Abstract
Identification of genes causing variation in daytime and nighttime respiration rates could advance our understanding of the basic molecular processes of human respiratory rhythmogenesis. This could also serve an important clinical purpose, because dysfunction of such processes has been identified as critically important in sleep disorders. We performed a sib-pair–based linkage analysis on ambulatory respiration rate, using the data from 270 sibling pairs who were genotyped at 374 markers on the autosomes, with an average distance of 9.65 cM. Uni- and multivariate variance-components–based multipoint linkage analyses were performed for respiration rate during three daytime periods (morning, afternoon, and evening) and during nighttime sleep. Evidence of linkage was found at chromosomal locations 3q27, 7p22, 10q26, and 22q12. The strongest evidence of linkage was found for respiration rate during sleep, with LOD scores of 2.36 at 3q27, 3.86 at 10q26, and 1.59 at 22q12. In a simultaneous analysis of these three loci, >50% of the variance in sleep respiration rate could be attributed to a quantitative-trait loci near marker D10S1248 at 10q. Genes in this area (GFRA1, ADORA2L, FGR2, EMX2, and HMX2) can be considered promising positional candidates for genetic association studies of respiratory control during sleep.
Introduction
Billions of mammals, including humans, depend on rhythmic breathing to regulate one of the foremost aspects of homeostasis: the appropriate exchange of oxygen and carbon dioxide. Although a number of theoretical models have been proposed, the actual mechanisms responsible for respiratory rhythmogenesis were largely misunderstood until recently, when important new insights were gained from in vitro studies of brain stem preparations from neonatal rodents. In the rostral ventrolateral medulla, a set of neurons known as the “preBötzinger complex” act as an inspiratory pacemaker that plays a vital role in respiratory rhythmogenesis (Rekling and Feldman 1998; Richter and Spyer 2001; Feldman et al. 2003). Bilateral outflow of the preBötzinger complex and its associated “distributed” network in the lower brain stem is transmitted to the spinal motor neurons (hypoglossal nerve and phrenic nerve) to produce rhythmic contraction.
Although in vitro models, with their elimination of the many complex peripheral inputs, allow reliable modeling of the neuronal pacemaker network in the preBötzinger complex, it has been widely recognized that they yield an oversimplified picture of the true in vivo generation of the respiratory rhythm (Richter and Spyer 2001; Hilaire and Pásaro 2003). An alternative way to access the molecular biology of respiration is to characterize the genetic variation involved in individual differences in the control of respiratory behavior. This genetic variation should generate clues to the signaling pathways in respiratory neurons by providing molecular targets for animal-genetics engineers to produce conditional knockouts or transgenic animals that would allow detailed anatomical tracing of the neural network involved in respiratory control. To ensure that the investigated pathways have a meaningful concomitant in humans, it would be advantageous to identify genes for respiration in a human population, preferably through measuring respiration under natural conditions.
Understanding the basic molecular processes in human respiratory rhythmogenesis serves an important clinical purpose, because such processes have been identified as critically important in sleep apnea (Hanly 1992; American Academy of Sleep Medicine Task Force 1999; Palmer and Redline 2003). Sleep apnea is defined as repetitive episodes of decreased or total cessation of respiratory airflow during sleep, leading to a >4% fall in oxygen saturation and to sleep fragmentation. Sleep apnea can be central or obstructive. Obstructive sleep apnea is caused by upper-airway collapse during inspiration and is accompanied by strenuous breathing efforts. In central sleep apnea, there are unknown primary defects in the central respiratory rhythmogenesis and/or the chemosensitive control mechanisms that lead to diminution or cessation of thorac-abdominal respiratory movements. Sleep apnea constitutes a major public health problem because of its high prevalence and its association with cardiovascular morbidity (Wolk et al. 2003).
Recently, we completed a large twin-sibling study to test the heritability of 24-h respiration rate (H. M. Kupper, G. Willemsen, D. Posthuma, D. De Boer, D. I. Boomsma, E. J. C. de Geus, unpublished data). We used ambulatory recording of the thorax impedance to obtain the respiration signal. This allows measurements in natural settings and, importantly, is sufficiently unobtrusive to allow recording during sleep (de Geus et al. 1995; de Geus and Van Doornen 1996). Heritability of respiration rate during the daytime was moderate (41%–50%), whereas heritability at night was high (81%).
Here, we report a whole-genome scan on ambulatory respiration rate, using the data from 270 sibling pairs who were genotyped at 374 marker loci on the autosomes, with an average distance of 9.65 cM (Kosambi). Variance-components–based, multipoint, model-free linkage analyses were performed separately on respiration-rate data obtained during three daytime periods (morning, afternoon, and evening) and during nighttime sleep.
Subjects and Methods
Subjects
In 1991, the Netherlands Twin Register started a longitudinal survey study of health and lifestyle (Boomsma et al. 2002). Questionnaires were sent out in 1991, 1993, 1995, and 1997 to adolescent and adult twins and their family members. Twin pairs were asked to participate in all years; parents were asked to participate in 1991, 1993, and 1995; and siblings were included in 1995 and 1997. On the basis of questionnaire data on anxiety and depression, a genetic-factor score was composed that was used for extreme discordant and concordant selection for a QTL study of anxious depression (Netherlands Twin Family Study of Anxious Depression; see Boomsma et al. [2000] for a detailed description). Primary ascertainment of families was through these extremely discordant or concordant sib pairs, but all other siblings in the family were invited to participate. The distribution of anxious depression in the resulting sample (N = 2,724), therefore, was near normal with only mild kurtosis, in comparison with that of the entire unselected sample (see fig. 2 in Boomsma et al. 2000).
All subjects and their parents were asked to provide a buccal swab for DNA isolation. Of the 1,962 subjects (72%) who returned a buccal swab, 917 (624 offspring and 293 parents) were genotyped at 379 markers on the autosomes. Subjects for whom <50% of the markers were successfully typed were removed from the sample. This resulted in a subsample of 558 offspring and 278 parents from 192 families for whom genotyping was successful. From the offspring who were successfully genotyped, 306 subjects from 123 families participated in 24-h ambulatory monitoring. For a total of 270 complete sib pairs, ambulatory respiratory signals and adequate marker data were available. Sib pairs could be full DZ twin pairs, an MZ or a DZ twin paired with a singleton sibling, or two singleton siblings. From MZ pairs, data from only one (randomly chosen) twin was used for genotyping and the linkage analyses.
