Abstract
U.S. populations are genetically admixed, but surprisingly little empirical data exists documenting the impact of such heterogeneity on type I and type II error in genetic-association studies of unrelated individuals. By applying several complementary analytical techniques, we characterize genetic background heterogeneity among 810 self-identified African American subjects sampled as part of a multisite cohort study of cardiovascular disease in older adults. On the basis of the typing of 24 ancestry-informative biallelic single-nucleotide–polymorphism markers, there was evidence of substantial population substructure and admixture. We used an allele-sharing–based clustering algorithm to infer evidence for four genetically distinct subpopulations. Using multivariable regression models, we demonstrate the complex interplay of genetic and socioeconomic factors on quantitative phenotypes related to cardiovascular disease and aging. Blood glucose level correlated with individual African ancestry, whereas body mass index was associated more strongly with genetic similarity. Blood pressure, HDL cholesterol level, C-reactive protein level, and carotid wall thickness were not associated with genetic background. Blood pressure and HDL cholesterol level varied by geographic site, whereas C-reactive protein level differed by occupation. Both ancestry and genetic similarity predicted the number and quality of years lived during follow-up, but socioeconomic factors largely accounted for these associations. When the 24 genetic markers were tested individually, there were an excess number of marker-trait associations, most of which were attenuated by adjustment for genetic ancestry. We conclude that the genetic demography underlying older individuals who self identify as African American is complex, and that controlling for both genetic admixture and socioeconomic characteristics will be required in assessing genetic associations with chronic-disease–related traits in African Americans. Complementary methods that identify discrete subgroups on the basis of genetic similarity may help to further characterize the complex biodemographic structure of human populations.
Introduction
Genetic-association studies are often performed in population samples of unrelated individuals to identify susceptibility loci for complex human traits. If subjects are sampled from two or more subpopulations for which the frequencies of marker alleles and traits differ, spurious associations may arise due to confounding by population substructure (Pritchard et al. 2000b; Schork et al. 2001; Risch et al. 2002). On the other hand, the increased extent of linkage disequilibrium between markers on the same chromosome, created by population admixture, may actually facilitate genome mapping of complex trait genes when exploited appropriately in the design of a study (Chakraborty and Weiss 1988; McKeigue 1998).
Prior studies assessing population stratification have primarily considered the impact of population subdivision and ancestral admixture proportions. Additional population genetic factors, however, may contribute to genetic background heterogeneity (Schork et al. 2001). Variation in allele frequencies as a result of genealogical differences between people in a sample may occur even in the absence of overt admixture. In addition, time-dependent population shifts, due to environmental or socioeconomic factors that influence migration or mating patterns, might create genetic heterogeneity across different age groups. These demographic movements may be especially relevant for studies of older adults in assessing complex diseases related to aging, as well as interindividual variation in life span or longevity (Yashin et al. 1999).
Biodemographic factors contributing to population heterogeneity and substructure are particularly important for genetic-association studies involving African Americans, among whom admixture with whites and Native Americans varies by geographic region (Parra et al. 1998, 2001; Pfaff et al. 2001; Smith et al. 2004). Among older adults in the United States, African Americans have a higher prevalence of cardiovascular disease (CVD) risk factors (Hutchinson et al. 1997; Kuller et al. 1998; Sundquist et al. 2001) and also greater clustering of CVD risk factors (Sharma et al. 2004), compared with non-Hispanic whites.
In light of the potential for confounding due to population stratification (Kittles et al. 2002; Freedman et al. 2004), as well as the opportunity for efficient genetic mapping of complex diseases by admixture linkage disequilibrium, we empirically evaluated the influence of population stratification on several common chronic-disease and aging-related phenotypes in a multicenter African American cohort. Our results show that there is substantial population admixture and substructure among the African American population, and that controlling for genetic ancestry not only may reduce false-positive associations but also may uncover a true association previously obscured by stratification. Our findings also show that controlling for social economic status, in addition to population stratification, is necessary in assessing genetic associations with chronic-disease–related traits in African American subjects.
Methods
Study Subjects
Study subjects were self-identified African American men and women aged ⩾65 years old who participated in the Cardiovascular Health Study (CHS) (Fried et al. 1991). CHS participants were recruited from lists of Medicare beneficiaries in four U.S. communities: Winston-Salem, NC; Pittsburgh, PA; Washington County, MD; and Sacramento, CA. The original CHS cohort, recruited from 1989 to 1990, included 246 African American participants. A second cohort of 678 African American participants was recruited from 1992 to 1993. Of 924 total African American participants, 810 are included in the present study. The reason for exclusion was either refusal of consent for genetic testing (n=62) or lack of an available DNA sample (n=52). All procedures were conducted under institutionally approved protocols for study of human subjects, and all subjects provided written informed consent.
Data Collection and Definition of Phenotypes Related to Vascular Disease and Aging
Data collection methods in the CHS have been described elsewhere (Fried et al. 1991). The baseline evaluation included demographic, lifestyle, and medical histories; physical examination; and fasting blood collection (Cushman et al. 1995). Quantitative phenotypes and CVD risk factors that were considered include systolic blood pressure (mm Hg), BMI (kg/m2), fasting blood glucose (mg/dL), HDL cholesterol level (mg/dL), and C-reactive protein (CRP) level (mg/liter). Carotid wall thickness, a quantitative measure of subclinical vascular disease, was defined as the mean maximal intimal-medial thickness of the near and far walls on both the left and right arteries, as determined by high-resolution ultrasonography (O’Leary et al. 1991). The outcome “years of life” (YOL) was defined as the number of years a participant was alive during 10 years of follow-up, and “years of healthy life” (YHL) was defined as the number of years the person reported being in excellent, very good, or good health during the 10 years of follow-up. This outcome was derived from standard information on self-rated health status (excellent/very good/good/fair/poor) collected at baseline and every 6 mo during follow-up (Diehr et al. 1998).
Selection of Ancestry-Informative Markers (AIMs) and Genotype Analysis
Twenty-four biallelic SNP markers (table 1) were chosen on the basis of known allele-frequency differences (δ values) between African, European, and Native American populations. A subset of these markers has been characterized and published by Mark Shriver and colleagues (Hoggart et al. 2003; Shriver et al. 2003). Additional markers were selected by identifying markers from dbSNP that had been typed in both European Americans and African Americans and that had a high allele-frequency difference between those populations (δ>0.5). The ancestral allele frequencies were then confirmed by genotyping the markers in populations collected from Sub-Saharan Africa (Nigeria, Central African Republic, and Sierra Leone [n=481]), Europe (Ireland, England, Germany, and Spain [n=243]), and Native American populations indigenous to the United States and Mexico (Maya, Pima, Cheyenne, and Pueblo [n=148]). The ancestral DNA samples were kindly provided by Dr. Mark Shriver. Detailed information regarding the markers characterized by Shriver and colleagues can be found at the dbSNP Web site under the submitter handle “PSU-ANTH,” or, for the newly identified markers, under the submitter handle “HapMap-UCSF-WU-FP-TDI.”
Table 1.
Marker Chromosomal Locations, Ancestral Population Allele Frequencies, and Allele Frequencies and Tests for Hardy-Weinberg Proportions in CHS African American Participants
|
Ancestral PopulationAllele 1 Frequencies |
Marker FST Values betweenAncestral Populationsa |
Findings for CHS African Americans |
||||||||
| Marker | Location | Alleles 1/2 | African | European | NativeAmerican | African/European | African/ NativeAmerican | European/ NativeAmerican | Allele 1Frequency | Hardy-WeinbergEquilibriumb |
| rs2814778 | 1q23.2 | A/G | .003 | .994 | .991 | .982 | .976 | .000 | .256 | .034 |
| rs930072 | 5p13 | C/T | .960 | .096 | .447 | .749 | .315 | .156 | .731 | .045 |
| rs7349 | 10p11.22 | C/T | .039 | .873 | .956 | .701 | .841 | .022 | .214 | .0004 |
| rs723632 | 1q32.3 | G/C | .100 | .919 | .674 | .671 | .347 | .093 | .277 | .822 |
| rs722098 | 21q21.1 | G/A | .902 | .177 | .717 | .529 | .055 | .295 | .702 | .038 |
| rs146026 | 13q13.1 | C/T | .256 | .917 | .826 | .450 | .327 | .018 | .377 | .132 |
| rs6003 | 1q31.3 | G/A | .702 | .083 | .031 | .402 | .485 | .013 | .570 | .674 |
| rs1985080 | 7p14.3 | G/A | .100 | .643 | .966 | .316 | .753 | .166 | .224 | .418 |
| rs518116 | 9q33.3 | G/A | .131 | .669 | .581 | .302 | .221 | .008 | .245 | .052 |
| rs3287 | 2p16.2 | G/A | .730 | .196 | .205 | .287 | .277 | .000 | .590 | .057 |
| rs1989486 | 19q13.42 | C/T | .045 | .578 | .404 | .331 | .185 | .030 | .219 | .120 |
| rs7041 | 4q13.3 | T/G | .928 | .413 | .451 | .300 | .266 | .001 | .815 | .223 |
| rs994174 | 10q23.1 | G/A | .758 | .246 | .264 | .262 | .244 | .000 | .667 | .062 |
| rs1800498 | 11q23.2 | T/C | .138 | .648 | .088 | .273 | .006 | .337 | .258 | .968 |
| rs2816 | 17p13.1 | T/C | .003 | .494 | .075 | .323 | .035 | .216 | .151 | .490 |
| rs2891 | 17p13.2 | G/A | .021 | .507 | .425 | .304 | .235 | .007 | .122 | .280 |
| rs3188520 | 20q11.22 | G/C | .828 | .349 | .439 | .237 | .163 | .008 | .747 | .156 |
| rss1042602 | 11q14.3 | A/C | .004 | .467 | .053 | .298 | .022 | .223 | .090 | .012 |
| rs326946 | 11q23.1 | G/T | .609 | .167 | .067 | .206 | .328 | .024 | .482 | .305 |
| rs2077863 | 18p11.21 | C/G | .511 | .925 | .926 | .212 | .213 | .000 | .660 | .861 |
| rs3188519 | 4q28.2 | C/T | .758 | .369 | .318 | .154 | .195 | .003 | .623 | .216 |
| rs594689 | 11q13.1 | A/G | .094 | .467 | .130 | .172 | .003 | .136 | .193 | .102 |
| rs2228478 | 16q24.3 | G/A | .508 | .136 | .043 | .158 | .271 | .027 | .393 | .458 |
| rs584059 | 3q23 | C/A | .494 | .140 | .467 | .145 | .001 | .126 | .419 | .917 |
Marker FST values (inverse of the variance of the estimated ancestral contributions) were calculated in accordance with Pfaff et al. (2004).
