Abstract
Generalized vitiligo is a common, autoimmune, familial-clustering depigmentary disorder of the skin and hair that results from selective destruction of melanocytes. Generalized vitiligo is likely a heterogeneous disease, with five susceptibility loci reported so far—on chromosomes 1p31, 6p21, 7q, 8p, and 17p13—in white populations. To investigate vitiligo susceptibility loci in the Chinese population, we performed a genomewide linkage analysis in 57 multiplex Chinese families, each with at least two affected siblings, and we identified interesting linkage evidence on 1p36, 4q13-q21, 6p21-p22, 6q24-q25, 14q12-q13, and 22q12. Subsequently, to extract more linkage information, we investigated our initial genomewide linkage findings in a follow-up analysis of 49 new families and additional markers. Our initial genomewide linkage analysis and our subsequent follow-up analysis have identified a novel linkage to vitiligo on 4q13-q21, with highly significant linkage evidence (a nonparametic LOD score of 4.62 [P=.000003] and a heterogeneity LOD score of 4.01, under a recessive inheritance model), suggesting that 4q13-q21 likely harbors a major susceptibility locus for vitiligo in the Chinese population. We observed a minimal overlap between the linkage results of our current genomewide analysis in the Chinese population and the results of previous analyses in white populations, and we thus hypothesize that, as a polygenic disorder, vitiligo may be associated with great genetic heterogeneity and a substantial difference in its genetic basis between ethnic populations.
Generalized vitiligo (MIM 193200) is a common acquired autoimmune disorder of skin and hair that results from selective destruction of melanocytes (Cho et al. 2000). It is characterized by the appearance of sharply delimited patches of white skin overlying hair, oral mucosa, and occasionally the eyes, which are due to noninflammatory loss of pigment-forming melanocytes in affected areas. Vitiligo is associated with autoimmune disorders such as hypothyroidism, diabetes mellitus, chronic active hepatitis, pernicious anemia, and adrenal insufficiency (Kovacs 1998). The population prevalence of vitiligo ranges from 0.1% to 2% and shows a wide variability among ethnic groups (Bolognia et al. 1998; Hann and Nordlund 2000). For example, whereas the estimated population prevalence of vitiligo is ∼0.38% for whites in the United States and northern Europe (Howitz et al. 1977), vitiligo affects only ∼0.19% of the population in China (Xu et al. 2002).
Genetic risk for vitiligo is well supported by multiple lines of evidence. Vitiligo is frequently associated with familial clustering (Mehta et al. 1973; Carnevale et al. 1980; Goudie et al. 1983; Hafez et al. 1983; Das et al. 1985; Majumder et al. 1993; Alkhateeb et al. 2003), and ∼20% of probands have at least one affected first-degree relative (Alkhateeb et al. 2003). The risk for first-degree relatives of patients with vitiligo to develop the disease is elevated by 7- to 10-fold (Nath et al. 1994) compared with the risk for the general population. Similarly, our recent study of the Chinese population indicated that ∼1.8% of patients’ first-degree relatives were affected with vitiligo, which was 9-fold higher than the prevalence rate in general population (Zhang et al. 2004b). In addition, segregation analysis suggested that vitiligo is a multifactorial and polygenic disorder that likely results from multiple genetic and environmental factors (Arcos-Burgos et al. 2002; Alkhateeb et al. 2003; Zhang et al. 2004b). However, no disease genes have been identified for vitiligo so far.
Several genomewide linkage analyses of vitiligo have been performed in the past few years, and multiple linkages to vitiligo have been identified (Nath et al. 2001; Alkhateeb et al. 2002; Fain et al. 2003; Spritz et al. 2004). Using 16 European American pedigrees with cosegregation of systemic lupus erythematosus and vitiligo, Nath et al. (2001) performed the first genomewide linkage analysis of vitiligo and identified a significant linkage on 17p13. Shortly afterwards, Spritz and his colleagues performed a series of genomewide linkage analyses of vitiligo in white families. Their initial genomewide scan was done in a three-generation multiplex family with cosegregation of vitiligo and Hashimoto thyroiditis, and they identified a candidate gene with highly significant linkage at a locus (named “AIS1”) on chromosome 1p32.2-p31.3 (Alkhateeb et al. 2002). Subsequently, they performed a follow-up genomewide linkage analysis by studying 70 additional white families (Fain et al. 2003). Their follow-up analysis confirmed the original linkage finding at the AIS1 locus with highly significant linkage evidence (nonparametric LOD [NPL] score of 5.56) and identified additional linkage evidence on chromosomes 1, 7, 8, 11, 19, and 22. More recently, the group performed an extended genomewide linkage analysis, using 102 multiplex families (including the 70 families used in the previous genomewide scan) (Spritz et al. 2004). The linkage results from the 102 families reinforced the strong support for the AIS1 locus and also confirmed the previously suggestive linkage findings on chromosomes 7q and 8p (AIS2 and AIS3). The study also provided supporting evidence for a disease locus on chromosome 17, which likely corresponds to the SLEV1 locus identified by Nath et al. (2001). Interestingly, by stratifying their 102 families into autoimmunity- and nonautoimmunity-associated groups, Spritz and colleagues (2004) found that, whereas the linkage evidence at the AIS1, AIS2, and SLEV1 loci was mainly from the autoimmunity-associated families, the evidence at the AIS3 locus was primarily from the nonautoimmunity-associated families, suggesting that generalized vitiligo might be divided into two distinct phenotypic subcategories that involve different disease loci or alleles. In addition to the genomewide linkage analyses, population-based association analyses were used to investigate several candidate genes for vitiligo—for example, CTLA-4 (Kemp et al. 1999; Blomhoff et al. 2005), CAT and TAP1 (Casp et al. 2002, 2003), MC1R and ASIP (Na et al. 2003), ACE (Jin et al. 2004), and HLA (Zamani et al. 2001; Tastan et al. 2004; Zhang et al. 2004a).
