Abstract
Numerous putative susceptibility loci have been described for psoriasis. Among the loci confirmed in the literature, PSORS1 (the major histocompatibility complex at 6p21.3) has the strongest effect. Recent studies have highlighted a 200-kb candidate region. However, this region has not been well delimited, mainly because of the strong linkage equilibrium among the associated alleles. To finely map PSORS1, we set up a study using 17 polymorphic markers in a 525-kb interval around the human leucocyte antigen C locus (HLA-C). The results uncovered five loci with alleles strongly associated with psoriasis (Sidak-corrected P [Pc] values from 1.8 × 10−7 to .003), all structured in a psoriasis-susceptibility haplotype (PSH). Subsequent analysis of extended haplotypes showed that the PSH was not only present on the traditional psoriasis-susceptibility extended haplotypes (HLA-Cw6-B57, HLA-Cw6-B37, and HLA-Cw6-B13) but also on a haplotype of Sardinian origin (HLA-Cw7-B58) found to be associated with psoriasis (Pc=.0009) because of an ancestral recombination with one of the susceptibility haplotypes carrying the HLA-Cw6 allele. Comparisons of the regions identical by descent among associated and nonassociated haplotypes highlighted a minimum region of 70 kb not recombinant with PSORS1, around the corneodesmosin (CDSN) gene.
Psoriasis (MIM 177900) is a chronic inflammatory skin disease with a prevalence of 2% in white populations. Skin lesions are characterized by angiogenesis, infiltrates of activated T cells, hyperproliferation of keratinocytes, and altered epidermal differentiation (Barker 1991). Clear evidence for a strong genetic component in susceptibility to psoriasis arises from strong familial clustering and the high concordance rate in MZ twins (Brandrup et al. 1982). Throughout the years, the search for genetic susceptibility factors has been hindered by the multifactorial nature of the disease. Early studies on different ethnic groups have repeatedly highlighted the strong association of the HLA-Cw6 allele with psoriasis (Tiilikainen et al. 1980; Roitberg-Tambur et al. 1994; Gonzaga et al. 1996). In more recent years, genomewide scans have made it possible to individuate new susceptibility loci and to provide documented evidence of the highly significant linkage for the 6p21.3 region, thus confirming the presence of a major psoriasis-susceptibility gene within the major histocompatibility complex (MHC) (Nair et al. 1997; Trembath et al. 1997; Jenisch et al. 1998). These findings have led to the construction of maps densely filled with genetic markers flanking the human leucocyte antigen C locus (HLA-C [MIM 142840]), all focused on the refinement of the localization of PSORS1 by analysis of linkage disequilibrium (Oka et al. 1999; Nair et al. 2000; Veal et al. 2002).
Although these studies have different conclusions, they highlight a region of 200 kb in which alleles of the HLA-C gene, the alpha-helix-coiled-coil-rod homolog gene (HCR [MIM 602593]), and the corneodesmosin gene (CDSN [MIM 602593]) have been found to be associated with the disease. Overall, these studies confirm the existence, in white populations, of a strong linkage disequilibrium region, between the HLA-C and CDSN genes, in which it is difficult to distinguish between the susceptibility alleles and the markers in linkage disequilibrium with these alleles. In a recent study of the Sardinian population performed by our group, psoriasis was found to be associated with an allele of the CDSN gene encoding the corneodesmosin protein, without any relationship to the HLA-C locus. Hypothetically, this decline in linkage disequilibrium may be attributable to ancestral recombination events in this population (Orrù et al. 2002). In an attempt to contribute to the fine mapping of PSORS1, we performed a case-control association study combined with a search for identical-by-descent (IBD) haplotypes by the use of 17 genetic markers spaced over a region of 525 kb.
For this study, we recruited 161 unrelated Sardinian patients with psoriasis vulgaris (90 females and 71 males; age at onset 30.4 ± 18.8 years) and 184 individuals belonging to 32 families with multiple cases of psoriasis. Among the families, 68 members were affected. A total of 160 unrelated individuals (84 females and 76 males; mean age 33.0 ± 7.7 years) were used as controls. Diagnosis of psoriasis vulgaris was based on clinical and histological findings. Family trees were used to document the Sardinian background in both patients and controls. After informed consent was obtained, genomic DNA of each individual was isolated from peripheral blood, in accordance with standard methods. This study was approved by the institutional review board and by the local public-health ethics committee.