P values <.05 for the test of Hardy-Weinberg equilibrium are shown in bold italics.
The 24 AIMs were distantly spaced throughout the genome so that they offer independent association about genetic background/ancestry. The average distance between adjacent markers on the same chromosome was 26 Mb (range 1–60 Mb). The mean δ value between African and European populations was 0.56 (range 0.36–0.99). The mean allele-frequency differential between African and Native American populations was 0.44 (range 0.03–0.99). The mean allele-frequency difference between European and Native American populations was 0.19 (range 0.001–0.56). Marker FST values were calculated as the inverse of the variance of the estimated ancestral contributions, in accordance with Pfaff et al. (2004), and are shown in table 1.
Genotyping assays were performed on blood drawn from 810 CHS African American participants who gave informed consent to DNA preparation and testing. Genotyping was performed using the AcycloPrime-FP (Perkin Elmer) method (Chen et al. 1999) under standard conditions: 5 μl PCR volume with Platinum Taq buffer, 2.5 mM MgCl2, 2.4–4.0 ng of genomic DNA, 50 μM dNTPs, 0.1 μM of primers, and 0.1 U of Platinum Taq (Invitrogen). Cycling conditions were 95°C for 2 min, followed by 35 cycles at 92°C for 10 s, 58°C for 20 s, and 68°C for 30 s, with a final extension at 68°C for 10 min. PCR products were purified enzymatically, and genotyping extension reactions were performed in accordance with kit directions. The primer sequences for PCR and genotyping extension reactions and any changes to standard conditions are presented in table A1 (online only).
Characterization of Population Structure, Admixture, and Genetic Background Similarity
Exact tests for Hardy-Weinberg equilibrium and linkage disequilibrium and Wright’s hierarchical F statistics (Wright 1951) as estimators of allele-frequency variation under a pure-drift model (Weir and Cockerham 1984) were computed using Genetic Data Analysis, version 1.1 (see Genetic Data Analysis Web site).
Group admixture proportions were estimated from the average coalescent times for a pair of alleles taken from within and between populations by use of the program ADMIX, version 2.0 (Dupanloup and Bertorelle 2001). SEs for the group admixture coefficients were calculated on the basis of 1,000 bootstraps. The proportion of African, European, and Native American ancestry for each individual was estimated by a maximum-likelihood method (Chakraborty et al. 1986) by use of the program IAE3 (Bonilla et al. 2004), kindly provided by Mark Shriver. This program also gives 1-SD support intervals for the estimated ancestral proportions.
We used two Bayesian Markov Chain–Monte Carlo methods to provide complementary information on genetic differentiation between and among populations under nonequilibrium conditions. Population structure and evidence for allelic association between linked markers caused by correlation in ancestry (i.e., “admixture linkage disequilibrium”) were evaluated by estimating the average recombination rate by use of the program STRUCTURE 2.1 (Pritchard et al. 2000a; Falush et al. 2003), with a burn-in of 50,000 iterations and 1,000,000 iterations. By relaxing the requirement for Hardy-Weinberg equilibrium within geographic subpopulations and by allowing for recent migration, local inbreeding coefficients were estimated using the program BayesAss 1.2 of Wilson and Rannala (2003), which was run for a total of 3,000,000 iterations, including an initial burn-in of 1,000,000 iterations.
A genetic-clustering algorithm based on pairwise, weighted allele-frequency sharing was used to assess genetic background similarity (Schork 2001; Schork et al. 2001). Allele-sharing matrices were constructed in accordance with the method of Lynch and Ritland (1999), as implemented in the program IDENTIX (Belkhir et al. 2002). The resulting similarity matrices were used in an agglomerative hierarchical cluster analysis with complete linkage, under the assumption of the existence of 2–15 genetically similar groups of individuals within the total sample. To determine the most likely number of groups in the sample, we assigned each individual to his or her most likely genetic subgroup on the basis of his or her allele-frequency profile, and we assessed phenotypic differences across the groups by performing standard ANOVA and nonparametric ANOVA or the Kruskall-Wallis test (Lehmann and D’Abrera 1998). To identify any genetic “outliers” whose genetic background is extremely different from the remaining cohort, we applied the multilocus genotype-based permutation test of Curtis et al. (2002), which was run for 10,000 iterations, with a significance threshold of P ⩽ 1 × 10−6.
Tests of Associations between Quantitative Traits and Biodemographic Variables
Associations between quantitative traits and biodemographic predictor variables (estimates of individual ancestry, genetic-cluster membership, socioeconomic status, clinic site, or individual AIM genotypes) were assessed by multiple linear regression, by use of the statistical package Stata 8.0. Levels of blood glucose, HDL cholesterol, and CRP were log transformed to reduce skewness and kurtosis. Individual marker genotypes were coded 0, 1, and 2, under the assumption of an additive genetic model. An individual's percentage of African ancestry was coded as a continuous variable, by use of his or her proportion of African ancestry estimated by maximum likelihood. Each clinic site was represented by an indicator variable, and the largest clinic (North Carolina) was omitted from the regression model as the reference group. Similarly, the four genetic-similarity clusters were coded as indicator variables, with cluster 1 as reference. We created categorical variables for education, income, and occupation type as proxies for socioeconomic status (SES). A three-level ordinal categorical variable for education was created by dividing the cohort on the basis of education level (from none to grade 9; high school or general equivalency diploma; or college, vocational, graduate, or professional training). Similarly, a three-level ordinal categorical variable was created on the basis of annual income levels <$8,000; $8,000–$35,000; and >$35,000. For type of occupation, we created three nonordered categories on the basis of a response card that indicated lifetime occupation: professional/technical/managerial/administrative positions and sales/clerical service were classified as “white-collar” occupations; craftsman/machine operator/laborer and farming/forestry work were grouped together as “blue-collar” occupations; and housewife, other occupation, or refusal to answer were combined into the category of “other” occupations.
Covariate-adjusted P values for associations between quantitative traits and population characteristics (clinic site, genetic similarity cluster, individual ancestry, or SES defined by education, income, or occupation) were determined by likelihood-ratio tests. The log likelihood of a “full” regression model containing the variable(s) for a particular characteristic was compared with a reduced model without the characteristic. We adjusted the nominal 5% significance level by the number of traits analyzed (n=8) and used a P value threshold of <.00625 for significance. Mean-adjusted trait values (and 95% CIs) for different levels of predictor variables were calculated from the linear-regression coefficients and SEs, with any additional covariates set to their respective mean values. All analyses were adjusted for age at baseline and for sex. Additionally, we adjusted some analyses for other clinical covariates known to be important for the particular quantitative trait (Hutchinson et al. 1997; Kuller et al. 1998; Sundquist et al. 2001; Sharma et al. 2004). Thus, systolic blood pressure was adjusted for treated hypertension; blood glucose level was adjusted for baseline diabetes; CRP level was adjusted for BMI, smoking, and diabetes; and carotid wall thickness was adjusted for smoking, hypertension, diabetes, BMI, and HDL cholesterol level. Both YOL and YHL were adjusted for hypertension, diabetes, current smoking, BMI, coronary heart disease, cancer, and self-reported health status.
Results
Population Substructure and Admixture in African American Cohort
Of the 24 AIMs tested, 6 (including 4 of the 5 markers having the largest allele-frequency differential between Africans and Europeans) deviated significantly from Hardy-Weinberg proportions (table 1). There was increased homozygosity, both overall (FIT=0.034; 95% CI 0.016–0.052) and within the four regional subpopulations (FIS=0.033; 95% CI 0.015–0.050). Even though the markers were unlinked or widely spaced throughout the genome, 170 (60%) of 285 pairwise combinations showed significant allelic association. Together, the excess homozygosity and association between unlinked markers suggest substantial population substructure and admixture in the CHS African American cohort due to continuous gene flow or nonrandom mating. The program STRUCTURE 2.1 showed there was a greater likelihood that the cohort descended from two ancestral populations (log likelihood −20,725) than three (−20,767) or four (−20,823) ancestral populations or than a single homogeneous population (−21,363). Under a linkage model with two ancestral populations, the presence of significant admixture linkage disequilibrium was confirmed (Falush et al. 2003).
The mean proportions (± SEs) of African, European, and Native American ancestry, estimated for the cohort as a whole, were 76.4 ± 0.6%, 20.9 ± 1.2%, and 2.7 ± 1.6%, respectively. We also estimated individual ancestry by using maximum-likelihood, but the mean SEs were much larger (15.6% for African, 17.9% for European, and 20.9% for Native American); these presumably reflect both the wide interindividual variation in degree of admixture and the lack of precision in distinguishing European from Native American ancestry by the current set of 24 markers. Individual African ancestral proportions estimated by use of STRUCTURE under a two-population admixture model were virtually identical to those estimated by maximum likelihood under a three-population model (correlation coefficient 0.98; P<.0001). We also conducted a principal-components analysis and compared the scores that individuals received for the principal components with ancestral proportions calculated by use of the maximum-likelihood model (table 2). We found a very high correlation between an individual’s score on the first principal component and estimated African ancestry (correlation coefficient 0.97; P<.0001). For European and Native American ancestry, the correlations with the first principal component were weaker. The second and third principal components were also weakly correlated with percentage European versus percentage Native American ancestry.
Table 2.