Here, we report the first genomewide linkage analysis of vitiligo in a Chinese population. Our family collection includes 106 Chinese multiplex families, each with at least two siblings affected with generalized vitiligo (table 1). All the families were recruited by experienced dermatologists from the Department of Dermatology at First Affiliated Hospital of Anhui Medical University at Hefei, Anhui, China, and from the Vitiligo Clinic of the Railway Hospital at Xiangfan, Hubei, China. The diagnosis of generalized vitiligo was made on the basis of the patient’s history and the presence of typical clinical features (discrete, well-circumscribed depigmented patches). Phenotypes were carefully determined by history, lesion maps, and, in most cases, physical examination and/or photographs. Any individual whose phenotype was questionable was excluded from the study. Only patients with clear signs of acquired patches on the extremities, trunk, genitalia, central face, or other areas were scored as affected. The mean age at onset of the 286 affected individuals (148 male [51.7%] and 138 female [48.3%]) from the 106 families was 18.6 years (range 1–63 years). Of the 106 families, 57 families were used in an initial genomewide linkage analysis, and 49 families were used in a follow-up linkage analysis in which the initial linkage findings from the genomewide scan were further investigated by use of dense marker coverage (table 1). For each family, blood samples were collected from all affected individuals, their parents, and additional family members connecting affected individuals. Among the 106 families, blood samples were collected from both parents in 81 families and from one parent in 17 families. Eight families were missing samples from both parents of affected individuals. When samples from one or both parents of an affected individual were unavailable, a blood sample was collected from at least one additional unaffected sibling. Informed consent was obtained from each recruited subject. The study was approved by the Ethical Committee of the Chinese National Human Genome Center at Shanghai.
Table 1.
Summary of Analyses of Families
|
Total No. of |
No. of Families by No. of Affecteds(Total No. of Affecteds) |
||||
| Analysis | Families | Affecteds | Two Affecteds | Three Affecteds | Four or More Affecteds |
| Initial genomewide scan | 57 | 172 | 23 (46) | 17 (51) | 17 (75) |
| Follow-up | 49 | 114 | 35 (70) | 12 (36) | 2 (8) |
| Combined | 106 | 286 | 58 (116) | 29 (87) | 19 (83) |
Genomic DNA was extracted from whole blood by use of a simple salting-out procedure, as described elsewhere (Miller et al. 1988). We performed a genomewide linkage analysis of vitiligo by genotyping 382 microsatellite markers from the ABI Prism Linkage Mapping Set (version 2) in 57 families. Average marker spacing is 8.85 cM (range 1–19 cM), and average marker information content is 0.72, on the basis of genotypes from the 57 families. All the marker positions were based on an interpolated genetic map that incorporates the information from a physical map (build 34.3) and from published deCODE and Marshfield genetic maps (see David Duffy’s QIMR Homepage). All the markers were genotyped in multiplex PCR, in accordance with the manufacturer's guidelines (Applied Biosystems). Before being used in linkage analysis, all genotyping data were subjected to quality checking by use of the programs PedCheck (O’Connell and Weeks 1998) and MERLIN (Abecasis et al. 2002). Problematic genotypes either were corrected by rechecking original allele callings or were removed if allele status could not be reliably determined.
