Abstract
Sotos syndrome (SoS) is a congenital dysmorphic disorder characterized by overgrowth in childhood, distinctive craniofacial features, and mental retardation. Haploinsufficiency of the NSD1 gene owing to either intragenic mutations or microdeletions is known to be the major cause of SoS. The common ∼2.2-Mb microdeletion encompasses the whole NSD1 gene and neighboring genes and is flanked by low-copy repeats (LCRs). Here, we report the identification of a 3.0-kb major recombination hotspot within these LCRs, in which we mapped deletion breakpoints in 78.7% (37/47) of patients with SoS who carry the common microdeletion. The deletion size was subsequently refined to 1.9 Mb. Sequencing of breakpoint fragments from all 37 patients revealed junctions between a segment of the proximal LCR (PLCR-B) and the corresponding region of the distal LCR (DLCR-2B). PLCR-B and DLCR-2B are the only directly oriented regions, whereas the remaining regions of the PLCR and DLCR are in inverted orientation. The PLCR, with a size of 394.0 kb, and the DLCR, with a size of of 429.8 kb, showed high overall homology (∼98.5%), with an increased sequence similarity (∼99.4%) within the 3.0-kb breakpoint cluster. Several recombination-associated motifs were identified in the hotspot and/or its vicinity. Interestingly, a 10-fold average increase of a translin motif, as compared with the normal distribution within the LCRs, was recognized. Furthermore, a heterozygous inversion of the interval between the LCRs was detected in all fathers of the children carrying a deletion in the paternally derived chromosome. The functional significance of these findings remains to be elucidated. Segmental duplications of the primate genome play a major role in chromosomal evolution. Evolutionary study showed that the duplication of the SoS LCRs occurred 23.3–47.6 million years ago, before the divergence of Old World monkeys.
Introduction
Sotos syndrome (SoS [MIM 117550]) is characterized by excessive growth, distinctive craniofacial features—such as macrodolichocephaly, a prominent forehead, downslanting palpebral fissures, and a pointed chin—and variable degrees of mental retardation (Cole and Hughes 1994). Intragenic mutations or submicroscopic whole-gene deletions of the nuclear-receptor–binding SET-domain–containing protein 1 (NSD1) gene at 5q35 are the main causes of SoS (Kurotaki et al. 2002, 2003; Douglas et al. 2003; Kamimura et al. 2003; Nagai et al. 2003; Rio et al. 2003; Turkmen et al. 2003; De Boer et al. 2004). Intragenic mutations prevail in white patients with SoS, whereas Japanese patients with SoS more frequently harbor a microdeletion (see review by Visser and Matsumoto [2003]). The common ∼2.2-Mb microdeletion includes NSD1 and adjacent genes (Kurotaki et al. 2002). Each deletion breakpoint is located in either of the two flanking low-copy repeats (LCRs) (Kurotaki et al. 2003). Most meiotic rearrangements seem to be of intrachromosomal origin and show a preference for the paternally derived chromosome (Miyake et al. 2003). In light of accumulating evidence, SoS was recently added to the list of genomic disorders (Kurotaki et al. 2003; Shaw and Lupski 2004). Genomic disorders are defined as pathological conditions in which the gain, loss, or disruption of dosage-sensitive gene(s) results in a recognized phenotype (Lupski 1998). Unequal rearrangement—so-called nonallelic homologous recombination—between regions of high homology (i.e., LCRs) is the most common mechanism (Lupski 1998; Stankiewicz and Lupski 2002a). This leads to duplication, deletion, inversion, or translocation of a genomic segment containing the dosage-sensitive gene(s) (Lupski 1998; Stankiewicz and Lupski 2002a). LCRs are thought to have been derived from a single original copy relatively recently in the primate evolution (Stankiewicz and Lupski 2002b). The architecture of LCRs and underlying mechanisms have been investigated in different genomic disorders (reviewed by Inoue and Lupski [2002]). Directly oriented LCRs are likely to result in either duplication or deletion, whereas inverted repeats lead to inversion of a DNA segment between the LCRs. Research on the identification of junction fragments and breakpoint locations by standard methods, such as pulsed-field gel electrophoresis (PFGE), Southern blot analysis, and construction of somatic hybrid cell lines (Shaffer and Lupski 2000; Bi et al. 2003; Shaw et al. 2004), is time consuming and labor intensive and is complicated by the high homology of the regions. Characterization of these breakpoint hotspots at the nucleotide level has proven, however, to be very important in understanding the underlying mechanism and in revealing candidate DNA structures that may stimulate strand exchange (Reiter et al. 1996, 1998; Lopez-Correa et al. 2001; Bayes et al. 2003; Bi et al. 2003; Shaw et al. 2004).
In the present study, we identified and characterized breakpoints of the common ∼2.2-Mb microdeletions at the nucleotide level in our series of patients with SoS. We used a BAC library constructed from the genomic DNA of a patient with SoS who carries the microdeletion, and we subsequently developed a PCR assay to screen other patients with SoS for the same breakpoint region. We also studied the SoS proximal and distal LCRs (PLCR and DLCR, respectively) in detail, by computational analysis using the published May 2004 human genome sequence, and we searched for recombination-associated elements in the identified breakpoint cluster and its neighboring regions. Furthermore, we screened the genomic segment between the LCRs for an inversion polymorphism in the parents of patients with SoS. For an evolutionary perspective, we determined the introduction of the duplicated SoS LCRs in primate/monkey evolution and compared the human SoS LCRs sequences with the draft of the chimpanzee genome sequence.
Material and Methods
Patients
This study included 47 Japanese patients with SoS who carry a common ∼2.2-Mb deletion, of whom 45 were reported elsewhere (Kurotaki et al. 2003). The control group consisted of 48 parents plus 4 patients with SoS and a proven smaller deletion (Kurotaki et al. 2003). With regard to evaluation of a genomic inversion polymorphism, 20 healthy Japanese individuals were also analyzed. Molecular confirmation of the microdeletion was performed in accordance with methods described elsewhere (Kurotaki et al. 2003). Genomic DNA for PCR study was obtained from peripheral blood cells or lymphoblastoid cell lines, by the use of standard methods. Experimental protocols were approved by the Committee for Ethical Issues on Human Genome and Gene Analysis at Nagasaki University and by the Committee for Ethical Issues at Yokohama City University School of Medicine.
Computational Analysis of LCRs
To characterize more precisely the identified LCRs, the computational method was used as described elsewhere (Estivill et al. 2002). A 6-Mb sequence covering both LCRs and adjacent proximal and distal regions was downloaded from the National Center for Biotechnology Information (NCBI) build 35 database (May 2004 version) available on the UCSC Genome Bioinformatics Web site. Repetitive sequences were masked by RepeatMasker (see RepeatMasker Web site). MegaBLAST2 running locally was used for comparison of the sequence with itself. A BLAST-report table was created, and parsing conditions for the results included alignment length ⩾80 bp, sequence identity ⩾90%, and an expected value of ⩽e−30. The results were copied into a Microsoft Office Excel worksheet and were analyzed. Identical hits were omitted from analysis to exclude overlapping alignments, and alignments separated by <10 kb were joined in contiguous alignments. Overall percentages of identity were calculated, and their orientations were determined.
Patient’s Genomic BAC Library and Clone Containing a Junction Fragment
A BAC library was constructed from a lymphoblastoid cell line from patient SoS 42 by GenoTechs. PCR-based library screening was performed with STS marker SHGC-16645 (GenBank accession number G17014). PCR was performed in a 10-μl mixture containing 0.5 μl of BAC-DNA mix (GenoTechs), 1μM of each primer, 0.5 units TaKaRa Ex Taq polymerase (Takara Bio), 0.2 mM of each dNTP, and 1 × Ex Taq Buffer (Takara Bio). PCR conditions included initial denaturation at 94°C for 2 min; 35 cycles at 94°C for 30 s, 50°C for 30 s, and 72°C for 30 s; followed by a final extension at 72°C for 7 min. PCR products were visualized on a 2% agarose gel by ethidium bromide staining. Identified BAC clones were cultured overnight in 275 ml of 1× Luria-Bertani medium containing 5% sucrose and 30 μg/ml chloramphenicol. BAC DNA was extracted by use of the Qiagen Midi Kit (Qiagen). BAC end sequences were determined by cycle sequencing with universal SP6 and T7 primers. The sequencing reaction was performed in a 40-μl mixture containing 1 μg of BAC DNA, 16 μl of BigDye (Applied Biosystems), and 0.2 μM of primer. Conditions were in accordance with the manufacturer’s guidelines, with an increased number of cycles (n=75). PCR products were electrophoresed in an ABI Prism 3100 DNA sequencer (Applied Biosystems) and were analyzed by AutoAssembler (Applied Biosystems). By use of a clone containing the junction fragment, screening of the paralogous sequence variant (PSV) (i.e., nucleotide difference between the PLCR and DLCR [Estivill et al. 2002], also called “cis morphism” [Boerkoel et al. 1999]) was performed by sequencing PCR products and the clone (primer sequences available on request). PCR was performed on 30 ng of purified clone DNA in a 20-μl mixture. Contents and conditions were identical to those described above. The PSVs were assigned to the PLCR or DLCR on the basis of the NCBI build 35 (May 2004) database. The breakpoint was approached in a walking manner with different primers from centromeric and telomeric sides. An 8.4-kb full segment covering the breakpoint region was sequenced.
