Abstract
d-2-hydroxyglutaric aciduria is a neurometabolic disorder with both a mild and a severe phenotype and with unknown etiology. Recently, a novel enzyme, d-2-hydroxyglutarate dehydrogenase, which converts d-2-hydroxyglutarate into 2-ketoglutarate, and its gene were identified. In the genes of two unrelated patients affected with d-2-hydroxyglutaric aciduria, we identified disease-causing mutations. One patient was homozygous for a missense mutation (c.1331T→C; p.Val444Ala). The other patient was compound heterozygous for a missense mutation (c.440T→G; p.Ile147Ser) and a splice-site mutation (IVS1-23A→G) that resulted in a null allele. Overexpression studies in HEK-293 cells of proteins containing the missense mutations showed a marked reduction of d-2-hydroxyglutarate dehydrogenase activity, proving that mutations in the d-2-hydroxyglutarate dehydrogenase gene cause d-2-hydroxyglutaric aciduria.
d-2-hydroxyglutaric aciduria (d-2-HGA) is a neurometabolic disorder, first described in 1980 (Chalmers et al. 1980). To date, >30 patients with d-2-HGA have been reported. The diagnosis is based on increased levels of d-2-hydroxyglutarate (d-2-HG) in body fluids, detected by analytic methods that separate and quantify individually the two isomers of 2-HG. Clinical symptoms of patients with d-2-HGA are developmental delay, epilepsy, hypotonia, and dysmorphic features. In an international survey of clinical data, both a mild and a severe phenotype could be distinguished (van der Knaap et al. 1999). The severe phenotype is homogeneous and is characterized by early-infantile-onset epileptic encephalopathy and, often, cardiomyopathy. Magnetic resonance imaging (MRI) reveals signs of disturbed cerebral maturation and white-matter abnormalities. The mild phenotype has a more variable clinical presentation and less-consistent MRI findings than the severe phenotype.
In the 25 years since the recognition of d-2-HGA, little progress has been made in understanding the biochemical and genetic basis of this disorder. Recently, the isolation and characterization of a dehydrogenase acting on d-2-HG was described (Achouri et al. 2004). This mitochondrial enzyme converts d-2-HG to 2-ketoglutarate with a Km value <10 μM, is homologous to flavin adenine dinucleotide–dependent d-lactate dehydrogenase, and most likely transfers its electrons to electron-transfer flavoprotein. Assays of d-2-hydroxyglutarate dehydrogenase in rat tissues indicate that this enzyme is most active in liver and kidney but is also active in heart and brain (E.V.S., unpublished data). Levels of d-2-HG in body fluids of healthy human individuals are at the low micromolar level, suggesting that d-2-hydroxyglutarate dehydrogenase is an important enzyme in the disposal of d-2-HG.
To determine whether mutations in the d-2-hydroxyglutarate dehydrogenase gene cause d-2-HGA, we investigated this gene by genomic and cDNA sequence analysis in two unrelated patients affected with the severe phenotype of d-2-HGA.
Patient 1 was a white Italian boy born to consanguineous healthy parents (first cousins). Clinical assessment revealed mild facial dysmorphia, with a reduced bitemporal diameter, a prominent forehead, and micrognathia. The patient exhibited psychomotor retardation, experienced episodes of vomiting, and suffered from tonic, tonic-clonic, and myoclonic seizures that were not responsive to antiepileptic treatment. He died at 2 years of age. Quantitative measurements of d-2-HG showed urine levels at 502 mM/M creatinine (normal, <18 mM/M creatinine), plasma levels at 26 μM/l (normal, <1μM/l), and cerebrospinal fluid levels at 6 μM/l (normal, <0.4 μM/l). l-2-HG was within the reference range, in all samples. Two siblings died with symptoms similar to those observed in the patient (no data available), and one sibling is unaffected.
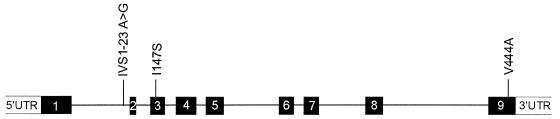
In the d-2-hydroxyglutarate dehydrogenase gene of patient 1, a homozygous T→C transition (c.1331T→C) that results in the substitution of alanine for valine at position 444 (p.Val444Ala) was found in exon 9 (see fig. 1). The parents were heterozygous for this mutation, proving homozygosity in their affected child. Valine and alanine belong to the same group of amino acids with aliphatic side chains, with a methyl group for alanine and a three-carbon unit for valine. In a multiple sequence alignment of available protein sequences, the valine 444 is conserved in the mouse d-2-hydroxyglutarate dehydrogenase and in the “actin-interacting protein 2” (NCBI accession number P46681) of Saccharomyces cerevisiae (Achouri et al. 2004). Overexpression of the mutant protein in HEK-293 cells showed that this amino acid change results in marked reduction of d-2-hydroxyglutarate dehydrogenase activity (table 1). These observations and the absence of the mutation in 210 control chromosomes indicate that the mutation is pathogenic.