Both nonparametric and parametric linkage analyses were performed using the program GENEHUNTER 2.1 (Kruglyak et al. 1996). For nonparametric analysis, the NPL score was calculated using the Sall statistic to capture the information about allele sharing between all affected individuals in a pedigree. For parametric analysis, the heterogeneity LOD (HLOD) score was calculated by assuming a dominant or recessive inheritance of the disease allele and a disease-allele frequency of 0.01, consistent with the population prevalence of 0.19% in China (Xu et al. 2002). Moreover, to reduce the impact of the nonpenetrant disease allele on linkage analysis, genomewide parametric analysis was performed by using an “affecteds-only” approach, in which all normal individuals were treated as “unknown” instead of “unaffected,” and by assuming a simple genetic model with high penetrance (0.99) and very low phenocopy rates (0.001). Marker-allele frequency was estimated on the basis of founders’ genotypes of either 57 families (for the genomewide analysis) or 106 families (for the follow-up analysis) by use of the program Linkage 5.10 (Lathrop and Lalouel 1984).
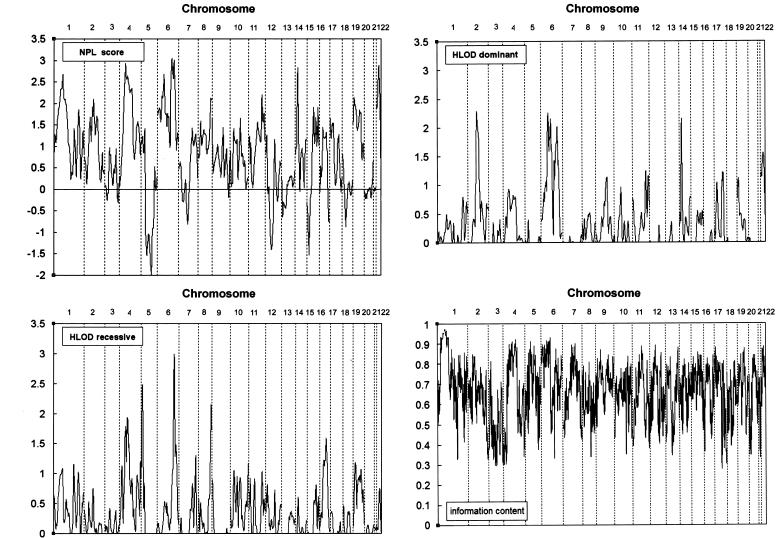
Multipoint nonparametric analysis identified linkage signals on 1p36, 4q13-q21, 6p21-p22, 6q24-q25, 14q12-q13, and 22q12 (fig. 1). Of these, the highest NPL score was 3.05 (P=.0017), identified at the marker interval D6S308–D6S441 on 6q24-q25; this was followed by NPL scores of 2.94 (P=.0021) at the marker interval D4S1592–D4S1534 on 4q13-q21, 2.87 (P=.0023) at the marker interval D22S280–D22S283 on 22q12, 2.83 (P=.0026) at the marker interval D14S275–D14S70 on 14q12-13, 2.68 (P=.0044) at the marker interval D6S422–D6S273 on 6p21-p22, and 2.67 (P=.0045) at the marker interval D1S234–D1S2885 on 1p36. Multipoint parametric linkage analysis under a recessive model of inheritance identified suggestive evidence for linkage (HLOD score of 1.86 or higher) (Lander and Kruglyak 1995) on 4q12-q13, 5p15, 6q24-q25, and 8q24 (fig. 1). The most significant HLOD score under a recessive model was 2.99 (α=36%) at the marker interval D6S308–D6S441, which was followed by multipoint HLOD scores of 2.48 (α=34%) at the marker interval D5S1981–D5S406, 2.15 (α=38%) at the marker interval D8S284–D8S272, and 1.94 (α=28%) at the marker interval D4S1592–D4S392. Under a dominant model of inheritance, multipoint parametric analysis identified suggestive linkage evidence on 2q13, 6p21-p22, 14q12-q13, and 22q12 (fig. 1). The highest HLOD score of 2.28 (α=59%) was identified at marker D2S160, which was followed by HLOD scores of 2.26 (α=37%) at the marker interval D6S422–D6S273, 2.15 (α=48%) at the marker interval D14S275–D14S70, and 2.07 (α=46%) at the marker interval D6S308–D6S441. For both nonparametric and parametric genomewide linkage analyses, two-point LOD scores (data not shown) were consistent with the multipoint LOD scores but were generally lower.
Figure 1.
Summary plot of the multipoint NPL and HLOD scores on 22 chromosomes from the genomewide linkage analysis of 382 microsatellite markers in 57 Chinese multiplex families with vitiligo.
