Abstract
Repeat length of the CAG microsatellite polymorphism in exon 1 of the androgen receptor (AR) gene has been associated with risk of prostate cancer in humans. This association has been the focus of >20 primary epidemiological publications and multiple review articles, but a consistent and reproducible association has yet to be confirmed. We systematically addressed possible causes of false-negative and false-positive association in >4,000 individuals from a multiethnic, prospective cohort study of prostate cancer, comprehensively studying genetic variation by microsatellite genotyping, direct resequencing of exons in advanced cancer cases, and haplotype analysis across the 180-kb AR genomic locus. These data failed to confirm that common genetic variation in the AR gene locus influences risk of prostate cancer. A systematic approach that assesses both coding and noncoding genetic variation in large and diverse patient samples can help clarify hypotheses about association between genetic variants and disease.
Introduction
Irreproducibility is a major issue in association studies (Lohmueller et al. 2003), and many factors can contribute to both false-negative and false-positive results. Possible explanations for irreproducibility include false-negative results due to inadequately powered studies, with sample sizes that are small relative to the true effect size (Cardon and Bell 2001) and inadequate characterization of genetic variation at the locus of interest; false-positive results due to inappropriately generous P value thresholds (Wacholder et al. 2004); and the possibility of ethnic heterogeneity in causal mutations (due to unmeasured genetic or environmental confounders). It is vital to distinguish true associations from false-positive results: a confirmed genetic association provides proof that a given pathway plays an etiological role in disease and, as such, lays the groundwork both for the development of drugs for treatment and/or prevention of disease and for the development of tests that could guide prediction, prognosis, and choice of therapy. Because downstream biological and clinical follow-up of positive findings demand substantial investments of time and money, however, it is essential to rigorously demonstrate the validity of putative associations, lest investment be wasted on the basis of spurious claims of association.
One important example is that of an association between genetic variation in the androgen receptor (AR) gene and risk of prostate cancer. Androgens play a critical role in development of the prostate gland, and blockade of androgen signaling is a mainstay of prostate cancer treatment. In the human population, the AR gene sequence varies in a manner that influences protein function: rare and dramatic expansions (>40 repeats) of a CAG microsatellite polymorphism in the coding region of AR cause spinal and bulbar muscular atrophy (MIM 313200), whereas common and more modest variation in the length of this same CAG repeat (population mean repeats ± SD equals 22±3) influences transactivation activity in vitro (Mhatre et al. 1993; Chamberlain et al. 1994; Beilin et al. 2000). For these reasons, the CAG polymorphism has been extensively studied for association with prostate cancer in cohorts of various sizes, many of which claim positive results (Irvine et al. 1995; Giovannucci et al. 1997; Ingles et al. 1997; Stanford et al. 1997; Bratt et al. 1999; Correa-Cerro et al. 1999; Edwards et al. 1999; Ekman et al. 1999; Hsing et al. 2000; Lange et al. 2000; Xue et al. 2000; Beilin et al. 2001; Latil et al. 2001; Modugno et al. 2001; Panz et al. 2001; Balic et al. 2002; Chen et al. 2002; Gsur et al. 2002; Mononen et al. 2002; Huang et al. 2003; Santos et al. 2003; Cicek et al. 2004)—albeit without emergence of a single model of genotype-phenotype correlation as consistently associated (for recent reviews, see Nelson and Witte [2002] and Simard et al. [2003]). Although not every study reported a positive association, inability to replicate has been attributed to inadequate sample size and/or to poor study design in reports of negative association (Kantoff et al. 1998; Buchanan et al. 2001; Coetzee and Irvine 2002; Nelson and Witte 2002).
To help clarify the relationship between inherited variation in the AR locus and prostate cancer, we systematically evaluated the common genetic variation at the AR locus in a large, multiethnic cohort of patients with sporadic prostate cancer.
Materials and Methods
The Multiethnic Cohort Study
Case and control subjects in the prostate cancer study were the male participants, from Hawaii and Los Angeles, in the Multiethnic Cohort Study (MEC). Details of the study have been published elsewhere (Kolonel et al. 2000). Briefly, >200,000 men and women, aged 45–75 years and residing in Hawaii and California, completed a questionnaire that included requests for data on demographic characteristics (including self-reported ancestry), lifestyle, health behaviors, and diagnoses, as well as a comprehensive dietary survey. Response rates were 72% for cases and 70% for controls.
Participants in the MEC are followed for incident cancers by computer linkage of the cohort with the Surveillance, Epidemiology, and End Results (SEER) cancer registries in Hawaii and Los Angeles, as well as with the California Cancer Registry. A total of 2,036 subjects with prostate cancer and 2,160 controls are included in the present study. Only incident cases from the African American, Japanese, Latino, and white sub-cohorts are included this study. A random sample of MEC participants was selected for blood collection, to serve as a cohort-based control for genetic analyses.
SNP Genotyping
SNPs were identified in both the public (dbSNP) and private (Celera) databases. The Sequenom MassArray system at Broad Institute was used to genotype the SNPs, as described elsewhere (Gabriel et al. 2002). In brief, primers and probes were designed for each SNP by the SpectroDesign software (FASTA sequences and primers and probes are available on request). Multiplex PCR was performed in 5-μl volumes that contain 0.1 U of Taq polymerase (Amplitaq Gold, Applied Biosystems [ABI]), 5 ng genomic DNA, 2.5 pmol of each PCR primer, and 2.5 μmol of dNTP. Thermocycling was at 95°C for 15 min, followed by 45 cycles of 95°C for 20 s, 56°C for 30 s, and 72°C for 30 s. Unincorporated dNTPs were deactivated using 0.3 U of shrimp alkaline phosphatase (Roche), followed by primer extension by use of 5.4 pmol of each primer extension probe, 50 μM of the appropriate dNTP/ddNTP combination, and 0.5 units of Thermosequenase (Amersham Pharmacia). Reactions were cycled at 94°C for 2 min, followed by 40 cycles of 94°C for 5 s, 50°C for 5 s, and 72°C for 5 s. After addition of a cation-exchange resin to remove residual salt from the reactions, ∼7 nl of the purified primer-extension reaction was loaded onto a matrix pad (3-hydroxypicolinic acid) of a SpectroCHIP (Sequenom). SpectroCHIPs were analyzed using a Bruker Biflex III MALDI-TOF mass spectrometer (SpectroREADER [Sequenom]) and were spectra processed using SpectroTYPER (Sequenom). Successful genotyping assays were defined as those in which 75% of all possible genotyping calls were obtained. Although we used 75% as a minimum threshold, we obtained an average of 90.5% for all genotypes attempted for each successful SNP.
