Abstract
The pattern of linkage disequilibrium (LD) is critical for association studies, in which disease-causing variants are identified by allelic association with adjacent markers. The aim of this study is to compare the LD patterns in several distinct European populations. We analyzed four genomic regions (in total, 749 kb) containing candidate genes for complex traits. Individuals were genotyped for markers that are evenly distributed at an average spacing of ∼2–4 kb in eight population-based samples from ongoing epidemiological studies across Europe. The Centre d'Etude du Polymorphisme Humain (CEPH) trios of the HapMap project were included and were used as a reference population. In general, we observed a conservation of the LD patterns across European samples. Nevertheless, shifts in the positions of the boundaries of high-LD regions can be demonstrated between populations, when assessed by a novel procedure based on bootstrapping. Transferability of LD information among populations was also tested. In two of the analyzed gene regions, sets of tagging single-nucleotide polymorphisms (tagSNPs) selected from the HapMap CEPH trios performed surprisingly well in all local European samples. However, significant variation in the other two gene regions predicts a restricted applicability of CEPH-derived tagging markers. Simulations based on our data set show the extent to which further gain in tagSNP efficiency and transferability can be achieved by increased SNP density.
Introduction
The efficiency of both candidate-gene and whole-genome approaches to identifying genetic loci associated with disease phenotypes relies on the minimization of SNP markers genotyped in a given population. For such a mapping approach, the selection process of markers to be genotyped is crucial (Chapman et al. 2003; Wang and Todd 2003). The observation that a significant fraction of the human genome is organized into a series of high–linkage disequilibrium (LD) regions that are separated by short segments in very low LD has led to the development of a number of algorithms that can be used to select informative markers for association studies (Cardon and Abecasis 2003). In Caucasians, approximately one-third to one-half of chromosomes are structured as high-LD regions, varying in length from a few kb to >300 kb (Gabriel et al. 2002; Phillips et al. 2003; Wall and Pritchard 2003; Ke et al. 2004). All marker-selection algorithms are based on the assumption that the complete set of sequence variants within a region of high background LD bears redundant information and can be significantly reduced to a selected subset of tagging markers. These markers can tag either neighboring markers or a set of common haplotypes within an LD block. There is an ongoing debate as to which tagging algorithm should be used, but little is known about the choice of reference populations to which such algorithms should be applied.
It has been suggested that the populations genotyped in the HapMap project may serve as reference populations for the selection of tagging markers in association studies (International HapMap Consortium 2003). In its first round, the HapMap project aims to genotype 600,000 SNPs at an average distance of 5 kb across the whole genome in four populations with African, Asian, and European ancestry (see HapMap Homepage). The European patterns are represented by 30 trios from a U.S. (Utah) population of northern and western European ancestry (CEPH sample [Dausset et al. 1990]).
As stated by the International HapMap Consortium (2003), the general applicability of the HapMap data has to be confirmed by samples from several local populations. Our study aims to describe the SNP allelic variation within candidate-gene regions in eight local European populations selected along a line from north to south. All samples represent population-based samples of ongoing epidemiological collections. The dense marker spacing of 2–4 kb over four autosomal regions (total size 749 kb) and a novel robust method to assess the reliability of LD block boundaries enables us to compare LD block boundaries and LD block content among these European populations. Although there was general agreement in the majority of LD patterns, detectable differences among study populations were found. In the context of association studies, we tested the performance of tagging SNPs (tagSNPs) that were defined in local population samples in comparison with tagSNPs that were defined in the HapMap sample, and we simulated the effect of an increased marker density.
Subjects and Methods
Population Samples
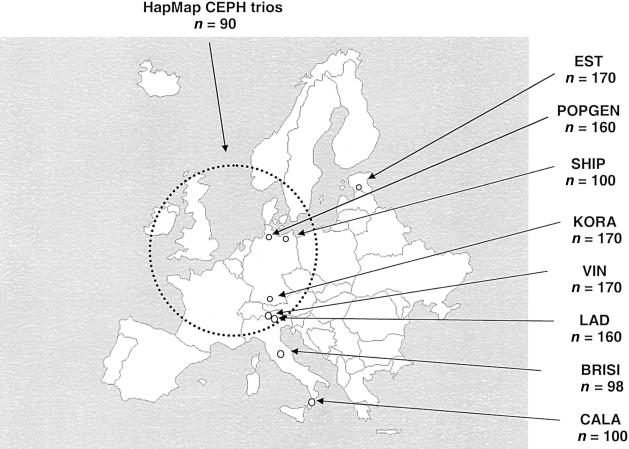
All population samples came from ongoing cross-sectional epidemiological surveys. Figure 1 shows the locations and sample sizes of eight regional population surveys. Samples were chosen randomly from the entire population. A ninth sample, 30 CEPH trios (Coriell Cell Repositories) used in the HapMap project, represents an emigrant population of northern and western European origin. The 170 individuals of Estonian ethnicity (EST) represent a random selection from ∼1.3 million Estonian inhabitants, excluding Russians. Randomly selected samples for the northern German population came from two epidemiological surveys, Study on Health in Pomerania (SHIP) (regional population size 212,000) and POPGEN (collected in Schleswig-Holstein [population size 1.15 million]) (see popgen Web site). The KORA samples were collected as part of a population-based, epidemiological project, KORA S2000 (Cooperative Health Research in the Region of Augsburg), and represent an urban region in the southern part of Germany with 610,000 inhabitants. Two Alpine populations were sampled: inhabitants of Vinschgau (VIN) in south Tyrol (population size 34,300), and members of the Ladin-speaking community (LAD) of Grödnertal and Gadertal (population size 16,800). The Brisighella sample (BRISI) represents a small town with 9,000 inhabitants from the region of Emilia-Romagna, Italy. A sample from Calabria (CALA) was sampled from a catchment area with 560,000 inhabitants. The sex ratio of all samples was ∼0.5, except for that of CALA, with 70 males and 30 females. Mean age was 55 years for all population samples except CALA and EST (mean 30 years). Prior to collection, we obtained approval from the relevant ethical committees/institutional review boards and informed consent from all participating subjects. When necessary, approval from data privacy oversight committees was obtained.
Figure 1.
Study populations and sample sizes (n)
SNP Selection and Genotyping
We selected four genomic regions, all containing candidate genes for different complex diseases. For each region, SNPs were evenly selected, covering the candidate gene and 76–174 kb of the upstream and downstream flanking regions (table 1). All information about the selected SNPs was extracted from the public dbSNP database. Genotyping of SNPs was achieved by primer extension of multiplex PCR products, with detection of the allele-specific extension products by matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF [Sequenom]) mass spectroscopy. The frequencies of genotypes from successfully typed SNPs (average call rate, 98%) were in Hardy-Weinberg equilibrium. The genotype data can be downloaded from our project Web site (see GSF European LD Pattern Project Web site).
Table 1.
Selected Gene Regions and SNPs
|
Size(in kb) of |
No. of SNPs |
Median Spacing(in kb) of |
|||||||||
| Gene | Disease | Chromosome Region | Gene | Region | Selecteda | Validatedb | Commonc | For HapMapComparisond | PopulationDifferentiatinge (%) | ValidatedSNPs | HapMapSNPs |
| SNCA | Parkinson | 4q21 | 112 | 188 | 97 | 78 | 73 | 33 | 5 (6) | 2.1 | 4.5 |
| LMNA | Cardiomyopathy | 1q21.2 | 23 | 177 | 37 | 29 | 27 | 17 | 4 (14) | 4.4 | 6.7 |
| FKBP5 | Depression | 6p21.31 | 115 | 289 | 76 | 44 | 37 | 37 | 10 (23) | 6.2f | 6.3 |
| PLAU | Alzheimer | 10q22.2 | 6.3 | 95 | 53 | 34 | 32 | 13 | 27 (79) | 2.2 | 6.0 |
Selected from public SNP databases.
Polymorphic in our sample set and in Hardy-Weinberg equilibrium.
Minor-allele frequency >5%.
For comparison, we selected a set of SNPs similar to the currently available set in the International HapMap Project (April 2004).
P<.001.
SNP spacing within the gene is 3.7 kb.
Statistics and LD-Pattern Analyses
Population differentiation was tested by permutation tests (10,000 permutations) based on F statistics, by use of the software package ARLEQUIN. FST values were calculated on three levels: for each marker separately, for each gene region separately, and for all four gene regions combined. FST values based on haplotype frequencies in each block were also tested. The standard expectation-maximization algorithm was used to estimate the haplotype frequencies.
To compare haplotype block boundaries among populations, it is critical to apply a relatively robust method for the definition of haplotype blocks on a constant set of common markers (Cardon and Abecasis 2003; Schwartz et al. 2003). We developed a simple bootstrap approach based on the standard algorithm of Gabriel et al. (2002), which invokes confidence bounds of pairwise D′ to define sequences of markers with little evidence of historical recombination. Bootstrapping, in the present context, means resampling the individual multilocus genotypes of a given population with replacement. The frequencies of block boundary positions across all 100 bootstrap runs represent confidence estimates for block borders. We plot boundary frequencies for the start and end of blocks separately, to be able to track individual blocks. In addition, we allowed blocks to overlap each other, which gives a more natural framework. Because most block definitions define haplotype blocks by block-internal characteristics, neighboring blocks may compete for the same intermediary markers, and there is no reason why one of the blocks (e.g., the larger of the two in a greedy algorithm) should win. Each block with at least one private marker is considered to be a block. Blocks completely nested within a larger block are not considered. An overall measure of similarity between two populations was calculated as the sum of cross-products of bootstrap frequencies, standardized by the sums of within-products of bootstrap frequencies, similar to the genetic identity of Nei (1972). An appropriate distance was given by the negative logarithm of this measure and was used in a multidimensional scaling algorithm to map the overall block similarity.
Selection and Efficiency Testing of tagSNPs
Either the CEPH trios or the local European populations, with varying numbers of randomly selected subsamples (100 replicates), were used as reference samples. For these reference samples, two different tagSNP selection algorithms were applied. The first algorithm finds SNPs that best tag other typed SNPs (i.e., tagSNPs); the second algorithm finds SNPs that represent common haplotypes within predefined blocks (i.e., haplotype-tagging SNPs [htSNPs]).
A greedy algorithm for the selection of tagSNPs (Carlson et al. 2004) was employed. In the first step, a SNP exceeding an r2 threshold of 0.8 with the maximum number of other SNP sites is identified. This SNP and all associated SNPs are grouped in one bin. A bin does not have to be a group of neighboring SNPs but rather can be split up in several regions. Any SNP exceeding the threshold r2 with all other sites in the bin is specified as a tagSNP. There may be more than one tagSNP, and we used only the one with the maximum average r2. This binning process is iterated, and all as-yet-unbinned SNPs are analyzed at each round, until all sites are binned or characterized as singleton bins. The efficiency of a given tagSNP set in other population samples is tested by the following criteria: an average r2 among all typed SNPs and the best SNP-specific tagSNP, a minimal r2 among all typed SNPs and the best SNP-specific tagSNP, and a ratio of SNPs above the threshold r2 to any tagSNP.
The htSNP selection method started with the definition of blocks, in accordance with the standard method of Gabriel et al. (2002). Because we wanted to compare the efficiency of htSNPs across several populations, we always used the block structure of the CEPH trios as the reference and forced the block structure of other populations to this reference structure. A single optimal set of htSNPs within each block was identified by sequential steps (Zhang and Jin 2003); these steps account for haplotype coverage (80% and 90% thresholds), optimal r2 among SNPs, and even spacing. To evaluate the selected htSNP set in any population and to compare between populations, we defined two statistics (chromosomal coverage of tagged haplotypes and ratio of nontagged common haplotypes), whereby common haplotypes are defined by a frequency >5%.
Results
Single SNPs
Allele frequencies of most single markers did not differ significantly among population samples. In three gene regions, the proportion of population-differentiating markers (defined by signficance level P<.001) varied between 6% and 23% (table 1). An exception was PLAU, with 79% population-differentiating SNPs. Maximum values for allele-frequency differences were ∼20% and were mostly seen between EST and CALA, thus indicating a geographical gradient between the northern and southern populations (table A1 [online only]). The pattern of population differentiation for each gene region is shown in table A2 (online only). Significant population differences in allele frequencies appeared mostly for the gene PLAU but also for FKBP5 and LMNA. The CEPH founders were significantly different from the southern Italian populations BRISI and CALA. EST and the northern German collections SHIP and POPGEN showed significant differences from all Italian populations. The Alpine populations of VIN and LAD differed both from the southern Italian populations and from the northern European populations. The overall pattern of genetic differentiation reflects well the geographical localization of the population samples (table A3 and fig. A1 [online only]).