Genotyping was performed on all individuals by PCR using microsatellite markers MIB (Grimaldi et al. 1996); C1_4_1, C1_2_5, C1_4_3, C_1_4_4, C1_2_6, C1_3_2, C2_4_4, C2_4_5, C4_2_7, and C4_4_9 (Oka et al. 1999); and M6S187 and M6S172 (Nair et al. 2000). Each PCR reaction mix was loaded onto an automated capillary sequencer (MegaBACE 1000 [Amersham Biosciences]) for electrophoretic separation and registration of electropherograms. The alleles were defined by comparisons with molecular markers, by use of the Genetic Profiler 1.1 software package (Amersham Biosciences) and by direct inspection of the electropherograms. The genotypes of SNP 9—located 4 kb centromeric to the HLA-C (Veal et al. 2002)—and a polymorphism of the OTF3 gene (MIM 164177) (Gonzalez et al. 2000) were determined by fluorescent primer extension, with the use of the MegaBACE SnuPe Genotyping Kit (Amersham Biosciences). The genotypes of the HLA-B, HLA-C, and CDSN loci were obtained as described by Orrù et al. (2002).
The differences in frequencies between patients and controls were evaluated by the two-tailed χ2 test with 1 df. The odds ratio (OR) was calculated by 2 × 2 contingency tables. Departures from Hardy-Weinberg equilibrium and the haplotype frequencies were calculated separately in patients and controls, by use of a Markov-chain simulation method and the expectation-maximization (EM) algorithm, respectively, as implemented in ARLEQUIN software (Schneider et al. 2000). To construct the complete haplotypes (17 loci), a maximum of 5–6 loci at a time were analyzed. Moving from the centromeric to the telomeric portion of the haplotype, the analysis was repeated, with the inclusion of one or two new loci each time, until the full haplotypes were generated. To estimate haplotype frequency, a maximum of nine loci at a time, including HLA-C and CDSN, were assessed. Only haplotypes confirmed in families and with a frequency >0.01 in at least one of the two groups (patients and controls) were considered for subsequent analysis. Less-frequent haplotypes with a variant attributable to the addition or loss of a single repeat, were added to the corresponding more-frequent haplotype. To estimate association between alleles of two different loci, standardized disequilibrium values were calculated: D′=D/Dmax=Pij-PiPj/Pi(1-Pj), where Pij corresponds to the observed frequency of the ij haplotype and Pi and Pj correspond to the frequencies of the i and j alleles, respectively. Association between haplotypes and psoriasis was also evaluated by the transmission-disequilibrium test (TDT) in 32 families with multiple cases of psoriasis, who were genotyped for all the markers included in the present study. In this study, P values were corrected using the Sidak test for multiple comparisons: Pc=1-(1-P)k, where k is the number of comparisons. Only Pc values <.01 were considered significant.
Initially, a single-marker case-control association study was performed on 161 cases of psoriasis and 160 controls by the use of 17 genetic markers in an MHC class I region of ∼525 kb. Statistical comparisons were made between the genotype frequencies of cases and controls. Those with the strongest deviation from the null hypothesis of no association are shown in table 1. After correction for multiple comparisons, five loci (M6S172, C1_3_2, CDSN, C2_4_4, and C2_4_5) showed highly significant values for positive association with psoriasis (Pc<.01; OR>2.5). Extremely high significant values were found for the allele CDSN*TTC (Pc=1.8×10-7), followed by those for the microsatellites C2_4_5 (Pc=.00038) and C1_3_2 (Pc=.0009); all were located in a region of 70 kb. Four loci centromeric to this region (C1_2_6, M6S172, C1_4_4, and C1_4_5) shared alleles negatively associated with psoriasis (Pc<.01; OR<0.39). The most centromeric and telomeric markers used in our study had no associated alleles or only revealed a feeble association with psoriasis. At the HLA-C locus, the allele HLA-Cw6 was not associated with psoriasis (Pc=.233). No significant deviation from the Hardy-Weinberg equilibrium was observed in the two groups (P>.05).
Table 1.