Subjects gave written informed consent, and both the DNA sampling and the ambulatory protocol were approved by the institutional review board of the Vrije Universiteit Medical Center.
Ambulatory Recording
Ambulatory recording of the thorax impedance (Z) was performed by the Vrije Universiteit Ambulatory Monitoring System (VU-AMS) with a six-spot electrode configuration (de Geus et al. 1995; de Geus and Van Doornen 1996). Two electrodes on the back were used to continuously send a high frequency current of 50 kHz 350 μA through the subject. Two electrodes on the chest were used to measure impedance. The upper measuring electrode was placed at the jugular notch of the sternum, between the collarbones. The lower measuring electrode was placed at the tip of the sternum (xiphoid process). The upper current electrode on the back was placed at least 3 cm above the horizontal plane of the upper measuring electrode. The lower current electrode on the back was placed at least 3 cm below the horizontal plane of the lower measuring electrode. The thoracic impedance signal was amplified and led to a precision rectifier. The rectified signal was filtered at 72 Hz (low pass) to give basal thoracic impedance, Z0. Filtering of Z0 at 0.1 Hz (high pass) supplies the dZ signal, which contains three major components: the high frequent impedance changes due to the ejection of blood into the aorta during systole; the low frequent impedance changes due to arm and upper-body movement; and, in between these frequencies, the thoracic impedance changes due to respiration. Fragments of 100-s dZ signal were band-pass filtered with 0.1 and 0.4 Hz cutoffs after being tapered with [sin(x)]2, yielding the respiration signal. VU-AMS software automatically detects the start of inspiration and expiration for each breath and displays the respiration signal with these markers for interactive visual inspection.
During the subject's awake time, the VU-AMS produces an audible alarm approximately every 30 min (±10 min randomized), to prompt the subject to fill out an activity diary. Subjects were instructed to write down the time and description of their activities and bodily postures during the past 30-min period, in chronological order. Information from the diary about physical activity and posture was combined with the body-movement signal from a built-in vertical accelerometer, to specify accurately the start and end times of any activity/posture changes of the subjects. We divided the entire recording into smaller fragments that were completely stationary with regard to physical activity and posture—for example, within each fragment, no shifts in posture occurred. Each fragment was coded for posture (lying, sitting, standing, walking, or bicycling), activity (e.g., deskwork, housekeeping, or watching TV), and location (e.g., at home, at work, or at a public place). The coded fragments were never <5 min or >1 h. On the basis of the reported times of awakening, lunch, dinner, and bedtime, data were aggregated across four periods: morning, afternoon, evening, and nighttime sleep. In 8% of the subjects, the exact time of awakening, lunch, dinner, or bedtime could not be extracted from either the diary or body movement. For these subjects, the missing time was imputed by use of the mean times of these events in the rest of the sample.
For the nighttime recording, the average respiration rate was aggregated across all valid breaths. For the three active periods of the day, the average respiration rates were computed using only the valid breaths obtained when subjects had been sitting. The use of sitting-only data eliminates the influence of individual differences in daytime physical-activity patterns, which are known to have a very strong effect on respiratory behavior. Total duration of sitting activities averaged 115 min in the morning, 152 min in the afternoon, and 137 min in the evening. In 34 subjects, one or more periods of the day were missing for the final analyses. This was because of equipment failure in a few cases, but it was mostly because the impedance signal quality was visually judged as insufficient for adequate respiration scoring. Consequently, the number of subjects varies slightly across the four periods (see table 1).
Table 1.
Ambulatory Respiration Rates in Male and Female Siblings during Three Daytime Periods and Nighttime Sleep[Note]
|
Respiration Rate (breaths/min) during |
|||||
| Subject Groupand Statistic | Age(years) | Morning | Afternoon | Evening | Night |
| Female: | |||||
| Mean | 32.14 | 16.40 | 16.79 | 17.36 | 15.98 |
| SD | 11.42 | 1.27 | 1.31 | 1.53 | 2.04 |
| n | 186 | 179 | 182 | 182 | 176 |
| Male: | |||||
| Mean | 32.09 | 16.11 | 16.66 | 17.37 | 15.10 |
| SD | 10.79 | 1.21 | 1.29 | 1.61 | 1.97 |
| n | 120 | 118 | 117 | 118 | 111 |
| All: | |||||
| Mean | 32.12 | 16.29 | 16.74 | 17.36 | 15.64 |
| SD | 11.16 | 1.25 | 1.30 | 1.56 | 2.06 |
| n | 306 | 297 | 299 | 300 | 287 |
Note.— To remove the effects of differences in physical-activity pattern during the awake periods, respiration rates were computed using data obtained only when subjects were sitting.
DNA Genotyping and Error Checking
Genotyping was conducted by the Marshfield Laboratory using the 10-cM spaced microsatellite screening set 10 (Yuan et al. 1997), with few alternative markers. Pedigrees were checked for Mendelian errors by use of the program Unknown (Schaffer 1996), and pedigree relationships in the entire sample were checked by use of the program GRR (Abecasis et al. 2001). Mendelian errors were removed by assigning missing values to the marker genotypes if the errors appeared incidental. Recombination likelihoods were checked using the Merlin program (Abecasis et al. 2002). Excessive recombinations were observed for 5 of the 379 autosomal markers that indicated potential problems: two markers on chromosome 1 (D1S160 and D1S1627-ATA25E0), two markers (D11S1985-GGAA5C04 and D11S2006-GATA46A12) in a group of five very closely or identically mapped markers on chromosome 11, and one marker on chromosome 20 (D20S159-UT1307). These five markers were excluded, leaving a final set of 374 markers for the analyses. For all other recombination problems, the data were cleaned using the default procedure of the Merlin program.
Marker distances in cM (Kosambi) were assigned by use of the deCODE map (Kong et al. 2002), when available. For markers not mapped by deCODE, the original distance provided on the Marshfield Web site (Broman et al. 1998) was transformed by linear interpolation from adjacent markers with known deCODE map values.