Sequencing
The exons of the AR gene were sequenced in DNA from 88 subjects with advanced prostate cancer, including 22 individuals from each of the four self-described ethnic groups—African American, white, Japanese, and Latino. Primers were designed to capture the full exonic sequence. First, amplification of the target site was performed. We used 22.5 ng of genomic DNA as template DNA. Each reaction consisted of a final volume of 30 μl; the final concentration for each reagent was 1.5 mM of MgCl2, 10× PCR Buffer II, 0.3 μl of 10mM dNTPs, 5 U/μl of AmpliTaq Gold, 0.017 μM mixed forward and reverse primers, and sterile water. This reaction was cycled under the following conditions: 96°C for 10 min, 35 cycles of 96°C for 30 s, 50°C for 2 min, and 72°C for 2 min, followed by 72°C for 2 min. We amplified sequences with PCR primers tailed with standard M13 sequencing sites (−21 forward and −28 reverse) and performed conventional dye-primer sequencing on ABI 377 sequencers. Sequences were base-called by the Phred program and assembled by the Phrap program, and polymorphism candidates were identified by the PolyPhred program. All results were visually inspected by at least two observers to confirm or refute the automated genotyping call.
Microsatellite Genotyping
Primers (5′-TCCAGAATCTGTTCCAGAGCGTGC-3′ and 5′-GCTGTGAAGGTTGCTGTTCCTCAT-3′) were used to PCR amplify the region flanking the CAG microsatellite repeat in exon 1 of the AR. One of the primers was 5′-modified with the fluorescent dye FAM. Products were analyzed by fragment analysis on an ABI 3730. The final reaction volume per reaction was 6 μl and comprised 5 ng of template DNA combined with 10× PCR buffer, 0.8 mM MgCl2, 0.2 mM dNTPs, 0.8 μM each of forward and reverse primers, 0.04 μl of HotStarTaq (Qiagen), and water (units given are for the final concentration). This mixture was placed in a thermal cycler (MJ Research) with the following settings: 92°C for 15 min; 94°C for 20 s, 56°C for 30 s, 72°C for 1 min, and 45 cycles at 72°C for 3 min. After cycling, each product was diluted 15-fold, and 2 μl of this dilution was added to each well. Then, 8 μl of Genescan 500-LIZ size standard:formamide mix (1:33) was added to each well. Products were electrophoresed on an ABI 3730 capillary sequencer. The data were analyzed by Genemapper software that automatically called fragment sizes relative to the ladder, to account for any migration artifact.
Known CAG lengths were sequenced for 60 individuals. The 60 individuals were split into two groups of 30—one group was used as a training set to derive the best fit for calculating the number of CAG repeats from the fragment length. The second group of 30 individuals was used as a test set to test the equation derived from the training set. We found that the equation y(number of CAG repeats)=0.3682 × (fragment length)−80.577 gave 100% accuracy.
Haplotype-Block Definition and Haplotype-Phase Determination
Haplotype blocks were determined using criteria described by Gabriel et al. (2002), on the basis of a composite of pairwise D′ values (Lewontin 1964). Empirical evidence shows that regions that meet the block definition of Gabriel et al. (2002) and that contain at least six SNPs have two properties: <5% of SNP pairs within such regions show strong evidence for recombination, and, within such regions, haplotype diversity is typically low.
Because the AR resides on the X chromosome and we studied men, haplotypes could be directly observed from genotype data without the need for statistical phasing. Missing data were resolved by use of the EM algorithm (Excoffier and Slatkin 1995).
Statistical Analysis
Unconditional logistic regression was used to analyze the resultant case and control data (SAS statistical software, version 8.0 [SAS Institute]). Analyses were conducted using the genetic data alone, adjusted for race/ethnicity as well as for stratification on race/ethnicity and disease severity. The haplotype analyses were performed by comparison of one haplotype with all others (reported in the “Results” section) as well as by use of the most common haplotype as a reference haplotype (data not shown). Both methods yielded similar nonsignificant results. Disease severity for cases was categorized into either “local” or “advanced” disease. “Local” was defined as “local disease with a well- or moderately differentiated grade”; “advanced” was used to mean “either local disease with a poorly differentiated grade” or “regional or distant disease.” Significance levels are reported two-sided and have not been corrected for multiple-hypothesis testing. The odds ratio (OR) is the ratio of the odds in favor of exposure among the cases to the odds in favor of exposure among the controls.
Among controls, analysis of variance was used to evaluate differences in CAG microsatellite repeat length across self-described ethnic groups. Unconditional logistic regression was used to model the association between repeat length and risk of prostate cancer. Repeat length was modeled using several approaches (continuous variable and cut point at <22 and <23 repeats) on the basis of previous literature. Analyses were adjusted for race/ethnicity, when combined. Analysis was also conducted to explore the role of disease severity and age with the association between repeat length and risk of prostate cancer. Both categorical and linear models were fit for the CAG repeat analysis; a categorical model did not provide a better fit to the data (P=.98).
For sequencing, the formula used for the power to detect a variant was: Power =1-(1-p)n, where p is the allele frequency and n is the number of chromosomes.
Results
We first studied the CAG-repeat polymorphism in exon 1 of the AR gene in 4,196 individuals from the prospectively collected MEC: 2,036 subjects with incident prostate cancer and 2,160 ethnically matched cohort controls (see tables A1 and A2 [online only] for descriptive characteristics). To our knowledge, this is the largest study of this variant in sporadic prostate cancer (nearly four times the size of the previous largest study) as well as the first to examine a large African American population, who suffer from particularly high rates of prostate cancer (Kolonel et al. 2000; Quinn and Babb 2002; SEER).