Our genomewide linkage analysis of vitiligo in the 57 Chinese families identified interesting linkage signals in six genomic regions. The most significant evidence for linkage was on 6q24-q25 and was supported by a maximum multipoint NPL score of 3.05 and a recessive HLOD score of 2.99. The second-most significant evidence was on 4q13-q21, with a maximum multipoint NPL score of 2.94 and a recessive HLOD score of 1.94, which was followed by linkage evidence on 6p21-p22, with a maximum multipoint NPL score of 2.67 and a dominant HLOD score of 2.26. Linkage evidence was also identified on 1p36, 14q12-q13, and 22q12, but the evidence was less significant (table 2). To further investigate these initial linkage findings, we analyzed an additional 49 Chinese families recruited from the same clinics in a follow-up analysis in which additional markers were genotyped to extract more linkage information. Instead of using the “affecteds-only” approach, we performed the follow-up parametric analysis of the initial linkage findings by assuming a disease-allele frequency of 0.01, a variable penetrance rate (range 0.5–0.99), and a fixed phenotype rate of 0.001 and by using the inheritance model under which the initial linkage evidence was identified in the genomewide analysis. Changing the penetrance rate made almost no impact on HLOD score results (data not shown); therefore, only HLOD scores under the penetrance rate of 0.99 were reported.
Table 2.
Summary of the Multipoint LOD Scores from the Initial Genomewide and Follow-up Linkage Analyses
|
Initial Analysisa |
Follow-Up Analysisb |
Combined Analysisc |
||||||||||||
| Chromosome | Marker or Interval | NPL | P | HLOD | α (%) | NPL | P | HLOD | α (%) | NPL | P | HLOD | α (%) | Genetic Modeld |
| 1p36 | D1S2674–D1S2885 | 2.67 | .0045 | 1.08 | 21 | .98 | .16 | .09 | 4.5 | 2.37 | .0093 | 1.17 | 11 | Rec |
| 4q13-q21 | D4S1592–D4S1534 | 2.94 | .0021 | 1.94 | 28 | 4.29 | .00001 | 2.74 | 35 | 4.62 | .000003 | 4.01 | 31 | Rec |
| 6p21-p22 | D6S289–D6S291 | 2.68 | .0044 | 2.26 | 37 | 2.17 | .016 | 1.32 | 45 | 3.16 | .00092 | 2.14 | 32 | Dom |
| 6q24-q25 | D6S308–D6S441 | 3.05 | .0017 | 2.99 | 36 | 1.27 | .11 | .31 | 11 | 2.98 | .0016 | 1.37 | 16 | Rec |
| 14q12-q13 | D14S70 | 2.83 | .0026 | 2.15 | 48 | .048 | .48 | 0 | 0 | 2.17 | .014 | 1.51 | 34 | Dom |
| 22q12 | D22S280–D22S283 | 2.87 | .0023 | 1.55 | 33 | .47 | .32 | .0066 | .038 | 1.75 | .039 | .72 | 19 | Dom |
Performed by analyzing 382 microsatellite markers in 57 multiplex Chinese families.
Performed by analyzing various numbers of markers in 49 multiplex Chinese families.
Performed by analyzing various numbers of markers in 106 multiplex Chinese families.
Rec = recessive inheritance; Dom = dominant inheritance.
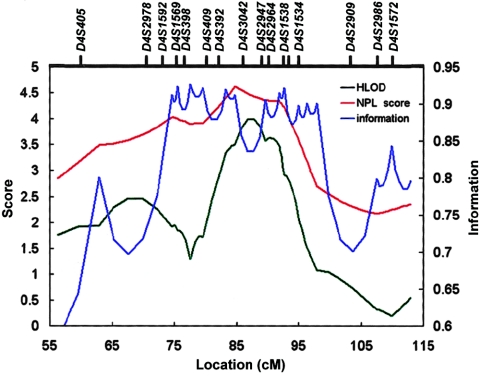
The initial linkage finding on 4q13-q21 was further investigated by genotyping 12 new and 5 original microsatellite markers in 106 families, which increased the marker density to an average marker spacing of 3.3 cM. We first performed a joint linkage analysis of 106 families, including 57 original and 49 new families. Multipoint linkage analysis of 17 markers yielded a maximum NPL score of 4.62 (P=.000003) at the marker interval D4S392–D4S3042 and a maximum HLOD score of 4.01 (α=31%) at the adjacent marker interval D4S3042–D4S2947, under a recessive model of inheritance (fig. 2 and table 2). The multipoint LOD score results were supported by a two-point analysis that yielded a maximum two-point NPL score of 4.53 (P=.000004) and a recessive HLOD score of 5.97 (α=42%) at the marker locus D4S392. Both nonparametric and parametric LOD scores of the joint linkage analysis of the 106 families on 4q13-q21 surpassed the suggested genomewide criteria for significant linkage evidence (Lander and Kruglyak 1995). In addition, we also performed an independent linkage analysis of the same 17 markers in 49 new families by using the same genetic model. The linkage analysis of the 49 new families provided independent supporting evidence for linkage, yielding a multipoint NPL score of 4.29 (P=.00001) and an HLOD score of 2.74 (α=35%) at the same genetic position. Therefore, our initial genomewide and subsequent fine-mapping linkage analyses have identified a novel linkage to vitiligo on 4q13-q21.