Linkage Analysis
Variation in respiration rate was decomposed into variation due to QTLs (σq2), variation due to additive influences (σa2), and variation due to nonshared environmental influences (σe2), by use of structural equation modeling as implemented in the Mx software package (see Mx software Web site). On the basis of genetic model fit in the full MZ and DZ twin samples, a model with additive genetic influences and unique environmental influences was known to best describe the twin covariance structure (H. M. Kupper, G. Willemsen, D. Posthuma, D. De Boer, D. I. Boomsma, E. J. C. de Geus, unpublished data). Age and sex were included as covariates in the model, to reduce residual variation in respiration rate and to increase power to detect linkage to a QTL. The formal model is represented by RR=μ+β1×sex+β2×age+ɛ, where RR is the observed respiration rate, μ is the intercept or grand mean, β1 is the deviation of males from females, β2 is the regression weight of age on respiration rate, and ɛ is the residual variance not explained by age or sex. The residual variance is divided into variance due to additive polygenetic variance; environmental variance, including measurement error; and variance associated with a putative QTL. Estimates of the variance component associated with a putative QTL were obtained by using the 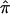 approach, in which the covariance due to the marker or trait locus for a sib pair is modeled as a function of the estimated proportion of alleles shared identical by descent (IBD). The general variance-covariance matrix for pair j,k of the ith family (Ωijk) is then given by
approach, in which the covariance due to the marker or trait locus for a sib pair is modeled as a function of the estimated proportion of alleles shared identical by descent (IBD). The general variance-covariance matrix for pair j,k of the ith family (Ωijk) is then given by
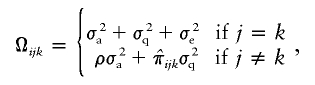 |
where ρ denotes the expected overall IBD proportion. The probabilities of sharing 0, 1, or 2 alleles IBD at every 1 cM (Haldane) over the genome were estimated by use of Merlin (Abecasis et al. 2002).
The effect of the QTLs was evaluated by the LOD score, computed as −2ln LR/4.6, where LR is the likelihood ratio. Descriptive significance levels for −2ln LR are obtainable from its asymptotic distribution, which is a 50:50 mixture of χ2 random variables with 1 and 0 df (Self and Liang 1987; Sham 1998, p. 265). For chromosomes with promising findings, however, empirical P values were determined by simulation, using the 1000× permutation algorithm as described by Lystig (2003).
In addition to the variance-components analyses discussed above, we reran the univariate linkage scans in Merlin Regress (Sham et al. 2002). Since virtually identical results were obtained from both procedures, only the Mx analyses will be presented here, for the sake of brevity. The Mx variance-components approach was chosen because it allowed additional multivariate and multilocus analyses.
Results
Table 1 shows the statistics of the genotyped sample of male and female offspring. Equating the means for males and females yielded a model with a significant loss of fit (χ24=20.76; P<.001). The main effect of sex shows a slightly slower respiration in males than in females. Respiration rate in all periods decreased with age, but the correlation was significant only for males in the morning sample (r=-0.20). Correlations between respiration rate and a summary factor score for anxious depression were nonsignificant (r values between −0.02 and 0.03). Heritabilities based on the sibling correlations were 29% in the morning, 37% in the afternoon, 43% in the evening, and 81% at night. Respiration rates from the four periods were significantly correlated (all values of r between 0.27 and 0.65).
Genomewide Linkage Analysis
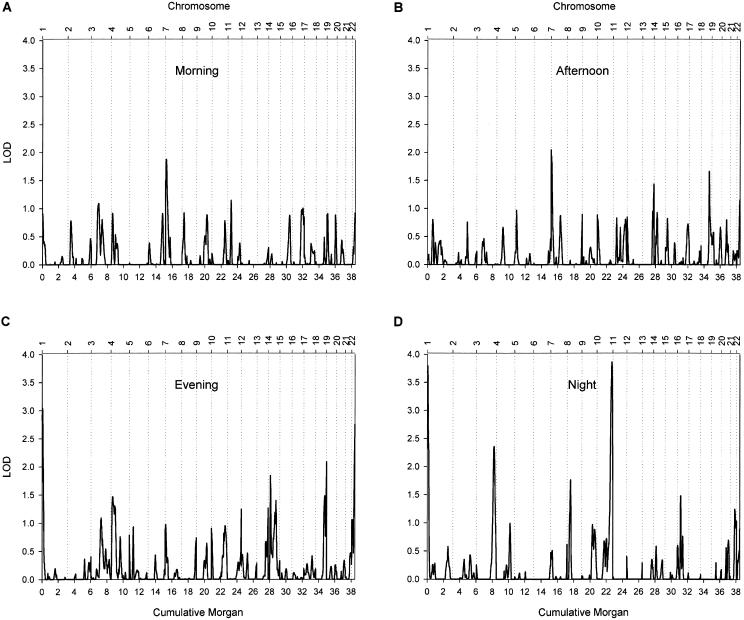
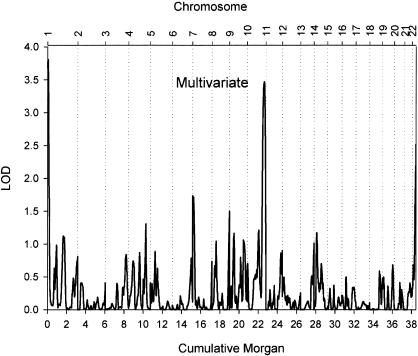
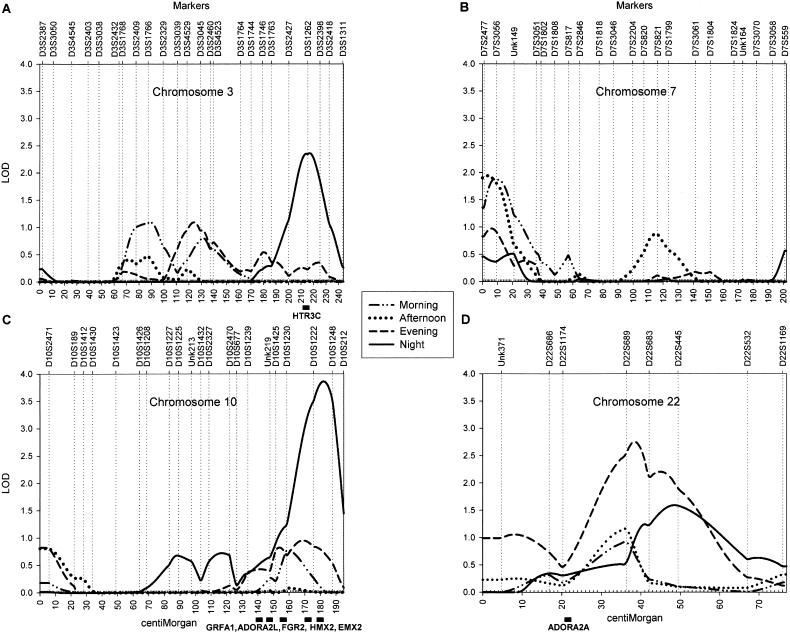
Figure 1 shows the results of the autosomal whole-genome scan. The highest peak (LOD score = 3.86) was found for respiration rate during sleep, on chromosome 10 at ∼182 cM from pter, between markers D10S1222 and D10S1248. A second locus had a suggestive influence on sleeping respiration rate (LOD score = 2.36); this locus was located on chromosome 3 at ∼217 cM from pter, between markers D3S1262 and D3S2398. Two additional regions were deemed of interest, because they yielded a LOD score >1.5 for respiration rate measured during at least two periods of the day. For morning and afternoon respiration rate, a suggestive peak was found on chromosome 7, near marker D7S3056. For evening and night respiration rate, suggestive peaks were found on chromosome 22; the peak for the evening respiration rate (LOD score = 2.75) was located at ∼39 cM from pter, between markers D22S689 and D22S683, and the peak for nighttime respiration rate (LOD score = 1.59) directly flanked it at ∼48 cM from pter, at marker D22S445. For the four regions with the strongest evidence of linkage, LOD scores for each of the periods are displayed in table 2. A multivariate analysis using all four periods confirmed the potential importance of these four loci (fig. 2), which are plotted in more detail in figure 3. The high LOD score on chromosome 1 did not have a flanking marker at the telomeric end and was considered to be an artifact.