As a primary analysis, we used unconditional logistic regression to examine the relationship between CAG-repeat length (as a continuous variable) and risk of prostate cancer. We found no evidence of heterogeneity among the ethnic groups, with regard to measures of association (P>.05) (table 1), and thus present data pooled across ethnic groups with adjustment for age and ethnicity as covariate.
Table 1.
Association Analysis of CAG Repeat Polymorphism (Continuous Variable), Stratified by Self-Reported Ethnicity
|
Findings by Ethnic Group |
||||||||||||
| African American |
Japanese |
Latino |
White |
|||||||||
| Subjects | ORa | 95% CI | P | ORa | 95% CI | P | ORa | 95% CI | P | ORa | 95% CI | P |
| All cases | 1.014 | .982–1.047 | .40 | 1.006 | .962–1.052 | .79 | 1.015 | .978–1.055 | .43 | 1.032 | .986–1.080 | .17 |
| Advanced cases | .983 | .937–1.032 | .50 | 1.064 | 1.001–1.130 | .05 | .993 | .943–1.047 | .8 | 1.055 | .987–1.126 | .11 |
| Local cases | 1.022 | .985–1.060 | .24 | .972 | .923–1.024 | .28 | 1.028 | .984–1.073 | .22 | 1.017 | .964–1.073 | .87 |
Adjusted for age.
We failed to detect a nominally significant association between CAG length and prostate cancer risk (OR 1.016 per CAG decrement; 95% CI 0.997–1.036; P=.11) (table 2). Because the previous literature on AR and prostate cancer provided a prior hypothesis as to the direction of the effect (that shorter repeat lengths are associated with increased risk), our result could be considered a one-sided test with a P value of .055 and thus borderline significant for replication. Because the study sample is large, however, our results provide very tight CIs on the estimate of risk: on the basis of our result, for example, we expect that, on repeated sampling, an OR between 0.99 and 1.036 would be observed 95% of the time.
Table 2.
Continuous and Cut Point Analyses of Prostate Cancer Risk with Decreased CAG-Repeat Length[Note]
|
Findings for CAG-Repeat Length |
||||||||||
| Continuous |
<22 versus ⩾22 |
<23 versus ⩾23 |
||||||||
| Subjects | No. of Controls/Cases | ORa | 95% CI | Pb | ORa | 95% CI | Pb | ORa | 95% CI | Pb |
| All cases | 2,160/2,036 | 1.016 | .997–1.036 | .11 | 1.084 | .954–1.231 | .22 | 1.115 | .981–1.267 | .09 |
| Advanced cases | 2,160/686 | 1.015 | .988–1.044 | .28 | 1.094 | .913–1.310 | .33 | 1.122 | .936–1.345 | .21 |
| Local cases | 2,160/1,239 | 1.013 | .991–1.036 | .25 | 1.068 | .922–1.238 | .38 | 1.087 | .937–1.261 | .27 |
Note.— There is no statistically significant association between the CAG-microsatellite polymorphism and prostate cancer risk for the entire population or when stratified by stage. Multiple analyses were performed, by using two cut points (22 and 23) as well as by modeling the CAG-repeat length as a continuous variable. These analyses demonstrate no significant association between prostate cancer risk and CAG-repeat length.
Adjusted for race/ethnicity and age.
Two-sided P value.
The literature that examines association of the AR and prostate cancer includes many different models for analysis of genotype (with consideration of repeat length as a threshold variable, for example, rather than as a continuous variable) and phenotype (with consideration of association analysis on a particular age at onset or status, such as advanced disease) (Giovannucci et al. 1997; Bratt et al. 1999). On the hypothesis that one of these models might provide a more reproducible and substantial association, we examined, as a secondary analysis, a variety of these models, including different cut points for repeat length (<22/22+ and <23/23+) and age at diagnosis. We found no significant evidence for association between the CAG polymorphism and cancer risk in any previously described model of genotype-phenotype correlation (tables 2 and A3 [online only]). The results in each ethnic subgroup (table 1) are neither nominally significant nor statistically distinguishable from the null result of the pooled sample. (On the causal hypothesis that the CAG repeat influences—either directly or by modification—the risk of prostate cancer, and absent any as-yet-uncharacterized gene-by-environment or gene-by-gene interaction, we would expect an effect of CAG length in all ethnic groups examined—and thus the best powered and most accurate estimate—to be that provided by all participants in the study adjusted for ethnicity.)
Given this very limited evidence for association between the CAG repeat and prostate cancer risk, we next considered the possibility that previous positive results had been due to linkage disequilibrium (LD) between the CAG repeat and some other (as-yet-undetected) causal variation in the AR gene region. To evaluate this possibility, we directly examined the coding region of the AR gene to search for protein-altering mutations, and we performed a detailed haplotype analysis spanning the AR gene region, on the hypothesis that noncoding (presumed regulatory) variants might play a role.
We resequenced each of the eight exons of the AR gene in 88 men with advanced prostate cancer: 22 each from the self-reported white, African American, Japanese, and Latino ethnic groups. Two synonymous changes (G213G in exon 1 and K581K in exon 2) were identified, but no novel amino acid–altering variants were found. This sample size provides 90% power to detect coding changes present in cases at a frequency of 3% or higher (in this pooled sample), which suggests that germline protein-altering mutations in the AR are rarely encountered, even in patients with advanced prostate cancer.