Figure 2.
Multipoint LOD scores on 4q13-q21 from the joint linkage analysis of 106 families. Multipoint NPL scores (red line); multipoint HLOD scores, under a recessive model of inheritance (black line); and marker information content (blue line) are plotted.
The two-point HLOD score at D4S392 is considerably higher than the multipoint HLOD score. This is not unexpected, because two-point linkage analysis is known to be more prone than multipoint analysis to inflated or deflated LOD scores. Marker information content at the D4S392 locus is 0.61 in our families, which is much lower than the marker information content of ∼0.9 that was achieved in our multipoint linkage analysis (fig. 2). A less informative marker, which is more likely to be present in two-point analysis than in multipoint analysis, can cause partial pedigrees to become uninformative at the marker locus. When partial linked or unlinked pedigrees become uninformative, the LOD score will be deflated or inflated, respectively. This problem is largely responsible for the phenomenon of having strong evidence for linkage to a single marker, without evidence, or with much weaker evidence, for the flanking markers.
The follow-up linkage analysis on 6p21-p22 was performed by analyzing nine new and six original markers in the 106 families and yielded a multipoint NPL score of 3.16 (P=.00092) at the marker interval D6S422–D6S1660 and a parametric HLOD score of 2.14 (α=32%) at the adjacent marker interval D6S1584– D6S422, under the dominant model of inheritance (table 2). Two-point nonparametric and parametric linkage analyses also provided supporting evidence (data not shown). Therefore, the follow-up linkage analysis of additional families and markers increased the NPL score from 2.67 to 3.16, and the enhanced linkage evidence was only slightly below the genomewide criteria for “suggestive linkage” (Lander and Kruglyak 1995; Nyholt 2000).
Our linkage finding on 6p21-p22 is consistent with the suggested autoimmune pathogenesis of vitiligo and the involvement of the major histocompatibility complex (MHC) locus in genetic predisposition to vitiligo. Autoimmune pathogenesis of vitiligo has been suggested by both clinical and immunological observations (Kovacs 1998; Ongenae et al. 2003; Le Poole et al. 2004). For example, vitiligo is associated with other autoimmune disorders in many patients. Immunological studies also demonstrated that autoantibodies to melanocytes can be detected, and depigmentation of skin is often accompanied by T-cell infiltration to the skin in many patients with vitiligo (Wankowicz-Kalinska et al. 2003). Moreover, repigmenting therapies are often associated with an immunosuppressive effect. All of these observations provide direct and indirect supporting evidence for an autoimmunity-mediated pathogenesis of vitiligo. Because of the extensive pathogenic association of the MHC locus with a large number of autoimmune disorders (Horton et al. 2004), the involvement of the MHC genes in the pathogenesis of vitiligo was suspected and was intensively investigated by genetic association analysis. Candidate gene–based genetic association analyses of vitiligo in Chinese (Zhang et al. 2004a) and white populations (Venneker et al. 1992; Zamani et al. 2001; Arcos-Burgos et al. 2002; de Vijlder et al. 2004; Tastan et al. 2004) have revealed multiple evidence for association between alleles of the MHC genes and vitiligo. Therefore, our linkage finding on 6p21-p22 was probably not unexpected, and it provides additional linkage evidence for the involvement of the MHC region in genetic risk of vitiligo. Moreover, our result also supports the suggestion that vitiligo shares some genetic risk factors with other autoimmune disorders (Spritz et al. 2004).
The linkage signal on 1p36 from the genomewide analysis was further investigated by analyzing six new and five original markers in 106 families. The follow-up linkage analysis of the 106 families yielded a multipoint NPL score of 2.37 (P=.009) and a multipoint recessive HLOD score of 1.17 at the marker interval D1S2734–D1S234 (table 2), which failed to strengthen the original linkage evidence on 1p36. Our moderate linkage evidence on chromosome 1p is at least 30 cM away from the AIS1 locus that was identified in the white population (Alkhateeb et al. 2002; Fain et al. 2003; Spritz et al. 2004). Therefore, our linkage analysis in Chinese families did not provide supporting evidence for this previously identified linkage on 1p in the white population. The linkage findings on 6q24-q25, 14q12-q13, and 22q12 were also further investigated in the 106 families, but none of these linkage findings was enhanced by the follow-up analysis of additional families (table 2).