Figure 1.
Univariate, multipoint, variance-components linkage of the 22 autosomes in 270 sib pairs. Linkage results are presented separately for the morning (A), afternoon (B), evening (C), and nighttime (D) and are adjusted for mean effects of sex and age on respiration rate. The X-axis plots genetic distance in cM (Haldane), and the Y-axis represents the LOD score.
Table 2.
Main Linkage Findings for Ambulatory Respiration Rate
| LOD Score (P)a for Period |
||||||
| Chromosome | Location(cM) | Morning | Afternoon | Evening | Night | Surrounding Markers |
| 3 | 217 | .00 | .00 | .26 | 2.36 (.080) | D3S1262, D3S2398 |
| 7 | 4 | 1.70 (.083) | 1.94 (.023) | .94 | .40 | D7S2477, D7S3056 |
| 10 | 182 | .04 | .00 | .74 | 3.86 (.004) | D10S1222, D10S1248 |
| 22 | 39 | .76 | .89 | 2.75 (.001) | .87 | D22S689, D22S683 |
For LOD scores >1.5, the empirical chromosome-wide P values are given (in parentheses) as obtained from 1,000 permutations of the data set.
Figure 2.
Multivariate, multipoint, variance-components linkage of the 22 autosomes in 270 sib pairs. Linkage results combine the four periods, each adjusted for mean effects of sex and age on respiration rate. The X-axis plots genetic distance in cM (Haldane), and the Y-axis represents the adjusted LOD score. The adjusted LOD score is determined as the 1-df LOD score required to give the same P value as the multivariate LOD score and can be compared directly with the univariate LOD scores.
Figure 3.
Best evidence of linkage to ambulatory respiration rate on chromosomes 3 (A), 7 (B), 10 (C), and 22 (D). Linkage results are presented for each of the four periods and are adjusted for mean effects of sex and age on respiration rate. The X-axis plots genetic distance in cM (Haldane), and the Y-axis represents the multipoint variance-components LOD score. Markers are arrayed in map order along the top of each plot.
Obstructive sleep apnea and obesity are highly comorbid, and the correlation between BMI and measures of sleep apnea has been shown to be attributable to genetic factors (Palmer et al. 2003). The correlations between respiration rate and BMI in our sample were all positive and significant (morning, 0.16; afternoon, 0.21; evening, 0.22; and night, 0.12; adjusted for age). The increased respiratory frequency with high BMI may reflect compensatory efforts against hypoventilation due to reduced chest-wall compliance or reduced nasopharyngeal caliber due to fat deposition in upper-airway tissues. To test this idea, we repeated all of the analyses described above with BMI as an additional covariate. Only neglible impact on the linkage signal was found, and the LOD scores for the four main loci (fig. 3) were essentially unchanged. Hence, linkage at these loci reflects direct genetic effects on respiratory regulation rather than indirect effects of genes influencing obesity.
Because respiration rate recorded during sleep showed the strongest linkage signals, the three loci with LOD scores >1.5 during sleep (on chromosomes 3 [217 cM], 10 [182 cM], and 22 [48 cM]) were modeled simultaneously as random effects in a three-loci linkage analysis. This simultaneous analysis is more robust to the well-known overestimation of variance attributable to markers in linkage analyses. In a single-locus analysis, the QTL effects may be overestimated and together sum to a percentage of explained genetic variance >100% (Göring et al. 2001). The three-locus analysis constrains the sum of the QTL variances to be not >100%. Table 3 shows a significant LOD score for all three loci simultaneously. For chromosome 22, the evidence of linkage disappeared in the multilocus analysis, but note that, for this marker, the largest linkage was found in the evening, not during sleep. For chromosomes 3 and 10, the evidence weakened somewhat in comparison with the single-locus analyses. In models that included the locus on either chromosome 3 or chromosome 10, however, the amount of genetic variance attributable to these loci was highly comparable to that found in the single-locus analyses. There is a slight discrepancy between the χ2 of a model leaving out three loci simultaneously and the summed χ2 of the models leaving them out one at a time, suggesting an interaction between loci.
Table 3.