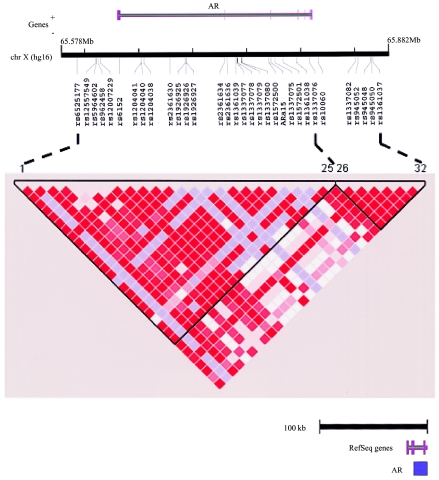
We next examined the hypothesis that there might be common noncoding variants that influence risk of prostate cancer (e.g., by alteration of gene expression). Because it is not yet possible to recognize regulatory variants from primary sequence data, and, given the large size of the AR gene region, complete resequencing in each case and control subject was impractical. For this reason, we used a haplotype-based approach to survey the 274 kb surrounding the AR gene locus. In total, 32 polymorphic SNPs (from the dbSNP and Celera databases) were genotyped in each of 1,756 people (882 cases; 874 controls) (fig. 1A). (Since the AR is on the X chromosome, and since our analysis considered males only, haplotype phase was directly observable from the genotype data.) The genotype data show two regions characterized by strong LD, which meets criteria for haplotype “blocks” as calibrated elsewhere (Gabriel et al. 2002) (fig. 1B). More than 93% of haplotypes matched one of the 4–8 haplotypes in each block, with a frequency of >1% in the multiethnic sample (data not shown). Given the number of markers typed, the strong LD observed, and the fact that we had directly resequenced the coding regions, we believe that the marker density achieved thoroughly captures the common genetic variation across these 274 kb around the AR gene locus.
Figure 1.
SNPs and LD across the AR locus. A, Physical relationship of SNPs to the AR locus (from University of California–Santa Cruz Genome Bioinformatics build hg16); 32 working polymorphic SNPs were genotyped across the AR locus, which spanned a total of 275 kb—the SNPs covered from 56 kb upstream to 37 kb downstream of the AR gene. Using previously defined criteria (see the “Materials and Methods” section), these 32 markers describe two strong blocks of LD (188 kb and 66 kb) with gaps between adjacent blocks (bounded by the markers selected) of 11.9 kb. B, LD plot for entire population across the AR locus. Each square in the plot represents the pairwise LD relationships among all 32 markers. The two blocks (see the “Materials and Methods” section) are highlighted and are bounded by markers 1–25 and 26–32. Red denotes strong LD (as measured by D′) with a high degree of statistical confidence, pale blue denotes a high D′ with relatively low statistical confidence, and white denotes low D′.
We examined association between prostate cancer risk and each SNP and common haplotype in this subset of men. No convincing association was observed (tables 3 and 4). The best results were two SNPs with a nominal P value of .01, a value not unexpected by chance, given 32 SNPs tested for association with two phenotypes (table 3). A combination of these results with the exon-resequencing data suggests that if common genetic variation at the AR influences risk, it would be found outside the coding region and demonstrate no LD with the well-defined haplotype structure examined.
Table 3.
OR, 95% CI, and P Value for the Minor Allele of Each AR SNP and Risk of Prostate Cancer (All Racial/Ethnic Groups Combined)[Note]
|
Findings for the Minor Allele of Each AR SNP and Risk of Prostate Cancer |
||||||
| All Cases |
Advanced-Stage Cases |
|||||
| SNP | ORa | 95% CI | Pb | ORa | 95% CI | Pb |
| rs6525177 | .93 | .61–1.42 | .75 | .89 | .50–1.57 | .69 |
| rs12557549 | 2.32 | .44–12.31 | .32 | 2.49 | .33–19.03 | .38 |
| rs5964602 | .90 | .60–1.36 | .61 | .85 | .49–1.48 | .56 |
| rs962458 | 1.22 | .79–1.89 | .36 | 1.40 | .80–2.46 | .24 |
| rs12007229 | .75 | .48–1.18 | .21 | .51 | .26–1.01 | .05 |
| rs6152 | 1.15 | .79–1.66 | .47 | 1.24 | .77–2.0 | .37 |
| rs1204041 | .61 | .36–1.03 | .07 | .42 | .20–.90 | .02 |
| rs1204040 | 1.20 | .80–1.79 | .37 | 1.09 | .64–1.86 | .76 |
| rs1204038 | .88 | .59–1.33 | .55 | .85 | .49–1.46 | .56 |
| rs2361630 | 1.17 | .81–1.71 | .41 | 1.29 | .79–2.12 | .31 |
| rs1926925 | .58 | .33–1.02 | .06 | .35 | .15–.86 | .02 |
| rs1926926 | .55 | .32–.96 | .04 | .29 | .11–.72 | .01 |
| rs1926927 | .54 | .31–.95 | .03 | .33 | .14–.81 | .01 |
| rs2361634 | .62 | .26–1.48 | .28 | 1.09 | .39–3.0 | .88 |
| rs2361636 | .46 | .25–.86 | .01 | .28 | .10–.75 | .01 |
| rs1361039 | 1.35 | .29–6.39 | .70 | 2.26 | .35–14.48 | .39 |
| rs1337077 | .63 | .36–1.10 | .11 | .41 | .18–.96 | .04 |
| rs1337078 | .58 | .33–1.02 | .06 | .40 | .17–.92 | .03 |
| rs1337079 | .57 | .32–.99 | .05 | .34 | .14–.81 | .02 |
| rs1337080 | .99 | .62–1.57 | .96 | .83 | .45–1.55 | .56 |
| rs1572500 | .54 | .30–.97 | .04 | .39 | .16–.96 | .04 |
| ARa15 | 4.63 | .47–45.9 | .19 | 6.18 | .52–74.03 | .15 |
| rs1337075 | .56 | .31–1.02 | .06 | .34 | .13–.86 | .02 |
| rs1572501 | 3.19 | .66–15.42 | .15 | .87 | .08–10.13 | .91 |
| rs1361038 | 1.12 | .57–2.19 | .75 | .85 | .34–2.11 | .73 |
| rs1337076 | 1.07 | .63–1.82 | .81 | .88 | .44–1.78 | .73 |
| rs10060 | 1.01 | .63–1.63 | .97 | .96 | .52–1.78 | .89 |
| rs1337082 | .96 | .65–1.41 | .84 | .90 | .54–1.49 | .68 |
| rs945052 | .98 | .59–1.61 | .93 | .85 | .44–1.64 | .62 |
| rs945048 | .94 | .57–1.54 | .80 | .77 | .39–1.50 | .43 |
| rs945050 | .87 | .61–1.25 | .45 | .81 | .51–1.31 | .40 |
| rs1361037 | 1.10 | .67–1.83 | .71 | .89 | .45–1.76 | .74 |
Note.— All 32 SNPs across the AR are individually tested for association with prostate cancer in the entire MEC population.