LD Structure
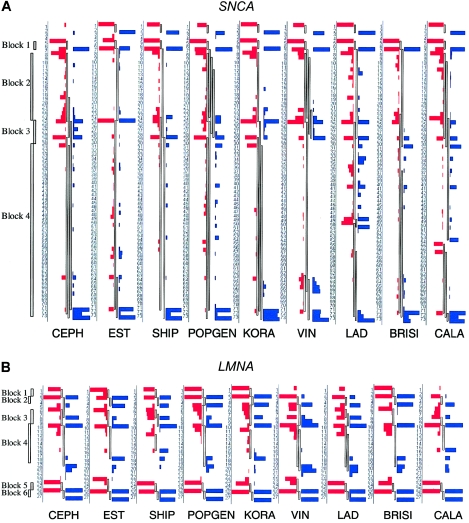
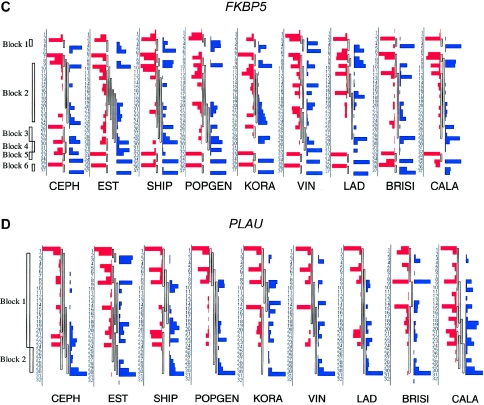
Standard plots of pairwise LD revealed similar patterns across samples (fig. A2 [online only]). To compare the LD structure across populations in a detailed and robust probability-based assessment, block overlaps were allowed and bootstrap frequencies of specific boundary positions were evaluated. The observed LD block structure and the bootstrap frequencies for each block start and block end are shown in figure 2. The calculations are based on a sample size of 100 individuals per population, to exclude variation in sample size as a potential confounder.
Figure 2.
Bootstrap frequencies of block starts and block ends in all population samples. All samples have an equal population size of 100 individuals (except BRISI, with 98 individuals). SNP markers are ordered vertically by their physical sequence. The length of red or blue bars indicates the bootstrap frequency of block starts or block ends, respectively, at the given position. Between the bars, the observed block structure is shown, with blocks allowed to overlap. To the left of each CEPH graph, the block structure of CEPH is shown, in accordance with the standard algorithm of Gabriel et al. (2002), without allowance for overlapping blocks . An example of a boundary shift can be seen at the end of block 4 in LMNA, which shows clear differences between the populations tested.
The general patterns of block structures are similar across samples, which is most prominent in the LMNA gene. Five of the six blocks in LMNA have nearly conserved block starts and ends across all study populations. Only the end of the largest block varied between positions 15 and 21, depending on the population studied. This represents a shift in the block extension in the range of 7–15 kb. Another example of differences in block boundaries is obvious for the SNCA region, where the largest block (between marker positions 30 and 73) has a tendency to break up into two pieces at different positions in the VIN (positions 63–67), LAD (positions 48–51), and CALA (positions 53–57) samples. Individual breakpoints of LD blocks for the Alpine populations VIN and LAD were also detected in the FKBP5 region between positions 14 and 18. PLAU also exhibited variable block structure.
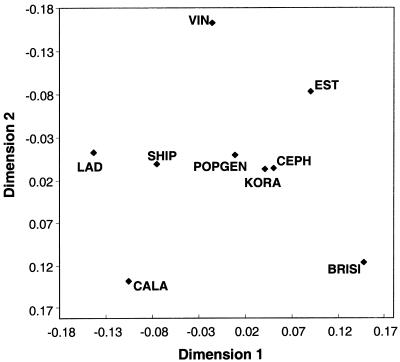
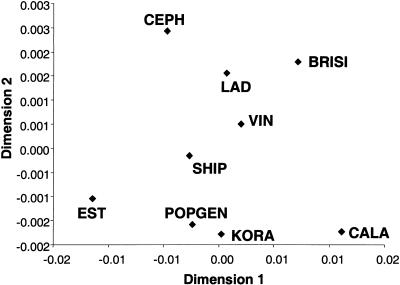
The overall variability in block structure among the populations is shown in figure 3. In a combined multidimensional scaling for all four gene regions, the most extreme and individual block structures were indicated for EST, LAD, VIN, BRISI, and CALA. The German populations, SHIP, POPGEN, and KORA, and the reference population CEPH appear in the center, indicating an intermediate block structure. Similar patterns were found when each gene was analyzed separately.
Figure 3.
Overall similarity of block boundaries across all four gene regions. The first two dimensions, after a multidimensional scaling of the dissimilarity measure of block boundaries, are shown. Sample sizes are adjusted to a size of 100 individuals. The Alpine and geographically peripheral populations (EST, LAD, VIN, BRISI, and CALA) differ the most from all other population samples.
Haplotypes
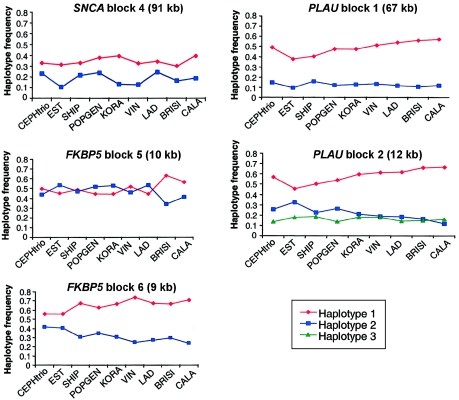
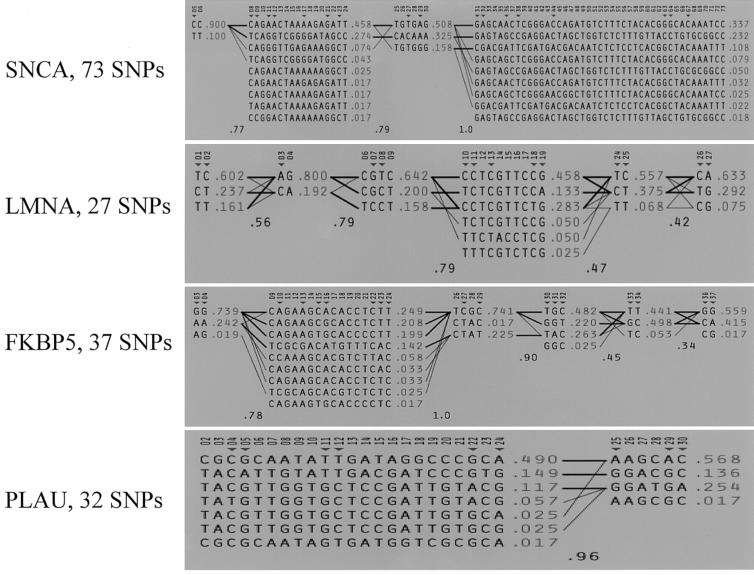
The standard algorithm of Gabriel et al. (2002), applied to the CEPH trios, allowed us to define four blocks in SNCA, six blocks in LMNA, six blocks in FKBP5, and two blocks in PLAU (see fig. 2, for reference of block positions, and fig. A3 [online only], for haplotype estimations). Significant haplotype frequency differences among populations were found only in block 4 of SNCA (P=.001), blocks 5 and 6 of FKBP5 (P=.03 and P=.02, respectively), and blocks 1 and 2 of PLAU (P=.001 and P=.01, respectively). Figure 4 shows the frequencies of all common haplotypes (frequency >10%) within the blocks that showed significant differences between populations. After Bonferroni correction for multiple testing, only the haplotype distributions of SNCA block 4 and PLAU block 1 remained significant. A clear geographic variation was evident with FKBP5 and PLAU but not with SNCA. In the PLAU gene region, haplotype 1 in block 1 showed the most extreme frequency values in EST (40%) and CALA (57%) and showed a gradient in between these two values in the remaining six populations. CEPH trios showed intermediate frequencies. A similar pattern was found for the haplotypes in block 2 of PLAU. There was also a gradient between EST and CALA in block 6 of FKBP5, but here CEPH trios are most similar to the EST sample. In block 5 of FKBP5, BRISI and CALA diverge from all other samples.
Figure 4.
Frequencies of common haplotypes (>10%) in all populations for the five haplotype blocks with significant population differentiation. For block numbers, see figure 2. Populations are arranged on the X-axis in a north-to-south localization. Geographical frequency gradients are prominent in blocks 1 and 2 of PLAU and in block 6 of FKBP5.
tagSNPs
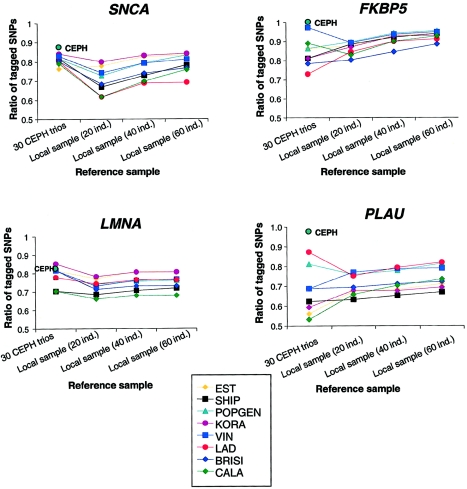
We first tested the efficiency of tagSNPs, which were defined to represent untagged SNPs with a high correlation coefficient (r2 > 0.8 [Carlson et al. 2004]). Figure 5 shows the performance of CEPH trios, as a reference for this tagSNP selection, in comparison with local population samples of different sizes. For each sample size, average values across 100 replicates are given. Only the criterion of a ratio of tagged SNPs above the threshold, which is the relative portion of SNPs correlated with any tagSNP by an r2 value >0.8, is shown. Other evaluation criteria (see the “Subjects and Methods” section) gave similar results (see table A4 [online only]). A reduced SNP set, which is comparable to the HapMap set, was used for the tagSNP selection (table 1). This reduced SNP set comprised ∼40% of SNPs identical to HapMap data. The selected tag SNPs were then tested on the full SNP set in all populations. The LD patterns appeared to be similar across all different SNP sets (see fig. A4 [online only]). There was no difference between the tagSNP set defined from CEPH trios and the tagSNP set defined from CEPH founders only, indicating that the additional phase information does not change the outcome.
Figure 5.
Performance of CEPH trios and local samples with different sample sizes used as references for tagSNP definition (by use of the method of Carlson et al. [2004]). The performance criterion shown is the ratio of tagged SNPs above the r2 threshold of 0.8. The tagSNP sets that were defined in the CEPH trios were tested on all populations, whereas the tagSNP sets of local samples were tested only on the same local population. CEPH trios performed relatively well as reference (ratio of tagged SNPs >0.7), except for the PLAU gene region.
The tagSNPs identified in the CEPH trios performed well for the genes SNCA, FKBP5, and LMNA. For >70% of typed SNPs, the r2 value was >0.8 with the best tagSNP. SNP allelic variation in KORA, for example, is well represented by CEPH tagSNPs for the genes SNCA and LMNA. In LMNA and SNCA, a local sample size of 20 individuals as a reference performs mostly worse than the 30 CEPH trios. Only a sample size of 40 or 60 individuals is comparable to CEPH trios. In FKBP5, most local samples of 20 individuals performed better as a reference than the CEPH sample, except for VIN and CALA. A different situation was seen in PLAU, where six populations showed a ratio of tagged SNPs of <70%, when the CEPH sample was used as reference—the worst ratio was from the CALA sample, with only 53%. For the same gene, data from 20 random individuals of most populations performed better as a reference than CEPH trios.
With the current HapMap SNP density as a reference, minimal r2 values between tagged and tagSNPs were as low as 0.036, even for the most conserved gene regions around SNCA and LMNA. We therefore tested the performance of tagSNP sets, when selected from the full SNP set in SNCA and PLAU, for which we had a >2-fold SNP density compared with that of the HapMap. The general pattern—a local sample with 20 individuals performs mostly better as a reference than the CEPH sample in PLAU, and CEPH performs better than local samples in SNCA—did not change, but the differences were less pronounced, and, even for PLAU, the ratio of tagged SNPs was >70% in all tested populations (table A4 [online only]).
Performance patterns were similar when the haplotype-based tagSNP selection method (i.e., the htSNP selection method) was used (Zhang and Jin 2003), but differences between CEPH and local references were weaker than those measured by the r2 method (table A5 [online only]). When CEPH trios were used as reference, chromosomal coverage of tagged haplotypes was below the intended threshold (80% and 90%, respectively) only for the PLAU gene (see population samples EST, SHIP, and KORA).