Simultaneous Linkage Analysis of Three Loci with a Single-Locus LOD Score >1.5 for Respiration Rate during Sleep[Note]
|
% Contribution to Variance for |
|||||||||
| Model | Significance Test for | A | E | Chr 3 | Chr 10 | Chr 22 | Δdf | χ2 | LOD |
| A+E+Chr 3, 10, and 22 | … | 0 | 11 | 33 | 56 | 0 | … | … | … |
| A+E+Chr 10 and 22 | Chr 3 | 10 | 20 | … | 60 | 9 | 1 | 9.02 | 1.96 |
| A+E+Chr 3 and 22 | Chr 10 | 9 | 23 | 40 | … | 28 | 1 | 12.91 | 2.81 |
| A+E+Chr 3 and 10 | Chr 22 | 0 | 11 | 33 | 56 | … | 1 | 0 | 0 |
| A+E | Chr 3, 10, and 22 | 81 | 19 | … | … | … | 3 | 27.08 | 4.49a |
Note.— The percentage contribution to the variance in respiration rate is given for background genetic influences (A), environmental influences (E), and the three loci on chromosomes 3 (217 cM), 10 (182 cM), and 22 (48 cM). The significance of the contribution of each locus separately and all three loci together derives from the contrast of the fit of the model with these loci (model row 1) and the fit of models without one (model rows 2–4) or all (model row 5) of these loci. Chr = chromosome(s); Δdf = change in degrees of freedom.
Adjusted LOD score, determined as the 1-df LOD score required to give the same P value as the multilocus LOD score.
Discussion
Using ambulatory recording of the thorax impedance, we obtained 24-h respiration rates in healthy human subjects under natural conditions. Four genomic regions were identified as having a high likelihood of harboring loci that influence respiration rate. Linkage of a locus on chromosome 10q to nighttime respiration rate exceeded the Lander-Kruglyak threshold for significance (Lander and Kruglyak 1995). A second locus on chromosome 3q was also suggestively linked to nighttime respiration, as was a third locus on chromosome 22q. A simultaneous three-locus analysis confirmed the importance of the 3q and 10q loci for respiration rate during sleep. The generally higher LOD scores for nighttime respiration may reflect the increased power of the genome scan with an increase in the heritability of respiration rate. In a recent twin family study (H. M. Kupper, G. Willemsen, D. Posthuma, D. De Boer, D. I. Boomsma, E. J. C. de Geus, unpublished data), heritability of respiration rate was found to be moderate during the daytime (41%–50%) but to sharply increase at night (81%) through a decrease in environmental variance coupled to a strong increase in genetic variance. This shift in genetic architecture suggests that respiration rate is under more genetic control during sleep than during awake periods. Neurobiologically, this makes good sense. Transcription of a number of genes appears to be selectively increased during sleep (Mackiewicz and Pack 2003). In addition, many environmental factors (speech, chewing, postural changes, and physical activity) impact respiration during the daytime, whereas, during sleep, respiratory frequency will be a more pure reflection of intrinsic rhythmogenesis by the brain stem.
Human and Animal Studies on the Genetics of Respiration
Several lines of evidence support an influence for genetic factors on respiratory control in humans. Most have focused on respiration during sleep because of its clinical relevance for sleep disorders, most prominently for obstructive sleep apnea (Gaultier et al. 2003; Palmer and Redline 2003). Attempts to find genes for respiration, therefore, have focused largely on the apnea-hypopnea index, the primary measure of obstructive sleep apnea. So far, candidate-gene association studies for apneic breathing have had limited success (Redline and Tishler 2000; Kadotani et al. 2001). In an authoritative review, Palmer and Redline (2003, table 2) compiled a list of the most plausible candidate genes for obstructive sleep apnea. None of these appeared to lie in the vicinity of the regions of (suggestive) linkage we found. In addition, our regions did not overlap with two loci (at 2p and 19p) for obstructive sleep apnea that were identified in a whole-genome scan of 66 pedigrees of American European origin (Palmer et al. 2003).
Congenital central hypoventilation syndrome (CCHS [MIM 209880]) is a second sleep disorder that may provide clues to positional candidates in the regions of our linkage peaks (Gaultier et al. 2003, 2004). This syndrome is characterized by deficient autonomic control over respiration and is hypothesized to account for some cases of sudden infant death syndrome (Gozal 1998). CCHS is caused by mutations of genes in the ret and endothelin pathways (see table 1 in Gaultier et al. 2004). The importance of some of these genes for respiration frequency has been confirmed in mice with loss-of-function mutations. Homozygous and heterozygous knockouts of gdnf, mash-1, and bdnf all significantly affected resting respiratory frequency (see table 2 in Gaultier et al. 2004). The human gene corresponding to one of these genes, the glial cell line–derived neurotrophic factor (GDNF) family receptor alpha-1 gene (GRFA1 [MIM 601496]), is mapped at ∼5 Mb before the start of our region on chromosome 10. Gene expression profiling has suggested deviations in GRFA1 receptor regulation in Hirschsprung disease (MIM 142623), a condition that is comorbid with CCHS (Iwashita et al. 2003), and at least one patient with CCHS showed a mutation in GFRA1 (Sasaki et al. 2003). Its proximity to our peak LOD score at 10q suggests that polymorphisms in this gene may affect variation in nighttime respiration rate in healthy subjects.
Animal models of respiratory rhythmogenesis and regulatory input to the brain stem have provided a number of further candidate genes. Hox paralogs and hoxregulating genes (e.g., hoxa, kreisler/mafB, c-jun, and Krox20) are involved in the primordial rhombomeric organization of the hindbrain. Knockout mutations in these genes resulted in phenotypes of decreased or increased respiratory frequency, compared with that of the wild type (Jacquin et al. 1996; Shirasawa et al. 2000; Domínguez del Toro et al. 2001; Chatonnet et al. 2002). Mice engineered to lack the bZIP transcriptional regulator gene MafB, prominently expressed in the preBötzinger complex, were shown to completely lack development of critical rhythm-generating neurons in the brain stem (Blanchi et al. 2003). Although none of these “rodent respiratory genes” had syntenic human genes in any of our four regions of linkage, at least two other homeobox genes, HMX2 (MIM 600647) and EMX2 (MIM 600035), with as-yet unknown relation to respiration, were found under our best linkage peak on 10q.