Adjusted for age and race/ethnicity.
P values are two-sided, with no correction for multiple testing.
Table 4.
Association of AR Haplotypes with Risk of Prostate Cancer[Note]
|
Frequency (%) among |
|||||
| LD Block (No. of Cases/Controls) and Haplotype | Cases | Controls | OR | 95% CI | P |
| 1 (843/805): | |||||
| GCCGCAGAATACAAGGGAAAGCCTA | 12.3 | 12.1 | 1.05 | .77–1.44 | .76 |
| GCCGCAGAATACAAGGGAAGGCCTA | 9.6 | 10.1 | .96 | .68–1.34 | .79 |
| GCCGCAAAATACAAGGGAAGGCCTG | 5.7 | 6.3 | .89 | .59–1.36 | .59 |
| GCCGCAAAATACAAGGGAAGGCCCG | 4.0 | 2.5 | 1.73 | .98–3.06 | .06 |
| GCCAAAGAATACAAGGGAAAGCCTA | 3.0 | 2.5 | 1.25 | .65–2.40 | .51 |
| GCCAAGACACTTGAAGAGGATCTTA | 9.4 | 12.2 | .75 | .54–1.04 | .09 |
| ACAACGGCGCACAAGGGAAAGCCTA | 49.4 | 47.1 | 1.12 | .85–1.48 | .43 |
| ACAACGGCGCACAGGGGAAAGCCTA | 1.2 | 1.5 | .73 | .29–1.81 | .49 |
| 2 (876/863): | |||||
| GAGCACA | 21.7 | 24.3 | .88 | .70–1.12 | .30 |
| GAACATA | 49.6 | 46.7 | 1.09 | .83–1.42 | .53 |
| GGGTTTG | 10.2 | 9.7 | 1.15 | .80–1.65 | .46 |
| TGGTTTG | 16.7 | 16.4 | 1.09 | .81–1.46 | .56 |
Note.— Haplotypes are compared against all other haplotypes (see the “Materials and Methods” section). No associations between common haplotype and prostate cancer are observed.
Discussion
One of the most vexing aspects of genetic-association studies is the interpretation of results that prove inconsistent on subsequent data collection and analysis. Because confirmed associations are of great value in understanding pathways of disease, initial claims often attract substantial attention. When subsequent studies fail to reproduce results, it is difficult for investigators in both the human genetic and biomedical communities to agree on an interpretation of the data.
The growing literature relating genetic variation in the AR gene to epidemiological risk of prostate cancer is one such example. The AR is a compelling biological candidate gene, and initial studies that reported association were well designed and highly suggestive of an effect of the CAG-repeat variation on prostate cancer risk. Since later reports collectively examined a variety of methods of analysis and failed to reach a single unified model of genotype-phenotype correlation, there has not been a clear and consistent picture of the impact of this gene on prostate cancer risk. We hypothesized that, by using a large sample and systematically examining genetic variation across the region, we might help clarify this association. The results were only marginally suggestive for association of the CAG repeat (treated as a continuous variable) and prostate cancer risk, with a maximal effect size smaller than previously suggested.
What might explain the differences between our results and those published elsewhere? Because our study is substantially larger than the studies that claimed more strongly positive effects (Giovannucci et al. 1997; Stanford et al. 1997; Mononen et al. 2002), it is unlikely that our study represents a false-negative result for a true association. Because we thoroughly searched coding regions (by direct resequencing) and noncoding regions (by haplotype analysis), it appears unlikely that we missed a true result because of an as-yet-undiscovered common variant in the AR gene. Thus, our data suggest either that the AR gene locus has (1) no effect on prostate cancer risk, (2) an effect that is extremely small (<4% per CAG repeat), or (3) an effect that is evident only in certain (and as yet unidentified) subgroups of patients that are based on an unmeasured genotypic, environmental, or behavioral modifier. If the effect is small but truly present, then even-larger samples (such as those of the National Cancer Institute Cohort Consortium for Breast and Prostate Cancer) will be needed to fully address the contribution of such variants to prostate cancer risk. If the effect is seen only in a subgroup of patients, it will be difficult to demonstrate unless and until the unmeasured confounding factors are identified.
Another class of a hypothesis for which the previous effect was true—and which can explain nonreplication—is that the definition and clinical characteristics of prostate cancer may have changed over time. This hypothesis is supported by the dramatic change in population screening brought on by widespread testing of serum levels of prostate-specific antigen (PSA). Since some of the positive studies were reported prior to widespread PSA testing, it is possible that failure of replication could be secondary to a shift in the spectrum and characteristics of prostate cancer cases detected by PSA, as compared with prostate cancer cases ascertained prior to PSA testing.
Obviously, we must consider the possibility that previous positive studies represent false-positive results due to statistical fluctuation or to unrecognized population stratification. Many of the previous studies showed only modest statistical significance, and no single model of genotype-phenotype correlation has consistently been observed across many studies. Given the low Bayesian prior probability that any one of the ∼30,000 genes in the human genome plays a role in disease (even a strong biological candidate like AR in prostate cancer) and the modest P values obtained elsewhere, false-positive results are not unexpected (Hirschhorn and Altshuler 2002; Lohmueller et al. 2003; Wacholder et al. 2004).