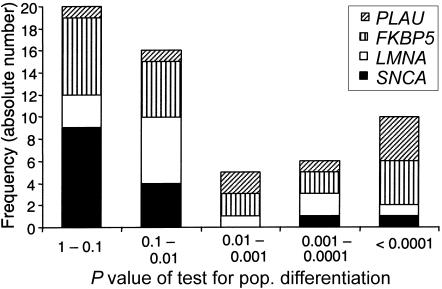
tagSNPs may also be seen as a set of relatively independent markers. To assess the probability of recruiting population-differentiating SNPs in a genomic approach, we plotted a histogram of the P values of tests for population differentiation for all tagSNPs defined by the method of Carlson et al. (2004) in CEPH trios (fig. 6). The majority of tagSNPs did not show strong population differences, underlining their universality. Most highly significant markers were found in the gene regions PLAU and FKBP5.
Figure 6.
Histogram of P values of tests for population differentiation, on the basis of 57 tagSNPs from all gene regions in the CEPH trios. The allele frequencies of most tagSNPs were similar across populations (P>.01). Exceptions were the tagSNPs of the PLAU gene.
Discussion
Individual population history and geographic variation may challenge the usefulness of a single European reference population for the selection of tagSNPs in association studies. Allele and haplotype frequencies show a clear geographic variation. Most dramatic frequency shifts lie in the upper range of values found in the survey of random loci done by Cavalli-Sforza et al. (1994), but these are still low compared with the values in strongly selected loci (e.g., cystic fibrosis variants [Lao et al. 2003]). The pattern of genetic differentiation corresponds relatively well to the European genetic variation described by Barbujani and Sokal (1990) and Cavalli-Sforza et al. (1994). The relatively strong genetic divergence of the Italian populations, CALA and BRISI, and the Alpine populations, LAD and VIN, from all other populations can be attributed to isolation resulting from linguistic differences (Germanic-Romance) and physical boundaries (the Alps). The two Italian populations, BRISI and CALA, also show significant differences from the CEPH sample and are therefore less well represented by this reference population.
Large-scale association studies—probably across different ethnic groups—are needed to detect small genetic effects on complex traits, and it is well known that even small amounts of cryptic population stratification can undermine such association studies (Marchini et al. 2004). However, high numbers of markers—in the range of several hundred microsatellite loci or a multitude of SNP loci—are required to detect genetic clusters of different ethnic origins in Europe (Rosenberg et al. 2002). Our results indicate that, in our total region of 749 kb, ∼28% (16/57) of the tagSNPs showed highly significant differences (P<.001) among our set of study populations. This rate indicates that the recruitment of population-differentiating SNPs for the purpose of genetic matching strategies in case-control studies is feasible (Hoggart et al. 2004).
Comparative analyses of the haplotype block structure revealed a high degree of concordance among European populations (Nejentsev et al. 2004; Ng et al. 2004; Stenzel et al. 2004), as well as among populations from different continents, such as Asia, Africa, and Europe (Gabriel et al. 2002; Wall and Pritchard 2003). This presumably reflects, in part, the shared ancestry of human populations or common variation patterns of recombination rates, but, to some extent, it also reflects the effect of uneven marker spacing in these studies. However, small, well-defined differences for block boundaries have been reported among Finnish subpopulations (Mannila et al. 2003). All the above-mentioned studies (except Mannila et al. 2003) compare the positions of LD block boundaries by use of a greedy algorithm—or just by inspection of pairwise LD measures—but do not account for the relative probabilities of specific boundary positions. With our method of evaluating the strength of block boundaries, which was applied to exactly the same set of common markers in each population, we were able to show clear examples of block boundary shifts and block fragmentation among European samples. The values of our similarity measure for block structure, which estimate the average probability that boundaries coincide, ranged from 0.72 among LAD and BRISI to 0.87 among SHIP and POPGEN. With the exception of the Alpine populations, the overall variation appeared in a pattern that was concordant with geography, indicating the usefulness of our similarity measure for population-genetic comparisons. The observed pattern suggests that demographic and/or biological factors shaping block boundaries vary in a geographical sense and differentiate in accordance with the level of presumed genetic isolation of populations.
The observed population differences in haplotype frequencies and LD structure may affect the power to detect phenotype-genotype associations. Association signals at markers, which are correlated with a true causal variant, may appear at different positions in populations with an individual LD structure, such as the VIN, LAD, and CALA populations. Repeated studies among such populations are likely to present different results and are problematic for finding positive replications. In contrast, population-specific fragmented LD blocks are useful for the fine-mapping of causal variants within the region.
We also tested the transferability of tagSNP sets among populations. It is often stated that tagSNPs are population specific and should be newly assessed in each local population or geographic area in which an association study is planned (Thompson et al. 2003; Weale et al. 2003; Carlson et al. 2004). On the other hand, the HapMap project claims that its data may be able to be used to define tagSNPs for related populations (International HapMap Consortium 2003). It has also been reported that tagSNPs can be effectively transferred among British, Norwegian, Finnish, and Romanian populations (Nejentsev et al. 2004). It is, however, not clear to what level of population differentiation tagSNPs are transferable between the HapMap data and local European populations. Our results indicate that tagSNPs defined in the HapMap CEPH trios perform relatively well for two of four candidate-gene regions, particularly in central European populations. For SNCA and LMNA, the data from CEPH trios perform even better as a reference than data from 20 local individuals. For two of the tested candidate genes (PLAU and FKBP5), CEPH is not such a good reference. A local sample size of only 20 individuals in most populations is more appropriate for determination of tagSNPs than the standard sample of 30 CEPH trios. By genotyping larger sample sizes (>20 individuals) in the population being studied, the advantage of a local reference will be stronger, but it appears that an increase in sample size beyond 40 individuals is not very effective. A substantial increase in tagSNP efficiency and transferability, however, is achieved by increasing the density of genotyped SNPs in the reference sample.
The surprisingly high performance of CEPH as a reference for tagSNP design in two gene regions was not due to an increased number of selected tagSNPs in CEPH or the additional phase information available from the trios (see table A4 [online only]). The special characteristic of CEPH being a multilocalized but panmictic European population probably confers the advantage to this sample collection. Our results suggest that future HapMap releases with a denser genotype data set will allow the sufficient selection of tagSNPs in the majority of gene regions in central European populations. However, for an as-yet-unknown proportion of genes, and especially for isolated and peripheral populations within Europe, the HapMap reference may not perform optimally, making it necessary to establish the LD pattern from a local sample.
Acknowledgments
This work was supported by the National Genome Research Network and the Bioinformatics for the Functional Analysis of Mammalian Genomes project from the German Federal Ministry of Education and Research. A.M. and E.L. were partially supported by Targeted Funding EMRE 0182582s03, and E.L. had a fellowship from the E.U. grants Mol Tools 503155 and “Genera” to Estonian Biocentre. M.R. and R.M. were supported by a core grant from the Estonian Ministry of Education and Research. The recruitment of the south Tyrolian samples VIN and LAD was supported by a grant from the Autonomous Province Bolzano and from the Südtiroler Sparkasse, Bolzano. The project POPGEN is supported by the Deutsche Forschungsgemeinschaft research group FOR 423 (“Polygenic Disorders”). The SHIP studies are funded by the German Federal Ministry for Education and Research (grant 01ZZ96030), by the Ministry for Education, Research, and Cultural Affairs, and by the Ministry for Social Affairs of the State of Mecklenburg-West Pomerania. We gratefully acknowledge the participation of all probands, as well as the review of the manuscript by Jack Favor.
Appendix A: Supplemental Material
Figure A1.
Multidimensional scaling plot based on Reynold distances (transformed FST values, linearized to population-divergence time). The FST values were calculated from allele frequencies of all four gene regions.
Figure A2.
LD structure (pairwise D′ values) across all nine population samples, with a minor-allele frequency (MAF) >5%.
Figure A3.
Estimated haplotypes with frequency >1% within each block of the four genomic regions for the CEPH trios. Blocks are defined by the standard Gabriel et al. (2002) algorithm (software used was Haploview).
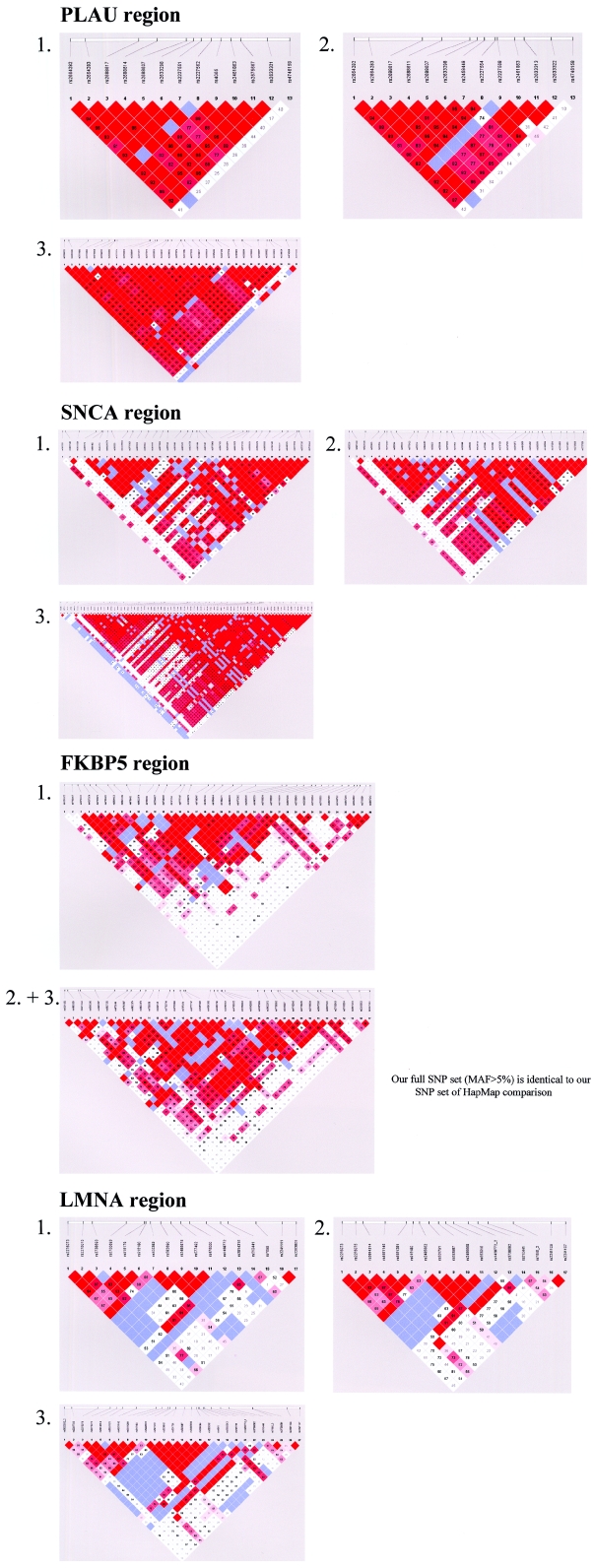
Figure A4.
LD structure in CEPH trios for all four gene regions. For each gene, comparisons of different SNP sets are shown. 1, Original HapMap SNP set. 2, Our SNP set for HapMap comparison. 3, Our full SNP set (minor-allele frequency [MAF] >5%).
Table A1.