Our linkage findings in the Chinese population show a minimal overlap with the previous linkage findings in white populations. Except for the 6p21-p22 region, our findings (highly significant linkage on 4q13-q21 and moderate linkage on 1p36, 6q24-q25, 14q12-q13, and 22q12) are all novel and were not reported in previous genomewide linkage analyses of white populations. Furthermore, previously identified strong linkage evidence on 1p31 (AIS1) (Alkhateeb et al. 2002; Fain et al. 2003; Spritz et al. 2004), 7q (AIS2), and 8p (AIS3) (Spritz et al. 2004) in the white population received little, if any, supporting evidence from our current linkage analysis of Chinese families. Such little overlap between the linkage findings in Chinese and white populations is very intriguing and might suggest that vitiligo is associated with strong genetic heterogeneity and that it involves different genetic risk factors in different ethnic populations. Meanwhile, we still need to keep in mind that it is possible that the current and previous linkage findings are largely false positives, which could account for the small amount of overlap in linkage findings. However, considering the high statistical significance of our linkage finding on 4q13-q21 in Chinese families and the previous linkage findings on 1p31 (AIS1), 7q (AIS2), and 8p (AIS3) in white families, we reason that this possibility is unlikely, although it can not be totally ruled out. Moreover, we hypothesize that there may be a significant difference in the genetic basis of vitiligo between Chinese and white populations. Such a difference in linkage results between Chinese and white populations can also be seen in studies of psoriasis, another autoimmune skin disorder (Gudjonsson et al. 2004), in which the 4q28-q31 region was recently recognized as a strong candidate region for hosting a susceptibility gene for psoriasis (Bowcock 2004). Linkage to psoriasis on 4q28-q31 was identified with highly significant evidence in a genomewide linkage analysis of Chinese families (Zhang et al. 2002) but not in any of five previous genomewide linkage analyses of white families. Independent supporting evidence for this linkage emerged only after the five white population–based genomewide linkage studies were pooled together in a meta-analysis (Sagoo et al. 2004). This suggests that, although the as-yet-unidentified susceptibility gene within the 4q28-q31 region is a major susceptibility gene that contributes significantly to genetic risk of psoriasis in the Chinese population, it probably contributes much less in the white population and thus can only be detected by the analysis of a very large number of families. An interesting question is whether the same is true for the linkage to vitiligo on 4q13-q21. Additional linkage studies in Chinese and white populations will likely shed light on this question.
In summary, we have performed the first genomewide linkage study of vitiligo in a Chinese population. We have identified linkage evidence in several genomic regions, and the evidence on 4q13-q21 has been confirmed by our follow-up analysis of additional new families. Not only has the overall linkage evidence from the joint linkage analysis of the 106 families surpassed the genomewide criteria for “significant linkage,” but the linkage results from the follow-up linkage analysis of the 49 new families also have provided independent evidence for linkage. Our linkage results strongly suggest that the 4q13-q21 region likely harbors a major susceptibility locus for vitiligo and should be targeted as a candidate for disease-gene discovery.
Acknowledgments
This work was funded by grants from the Chinese High Tech Program (863) (2003AA227030, 2002BA711A10, 2001AA227031, and 2001AA224021), the Shanghai Science and Technology Committee (03DJ14008), and the Chinese Ministry of Education (2003). We are grateful to all the families who participated in this study.
Electronic-Database Information
The URLs for data presented herein are as follows:
- David Duffy’s QIMR Homepage, http://www2.qimr.edu.au/davidD/
- MERLIN, http://www.sph.umich.edu/csg/abecasis/Merlin/index.html
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for vitiligo)
References
- Abecasis GR, Cherny SS, Cookson WO, Cardon LR (2002) Merlin-rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet 30:97–101 10.1038/ng786 [DOI] [PubMed] [Google Scholar]
