736936 | .065 | .144 | .046 | … | … |
| HLA-G*-964 | 29.90 | G/A | 1632947 | .051 | .060 | .046 | .0040 | .151 |
| HLA-G*-725 | 29.90 | C/G/T | 12233334 | .668 | .514 | .60 | .62 | .060 |
| HLA-G*-689 | 29.90 | A/G | 2735022 | .025 | .105 | .076 | … | … |
| HLA-G*-666 | 29.90 | T/G | 1632945 | .121 | .170 | .076 | … | … |
| HLA-G*-201 | 29.90 | G/A | 1631950 | .105 | .034 | .046 | … | … |
| HLA-G*1489 | 29.90 | C/T (H93H) | 1624278 | .182 | 1.0 | .207 | … | … |
| HLA-G*1538 | 29.90 | C/T (L110I) | .495 | 1.0 | .201 | … | … | |
| HLA-G*3741ins | 29.90 | 14 bp in/del | 16375 | .013 | .647 | .16 | .150 | .720 |
| IG.6-1 | 29.93 | G/A | 2844826 | .166 | .516 | … | … | … |
| IG.6-2 | 29.96 | G/C/Δ | 1054175 | .069 | .523 | … | … | … |
| HCGIV.9*2 | 29.99 | A/G/Δ | 2517782 | .614 | .027 | … | … | … |
| HCGIV.9*1 | 30.00 | A/G/Δ | 1056242 | .462 | .444 | … | … | … |
| IG.6-3 | 30.00 | C/T/Δ | 2517753 | .310 | .258 | … | … | … |
| IG.6-4 | 30.05 | A/G | 2394255 | .787 | 1.00 | … | … | … |
Note.— P values < .05 are shown in bold italics. TDT was performed for trios with asthma from CSGA families and trios and for Dutch families; a case-control test (Bourgain et al. 2003) was performed for Hutterites.
Table A4.
Methods Used for Genotyping[Note]
| Locus | dbSNP rs# | Genotyping Method |
| RFP*2 | 209131 | SBE-FP |
| RFP*1 | 1237485 | SBE-FP |
| HS6M1-4P*1 | 3131088 | SBE-FP |
| OR2J3*1 | 3129157 | DNAPrint |
| OR2J2*1 | 3130743 | SBE-FP |
| OR5V1*1 | 6930033 | SBE-FP |
| OR5V1*2 | 4713210 | DNAPrint |
| OR12D3*3 | 4713211 | DNAPrint |
| OR12D3*2 | 2354502 | DNAPrint |
| OR12D3*1 | 238881 | SBE-FP |
| OR12D2*3 | SBE-FP | |
| OR12D2*2 | 4987411 | DNAPrint |
| OR12D2*1 | 2073151 | DNAPrint |
| OR11A1*1 | 7770592 | DNAPrint |
| OR10C1*1 | 2074469 | DNAPrint |
| OR10C1*2 | 2074464 | DNAPrint |
| OR2H1*1 | 2021729 | DNAPrint |
| OR2H1*2 | 3128854 | DNAPrint |
| MRG*1 | 1233492 | SBE-FP |
| UBD*1 | 444013 | SBE-FP |
| UBD*2 | 362513 | DNAPrint |
| OR2H3*1 | 3129034 | SBE-FP |
| OR2H3*2 | 1233387 | SBE-FP |
| GABBR1*8 | Seq | |
| GABBR1*7 | Seq | |
| GABBR1*1 | 881284 | SBE-FP |
| GABBR1*19 | Seq | |
| GABBR1*6 | 29261 | Seq |
| GABBR1*5 | Seq | |
| GABBR1*2 | 29230 | SBE-FP |
| GABBR1*21 | SBE-FP | |
| GABBR1*22 | 29257 | SBE-FP |
| GABBR1*4 | In/del | |
| GABBR1*3 | 29221 | SBE-FP |
| GABBR1*24 | 29242 | DNAPrint |
| GABBR1*23 | DNAPrint | |
| MOG*3 | 3130250 | SBE-FP |
| MOG*2 | 1318631 | SBE-FP |
| MOG*4 | 2535246 | TM |
| LOC222567*1 | 2535238 | TM |
| HLA-F*2 | 136126 | DNAPrint |
| HLA-F*1 | 176925 | SBE-FP |
| HCGIV.9*1 | 1610718 | SBE-FP |
| HLA-G*-1306 | 1736936 | Dot blot |
| HLA-G*-964 | 1632947 | Dot blot |
| HLA-G*-725 | 12233334 | SBE-FP |
| HLA-G*-689 | 2735022 | Dot blot |
| HLA-G*-666 | 1632945 | Dot blot |
| HLA-G*-201 | 1631950 | Dot blot |
| HLA-G*1489 | 1624278 | Dot blot |
| HLA-G*1538 | Dot blot | |
| HLA-G*3741ins | 16375 | In/del |
| IG.6-1 | 2844826 | TM |
| IG.6-2 | 1054175 | TM/RT |
| HCGIV.9*2 | 2517782 | TM/RT |
| HCGIV.9*1 | 1056242 | TM/RT |
| IG.6-3 | 2517753 | TM/RT |
| HLA-A | 2394255 | LAS |
| IG.6-4 | 2394255 | TM |
Note.— SBE-FP, single-base extension with fluorescent polarization (Chen et al. 1999); DNAPrint, DNAPrint Genomics (see DNAPrint Genomics Web site); Seq, direct sequencing on ABI 3100 (Applied Biosystems); in/del, insertion/deletion polymorphism detected by size separation; TM, Taqman Assays-on-Demand or Assays-by-Design (Applied Biosystems); dot blot, hybridization with allele-specific probes (Aldrich et al. 2001); RT, RT-PCR to determine presence or absence of deletion (third allele) (Geraghty et al. 1992); and LAS, immobilized probe linear array system (Roche Molecular Systems) (Mirel et al. 2002).