Correlations between Individual Ancestry Estimated by Maximum Likelihood and Principal-Component Analysis
|
Ancestral Proportions Estimated by Maximum Likelihood |
||||||
| African |
European |
Native American |
||||
| PrincipalComponent | CorrelationCoefficienta | P Valueb | CorrelationCoefficienta | P Valueb | CorrelationCoefficienta | P Valueb |
| 1st | .9718 | .0000 | −.8523 | .0000 | −.2884 | .0000 |
| 2nd | −.0085 | 1.0000 | −.0656 | 1.0000 | .1018 | .3458 |
| 3rd | −.0589 | 1.0000 | −.1619 | .0027 | .3076 | .0000 |
| 4th | .0048 | 1.0000 | .1034 | .3102 | −.1477 | .0101 |
Pearson correlation coefficient for the comparison of ancestry with principal component.
Bonferroni-corrected P value.
Genetic Differentiation among Geographic Subpopulations
The coancestry coefficient estimator of FST was 0.0013 (95% CI 0.0003–0.0026), suggesting a small but significant amount of genetic differentiation among the four regions of the United States from which the CHS participants were sampled. Mean age- and sex-adjusted individual ancestry estimates differed across the four CHS clinic sites (P=.005). Group admixture estimates by clinic site are shown in table 3. Exclusion of the Maryland African American residents did not appreciably alter the variation in admixture (P=.01) but did attenuate the allele-frequency differences among the three larger population samples (FST=0.0007; 95% CI −0.00009 to 0.0017). Potential local inbreeding effects for the North Carolina, California, Maryland, and Pennsylvania African American populations were estimated at 0.025 ± 0.014, 0.092 ± 0.065, −0.006 ± 0.037, and −0.035 ± 0.079, respectively. Together, these results suggest that local population differences may play a role in shaping the overall genetic heterogeneity and structure of the entire CHS African American cohort.
Table 3.
Group Admixture Estimates by CHS Clinic Site or by Clusters Inferred on the Basis of Genetic Similarity[Note]
|
Estimated Ancestral Proportions(% ± SE) |
||||
| BiodemographicCharacteristic | N | African | European | Native American |
| Clinical centera: | ||||
| Winston-Salem | 299 | 79.1 ± .9 | 17.0 ± 1.7 | 3.9 ± 2.1 |
| Sacramento | 214 | 74.4 ± 1.0 | 20.6 ± 2.0 | 4.9 ± 2.4 |
| Pittsburgh | 285 | 75.2 ± .9 | 23.9 ± 1.8 | .9 ± 2.2 |
| Genetic similarity: | ||||
| Cluster 1 | 467 | 86.4 ± .7 | 12.1 ± 1.3 | 1.6 ± 1.7 |
| Cluster 2 | 32 | 76.1 ± 2.3 | 25.0 ± 5.0 | .0 ± 6.0 |
| Cluster 3 | 74 | 41.4 ± 1.6 | 37.1 ± 3.6 | 21.5 ± 4.2 |
| Cluster 4 | 236 | 67.2 ± 1.0 | 33.2 ± 2.1 | .0 ± 2.6 |
Population Subdivision Due to Genetic Background Similarity
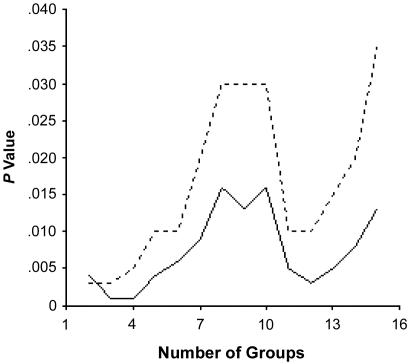
As a complementary approach to population-structure assessment, discrete clusters of genetically similar individuals were identified through the use of pairwise, allele-frequency–weighted, identity-by-state, allele-sharing matrices. The most likely number of genetically similar clusters of individuals within the total sample was determined by testing allele frequency and phenotypic differences within the total sample of 810 individuals. As shown in figure 1, the most significant differences in male BMI involved the assumption of four groups of individuals (P<.0001). These and other data (N.J.S., unpublished data) suggest that, within the cohort, there are likely four genetically distinct subgroups. The distribution of these four subpopulations, identified on the basis of genetic background similarity, did not differ among the four geographic subregions (P=.20) but did differ with respect to individual admixture proportions (P<.001) and group admixture estimates (table 3). These results suggest that the empirically determined clusters actually reflect the differences in degrees of admixture among the study subjects.
Figure 1.
Differences in BMI for males across groups, determined by the use of cluster analyses involving allele-similarity matrices. The solid line represents the P values associated with standard ANOVA, and the dotted line represents the P values associated with nonparametric ANOVA.
Two participants, one from North Carolina and the other from California, had multilocus genetic backgrounds that were extremely different from the remaining cohort. These two genetic “outliers” were confirmed to have 0% and 3% African ancestry, respectively, and 100% and 97% European ancestry, respectively. Both belonged to genetic similarity cluster 4. These two individuals were excluded from further analyses. Additional investigation did not reveal any evidence that either individual had been inadvertently misclassified within the CHS data set.
Relationships among Population Genetic, Demographic, and SES Variables
The mean age at baseline of the CHS African American participants was 73 years (range 65–93 years). Individual African ancestry averaged 74% among the 65–69-year-olds (n=269), 72% among the 70–74-year-olds (n=279), 74% among the 75–79-year-olds (n=160), and 78% among the participants aged ⩾80 years (n=102). These differences were not significant (P=.10). Education, income, and occupation type all differed strongly by individual admixture proportions and genetic background similarity (all P values <.001). Moreover, there were differences in education (P<.001), income (P<.001), and occupation (P=.02) among clinic sites. These data highlight the complex interrelationships that exist between genetic or ancestral background and current social, economic, and environmental conditions in human populations.
Associations between Population-Structure Characteristics and Chronic-Disease Phenotypes
To address the influence of different forms of population structure on traits related to CVD and aging, we performed direct tests of association between several quantitative phenotypes and estimates of individual ancestry, genetic similarity, SES, and clinic site (tables 4, 5, 6, and 7). Each biodemographic characteristic was examined separately and in a multivariable model that was simultaneously adjusted for other characteristics. Higher fasting blood glucose levels were associated most strongly with African ancestry. Mean glucose levels, adjusted for age, sex, and baseline diabetes status, were 19 mg/dL higher (95% CI 5–33) among subjects with 100% African ancestry compared with those with 0% African ancestry. The glucose-ancestry association was altered only minimally by additional adjustment for SES and clinic site. Systolic blood pressure and HDL cholesterol levels were higher among African Americans sampled from the Sacramento area than among those sampled from Winston-Salem or Pittsburgh. In multivariable-adjusted models, genetic background or SES did not seem to account appreciably for these geographic differences (tables 4 and 5). CRP levels were influenced by type of occupation (table 6). Blue-collar workers had 23% higher (range 5%–41% higher) CRP levels relative to those of white-collar workers. Carotid arterial wall thickness did not vary significantly by any of the biodemographic indicators in table 6.
Table 4.
Blood Glucose and Systolic Blood Pressure by Clinic Site, Genetic Background Similarity, Admixture, and SES[Note]
|
Mean Blood Glucose (95% CI)[mg/dl] |
Mean Systolic BloodPressure (95% CI)[mm Hg] |
||||
| BiodemographicCharacteristic | N | Minimally Adjusted | Fully Adjusted | Minimally Adjusted | Fully Adjusted |
| Clinical centera: | |||||
| Winston-Salem | 299 | 114 (111–117) | 114 (111–117) | 141 (138–143) | 141 (138–143) |
| Sacramento | 213 | 109 (106–112) | 110 (106–114) | 146 (143–149) | 145 (142–148) |
| Pittsburgh | 285 | 114 (111–116) | 113 (110–116) | 140 (138–143) | 141 (138–143) |
| Genetic similarityb: | |||||
| Cluster 1 | 467 | 114 (111–116) | 114 (112–117) | 141 (139–143) | 141 (139–144) |
| Cluster 2 | 32 | 105 (97–114) | 105 (97–114) | 144 (137–152) | 144 (136–152) |
| Cluster 3 | 74 | 107 (101–113) | 108 (102–114) | 142 (136–147) | 141 (136–147) |
| Cluster 4 | 234 | 112 (109–115) | 111 (108–115) | 142 (140–145) | 142 (139–145) |
| Genetic ancestryc: | |||||
| 0% African (estimated) | 102 (96–108) | 104 (98–111) | 139 (133–145) | 138 (132–144) | |
| 100% African (estimated) | 116 (113–119) | 116 (112–119) | 143 (140–145) | 143 (141–146) | |
| Educationd: | |||||
| None–grade 9 | 237 | 116 (112–119) | 114 (111–117) | 140 (138–143) | 140 (139–143) |
| High school | 294 | 110 (107–113) | 112 (111–114) | 142 (140–143) | 142 (140–143) |
| Professional/vocational | 274 | 111 (108–114) | 111 (108–114) | 143 (141–145) | 143 (140–145) |
| Annual incomee: | |||||
| <$8,000 | 285 | 115 (112–118) | 114 (111–117) | 143 (140–145) | 143 (140–145) |
| $8,000–$35,000 | 398 | 111 (109–114) | 112 (110–114) | 142 (140–143) | 141 (140–143) |
| >$35,000 | 77 | 109 (103–115) | 110 (105–114) | 141 (137–144) | 140 (136–144) |
| Occupation typef: | |||||
| White collar | 300 | 111 (108–114) | 112 (109–115) | 142 (139–144) | 142 (139–144) |
| Blue collar | 232 | 111 (108–115) | 110 (107–114) | 139 (136–142) | 140 (137–143) |
| Housewife/other | 275 | 114 (111–118) | 115 (111–118) | 144 (141–146) | 144 (141–146) |
Note.— Likelihood-ratio tests of association were performed by multiple linear regression of each phenotypic trait on biodemographic characteristics. Minimally adjusted models were adjusted for age, sex, and any clinically relevant covariates, as described in the “Methods” section. Fully adjusted models additionally contained variables for remaining biodemographic characteristics. In the footnotes below, P values in bold italics are less than the nominal significance level of 5% adjusted for the number of traits assessed (n=8; P<.00625).
For measurements by clinical center, the minimally adjusted P value was .05 and the fully adjusted P value was .25 for blood glucose, and the minimally adjusted P value was .005 and the fully adjusted P value was .04 for systolic blood pressure.
For measurements by genetic similarity, the minimally adjusted P value was .06 and the fully adjusted P value was .09 for blood glucose, and the minimally adjusted P value was .86 and the fully adjusted P value was .90 for systolic blood pressure.