Minor-Allele Frequency for All Four Gene Regions
|
Minor-Allele Frequency for |
||||||||||||
| Gene Region and SNP | NCBI Build 34 (hg16) | Major Allele | Minor Allele | CEPH Trio Founders | EST | SHIP | POPGEN | KORA | VIN | LAD | BRISI | CALA |
| SNCA: | ||||||||||||
| rs4122859 | 91054570 | A | G | .042 | .121 | .05 | .075 | .08 | .091 | .081 | .092 | .09 |
| rs3857046 | 91059165 | G | A | .042 | .089 | .05 | .066 | .068 | .077 | .081 | .031 | .076 |
| rs3857047 | 91064912 | T | G | .058 | .156 | .14 | .145 | .176 | .138 | .128 | .138 | .21 |
| rs356229 | 91064991 | A | G | .45 | .379 | .419 | .359 | .396 | .385 | .4 | .378 | .328 |
| rs3857048 | 91067353 | C | T | .1 | .109 | .08 | .116 | .136 | .106 | .11 | .117 | .143 |
| rs3906628 | 91076920 | C | T | .1 | .109 | .081 | .116 | .136 | .112 | .111 | .117 | .143 |
| rs356183 | 91084492 | G | C | .467 | .485 | .434 | .44 | .435 | .422 | .439 | .44 | .422 |
| rs356180 | 91086521 | C | T | .35 | .324 | .354 | .3 | .349 | .339 | .347 | .322 | .27 |
| rs356169 | 91091162 | A | C | .342 | .362 | .345 | .334 | .376 | .355 | .394 | .335 | .345 |
| rs2572323 | 91092946 | G | A | .325 | .323 | .35 | .3 | .339 | .338 | .352 | .299 | .305 |
| rs356215 | 91094955 | A | G | .466 | .449 | .442 | .545 | .398 | .399 | .436 | .387 | .47 |
| rs356219 | 91095995 | A | G | .425 | .407 | .404 | .375 | .353 | .377 | .368 | .347 | .343 |
| rs356220 | 91099734 | C | T | .433 | .413 | .308 | .373 | .35 | .371 | .296 | .353 | .259 |
| rs356222 | 91101517 | T | C | .322 | .312 | .308 | .294 | .325 | .321 | .336 | .293 | .253 |
| rs356165 | 91105280 | A | G | .433 | .41 | .399 | .375 | .35 | .371 | .355 | .358 | .34 |
| rs3775422 | 91113058 | G | A | .1 | .072 | .062 | .075 | .021 | .033 | .019 | .048 | .078 |
| rs356205 | 91115580 | A | G | .336 | .29 | .32 | .303 | .318 | .321 | .34 | .301 | .245 |
| rs3822086 | 91123188 | C | T | .1 | .093 | .065 | .075 | .021 | .042 | .019 | .059 | .086 |
| rs356203 | 91124435 | A | G | .441 | .414 | .394 | .375 | .385 | .393 | .355 | .35 | .34 |
| rs356202 | 91124685 | A | G | .342 | .321 | .328 | .3 | .327 | .334 | .336 | .289 | .25 |
| rs356200 | 91127008 | G | A | .481 | .518 | .455 | .482 | .457 | .454 | .453 | .432 | .445 |
| rs2736991 | 91130064 | A | T | .312 | .352 | .33 | .294 | .339 | .356 | .334 | .306 | .237 |
| rs356167 | 91132164 | G | A | .298 | .296 | .273 | .28 | .269 | .307 | .263 | .265 | .209 |
| rs356168 | 91132825 | A | G | .483 | .515 | .45 | .475 | .45 | .459 | .453 | .426 | .445 |
| rs2736990 | 91136935 | T | C | .483 | .506 | .449 | .472 | .436 | .443 | .453 | .418 | .439 |
| rs2572324 | 91137192 | T | C | .333 | .315 | .32 | .299 | .33 | .329 | .334 | .292 | .253 |
| rs356199 | 91140721 | T | C | .322 | .293 | .293 | .304 | .306 | .308 | .334 | .301 | .258 |
| rs356195 | 91141562 | G | A | .34 | .295 | .298 | .309 | .313 | .298 | .334 | .305 | .25 |
| rs356192 | 91146321 | T | C | .325 | .295 | .3 | .3 | .301 | .31 | .334 | .304 | .25 |
| rs356189 | 91149526 | G | A | .331 | .29 | .295 | .304 | .296 | .314 | .333 | .301 | .25 |
| rs356188 | 91149931 | A | G | .158 | .197 | .215 | .166 | .21 | .219 | .216 | .214 | .175 |
| rs356187 | 91150862 | G | A | .339 | .289 | .295 | .299 | .304 | .317 | .334 | .301 | .25 |
| rs356164 | 91151870 | G | C | .117 | .185 | .146 | .103 | .14 | .141 | .154 | .156 | .066 |
| rs356162 | 91155551 | A | G | .158 | .203 | .217 | .166 | .207 | .216 | .216 | .222 | .175 |
| rs4031753 | 91158555 | G | C | .1 | .087 | .066 | .081 | .021 | .046 | .019 | .056 | .085 |
| rs356184 | 91161627 | G | A | .322 | .297 | .295 | .3 | .296 | .31 | .334 | .307 | .255 |
| rs356186 | 91163758 | C | T | .136 | .173 | .2 | .152 | .189 | .195 | .216 | .196 | .17 |
| rs2737033 | 91166341 | A | G | .325 | .292 | .295 | .297 | .313 | .315 | .341 | .299 | .255 |
| rs2737029 | 91170164 | A | G | .45 | .449 | .39 | .428 | .38 | .405 | .394 | .399 | .38 |
| rs2737028 | 91175410 | C | T | .149 | .203 | .217 | .161 | .205 | .211 | .216 | .213 | .175 |
| rs2737025 | 91177586 | C | T | .118 | .219 | .018 | .028 | .213 | .217 | .048 | .212 | .011 |
| rs2737024 | 91179954 | T | C | .317 | .282 | .282 | .287 | .304 | .31 | .337 | .304 | .237 |
| rs2583959 | 91180031 | C | G | .328 | .284 | .295 | .297 | .302 | .308 | .334 | .301 | .25 |
| rs2619373 | 91180827 | G | A | .317 | .285 | .308 | .3 | .303 | .312 | .321 | .301 | .234 |
| rs2197120 | 91187996 | G | A | .161 | .203 | .215 | .166 | .206 | .214 | .216 | .224 | .175 |
| rs2619368 | 91188141 | G | T | .15 | .175 | .212 | .167 | .18 | .183 | .214 | .194 | .173 |
| rs2619369 | 91192063 | A | G | .042 | .04 | .02 | .034 | .027 | .024 | .019 | .036 | .01 |
| rs748849 | 91193355 | A | G | .147 | .155 | .214 | .154 | .193 | .191 | .212 | .198 | .177 |
| rs1837890 | 91194400 | C | A | .192 | .29 | .255 | .23 | .249 | .269 | .266 | .258 | .235 |
| rs2619370 | 91194979 | C | T | .325 | .279 | .291 | .276 | .298 | .314 | .32 | .312 | .253 |
| rs1442145 | 91195317 | A | G | .173 | .303 | .25 | .216 | .217 | .259 | .252 | .273 | .239 |
| rs972880 | 91197649 | G | A | .164 | .207 | .203 | .174 | .219 | .233 | .217 | .223 | .159 |
| rs1812923 | 91197933 | C | A | .492 | .429 | .421 | .474 | .446 | .428 | .389 | .422 | .495 |
| rs2737021 | 91198386 | T | A | .153 | .205 | .217 | .166 | .212 | .211 | .253 | .224 | .192 |
| rs2619341 | 91200167 | G | A | .147 | .204 | .22 | .166 | .205 | .214 | .214 | .224 | .175 |
| rs2737014 | 91203850 | T | G | .316 | .281 | .29 | .297 | .311 | .304 | .333 | .302 | .255 |
| rs2737012 | 91204101 | C | T | .33 | .284 | .298 | .296 | .301 | .301 | .333 | .305 | .255 |
| rs2583969 | 91204527 | T | C | .325 | .28 | .288 | .294 | .277 | .298 | .331 | .292 | .258 |
| rs2737010 | 91204860 | T | C | .164 | .208 | .188 | .16 | .218 | .236 | .213 | .23 | .151 |
| rs2737009 | 91205230 | T | C | .322 | .298 | .29 | .297 | .293 | .316 | .331 | .324 | .255 |
| rs2737008 | 91205577 | C | T | .325 | .274 | .29 | .297 | .289 | .299 | .331 | .293 | .255 |
| rs1811442 | 91206145 | T | C | .147 | .202 | .22 | .166 | .213 | .223 | .217 | .224 | .175 |
| rs920624 | 91206589 | A | T | .483 | .486 | .531 | .472 | .517 | .544 | .56 | .575 | .432 |
| rs1442146 | 91207040 | C | G | .325 | .284 | .288 | .299 | .295 | .31 | .329 | .301 | .253 |
| rs1442149 | 91207526 | A | T | .322 | .287 | .29 | .296 | .301 | .304 | .331 | .3 | .255 |
| rs2737001 | 91211871 | C | T | .325 | .282 | .293 | .3 | .288 | .298 | .331 | .3 | .255 |
| rs1372516 | 91214124 | G | A | .328 | .283 | .29 | .303 | .291 | .308 | .331 | .301 | .255 |
| rs2028535 | 91214815 | G | C | .319 | .218 | .286 | .289 | .261 | .258 | .326 | .294 | .25 |
| rs2301135 | 91216783 | C | G | .483 | .506 | .49 | .525 | .471 | .462 | .447 | .443 | .555 |
| rs2301134 | 91217339 | T | C | .483 | .509 | .495 | .525 | .47 | .471 | .446 | .438 | .56 |
| rs2619364 | 91218281 | A | G | .317 | .284 | .283 | .297 | .291 | .308 | .33 | .294 | .265 |
| rs2583988 | 91219222 | C | T | .258 | .132 | .242 | .278 | .145 | .15 | .287 | .172 | .217 |
| rs2619366 | 91221654 | A | G | .317 | .284 | .288 | .297 | .298 | .304 | .328 | .291 | .265 |
| rs2619367 | 91225489 | A | C | .317 | .285 | .293 | .302 | .298 | .312 | .326 | .291 | .27 |
| rs2583989 | 91227816 | A | G | .317 | .278 | .286 | .296 | .298 | .308 | .326 | .28 | .263 |
| rs2736993 | 91231245 | T | G | .317 | .283 | .288 | .299 | .294 | .312 | .328 | .291 | .27 |
| rs2737026 | 91238217 | C | T | .186 | .177 | .22 | .186 | .225 | .227 | .225 | .239 | .205 |
| rs2736994 | 91242922 | C | T | .183 | .207 | .2 | .163 | .208 | .205 | .216 | .26 | .19 |
| LMNA: | ||||||||||||
| rs3820592_2 | 153222805 | T | C | .23 | .24 | .21 | .17 | .23 | .18 | .18 | .22 | .12 |
| rs2297792 | 153228236 | C | T | .4 | .34 | .31 | .32 | .41 | .35 | .33 | .41 | .35 |
| rs2275073 | 153247612 | A | C | .2 | .16 | .2 | .19 | .22 | .19 | .13 | .26 | .2 |
| rs2275075 | 153257102 | G | A | .19 | .14 | .18 | .17 | .19 | .15 | .12 | .23 | .18 |
| rs3814314 | 153261975 | T | A | .21 | .21 | .21 | .27 | .23 | .24 | .25 | .23 | .14 |
| rs6691151 | 153266330 | C | T | .15 | .1 | .13 | .13 | .13 | .14 | .09 | .23 | .19 |
| rs4661146 | 153276942 | G | C | .16 | .1 | .13 | .13 | .13 | .14 | .09 | .24 | .19 |
| rs6661281 | 153291637 | T | C | .36 | .32 | .35 | .4 | .37 | .41 | .33 | .46 | .34 |
| rs915180 | 153295875 | C | T | .36 | .33 | .37 | .4 | .38 | .39 | .35 | .46 | .34 |
| rs2485662 | 153300260 | C | T | .26 | .27 | .25 | .28 | .28 | .34 | .3 | .29 | .24 |
| LMNA_S17S | 153301572 | C | T | .01 | 0 | .01 | .01 | .01 | .01 | 0 | .03 | .01 |
| rs547915 | 153302167 | C | T | .08 | .09 | .04 | .05 | .09 | .12 | .04 | .07 | .08 |
| rs503815 | 153306401 | T | C | .05 | .04 | .04 | .04 | .07 | .1 | .05 | .06 | .1 |
| rs501791 | 153306665 | C | T | .05 | .04 | .04 | .05 | .06 | .1 | .05 | .06 | .09 |
| rs593987 | 153313179 | G | A | .05 | .04 | .03 | .05 | .07 | .1 | .04 | .06 | .09 |
| LMNAR119R | 153317223 | C | T | 0 | .01 | 0 | 0 | .01 | .01 | 0 | 0 | 0 |
| rs2485668 | 153318341 | T | C | .05 | .04 | .04 | .05 | .07 | .1 | .05 | .06 | .1 |
| rs538089 | 153321820 | T | C | .08 | .09 | .05 | .04 | .09 | .12 | .05 | .08 | .06 |