Electronic-Database Information
The URLs for data presented herein are as follows:
- dbSNP Home Page, http://www.ncbi.nlm.nih.gov/SNP/
- DNAPrint Genomics, http://www.dnaprint.com/genotyping.html
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for asthma, HLA-G, BHR, and atopy)
References
- Adams KM, Nelson JL (2004) Microchimerism: an investigative frontier in autoimmunity and transplantation. JAMA 291:1127–1131 [DOI] [PubMed] [Google Scholar]
- Begovich AB, McClure GR, Suraj VC, Helmuth RC, Fildes N, Bugawan TL, Erlich HA, Klitz W (1992) Polymorphism, recombination, and linkage disequilibrium within the HLA class II region. J Immunol 148:249–258 [PubMed] [Google Scholar]
- Bourgain C, Hoffjan S, Nicolae R, Newman D, Steiner L, Walker K, Reynolds R, Ober C, McPeek MS (2003) Novel case-control test in a founder population identifies p-selectin as an atopy-susceptibility locus. Am J Hum Genet 73:612–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collaborative Study on the Genetics of Asthma (1997) A genome-wide search for asthma susceptibility loci in ethnically diverse populations. Nat Genet 15:389–392 [DOI] [PubMed] [Google Scholar]
- Fujii T, Ishitani A, Geraghty DE (1994) A soluble form of the HLA-G antigen is encoded by a messenger ribonucleic acid containing intron 4. J Immunol 153:5516–5524 [PubMed] [Google Scholar]
- GAMES, Transatlantic Multiple Sclerosis Genetics Cooperative (2003) A meta-analysis of whole genome linkage screens in multiple sclerosis. J Neuroimmunol 143:39–46 [DOI] [PubMed] [Google Scholar]
- Hunt JS, Petroff MG, Morales P, Sedlmayr P, Geraghty DE, Ober C (2000) HLA-G in reproduction: studies on the maternal-fetal interface. Hum Immunol 61:1113–1117 [DOI] [PubMed] [Google Scholar]
- Jones CA, Holloway JA, Warner JO (2000) Does atopic disease start in foetal life? Allergy 55:2–10 [DOI] [PubMed] [Google Scholar]
- Kapasi K, Albert SE, Yie S, Zavazava N, Librach CL (2000) HLA-G has a concentration-dependent effect on the generation of an allo-CTL response. Immunology 101:191–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khosrotehrani K, Le Danff C, Reynaud-Mendel B, Dubertret L, Carosella ED, Aractingi S (2001) HLA-G expression in atopic dermatitis. J Invest Dermatol 117:750–752 [DOI] [PubMed] [Google Scholar]
- Koppelman GH, Stine OC, Xu J, Howard TD, Zheng SL, Kauffman HF, Bleecker ER, Meyers DA, Postma DS (2002) Genome-wide search for atopy susceptibility genes in Dutch families with asthma. J Allergy Clin Immunol 109:498–506 [DOI] [PubMed] [Google Scholar]
- Le Bouteiller P, Legrand-Abravanel F, Solier C (2003) Soluble HLA-G1 at the materno-foetal interface—a review. Placenta Suppl 24:S10–S15 [DOI] [PubMed] [Google Scholar]
- Lester LA, Rich SS, Blumenthal MN, Togias A, Murphy S, Malveaux F, Miller ME, Dunston GM, Solway J, Wolf RL, Samet JM, Marsh DG, Meyers DA, Ober C, Bleecker ER (2001) Ethnic differences in asthma and associated phenotypes: Collaborative Study on the Genetics of Asthma. J Allergy Clin Immunol 108:357–362 [DOI] [PubMed] [Google Scholar]
- Lin H, Mosmann TR, Guilbert L, Tuntipopipat S, Wegmann TG (1993) Synthesis of T helper 2-type cytokines at the maternal-fetal interface. J Immunol 151:4562–4573 [PubMed] [Google Scholar]