For measurements by genetic ancestry, the minimally adjusted P value was .002 and the fully adjusted P value was .01 for blood glucose, and the minimally adjusted P value was .29 and the fully adjusted P value was .23 for systolic blood pressure.
For measurements by education, the minimally adjusted P value was .08 and the fully adjusted P value was .26 for blood glucose, and the minimally adjusted P value was .16 and the fully adjusted P value was .24 for systolic blood pressure.
For measurements by annual income, the minimally adjusted P value was .04 and the fully adjusted P value was .20 for blood glucose, and the minimally adjusted P value was .43 and the fully adjusted P value was .32 for systolic blood pressure.
For measurements by occupation type, the minimally adjusted P value was .30 and the fully adjusted P value was .21 for blood glucose, and the minimally adjusted P value was .14 and the fully adjusted P value was .16 for systolic blood pressure.
Table 5.
HDL Cholesterol and BMI by Clinic Site, Genetic Background Similarity, Admixture, and SES[Note]
|
Mean HDL Cholesterol (95% CI)[mg/dl] |
Mean BMI (95% CI)[kg/m2] |
||||
| BiodemographicCharacteristic | N | Minimally Adjusted | Fully Adjusted | Minimally Adjusted | Fully Adjusted |
| Clinical centera: | |||||
| Winston-Salem | 299 | 55 (53–56) | 55 (53–56) | 28.3 (27.7–28.9) | 28.3 (27.7–29.0) |
| Sacramento | 213 | 59 (57–61) | 59 (57–61) | 29.3 (27.9–29.3) | 28.9 (28.1–29.6) |
| Pittsburgh | 285 | 55 (53–56) | 55 (53–57) | 28.5 (27.9–29.2) | 28.5 (27.9–29.1) |
| Genetic similarityb: | |||||
| Cluster 1 | 467 | 56 (55–57) | 56 (55–57) | 28.7 (28.2–29.2) | 28.7 (28.2–29.2) |
| Cluster 2 | 32 | 56 (51–61) | 55 (51–60) | 26.4 (24.5–28.2) | 26.5 (24.4–28.4) |
| Cluster 3 | 74 | 54 (51–57) | 53 (50–56) | 29.4 (28.2–30.6) | 29.7 (28.4–31.0) |
| Cluster 4 | 234 | 57 (55–59) | 56 (55–58) | 28.1 (27.4–28.8) | 28.2 (27.5–28.9) |
| Genetic ancestryc: | |||||
| 0% African (estimated) | 55 (52–59) | 54 (51–58) | 27.8 (26.4–29.1) | 28.0 (26.5–29.5) | |
| 100% African (estimated) | 56 (54–58) | 56 (55–58) | 28.8 (28.2–29.4) | 28.7 (28.1–29.4) | |
| Educationd: | |||||
| None–grade 9 | 237 | 55 (54–57) | 56 (54–57) | 28.6 (28.0–29.2) | 28.6 (28.0–29.3) |
| High school | 294 | 56 (55–57) | 56 (55–57) | 28.5 (28.1–28.9) | 28.5 (28.1–28.9) |
| Professional/vocational | 274 | 56 (55–58) | 56 (54–58) | 28.4 (27.8–28.9) | 28.4 (27.8–29.0) |
| Annual incomee: | |||||
| <$8,000 | 285 | 55 (53–56) | 55 (53–56) | 28.9 (28.3–29.5) | 28.9 (28.3–29.6) |
| $8,000– $35,000 | 398 | 56 (55–57) | 56 (55–57) | 28.5 (28.0–28.9) | 28.4 (28.0–28.9) |
| >$35,000 | 77 | 58 (56–61) | 58 (55–60) | 28.0 (27.1–28.9) | 28.0 (27.0–28.9) |
| Occupation typef: | |||||
| White collar | 300 | 58 (56–59) | 58 (56–59) | 28.0 (27.4–28.6) | 28.0 (27.4–28.6) |
| Blue collar | 232 | 54 (52–56) | 54 (53–56) | 28.7 (28.0–29.4) | 28.7 (28.0–29.5) |
| Housewife/other | 275 | 55 (53–57) | 55 (53–57) | 28.8 (28.2–29.5) | 28.8 (28.2–29.5) |
Note.— Likelihood-ratio tests of association were performed by multiple linear regression of each phenotypic trait on biodemographic characteristics. Minimally adjusted models were adjusted for age, sex, and any clinically relevant covariates, as described in the “Methods” section. Fully adjusted models additionally contained variables for remaining biodemographic characteristics. In the footnotes below, P values in bold italics are less than the nominal significance level of 5% adjusted for the number of traits assessed (n=8; P<.00625).
For measurements by clinical center, the minimally adjusted P value was .0005 and the fully adjusted P value was .005 for HDL cholesterol, and the minimally adjusted P value was .61 and the fully adjusted P value was .57 for BMI.
For measurements by genetic similarity, the minimally adjusted P value was .45 and the fully adjusted P value was .28 for HDL cholesterol, and the minimally adjusted P value was .03 and the fully adjusted P value was .02 for BMI.
For measurements by genetic ancestry, the minimally adjusted P value was .84 and the fully adjusted P value was .43 for HDL cholesterol, and the minimally adjusted P value was .28 and the fully adjusted P value was .46 for BMI.
For measurements by education, the minimally adjusted P value was .47 and the fully adjusted P value was .90 for HDL cholesterol, and the minimally adjusted P value was .57 and the fully adjusted P value was .64 for BMI.
For measurements by annual income, the minimally adjusted P value was .05 and the fully adjusted P value was .10 for HDL cholesterol, and the minimally adjusted P value was .20 and the fully adjusted P value was .16 for BMI.
For measurements by occupation type, the minimally adjusted P value was .008 and the fully adjusted P value was .02 for HDL cholesterol, and the minimally adjusted P value was .13 and the fully adjusted P value was .14 for BMI.
Table 6.
CRP Levels and Carotid Wall Thickness by Clinic Site, Genetic Background Similarity, Admixture, and SES[Note]
|
Mean CRP Levels (95% CI)[mg/liter] |
Mean Carotid WallThickness (95% CI)[mm] |
||||
| BiodemographicCharacteristic | N | Minimally Adjusted | Fully Adjusted | Minimally Adjusted | Fully Adjusted |
| Clinical centera: | |||||
| Winston-Salem | 299 | 2.82 (2.51–3.17) | 2.81 (2.49–3.17) | 1.12 (1.09–1.14) | 1.12 (1.10–1.15) |
| Sacramento | 213 | 2.43 (2.11–2.78) | 2.54 (2.20–2.93) | 1.12 (1.10–1.15) | 1.13 (1.10–1.16) |
| Pittsburgh | 285 | 2.57 (2.28–2.88) | 2.58 (2.29–2.90) | 1.12 (1.10–1.15) | 1.12 (1.10–1.15) |
| Genetic similarityb: | |||||
| Cluster 1 | 467 | 2.66 (2.42–2.91) | 2.66 (2.41–2.92) | 1.13 (1.11–1.15) | 1.13 (1.11–1.15) |
| Cluster 2 | 32 | 2.67 (1.88–3.80) | 2.57 (1.80–3.67) | 1.09 (1.01–1.16) | 1.08 (1.00–1.15) |
| Cluster 3 | 74 | 2.35 (1.86–2.96) | 2.41 (1.89–3.08) | 1.09 (1.04–1.14) | 1.09 (1.04–1.14) |
| Cluster 4 | 234 | 2.58 (2.27–2.94) | 2.66 (2.33–3.03) | 1.13 (1.10–1.15) | 1.13 (1.10–1.16) |
| Genetic ancestryc: | |||||
| 0% African (estimated) | 2.12 (1.63–2.76) | 2.37 (1.79–3.14) | 1.09 (1.03–1.14) | 1.09 (1.03–1.15) | |
| 100% African (estimated) | 2.80 (2.50–3.14) | 2.72 (2.42–3.07) | 1.14 (1.11–1.16) | 1.13 (1.11–1.16) | |
| Educationd: | |||||
| None–grade 9 | 237 | 2.80 (2.48–3.14) | 2.75 (2.44–3.11) | 1.14 (1.11–1.16) | 1.14 (1.11–1.16) |
| High school | 294 | 2.62 (2.44–2.81) | 2.62 (2.44–2.81) | 1.12 (1.11–1.14) | 1.12 (1.11–1.14) |
| Professional/vocational | 274 | 2.45 (2.20–2.74) | 2.49 (2.23–2.79) | 1.11 (1.09–1.13) | 1.11 (1.09–1.14) |
| Annual incomee: | |||||
| <$8,000 | 285 | 2.81 (2.50–3.15) | 2.77 (2.46–3.11) | 1.13 (1.10–1.15) | 1.13 (1.10–1.15) |
| $8,000– $35,000 | 398 | 2.55 (2.36–3.77) | 2.57 (2.37–3.79) | 1.12 (1.11–1.14) | 1.13 (1.11–1.14) |
| >$35,000 | 77 | 2.48 (2.25–2.75) | 2.39 (2.00–2.86) | 1.12 (1.08–1.15) | 1.12 (1.08–1.16) |
| Occupation typef: | |||||
| White collar | 300 | 2.52 (2.25–2.83) | 2.54 (2.26–2.86) | 1.12 (1.10–1.15) | 1.12 (1.10–1.15) |
| Blue collar | 232 | 3.15 (2.75–3.62) | 3.12 (2.70–3.59) | 1.12 (1.09–1.15) | 1.12 (1.09–1.15) |
| Housewife/other | 275 | 2.30 (2.03–2.60) | 2.31 (2.03–2.62) | 1.12 (1.10–1.15) | 1.12 (1.09–1.15) |
Note.— Likelihood-ratio tests of association were performed by multiple linear regression of each phenotypic trait on biodemographic characteristics. Minimally adjusted models were adjusted for age, sex, and any clinically relevant covariates, as described in the “Methods” section. Fully adjusted models additionally contained variables for remaining biodemographic characteristics. In the footnotes below, the P value in bold italics is less than the nominal significance level of 5% adjusted for the number of traits assessed (n=8; P<.00625).