| rs553016 | 153323655 | C | T | .08 | .09 | .07 | .05 | .08 | .12 | .06 | .08 | .11 |
| rs4641 | 153324326 | C | T | .28 | .17 | .25 | .26 | .24 | .24 | .25 | .27 | .25 |
| rs520973 | 153324811 | G | A | 0 | .04 | .03 | 0 | .08 | .09 | .01 | .06 | .05 |
| rs6669212 | 153327105 | G | A | .13 | .16 | .17 | .18 | .15 | .18 | .23 | .16 | .11 |
| rs545731 | 153328082 | G | A | .06 | .04 | .06 | .07 | .06 | .1 | .07 | .09 | .08 |
| rs1468772_2 | 153333280 | T | G | .28 | .33 | .35 | .26 | .26 | .27 | .26 | .3 | .29 |
| rs3738582 | 153340022 | C | G | .22 | .18 | .17 | .19 | .25 | .26 | .17 | .24 | .25 |
| rs510441 | 153346760 | A | G | .22 | .25 | .31 | .21 | .25 | .27 | .31 | .24 | .31 |
| rs7695_2 | 153364118 | T | C | .38 | .39 | .4 | .37 | .37 | .42 | .43 | .39 | .36 |
| rs3738581 | 153364350 | C | T | .44 | .44 | .44 | .38 | .4 | .45 | .47 | .41 | .43 |
| rs2241109 | 153389874 | C | T | .3 | .34 | .22 | .3 | .33 | .21 | .22 | .26 | .29 |
| rs2241107 | 153399502 | A | G | .37 | .46 | .32 | .36 | .41 | .28 | .32 | .34 | .38 |
| FKBP5: | ||||||||||||
| rs1051952 | 35467201 | A | C | .45 | .44 | .49 | .48 | .43 | .41 | .4 | .34 | .38 |
| rs1883636 | 35470074 | A | G | .12 | .11 | .17 | .15 | .13 | .13 | .11 | .08 | .11 |
| rs2273000 | 35479883 | G | A | .27 | .29 | .32 | .33 | .32 | .29 | .24 | .33 | .37 |
| rs1540910 | 35481677 | G | A | .24 | .26 | .28 | .28 | .3 | .26 | .2 | .29 | .33 |
| rs1883637 | 35496014 | C | T | .08 | .08 | .11 | .09 | .08 | .09 | .06 | .03 | .04 |
| rs3807050 | 35514341 | C | T | .34 | .25 | .25 | .21 | .19 | .23 | .29 | .12 | .11 |
| rs873941 | 35526292 | A | T | .26 | .31 | .26 | .33 | .34 | .27 | .34 | .36 | .42 |
| rs4713897 | 35529985 | G | A | .17 | .2 | .18 | .21 | .2 | .17 | .15 | .22 | .26 |
| rs3800374 | 35538821 | C | T | .17 | .18 | .2 | .21 | .18 | .17 | .18 | .22 | .3 |
| rs3800373 | 35543891 | A | C | .23 | .22 | .29 | .28 | .28 | .25 | .25 | .27 | .31 |
| rs755658 | 35551085 | G | A | .05 | .08 | .11 | .09 | .09 | .08 | .06 | .04 | .04 |
| rs992105 | 35556598 | A | C | .14 | .14 | .16 | .18 | .16 | .15 | .16 | .23 | .25 |
| rs7753746 | 35566837 | A | G | .15 | .14 | .14 | .16 | .19 | .15 | .18 | .28 | .28 |
| rs4713899 | 35570696 | G | A | .15 | .14 | .14 | .16 | .19 | .14 | .17 | .26 | .27 |
| rs737054 | 35576902 | C | T | .23 | .3 | .27 | .27 | .3 | .32 | .3 | .32 | .29 |
| rs3777747 | 35580417 | A | G | .42 | .49 | .46 | .47 | .45 | .49 | .42 | .53 | .52 |
| rs6457836 | 35581713 | C | T | .15 | .15 | .14 | .16 | .2 | .14 | .18 | .29 | .28 |
| rs7747121 | 35595398 | A | G | .01 | .01 | .01 | .01 | .03 | .01 | .01 | .03 | .02 |
| rs1591365 | 35605522 | A | G | .25 | .25 | .29 | .29 | .3 | .24 | .28 | .34 | .34 |
| rs1360780 | 35608986 | C | T | .24 | .25 | .28 | .29 | .3 | .24 | .28 | .33 | .33 |
| rs2143404 | 35612096 | C | T | .15 | .14 | .13 | .15 | .18 | .14 | .2 | .24 | .27 |
| rs4713902 | 35615441 | T | C | .21 | .29 | .26 | .28 | .29 | .32 | .27 | .31 | .31 |
| rs1334894 | 35616545 | C | T | .06 | .09 | .11 | .09 | .08 | .08 | .06 | .03 | .04 |
| rs6912833 | 35619000 | T | A | .25 | .27 | .27 | .28 | .31 | .27 | .29 | .34 | .32 |
| rs1475774 | 35620969 | G | A | .01 | .01 | .02 | .02 | .02 | .01 | .02 | .02 | .02 |
| rs2092427 | 35623622 | G | A | .01 | .01 | .02 | .02 | .02 | .01 | .02 | .02 | .02 |
| rs7747647 | 35630615 | A | C | 0 | .03 | .05 | .06 | .09 | .03 | .05 | .06 | .1 |
| rs4713907 | 35644490 | G | A | 0 | 0 | .01 | .02 | .01 | 0 | .01 | .02 | .01 |
| rs4713908 | 35648725 | A | G | .01 | .01 | .02 | .02 | .02 | .01 | .02 | .02 | .02 |
| rs6457839 | 35650245 | T | C | .31 | .34 | .29 | .31 | .34 | .33 | .29 | .41 | .32 |
| rs3800372 | 35656660 | T | C | .22 | .26 | .31 | .32 | .18 | .23 | .3 | .36 | .34 |
| rs7759392 | 35663234 | T | C | .24 | .28 | .29 | .29 | .3 | .27 | .29 | .32 | .31 |
| rs943297 | 35669275 | C | T | .24 | .29 | .27 | .28 | .3 | .27 | .28 | .33 | .31 |
| rs4713916 | 35671398 | G | A | .25 | .28 | .28 | .28 | .29 | .27 | .28 | .32 | .31 |
| rs4713921 | 35683192 | C | T | .23 | .31 | .29 | .28 | .31 | .28 | .29 | .34 | .32 |
| rs2766534 | 35687129 | T | G | .24 | .21 | .22 | .21 | .19 | .16 | .13 | .21 | .2 |
| rs2817035 | 35697778 | G | A | .26 | .28 | .29 | .28 | .3 | .28 | .28 | .32 | .32 |
| rs2817041 | 35707307 | C | T | .23 | .2 | .19 | .21 | .15 | .15 | .11 | .24 | .19 |
| rs2766543 | 35710049 | T | G | .51 | .45 | .49 | .45 | .45 | .52 | .44 | .64 | .57 |
| rs2766554 | 35720742 | C | T | .45 | .53 | .49 | .52 | .53 | .47 | .53 | .36 | .41 |
| rs2817054 | 35735114 | G | A | .5 | .44 | .39 | .44 | .39 | .27 | .36 | .31 | .22 |
| rs2296662 | 35746184 | G | C | .43 | .44 | .32 | .37 | .32 | .25 | .32 | .34 | .27 |
| rs2817010 | 35755964 | G | A | .42 | .41 | .31 | .35 | .31 | .25 | .28 | .3 | .26 |
| rs2766597 | 35766458 | A | G | .03 | .01 | .02 | .01 | .01 | .01 | .01 | .02 | .01 |
| PLAU: | ||||||||||||
| rs2688626 | 74955693 | A | G | .25 | .27 | .2 | .27 | .19 | .15 | .18 | .12 | .12 |
| rs2250140 | 74957484 | C | T | .43 | .51 | .51 | .46 | .42 | .39 | .39 | .33 | .35 |
| rs2664282 | 74965360 | G | A | .43 | .44 | .51 | .46 | .39 | .33 | .38 | .28 | .32 |
| rs2664283 | 74970976 | C | T | .08 | .07 | .06 | .06 | .04 | .03 | .03 | .02 | .02 |
| rs4746154 | 74973999 | G | A | .18 | .2 | .3 | .18 | .21 | .22 | .19 | .18 | .22 |
| rs2688625 | 74976151 | C | T | .43 | .51 | .51 | .46 | .42 | .37 | .4 | .33 | .35 |
| rs2633312 | 74976358 | A | T | .43 | .47 | .5 | .46 | .45 | .39 | .4 | .32 | .35 |
| rs2675671 | 74977363 | A | G | .43 | .51 | .52 | .46 | .43 | .38 | .39 | .33 | .36 |
| rs2633303 | 74990485 | T | G | .26 | .33 | .25 | .28 | .21 | .17 | .19 | .15 | .11 |
| rs2688617 | 74990750 | A | T | .26 | .33 | .24 | .28 | .21 | .17 | .19 | .15 | .11 |
| rs2675677 | 74992852 | T | G | .27 | .33 | .26 | .3 | .21 | .17 | .2 | .17 | .13 |
| rs2675675 | 74993651 | T | C | .26 | .33 | .24 | .24 | .2 | .17 | .18 | .16 | .11 |
| rs2633306 | 74997050 | G | T | .26 | .34 | .26 | .29 | .22 | .17 | .21 | .16 | .12 |
| rs2688611 | 74997975 | A | C | .27 | .37 | .25 | .28 | .28 | .22 | .19 | .19 | .11 |
| rs2688610 | 74999534 | T | C | .46 | .54 | .49 | .45 | .45 | .41 | .39 | .38 | .36 |
| rs2675679 | 75003184 | A | G | .47 | .55 | .49 | .47 | .43 | .38 | .39 | .34 | .4 |
| rs2675680 | 75003467 | G | A | .43 | .52 | .49 | .45 | .43 | .38 | .39 | .32 | .35 |
| rs2675663 | 75004873 | G | T | .46 | .54 | .51 | .47 | .44 | .39 | .4 | .35 | .4 |
| rs2688607 | 75008339 | C | T | .24 | .32 | .27 | .26 | .2 | .2 | .22 | .18 | .14 |
| rs2633298 | 75010942 | C | G | .26 | .37 | .3 | .32 | .25 | .21 | .23 | .2 | .19 |
| rs2459449 | 75013616 | C | T | .26 | .32 | .29 | .31 | .22 | .2 | .23 | .19 | .14 |
| rs2227553 | 75014546 | T | G | 0 | .02 | .01 | .01 | .02 | .01 | .02 | .01 | .02 |
| rs2227564 | 75017704 | G | A | .21 | .3 | .24 | .26 | .2 | .19 | .21 | .17 | .13 |
| rs2227568 | 75018482 | C | T | .15 | .16 | .18 | .14 | .11 | .15 | .13 | .14 | .13 |
| rs2227583 | 75019822 | T | C | .02 | .01 | .01 | .02 | .01 | 0 | .01 | .02 | .03 |
| rs2461863 | 75024839 | A | G | .4 | .51 | .45 | .43 | .38 | .38 | .35 | .33 | .28 |
| rs2633314 | 75028193 | A | G | .42 | .54 | .47 | .46 | .4 | .38 | .37 | .33 | .31 |
| rs2633313 | 75028468 | A | G | .41 | .52 | .47 | .44 | .39 | .37 | .36 | .34 | .29 |
| rs2633317 | 75034871 | G | A | .4 | .52 | .46 | .42 | .39 | .37 | .37 | .33 | .29 |
| rs2633322 | 75038535 | C | T | .26 | .34 | .26 | .28 | .21 | .19 | .21 | .17 | .13 |
| rs2633323 | 75039429 | A | G | .42 | .54 | .46 | .45 | .4 | .38 | .36 | .33 | .33 |
| rs2688624 | 75040327 | C | A | .25 | .33 | .24 | .28 | .21 | .19 | .19 | .17 | .12 |
| rs4746158 | 75046458 | A | G | .23 | .33 | .29 | .27 | .28 | .25 | .28 | .36 | .39 |
| rs2675661 | 75050580 | A | C | .04 | .11 | .08 | .08 | .06 | .05 | .06 | .09 | .14 |
Table A2.
Symmetric Matrix of Significant Population Pairwise FST Values for Each Gene Region[Note]
| Population | CEPH | EST | SHIP | POPGEN | KORA | VIN | LAD | BRISI | CALA |
| CEPH | F, P | F, P | |||||||
| EST | P | L, P | P | F, L, P | F, P | ||||
| SHIP | P | P | P | P | |||||
| POPGEN | P | F, P | P | ||||||
| KORA | P | ||||||||
| VIN | L, P | P | P | F | |||||
| LAD | P | P | F, L | L | |||||
| BRISI | F, P | F, L, P | P | F, P | F | F, L | |||
| CALA | F, P | F, P | P | P | L |
Note.— The gene regions with the significant (P<.01 [permutation tests]) differentiations are indicated as follows: P = PLAU; F = FKBP5; L = LMNA; and S = SNCA.
Table A3.
Population Pairwise FST Values, on the Basis of All Four Gene Regions
| Population | CEPH | EST | SHIP | POPGEN | KORA | VIN | LAD | BRISI | CALA |
| CEPH | .0000 | ||||||||
| EST | .0022 | .0000 | |||||||
| SHIP | −.0011 | .0000 | .0000 | ||||||
| POPGEN | −.0007 | .0008 | −.0019 | .0000 | |||||
| KORA | .0017 | .0048 | −.0010 | .0001 | .0000 | ||||
| VIN | .0039 | .0107 | .0017 | .0040 | −.0003 | .0000 | |||
| LAD | .0024 | .0095 | .0023 | .0030 | −.0006 | −.0007 | .0000 | ||
| BRISI | .0101 | .0180 | .0080 | .0083 | .0026 | .0018 | .0037 | .0000 | |
| CALA | .0156 | .0219 | .0120 | .0109 | .0053 | .0053 | .0084 | .0000 | .0000 |
Table A4.