Homeobox genes have also been prominently included in the list of possible candidate genes for obstructive sleep apnea (Palmer and Redline 2003), because they can have an effect on craniofacial form and upper-airway anatomy. The >100 craniofacial malformation (“craniosynostosis”) syndromes may well be regarded as the third “disorder” that can affect ventilation. Craniofacial development from skeletogenic differentiation of the cranial neural crest is governed almost entirely by the fibroblast growth factors (Wilkie and Morriss-Kay 2001). A major role in craniosynostosis syndromes is suggested for fibroblast growth factor receptor 2 gene (FGFR2 [MIM 176943]), which is directly under our best linkage peak at 10q26. Deviant respiratory control has frequently been reported in craniosynostosis syndromes, often severe enough to require surgical correction or prolonged continuous positive airway pressure therapy (Gonsalez et al. 1996; Perkins et al. 1997). Milder nonmorbid mutations in FGFR2 may well affect population variation in craniofacial build and basal respiration rate.
Positional Candidates
In a second strategy to confirm our linkage-peak results, we scanned the Ensembl human genome map (version 19.34b.2 of NCBI assembly 34 [July 2004 freeze]; see Ensemble Web site) for genes in these regions that could be plausibly linked to respiratory behavior. “Broad” peaks were used, spanning between the markers that defined the upstroke and incisura of the LOD score peaks (D3S2427–D3S2418, D7S2477–D7S3051, D10S1230–D10S212, and D22S1174–D22S532). By necessity, successful recognition of potentially relevant genes in the Ensembl-generated lists of genes in the linkage regions is limited by current understanding of the molecular biology of respiration. At least five positional candidate genes, however, could be plausibly connected to the regulation of nighttime respiratory frequency.
Adenosine and its analogues have been shown to increase respiratory ventilation in a dose-dependent manner (Monteiro and Ribeiro 1987) and to modulate the incidence of sleep apneas in rats (Monti et al. 1995). In a sheep model of fetal breathing, A1 receptors were found to tonically inhibit respiratory drive, A2A receptors to tonically inhibit REM sleep, and both A1 and A2A receptors to mediate the depressant effects of adenosine on REM sleep and breathing (Koos et al. 2001). Adenosine mediates its effects through four receptor subtypes: the A1, A2A, A2B, and A3 receptors (Fredholm et al. 1994). MacCollin et al. (1994) localized the ADORA2A gene (MIM 102776) to chromosome 22q11.23, just outside our region on 22q that showed linkage to respiration rate during the evening and sleep. At 10q25.3-q26.3 (ADORA2L [MIM 102777]), another gene for an A2 adenosine receptor subtype is suspected, the function of which is currently unknown. Given the convergence of the loci at chromosomes 10 and 22 in a single biological pathway and the importance of adenosine in respiration, we suggest that ADORA2A and ADORA2L may be promising positional candidate genes for respiration rate.
Serotonin and serotonergic drugs have significant effects on respiration, and serotonin has been implicated in the pathogenesis of sleep disorders (Richerson 2004). Serotonergic neurons in the Raphé nucleus act as chemoreceptors and enhance respiratory rhythm generation in the preBötzinger complex. These effects have been shown to be mediated through serotonin receptors type 1A, 2A , 4A, and 7 (Richter et al. 2003; Richerson 2004). However, a role for serotonin type 3 (5-HT3) receptors can also be assumed. Serotonin antagonists selective for the 5-HT3 receptor suppress sleep-related central apneas in rats (Radulovacki et al. 1998) and obstructive sleep apnea in the English bulldog (Veasey et al. 2001). A recently identified cluster of three novel serotonin type 3 receptor genes (HTR3C, HTR3D, and HTR3E) is localized at 3q27 (Karnovsky et al. 2003), directly in our area of linkage. Comparative expression analysis suggested that HTR3D and HTR3E expression were limited to colon, kidney, liver, and intestine, whereas the HTR3C gene is widely expressed in many tissues, including the brain (Niesler et al. 2003). These findings lead us to suggest that HTR3C, a gene homologous to 5-HT3A and 5-HT3B receptors, qualifies as a potential candidate gene for respiration rate.
In summary, evidence of linkage was found for respiration rate during sleep at 3q27, 7p22, 10q26, and 22q12. Strongest evidence of linkage was found between markers D10S1222 and D10S1248 on chromosome 10. From the Ensembl database, we identified GFRA1, ADORA2L, FGR2, EMX2, and HMX2 as biologically plausible candidate genes harbored by this linkage region. Further candidates suggested by our linkage findings are the ADORA2A adenosine receptor gene on 22q and the HTRC3 serotonin receptor gene at 3q. Identification of the genetic variation influencing human respiratory phenotypes would serve an important clinical purpose. It could increase our understanding of the molecular and cellular bases of disorders of rhythmogenesis such as sleep apnea, unexplained stillbirth, and certain cases of sudden infant death syndrome.
Acknowledgments
This work was supported by grants 575-25-006 and 904-61-090 from the Netherlands Organization for Scientific Research (NWO) and by the use of their supercomputer facilities (NCF 2004/00931). Genotyping was performed by the Marshfield Center for Medical Genetics. D.P. was supported by the GenomeEUtwin project (European Union contract QLG2-CT-2002-01254). We would like to thank E. Suchiman and N. Lakenberg, for DNA isolation.