Single-Marker Association Study
|
Frequency in |
P Valuec |
||||
| Locus (Locationa)and Alleleb | Cases | Controls | P | Pc | OR |
| MIB (0): | |||||
| 1 | .273 | .413 | .01207 | .23453 | .54 |
| C1_4_1 (18): | |||||
| 2 | .522 | .675 | .00722 | .14737 | .53 |
| C1_2_5 (72): | |||||
| 12 | .286 | .144 | .00308 | .06554 | 2.38 |
| SNP 9 (15): | |||||
| 2 | .379 | .250 | .01798 | .32909 | 1.83 |
| HLA-C (4): | |||||
| 5 | .168 | .338 | .00074 | .01620 | .40 |
| 7 | .528 | .350 | .00194 | .04178 | 2.08 |
| C1_4_3 (29): | |||||
| 4 | .130 | .325 | 5.70×10−5 | .00125 | .31 |
| 8 | .422 | .269 | .00551 | .11438 | 1.99 |
| C1_4_4 (5): | |||||
| 2 | .447 | .281 | .00295 | .06292 | 2.07 |
| 7 | .161 | .356 | .00011 | .00252 | .35 |
| M6S172 (16): | |||||
| 1 | .714 | .481 | 3.41×10−5 | .00075 | 2.69 |
| 4 | .311 | .538 | 6.30×10−5 | .00139 | .39 |
| C1_2_6 (55): | |||||
| 2 | .447 | .694 | 1.37×10−5 | .00030 | .36 |
| OTF-3 (8): | |||||
| 2 | .752 | .606 | .00761 | .15473 | 1.96 |
| C1_3_2 (46): | |||||
| 1 | .596 | .363 | .00005 | .00099 | 2.60 |
| CDSN (7): | |||||
| TTC | .634 | .306 | 8.30×10−9 | 1.83×10−7 | 3.92 |
| TGT | .385 | .563 | .00213 | .04587 | .49 |
| C2_4_4 (50): | |||||
| 2 | .745 | .538 | .00017 | .00363 | 2.52 |
| C2_4_5 (14): | |||||
| 4 | .646 | .400 | 1.71×10−5 | .00038 | 2.74 |
| M6S187 (73): | |||||
| 6 | .230 | .394 | .00228 | .04907 | .46 |
| C4_2_7 (95): | |||||
| 13 | .118 | .206 | .04617 | .64649 | .51 |
| C4_4_9 (19): | |||||
| 5 | .683 | .725 | .48551 | 1.00000 | .82 |
Distance (in kb) from the preceding locus.
Allele with the largest deviation from the null hypothesis of no association.
P is determined by the χ2 two-tailed test; Pc is the P value corrected by the Sidak test for multiple comparisons. All statistically significant values are indicated in bold italics.
To understand the relationship between the associated loci, we performed a haplotype study, using the maximum-likelihood method calculated by the EM algorithm and the reconstruction of haplotype segregation within families followed by the TDT. Table 2 shows the haplotypes obtained with psoriasis-associated loci plus HLA-C and shows their distribution in the two groups—patients and controls. The single alleles found to be associated with psoriasis were structured on three different haplotypes, which, in turn, showed significant association with the disease, both in the case-control association study and in the TDT. However, whereas the negatively associated alleles were found on a single haplotype that exclusively carried the HLA-Cw5 alleles (haplotype D), the positively associated alleles were present on two different haplotypes (haplotypes E and F) that carried the HLA-Cw6 and HLA-Cw7 alleles, respectively, and different alleles at the C1_4_4 and C1_2_6 loci. The common portion of these haplotypes (M6S172*1–C1_3_2*1–CDSN*TTC–C2_4_4*2–C_2_4_5*4) was present in 103 (32%) of the psoriatic chromosomes, compared with 42 (13%) of the control chromosomes (Pc=2.5×10-7; OR=3.1), and was overtransmitted within families (transmitted = 26; not transmitted = 5; P=.0002). This haplotype, which we defined as the “psoriasis-susceptibility haplotype” (PSH), was almost exclusively carried by the E and F haplotypes—since its presence in other haplotypes was rare (four in patients and three in controls)—and, even so, was not correlated to a specific haplotype. As is summarized in table 3, the increased frequency of PSH observed in patients did not depend on a decreased frequency of the D haplotype (HLA-Cw5 and C1_4_4*7 genotypes excluded) or an increased frequency of the E (HLA-Cw6) and F (HLA-Cw7) haplotypes.