For measurements by clinical center, the minimally adjusted P value was .23 and the fully adjusted P value was .49 for CRP level, and the minimally adjusted P value was .95 and the fully adjusted P value was .94 for carotid wall thickness.
For measurements by genetic similarity, the minimally adjusted P value was .80 and the fully adjusted P value was .91 for CRP level, and the minimally adjusted P value was .44 and the fully adjusted P value was .32 for carotid wall thickness.
For measurements by genetic ancestry, the minimally adjusted P value was .11 and the fully adjusted P value was .46 for CRP level, and the minimally adjusted P value was .19 and the fully adjusted P value was .31 for carotid wall thickness.
For measurements by education, the minimally adjusted P value was .16 and the fully adjusted P value was .31 for CRP level, and the minimally adjusted P value was .20 and the fully adjusted P value was .26 for carotid wall thickness.
For measurements by annual income, the minimally adjusted P value was .13 and the fully adjusted P value was .27 for CRP level, and the minimally adjusted P value was .70 and the fully adjusted P value was .93 for carotid wall thickness.
For measurements by occupation type, the minimally adjusted P value was .004 and the fully adjusted P value was .01 for CRP level, and the minimally adjusted P value was .99 and the fully adjusted P value was .97 for carotid wall thickness.
Table 7.
YOL and YHL by Clinic Site, Genetic Background Similarity, Admixture, and SES[Note]
|
Mean YOL (95% CI)[years] |
Mean YHL (95% CI)[years] |
||||
| BiodemographicCharacteristic | N | Minimally Adjusted | Fully Adjusted | Minimally Adjusted | Fully Adjusted |
| Clinical centera: | |||||
| Winston-Salem | 299 | 8.13 (7.83–8.42) | 8.11 (7.80–8.42) | 5.07 (4.76–5.38) | 5.10 (4.77–5.43) |
| Sacramento | 213 | 8.54 (8.19–8.90) | 8.47 (8.10–8.83) | 5.63 (5.26–6.01) | 5.45 (5.06–5.84) |
| Pittsburgh | 285 | 8.25 (7.95–8.55) | 8.26 (7.95–8.56) | 4.95 (4.63–5.27) | 4.93 (4.61–5.26) |
| Genetic similarityb: | |||||
| Cluster 1 | 467 | 7.98 (7.74–8.21) | 7.97 (7.72–8.21) | 4.93 (4.68–5.18) | 4.94 (4.68–5.20) |
| Cluster 2 | 32 | 7.99 (7.09–8.89) | 7.96 (7.04–8.88) | 5.05 (4.09–6.00) | 5.07 (4.10–6.05) |
| Cluster 3 | 74 | 8.69 (8.10–9.28) | 8.64 (8.01–9.26) | 5.90 (5.27–6.53) | 5.79 (5.13–6.45 |
| Cluster 4 | 234 | 8.76 (8.43–9.09) | 8.71 (8.37–9.05) | 5.48 (5.13–5.83) | 5.34 (4.98–5.69) |
| Genetic ancestryc: | |||||
| 0% African (estimated) | 9.15 (8.48–9.82) | 9.02 (8.30–9.75) | 6.08 (5.37–6.80) | 5.59 (4.82–6.36) | |
| 100% African (estimated) | 7.97 (7.68–8.26) | 7.98 (7.67–8.28) | 4.86 (4.55–5.17) | 4.98 (4.65–5.30) | |
| Educationd: | |||||
| None–grade 9 | 237 | 8.16 (7.85–8.46) | 8.28 (7.97–8.59) | 4.69 (4.37–5.01) | 4.77 (4.44–5.10) |
| High school | 294 | 8.26 (8.09–8.44) | 8.27 (8.09–8.44) | 5.15 (4.96–5.33) | 5.15 (4.96–5.34) |
| Professional/vocational | 274 | 8.37 (8.09–8.65) | 8.26 (7.97–8.55) | 5.60 (5.30–5.90) | 5.53 (5.22–5.84) |
| Annual incomee: | |||||
| <$8,000 | 285 | 8.13 (7.83–8.42) | 8.25 (7.95–8.55) | 4.83 (4.51–5.14) | 4.94 (4.62–5.27) |
| $8,000– $35,000 | 398 | 8.30 (8.10–8.50) | 8.25 (8.05–8.45) | 5.27 (5.05–5.48) | 5.22 (5.01–5.44) |
| >$35,000 | 77 | 8.48 (8.04–8.92) | 8.25 (7.79–8.70) | 5.70 (5.24–6.17) | 5.50 (5.02–5.99) |
| Occupation typef: | |||||
| White collar | 300 | 8.41 (8.11–8.71) | 8.32 (8.02–8.62) | 5.63 (5.32–5.95) | 5.48 (5.11–5.85) |
| Blue collar | 232 | 8.07 (7.71–8.42) | 8.13 (7.77–8.49) | 4.60 (4.23–4.98) | 4.75 (4.33–5.17) |
| Housewife/other | 275 | 8.30 (7.98–8.62) | 8.34 (8.02–8.66) | 5.17 (4.83–5.51) | 5.09 (4.72–5.47) |
Note.— Likelihood-ratio tests of association were performed by multiple linear regression of each phenotypic trait on biodemographic characteristics. Minimally adjusted models were adjusted for age, sex, and any clinically relevant covariates, as described in the “Methods” section. Fully adjusted models additionally contained variables for remaining biodemographic characteristics. In the footnotes below, P values in bold italics are less than the nominal significance level of 5% adjusted for the number of traits assessed (n=8; P<.00625).
For measurements by clinical center, the minimally adjusted P value was .32 and the fully adjusted P value was .36 for YOL, and the minimally adjusted P value was .04 and the fully adjusted P value was .14 for YHL.
For measurements by genetic similarity, the minimally adjusted P value was .0007 and the fully adjusted P value was .003 for YOL, and the minimally adjusted P value was .008 and the fully adjusted P value was .08 for YHL.
For measurements by genetic ancestry, the minimally adjusted P value was .008 and the fully adjusted P value was .03 for YOL, and the minimally adjusted P value was .01 and the fully adjusted P value was .23 for YHL.
For measurements by education, the minimally adjusted P value was .37 and the fully adjusted P value was .93 for YOL, and the minimally adjusted P value was .003 and the fully adjusted P value was .004 for YHL.
For measurements by annual income, the minimally adjusted P value was .27 and the fully adjusted P value was .99 for YOL, and the minimally adjusted P value was .01 and the fully adjusted P value was .12 for YHL.
For measurements by occupation type, the minimally adjusted P value was .36 and the fully adjusted P value was .66 for YOL, and the minimally adjusted P value was .0003 and the fully adjusted P value was .002 for YHL.
BMI appeared to be influenced more by genetic similarity than by ancestral proportions (table 5). Moreover, the associations differed by sex (P value for sex-genetic similarity cluster interaction on BMI was .03). The mean age-, clinic-, and SES-adjusted BMI was highest among men in genetic similarity cluster 3 (28.7 kg/m2; 95% CI 27.1–30.3) and was lowest among men in genetic similarity cluster 2 (24.6 kg/m2; 95% CI 22.6–26.5). In contrast, income level remained the only significant predictor of BMI among women, after multivariable adjustment (P=.03). The mean-adjusted BMI was 30.3 kg/m2 (95% CI 29.6–31.1) for women in the lowest income group, compared with 28.3 kg/m2 (95% CI 26.8–29.8) for women in the highest income group.
African ancestry and genetic similarity, even when adjusted for baseline age, BMI, sex, self-rated health status, smoking, hypertension, diabetes, coronary heart disease, and cancer, were associated with both YOL and YHL during follow-up (table 7). For the YHL outcome, SES adjustment attenuated these associations. Moreover, when all of the biodemographic covariates in table 7 were included together simultaneously, income, education, and occupation (P=.005), rather than individual ancestry (P=.50) or genetic similarity (P=.12), remained the only significant predictor of YHL. In contrast, genetic background similarity (P=.02) remained the only significant biodemographic predictor of the outcome YOL in a multivariable model; individuals belonging to genetic similarity cluster 4 lived, on average, an additional 9 mo (95% CI 3–14), compared with genetic similarity cluster 1.
Associations of Phenotypic Traits with Individual AIMs
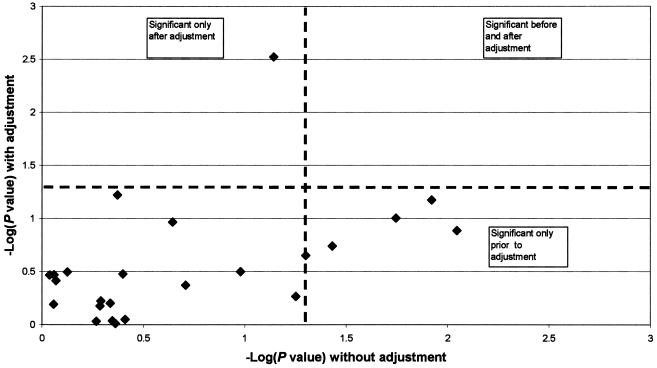
Tests of association for each AIM with each phenotypic trait are shown in table 8. In general, trait-associated markers tended to be among those with the highest African/European allele-frequency differential. Under the null hypothesis, ∼1/24 markers would be expected by chance to be associated with any single trait (with the significance threshold of P<.05). By comparison, blood glucose was associated with five markers. Adjustment for individual ancestry attenuated each of the five marker–blood glucose associations (fig. 2). These findings strongly suggest false-positive associations due to population stratification. Note also in figure 2 that an additional marker (rs722098) showed no association before adjustment but was associated with glucose level (P=.002) only after correction for African ancestry.
Table 8.