PCR Assay for Detection of Recombined Deletion-Junction Fragment in Patients with SoS
After a clone containing the junction fragment in patient SoS 42 was identified, we hypothesized that breakpoints in other patients with SoS have occurred in the same region. Forward and reverse primers specific to the PLCR and DLCR, respectively, were designed using the online version of Primer3 (Rozen and Skaletsky 2000; see Primer3 Web site), to amplify the junction fragment. In the forward and reverse primer of set 1, the penultimate or third nucleotide from the 3′ end was mismatched, to increase specificity for a targeted LCR (Ayyadevara et al. 2000; Pettersson et al. 2003). The optimal PCR conditions were determined experimentally. PCR was performed using the GeneAmp XL PCR Kit (Applied Biosystems) in a 100-μl reaction mixture containing 2 U of rTth DNA polymerase, 30 μl of 3.3 × XL Buffer II, 200 μM of each dNTP, and a final primer concentration of 0.075 μM each. The conditions for primer set 1 included initial denaturation at 94°C for 1 min, 29 cycles at 94°C for 15 s and 69°C for 6 min, and final extension for 15 min at 72°C. Only the extension conditions in the PCR for primer set 2—at 68°C for 6 min—differed from the conditions for set 1. The products, 6.8 kb for set 1 and 6.9 kb for set 2, were visualized on a 1% ethidium bromide–stained gel. Nested PCR was subsequently performed using the first PCR product as a template (PCR primers and conditions available on request). PCR products were purified with ExoSAP-IT (USB Corporation), and the sequence was determined as described above.
Characterization of the SoS Recombination Hotspot Region
The identified 3.0-kb hotspot and 1-kb flanking DNA sequences were characterized by RepeatMasker and by a search for known recombination- and replication-associated motifs, as described elsewhere (Badge et al. 2000; Abeysinghe et al. 2003). The motifs and corresponding sequences are presented in table 1. The motif search and calculation of the GC percentage were performed with DNASIS Pro software (Hitachi Software Engineering).
Table 1.
Recombination-Associated Motifs, Searched for in the 3.0-kb Breakpoint Cluster and Adjacent Regions
| Motifa and Sequences (5′→3′) | No. in Hotspot |
| χ element from Escherichia coli: | |
| GCTGGTGG | 0 |
| Ade6-M26 heptamer from Schizosaccharomyces pombe: | |
| ATGACGT | 0 |
| ARSb consensus from Saccharomyces cerevisiae: | |
| WTTTATRTTTW | 0 |
| ARSb consensus from Saccharomyces pombe: | |
| WRTTTATTTAW | 0 |
| Consensus scaffold attachment regions: | |
| AATAAAYAAA | 0 |
| TTWTWTTWTT | 0 |
| WADAWAYAWW | 0 |
| TWWTDTTWWW | 0 |
| Deletion hotspot consensus: | |
| TGRRKM | 11 |
| DNA polymerase arrest site: | |
| WGGAG | 5 |
| DNA polymerase α frameshift hotspots: | |
| TCCCCC | 1 |
| CTGGCG | 0 |
| DNA polymerase β frameshift hotspots: | |
| ACCCWR | 1 |
| TTTT | 12 |
| DNA polymerase α/β frameshift hotspots: | |
| ACCCCA | 1 |
| TGGNGT | 4 |
| Drosophila topoisomerase II consensus: | |
| GTNWAYATTNATNNR | 0 |
| Heptamer recombination signal: | |
| CACAGTG | 2 |
| Human hypervariable minisatellite sequences: | |
| GGAGGTGGGCAGGARG | 0 |
| AGAGGTGGGCAGGTGG | 0 |
| Human minisatellite core sequence: | |
| GGGCAGGARG | 0 |
| Human replication origin consensus: | |
| WAWTTDDWWWDHWGWHMAWTT | 0 |
| Human minisatellite conserved sequence/χ-like element: | |
| GCWGGWGG | 0 |
| Immunoglobulin heavy chain class switch repeats: | |
| GAGCT | 1 |
| GGGCT | 8 |
| GGGGT | 3 |
| TGGGG | 9 |
| TGAGC | 7 |
| Long terminal repeat (LTR-IS) motif: | |
| TGGAAATCCCC | 0 |
| Mariner transposon-like element (3′ end): | |
| GAAAATGAAGCTATTTACCCAGGA | 0 |
| Murine MHC recombination hotspot: | |
| CAGRCAGR | 0 |
| Murine parvovirus recombination hotspot: | |
| CTWTTY | 2 |
| Nonamer recombination signal: | |
| ACAAAAACC | 0 |
| Pur binding site: | |
| GGNNGAGGGAGARRRR | 0 |
| Retrotransposon long terminal repeat sequence: | |
| TCATACACCACGCAGGGGTAGAGGACT | 0 |
| Translin binding sites: | |
| ATGCAG | 1 |
| GCCCWSSW | 14 |
| Vaccinia topoisomerase I consensus: | |
| YCCTT | 12 |
| Vertebrate topoisomerase II consensus: | |
| RNYNNCNNGYNGKTNYNY | 0 |
| XY32 homopurine-pyrimidine H-palindrome motif: | |
| AAGGGAGAARGGGTATAGGGRAAGAGGGAA | 0 |
Inversion-Polymorphism Screening of the Genomic Interval between the LCRs
Interphase nuclei were prepared for FISH from peripheral blood lymphocytes or immortalized lymphoblastoid cell lines from 40 parents of 20 patients with SoS and a microdeletion (17 common and 3 smaller microdeletions [Kurotaki et al. 2003]) and from 20 healthy Japanese controls, in accordance with standard protocols described elsewhere (Shimokawa et al. 2004). The parental origin of the chromosome harboring the microdeletion was reported elsewhere (Miyake et al. 2003). BAC clones CTB-162J7 (GenBank accession number AC010297) and CTB-22D11 (GenBank accession number AC090063) were selected from the UCSC Genome Browser NCBI build 35 (May 2004), and PAC clone GS-240G13 (5q subtelomeric clone) was described elsewhere (Knight et al. 2000). These three clones were labeled with SpectrumGreen-11-dUTP (green) (Vysis), with SpectrumOrange-11-dUTP (red) (Vysis), and with both (yellow), respectively. Authors R.V. and O.S. blindly evaluated the FISH slides, and 30–50 interphase nuclei were scored by each author. Only concordant results (i.e., one specific pattern observed in >50% of cells by both R.V. and O.S.) were regarded as conclusive.
Evolutionary Study
FISH was performed on interphase and metaphase chromosomes of lymphoblastoid cell lines from the chimpanzee (Pan troglodytes), orangutan (Pongo pygmaeus), gibbon (Hylobates lar), Old World monkey (Macaca fuscata), and the New World monkey marmoset (Callithrix jacchus), in accordance with the methods described elsewhere (Sugawara et al. 2003). BAC clones RP11-546L14 (GenBank accession number AC108509; which mapped to PLCR-A, -B, and -C, with a higher homology to the DLCR [98.1%–98.9%]) and CTD-2272F9 (GenBank accession number AC124851; which mapped to approximately the region covering DLCR-C and DLCR-H, with a somewhat lower homology to the PLCR [96.1%–99.1%]) were labeled with SpectrumOrange-11-dUTP and SpectrumGreen-11-dUTP, respectively.