Performance of tagSNPs in All Four Gene Regions[Note]
|
Results for |
||||||||
| SNP Set Similar to HapMap |
Full SNP Set, Using |
|||||||
| Local Sample |
Local Population |
|||||||
| Region, Tested Sample, and Evaluation criterion | CEPH Trios | n=20 | n=40 | n=60 | CEPH Trios Phased | CEPH Founders (n=60) | n=20 | n=60 |
| SNCAa: | ||||||||
| EST: | ||||||||
| No. of tagSNPs | 9 | 10.48 | 10.76 | 10.85 | 15 | 15 | 18.12 | 18.78 |
| Ratio of tagged SNPsb | .763 | .777 | .827 | .831 | .921 | .921 | .823 | .836 |
| Mean r2c | .874 | .872 | .889 | .891 | .918 | .920 | .894 | .900 |
| Minimal r2d | .145 | .145 | .145 | .145 | .660 | .660 | .345 | .339 |
| SHIP: | ||||||||
| No. of tagSNPs | 9 | 9.09 | 9.15 | 9.47 | 15 | 15 | 15.45 | 16.25 |
| Ratio of tagged SNPs | .816 | .669 | .728 | .781 | .934 | .947 | .838 | .861 |
| Mean r2 | .896 | .847 | .867 | .884 | .939 | .951 | .909 | .917 |
| Minimal r2 | .064 | .064 | .064 | .064 | .651 | .740 | .364 | .382 |
| POPGEN: | ||||||||
| No. of tagSNPs | 9 | 9.6 | 9.74 | 9.93 | 15 | 15 | 15.98 | 16.57 |
| Ratio of tagged SNPs | .816 | .727 | .793 | .831 | .934 | .934 | .822 | .857 |
| Mean r2 | .895 | .857 | .883 | .894 | .945 | .947 | .902 | .922 |
| Minimal r2 | .096 | .095 | .096 | .096 | .444 | .444 | .399 | .418 |
| KORA: | ||||||||
| No. of tagSNPs | 9 | 10.95 | 10.43 | 10.34 | 15 | 15 | 17.56 | 17.58 |
| Ratio of tagged SNPs | .842 | .798 | .833 | .841 | .895 | .895 | .800 | .758 |
| Mean r2 | .867 | .871 | .883 | .886 | .929 | .930 | .875 | .838 |
| Minimal r2 | .125 | .124 | .125 | .125 | .700 | .700 | .343 | .282 |
| VIN: | ||||||||
| No. of tagSNPs | 9 | 10.21 | 10.05 | 9.9 | 15 | 15 | 18 | 17.61 |
| Ratio of tagged SNPs | .829 | .743 | .793 | .812 | .961 | .974 | .825 | .822 |
| Mean r2 | .871 | .852 | .869 | .874 | .915 | .918 | .884 | .884 |
| Minimal r2 | .101 | .099 | .101 | .101 | .740 | .740 | .342 | .339 |
| LAD: | ||||||||
| No. of tagSNPs | 9 | 9.78 | 9.91 | 9.71 | 15 | 15 | 16.02 | 17.83 |
| Ratio of tagged SNPs | .803 | .618 | .689 | .691 | .855 | .855 | .788 | .830 |
| Mean r2 | .904 | .850 | .872 | .874 | .932 | .935 | .894 | .913 |
| Minimal r2 | .160 | .160 | .160 | .160 | .651 | .651 | .383 | .414 |
| BRISI: | ||||||||
| No. of tagSNPs | 9 | 10.31 | 10.73 | 10.76 | 15 | 15 | 17.92 | 18.68 |
| Ratio of tagged SNPs | .803 | .682 | .740 | .767 | .908 | .908 | .805 | .808 |
| Mean r2 | .870 | .842 | .864 | .872 | .908 | .906 | .897 | .894 |
| Minimal r2 | .036 | .034 | .035 | .035 | .201 | .201 | .348 | .345 |
| CALA: | ||||||||
| No. of tagSNPs | 9 | 9.75 | 10.31 | 10.41 | 15 | 15 | 17.87 | 19.63 |
| Ratio of tagged SNPs | .789 | .616 | .698 | .756 | .882 | .895 | .799 | .842 |
| Mean r2 | .872 | .812 | .836 | .855 | .921 | .923 | .887 | .912 |
| Minimal r2 | .075 | .074 | .074 | .075 | .584 | .584 | .296 | .325 |
| CEPH founders: | ||||||||
| No. of tagSNPs | 9 | 10.7 | 10.1 | |||||
| Ratio of tagged SNPs | .87 | .82 | .87 | |||||
| Mean r2 | .9 | .88 | .9 | |||||
| Minimal r2 | .08 | .08 | .08 | |||||
| LMNAe: | ||||||||
| EST: | ||||||||
| No. of tagSNPs | 13 | 13.22 | 13.5 | 13.47 | ||||
| Ratio of tagged SNPs | .815 | .769 | .808 | .806 | ||||
| Mean r2 | .871 | .851 | .871 | .870 | ||||
| Minimal r2 | .185 | .187 | .188 | .189 | ||||
| SHIP: | ||||||||
| No. of tagSNPs | 13 | 13.25 | 13.48 | 13.44 | ||||
| Ratio of tagged SNPs | .704 | .684 | .707 | .718 | ||||
| Mean r2 | .836 | .818 | .829 | .835 | ||||
| Minimal r2 | .188 | .186 | .186 | .188 | ||||
| POPGEN: | ||||||||
| No. of tagSNPs | 13 | 12.7 | 12.68 | 12.73 | ||||
| Ratio of tagged SNPs | .778 | .743 | .756 | .762 | ||||
| Mean r2 | .846 | .832 | .836 | .840 | ||||
| Minimal r2 | .148 | .146 | .146 | .148 | ||||
| KORA: | ||||||||
| No. of tagSNPs | 13 | 12.75 | 12.93 | 12.77 | ||||
| Ratio of tagged SNPs | .852 | .781 | .807 | .805 | ||||
| Mean r2 | .873 | .853 | .862 | .861 | ||||
| Minimal r2 | .159 | .158 | .159 | .159 | ||||
| VIN: | ||||||||
| No. of tagSNPs | 13 | 12.51 | 12.77 | 12.92 | ||||
| Ratio of tagged SNPs | .815 | .73 | .760 | .767 | ||||
| Mean r2 | .867 | .845 | .852 | .855 | ||||
| Minimal r2 | .153 | .146 | .149 | .150 | ||||
| LAD: | ||||||||
| No. of tagSNPs | 13 | 12.37 | 12.53 | 12.51 | ||||
| Ratio of tagged SNPs | .778 | .742 | .763 | .765 | ||||
| Mean r2 | .865 | .841 | .850 | .850 | ||||
| Minimal r2 | .156 | .155 | .155 | .156 | ||||
| BRISI: | ||||||||
| No. of tagSNPs | 13 | 12.14 | 12.19 | 12.22 | ||||
| Ratio of tagged SNPs | .815 | .713 | .729 | .729 | ||||
| Mean r2 | .870 | .848 | .852 | .853 | ||||
| Minimal r2 | .264 | .262 | .260 | .262 | ||||
| CALA: | ||||||||
| No. of tagSNPs | 13 | 11.9 | 12.34 | 12.36 | ||||
| Ratio of tagged SNPs | .704 | .661 | .679 | .68 | ||||
| Mean r2 | .859 | .831 | .840 | .841 | ||||
| Minimal r2 | .253 | .231 | .242 | .247 | ||||
| CEPH founders: | ||||||||
| No. of tagSNPs | 13 | 12.2 | 12.5 | |||||
| Ratio of tagged SNPs | .81 | .77 | .77 | |||||
| Mean r2 | .88 | .87 | .87 | |||||
| Minimal r2 | .23 | .23 | .23 | |||||
| FKBP5f: | ||||||||
| EST: | ||||||||
| No. of tagSNPs | 20 | 19.21 | 19.72 | 19.81 | ||||
| Ratio of tagged SNPs | .865 | .875 | .934 | .938 | ||||
| Mean r2 | .928 | .929 | .889 | .950 | ||||
| Minimal r2 | .536 | .620 | .713 | .746 | ||||
| SHIP: | ||||||||
| No. of tagSNPs | 20 | 18.95 | 19.16 | 19.24 | ||||
| Ratio of tagged SNPs | .811 | .886 | .92 | .939 | ||||
| Mean r2 | .936 | .934 | .944 | .949 | ||||
| Minimal r2 | .607 | .649 | .712 | .753 | ||||
| POPGEN: | ||||||||
| No. of tagSNPs | 20 | 18.55 | 18.9 | 19.26 | ||||
| Ratio of tagged SNPs | .865 | .896 | .942 | .956 | ||||
| Mean r2 | .936 | .929 | .942 | .947 | ||||
| Minimal r2 | .607 | .622 | .711 | .749 | ||||
| KORA: | ||||||||
| No. of tagSNPs | 20 | 20.12 | 20.79 | 20.66 | ||||
| Ratio of tagged SNPs | .811 | .870 | .928 | .931 | ||||
| Mean r2 | .923 | .930 | .953 | .954 | ||||
| Minimal r2 | .513 | .586 | .703 | .721 | ||||
| VIN: | ||||||||
| No. of tagSNPs | 20 | 18.96 | 19.27 | 19.32 | ||||
| Ratio of tagged SNPs | .973 | .894 | .936 | .949 | ||||
| Mean r2 | .948 | .930 | .943 | .949 | ||||
| Minimal r2 | .541 | .609 | .695 | .726 | ||||
| LAD: | ||||||||
| No. of tagSNPs | 20 | 20.7 | 21.23 | 21.34 | ||||
| Ratio of tagged SNPs | .730 | .845 | .901 | .913 | ||||
| Mean r2 | .904 | .930 | .950 | .953 | ||||
| Minimal r2 | .499 | .577 | .700 | .725 | ||||
| BRISI: | ||||||||
| No. of tagSNPs | 20 | 20.38 | 20.85 | 21.07 | ||||
| Ratio of tagged SNPs | .784 | .803 | .846 | .884 | ||||
| Mean r2 | .917 | .921 | .935 | .944 | ||||
| Minimal r2 | .408 | .609 | .676 | .721 | ||||
| CALA: | ||||||||
| No. of tagSNPs | 20 | 18.76 | 19.06 | 19.55 | ||||
| Ratio of tagged SNPs | .892 | .831 | .901 | .928 | ||||
| Mean r2 | .934 | .912 | .937 | .945 | ||||
| Minimal r2 | .539 | .591 | .689 | .738 | ||||
| CEPH founders | ||||||||
| No. of tagSNPs | 20 | 19.9 | 20.3 | |||||
| Ratio of tagged SNPs | 1 | .88 | .94 | |||||
| Mean r2 | .96 | .95 | .96 | |||||
| Minimal r2 | .82 | .65 | .77 | |||||
| PLAUg: | ||||||||
| EST: | ||||||||
| No. of tagSNPs | 8 | 8.29 | 8.25 | 8.76 | 9 | 9 | 12.04 | 12.31 |
| Ratio of tagged SNPs | .563 | .673 | .682 | .713 | .844 | .844 | .803 | .858 |
| Mean r2 | .827 | .844 | .845 | .855 | .904 | .903 | .909 | .922 |
| Minimal r2 | .24 | .24 | .24 | .24 | .457 | .457 | .670 | .726 |
| SHIP: | ||||||||
| No. of tagSNPs | 8 | 8.14 | 8.34 | 8.58 | 9 | 9 | 11.61 | 12.27 |
| Ratio of tagged SNPs | .625 | .633 | .652 | .672 | .750 | .750 | .792 | .881 |
| Mean r2 | .830 | .839 | .845 | .850 | .896 | .897 | .910 | .930 |
| Minimal r2 | .074 | .07 | .07 | .07 | .511 | .511 | .681 | .739 |
| POPGEN: | ||||||||
| No. of tagSNPs | 8 | 7.11 | 7.09 | 7.35 | 9 | 9 | 10.29 | 10.07 |
| Ratio of tagged SNPs | .813 | .758 | .780 | .814 | .844 | .844 | .845 | .900 |
| Mean r2 | .870 | .856 | .860 | .867 | .896 | .896 | .903 | .914 |
| Minimal r2 | .130 | .130 | .130 | .130 | .528 | .528 | .689 | .759 |
| KORA: | ||||||||
| No. of tagSNPs | 8 | 8.41 | 8.34 | 8.44 | 9 | 9 | 11.03 | 11.77 |
| Ratio of tagged SNPs | .594 | .678 | .676 | .694 | .875 | .875 | .812 | .904 |
| Mean r2 | .831 | .854 | .854 | .857 | .918 | .918 | .906 | .932 |
| Minimal r2 | .165 | .165 | .165 | .165 | .563 | .563 | .623 | .738 |
| VIN: | ||||||||
| No. of tagSNPs | 8 | 8.1 | 8.14 | 8.23 | 9 | 9 | 10.89 | 10.8 |
| Ratio of tagged SNPs | .688 | .771 | .788 | .792 | .906 | .938 | .846 | .911 |
| Mean r2 | .848 | .867 | .872 | .873 | .926 | .921 | .909 | .928 |
| Minimal r2 | .184 | .184 | .184 | .184 | .604 | .604 | .660 | .734 |
| LAD: | ||||||||
| No. of tagSNPs | 8 | 6.74 | 6.88 | 6.77 | 9 | 9 | 9.98 | 9.77 |
| Ratio of tagged SNPs | .875 | .754 | .793 | .821 | .969 | .969 | .886 | .919 |
| Mean r2 | .879 | .861 | .866 | .868 | .933 | .933 | .920 | .928 |
| Minimal r2 | .091 | .091 | .091 | .091 | .624 | .624 | .707 | .763 |
| BRISI: | ||||||||
| No. of tagSNPs | 8 | 7.98 | 8.26 | 8.39 | 9 | 9 | 11.43 | 12 |
| Ratio of tagged SNPs | .688 | .695 | .711 | .724 | .750 | .750 | .838 | .903 |
| Mean r2 | .833 | .841 | .845 | .849 | .897 | .897 | .904 | .920 |
| Minimal r2 | .197 | .197 | .197 | .197 | .464 | .464 | .610 | .715 |
| CALA: | ||||||||
| No. of tagSNPs | 8 | 8.2 | 8.78 | 9.16 | 9 | 9 | 12.44 | 12.64 |
| Ratio of tagged SNPs | .531 | .659 | .703 | .734 | .813 | .813 | .822 | .913 |
| Mean r2 | .791 | .824 | .840 | .851 | .903 | .901 | .902 | .926 |
| Minimal r2 | .113 | .113 | .113 | .113 | .504 | .509 | .628 | .724 |
| CEPH founders: | ||||||||
| No. of tagSNPs | 8 | 7 | 7.2 | |||||
| Ratio of tagged SNPs | .97 | .78 | .82 | |||||
| Mean r2 | .93 | .89 | .89 | |||||
| Minimal r2 | .26 | .26 | .26 | |||||
Note.— tagSNPs were defined in accordance with the r2 method of Carlson et al. (2004).