Tests of Association between Each AIM and the Cardiovascular Risk Factors and Longevity Outcomes[Note]
|
P Values for |
||||||||
| Markera | BloodGlucose | SystolicBlood Pressure | HDLCholesterol | BMI | CRP Levels | Carotid WallThickness | YOL | YHL |
| rs2814778 | .37 | .46 | .65 | .89 | .004 | .55 | .01 | .23 |
| rs930072 | .005 | .18 | .49 | .16 | .05 | .02 | .03 | .02 |
| rs7349 | .04 | .28 | .75 | .18 | .97 | .91 | .10 | .01 |
| rs723632 | .07 | .94 | .25 | .05 | .95 | .85 | .65 | .40 |
| rs722098 | .56 | .58 | .16 | .28 | .29 | .87 | .05 | .09 |
| rs146026 | .04 | .21 | .02 | .93 | .54 | .02 | .41 | .58 |
| rs6003 | .12 | .33 | .18 | .95 | .07 | .82 | .43 | .57 |
| rs1985080 | .91 | .92 | .74 | .86 | .81 | .56 | .94 | .78 |
| rs518116 | .01 | .28 | .68 | .76 | .60 | .38 | .08 | .07 |
| rs3287 | .01 | .49 | .15 | .73 | .50 | .69 | .49 | .25 |
| rs1989486 | .52 | .43 | .08 | .32 | .32 | .99 | .16 | .99 |
| rs7041 | .07 | .23 | .72 | .38 | .66 | .10 | .06 | .22 |
| rs994174 | .45 | .18 | .44 | .76 | .30 | .57 | .33 | .18 |
| rs1800498 | .67 | .56 | .15 | .71 | .86 | .75 | .03 | .15 |
| rs2816 | .77 | .36 | .38 | .59 | .15 | .21 | .82 | .54 |
| rs2891 | .15 | .41 | .59 | .18 | .28 | .98 | .19 | .16 |
| rs3188520 | .77 | .62 | .49 | .07 | .19 | .10 | .61 | .13 |
| rss1042602 | .96 | .04 | .13 | .95 | .62 | .72 | .22 | .18 |
| rs326946 | .10 | .67 | .87 | .21 | .23 | .64 | .27 | .10 |
| rs2077863 | .23 | .40 | .99 | .38 | .63 | .35 | .17 | .91 |
| rs3188519 | .92 | .32 | .96 | .16 | .84 | .08 | .73 | .48 |
| rs594689 | .24 | .62 | .48 | .20 | .44 | .07 | .29 | .93 |
| rs2228478 | .45 | .37 | .26 | .03 | .04 | .81 | .01 | .21 |
| rs584059 | .43 | .04 | .41 | .11 | .27 | .45 | .71 | .86 |
Note.— Likelihood-ratio tests of association were performed using multiple linear regression, adjusted for age, sex, and any clinically relevant covariates, as described in the “Methods” section. P values ⩽.05 are indicated in bold italics.
Markers are listed in decreasing order of African/European FST values.
Figure 2.
Associations of markers with glucose, before and after adjustment for African ancestry proportions
Discussion
Our results suggest a complex relationship between aging-related traits, genetic background heterogeneity, and population structure among older African American adults. Overall, there was evidence of substantial population subdivision and genetic admixture, as demonstrated by decreased marker heterozygosity and excess allelic association of unlinked markers. Remarkably, ∼60% of pairs of markers in this data set were associated, despite the fact that the markers were randomly scattered throughout the genome. The association between markers that are not in physical proximity indicates that the rate of spurious associations without adjustment for population stratification is likely to be particularly high in this population, since association cannot be a reliable measure of genomic position. However, the rate of association between these markers is much higher than the rate expected with random markers, since there is a linear relationship between the strength of allelic association and marker informativeness in admixed populations (Chakraborty and Weiss 1988).
All of the phenotypic traits under study are known to be influenced by environmental factors, some of which are related to SES. In our analyses, SES adjustment seemed to weaken the association of genetic background with some traits but not others. However, education, income, and occupation type likely represent only crude proxies for current SES (Kaufman et al. 1997), given the retirement status of the participants. Moreover, clinical disease is common among older cohorts such as the CHS cohort (Kuller et al. 1998). Although we additionally adjusted our analyses for known clinical confounders, residual nonrandom associations between health care access or adequacy of treatment and social characteristics may persist. Therefore, we cannot exclude residual confounding by environmental determinants as a possible explanation for the observed genetic ancestry-trait associations (Risch et al. 2002; Kittles and Weiss 2003). Ultimately, proof that association between genetic ancestry and a particular phenotype is due to genetic etiology lies in the identification of a specific genomic region or regions that account for the association. This would require a whole-genome admixture mapping survey, which depends on the typing of hundreds to thousands of markers (Smith et al. 2004; McKeigue 2005).
The observed association of increased blood glucose levels with African ancestry is consistent with reports of higher fasting glucose and increased prevalence of diabetes in older African Americans (Haffner et al. 1996). A similar association between insulin resistance and African ancestry, independent of SES, was recently reported in a study of children residing in the southern United States, which included individuals whose self-reported race/ethnicity was white and African American (Gower et al. 2003). Our analysis included only individuals who self reported as African American and was also adjusted for education and income levels.
The sex-dependent associations we observed for BMI are noteworthy in light of the higher prevalence of female obesity but lower levels of male obesity among older African Americans compared with older whites (Hutchinson et al. 1997; Kuller et al. 1998; Sundquist et al. 2001). The propensity for weight gain and obesity in black women has been associated with lower SES and higher physical inactivity (Fernandez et al. 2003). Our findings suggest a possible contribution of either genetic background or other distinct environmental factors correlated with genetic background to the lower rates of obesity in African American men.
SES has a major impact on all-cause mortality within and among age, sex, and race strata (Lin et al. 2003). In the CHS African American cohort, longevity appeared to be influenced by various indicators of population and social structure. The association with SES indicators was particularly strong for YHL, a measure that incorporates both length and quality of life. For YOL, which more objectively quantifies survival time or all-cause mortality, the association with genetic background appeared stronger but was attenuated somewhat by SES adjustment. Genetic factors may influence mortality in older adults, particularly at very advanced ages (Perls et al. 2002). It is important to recognize, however, that genetic similarity or shared ancestry are likely correlated with a range of social, cultural, and/or environmental variables that influence disease occurrence and mortality yet remain unmeasured or not adequately accounted for in our analysis (Risch et al. 2002; Kittles and Weiss 2003). The substantial effect of SES on the genetic associations with longevity highlights an important principle: excess type I error can occur in admixed populations even as a result of environmental factors (Risch et al. 2002; Cardon and Palmer 2003). In this case, SES is associated with genetic ancestry, leading to confounding in tests for individual markers.
Our findings for CHS strongly suggest that controlling for population structure/admixture will be required in large, multicenter genetic-association studies that assess common chronic-disease–related traits in African American population samples. Individual ancestry can be estimated in African American samples by typing a reasonable number of markers that are highly differentiated in allele frequency across parental populations. Conditioning on admixture proportions in a multilocus analysis can control for confounding due to population stratification (McKeigue et al. 2000; Pfaff et al. 2001; Hoggart et al. 2003). As illustrated in figure 2, controlling for genetic ancestry should not only reduce false-positive associations but may also uncover a true association previously obscured by stratification. On the basis of dynamic relationships among various genetic and environmental determinants of disease susceptibility, additional multilocus methods—such as those that detect genetic similarity, cryptic relatedness, or rates of migration under nonequilibrium conditions—may help to characterize the complex genetic demography of an epidemiologic sample (Overall and Nichols 2001; Schork 2001; Schork et al. 2001; Curtis et al. 2002; Wilson and Rannala 2003) and thereby provide additional information about the genetic architecture of common diseases of aging in heterogeneous outbred populations.
The limited number of markers we used may have resulted in imprecise estimates of individual ancestry or genetic background similarity. However, several different statistical methods of differentiating individuals, including the Bayesian algorithm in the program STRUCTURE and the results of the principal-components method, demonstrated a very high degree of correlation with our estimates of African ancestry from the maximum-likelihood model. This is not unexpected, since the markers were chosen primarily on the basis of frequency differential between African and European parental populations.
Since the markers we used had less ability to distinguish Native American ancestry from European ancestry, the correlations were less robust for Native American or European ancestry estimated by maximum likelihood, STRUCTURE, and principal-components analysis. This is also reflected by the wide CIs associated with our estimates of Native American ancestry. Our results are not inconsistent with previous studies, such as those of Parra et al. (1998) and Smith et al. (2004), who estimated the Native American ancestry of African American populations at 1%–2%. Interestingly, there appeared to be a somewhat higher proportion of Native American ancestry among individuals within genetic similarity cluster 3. Typing more markers informative for Native American ancestry will be necessary to confirm these findings, which might lead to greater precision in controlling for admixture in association studies.
Our results also are in agreement with other studies showing ∼20% European admixture among African Americans, with somewhat higher contributions of European ancestry in northern or western U.S. populations (Chakraborty et al. 1986; Parra et al. 1998; McKeigue et al. 2000; Pfaff et al. 2001; Hoggart et al. 2003). Whether genetic heterogeneity among the African parental source populations has contributed to local variations in admixture among modern African American populations remains uncertain. Despite earlier studies suggesting genetic heterogeneity within continental Africa (reviewed by Tishkoff and Williams 2002; Kittles and Weiss 2003), markers such as the ones we typed, which have large frequency differences between European and African populations, appear to have much smaller variations within continental Africa (Collins-Schramm et al. 2002).
Our analysis excluded the individuals who self reported as white in the CHS cohort. Since other studies report that the proportion of African ancestry among U.S. non-Hispanic whites is <5%, the exclusion of self-identified white CHS participants from our study sample is unlikely to have impacted our findings in a substantial way. Our study does not address the question of population stratification among European Americans. Since allele-frequency differences between European subpopulations are likely smaller than those between European and African ancestral populations, a larger of number of markers will be required to assess the impact of stratification within non-Hispanic white populations.
Individuals who self identify as African American are culturally, socially, and genetically heterogeneous. SES and related factors, such as access to health care, play a major role in healthy aging. In some instances, these nongenetic factors may account for all or part of the association between a phenotype and ancestry. Nevertheless, from a public health and epidemiologic standpoint, an objective assessment of genetic background may provide additional information relevant to potential nongenetic confounders and predictors of disease risk as well as insight into genetic contributions. These considerations highlight the need for further investigation of the various SES and biodemographic factors that influence life span or quality of life in older adults.