False-positive association due to population stratification is another possibility, and it is particularly relevant in studies of prostate cancer (Kittles et al. 2002; Freedman et al. 2004). Prostate cancer is more common in African Americans than in Americans of self-described European ancestry, and African Americans have a lower average repeat length at the exon 1 CAG than do whites (Edwards et al. 1992; Sartor et al. 1999; the present study). Thus, unrecognized population substructure could lead to spurious association between CAG-repeat length and prostate cancer risk (Pritchard and Rosenberg 1999; Kittles et al. 2002; Freedman et al. 2004). Whether the samples studied in the past contained cryptic substructure—and whether any such effect would have been large enough to have influenced the prior literature—has yet to be determined.
The causes of irreproducibility in any particular association of genotype and phenotype are difficult to identify and can be attributed to biological, statistical, or technical reasons. From a practical point of view, however, those associations that are robust and reproducible are the most certain to be biologically valid and of relevance to patients in the average clinical setting. The combination of a large, diverse patient sample and systematic evaluation of genetic variation in exons (by resequencing) and surrounding genomic regions (by haplotype analysis) provides a route for clarification of correlations between genotype and phenotype and addresses each possible cause of false-positive and false-negative association. Development of a set of standards for performing such validation studies will be increasingly crucial as reports of genetic association proliferate in the years to come.
Acknowledgments
We thank our colleagues at Massachusetts General Hospital and in the Program in Medical and Population Genetics at the Broad Institute, particularly Mark Daly, Stacey Gabriel, and the genotyping team. D.A. is a Charles E. Culpeper Scholar of the Rockefeller Brothers Fund and a Burroughs Wellcome Fund Clinical Scholar in Translational Research. J.N.H. is a recipient of a Burroughs Wellcome Career Award in the Biomedical Sciences. M.L.F. is supported by a Howard Hughes Medical Institute physician postdoctoral fellowship and Department of Defense Health Disparity Training–Prostate Scholar Award DAMD 17-02-1-0246. This work was supported by National Cancer Institute grant 5 R01 CA 63464: Genetic Susceptibility to Cancer in Multiethnic Cohorts.
Appendix A
Table A1.
Descriptive Characteristics of the MEC
|
No. of MEC Subjects |
||
| Characteristic | Controls | Cases |
| Ethnicity: | ||
| African American | 664 | 635 |
| Japanese | 476 | 422 |
| Latino | 574 | 576 |
| White | 446 | 403 |
| Age (in years): | ||
| <60 | 246 | 223 |
| 60–64 | 305 | 285 |
| 65–70 | 608 | 578 |
| >70 | 1,001 | 950 |
| Disease severity: | ||
| High | 686 | |
| Low | 1,239 | |
Table A2.
Distribution of AR CAG-Repeat Length for Subjects with Prostate Cancer and Control Individuals
|
Findings for |
||||||||||
| African Americans |
Japanese |
Latinos |
Whites |
All |
||||||
| CAG-Repeat Length | Cases(n=637) | Controls(n=664) | Cases(n=431) | Controls(n=476) | Cases(n=576) | Controls(n=574) | Cases(n=405) | Controls(n=446) | Cases(n=2,049) | Controls(n=2,160) |
| Mean | 19.95 | 20.11 | 22.72 | 22.8 | 22.53 | 22.67 | 21.74 | 22.02 | 21.61 | 21.78 |
| SD | 3.39 | 3.39 | 3.03 | 2.98 | 2.88 | 3.21 | 3.03 | 2.93 | 3.31 | 3.36 |
| Median | 19 | 20 | 23 | 23 | 23 | 23 | 21 | 22 | 22 | 22 |
| Range | 12–33 | 11–41 | 12–38 | 12–33 | 12–33 | 12–38 | 12–34 | 12–35 | 12–38 | 11–41 |
Table A3.
Analyses of Continuous and Cut Point CAG-Repeat Length, Stratified by Age at Diagnosis of Prostate Cancer
|
Findings for CAG-Repeat Length as Variable |
||||||
| Continuous |
<22 versus ⩾22 |
<23 versus ⩾23 |
||||
| Age at Diagnosis(years) | ORa | 95% CI | ORa | 95% CI | ORa | 95% CI |
| 40–64 | 1.009 | .971–1.048 | 1.042 | .810–1.340 | 1.037 | .805–1.337 |
| ⩾65 | 1.019 | .996–1.042 | 1.098 | .947–1.272 | 1.142 | .985–1.323 |
Adjusted for race/ethnicity and age.