To determine the homology between the SoS LCRs in the human genome and the chimpanzee (P. troglodytes) genome, orthologous sequences spanning 6 Mb and covering NSD1 and flanking regions on the chimpanzee chromosome 4 were downloaded from the NCBI build 1 database (November 2003) of the UCSC Chimp Genome Browser and were masked for repetitive sequences by RepeatMasker. The masked human PLCR, DLCR, and interval sequences were each compared with the chimp sequence by MegaBlast2 and were analyzed in accordance with the conditions described in the section “Computational Analysis of LCRs” above.
Results
Computational Analysis of LCRs
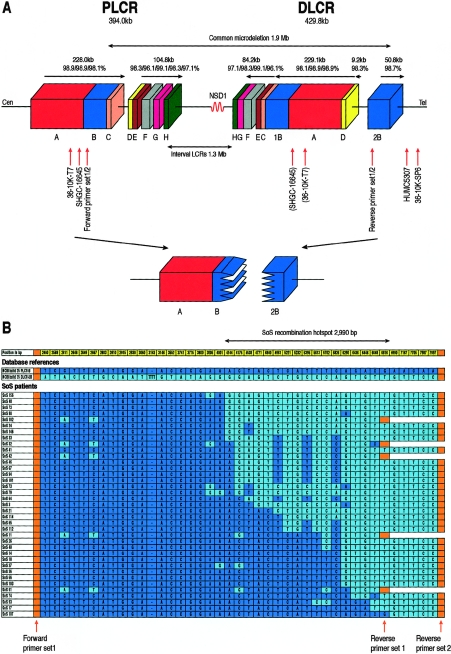
Results of the computational analysis are shown in figure 1. The sizes of the PLCR (UCSC Genome Browser NCBI build 35 [May 2004] nucleotide coordinates 175263255–175657241) and DLCR (UCSC coordinates 176984723–177414477) were found to be 394.0 kb and 429.8 kb, respectively, and they are separated by an ∼1.3-Mb interval. The identity between the PLCR and DLCR is at least 96.1%, although the largest part (regions A, B, and C) has an identity of 98.1%–98.9% (overall SoS LCR homology is ∼98.5%). The DLCR is mainly inversely oriented with regard to the PLCR, except for the PLCR-B region (UCSC coordinates 175417987–175481486), which is represented twice in the DLCR: once in inverted orientation (DLCR-1B [UCSC coordinates 177075889–177145310]) and once in direct orientation (DLCR-2B [UCSC coordinates 177363698–177414477]) (see fig. 1A). In the proximal and distal regions adjacent to the LCRs of at least 1.3 Mb, no other similar LCRs were found.
Figure 1.
A, Schematic presentation of the structure of the two LCRs flanking microdeletions in SoS. Blocks with the same color and letter represent corresponding regions with sequence homology to each other. The size of (groups of) blocks and homology percentage of each block are shown above the horizontal arrows, which indicate the genomic orientation. 36-10K-SP6 and 36-10K-T7 show the position of the BAC end sequences found in clone 36-10K. STS markers SHGC-16645 and HUMC5307 could be amplified from clone 36-10K, and “(SHGC-16645)” and “(36-10K-T7)” indicate the possible corresponding sequences located in DLCR-A. The relative position of primer sets 1 and 2 are shown. The lower part shows a deletion resulting in the formation of a junction fragment (as identified in the present study) occuring between PLCR-B and the corresponding DLCR-2B. Cen = centromere; Tel = telomere. B, PSVs found in and around the breakpoint region. Dark blue and light blue represent PSVs of PLCR-B and DLCR-2B deposited in the NCBI build 35, respectively. The position of each PSV (in bp) is numbered, starting from the forward primer of set 1. PSVs in the first ∼2.46-kb and the location of the forward primer of set 2 are not shown. The position of the forward primer of set 1 and the reverse primers of sets 1 and 2 are indicated by orange arrows and blocks. Patients with SoS are arranged in order on the basis of the location of their breakpoints. The size of the 2,990-bp hotspot is indicated with a bidirectional arrow above the figure.
BAC Library of SoS 42 and Clone 36-10K Containing the Junction Fragment
The BAC library had a 100–150-kb insert DNA, on the basis of the size of sampled clones, and an ∼1–2-fold coverage of the whole genome. A BAC clone, 36-10K, was identified in the library and had BAC end sequences similar to PLCR-A (the T7 end [GenBank accession number AY753210; UCSC coordinates 175374945–175375414]) and distal to the DLCR (the SP6 end [GenBank accession number AY753209; UCSC coordinates 177451285–177451735]), as shown in figure 1A. The size of inserted DNA is estimated to be 131 kb (NCBI build 35 [May 2004] database). STS markers SHGC-16645, located at the PLCR, and HUMC5307 (GenBank accession number L28294), distal to the DLCR, could be amplified from this clone and were also confirmed by BLAST. This indicated a junction fragment containing segments of both the PLCR and DLCR. PSVs of nested PCR products were assigned to the PLCR or DLCR by BLAST and were used to delimit the breakpoint region. A region of ∼2.1 kb between positions 4144 and 6290 (see SoS 42 in fig. 1B) was identified, showing a transition from the PLCR to DLCR, on the basis of PSVs. No deletions or insertions were found in the junction fragment. In both centromeric PLCR and telomeric DLCR segments of the breakpoint region (see SoS 42 positions 2611, 2667, 4961, 5395, and 5826 in fig. 1B), the presumed PSVs were polymorphic, as was confirmed in other individuals (fig. 1B).
PCR Assay to Detect Breakpoint Junctions in Other Patients with SoS
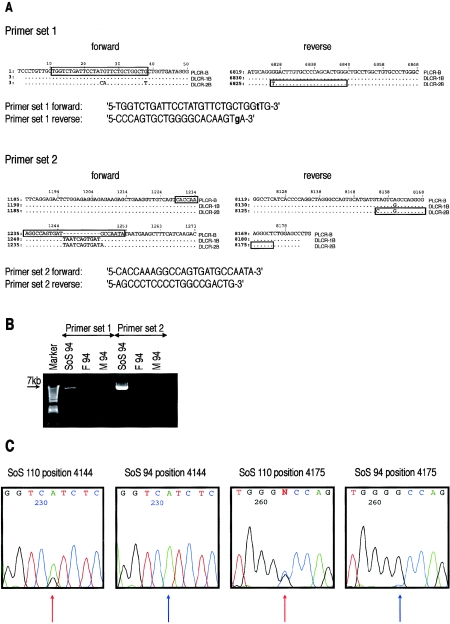
Initially, primer set 1 was designed to screen other patients with SoS for the same breakpoint region as patient SoS 42 (see fig. 2A and 2B). The overall results are shown in table 2, and the results for each individual patient are shown in table A1 (online only). Of a total of 47 patients, 33 (70.2%) showed an amplified PCR product. This was checked by sequence analysis, which confirmed the breakpoint in 30 patients (63.8%). Three patients (6.4%)—SoS 5, SoS 107, and SoS 110—showed a heterozygous appearance of the PSVs, on the basis of their sequence electropherograms (fig. 2C), and were therefore regarded as showing false-positive results. This indicates that the primer set 1 may have also amplified products from normal PLCR-B and DLCR-1B. The PCR assay was negative in 14 (29.8%) of 47 patients and in 47 (90.4%) of 52 controls, although 5 controls (9.6%) gave a slightly positive PCR product. Patterns of the sequence electropherograms in these five controls were similar to those of patients SoS 5, SoS 107, and SoS 110 (fig. 2C). Changing the PCR conditions and concentrations did not improve PCR yield or specificity.
Figure 2.
A, Alignments of the PLCR and DLCR at the primer sites, as generated by MultiPipMaker (Schwartz et al. 2003). PLCR-B, DLCR-1B, and DLCR-2B are the regions also shown in figure 1A. Dots in the alignments indicate identical nucleotides; PSVs are shown with their respective nucleotides. Boxes represent the position of primer sequences. Nucleotides in lowercase bold letters are mismatched nucleotides introduced to increase PCR specificity. The position is shown on the left of the alignments and starts 10 bp proximal to the forward primer of set 1. B, PCR results in patient SoS 94 and parents, for primer sets 1 and 2. Left lane, a 1-kb plus DNA ladder (Invitrogen). F 94 = father of SoS 94; M 94 = mother of SoS 94. C, Electropherograms of PSVs at positions 4144 and 4175. SoS 110 shows heterozygous patterns of PSVs (red arrows) in the PCR assay with primer set 1 and is therefore regarded as having a false-positive PCR result. SoS 94, in whom the junction fragment was identified, shows no heterozygous patterns of the PSVs (blue arrows).