In summary, there is evidence of substantial substructure and admixture among the CHS African American population. In addition, our analyses have shown that nongenetic factors may, in fact, confound genetic associations among populations with recent admixture and population substructure. Therefore, both controlling for population admixture by use of genetic markers and controlling for sociodemographic measures will be required in assessing genetic associations with complex chronic-disease traits in African American subjects.
Acknowledgments
We thank Mark D. Shriver for providing the program used for maximum-likelihood estimation and for providing DNA samples of the three ancestral populations. The research reported in this article was supported by National Heart, Lung, and Blood Institute contracts N01-HC-85079 through N01-HC-85086, N01-HC-35129, and N01 HC-15103. A full list of participating CHS investigators and institutions can be found at the CHS Web site.
Appendix A
Table A1.
PCR Assay Conditions and Oligonucleotide Primers for the 24 AIMs[Note]
|
PCR Primer (5′→3′) |
||||||||
| Marker | Alternate Namea | Conditionsb | Tm(°C) | Strandc | Dye Terminator Mixd | Forward | Reverse | Genotyping Primer (5′→3′) |
| rs2814778 | FY-NULL | .2 μM primers, 100 μM dNTPs, 5% DMSO | 58 | Reverse | CT | GCATAGGGATAAGGGACTCT | AACCTCAAAACAGGAAGACC | GCCCTCATTAGTCCTTGGCTCTTA |
| rs930072 | Standard | 58 | Reverse | GA | TGCCACTGTCTCTTAGATTACAT | CAAAGAAAGTCACTTCAAATCTC | CTTCTGGCCCCTTTATAGACTAGCTT | |
| rs7349 | Standard | 58 | Reverse | GA | TAACATTTATACTTGCCTTGGAC | ATTGTTCCCAACACTGTTTATAG | GGAAATGAGAGTTGTATGGTTAGGCT | |
| rs723632 | Standard | 58 | Forward | GC | ATATTTGAACCTTGTGCTTCAG | TAAGAGAGTTTATGATGGGTGTC | CCCATCCCCAGGCCTCTTTA | |
| rs722098 | Standard | 58 | Reverse | CT | TAATGAACACAGCCATCTAGTCT | AAATATTCAGCACATCCAAAAT | GAAATATTCAGCACATCCAAAATTTAAATCCTTA | |
| rs146026 | Standard | 58 | Reverse | GA | TGAGCACTGGCTTTATTATTATT | CAACTGCTTTAAAAAGAGAGAGA | TGAAACACACAAAATGCATTACAATCGTA | |
| rs6003 | F13B | Standard | 58 | Reverse | CT | ATGAAATCGCCAATAATAACAT | AGAGAGACATTGAACCACTTCTT | CCCTGAAGCGCAACCATAA |
| rs1985080 | Standard | 58 | Reverse | CT | TTTTTAAAATGGTATCAAACACC | GTCATCCCTATACCTTCTGACTT | GTGGATTGTACAGTGAGCCACTTA | |
| rs518116 | Standard | 58 | Reverse | CT | TTACATCATGAGAATCACTTTTTC | GTCATCTACTACACAGGGAGGTA | TTCAGTTTGGGCAAATTAGTTTGCAC | |
| rs3287 | WI-16857 | 5% DMSO | 57 | Reverse | CT | TATAATCCATCCTCCAACACA | AAGAGCAGATTTCTCCACATTAT | TCTTGTGAATTGGGAAGACCACTAG |
| rs1989486 | Standard | 58 | Reverse | GA | AAATGGACACAAAAATAAAGATG | GTTTAAAATTGATTGCTGTTCTC | CCCTTGATGAACAAGGCTAACCT | |
| rs7041 | 5% DMSO | 58 | Forward | GT | AATGGCTATTATTTTGCATTAGA | TAAAAGATTCTGCCATGTTAAGT | GCGACTAAAAGCAAAATTGCCTGA | |
| rs994174 | Standard | 58 | Reverse | CT | ATTGTGAAGTACAGGTGTGGTAT | TTTGGAGTCTCAGTGTTCTTATC | GTACTGCTTCTAATAGTACTCAGTGCCA | |
| rs1800498 | DRD2Taq_D | .4 μM primers, 3.5mM MgCl2 | 58 | Reverse | GA | TATAAGCATCAAGTGTTTGGAAC | GAGAAGAAAACAGAACAAGATCA | GCCCCAGGTTCCCTAGTC |
| rs2816 | WI-7423 | Standard | 58 | Forward | CT | CTCATAGCTGCTGCTTCAAT | ACAGCATGCCTTTATTTCAC | CATTGCTGGGCTGTGTTCC |
| rs2891 | WI-14867 | Standard | 58 | Forward | GA | CAAAGTGTTGGAAATGTCATC | GAACAACTGATGAACTGAATTTT | ATGGGGCTGCAGACACTC |
| rs3188520 | Standard | 61 | Reverse | GC | CTCTGACTGAGAAACTGAACAAT | AGGGTACCTAGCACCTTGTATT | AAAACAGGCAATCCTCCTAAGTCT | |
| rs1042602 | TYR-192 | Standard | 58 | Reverse | GT | CTACCTCACTTTAGCAAAGCATA | ACCGCAACAAGAAGAGTCTAT | GCAAAATCAATGTCTCTCCAGATTTCA |
| rs326946 | Standard | 58 | Reverse | CA | AACCCCTGAGATAGAACATAGAT | GGTAAGAAATGACTCAGAAGACA | TTTTTGCAAAGTGGCAGGATTTAATAATAATAAT | |
| rs2077863 | Standard | 58 | Reverse | GC | AGGTCCAGAAATGGCTCTAT | GACTGATAGTTGTTCCTGCTG | GCATCTATCCTAGGTTTGTGGATAGGA | |
| rs3188519 | Standard | 58 | Forward | CT | TAGGTCTCAGGTAGCAAGAGTC | CAGATGATTCATACACACCCTAT | CCAGCCACCAGTCACCC | |
| rs594689 | D11S429 | 5% DMSO | 58 | Reverse | CT | GATCTGAAAGGCATTGTAGG | CTTTGTAGGGTAGGAGAAATTG | GGGGACGGGCAAGGAG |
| rs2228478 | MC1R-314 | Standard | 58 | Reverse | CT | CAAGAACTTCAACCTCTTTCTC | AGATCATTTAGTCCATCCTCTTT | GCGCTCACCAGGAGCA |
| rs584059 | Standard | 58 | Forward | CA | TCATCAGGATCCTCCAATACT | CTCCACTTCTCTGGAATTTTT | CAACCCAGGACCCACAGAAG | |
Note.— Tm = the annealing temperature used in PCR cycles.
Additional information on markers with an alternate name can be found in dbSNP under the handle PSU-ANTH.
Standard PCR conditions were 5 μl PCR volume with 2.4 ng genomic DNA, Platinum Taq buffer, 2.5 mM MgCl2, 50 μM dNTPs, 0.1μM PCR primers, 0.1 unit Platinum Taq, and 1 μl of extra water to counteract evaporation.
The strand of DNA for which the genotyping primer will give results.
The mix of dye terminators used in the genotyping reaction (based on the strand used for the genotyping primer).
Electronic-Database Information
The URLs for data presented herein are as follows:
- CHS, http://www.chs-nhlbi.org
- dbSNP, http://www.ncbi.nlm.nih.gov/SNP/
- Genetic Data Analysis, http://hydrodictyon.eeb.uconn.edu/people/plewis/software.php
References
- Belkhir K, Castric V, Bonhomme F (2002) IDENTIX , a software to test for relatedness in a population using permutation methods. Mol Ecol Notes 2:611–614 10.1046/j.1471-8286.2002.00273.x [DOI] [Google Scholar]
- Bonilla C, Parra EJ, Pfaff CL, Dios S, Marshall JA, Hamman RF, Ferrell RE, Hoggart CL, McKeigue PM, Shriver MD (2004) Admixture in the Hispanics of the San Luis Valley, Colorado, and its implications for complex trait gene mapping. Ann Hum Genet 68:139–153 10.1046/j.1529-8817.2003.00084.x [DOI] [PubMed] [Google Scholar]
- Cardon LR, Palmer LJ (2003) Population stratification and spurious allelic association. Lancet 361:598–604 10.1016/S0140-6736(03)12520-2 [DOI] [PubMed] [Google Scholar]
- Chakraborty R, Ferrell RE, Stern MP, Haffner SM, Hazuda HP, Rosenthal M (1986) Relationship of prevalence of non-insulin dependent diabetes mellitus to Amerindian admixture in the Mexican Americans of San Antonio, Texas. Genet Epidemiol 3:435–454 [DOI] [PubMed] [Google Scholar]
- Chakraborty R, Weiss KM (1988) Admixture as a tool for finding linked genes and detecting that difference from allelic association between loci. Proc Natl Acad Sci USA 85:9119–9123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Levine L, Kwok PY (1999) Fluorescence polarization in homogeneous nucleic acid analysis. Genome Res 9:492–498 [PMC free article] [PubMed] [Google Scholar]
- Collins-Schramm HE, Kittles RA, Operario DJ, Weber JL, Criswell LA, Cooper RS, Seldin MF (2002) Markers that discriminate between European and African ancestry show limited variation within Africa. Hum Genet 111:566–569 10.1007/s00439-002-0818-z [DOI] [PubMed] [Google Scholar]
- Curtis D, North BV, Gurling HM, Blaveri E, Sham PC (2002) A quick and simple method for detecting subjects with abnormal genetic background in case-control samples. Ann Hum Genet 66:235–244 10.1046/j.1469-1809.2002.00109.x [DOI] [PubMed] [Google Scholar]
- Cushman M, Cornell E, Howard P, Bovill E, Tracy R (1995) Laboratory methods and quality assurance in the Cardiovascular Health Study. Clin Chem 41:264–270 [PubMed] [Google Scholar]
- Diehr P, Patrick DL, Bild DE, Burke GL, Williamson JD (1998) Predicting future years of healthy life for older adults. J Clin Epidemiol 51:343–353 10.1016/S0895-4356(97)00298-9 [DOI] [PubMed] [Google Scholar]
- Dupanloup I, Bertorelle G (2001) Inferring admixture proportions from molecular data: extension to any number of parental populations. Mol Biol Evol 18:672–675 [DOI] [PubMed] [Google Scholar]
- Falush D, Stephens M, Pritchard JK (2003) Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164:1567–1587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez JR, Shriver MD, Beasley TM, Rafla-Demetrious N, Parra E, Albu J, Nicklas B, Ryan AS, McKeigue PM, Hoggart CL, Weinsier RL, Allison DB (2003) Association of African genetic admixture with resting metabolic rate and obesity among women. Obes Res 11:904–911 [DOI] [PubMed] [Google Scholar]
- Freedman ML, Reich D, Penney KL, McDonald GJ, Mignault AA, Patterson N, Gabriel SB, Topol EJ, Smoller JW, Pato CN, Pato MT, Petryshen TL, Kolonel LN, Lander ES, Sklar P, Henderson B, Hirschhorn JN, Altshuler D (2004) Assessing the impact of population stratification on genetic association studies. Nat Genet 36:388–393 10.1038/ng1333 [DOI] [PubMed] [Google Scholar]