Table 2.
Haplotype Association Study in the Sardinian Population[Note]
|
Allele at Locus |
Cases |
Controls |
P Valueb |
TDTc |
|||||||||||||||
| Haplotype | HLA-C | C1_4_3 | C1_4_4 | M6S172 | C1_2_6 | C1_3_2 | CDSN | C2_4_4 | C2_4_5 | n | Frequencya | n | Frequencya | P | Pc | OR | T | NT | P |
| A | 12 | 2 | 9 | 3 | 7 | 8 | TGT | 9 | 10 | 14 | .043 | 11 | .034 | .69496 | .99993 | 1.28 | 8 | 5 | .40540 |
| B | 12 | 6 | 1 | 5 | – | – | TTT | 5 | 12 | 4 | .012 | 1 | .003 | .37296 | .97610 | 4.01 | 1 | 0 | – |
| C | 4 | 7 | 1 | 5 | 2 | 7 | TTT | 8 | 3 | 26 | .081 | 27 | .084 | .98111 | 1.00000 | .95 | 15 | 7 | .00881 |
| D | 5 | 4 |
7 |
4 |
2 |
9 | TGT | 11 | 10 | 14 | .043 | 40 | .125 | .00035 | .00276 | .32 | 2 | 19 | .00020 |
| E | 6 | 8 | 2 | 1 |
8 | 1 |
TTC |
2 |
4 |
44 | .137 | 17 | .053 | .00051 | .00410 | 2.82 | 12 | 2 | .00750 |
| F | 7 | 9 | 11 | 1 |
5 | 1 |
TTC |
2 |
4 |
55 | .171 | 22 | .069 | .00011 | .00091 | 2.79 | 14 | 3 | .00760 |
| G | 7 | 3 | 8 | 4 | 2 | 7 | CTC | 2 | 2 | 13 | .040 | 8 | .025 | .38266 | .97891 | 1.64 | 4 | 9 | .16550 |
| H | 8 | 8 | 2 | 1 | 9 | 8 | CTC | 10 | 11 | 21 | .065 | 20 | .063 | .98355 | 1.00000 | 1.05 | 9 | 10 | .81850 |
Note.— The single alleles found to be associated with psoriasis are underlined. The alleles not always present on all haplotypes are shown in bold italics. “–” indicates ⩾3 variable alleles.
Estimated by the EM method.
P is determined by the χ2 two-tailed test; Pc is the P value corrected by the Sidak test for multiple comparisons. All statistically significant values are indicated in bold italics.
T = transmitted; N = not transmitted.
Table 3.
Distribution of PSH Haplotypes in Selected Patients with Psoriasis and Controls
|
Distribution ofHaplotype in |
P Valuea |
||||
| Excluded Genotype | Cases | Controls | P | Pc | OR |
| HLA-Cw5 | 87/268 | 31/212 | 1.08×10-5 | .00012 | 2.81 |
| C1_4_4*7 | 86/270 | 24/206 | 3.96×10-7 | 4.76×10-6 | 3.50 |
| HLA-Cw6 | 49/222 | 19/260 | 6.47×10-6 | 7.77×10-5 | 3.59 |
| HLA-Cw7 | 36/152 | 12/208 | 1.73×10-6 | 2.08×10-5 | 5.07 |
P is determined by the χ2 two-tailed test; Pc is the P value corrected by the Sidak test for multiple comparisons.
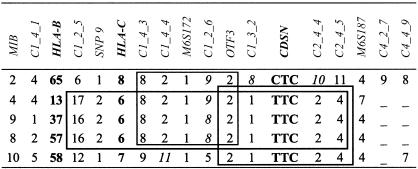
Finally, the construction of the haplotypes, by use of all loci analyzed in the present study (fig. 1), made it possible to search for alleles shared by different haplotypes and to further clarify the nature of the association of PSH with psoriasis. As concerns the two haplotypes with PSH, the E haplotype was found to be the sum of three extended haplotypes (EHs), Cw6-B13 (EH13.1), Cw6-B37 (EH37.1), and Cw6-B57 (EH57.1), whereas the F haplotype corresponded to the extended haplotype HLA-Cw7-B58 (EH58.1).
Figure 1.
Haplotypes with IBD regions. Loci and markers are listed across the top, with alleles below. The IBD regions are shown within frames. The alleles not always present on all haplotypes are given in italics, and “–” indicates ⩾3 variable alleles.