Electronic-Database Information
The URLs for data presented herein are as follows:
- Ensembl, http://www.ensembl.org/ (for human genome map version 19.34b.2 of NCBI assembly 34 [July 2004 freeze])
- Marshfield Center for Medical Genetics, http://www.marshfieldclinic.org/research/genetics/
- Mx software, http://www.vcu.edu/mx
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for CCHS, GRFA1, Hirschsprung disease, HMX2, EMX2, FGFR2, ADORA2A, and ADORA2L)
References
- Abecasis GR, Cherny SS, Cookson WO, Cardon LR (2001) GRR: graphical representation of relationship errors. Bioinformatics 17:742–743 10.1093/bioinformatics/17.8.742 [DOI] [PubMed] [Google Scholar]
- ——— (2002) Merlin—rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet 30:97–101 10.1038/ng786 [DOI] [PubMed] [Google Scholar]
- American Academy of Sleep Medicine Task Force (1999) Sleep-related breathing disorders in adults: recommendations for syndrome definition and measurement techniques in clinical research. Sleep 22:667–689 [PubMed] [Google Scholar]
- Blanchi B, Kelly LM, Viemari JC, Lafon I, Burnet H, Bévengut M, Tillmanns S, Daniel L, Graf T, Hilaire G, Sieweke MH (2003) MafB deficiency causes defective respiratory rhythmogenesis and fatal central apnea at birth. Nat Neurosci 6:1091–1100 10.1038/nn1129 [DOI] [PubMed] [Google Scholar]
- Boomsma DI, Beem AL, van den Berg M, Dolan CV, Koopmans JR, Vink JM, de Geus EJ, Slagboom PE (2000) Netherlands twin family study of anxious depression (NETSAD). Twin Res 3:323–334 10.1375/136905200320565300 [DOI] [PubMed] [Google Scholar]
- Boomsma DI, Vink JM, Van Beijsterveldt TC, de Geus EJ, Beem AL, Mulder EJ, Derks EM, Riese H, Willemsen GA, Bartels M, van den Berg M, Kupper NH, Polderman TJ, Posthuma D, Rietveld MJ, Stubbe JH, Knol LI, Stroet T, van Baal GC (2002) Netherlands Twin Register: a focus on longitudinal research. Twin Res 5:401–406 10.1375/136905202320906174 [DOI] [PubMed] [Google Scholar]
- Broman KW, Murray JC, Sheffield VC, White RL, Weber JL (1998) Comprehensive human genetic maps: individual and sex-specific variation in recombination. Am J Hum Genet 63:861–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatonnet F, Thoby-Brisson M, Abadie V, Domínguez del Toro E, Champagnat J, Fortin G (2002) Early development of respiratory rhythm generation in mouse and chick. Respir Physiol Neurobiol 131:5–13 10.1016/S1569-9048(02)00033-2 [DOI] [PubMed] [Google Scholar]
- de Geus EJC, Van Doornen LJP (1996) Ambulatory assessment of parasympathetic/sympathetic balance by impedance cardiography. In: Fahrenberg J, Myrtek M (eds) Computer-assisted psychological and psychophysiological methods in monitoring and field studies. Hogrefe and Huber Publishers, Seattle, pp 141–163 [Google Scholar]
- de Geus EJC, Willemsen AHM, Klaver CH, Van Doornen LJ (1995) Ambulatory measurement of respiratory sinus arrhythmia and respiration rate. Biol Psychol 41:205–227 10.1016/0301-0511(95)05137-6 [DOI] [PubMed] [Google Scholar]
- Domínguez del Toro E, Borday V, Davenne M, Neun R, Rijli FM, Champagnat J (2001) Generation of a novel functional neuronal circuit in Hoxa1 mutant mice. J Neurosci 21:5637–5642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman JL, Mitchell GS, Nattie EE (2003) Breathing: rhythmicity, plasticity, chemosensitivity. Annu Rev Neurosci 26:239–266 10.1146/annurev.neuro.26.041002.131103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredholm BB, Abbracchio MP, Burnstock G, Daly JW, Harden TK, Jacobson KA, Leff P, Williams M (1994) Nomenclature and classification of purinoceptors. Pharmacol Rev 46:143–156 [PMC free article] [PubMed] [Google Scholar]
- Gaultier C, Amiel J, Dauger S, Trang H, Lyonnet S, Gallego J, Simonneau M (2004) Genetics and early disturbances of breathing control. Pediatr Res 55:729–733 10.1203/01.PDR.0000115677.78759.C5 [DOI] [PubMed] [Google Scholar]
- Gaultier C, Dauger S, Simonneau M, Gallego J (2003) Genes modulating chemical breathing control: lessons from mutant animals. Respir Physiol Neurobiol 136:105–114 10.1016/S1569-9048(03)00075-2 [DOI] [PubMed] [Google Scholar]
- Gonsalez S, Thompson D, Hayward R, Lane R (1996) Treatment of obstructive sleep apnoea using nasal CPAP in children with craniofacial dysostoses. Childs Nerv Syst 12:713–719 10.1007/BF00366156 [DOI] [PubMed] [Google Scholar]
- Göring HH, Terwilliger JD, Blangero J (2001) Large upward bias in estimation of locus-specific effects from genomewide scans. Am J Hum Genet 69:1357–1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gozal D (1998) Congenital central hypoventilation syndrome: an update. Pediatr Pulmonol 26:273–282 [DOI] [PubMed] [Google Scholar]
- Hanly PJ (1992) Mechanisms and management of central sleep apnea. Lung 170:1–17 10.1007/BF00164751 [DOI] [PubMed] [Google Scholar]
- Hilaire G, Pásaro R (2003) Genesis and control of the respiratory rhythm in adult mammals. News Physiol Sci 18:23–28 [DOI] [PubMed] [Google Scholar]
- Iwashita T, Kruger GM, Pardal R, Kiel MJ, Morrison SJ (2003) Hirschsprung disease is linked to defects in neural crest stem cell function. Science 301:972–976 10.1126/science.1085649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquin TD, Borday V, Schneider-Maunoury S, Topilko P, Ghilini G, Kato F, Charnay P, Champagnat J (1996) Reorganization of pontine rhythmogenic neuronal networks in Krox-20 knockout mice. Neuron 17:747–758 10.1016/S0896-6273(00)80206-8 [DOI] [PubMed] [Google Scholar]
- Kadotani H, Kadotani T, Young T, Peppard PE, Finn L, Colrain IM, Murphy GM Jr, Mignot E (2001) Association between apolipoprotein E ε4 and sleep-disordered breathing in adults. JAMA 285:2888–2890 10.1001/jama.285.22.2888 [DOI] [PubMed] [Google Scholar]