Table 2.
Results of PCR and Sequencing Using Two Primer Sets[Note]
|
PCR Results |
Sequence Results ofPositive PCR Products |
|||
| Primer Setand Subjects | No. (%) Positive | No. (%) Negative | No. (%) Positive | No. (%) Negative |
| Set 1: | ||||
| Patients (n=47) | 33 (70.2) | 14 (29.8) | 30 (63.8) | 3 (6.4) |
| Controls (n=52) | 5 (9.6) | 47 (90.4) | 0 (0) | 5 (9.6) |
| Set 2: | ||||
| Patients (n=47) | 32 (68.1) | 15 (31.9) | 32 (68.1) | 0 (0) |
| Controls (n=52) | 0 (0) | 52 (100) | 0 (0) | 0 (0) |
Note.— Overall results for patients (n=47): identified junction, 37 (78.7%); not-identified junction, 10 (21.3%).
Primer set 2 was subsequently designed on the basis of the sequence results of set 1 (fig. 2A and 2B). The results are reported in table 2. Thirty-two (68.1%) of the 47 patients showed positive results, and this was confirmed by sequence analysis. All 48 available parental samples and the 4 cases with a smaller deletion were negative. Samples from patients SoS 5 and SoS 110 and five controls, assumed to be false positives by the results of set 1, were negative for set 2. However, sample SoS 107 was positive for set 2, which was confirmed by sequence analysis. Additionally, samples from five patients (SoS 11, SoS 32, SoS 42, SoS 61, and SoS 102) were negative for set 2, as the result of an insertional polymorphism (TAATCAGTGAT) in their genome at the site where the forward primer of set 2 was designed (fig. 2A). On the other hand, patients SoS 40, SoS 66, SoS 67, SoS 72, SoS 74, and SoS 103 showed positive results, which were confirmed by sequencing. Overall, we were able to map the breakpoint in 78.7% (37/47) of the patients with the common microdeletion within a 2,990-bp recombination hotspot, whereas breakpoints of 10 patients (21.3%) were suspected to be located elsewhere.
Characterization of SoS Recombination Hotspot
On the basis of the study results, the size of deletions with breakpoints clustering at the 3.0-kb region was calculated to be 1.9 Mb, close to the ∼2.2 Mb estimated elsewhere (Kurotaki et al. 2002). No inserted or deleted nucleotides were identified within the breakpoint-cluster region. At PSV positions 2611, 2667, 3905, 4001, 4175, 5613, and 5702 in the PLCR part, nucleotides assigned to the DLCR on the basis of the human genome database were found. Also, in the ∼2.46-kb region centromeric to position 2460, several similar polymorphisms were identified (data not shown). On the other hand, nucleotides of the PLCR were found in the DLCR portion, at positions 4538, 4961, 5395, 5826, 6290, and 6648.
Higher homology (99.4%) and higher GC content (55.0%–55.2%) were observed in the hotspot regions, compared with 98.7% homology in the remaining PLCR-B and DLCR-2B regions, 44.7% GC content in PLCR-B (44.2% in PLCR), and 44.6% GC content in DLCR-2B (44.9% in DLCR).
The search for sequence motifs prone to recombination and replication revealed several motifs within the 3.0-kb hotspot region (table 1). The frequency of these motifs is in concordance with the distribution throughout the LCRs, except for the translin target-site (5′-GCCCWSSW-3′) motif that showed a 9-fold and an 11-fold increase over the DLCR and PLCR, respectively. In the 1-kb flanking regions, several similar motifs were identified. Furthermore, in the centromeric region, two scaffold-attachment regions (SARs) of 5′-TTWTWTTWTT-3′ and four SARs of 5′-TWWTDTTWWW-3′ were identified. At 559 bp and 702 bp proximal to the hotspot and at 733 bp distal to the breakpoint region, a 302-bp Alu repeat and 85-bp and 210-bp mammalian interspersed repeats were found, respectively.
Inversion-Polymorphism Screening of the 1.3-Mb Genomic Interval
Results of inversion-polymorphism screening are presented in table 3. An example of an inverted and normal-oriented interval segment, as detected by three-color FISH, is shown in figure 3A. The FISH results were concordant after single-time evaluation for 31 parents (77.5%) and 10 controls (50%). In the parents of patients with a paternal microdeletion, 14 (100%) of the 14 fathers and 8 (88.9%) of the 9 mothers showed a heterozygous inversion of the 1.3-Mb interval. In the two patients with a deletion in the maternally derived chromosome, all four parents were heterozygous for the inversion. Additionally, in two cases in which the parental deletion origin was not determined, both of the fathers and one mother carried a heterozygous inversion and the other mother had a normal status. A heterozygous pattern was observed in the control group in 4 (66.7%) of the 6 males and in 3 (75%) of the 4 females.
Table 3.
Results of Heterozygosity Screening of the 1.3-Mb Interval
|
Finding in Fathers |
Finding in Mothers |
|||||||
| Informative (18/20 [90%]) |
Informative (13/20 [65%]) |
|||||||
| Subject: | HomozygousInversion | HeterozygousInversion | Normal | Not Informative (2/20 [10%]) | HomozygousInversion | HeterozygousInversion | Normal | Not Informative (7/20 [35%]) |
| Patients with SoS and: | ||||||||
| Paternal deletion (n=16) | 0 | 14/14a (100%) | 0 | … | 0 | 8/9b (88.9%) | 1/9 (11%) | … |
| Maternal deletion (n=2) | 0 | 2/2 (100%) | 0 | … | 0 | 2/2 (100%) | 0 | … |
| Deletion not determined (n=2) | 0 |
2/2 (100%) |
0 |
… |
0 |
1/2 (50%) |
1/2 (50%) |
… |
|
Finding in Males |
Finding in Females |
|||||||
| Informative (6/10 [60%]) |
Informative (4/10 [40%]) |
|||||||
| HomozygousInversion |
HeterozygousInversion |
Normal |
Not Informative (4/10 [40%]) |
HomozygousInversion |
HeterozygousInversion |
Normal |
Not Informative (6/10 [60%]) |
|
| Controls (n=20) | 0 | 4/6 (66.7%) | 2/6 (33.3%) | … | 0 | 3/4 (75%) | 1/4 (25%) | … |
Includes three fathers of patients with SoS and a smaller deletion.
Includes two mothers of patients with SoS and a smaller deletion.
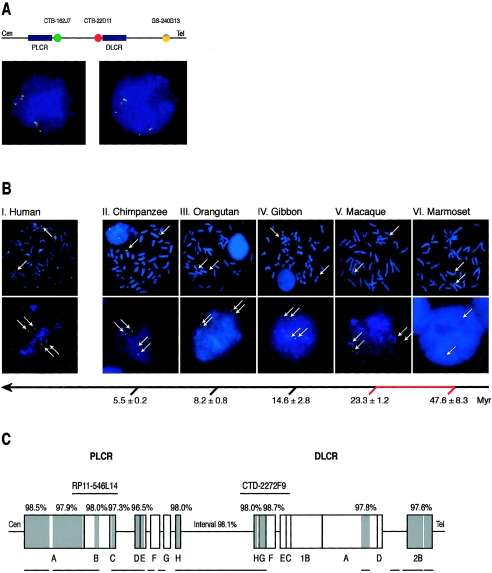
Figure 3.