Thorough examination of the haplotypes from locus C1_2_5 to locus C2_4_5 revealed three IBD regions. These regions are represented within frames in figure 1. The largest frame corresponds to the cluster of Cw6 haplotypes and includes the alleles from C1_2_5 to C2_4_5 (250 kb). The two allelic variants in the C1_2_5 and C1_2_6 loci may be the result of slippage-mispairing mutations. This block of alleles has been reported elsewhere (Nair et al. 2000), and the markers in common that are used in the present study confirm its structure. The smallest frame coincided with OTF3*2 and included four of the five PSH alleles. However, it is possible that the recombination event did not involve the centromeric end of PSH but that it occurred between the C1_2_6 and OTF3 loci as the result of a noncontiguous allele at the C1_2_6 locus. The HLA-Cw8-B65 (EH65.1) haplotype was included in the table, because it contained a region of 85 kb IBD to the cluster of Cw6 haplotypes. On the basis of the association data found for this haplotype (haplotype H; see table 2), it seemed possible to rule out this region. Hence, the minimum region nonrecombinant with PSH can be restricted to the loci C1_3_2–C2_4_5. We conclude that this region has the highest probability of containing PSORS1.
Of the 13 microsatellite markers investigated in the present study, 10 corresponded to those studied by Nair et al. (2000). This enabled us to search for PSH alleles (CDSN excluded) among the 66 clusters of haplotypes identified by those authors in Northern European white families. The results of our search showed that PSH—or the portion C1_3_2–C2_4_5, in which we mapped PSORS1—was exclusively present on haplotypes 19–25, corresponding to the cluster of haplotypes HLA-Cw6.
The first important finding emerging from this data is that the genetic background of the Sardinian population, with respect to PSORS1, is the same as that of other white populations and that the recently reported lack of association between the HLA-C alleles and psoriasis (Orrù et al. 2002) is not the result of the genetic heterogeneity of the population but is, instead, a likely consequence of an ancestral recombination between one of the Cw6 haplotypes and the HLA-Cw7-B58 haplotype. Another important outcome of our study is the definitive exclusion of the loci centromeric to the M6S172 marker—including the HLA-C locus—from the PSORS1 candidate region.
The HCR and CDSN genes map inside the PSORS1 critical region defined in the present study. Both genes have been investigated recently in numerous association studies, because, at a functional level, they are good candidates for psoriasis susceptibility (Simon et al. 1997; Asumalahti et al. 2002; Elomaa et al. 2004). In addition, SPR1, SEEK1, and STG map to this region, and, although functional data is lacking for these genes, their specific expression in the skin (Holm et al. 2003) has aroused considerable interest. Some polymorphisms of the CDSN gene have been found to be associated with psoriasis, but these associations have been attributed to linkage disequilibrium with HLA-Cw6 (Jenisch et al. 1999; Guerrin et al. 2001; Asumalahti et al. 2002). This can be explained by the block of IBD alleles carried by haplotypes of the Cw6 cluster in an ∼250-kb region from the C1_2_5 marker (19 kb centromeric to HLA-C) to the C2_4_5 marker (60 kb telomeric to CDSN), as documented in this and other reports (Nair et al. 2000). In the Sardinian population, an ancestral recombination has confined this block of linkage disequilibrium to the PSH region.
Other studies do not confirm the association of the CDSN gene with psoriasis (Gonzalez et al. 2000; Chang et al. 2003), probably because the association of CDSN and HCR is mostly linked to intragenic haplotypes of SNPs (Jenisch et al. 1999; Asumalahti et al. 2002) and is not always uncovered when the SNPs are considered separately. Our findings support the hypothesis that the CDSN*TTC allele is involved in the disease. Major opposition to this hypothesis arises from the fact that this allele was not found to be associated with psoriasis in Thai (Romphruk et al. 2003) and Japanese (Hui et al. 2002) populations. On the basis of the recent identification of a psoriasis-risk haplotype that conserves the SNP9*2 and CDSN*TTC alleles, an equal role for HLA-C and CDSN in psoriasis susceptibility has been suggested (Capon et al. 2003). Since the CDSN*TTC allele had the strongest association in our study, we decided to investigate the effect of HLA-C alleles on this association. The results summarized in table 4 show that the influence exerted by these alleles was irrelevant. The strength of association found for the CDSN*TTC allele was only slightly decreased in patients and controls negatively selected for the alleles HLA-Cw6 and SNP9*2 (OR=3.3 and 3.7, respectively), compared with the total sampling (OR=3.9), and was even increased in samples negatively selected for HLA-Cw7 (OR=4.4). In contrast, HLA-C alleles are completely dependent on CDSN*TTC, since they lose any tendency whatsoever toward association with psoriasis when this allele is excluded from the sampling.