- Alkhateeb A, Fain PR, Thody T, Bennett DC, Spritz RA (2003) Vitiligo and associated autoimmune diseases in Caucasian probands and their families. Pigment Cell Res 16:208–214 10.1034/j.1600-0749.2003.00032.x [DOI] [PubMed] [Google Scholar]
- Alkhateeb A, Stetler GL, Old W, Talbert J, Uhlhorn C, Taylor M, Fox A, Miller C, Dills DG, Ridgway EC, Bennett DC, Fain PR, Spritz RA (2002) Mapping of an autoimmunity susceptibility locus (AIS1) to chromosome 1p31.3-p32.2. Hum Mol Genet 11:661–667 10.1093/hmg/11.6.661 [DOI] [PubMed] [Google Scholar]
- Arcos-Burgos M, Parodi E, Salgar M, Bedoya E, Builes JJ, Jaramillo D, Ceballos G, Uribe A, Rivera N, Rivera D, Fonseca I, Camargo M, Palacio LG (2002) Vitiligo: complex segregation and linkage disequilibrium analyses with respect to microsatellite loci spanning the HLA. Hum Genet 110:334–342 10.1007/s00439-002-0687-5 [DOI] [PubMed] [Google Scholar]
- Blomhoff A, Helen Kemp E, Gawkrodger DJ, Weetman AP, Husebye ES, Akselsen HE, Lie BA, Undlien DE (2005) CTLA4 polymorphisms are associated with vitiligo, in patients with concomitant autoimmune diseases. Pigment Cell Res 18:55–58 10.1111/j.1600-0749.2004.00196.x [DOI] [PubMed] [Google Scholar]
- Bolognia JL, Nordlund JJ, Ortonne J-P (1998) Vitiligo vulgaris. In: Nordlund JJ, Boissy RE, Hearing VJ, King RA, Ortonne J-P (eds) The pigmentary system. Oxford University Press, New York, pp 513–551 [Google Scholar]
- Bowcock AM (2004) Psoriasis genetics: the way forward. J Invest Dermatol 122:xv–xvii 10.1111/j.0022-202X.2004.22627.x [DOI] [PubMed] [Google Scholar]
- Carnevale A, Zavala C, Castillo VD, Maldonado RR, Tamayo L (1980) Analisis genetico de 127 families con vitiligo. Rev Invest Clin 32:37–41 [PubMed] [Google Scholar]
- Casp CB, She JX, McCormack WT (2002) Genetic association of the catalase gene (CAT) with vitiligo susceptibility. Pigment Cell Res 15:62–66 10.1034/j.1600-0749.2002.00057.x [DOI] [PubMed] [Google Scholar]
- ——— (2003) Genes of the LMP/TAP cluster are associated with the human autoimmune disease vitiligo. Genes Immun 4:492–499 10.1038/sj.gene.6364016 [DOI] [PubMed] [Google Scholar]
- Cho S, Kang HC, Hahm JH (2000) Characteristics of vitiligo in Korean children. Pediatr Dermatol 17:189–193 10.1046/j.1525-1470.2000.01749.x [DOI] [PubMed] [Google Scholar]
- Das SK, Majumder PP, Majumder TK, Haldar B (1985) Studies on vitiligo. II. Familial aggregation and genetics. Genet Epidemiol 2:255–262 [DOI] [PubMed] [Google Scholar]
- de Vijlder HC, Westerhof W, Schreuder GM, de Lange P, Claas FH (2004) Difference in pathogenesis between vitiligo vulgaris and halo nevi associated with vitiligo is supported by an HLA association study. Pigment Cell Res 17:270–274 10.1111/j.1600-0749.2004.00145.x [DOI] [PubMed] [Google Scholar]
- Fain PR, Gowan K, LaBerge GS, Alkhateeb A, Stetler GL, Talbert J, Bennett DC, Spritz RA (2003) A genomewide screen for generalized vitiligo: confirmation of AIS1 on chromosome 1p31 and evidence for additional susceptibility loci. Am J Hum Genet 72:1560–1564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goudie BM, Wilkinson C, Goudie RB (1983) A family study of vitiligo patterns. Scott Med J 28:338–342 [DOI] [PubMed] [Google Scholar]
- Gudjonsson JE, Johnston A, Sigmundsdottir H, Valdimarsson H (2004) Immunopathogenic mechanisms in psoriasis. Clin Exp Immunol 135:1–8 10.1111/j.1365-2249.2004.02310.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafez M, Sharaf L, Abd el-Nabi SM (1983) The genetics of vitiligo. Acta Derm Venereol 63:249–251 [PubMed] [Google Scholar]
- Hann S-K, Nordlund J (2000) Vitiligo: a comprehensive monograph on basic and clinical science. Blackwell Science, Oxford, United Kingdom [Google Scholar]
- Horton R, Wilming L, Rand V, Lovering RC, Bruford EA, Khodiyar VK, Lush MJ, Povey S, Talbot CC Jr, Wright MW, Wain HM, Trowsdale J, Ziegler A, Beck S (2004) Gene map of the extended human MHC. Nat Rev Genet 5:889–899 10.1038/nrg1489 [DOI] [PubMed] [Google Scholar]
- Howitz J, Brodthagen H, Schwartz M, Thompsen K (1977) Prevalence of vitiligo: epidemiological survey of the Isle of Bornholm, Denmark. Arch Dermatol 113:47–52 10.1001/archderm.113.1.47 [DOI] [PubMed] [Google Scholar]