A, Three-color FISH analysis for confirmation of the inversion polymorphism of the 1.3-Mb interval between the LCRs. Clones CTB-162J7 and CTB-22D11 were located in the interval, and GS-240G13 was located at the 5q subtelomeric region, as shown above the interphase FISH photographs. CTB-162J7, CTB-22D11, and GS-240G13 were labeled with SpectrumGreen (green), SpectrumOrange (red), and a 50-50 combination of SpectrumGreen and SpectrumOrange (yellow), respectively. The left photograph indicates a normal homozygous state (green-red-yellow) of the interval in both chromosomes. The right photograph shows a heterozygous inversion pattern (red-green-yellow) in one chromosome and a normal pattern (green-red-yellow) in the other. B, FISH signals (arrows) of BAC clone CTD-2272F9 (SpectrumGreen-11-dUTP) on metaphase (upper row) and interphase (lower row) cells from human (Homo sapiens) (I), chimpanzee (Pan troglodytes) (II), orangutan (Pongo pygmaeus) (III), gibbon (Hylobates lar) (IV), Old World monkey (Macaca fuscata) (V), and the New World monkey marmoset (Callithrix jacchus) (VI). The BAC clone represents the region of DLCR between DLCR-C and DLCR-H (see panel C). Duplicated signals are observed in interphase cells in photographs I–V; a single signal is seen in VI. The arrow with horizontal branches indicates the molecular timescale, as described elsewhere (Kumar and Hedges 1998), with estimated time (±1 SE) showing the divergence of the human and other species. The red portion of the branched arrow shows the introduction of the SoS LCR duplication in the primate/monkey evolution. Myr = million years ago. C, Schematic presentation of the human LCRs (blocks) and intervals (lines) and their orthologous chimpanzee genome sequences. Gray blocks indicate regions homologous to the chimpanzee sequence. The similarity percentages of these regions (i.e., similarity between the human and chimpanzee sequence) are shown above the blocks. Lines below the LCRs depict the available chimpanzee sequences, and interruptions in lines indicate gaps (>5 kb) in the present NCBI build 1 (November 2003) UCSC Chimp Genome Browser. RP11-546L14 and CTD-2272F9 show the position of the FISH probes, as used in the evolutionary study (see panel B).
Evolutionary Study
Results of the evolutionary study are presented in figure 3B. A duplicated FISH signal for probe CTD-2272F9 (SpectrumGreen) (fig. 3) and probe RP11-546L14 (SpectrumOrange) (results not shown) was clearly observed in interphase cells of the chimpanzee, orangutan, gibbon, and Old World monkey, whereas a single FISH signal of CTD-2272F9 was observed in the New World monkey marmoset. The signal of probe RP11-546L14 could not be recognized in the marmoset, either because of very low sequence homology or because of poor quality of the chromosomes and interphases despite different preparations. The results suggest that the duplication of SoS LCRs occurred at least after the divergence of the New World monkeys and before the divergence of Old World monkeys. This is estimated to be ∼23.3–47.6 million years ago (Kumar and Hedges 1998).
The sequence comparison between the human SoS LCRs and the corresponding chimpanzee sequences is shown in figure 3C. Parts of both LCRs could be identified in the chimpanzee sequences, as well as the 1.3-Mb interval. Similarity varied from ∼96.5% to ∼98.5% for the sequences available. These results support an introduction of the LCRs before the divergence of the chimpanzee. With an ∼95% stated coverage of the NCBI build 1 (November 2003) draft chimpanzee genome sequence, sequence gaps in the chimpanzee region homologous to the PLCR and DLCR accounted for 38.4% and 68.6%, respectively.
Discussion
The common-size microdeletions in SoS and the existence of flanking LCRs harboring the deletion breakpoints are lines of evidence for nonrandom recurrent events of these deletions (Kurotaki et al. 2003). The identification in the present study of a 3.0-kb region that harbors 78.7% (37/47) of the breakpoints is the first evidence to show that unequal strand exchanges occur at a specific, nonrandom recombination hotspot in the majority of patients with SoS. Deletions with determined junction boundaries in the hotspot region were calculated to be 1.9 Mb in size. The detailed structure of the LCRs in our study is based on in silico characterization. Although in silico analysis might not perfectly represent the in vivo situation, its usefulness has already been proven in the analysis of chromosomal segment duplications (Estivill et al. 2002; Cheung et al. 2003; Gimelli et al. 2003). Furthermore, the results were in concordance with past and present experimental data (Kurotaki et al. 2003). Our schematic presentation is, therefore, the most detailed construction of the SoS LCR region.
Standard techniques to identify junction fragments are usually PFGE, Southern blot hybridization, and construction of hybrid cell lines (Shaffer and Lupski 2000; Bi et al. 2003; Shaw et al. 2004). In the present study, a BAC library was constructed from genomic DNA of a patient with SoS who carried a common microdeletion, and PCR-based screening of the library was performed. This strategy turned out to be very effective for the identification of a BAC clone containing a junction fragment, as well as other clones of the PLCR and DLCR (data not shown).
The T7-end sequence of BAC clone 36-10K and the STS marker SHGC-16645 were mapped to PLCR-A, with only a slightly higher identity than that to the corresponding DLCR-A portion (fig. 1A). This might suggest that the clone 36-10K contains a DNA fragment from DLCR-A to its neighboring telomeric portion, the SP6-end sequence. In this case, the clone would have contained ∼264 kb, and the genomic orientation would have been incorrect. Furthermore, markers from the unique region in the DLCR could not be amplified (data not shown), and the PSVs showed no pattern in concordance with the DLCR-A and -1B regions. Therefore, the clone 36-10K contains a junction fragment of PLCR-B and the directly oriented part of the DLCR (DLCR-2B) and has an estimated total length of 131 kb.
The PCR systems we developed successfully defined deletion breakpoints in 37 (78.7%) of the 47 patients with SoS in our study. Very high sequence similarity between the two LCRs forced us to design specific primers. Although our first set (set 1) of primers had a false-positive rate of 9.6% in the control group, this could be verified by sequence electropherogram analysis. The second primer set (set 2) seemed highly specific in our patient samples but failed to detect the junction fragments in 13.5% (5/37) of samples because of the presence of an insertional polymorphism at the forward primer site. Stringent conditions for set 1 also caused a small yield with a false-negative rate of 16.2% (6/37). However, the breakpoints of all these patients were detected by set 2. The combination of both sets enabled us to identify the junction fragment in 78.7% of the patient samples. The patients who showed negative PCR results are likely to have breakpoints in a different region or different hotspot(s). Alternatively, low quality of their DNA may have interfered with the long-PCR assay, or polymorphic PSVs may have occurred at the primer sites. Because of polymorphic PSVs, false-positive results by PCR could also appear in the normal population without any junction fragment. In our control samples, we did not find any such incorrect results, although investigations of a larger population are necessary to determine the sensitivity and specificity of the primer sets and their clinical applicability.
The ∼7-kb fragments amplified by both primer sets provided information about the region proximal and distal to the breakpoint cluster. PSVs, which turned out to be polymorphic, were possibly caused by gene-conversion events. In a recombination hotspot for neurofibromatosis type 1 (NF1 [MIM 162200]) and for some other genomic disorders, gene conversion is also documented (Reiter et al. 1998; Lopes et al. 1999; Han et al. 2000; Lopez-Correa et al. 2001; Bi et al. 2003). Highly frequent gene conversions were recently confirmed in relation to hotspots for meiotic equal recombination, suggesting similar causative mechanisms between equal and unequal recombinations (Jeffreys and May 2004). However, Han et al. (2000) found no increased recombination frequency in a recombination hotspot for Charcot-Marie-Tooth disease type 1A (CMT1A [MIM 118220]) and defined it as a hotspot with positional preference for recombination (Han et al. 2000). Therefore, until the recombination frequency in the SoS breakpoint cluster is determined, the 3.0-kb hotspot indicates a positional preference for recombination.
The absence of loss or gain of sequences at the breakpoints, such as nucleotide deletions or insertions and junctions between homologous PLCR-B and DLCR-2B, is supportive evidence of nonallelic homologous recombination as the general underlying mechanism in SoS. Nevertheless, specific interactions stimulating strand breaks and unequal crossovers at recombination hotspots in genomic disorders remain unknown.
We found an elevated GC content (∼55% vs. an average of ∼44.5% in the LCRs) in the SoS breakpoint hotspot region. A raised GC level was also noted in the recombination hotspot for NF1 (Lopez-Correa et al. 2001) and in the 12-kb hotspot on chromosome 17p11.2, which is associated with deletions in Smith-Magenis syndrome (SMS [MIM 182290]) and with reciprocal duplications in patients with dup(17)(p11.2p11.2) (Bi et al. 2003). However, deletion breakpoints are, in general, rather AT rich, compared with GC-rich translocation breakpoints (Abeysinghe et al. 2003).