Table 4.
Distribution of Genotypes in Selected Individuals
| Subject Group and Characteristic | CDSN*TTC | SNP 9*2 | HLA-Cw6 | HLA-Cw7 |
| CDSN*TTC negative: | ||||
| No. of cases with genotype (n=59) | - | 14 | 3 | 23 |
| No. of controls with genotype (n=111) | - | 21 | 11 | 28 |
| Pc | - | .99998 | .99872 | .68380 |
| OR | - | 1.33 | .49 | 1.89 |
| SNP 9*2 negative: | ||||
| No. of cases with genotype (n=100) | 55 | - | 7 | 68 |
| No. of controls with genotype (n=120) | 30 | - | 7 | 46 |
| Pc | .00012 | - | 1 | .00026 |
| OR | 3.67 | - | .93 | 3.42 |
| HLA-Cw6 negative: | ||||
| No. of cases with genotype (n=111) | 55 | 18 | - | 71 |
| No. of controls with genotype (n=130) | 30 | 19 | - | 48 |
| Pc | .00039 | 1 | - | .00059 |
| OR | 3.27 | 1.3 | - | 3.03 |
| HLA-Cw7 negative: | ||||
| No. of cases with genotype (n=76) | 40 | 44 | 36 | - |
| No. of controls with genotype (n=104) | 21 | 30 | 22 | - |
| Pc | .00014 | .00205 | .00451 | - |
| OR | 4.39 | 3.39 | 3.35 | - |
Although our data demonstrate the independence of the association with the CDSN*TTC allele, they do not make it possible to differentiate the effect that this allele has on the rest of the region in which we map PSORS1. However, it is likely that minor loci interact with PSORS1 in the predisposition to psoriasis. The existence of IBD regions in different HLA psoriasis-susceptibility haplotypes implies that such haplotypes have the same susceptibility allele and therefore confer an identical risk of psoriasis. The data obtained in this study seem to indicate that haplotypes of the HLA-Cw6 cluster confer a higher risk of psoriasis than the HLA-Cw7-B58 haplotype (OR=5.1 vs. 3.6). The three haplotypes of the Cw6 cluster were elsewhere shown to confer different levels of risk for psoriasis (Jenisch et al. 1998), suggesting that other genetic factors mapping in the MHC region may interact with PSORS1. A precious tool to enlighten this aspect will be comparisons among recombinant portions of psoriasis-risk haplotypes. The discovery of the recombinant HLA-Cw7-B58 haplotype should contribute toward unraveling the etiology of psoriasis.
Acknowledgments
The authors wish to thank all the patients with psoriasis whose participation made this project possible. The authors would also like to thank Anna Koopmans for the preparation of the manuscript. This research was supported by grant 1710 from the Sardinian Regional Government.