Figure 1.
Schematic presentation of the d-2-hydroxyglutarate dehydrogenase gene, showing exons (numbered blackened boxes) and introns (see Genbank for genomic DNA [accession number 27465811] and cDNA [accession number 22477763]). The three pathogenic mutations that were found in two patients affected with d-2-HGA are shown. The IVS1-23A→G mutation creates an alternative splice-acceptor site located 19 nt upstream of the wild-type splice-acceptor site (r.295_296ins19). Figure is drawn to scale.
Table 1.
d-2-Hydroxyglutarate Dehydrogenase Activity in HEK-293 Cells Overexpressing the Wild-Type or Mutated Human Enzyme[Note]
| Transfectedd-2-Hydroxyglutarate Dehydrogenase | Activitya(pM/min/mg protein) |
| Wild type | 2.00 ± .3 |
| Val444Ala mutant | .5 ± .1 |
| Ile147Ser mutant | 0 |
Note.— Transient transfections were performed as described elsewhere (Achouri et al. 2004). The nucleic acid sequences of the mutated plasmids were confirmed by sequence analysis. The enzymatic activity was assayed by use of the radiochemical assay in the presence of 2 μM d-2-hydroxyglutarate without added Co2+.
Activities are expressed as the activity observed after subtraction of the value in nontransfected HEK cells (0.16 ± 0.02 pM/min/mg protein). Data are mean ± SEM of three or four transfections.
Patient 2 was a white American girl born to nonconsanguineous parents of mixed ethnic ancestry. Her clinical details have been described elsewhere (Gibson et al. 1993). She presented with generalized tonic-clonic seizures, infantile spasms with hypsarrhythmia, hypotonia, a movement disorder, cortical blindness, and developmental delay. Quantitative measurements of d-2-HG showed urine levels at 1,747 mM/M creatinine, plasma levels at 73 μM/l, and cerebrospinal fluid levels at 25 μM/l. l-2-HG was within the reference range, in all samples.
In the d-2-hydroxyglutarate dehydrogenase gene of this patient, a heterozygous missense mutation in exon 2 and a heterozygous mutation in intron 1 were found (see fig. 1). The missense mutation consists of a T→G transversion at position 440 (c.440T→G), resulting in the substitution of serine for isoleucine (p.Ile147Ser). Isoleucine is conserved in the mouse d-2-hydroxyglutarate dehydrogenase protein and in the actin-interacting protein 2 of S. cerevisiae (Achouri et al. 2004). The conservation of isoleucine, the differences in the chemical properties of serine versus isoleucine, and the absence of this mutation in 210 control chromosomes suggest that this substitution is pathogenic. Indeed, overexpression in HEK-293 cells of the mutant protein containing p.Ile147Ser proved that the amino acid change results in marked reduction of d-2-hydroxyglutarate dehydrogenase activity (table 1).
The heterozygous mutation in intron 1, IVS1-23A→G, was not found in 210 control chromosomes and thus could represent a pathogenic mutation. RT-PCR was performed on RNA isolated from cultured fibroblasts of the patient and showed that only the allele containing the missense mutation was present, which indicates that the second allele is a null allele. mRNA was isolated from fibroblasts that were incubated for 5 h with cycloheximide (0.25 mg/ml), an inhibitor of protein synthesis, which avoids nonsense-mediated decay. RT-PCR analysis using this mRNA identified the structure of the second mutant allele. It arises as a result of the creation of an alternative splice-acceptor site located 19 nt upstream of the wild-type splice-acceptor site, leading to the insertion of 19 nt into the mature mRNA (r.295_296ins19), and it involves the IVS1-23A→G site. This insertion results in a frameshift that causes a premature stop (p.Cys100fsX9). The combination of a null allele and a pathogenic missense mutation indicates that, in this patient, d-2-HGA is caused by a deficiency of d-2-hydroxyglutarate dehydrogenase.