Electronic-Database Information
The URLs for data presented herein are as follows:
- Celera Discovery System, http://www.celeradiscoverysystem.com/index.cfm
- dbSNP Database, http://www.ncbi.nlm.nih.gov/SNP/
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for spinal and bulbar muscular atrophy)
- Surveillance, Epidemiology, and End Results (SEER), http://seer.cancer.gov/faststats/html/inc_prost.html
- University of California–Santa Cruz Genome Bioinformatics, http://genome.ucsc.edu/
References
- Balic I, Graham ST, Troyer DA, Higgins BA, Pollock BH, Johnson-Pais TL, Thompson IM, Leach RJ (2002) Androgen receptor length polymorphism associated with prostate cancer risk in Hispanic men. J Urol 168:2245–2248 10.1097/00005392-200211000-00107 [DOI] [PubMed] [Google Scholar]
- Beilin J, Ball EM, Favaloro JM, Zajac JD (2000) Effect of the androgen receptor CAG repeat polymorphism on transcriptional activity: specificity in prostate and non-prostate cell lines. J Mol Endocrinol 25:85–96 [DOI] [PubMed] [Google Scholar]
- Beilin J, Harewood L, Frydenberg M, Mameghan H, Martyres RF, Farish SJ, Yue C, Deam DR, Byron KA, Zajac JD (2001) A case-control study of the androgen receptor gene CAG repeat polymorphism in Australian prostate carcinoma subjects. Cancer 92:941–949 [DOI] [PubMed] [Google Scholar]
- Bratt O, Borg A, Kristoffersson U, Lundgren R, Zhang QX, Olsson H (1999) CAG repeat length in the androgen receptor gene is related to age at diagnosis of prostate cancer and response to endocrine therapy, but not to prostate cancer risk. Br J Cancer 81:672–676 10.1038/sj.bjc.6690746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchanan G, Irvine RA, Coetzee GA, Tilley WD (2001) Contribution of the androgen receptor to prostate cancer predisposition and progression. Cancer Metastasis Rev 20:207–223 10.1023/A:1015531326689 [DOI] [PubMed] [Google Scholar]
- Cardon LR, Bell JI (2001) Association study designs for complex diseases. Nat Rev Genet 2:91–99 10.1038/35052543 [DOI] [PubMed] [Google Scholar]
- Chamberlain NL, Driver ED, Miesfeld RL (1994) The length and location of CAG trinucleotide repeats in the androgen receptor N-terminal domain affect transactivation function. Nucleic Acids Res 22:3181–3186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Lamharzi N, Weiss NS, Etzioni R, Dightman DA, Barnett M, DiTommaso D, Goodman G (2002) Androgen receptor polymorphisms and the incidence of prostate cancer. Cancer Epidemiol Biomarkers Prev 11:1033–1040 [PubMed] [Google Scholar]
- Cicek MS, Conti DV, Curran A, Neville PJ, Paris PL, Casey G, Witte JS (2004) Association of prostate cancer risk and aggressiveness to androgen pathway genes: SRD5A2, CYP17, and the AR. Prostate 59:69–76 10.1002/pros.10358 [DOI] [PubMed] [Google Scholar]
- Coetzee G, Irvine R (2002) Size of the androgen receptor CAG repeat and prostate cancer: does it matter? J Clin Oncol 20:3572–3573 [DOI] [PubMed] [Google Scholar]
- Correa-Cerro L, Wohr G, Haussler J, Berthon P, Drelon E, Mangin P, Fournier G, Cussenot O, Kraus P, Just W, Paiss T, Cantu JM, Vogel W (1999) (CAG)nCAA and GGN repeats in the human androgen receptor gene are not associated with prostate cancer in a French-German population. Eur J Hum Genet 7:357–362 10.1038/sj.ejhg.5200298 [DOI] [PubMed] [Google Scholar]
- Edwards A, Hammond HA, Jin L, Caskey CT, Chakraborty R (1992) Genetic variation at five trimeric and tetrameric tandem repeat loci in four human population groups. Genomics 12:241–253 10.1016/0888-7543(92)90371-X [DOI] [PubMed] [Google Scholar]
- Edwards SM, Badzioch MD, Minter R, Hamoudi R, Collins N, Ardern-Jones A, Dowe A, Osborne S, Kelly J, Shearer R, Easton DF, Saunders GF, Dearnaley DP, Eeles RA (1999) Androgen receptor polymorphisms: association with prostate cancer risk, relapse and overall survival. Int J Cancer 84:458–465 [DOI] [PubMed] [Google Scholar]
- Ekman P, Gronberg H, Matsuyama H, Kivineva M, Bergerheim US, Li C (1999) Links between genetic and environmental factors and prostate cancer risk. Prostate 39:262–268 [DOI] [PubMed] [Google Scholar]
- Excoffier L, Slatkin M (1995) Maximum-likelihood estimation of molecular haplotype frequencies in a diploid population. Mol Biol Evol 12:921–927 [DOI] [PubMed] [Google Scholar]
- Freedman ML, Reich D, Penney KL, McDonald GJ, Mignault AA, Patterson N, Gabriel SB, Topol EJ, Smoller JW, Pato CN, Pato MT, Petryshen TL, Kolonel LN, Lander ES, Sklar P, Henderson B, Hirschhorn JN, Altshuler D (2004) Assessing the impact of population stratification on genetic association studies. Nat Genet 36:388–393 10.1038/ng1333 [DOI] [PubMed] [Google Scholar]
- Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, Higgins J, DeFelice M, Lochner A, Faggart M, Liu-Cordero SN, Rotimi C, Adeyemo A, Cooper R, Ward R, Lander ES, Daly MJ, Altshuler D (2002) The structure of haplotype blocks in the human genome. Science 296:2225–2229 10.1126/science.1069424 [DOI] [PubMed] [Google Scholar]
- Giovannucci E, Stampfer MJ, Krithivas K, Brown M, Dahl D, Brufsky A, Talcott J, Hennekens CH, Kantoff PW (1997) The CAG repeat within the androgen receptor gene and its relationship to prostate cancer. Proc Natl Acad Sci USA 94:3320–3323 10.1073/pnas.94.7.3320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gsur A, Preyer M, Haidinger G, Zidek T, Madersbacher S, Schatzl G, Marberger M, Vutuc C, Micksche M (2002) Polymorphic CAG repeats in the androgen receptor gene, prostate-specific antigen polymorphism and prostate cancer risk. Carcinogenesis 23:1647–1651 10.1093/carcin/23.10.1647 [DOI] [PubMed] [Google Scholar]
- Hirschhorn JN, Altshuler D (2002) Once and again: issues surrounding replication in genetic association studies. J Clin Endocrinol Metab 87:4438–4441 10.1210/jc.2002-021329 [DOI] [PubMed] [Google Scholar]