Electronic-Database Information
The URL for data presented herein is as follows:
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for HLA-C, CDSN, OTF3, HCR, and psoriasis)
References
- Asumalahti K, Veal C, Laitinen T, Suomela S, Allen M, Elomaa O, Moser M, de Cid R, Ripatti S, Vorechovsky I, Marcusson JA, Nakagawa H, Lazaro C, Estivill X, Capon F, Novelli G, Saarialho-Kere U, Barker J, Trembath R, Kere J; Psoriasis Consortium (2002) Coding haplotype analysis supports HCR as the putative susceptibility gene for psoriasis at the MHC PSORS1 locus. Hum Mol Genet 11:589–597 10.1093/hmg/11.5.589 [DOI] [PubMed] [Google Scholar]
- Barker JN (1991) The pathophysiology of psoriasis. Lancet 338:227–230 10.1016/0140-6736(91)90357-U [DOI] [PubMed] [Google Scholar]
- Brandrup F, Holm N, Grunnet N, Henningsen K, Hansen HE (1982) Psoriasis in homozygotic twins: variations in expression in individuals with identical genetic constitution. Acta Derm Venerol 62:229–236 [PubMed] [Google Scholar]
- Capon F, Toal IK, Evans JC, Allen MH, Patel S, Tillman D, Burden D, Barker JN, Trembath RC (2003) Haplotype analysis of distantly related populations implicates corneodesmosin in psoriasis susceptibility. J Med Genet 40:447–452 10.1136/jmg.40.6.447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YT, Tsai SF, Lee DD, Shiao YM, Huang CY, Liu HN, Wang WJ, Wong CK (2003) A study of candidate genes for psoriasis near HLA-C in Chinese patients with psoriasis. Br J Dermatol 148:418–423 10.1046/j.1365-2133.2003.05166.x [DOI] [PubMed] [Google Scholar]
- Elomaa O, Majuri I, Suomela S, Asumalahti K, Jiao H, Mirzaei Z, Rozell B, Dahlman-Wright K, Pispa J, Kere J, Saarialho-Kere U (2004) Transgenic mouse models support HCR as an effector gene in the PSORS1 locus. Hum Mol Genet 13:1551–1561 10.1093/hmg/ddh178 [DOI] [PubMed] [Google Scholar]
- Gonzaga HF, Torres EA, Alchorne MM, Gerbase-Delima M (1996) Both psoriasis and benign migratory glossitis are associated with HLA-Cw6. Br J Dermatol 135:368–370 10.1046/j.1365-2133.1996.d01-1006.x [DOI] [PubMed] [Google Scholar]
- Gonzalez S, Martinez Borra J, Del Rio JS, Santos-Juanes J, Lopez-Vazquez A, Blanco-Gelaz M, Lopez-Larrea C (2000) The OTF3 gene polymorphism confers susceptibility to psoriasis independent of the association of HLA-Cw*0602. J Invest Dermatol 115:824–828 10.1046/j.1523-1747.2000.00133.x [DOI] [PubMed] [Google Scholar]
- Grimaldi MC, Clayton J, Pontarotti P, Cambon-Thomsen A, Crouau-Roy B (1996) New highly polymorphic microsatellite marker in linkage disequilibrium with HLA-B. Hum Immunol 51:89–94 10.1016/S0198-8859(96)00228-5 [DOI] [PubMed] [Google Scholar]
- Guerrin M, Vincent C, Simon M, Tazi Ahnini R, Fort M, Serre G (2001) Identification of six novel polymorphisms in the human corneodesmosin gene. Tissue Antigens 57:32–38 10.1034/j.1399-0039.2001.057001032.x [DOI] [PubMed] [Google Scholar]
- Holm SJ, Carlen LM, Mallbris L, Stahle-Backdahl M, O’Brien KP (2003) Polymorphisms in the SEEK1 and SPR1 genes on 6p21.3 associate with psoriasis in the Swedish population. Exp Dermatol 12:435–444 10.1034/j.1600-0625.2003.00048.x [DOI] [PubMed] [Google Scholar]
- Hui J, Oka A, Tamiya G, Tomizawa M, Kulski JK, Penhale WJ, Tay GK, Iizuka M, Ozawa A, Inoko H (2002) Corneodesmosin DNA polymorphisms in MHC haplotypes and Japanese patients with psoriasis. Tissue Antigens 60:77–83 10.1034/j.1399-0039.2002.600110.x [DOI] [PubMed] [Google Scholar]