tagSNPs defined in CEPH trios (HapMap comparison SNP set) for SNCA are rs2583969, rs2572323, rs356188, rs356200, rs4031753, rs3857048, rs2736994, rs356229, and rs1812923.
Ratio of SNPs above the r2 threshold of 0.8 to any tagSNP.
Mean r2 among all typed SNPs and the best SNP-specific tagSNP.
Minimal r2 among all typed SNPs and the best SNP-specific tagSNP.
tagSNPs defined in CEPH trios (HapMap comparison SNP set) for LMNA are rs501791, rs2275073, rs6661281, rs510441, rs7695_2, rs2241109, rs2241107, rs2485662, rs553016, rs1468772_2, rs4661146, rs3814314, and rs3738582.
tagSNPs defined in CEPH trios (HapMap comparison SNP set) for FKBP5 are rs992105, rs943297, rs1591365, rs2273000, rs1334894, rs4713902, rs2296662, rs2766534, rs1051952, rs2766554, rs2766543, rs3800372, rs2817054, rs3777747, rs873941, rs3807050, rs1883637, rs6457839, rs2817035, and rs1883636.
tagSNPs defined in CEPH trios (HapMap comparison SNP set) for PLAU are rs2688607, rs2633313, rs2664282, rs2227568, rs2227564, rs2633322, rs4746158, and rs2664283.
Table A5.
Performance of htSNPs in Four Gene Regions[Note]
|
Performance of htSNPs Defined in |
||||||||
| CEPH Trios |
Local Population |
|||||||
|
n=20 |
n=40 |
n=60 |
||||||
| Region, Tested Sample, and Evaluation Criterion | 90% | 80% | 90% | 80% | 90% | 80% | 90% | 80% |
| SNCA: | ||||||||
| CEPH: | ||||||||
| No. of htSNPs | 6 | 5 | ||||||
| Coverage of tagged haplotypes | .952 | .906 | ||||||
| Ratio of nontagged haplotypes (>5%) | 0 | .182 | ||||||
| EST: | ||||||||
| No. of htSNPs | 6 | 5 | 8.3 | 6.3 | 8.2 | 6.2 | 7.7 | 5.7 |
| Coverage of tagged haplotypes | .923 | .887 | .946 | .904 | .956 | .915 | .943 | .900 |
| Ratio of nontagged haplotypes (>5%) | .154 | .308 | .092 | .223 | .046 | .192 | .077 | .262 |
| SHIP: | ||||||||
| No. of htSNPs | 6 | 5 | 7.5 | 5.5 | 8 | 5.6 | 6.7 | 5.2 |
| Coverage of tagged haplotypes | .938 | .907 | .934 | .911 | .953 | .918 | .938 | .910 |
| Ratio of nontagged haplotypes (>5%) | 0 | .182 | .073 | .127 | .027 | .118 | .036 | .164 |
| POPGEN: | ||||||||
| No. of htSNPs | 6 | 5 | 8 | 5.6 | 7.4 | 5.4 | 7.2 | 5.2 |
| Coverage of tagged haplotypes | .941 | .905 | .942 | .911 | .953 | .915 | .957 | .908 |
| Ratio of nontagged haplotypes (>5%) | .083 | .25 | .075 | .2 | .04 | .2 | .025 | .225 |
| KORA: | ||||||||
| No. of htSNPs | 6 | 5 | 7 | 5.4 | 7.3 | 5.2 | 6.6 | 5.1 |
| Coverage of tagged haplotypes | .921 | .913 | .932 | .917 | .937 | .913 | .932 | .914 |
| Ratio of nontagged haplotypes (>5%) | .1 | .1 | .03 | .08 | .02 | .09 | .02 | .09 |
| VIN: | ||||||||
| No. of htSNPs | 6 | 5 | 7 | 5.2 | 7.2 | 5.2 | 6.7 | 5 |
| Coverage of tagged haplotypes | .935 | .918 | .937 | .916 | .942 | .921 | .938 | .918 |
| Ratio of nontagged haplotypes (>5%) | .1 | .1 | .03 | .1 | .03 | .08 | .04 | . 1 |
| LAD: | ||||||||
| No. of htSNPs | 6 | 5 | 7.1 | 4.9 | 6.7 | 4.9 | 7 | 5.3 |
| Coverage of tagged haplotypes | .934 | .926 | .948 | .912 | .946 | .916 | .951 | .929 |
| Ratio of nontagged haplotypes (>5%) | .1 | .1 | .03 | .11 | .02 | .11 | .02 | .08 |
| BRISI: | ||||||||
| No. of htSNPs | 6 | 5 | 7.9 | 5.6 | 7.3 | 5.2 | 7 | 5.1 |
| Coverage of tagged haplotypes | .930 | .909 | .941 | .913 | .936 | .912 | .931 | .910 |
| Ratio of nontagged haplotypes (>5%) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| CALA: | ||||||||
| No. of htSNPs | 6 | 5 | 7.5 | 5.9 | 7.6 | 5.6 | 7.2 | 4.3 |
| Coverage of tagged haplotypes | .921 | .899 | .918 | .901 | .940 | .910 | .937 | .859 |
| Ratio of nontagged haplotypes (>5%) | .167 | .25 | .133 | .208 | .058 | .192 | .075 | .292 |
| LMNA: | ||||||||
| CEPH: | ||||||||
| No. of htSNPs | 5 | 4 | ||||||
| Coverage of tagged haplotypes | .977 | .938 | ||||||
| Ratio of nontagged haplotypes (>5%) | .1 | .2 | ||||||
| EST: | ||||||||
| No. of htSNPs | 5 | 4 | 5.2 | 4.1 | 5.3 | 4 | 5.2 | 4 |
| Coverage of tagged haplotypes | .963 | .937 | .969 | .938 | .971 | .936 | .969 | .935 |
| Ratio of nontagged haplotypes (>5%) | .11 | .22 | .09 | .211 | .08 | .22 | .09 | .22 |
| SHIP: | ||||||||
| No. of htSNPs | 5 | 4 | 5.6 | 4.3 | 5.3 | 4 | 5.3 | 4 |
| Coverage of tagged haplotypes | .965 | .934 | .971 | .937 | .971 | .934 | .969 | .933 |
| Ratio of nontagged haplotypes (>5%) | .11 | .22 | .044 | .189 | .08 | .22 | .078 | .22 |
| POPGEN: | ||||||||
| No. of htSNPs | 5 | 4 | 4.6 | 4.2 | 4.9 | 4 | 4.9 | 4 |
| Coverage of tagged haplotypes | .97 | .938 | .954 | .941 | .967 | .938 | .965 | .938 |
| Ratio of nontagged haplotypes (>5%) | .11 | .22 | .156 | .2 | .122 | .22 | .122 | .22 |
| KORA: | ||||||||
| No. of htSNPs | 5 | 4 | 5 | 4 | 4.1 | 4 | 5.2 | 4 |
| Coverage of tagged haplotypes | .969 | .937 | .965 | .937 | .971 | .937 | .972 | .937 |
| Ratio of nontagged haplotypes (>5%) | .1 | .2 | .1 | .2 | .09 | .2 | .08 | .2 |
| VIN: | ||||||||
| No. of htSNPs | 5 | 4 | 5 | 4.3 | 5.3 | 4 | 5 | 4.1 |
| Coverage of tagged haplotypes | .965 | .932 | .964 | .941 | .970 | .932 | .966 | .935 |
| Ratio of nontagged haplotypes (>5%) | .1 | .2 | .1 | .17 | .08 | .2 | .1 | .19 |
| LAD: | ||||||||
| No. of htSNPs | 5 | 4 | 5 | 4.1 | 5.1 | 4.1 | 5.4 | 4 |
| Coverage of tagged haplotypes | .964 | .943 | .966 | .945 | .971 | .946 | .977 | .942 |
| Ratio of nontagged haplotypes (>5%) | .11 | .22 | .122 | .211 | .1 | .211 | .067 | .22 |
| BRISI: | ||||||||
| No. of htSNPs | 5 | 4 | 5.1 | 4.4 | 5.1 | 4.3 | 5.3 | 4.6 |
| Coverage of tagged haplotypes | .972 | .912 | .967 | .939 | .974 | .934 | .978 | .950 |
| Ratio of nontagged haplotypes (>5%) | .1 | .2 | .09 | .16 | .09 | .17 | .07 | .14 |
| CALA: | ||||||||
| No. of htSNPs | 5 | 4 | 5.3 | 4.4 | 5 | 4 | 5.2 | 4.1 |
| Coverage of tagged haplotypes | .969 | .926 | .970 | .946 | .971 | .929 | .974 | .933 |
| Ratio of nontagged haplotypes (>5%) | .1 | .2 | .09 | .16 | .1 | .2 | .08 | .19 |
| FKBP5: | ||||||||
| CEPH: | ||||||||
| No. of htSNPs | 12 | 10 | ||||||
| Coverage of tagged haplotypes | .967 | .948 | ||||||
| Ratio of nontagged haplotypes (>5%) | .059 | .059 | ||||||
| FKBP5: | ||||||||
| EST: | ||||||||
| No. of htSNPs | 12 | 10 | 11.9 | 10.3 | 11.7 | 10 | 11.4 | 9.9 |
| Coverage of tagged haplotypes | .958 | .937 | .935 | .915 | .948 | .931 | .951 | .932 |
| Ratio of nontagged haplotypes (>5%) | 0 | 0 | .04 | .075 | .006 | .038 | 0 | .019 |
| SHIP: | ||||||||
| No. of htSNPs | 12 | 10 | 11.7 | 9.2 | 10.9 | 9.4 | 10.4 | 9.2 |
| Coverage of tagged haplotypes | .953 | .942 | .944 | .919 | .948 | .926 | .944 | .923 |
| Ratio of nontagged haplotypes (>5%) | .059 | .059 | .065 | .118 | .047 | .1 | .053 | .106 |
| POPGEN: | ||||||||
| No. of htSNPs | 12 | 10 | 10.9 | 9.3 | 11 | 9.6 | 10.8 | 9.2 |
| Coverage of tagged haplotypes | .954 | .939 | .942 | .919 | .947 | .929 | .945 | .922 |
| Ratio of nontagged haplotypes (>5%) | .059 | .059 | .071 | .118 | .065 | .088 | .059 | .106 |
| KORA: | ||||||||
| No. of htSNPs | 12 | 10 | 11.8 | 9.6 | 11.6 | 9.9 | 10.9 | 9.2 |
| Coverage of tagged haplotypes | .943 | .932 | .933 | .911 | .943 | .921 | .938 | .916 |
| Ratio of nontagged haplotypes (>5%) | 0 | 0 | .019 | .069 | 0 | .031 | 0 | .05 |
| Tested samples: | ||||||||
| VIN: | ||||||||
| No. of htSNPs | 12 | 10 | 11.3 | 9.7 | 11.5 | 9.6 | 11.3 | 9.4 |
| Coverage of tagged haplotypes | .959 | .940 | .939 | .924 | .951 | .930 | .950 | .929 |
| Ratio of nontagged haplotypes (>5%) | 0 | 0 | .031 | .056 | .006 | .044 | 0 | .038 |
| LAD: | ||||||||
| No. of htSNPs | 12 | 10 | 11.9 | 9 | 11.9 | 9.1 | 11.3 | 8.8 |
| Coverage of tagged haplotypes | .931 | .927 | .924 | .899 | .937 | .902 | .932 | .908 |
| Ratio of nontagged haplotypes (>5%) | .059 | .059 | .106 | .165 | .047 | .147 | .059 | .135 |
| BRISI: | ||||||||
| No. of htSNPs | 12 | 10 | 12.1 | 9.6 | 12.7 | 9.9 | 11.9 | 10 |
| Coverage of tagged haplotypes | .918 | .913 | .933 | .895 | .940 | .911 | .940 | .913 |
| Ratio of nontagged haplotypes (>5%) | .167 | .111 | .056 | .156 | .05 | .144 | .039 | .144 |
| CALA: | ||||||||
| No. of htSNPs | 12 | 10 | 11.6 | 8.7 | 11.2 | 9.4 | 10.7 | 8.9 |
| Coverage of tagged haplotypes | .935 | .932 | .935 | .907 | .939 | .926 | .935 | .923 |
| Ratio of nontagged haplotypes (>5%) | 0 | 0 | .013 | .047 | 0 | 0 | 0 | .007 |
| PLAU: | ||||||||
| CEPH: | ||||||||
| No. of htSNPs | 6 | 4 | ||||||
| Coverage of tagged haplotypes | .965 | .857 | ||||||
| Ratio of nontagged haplotypes (>5%) | 0 | .25 | ||||||
| EST: | ||||||||