A mariner-like transposon element was mapped at ∼700 bp centromeric to the hotspot associated with deletions in CMT1A and with duplications in hereditary neuropathy with liability to pressure palsies (HNPP [MIM 162500]) (Kiyosawa and Chance 1996). No evidence was found, however, that this element is functionally active. Furthermore, five human minisatellite-like sequence clusters and one Escherichia coli χ-like element were found within a 741-bp recombination interval in a CMT1A LCR (Lopes et al. 1998). In a 12-kb hotspot in the SMS LCRs, a human minisatellite core sequence and four χ-sites were identified (Bi et al. 2003). In a 2-kb recombination hotspot for NF1, a χ-like element was also found (Lopez-Correa et al. 2001). Several recombination-prone motifs were recently identified in a 524-bp hotspot of uncommon deletions in SMS, and four of the six junctions were mapped within an AluSq/x element (Shaw et al. 2004). Since all the elements mentioned above show a distribution throughout the human genome, the significance of these findings in LCRs of genomic disorders has yet to be determined. We also identified multiple motifs within the 3.0-kb hotspot interval and adjacent regions. Interestingly, a 10-fold increase of the translin target site (5′-GCCCWSSW-3′) was found, compared with the overall distribution within the LCRs. Translin target sequences are found in a significantly higher concentration in or around regions of translocation and deletion breakpoint clusters (Abeysinghe et al. 2003). The translin protein is thought to recognize single-stranded DNA ends of staggered breaks that may occur at recombination hotspots (Kasai et al. 1997). The role of the translin target sites and other motifs in stimulating unequal strand exchanges in the 3.0-kb hotspot in SoS remains to be elucidated.
The prevalence of common deletions in the Japanese patients with SoS, compared with the few cases of NSD1 deletions in the white patients with SoS, is remarkable. Furthermore, the identification of a deletion hotspot raises the likelihood of a nonrandom underlying causative mechanism. Consequently, these two arguments seem to be suggestive of a genomic variation, which would be more or less specific to the Japanese population and would predispose to unequal meiotic recombination between the SoS LCRs. A genomic inversion of the DNA segment between LCRs is thought to be associated with abnormal meiotic pairing and would therefore increase susceptibility to unequal recombination (Osborne et al. 2001). For example, in a study of Angelman syndrome (AS [MIM 105830]), a heterozygous inversion of 15q11-q13 was observed in four (66.7%) of six mothers of patients with AS carrying a type II 15q11-q13 microdeletion of maternal origin, compared with 9% in the normal population (Gimelli et al. 2003). In studies of Williams-Beuren syndrome (WBS [MIM 194050]), ∼30% of the parents transmitting the chromosome in which the microdeletion subsequently occurred carried a heterozygous inversion (Osborne et al. 2001; Bayes et al. 2003), whereas this inversion was not found in 26 healthy individuals (Osborne et al. 2001). We screened for an inversion including at least the 1.3-Mb interval flanked by LCRs. We observed a heterozygous state in 100% of the fathers of patients with SoS and a microdeletion of paternal origin. Also, in the control group, a high percentage of heterozygosity, 66.7% in males and 75% in females, was observed.
Although the high percentage of heterozygosity could indicate a common polymorphism in the Japanese population and, consequently, a higher susceptibility to microdeletions in SoS, these results should be evaluated carefully. First, a relatively high percentage of interpersonal difference was noted, and nonconcordant results were discarded from conclusive data, creating a possible bias of the final results. High complexity of three-colored clones ordered in an ∼5 Mb (1.3 Mb plus ∼3.7 Mb) genomic region might have influenced the results. Additional techniques, such as fiber FISH, are therefore required to confirm the inversion status, as well as the physical size. Second, large-scale studies of Japanese and non-Japanese individuals are necessary for confirmation of our results on a population level. Third, a group of 24 cases of whole NSD1 gene deletions, comprising patients from the United Kingdom, the United States, Europe, and Australia, was presented recently (Tatton-Brown et al. 2004). The deletion found was of paternal origin in 15 of 17 patients, and 14 of these patients showed a similar-sized deletion, a result apparently similar to our findings in Japanese patients, although the deletion size was not clearly stated. Further research in different SoS populations is therefore necessary to delineate a possible racial difference in relation to genomic variants in SoS.
The major forces of the primate evolution are single base-pair mutations, sequence duplications, and chromosomal rearrangements (Samonte and Eichler 2002). The introduction of segmental duplications, derived from an ancestral copy, is generally dated within the past 35 million years ago (Eichler 2001). A more precise timescale for different genomic disorders has been reviewed by Stankiewicz and Lupski (2002b). Our study suggested that duplication of the SoS LCRs may have happened ∼23.3–47.6 million years ago. Since clone RP11-546L14 could not be evaluated in the New World monkey, we are unable to completely exclude the possibility of multiple duplication events within the SoS LCRs at different times in evolution. Sequence homology in the LCRs is slightly higher in the A, B, and C regions, which could indicate a more recent duplication in comparison with the D, E, F, G, and H regions, which showed a slightly lower homology. On the other hand, the difference in similarity might also be caused by a higher recombination frequency in the D, E, F, and G regions and/or a higher gene-conversion rate. Comparison of the SoS LCRs with the orthologous draft chimpanzee genome sequence showed a homology of ∼96.5%–98.5% for the sequences available. However, the draft chimpanzee genome sequence still contains too many gaps to allow us to perform extensive analysis to determine possible recombination and gene-conversion rates. Further updating of this draft genome sequence, as well as the publication of the orangutan genome sequence, will undoubtedly create conditions for a more detailed analysis of the evolution of SoS LCRs.
In conclusion, the identification of a 3.0-kb hotspot harboring 78.7% of breakpoints of the common 1.9-Mb deletions in patients with SoS gives detailed information about the crossover location and provides insight into the underlying mechanism of unequal recombination. Identification of additional hotspots will be important for further elucidation of the causative mechanisms.
Acknowledgments
We kindly express our gratitude to the patients, their parents, and the referring physicians, for their participation in this study. Furthermore, we thank Ms. Yasuko Noguchi, Ms. Kazumi Miyazaki, and Ms. Naoko Yanai, for their excellent technical assistance. This study was supported by the Japan Science and Technology Agency (Core Research for Evolutional Science and Technology) and by the International Consortium for Medical Care of Hibakusha and Radiation Life Science, The 21st Century Center of Excellence.
Appendix A: Supplemental Data
Table A1.
Results of PCR and Sequencing Using Two Primer Sets, in Patients with SoS[Note]
|
Finding in Patienta |
|||||
| Patient | PCR Set 1 | Sequence Set 1 | PCR Set 2 | Sequence Set 2 | Conclusionb |
| SoS 3 | + | + | + | + | + |
| SoS 14 | + | + | + | + | + |
| SoS 17 | + | + | + | + | + |
| SoS 18 | + | + | + | + | + |
| SoS 21 | + | + | + | + | + |
| SoS 24 | + | + | + | + | + |
| SoS 26 | + | + | + | + | + |
| SoS 27 | + | + | + | + | + |
| SoS 33 | + | + | + | + | + |
| SoS 36 | + | + | + | + | + |
| SoS 41 | + | + | + | + | + |
| SoS 46 | + | + | + | + | + |
| SoS 48 | + | + | + | + | + |
| SoS 64 | + | + | + | + | + |
| SoS 65 | + | + | + | + | + |
| SoS 73 | + | + | + | + | + |
| SoS 79 | + | + | + | + | + |
| SoS 80 | + | + | + | + | + |
| SoS 93 | + | + | + | + | + |
| SoS 94 | + | + | + | + | + |
| SoS 101 | + | + | + | + | + |
| SoS 112 | + | + | + | + | + |
| SoS 114 | + | + | + | + | + |
| SoS 146 | + | + | + | + | + |
| Sos 156 | + | + | + | + | + |
| SoS 11 | + | + | − | + | |
| SoS 32 | + | + | − | + | |
| SoS 42 | + | + | − | + | |
| SoS 61 | + | + | − | + | |
| SoS 102 | + | + | − | + | |
| SoS 40 | − | + | + | + | |
| SoS 66 | − | + | + | + | |
| SoS 67 | − | + | + | + | |
| SoS 72 | − | + | + | + | |
| SoS 74 | − | + | + | + | |
| SoS 103 | − | + | + | + | |
| SoS 107 | + | − | + | + | + |
| SoS 5 | + | − | − | − | |
| SoS 110 | + | − | − | − | |
| SoS 4 | − | − | − | ||
| SoS 12 | − | − | − | ||
| SoS 31 | − | − | − | ||
| SoS 69 | − | − | − | ||
| SoS 71 | − | − | − | ||
| SoS 85 | − | − | − | ||
| SoS 96 | − | − | − | ||
| SoS 100 | − | − | − | ||
Note.— Blank spaces in the appendix indicate that sequences could not be performed (because of a negative PCR result).