- Martinez FD (1997) Maternal risk factors in asthma. Ciba Found Symp 206:233–219 [PubMed] [Google Scholar]
- Mathew CG, Lewis CM (2004) Genetics of inflammatory bowel disease: progress and prospects. Hum Mol Genet Suppl 13 Spec No 1:R161–R168 [DOI] [PubMed] [Google Scholar]
- Morales PJ, Pace JL, Platt JS, Phillips TA, Morgan K, Fazleabas AT, Hunt JS (2003) Placental cell expression of HLA-G2 isoforms is limited to the invasive trophoblast phenotype. J Immunol 171:6215–6224 [DOI] [PubMed] [Google Scholar]
- Moreau P, Mouillot G, Rousseau P, Marcou C, Dausset J, Carosella ED (2003) HLA-G gene repression is reversed by demethylation. Proc Natl Acad Sci USA 100:1191–1196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mungall AJ, Palmer SA, Sims SK, Edwards CA, Ashurst JL, Wilming L, Jones MC, et al (2003) The DNA sequence and analysis of human chromosome 6. Nature 425:805–811 [DOI] [PubMed] [Google Scholar]
- Nicolae DL, Kong A (2004) Measuring the relative information in allele-sharing linkage studies. Biometrics 60:368–375 [DOI] [PubMed] [Google Scholar]
- Ober C, Aldrich A (1997) HLA-G polymorphisms: neutral evolution or novel function? J Reprod Immunol 36:1–21 [DOI] [PubMed] [Google Scholar]
- Ober C, Aldrich CL, Chervoneva I, Billstrand C, Rahimov F, Gray HL, Hyslop T (2003) Variation in the HLA-G promoter region influences miscarriage rates. Am J Hum Genet 72:1425–1435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ober C, Rosinsky B, Grimsley C, van der Ven K, Robertson A, Runge A (1996) Population genetics studies of HLA-G: allele frequencies and linkage disequilibrium with HLA-A. J Reprod Immunol 32:111–123 [DOI] [PubMed] [Google Scholar]
- Ober C, Tsalenko A, Parry R, Cox NJ (2000) A second generation genome-wide screen for asthma susceptibility alleles in a founder population. Am J Hum Genet 67:1154–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onno M, Amiot L, Bertho N, Drenou B, Fauchet R (1997) CpG methylation patterns in the 5′ part of the nonclassical HLA-G gene in peripheral blood CD34+ cells and CD2+ lymphocytes. Tissue Antigens 49:356–364 [DOI] [PubMed] [Google Scholar]
- Osmond C, Barker DJ (2000) Fetal, infant, and childhood growth are predictors of coronary heart disease, diabetes, and hypertension in adult men and women. Environ Health Perspect Suppl 108:545–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pangault C, Le Friec G, Caulet-Maugendre S, Lena H, Amiot L, Guilloux B, Onno M, Fauchet R (2002) Lung macrophages and dendritic cells express HLA-G molecules in pulmonary diseases. Hum Immunol 63:83–90 [DOI] [PubMed] [Google Scholar]
- Panhuysen CI, Bleecker ER, Koeter GH, Meyers DA, Postma DS (1995) Dutch approach to the study of the genetics of asthma. Clin Exp Allergy Suppl 25:35–38 [DOI] [PubMed] [Google Scholar]
- Rouas-Freiss N, LeMaoult J, Moreau P, Dausset J, Carosella ED (2003) HLA-G in transplantation: a relevant molecule for inhibition of graft rejection? Am J Transplant 3:11–16 [DOI] [PubMed] [Google Scholar]
- Soderhall C, Bradley M, Kockum I, Wahlgren CF, Luthman H, Nordenskjold M (2001) Linkage and association to candidate regions in Swedish atopic dermatitis families. Hum Genet 109:129–135 [DOI] [PubMed] [Google Scholar]