Our data show, for the first time, that mutations in the d-2-hydroxyglutarate dehydrogenase gene can cause d-2-HGA. This result proves that d-2-HGA is an inborn error of metabolism with autosomal recessive inheritance and is consistent with our previous studies in which we found increased levels of d-2-HG in culture media of fibroblasts from patients with d-2-HGA (Struys et al. 2003). The low activities of d-2-hydroxyglutarate dehydrogenase in fibroblasts, leukocytes, and lymphocytes, however, do not allow accurate measurement by radiochemical assay (Achouri et al. 2004), which is the only available method for this enzyme to date. In this procedure, racemic [2-3H]d/l-2-HG is used as the substrate, requiring the separation of d-2-hydroxyglutarate dehydrogenase and l-2-hydroxyglutarate dehydrogenase (Rzem et al. 2004) prior to the enzyme assay. This is obligatory, since both d-2-hydroxyglutarate dehydrogenase and l-2-hydroxyglutarate dehydrogenase act on [2-3H]d/l-2-HG, yielding titrated water, which makes the assay nonspecific. Alternatively, if the racemic substrate [2-3H]d/l-2-HG could be separated into enantiomeric pure substrates by chromatographic techniques, then the assay would become specific for the corresponding hydroxyglutarate dehydrogenase. Unfortunately, both the separation of the enzymes from fibroblasts, leukocytes, and lymphocytes and the separation of the racemic substrate are not feasible at this time.
Although the pathophysiologic mechanism by which d-2-HGA causes neurologic dysfunction remains unknown, the elucidation of disease-causing mutations in the d-2-hydroxyglutarate dehydrogenase gene extends prenatal diagnoses from the established measurement of d-2-HG in amniotic fluid to DNA analysis during the first trimester for families with proven pathogenic mutations.
Acknowledgments
We acknowledge Patricia S. Darmin for her outstanding technical support (PCR and DNA sequence analysis). This study was supported by Interuniversity Attraction Poles Programme—Belgian Science Policy.
Electronic-Database Information
The accession numbers and URLs for data presented herein are as follows:
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for genomic DNA [accession number 27465811] and cDNA [accession number 22477763])
- NCBI Protein Database, http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=Protein (for the actin-interacting protein 2 of S. cerevisiae [accession number P46681])
References
- Achouri Y, Noël G, Vertommen D, Rider MH, Veiga-da-Cunha M, Van Schaftingen E (2004) Identification of a dehydrogenase acting on d-2-hydroxyglutarate. Biochem J 381:35–42 10.1042/BJ20031933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalmers RA, Lawson AM, Watts RWE, Tavill AS, Kamerling JP, Hey E, Ogilve D (1980) d-2-hydroxyglutaric aciduria: case report and biochemical studies. J Inherit Metab Dis 3:11–15 [DOI] [PubMed] [Google Scholar]
- Gibson KM, Craigen W, Herman GE, Jakobs C (1993) d-2-hydroxyglutaric acidemia in a newborn with neurological abnormalities: a new neurometabolic disorder. J Inherit Metab Dis 16:497–500 10.1007/BF00711664 [DOI] [PubMed] [Google Scholar]
- Rzem R, Veiga-da-Cunha M, Noël G, Gofette S, Nassogne M-C, Tabarki B, Schöller C, Marquardt T, Vikkula M, Van Schaftingen E (2004) A gene encoding a putative FAD-dependent l-2-hydroxyglutarate dehydrogenase is mutated in l-2-hydroxyglutaric aciduria. Proc Natl Acad Sci USA 101:16849–16854 10.1073/pnas.0404840101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struys EA, Verhoeven NM, Roos B, Jakobs C (2003) Disease-related metabolites in culture medium of fibroblasts from patients with d-2-hydroxyglutaric aciduria, l-2-hydroxyglutaric aciduria, and d/l-2-hydroxyglutaric aciduria. Clin Chem 49:1133–1138 10.1373/49.7.1133 [DOI] [PubMed] [Google Scholar]
- van der Knaap MS, Jakobs C, Hoffmann GF, Nyhan WL, Renier WO, Smeitink JA, Catsman-Berrevoets CE, Hjalmarson O, Vallance H, Sugita K, Bowe CM, Herrin JT, Craigen WJ, Buist NR, Brookfield DS, Chalmers RA (1999) d-2-hydroxyglutaric aciduria: biochemical marker or clinical disease entity? Ann Neurol 45:111–119 [DOI] [PubMed] [Google Scholar]