+ = positive result; − = negative result.
Refers to whether or not the breakpoint is identified on the basis of the PCR and sequence results of sets 1 and 2.
Electronic-Database Information
Accession numbers and URLs for data presented herein are as follows:
- BLAST, http://www.ncbi.nlm.nih.gov/BLAST/
- GenBank, http://www.ncbi.nih.gov/GenBank/ (for SHGC-16645 [accession number G17014], CTB-162J7 [accession number AC010297], CTB-22D11 [accession number AC090063], RP11-546L14 [accession number AC108509], CTD-2272F9 [accession number AC124851], 36-10K-T7 [accession number AY753210], 36-10K-SP6 [accession number AY753209], and HUMC5307 [accession number L28294])
- MultiPipMaker, http://pipmaker.bx.psu.edu/pipmaker/
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for AS, CMT1A, HNPP, NF1, SMS, SoS, and WBS)
- Primer3, http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi
- RepeatMasker, http://www.repeatmasker.org/
- UCSC Genome Bioinformatics, http://genome.ucsc.edu/ (for human genome browser and chimpanzee genome browser)
References
- Abeysinghe SS, Chuzhanova N, Krawczak M, Ball EV, Cooper DN (2003) Translocation and gross deletion breakpoints in human inherited disease and cancer. I: Nucleotide composition and recombination-associated motifs. Hum Mutat 22:229–244 10.1002/humu.10254 [DOI] [PubMed] [Google Scholar]
- Ayyadevara S, Thaden JJ, Shmookler Reis RJ (2000) Discrimination of primer 3′-nucleotide mismatch by taq DNA polymerase during polymerase chain reaction. Anal Biochem 284:11–18 10.1006/abio.2000.4635 [DOI] [PubMed] [Google Scholar]
- Badge RM, Yardley J, Jeffreys AJ, Armour JA (2000) Crossover breakpoint mapping identifies a subtelomeric hotspot for male meiotic recombination. Hum Mol Genet 9:1239–1244 10.1093/hmg/9.8.1239 [DOI] [PubMed] [Google Scholar]
- Bayes M, Magano LF, Rivera N, Flores R, Perez Jurado LA (2003) Mutational mechanisms of Williams-Beuren syndrome deletions. Am J Hum Genet 73:131–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi W, Park SS, Shaw CJ, Withers MA, Patel PI, Lupski JR (2003) Reciprocal crossovers and a positional preference for strand exchange in recombination events resulting in deletion or duplication of chromosome 17p11.2. Am J Hum Genet 73:1302–1315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boerkoel CF, Inoue K, Reiter LT, Warner LE, Lupski JR (1999) Molecular mechanisms for CMT1A duplication and HNPP deletion. Ann NY Acad Sci 883:22–35 [PubMed] [Google Scholar]
- Cheung J, Estivill X, Khaja R, MacDonald JR, Lau K, Tsui LC, Scherer SW (2003) Genome-wide detection of segmental duplications and potential assembly errors in the human genome sequence. Genome Biol 4:R25 10.1186/gb-2003-4-4-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole TRP, Hughes HE (1994) Sotos syndrome: a study of the diagnostic criteria and natural history. J Med Genet 31:20–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Boer L, Kant S, Karperien M, van Beers L, Tjon Y, Vink G, van Tol D, Dauwerse H, Le Cessie S, Beemer FA, van der Burgt I, Hamel BCJ, Hennekam RC, Kuhnle U, Mathijssen IB, Veenstra-Knol HE, Stumpel CT, Breuning MH, Wit JM (2004) Genotype-phenotype correlation in patients suspected of having Sotos syndrome. Horm Res 62:197–207 10.1159/000081063 [DOI] [PubMed] [Google Scholar]
- Douglas J, Hanks S, Temple IK, Davies S, Murray A, Upadhyaya M, Tomkins S, Hughes HE, Cole TR, Rahman N (2003) NSD1 mutations are the major cause of Sotos syndrome and occur in some cases of Weaver syndrome but are rare in other overgrowth phenotypes. Am J Hum Genet 72:132–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichler EE (2001) Recent duplication, domain accretion and the dynamic mutation of the human genome. Trends Genet 17:661–669 10.1016/S0168-9525(01)02492-1 [DOI] [PubMed] [Google Scholar]
- Estivill X, Cheung J, Pujana MA, Nakabayashi K, Scherer SW, Tsui LC (2002) Chromosomal regions containing high-density and ambiguously mapped putative single nucleotide polymorphisms (SNPs) correlate with segmental duplications in the human genome. Hum Mol Genet 11:1987–1995 10.1093/hmg/11.17.1987 [DOI] [PubMed] [Google Scholar]
- Gimelli G, Pujana MA, Patricelli MG, Russo S, Giardino D, Larizza L, Cheung J, Armengol L, Schinzel A, Estivill X, Zuffardi O (2003) Genomic inversions of human chromosome 15q11-q13 in mothers of Angelman syndrome patients with class II (BP2/3) deletions. Hum Mol Genet 12:849–858 10.1093/hmg/ddg101 [DOI] [PubMed] [Google Scholar]
- Han LL, Keller MP, Navidi W, Chance PF, Arnheim N (2000) Unequal exchange at the Charcot-Marie-Tooth disease type 1A recombination hot-spot is not elevated above the genome average rate. Hum Mol Genet 9:1881–1889 10.1093/hmg/9.12.1881 [DOI] [PubMed] [Google Scholar]
- Inoue K, Lupski JR (2002) Molecular mechanisms for genomic disorders. Annu Rev Genomics Hum Genet 3:199–242 10.1146/annurev.genom.3.032802.120023 [DOI] [PubMed] [Google Scholar]
- Jeffreys AJ, May CA (2004) Intense and highly localized gene conversion activity in human meiotic crossover hot spots. Nat Genet 36:151–156 10.1038/ng1287 [DOI] [PubMed] [Google Scholar]
- Kamimura J, Endo Y, Kurotaki N, Kinoshita A, Miyake N, Shimokawa O, Harada N, Visser R, Ohashi H, Miyakawa K, Gerritsen J, Innes AM, Lagace L, Frydman M, Okamoto N, Puttinger R, Raskin S, Resic B, Culic V, Yoshiura K, Ohta T, Kishino T, Ishikawa M, Niikawa N, Matsumoto N (2003) Identification of eight novel NSD1 mutations in Sotos syndrome. J Med Genet 40:e126 10.1136/jmg.40.11.e126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasai M, Matsuzaki T, Katayanagi K, Omori A, Maziarz RT, Strominger JL, Aoki K, Suzuki K (1997) The translin ring specifically recognizes DNA ends at recombination hot spots in the human genome. J Biol Chem 272:11402–11407 10.1074/jbc.272.17.11402 [DOI] [PubMed] [Google Scholar]
- Kiyosawa H, Chance PF (1996) Primate origin of the CMT1A-REP repeat and analysis of a putative transposon-associated recombinational hotspot. Hum Mol Genet 5:745–753 10.1093/hmg/5.6.745 [DOI] [PubMed] [Google Scholar]
- Knight SJ, Lese CM, Precht KS, Kuc J, Ning Y, Lucas S, Regan R, Brenan M, Nicod A, Lawrie NM, Cardy DL, Nguyen H, Hudson TJ, Riethman HC, Ledbetter DH, Flint J (2000) An optimized set of human telomere clones for studying telomere integrity and architecture. Am J Hum Genet 67:320–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Hedges SB (1998) A molecular timescale for vertebrate evolution. Nature 392:917–920 10.1038/31927 [DOI] [PubMed] [Google Scholar]
- Kurotaki N, Harada N, Shimokawa O, Miyake N, Kawame H, Uetake K, Makita Y, et al (2003) Fifty microdeletions among 112 cases of Sotos syndrome: low copy repeats possibly mediate the common deletion. Hum Mutat 22:378–387 10.1002/humu.10270 [DOI] [PubMed] [Google Scholar]