- Fried LP, Borhani NO, Enright P, Furberg CD, Gardin JM, Kronmal RA, Kuller LH, Manolio TA, Mittelmark MB, Newman A, O’Leary DH, Psaty BM, Rautaharju P, Tracy RP, Weiler PG (1991) The Cardiovascular Health Study: design and rationale. Ann Epidemiol 1:263–276 [DOI] [PubMed] [Google Scholar]
- Gower BA, Fernandez JR, Beasley TM, Shriver MD, Goran MI (2003) Using genetic admixture to explain racial differences in insulin-related phenotypes. Diabetes 52:1047–1051 [DOI] [PubMed] [Google Scholar]
- Haffner SM, D'Agostino R, Saad MF, Rewers M, Mykkanen L, Selby J, Howard G, Savage PJ, Hamman RF, Wagenknecht LE, Bergman RN (1996) Increased insulin resistance and insulin secretion in nondiabetic African Americans and Hispanics compared with non-Hispanic whites: The Insulin Resistance Atherosclerosis Study. Diabetes 45:742–748 [DOI] [PubMed] [Google Scholar]
- Hoggart CJ, Parra EJ, Shriver MD, Bonilla C, Kittles RA, Clayton DG, McKeigue PM (2003) Control of confounding of genetic associations in stratified populations. Am J Hum Genet 72:1492–1504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchinson RG, Watson RL, Davis CE, Barnes R, Brown S, Romm F, Spencer JM, Tyroler HA, Wu K (1997) Racial differences in risk factors for atherosclerosis. The ARIC Study: Atherosclerosis Risk in Communities. Angiology 48:279–290 [DOI] [PubMed] [Google Scholar]
- Kaufman JS, Cooper RS, McGee DL (1997) Socioeconomic status and health in blacks and whites: the problem of residual confounding and the resiliency of race. Epidemiology 8:621–628 10.1097/00001648-199710000-00002 [DOI] [PubMed] [Google Scholar]
- Kittles RA, Chen W, Panguluri RK, Ahaghotu C, Jackson A, Adebamowo CA, Griffin R, Williams T, Ukoli F, Adams-Campbell L, Kwagyan J, Isaacs W, Freeman V, Dunston GM (2002) CYP3A4-V and prostate cancer in African Americans: causal or confounding association because of population stratification? Hum Genet 110:553–560 10.1007/s00439-002-0731-5 [DOI] [PubMed] [Google Scholar]
- Kittles RA, Weiss KM (2003) Race, ancestry, and genes: implications for defining disease risk. Annu Rev Genomics Hum Genet 4:33–67 10.1146/annurev.genom.4.070802.110356 [DOI] [PubMed] [Google Scholar]
- Kuller L, Fisher L, McClelland R, Fried L, Cushman M, Jackson S, Manolio T (1998) Differences in prevalence of and risk factors for subclinical vascular disease among black and white participants in the Cardiovascular Health Study. Arterioscler Thromb Vasc Biol 18:283–293 [DOI] [PubMed] [Google Scholar]
- Lehmann EL, D’Abrera HJM (1998) Nonparametrics: statistical methods based on ranks. Prentice-Hall, Englewood Cliffs, NJ [Google Scholar]
- Lin CC, Rogot E, Johnson NJ, Sorlie PD, Arias E (2003) A further study of life expectancy by socioeconomic factors in the National Longitudinal Mortality Study. Ethn Dis 13:240–247 [PubMed] [Google Scholar]
- Lynch M, Ritland K (1999) Estimation of pairwise relatedness with molecular markers. Genetics 152:1753–1766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKeigue PM (1998) Mapping genes that underlie ethnic differences in disease risk: methods for detecting linkage in admixed populations, by conditioning on parental admixture. Am J Hum Genet 63:241–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ——— (2005) Prospects for admixture mapping of complex traits. Am J Hum Genet 76:1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKeigue PM, Carpenter J, Parra EJ, Shriver MD (2000) Estimation of admixture and detection of linkage in admixed populations by a Bayesian approach using Markov chain simulation: application to African American populations. Ann Hum Genet 64:171–186 10.1046/j.1469-1809.2000.6420171.x [DOI] [PubMed] [Google Scholar]
- O’Leary DH, Polak JF, Wolfson SK Jr, Bond MG, Bommer W, Sheth S, Psaty BM, Sharrett AR, Manolio TA (1991) Use of ultrasonography to evaluate carotid atherosclerosis in the elderly: the Cardiovascular Health Study. Stroke 22:1155–1163 [DOI] [PubMed] [Google Scholar]
- Overall AD, Nichols RA (2001) A method for distinguishing consanguinity and population substructure using multilocus genotype data. Mol Biol Evol 18:2048–2056 [DOI] [PubMed] [Google Scholar]
- Parra EJ, Kittles RA, Argyropoulos G, Pfaff CL, Hiester K, Bonilla C, Sylvester N, Parrish-Gause D, Garvey WT, Jin L, McKeigue PM, Kamboh MI, Ferrell RE, Pollitzer WS, Shriver MD (2001) Ancestral proportions and admixture dynamics in geographically defined African Americans living in South Carolina. Am J Phys Anthropol 114:18–29 [DOI] [PubMed] [Google Scholar]
- Parra EJ, Marcini A, Akey J, Martinson J, Batzer MA, Cooper R, Forrester T, Allison DB, Deka R, Ferrell RE, Shriver MD (1998) Estimating African American admixture proportions by use of population-specific alleles. Am J Hum Genet 63:1839–1851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perls TT, Wilmoth J, Levenson R, Drinkwater M, Cohen M, Bogan H, Joyce E, Brewster S, Kunkel L, Puca A (2002) Life-long sustained mortality advantage of siblings of centenarians. Proc Natl Acad Sci USA 99:8442–8447 10.1073/pnas.122587599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaff CL, Barnholtz-Sloan J, Wagner JK, Long JC (2004) Information on ancestry from genetic markers. Genet Epidemiol 26:305–315 10.1002/gepi.10319 [DOI] [PubMed] [Google Scholar]
- Pfaff CL, Parra EJ, Bonilla C, Hiester K, McKeigue PM, Kamboh MI, Hutchinson RG, Ferrell RE, Boerwinkle E, Shriver MD (2001) Population structure in admixed populations: effect of admixture dynamics on the pattern of linkage disequilibrium. Am J Hum Genet 68:198–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P (2000a) Inference of population structure from multilocus genotype data. Genetics 155:945–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Rosenberg NA, Donnelly P (2000b) Association mapping in structured populations. Am J Hum Genet 67:170–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risch N, Burchard E, Ziv E, Tang H (2002) Categorization of humans in biomedical research: genes, race and disease. Genome Biol 3:1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schork NJ (2001) Genome partitioning and whole-genome analysis. Adv Genet 42:299–322 [DOI] [PubMed] [Google Scholar]
- Schork NJ, Fallin D, Thiel B, Xu X, Broeckel U, Jacob HJ, Cohen D (2001) The future of genetic case-control studies. Adv Genet 42:191–212 [DOI] [PubMed] [Google Scholar]
- Sharma S, Malarcher AM, Giles WH, Myers G (2004) Racial, ethnic and socioeconomic disparities in the clustering of cardiovascular disease risk factors. Ethn Dis 14:43–48 [PubMed] [Google Scholar]
- Shriver MD, Parra EJ, Dios S, Bonilla C, Norton H, Jovel C, Pfaff C, Jones C, Massac A, Cameron N, Baron A, Jackson T, Argyropoulos G, Jin L, Hoggart CJ, McKeigue PM, Kittles RA (2003) Skin pigmentation, biogeographical ancestry and admixture mapping. Hum Genet 112:387–399 [DOI] [PubMed] [Google Scholar]
- Smith MW, Patterson N, Lautenberger JA, Truelove AL, McDonald GJ, Waliszewska A, Kessing BD, et al (2004) A high-density admixture map for disease gene discovery in African Americans. Am J Hum Genet 74:1001–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundquist J, Winkleby MA, Pudaric S (2001) Cardiovascular disease risk factors among older black, Mexican-American, and white women and men: an analysis of NHANES III, 1988–1994. Third National Health and Nutrition Examination Survey. J Am Geriatr Soc 49:109–116 10.1046/j.1532-5415.2001.49030.x [DOI] [PubMed] [Google Scholar]
- Tishkoff SA, Williams SM (2002) Genetic analysis of African populations: human evolution and complex disease. Nat Rev Genet 3:611–621 [DOI] [PubMed] [Google Scholar]
- Weir BS, Cockerham CC (1984) Estimating F-statistics for the analysis of population structure. Evolution 38:1358–1370 [DOI] [PubMed] [Google Scholar]
- Wilson GA, Rannala B (2003) Bayesian inference of recent migration rates using multilocus genotypes. Genetics 163:1177–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S (1951) The genetical structure of populations. Ann Eugen 15:323–354 [DOI] [PubMed] [Google Scholar]
- Yashin AI, De Benedictis G, Vaupel JW, Tan Q, Andreev KF, Iachine IA, Bonafe M, DeLuca M, Valensin S, Carotenuto L, Franceschi C (1999) Genes, demography, and life span: the contribution of demographic data in genetic studies on aging and longevity. Am J Hum Genet 65:1178–1193 [DOI] [PMC free article] [PubMed] [Google Scholar]