- Karnovsky AM, Gotow LF, McKinley DD, Piechan JL, Ruble CL, Mills CJ, Schellin KA, Slightom JL, Fitzgerald LR, Benjamin CW, Roberds SL (2003) A cluster of novel serotonin receptor 3-like genes on human chromosome 3. Gene 319:137–148 10.1016/S0378-1119(03)00803-5 [DOI] [PubMed] [Google Scholar]
- Kong A, Gudbjartsson DF, Sainz J, Jonsdottir GM, Gudjonsson SA, Richardsson B, Sigurdardottir S, Barnard J, Hallbeck B, Masson G, Shlien A, Palsson ST, Frigge ML, Thorgeirsson TE, Gulcher JR, Stefansson K (2002) A high-resolution recombination map of the human genome. Nat Genet 31:241–247 [DOI] [PubMed] [Google Scholar]
- Koos BJ, Maeda T, Jan C (2001) Adenosine A1 and A2A receptors modulate sleep state and breathing in fetal sheep. J Appl Physiol 91:343–350 [DOI] [PubMed] [Google Scholar]
- Lander E, Kruglyak L (1995) Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet 11:241–247 10.1038/ng1195-241 [DOI] [PubMed] [Google Scholar]
- Lystig TC (2003) Adjusted P values from genome-wide scans. Genetics 164:1683–1687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacCollin M, Peterfreund R, MacDonald M, Fink JS, Gusella J (1994) Mapping of a human A2a adenosine receptor (ADORA2) to chromosome 22. Genomics 20:332–333 10.1006/geno.1994.1181 [DOI] [PubMed] [Google Scholar]
- Mackiewicz M, Pack AI (2003) Functional genomics of sleep. Respir Physiol Neurobiol 135:207–220 10.1016/S1569-9048(03)00045-4 [DOI] [PubMed] [Google Scholar]
- Monteiro EC, Ribeiro JA (1987) Ventilatory effects of adenosine mediated by carotid body chemoreceptors in the rat. Naunyn Schmiedebergs Arch Pharmacol 335:143–148 [DOI] [PubMed] [Google Scholar]
- Monti D, Carley DW, Radulovacki M (1995) Adenosine analogues modulate the incidence of sleep apneas in rats. Pharmacol Biochem Behav 51:125–131 10.1016/0091-3057(94)00395-Y [DOI] [PubMed] [Google Scholar]
- Niesler B, Frank B, Kapeller J, Rappold GA (2003) Cloning, physical mapping and expression analysis of the human 5-HT3 serotonin receptor-like genes HTR3C, HTR3D and HTR3E. Gene 310:101–111 10.1016/S0378-1119(03)00503-1 [DOI] [PubMed] [Google Scholar]
- Palmer LJ, Buxbaum SG, Larkin E, Patel SR, Elston RC, Tishler PV, Redline S (2003) A whole-genome scan for obstructive sleep apnea and obesity. Am J Hum Genet 72:340–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer LJ, Redline S (2003) Genomic approaches to understanding obstructive sleep apnea. Respir Physiol Neurobiol 135:187–205 10.1016/S1569-9048(03)00044-2 [DOI] [PubMed] [Google Scholar]
- Perkins JA, Sie KC, Milczuk H, Richardson MA (1997) Airway management in children with craniofacial anomalies. Cleft Palate Craniofac J 34:135–140 [DOI] [PubMed] [Google Scholar]
- Radulovacki M, Trbovic SM, Carley DW (1998) Serotonin 5-HT3-receptor antagonist GR 38032F suppresses sleep apneas in rats. Sleep 21:131–136 [DOI] [PubMed] [Google Scholar]
- Redline S, Tishler PV (2000) The genetics of sleep apnea. Sleep Med Rev 4:583–602 10.1053/smrv.2000.0120 [DOI] [PubMed] [Google Scholar]
- Rekling JC, Feldman JL (1998) PreBötzinger complex and pacemaker neurons: hypothesized site and kernel for respiratory rhythm generation. Annu Rev Physiol 60:385–405 10.1146/annurev.physiol.60.1.385 [DOI] [PubMed] [Google Scholar]
- Richerson GB (2004) Serotonergic neurons as carbon dioxide sensors that maintain pH homeostasis. Nat Rev Neurosci 5:449–461 10.1038/nrn1409 [DOI] [PubMed] [Google Scholar]
- Richter DW, Manzke T, Wilken B, Ponimaskin E (2003) Serotonin receptors: guardians of stable breathing. Trends Mol Med 9:542–548 10.1016/j.molmed.2003.10.010 [DOI] [PubMed] [Google Scholar]
- Richter DW, Spyer KM (2001) Studying rhythmogenesis of breathing: comparison of in vivo and in vitro models. Trends Neurosci 24:464–472 10.1016/S0166-2236(00)01867-1 [DOI] [PubMed] [Google Scholar]
- Sasaki A, Kanai M, Kijima K, Akaba K, Hashimoto M, Hasegawa H, Otaki S, Koizumi T, Kusuda S, Ogawa Y, Tuchiya K, Yamamoto W, Nakamura T, Hayasaka K (2003) Molecular analysis of congenital central hypoventilation syndrome. Hum Genet 114:22–26 10.1007/s00439-003-1036-z [DOI] [PubMed] [Google Scholar]
- Schaffer AA (1996) Faster linkage analysis computations for pedigrees with loops or unused alleles. Hum Hered 46:226–235 [DOI] [PubMed] [Google Scholar]
- Self SG, Liang K-Y (1987) Asymptotic properties of maximum likelihood estimators and likelihood ratio tests under nonstandard conditions. J Am Stat Assoc 82:605–610 [Google Scholar]
- Sham P (1998) Statistics in human genetics. Arnold Publishers, London [Google Scholar]
- Sham PC, Purcell S, Cherny SS, Abecasis GR (2002) Powerful regression-based quantitative-trait linkage analysis of general pedigrees. Am J Hum Genet 71:238–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirasawa S, Arata A, Onimaru H, Roth KA, Brown GA, Horning S, Arata S, Okumura K, Sasazuki T, Korsmeyer SJ (2000) Rnx deficiency results in congenital central hypoventilation. Nat Genet 24:287–290 10.1038/73516 [DOI] [PubMed] [Google Scholar]
- Veasey SC, Chachkes J, Fenik P, Hendricks JC (2001) The effects of ondansetron on sleep-disordered breathing in the English bulldog. Sleep 24:155–160 [DOI] [PubMed] [Google Scholar]
- Wilkie AO, Morriss-Kay GM (2001) Genetics of craniofacial development and malformation. Nat Rev Genet 2:458–468 10.1038/35076601 [DOI] [PubMed] [Google Scholar]
- Wolk R, Kara T, Somers VK (2003) Sleep-disordered breathing and cardiovascular disease. Circulation 108:9–12 10.1161/01.CIR.0000072346.56728.E4 [DOI] [PubMed] [Google Scholar]
- Yuan B, Vaske D, Weber JL, Beck J, Sheffield VC (1997) Improved set of short-tandem-repeat polymorphisms for screening the human genome. Am J Hum Genet 60:459–460 [PMC free article] [PubMed] [Google Scholar]