- Kurotaki N, Imaizumi K, Harada N, Masuno M, Kondoh T, Nagai T, Ohashi H, Naritomi K, Tsukahara M, Makita Y, Sugimoto T, Sonoda T, Hasegawa T, Chinen Y, Tomita H, Kinoshita A, Mizuguchi T, Yoshiura K, Ohta T, Kishino T, Fukushima Y, Niikawa N, Matsumoto N (2002) Haploinsufficiency of NSD1 causes Sotos syndrome. Nat Genet 30:365–366 10.1038/ng863 [DOI] [PubMed] [Google Scholar]
- Lopes J, Ravise N, Vandenberghe A, Palau F, Ionasescu V, Mayer M, Levy N, Wood N, Tachi N, Bouche P, Latour P, Ruberg M, Brice A, LeGuern E (1998) Fine mapping of de novo CMT1A and HNPP rearrangements within CMT1A-REPs evidences two distinct sex-dependent mechanisms and candidate sequences involved in recombination. Hum Mol Genet 7:141–148 10.1093/hmg/7.1.141 [DOI] [PubMed] [Google Scholar]
- Lopes J, Tardieu S, Silander K, Blair I, Vandenberghe A, Palau F, Ruberg M, Brice A, LeGuern E (1999) Homologous DNA exchanges in humans can be explained by the yeast double-strand break repair model: a study of 17p11.2 rearrangements associated with CMT1A and HNPP. Hum Mol Genet 8:2285–2292 10.1093/hmg/8.12.2285 [DOI] [PubMed] [Google Scholar]
- Lopez-Correa C, Dorschner M, Brems H, Lazaro C, Clementi M, Upadhyaya M, Dooijes D, Moog U, Kehrer-Sawatzki H, Rutkowski JL, Fryns JP, Marynen P, Stephens K, Legius E (2001) Recombination hotspot in NF1 microdeletion patients. Hum Mol Genet 10:1387–1392 10.1093/hmg/10.13.1387 [DOI] [PubMed] [Google Scholar]
- Lupski JR (1998) Genomic disorders: structural features of the genome can lead to DNA rearrangements and human disease traits. Trends Genet 14:417–422 10.1016/S0168-9525(98)01555-8 [DOI] [PubMed] [Google Scholar]
- Miyake N, Kurotaki N, Sugawara H, Shimokawa O, Harada N, Kondoh T, Tsukahara M, Ishikiriyama S, Sonoda T, Miyoshi Y, Sakazume S, Fukushima Y, Ohashi H, Nagai T, Kawame H, Kurosawa K, Touyama M, Shiihara T, Okamoto N, Nishimoto J, Yoshiura K, Ohta T, Kishino T, Niikawa N, Matsumoto N (2003) Preferential paternal origin of microdeletions caused by prezygotic chromosome or chromatid rearrangements in Sotos syndrome. Am J Hum Genet 72:1331–1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai T, Matsumoto N, Kurotaki N, Harada N, Niikawa N, Ogata T, Imaizumi K, Kurosawa K, Kondoh T, Ohashi H, Tsukahara M, Makita Y, Sugimoto T, Sonoda T, Yokoyama T, Uetake K, Sakazume S, Fukushima Y, Naritomi K (2003) Sotos syndrome and haploinsufficiency of NSD1: clinical features of intragenic mutations and submicroscopic deletions. J Med Genet 40:285–289 10.1136/jmg.40.4.285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne LR, Li M, Pober B, Chitayat D, Bodurtha J, Mandel A, Costa T, Grebe T, Cox S, Tsui LC, Scherer SW (2001) A 1.5 million–base pair inversion polymorphism in families with Williams-Beuren syndrome. Nat Genet 29:321–325 10.1038/ng753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersson M, Bylund M, Alderborn A (2003) Molecular haplotype determination using allele-specific PCR and pyrosequencing technology. Genomics 82:390–396 10.1016/S0888-7543(03)00177-0 [DOI] [PubMed] [Google Scholar]
- Reiter LT, Hastings PJ, Nelis E, De Jonghe P, Van Broeckhoven C, Lupski JR (1998) Human meiotic recombination products revealed by sequencing a hotspot for homologous strand exchange in multiple HNPP deletion patients. Am J Hum Genet 62:1023–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter LT, Murakami T, Koeuth T, Pentao L, Muzny DM, Gibbs RA, Lupski JR (1996) A recombination hotspot responsible for two inherited peripheral neuropathies is located near a mariner transposon-like element. Nat Genet 12:288–297 10.1038/ng0396-288 [DOI] [PubMed] [Google Scholar]
- Rio M, Clech L, Amiel J, Faivre L, Lyonnet S, Le Merrer M, Odent S, Lacombe D, Edery P, Brauner R, Raoul O, Gosset P, Prieur M, Vekemans M, Munnich A, Colleaux L, Cormier-Daire V (2003) Spectrum of NSD1 mutations in Sotos and Weaver syndromes. J Med Genet 40:436–440 10.1136/jmg.40.6.436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H (2000) Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol 132:365–386 [DOI] [PubMed] [Google Scholar]
- Samonte RV, Eichler EE (2002) Segmental duplications and the evolution of the primate genome. Nat Rev Genet 3:65–72 10.1038/nrg705 [DOI] [PubMed] [Google Scholar]
- Schwartz S, Elnitski L, Li M, Weirauch M, Riemer C, Smit A, Green ED, Hardison RC, Miller W (2003) MultiPipMaker and supporting tools: alignments and analysis of multiple genomic DNA sequences. Nucleic Acids Res 31:3518–3524 10.1093/nar/gkg579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer LG, Lupski JR (2000) Molecular mechanisms for constitutional chromosomal rearrangements in humans. Annu Rev Genet 34:297–329 10.1146/annurev.genet.34.1.297 [DOI] [PubMed] [Google Scholar]
- Shaw CJ, Lupski JR (2004) Implications of human genome architecture for rearrangement-based disorders: the genomic basis of disease. Hum Mol Genet Suppl 13:R57–R64 10.1093/hmg/ddh073 [DOI] [PubMed] [Google Scholar]
- Shaw CJ, Withers MA, Lupski JR (2004) Uncommon deletions of the Smith-Magenis syndrome region can be recurrent when alternate low-copy repeats act as homologous recombination substrates. Am J Hum Genet 75:75–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimokawa O, Kurosawa K, Ida T, Harada N, Kondoh T, Miyake N, Yoshiura K, Kishino T, Ohta T, Niikawa N, Matsumoto N (2004) Molecular characterization of inv dup del(8p): analysis of five cases. Am J Med Genet 128A:133–137 [DOI] [PubMed] [Google Scholar]
- Stankiewicz P, Lupski JR (2002a) Genome architecture, rearrangements and genomic disorders. Trends Genet 18:74–82 10.1016/S0168-9525(02)02592-1 [DOI] [PubMed] [Google Scholar]
- ——— (2002b) Molecular-evolutionary mechanisms for genomic disorders. Curr Opin Genet Dev 12:312–319 10.1016/S0959-437X(02)00304-0 [DOI] [PubMed] [Google Scholar]
- Sugawara H, Harada N, Ida T, Ishida T, Ledbetter DH, Yoshiura K, Ohta T, Kishino T, Niikawa N, Matsumoto N (2003) Complex low-copy repeats associated with a common polymorphic inversion at human chromosome 8p23. Genomics 82:238–244 10.1016/S0888-7543(03)00108-3 [DOI] [PubMed] [Google Scholar]
- Tatton-Brown K, Douglas J, Coleman K, Cole T, Rahman N (2004) Clinical and molecular features of Sotos syndrome caused by microdeletions encompassing NSD1. Paper presented at the Clinical Genetics Society, Spring Conference, Oxford, March 24 [Google Scholar]
- Turkmen S, Gillessen-Kaesbach G, Meinecke P, Albrecht B, Neumann LM, Hesse V, Palanduz S, Balg S, Majewski F, Fuchs S, Zschieschang P, Greiwe M, Mennicke K, Kreuz FR, Dehmel HJ, Rodeck B, Kunze J, Tinschert S, Mundlos S, Horn D (2003) Mutations in NSD1 are responsible for Sotos syndrome, but are not a frequent finding in other overgrowth phenotypes. Eur J Hum Genet 11:858–865 10.1038/sj.ejhg.5201050 [DOI] [PubMed] [Google Scholar]
- Visser R, Matsumoto N (2003) Genetics of Sotos syndrome. Curr Opin Pediatr 15:598–606 10.1097/00008480-200312000-00010 [DOI] [PubMed] [Google Scholar]