- Hsing AW, Gao YT, Wu G, Wang X, Deng J, Chen YL, Sesterhenn IA, Mostofi FK, Benichou J, Chang C (2000) Polymorphic CAG and GGN repeat lengths in the androgen receptor gene and prostate cancer risk: a population-based case-control study in China. Cancer Res 60:5111–5116 [PubMed] [Google Scholar]
- Huang SP, Chou YH, Chang WS, Wu MT, Yu CC, Wu T, Lee YH, Huang JK, Wu WJ, Huang CH (2003) Androgen receptor gene polymorphism and prostate cancer in Taiwan. J Formos Med Assoc 102:680–686 [PubMed] [Google Scholar]
- Ingles SA, Ross RK, Yu MC, Irvine RA, La Pera G, Haile RW, Coetzee GA (1997) Association of prostate cancer risk with genetic polymorphisms in vitamin D receptor and androgen receptor. J Natl Cancer Inst 89:166–170 10.1093/jnci/89.2.166 [DOI] [PubMed] [Google Scholar]
- Irvine RA, Yu MC, Ross RK, Coetzee GA (1995) The CAG and GGC microsatellites of the androgen receptor gene are in linkage disequilibrium in men with prostate cancer. Cancer Res 55:1937–1940 [PubMed] [Google Scholar]
- Kantoff P, Giovannucci E, Brown M (1998) The androgen receptor CAG repeat polymorphism and its relationship to prostate cancer. Biochim Biophys Acta 1378:C1–C5 [DOI] [PubMed] [Google Scholar]
- Kittles RA, Chen W, Panguluri RK, Ahaghotu C, Jackson A, Adebamowo CA, Griffin R, Williams T, Ukoli F, Adams-Campbell L, Kwagyan J, Isaacs W, Freeman V, Dunston GM (2002) CYP3A4-V and prostate cancer in African Americans: causal or confounding association because of population stratification? Hum Genet 110:553–560 10.1007/s00439-002-0731-5 [DOI] [PubMed] [Google Scholar]
- Kolonel LN, Henderson BE, Hankin JH, Nomura AM, Wilkens LR, Pike MC, Stram DO, Monroe KR, Earle ME, Nagamine FS (2000) A multiethnic cohort in Hawaii and Los Angeles: baseline characteristics. Am J Epidemiol 151:346–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange EM, Chen H, Brierley K, Livermore H, Wojno KJ, Langefeld CD, Lange K, Cooney KA (2000) The polymorphic exon 1 androgen receptor CAG repeat in men with a potential inherited predisposition to prostate cancer. Cancer Epidemiol Biomarkers Prev 9:439–442 [PubMed] [Google Scholar]
- Latil AG, Azzouzi R, Cancel GS, Guillaume EC, Cochan-Priollet B, Berthon PL, Cussenot O (2001) Prostate carcinoma risk and allelic variants of genes involved in androgen biosynthesis and metabolism pathways. Cancer 92:1130–1137 [DOI] [PubMed] [Google Scholar]
- Lewontin RC (1964) The interaction of selection and linkage. I. General considerations; heterotic models. Genetics 49:49–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohmueller KE, Pearce CL, Pike M, Lander ES, Hirschhorn JN (2003) Meta-analysis of genetic association studies supports a contribution of common variants to susceptibility to common disease. Nat Genet 33:177–182 10.1038/ng1071 [DOI] [PubMed] [Google Scholar]
- Mhatre AN, Trifiro MA, Kaufman M, Kazemi-Esfarjani P, Figlewicz D, Rouleau G, Pinsky L (1993) Reduced transcriptional regulatory competence of the androgen receptor in X-linked spinal and bulbar muscular atrophy. Nat Genet 5:184–188 10.1038/ng1093-184 [DOI] [PubMed] [Google Scholar]
- Modugno F, Weissfeld JL, Trump DL, Zmuda JM, Shea P, Cauley JA, Ferrell RE (2001) Allelic variants of aromatase and the androgen and estrogen receptors: toward a multigenic model of prostate cancer risk. Clin Cancer Res 7:3092–3096 [PubMed] [Google Scholar]
- Mononen N, Ikonen T, Autio V, Rokman A, Matikainen MP, Tammela TL, Kallioniemi OP, Koivisto PA, Schleutker J (2002) Androgen receptor CAG polymorphism and prostate cancer risk. Hum Genet 111:166–171 10.1007/s00439-002-0776-5 [DOI] [PubMed] [Google Scholar]
- Nelson KA, Witte JS (2002) Androgen receptor CAG repeats and prostate cancer. Am J Epidemiol 155:883–890 10.1093/aje/155.10.883 [DOI] [PubMed] [Google Scholar]
- Panz VR, Joffe BI, Spitz I, Lindenberg T, Farkas A, Haffejee M (2001) Tandem CAG repeats of the androgen receptor gene and prostate cancer risk in black and white men. Endocrine 15:213–216 10.1385/ENDO:15:2:213 [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Rosenberg NA (1999) Use of unlinked genetic markers to detect population stratification in association studies. Am J Hum Genet 65:220–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn M, Babb P (2002) Patterns and trends in prostate cancer incidence, survival, prevalence and mortality. II. Individual countries. BJU Int 90:174–184 10.1046/j.1464-410X.2002.02823.x [DOI] [PubMed] [Google Scholar]
- Santos ML, Sarkis AS, Nishimoto IN, Nagai MA (2003) Androgen receptor CAG repeat polymorphism in prostate cancer from a Brazilian population. Cancer Detect Prev 27:321–326 10.1016/S0361-090X(03)00106-5 [DOI] [PubMed] [Google Scholar]
- Sartor O, Zheng Q, Eastham JA (1999) Androgen receptor gene CAG repeat length varies in a race-specific fashion in men without prostate cancer. Urology 53:378–380 10.1016/S0090-4295(98)00481-6 [DOI] [PubMed] [Google Scholar]
- Simard J, Dumont M, Labuda D, Sinnett D, Meloche C, El-Alfy M, Berger L, Lees E, Labrie F, Tavtigian SV (2003) Prostate cancer susceptibility genes: lessons learned and challenges posed. Endocr Relat Cancer 10:225–259 [DOI] [PubMed] [Google Scholar]
- Stanford JL, Just JJ, Gibbs M, Wicklund KG, Neal CL, Blumenstein BA, Ostrander EA (1997) Polymorphic repeats in the androgen receptor gene: molecular markers of prostate cancer risk. Cancer Res 57:1194–1198 [PubMed] [Google Scholar]
- Wacholder S, Chanock S, Garcia-Closas M, El Ghormli L, Rothman N (2004) Assessing the probability that a positive report is false: an approach for molecular epidemiology studies. J Natl Cancer Inst 96:434–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue W, Irvine RA, Yu MC, Ross RK, Coetzee GA, Ingles SA (2000) Susceptibility to prostate cancer: interaction between genotypes at the androgen receptor and prostate-specific antigen loci. Cancer Res 60:839–841 [PubMed] [Google Scholar]