| No. of htSNPs | 6 | 4 | 7.4 | 5 | 6.6 | 4.8 | 6.4 | 3.7 |
| Coverage of tagged haplotypes | .872 | .756 | .899 | .855 | .905 | .833 | .905 | .786 |
| Ratio of nontagged haplotypes (>5%) | 0 | .167 | 0 | .03 | .017 | .117 | 0 | .167 |
| SHIP: | ||||||||
| No. of htSNPs | 6 | 4 | 6.7 | 4.9 | 6.7 | 4.6 | 6.7 | 4.5 |
| Coverage of tagged haplotypes | .876 | .769 | .898 | .838 | .909 | .826 | .912 | .828 |
| Ratio of nontagged haplotypes (>5%) | .143 | .286 | .029 | .1 | .014 | .114 | 0 | .129 |
| POPGEN: | ||||||||
| No. of htSNPs | 6 | 4 | 6.1 | 4.1 | 6.7 | 4.1 | 6.1 | 3.5 |
| Coverage of tagged haplotypes | .913 | .827 | .894 | .825 | .923 | .839 | .924 | .817 |
| Ratio of nontagged haplotypes (>5%) | 0 | .167 | .033 | .167 | .017 | .15 | 0 | .167 |
| KORA: | ||||||||
| No. of htSNPs | 6 | 4 | 5.5 | 3.8 | 6.4 | 3.4 | 5.5 | 4 |
| Coverage of tagged haplotypes | .891 | .794 | .883 | .817 | .910 | .788 | .901 | .823 |
| Ratio of nontagged haplotypes (>5%) | 0 | .167 | .033 | .133 | .033 | .2 | .017 | .117 |
| VIN: | ||||||||
| No. of htSNPs | 6 | 4 | 5.4 | 3.4 | 5.6 | 3.4 | 5.2 | 3.5 |
| Coverage of tagged haplotypes | .918 | .810 | .915 | .819 | .925 | .813 | .919 | .829 |
| Ratio of nontagged haplotypes (>5%) | 0 | .167 | .017 | .167 | 0 | .167 | 0 | .133 |
| LAD: | ||||||||
| No. of htSNPs | 6 | 4 | 5.1 | 3.1 | 4.9 | 3.1 | 5.1 | 3.3 |
| Coverage of tagged haplotypes | .921 | .844 | .903 | .844 | .923 | .847 | .924 | .857 |
| Ratio of nontagged haplotypes (>5%) | 0 | .167 | .05 | .15 | 0 | .15 | 0 | .117 |
| BRISI: | ||||||||
| No. of htSNPs | 6 | 4 | 6 | 3.6 | 6.3 | 3.7 | 5.3 | 3.1 |
| Coverage of tagged haplotypes | .915 | .817 | .908 | .824 | .936 | .854 | .921 | .824 |
| Ratio of nontagged haplotypes (>5%) | 0 | .167 | .067 | .217 | .017 | .117 | .017 | .167 |
| CALA: | ||||||||
| No. of htSNPs | 6 | 4 | 4.9 | 3.3 | 5.5 | 3.6 | 5 | 3.1 |
| Coverage of tagged haplotypes | .907 | .825 | .897 | .845 | .915 | .852 | .910 | .842 |
| Ratio of nontagged haplotypes (>5%) | .143 | .286 | .114 | .243 | .057 | .229 | .057 | .271 |
Note.— Zhang and Jin's (2003) htSNP selection method was used; htSNPs were selected by use of two different thresholds (80% and 90%) for coverage of common haplotypes.
Electronic-Database Information
The URLs for data presented herein are as follows:
- GSF European LD Pattern Project, http://ihg.gsf.de/LD/ (for a downloadable version of the genotype data presented in this study)
- HapMap Homepage, http://www.hapmap.org/ (for the International HapMap Project)
- popgen, http://www.popgen.de/
References
- Barbujani G, Sokal RR (1990) Zones of sharp genetic change in Europe are also linguistic boundaries. Proc Natl Acad Sci USA 87:1816–1819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardon LR, Abecasis GR (2003) Using haplotype blocks to map human complex trait loci. Trends Genet 19:135–140 10.1016/S0168-9525(03)00022-2 [DOI] [PubMed] [Google Scholar]
- Carlson CS, Eberle MA, Rieder MJ, Yi Q, Kruglyak L, Nickerson DA (2004) Selecting a maximally informative set of single-nucleotide polymorphisms for association analyses using linkage disequilibrium. Am J Hum Genet 74:106–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalli-Sforza LL, Menozzi P, Piazza A (1994) The history and geography of human genes. Princeton University Press, Princeton, NJ [Google Scholar]
- Chapman JM, Cooper JD, Todd JA, Clayton DG (2003) Detecting disease associations due to linkage disequilibrium using haplotype tags: a class of tests and the determinants of statistical power. Hum Hered 56:18–31 10.1159/000073729 [DOI] [PubMed] [Google Scholar]
- Dausset J, Cann H, Cohen D, Lathrop M, Lalouel JM, White R (1990) Centre d’etude du polymorphisme humain (CEPH): collaborative genetic mapping of the human genome. Genomics 6:575–577 10.1016/0888-7543(90)90491-C [DOI] [PubMed] [Google Scholar]
- Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, Higgins J, DeFelice M, Lochner A, Faggart M, Liu-Cordero SN, Rotimi C, Adeyemo A, Cooper R, Ward R, Lander ES, Daly MJ, Altshuler D (2002) The structure of haplotype blocks in the human genome. Science 296:2225–2229 10.1126/science.1069424 [DOI] [PubMed] [Google Scholar]
- Hoggart CJ, Shriver MD, Kittles RA, Clayton DG, McKeigue PM (2004) Design and analysis of admixture mapping studies. Am J Hum Genet 74:965–978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- International HapMap Consortium (2003) The International HapMap Project. Nature 426:789–796 10.1038/nature02168 [DOI] [PubMed] [Google Scholar]
- Ke X, Hunt S, Tapper W, Lawrence R, Stavrides G, Ghori J, Whittaker P, Collins A, Morris AP, Bentley D, Cardon LR, Deloukas P (2004) The impact of SNP density on fine-scale patterns of linkage disequilibrium. Hum Mol Genet 13:577–588 10.1093/hmg/ddh060 [DOI] [PubMed] [Google Scholar]
- Lao O, Andres AM, Mateu E, Bertranpetit J, Calafell F (2003) Spatial patterns of cystic fibrosis mutation spectra in European populations. Eur J Hum Genet 11:385–394 10.1038/sj.ejhg.5200970 [DOI] [PubMed] [Google Scholar]
- Mannila H, Koivisto M, Perola M, Varilo T, Hennah W, Ekelund J, Lukk M, Peltonen L, Ukkonen E (2003) Minimum description length block finder, a method to identify haplotype blocks and to compare the strength of block boundaries. Am J Hum Genet 73:86–94 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Marchini J, Cardon LR, Phillips MS, Donnelly P (2004) The effects of human population structure on large genetic association studies. Nat Genet 36:512–517 10.1038/ng1337 [DOI] [PubMed] [Google Scholar]
- Nei M (1972) Genetic distance between populations. Am Nat 106:283–292 [Google Scholar]
- Nejentsev S, Godfrey L, Snook H, Rance H, Nutland S, Walker NM, Lam AC, Guja C, Ionescu-Tirgoviste C, Undlien DE, Ronningen KS, Tuomilehto-Wolf E, Tuomilehto J, Newport MJ, Clayton DG, Todd JA (2004) Comparative high-resolution analysis of linkage disequilibrium and tag single nucleotide polymorphisms between populations in the vitamin D receptor gene. Hum Mol Genet 13:1633–1639 10.1093/hmg/ddh169 [DOI] [PubMed] [Google Scholar]
- Ng MCY, Wang Y, So WY, Cheng S, Visvikis S, Zee RYL, Fernandez-Cruz A, Lindpaintner K, Chan JCN (2004) Ethnic differences in the linkage disequilibrium and distribution of single-nucleotide polymorphisms in 35 candidate genes for cardiovascular diseases. Genomics 83:559–565 10.1016/j.ygeno.2003.09.008 [DOI] [PubMed] [Google Scholar]
- Phillips MS, Lawrence R, Sachidanandam R, Morris AP, Balding DJ, Donaldson MA, Studebaker JF, et al (2003) Chromosome-wide distribution of haplotype blocks and the role of recombination hot spots. Nat Genet 33:382–387 10.1038/ng1100 [DOI] [PubMed] [Google Scholar]
- Rosenberg NA, Pritchard JK, Weber JL, Cann HM, Kidd KK, Zhivotovsky LA, Feldman MW (2002) Genetic structure of human populations. Science 298:2381–2385 10.1126/science.1078311 [DOI] [PubMed] [Google Scholar]
- Schwartz R, Halldorsson BV, Bafna V, Clark AG, Istrail S (2003) Robustness of inference of haplotype block structure. J Comput Biol 10:13–19 10.1089/106652703763255642 [DOI] [PubMed] [Google Scholar]
- Stenzel A, Lu T, Koch WA, Hampe J, Guenther SM, De La Vega FM, Krawczak M, Schreiber S (2004) Patterns of linkage disequilibrium in the MHC region on human chromosome 6p. Hum Genet 114:377–385 10.1007/s00439-003-1075-5 [DOI] [PubMed] [Google Scholar]
- Thompson D, Stram D, Goldgar D, Witte JS (2003) Haplotype tagging single nucleotide polymorphisms and association studies. Hum Hered 56:48–55 10.1159/000073732 [DOI] [PubMed] [Google Scholar]
- Wall JD, Pritchard JK (2003) Haplotype blocks and linkage disequilibrium in the human genome. Nat Rev Genet 4:587–597 10.1038/nrg1123 [DOI] [PubMed] [Google Scholar]
- Wang WYS, Todd JA (2003) The usefulness of different density SNP maps for disease association studies of common variants. Hum Mol Genet 12:3145–3149 10.1093/hmg/ddg337 [DOI] [PubMed] [Google Scholar]
- Weale ME, Depondt C, Macdonald SJ, Smith A, Lai PS, Shorvon SD, Wood NW, Goldstein DB (2003) Selection and evaluation of tagging SNPs in the neuronal-sodium-channel gene SCN1A: implications for linkage-disequilibrium gene mapping. Am J Hum Genet 73:551–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Jin L (2003) HaploBlockFinder: haplotype block analyses. Bioinformatics 19:1300–1301 10.1093/bioinformatics/btg142 [DOI] [PubMed] [Google Scholar]