- Stewart CA, Horton R, Allcock RJ, Ashurst JL, Atrazhev AM, Coggill P, Dunham I, et al (2004) Complete MHC haplotype sequencing for common disease gene mapping. Genome Res 14:1176–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Cox NJ, McPeek MS (2002) A statistical method for identification of polymorphisms that explain a linkage result. Am J Hum Genet 70:399–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres MI, Le Discorde M, Lorite P, Rios A, Gassull MA, Gil A, Maldonado J, Dausset J, Carosella ED (2004) Expression of HLA-G in inflammatory bowel disease provides a potential way to distinguish between ulcerative colitis and Crohn’s disease. Int Immunol 16:579–583 [DOI] [PubMed] [Google Scholar]
- Wiendl H, Mitsdoerffer M, Weller M (2003) Express and protect yourself: the potential role of HLA-G on muscle cells and in inflammatory myopathies. Hum Immunol 64:1050–1056 [DOI] [PubMed] [Google Scholar]
- Wright P, Nimgaonkar VL, Donaldson PT, Murray RM (2001) Schizophrenia and HLA: a review. Schizophr Res 47:1–12 [DOI] [PubMed] [Google Scholar]
- Xu J, Meyers DA, Ober C, Blumenthal MN, Mellen B, Barnes KC, King RA, Lester LA, Howard TD, Solway J, Langefeld CD, Beaty TH, Rich SS, Bleecker ER, Cox NJ (2001) Genomewide screen and identification of gene-gene interactions for asthma-susceptibility loci in three U.S. populations: Collaborative Study on the Genetics of Asthma. Am J Hum Genet 68:1437–1446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Chu W, Geraghty DE, Hunt JS (1996) Expression of HLA-G in human mononuclear phagocytes and selective induction by IFN-γ. J Immunol 156:4224–4231 [PubMed] [Google Scholar]
- Zhang W, Collins A, Maniatis N, Tapper W, Morton NE (2002a) Properties of linkage disequilibrium (LD) maps. Proc Natl Acad Sci USA 99:17004–17007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XJ, He PP, Wang ZX, Zhang J, Li YB, Wang HY, Wei SC, Chen SY, Xu SJ, Jin L, Yang S, Huang W (2002b) Evidence for a major psoriasis susceptibility locus at 6p21(PSORS1) and a novel candidate region at 4q31 by genome-wide scan in Chinese Hans. J Invest Dermatol 119:1361–1366 [DOI] [PubMed] [Google Scholar]
Supplemental References
- Aldrich CL, Stephenson MD, Karrison T, Odem RR, Branch DW, Scott JR, Schreiber JR, Ober C (2001) HLA-G genotypes and pregnancy outcome in couples with unexplained recurrent miscarriage. Mol Hum Reprod 7:1167–1172. [DOI] [PubMed] [Google Scholar]
- Chen X, Levine L, Kwok P-Y (1999) Fluorescence polarization in homogeneous nucleic acid analysis. Genome Res 9:492-498 [PMC free article] [PubMed] [Google Scholar]
- Geraghty DE, Pei J, Lipsky B, Hansen JA, Taillon-Miller P, Bronson SK, Chaplin DD (1992) Cloning and physical mapping of the HLA class I region spanning the HLA- E-to-HLA-F interval by using yeast artificial chromosomes. PNAS 89:2669–2673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudbjartsson DF, Jonasson K, Frigge ML, Kong A (2000) Allegro, a new computer program for multipoint linkage analysis. Nat Genet 25:12–13 [DOI] [PubMed] [Google Scholar]
- Kong A, Cox NJ (1997) Allele-sharing models: LOD scores and accurate linkage tests. Am J Hum Genet 61:1179–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirel DB, Valdes AM, Lazzeroni LC, Reynolds RL, Erlich HA, Noble JA (2002) Association of IL4R haplotypes with type 1 diabetes. Diabetes 51:3336–3341 [DOI] [PubMed] [Google Scholar]
- Spielman RS, McGinnis RE, Ewens WJ (1993) Transmission test for linkage disequilibrium: the insulin gene region and insulin dependent diabetes mellitus (IDDM). Am J Hum Genet 52:506–516 [PMC free article] [PubMed] [Google Scholar]