- Jenisch S, Henseler T, Nair RP, Guo SW, Westphal E, Stuart P, Kronke M, Voorhees JJ, Christophers E, Elder JT (1998) Linkage analysis of human leucocyte antigen (HLA) markers in familial psoriasis: strong disequilibrium effects provide evidence for a major determinant in the HLA-B/-C region. Am J Hum Genet 63:191–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenisch S, Koch S, Henseler T, Nair RP, Elder JT, Watts CE, Westphal E, Voorhees JJ, Christophers E, Kronke M (1999) Corneodesmosin gene polymorphism demonstrates strong linkage disequilibrium with HLA and association with psoriasis vulgaris. Tissue Antigens 54:439–449 10.1034/j.1399-0039.1999.540501.x [DOI] [PubMed] [Google Scholar]
- Nair RP, Henseler T, Jenisch S, Stuart P, Bichakjian CK, Lenk W, Westphal E, Guo SW, Christophers E, Voorhees JJ, Elder JT (1997) Evidence for two psoriasis susceptibility loci (HLA and 17q) and two novel candidate regions (16q and 20p) by genome-wide scan. Hum Mol Genet 6:1349–1356 10.1093/hmg/6.8.1349 [DOI] [PubMed] [Google Scholar]
- Nair RP, Stuart P, Henseler T, Jenisch S, Chia NV, Westphal E, Schork NJ, Kim J, Lim HW, Christophers E, Voorhees JJ, Elder JT (2000) Localization of psoriasis-susceptibility locus PSORS1 to a 60-kb interval telomeric to HLA-C. Am J Hum Genet 66:1833–1844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka A, Tamiya G, Tomizawa M, Ota M, Katsuyama Y, Makino S, Shiina T, Yoshitome M, Iizuka M, Sasao Y, Iwashita K, Kawakubo Y, Sugai J, Ozawa A, Ohkido M, Kimura M, Bahram S, Inoko H (1999) Association analysis using refined microsatellite markers localizes a susceptibility locus for psoriasis vulgaris within a 111 kb segment telomeric to the HLA-C gene. Hum Mol Genet 8:2165–2170 10.1093/hmg/8.12.2165 [DOI] [PubMed] [Google Scholar]
- Orrù S, Giuressi E, Casula M, Loizedda A, Murru R, Mulargia M, Masala MV, Cerimele D, Zucca M, Aste N, Biggio P, Carcassi C, Contu L (2002) Psoriasis is associated with a SNP haplotype of the corneodesmosin gene (CDSN). Tissue Antigens 60:292–298 10.1034/j.1399-0039.2002.600403.x [DOI] [PubMed] [Google Scholar]
- Roitberg-Tambur A, Friedmann A, Tzfoni EE, Battat S, Ben Hammo R, Safirman C, Tokunaga K, Asahina A, Brautbar C (1994) Do specific pockets of HLA-C molecules predispose Jewish patients to psoriasis vulgaris? J Am Acad Dermatol 31:964–968 [DOI] [PubMed] [Google Scholar]
- Romphruk AV, Oka A, Romphruk A, Tomizawa M, Choonhakarn C, Naruse TK, Puapairoj C, Tamiya G, Leelayuwat C, Inoko H (2003) Corneodesmosin gene: no evidence for PSORS 1 gene in North-eastern Thai psoriasis patients. Tissue Antigens 62:217–224 10.1034/j.1399-0039.2003.00056.x [DOI] [PubMed] [Google Scholar]
- Schneider S, Roessli D, Excoffier L (2000) Arlequin version 2.0: a software for population genetics data analysis. Genetics and Biometry Laboratory, University of Geneva, Switzerland [Google Scholar]
- Simon M, Montézin M, Guerrin M, Durieux JJ, Serre G (1997) Characterization and purification of human corneodesmosin, an epidermal basic glycoprotein associated with corneocyte-specific modified desmosomes. J Biol Chem 272:31770–31776 10.1074/jbc.272.50.31770 [DOI] [PubMed] [Google Scholar]
- Tiilikainen A, Lassus A, Karvonen J, Vartiainen P, Julin M (1980) Psoriasis and HLA-Cw6. Br J Dermatol 102:179–184 [DOI] [PubMed] [Google Scholar]
- Trembath RC, Clough RL, Rosbotham JL, Jones AB, Camp RD, Frodsham A, Browne J, Barber R, Terwilliger J, Lathrop GM, Barker JN (1997) Identification of a major susceptibility locus on chromosome 6p and evidence for further disease loci revealed by a two stage genome-wide search in psoriasis. Hum Mol Genet 6:813–820 10.1093/hmg/6.5.813 [DOI] [PubMed] [Google Scholar]
- Veal CD, Capon F, Allen MH, Heath EK, Evans JC, Jones A, Patel S, Burden D, Tillman D, Barker JN, Trembath RC (2002) Family-based analysis using a dense single-nucleotide polymorphism–based map defines genetic variation at PSORS1, the major psoriasis-susceptibility locus. Am J Hum Genet 71:554–564 [DOI] [PMC free article] [PubMed] [Google Scholar]