- Jin SY, Park HH, Li GZ, Lee HJ, Hong MS, Hong SJ, Park HK, Chung JH, Lee MH (2004) Association of angiotensin converting enzyme gene I/D polymorphism of vitiligo in Korean population. Pigment Cell Res 17:84–86 10.1046/j.1600-0749.2003.00105.x [DOI] [PubMed] [Google Scholar]
- Kemp EH, Ajjan RA, Waterman EA, Gawkrodger DJ, Cork MJ, Watson PF, Weetman AP (1999) Analysis of a microsatellite polymorphism of the cytotoxic T-lymphocyte antigen-4 gene in patients with vitiligo. Br J Dermatol 140:73–78 10.1046/j.1365-2133.1999.02610.x [DOI] [PubMed] [Google Scholar]
- Kovacs SO (1998) Vitiligo. J Am Acad Dermatol 38:647–666 [DOI] [PubMed] [Google Scholar]
- Kruglyak L, Daly MJ, Reeve-Daly, MP, Lander ES (1996) Parametric and nonparametric linkage analysis: a unified multipoint approach. Am J Hum Genet 58:1347–1363 [PMC free article] [PubMed] [Google Scholar]
- Lander E, Kruglyak L (1995) Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet 11:241–247 10.1038/ng1195-241 [DOI] [PubMed] [Google Scholar]
- Lathrop GM, Lalouel JM (1984) Easy calculations of lod scores and genetic risks on small computers. Am J Hum Genet 36:460–465 [PMC free article] [PubMed] [Google Scholar]
- Le Poole IC, Wankowicz-Kalinska A, van den Wijngaard RM, Nickoloff BJ, Das PK (2004) Autoimmune aspects of depigmentation in vitiligo. J Investig Dermatol Symp Proc 9:68–72 10.1111/j.1087-0024.2004.00825.x [DOI] [PubMed] [Google Scholar]
- Majumder PP, Nordlund JJ, Nath SK (1993) Pattern of familial aggregation of vitiligo. Arch Dermatol 129:994–998 10.1001/archderm.129.8.994 [DOI] [PubMed] [Google Scholar]
- Mehta NR, Shah KC, Theodore C, Vyas VP, Patel AB (1973) Epidemiological study of vitiligo in Surat area. Indian J Med Res 61:145–154 [PubMed] [Google Scholar]
- Miller S, Dykes D, Polesky H (1988) A simple salting out procedure for extraction of high molecular weight DNA from human nucleated cells. Nucl Acids Res 16:1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Na GY, Lee KH, Kim MK, Lee SJ, Kim do W, Kim JC (2003) Polymorphisms in the melanocortin-1 receptor (MC1R) and agouti signaling protein (ASIP) genes in Korean vitiligo patients. Pigment Cell Res 16:383–387 10.1034/j.1600-0749.2003.00062.x [DOI] [PubMed] [Google Scholar]
- Nath SK, Kelly JA, Namjou B, Lam T, Bruner GR, Scofield RH, Aston CE, Harley JB (2001) Evidence for a susceptibility gene, SLEV1, on chromosome 17p13 in families with vitiligo-related systemic lupus erythematosus. Am J Hum Genet 69:1401–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nath SK, Majumder PP, Nordlund JJ (1994) Genetic epidemiology of vitiligo: multilocus recessivity cross-validated. Am J Hum Genet 55:981–990 [PMC free article] [PubMed] [Google Scholar]
- Nyholt DR (2000) All LODs are not created equal. Am J Hum Genet 67:282–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell JR, Weeks DE (1998) PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet 63:259–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ongenae K, Van Geel N, Naeyaert JM (2003) Evidence for an autoimmune pathogenesis of vitiligo. Pigment Cell Res 16:90–100 10.1034/j.1600-0749.2003.00023.x [DOI] [PubMed] [Google Scholar]
- Sagoo GS, Tazi-Ahnini R, Barker JW, Elder JT, Nair RP, Samuelsson L, Traupe H, Trembath RC, Robinson DA, Iles MM (2004) Meta-analysis of genome-wide studies of psoriasis susceptibility reveals linkage to chromosomes 6p21 and 4q28-q31 in Caucasian and Chinese Hans population. J Invest Dermatol 122:1401–1405 10.1111/j.0022-202X.2004.22607.x [DOI] [PubMed] [Google Scholar]
- Spritz RA, Gowan K, Bennett DC, Fain PR (2004) Novel vitiligo susceptibility loci on chromosomes 7 (AIS2) and 8 (AIS3), confirmation of SLEV1 on chromosome 17, and their roles in an autoimmune diathesis. Am J Hum Genet 74:188–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tastan HB, Akar A, Orkunoglu FE, Arca E, Inal A (2004) Association of HLA class I antigens and HLA class II alleles with vitiligo in a Turkish population. Pigment Cell Res 17:181–184 10.1111/j.1600-0749.2004.00141.x [DOI] [PubMed] [Google Scholar]
- Venneker GT, Westerhof W, de Vries IJ (1992) Molecular heterogeneity of the fourth component of complement (C4) and its genes in vitiligo. J Invest Dermatol 99:853–858 10.1111/1523-1747.ep12614826 [DOI] [PubMed] [Google Scholar]
- Wankowicz-Kalinska A, van den Wijngaard RM, Tigges BJ, Westerhof W, Ogg GS, Cerundolo V, Storkus WJ, Das PK (2003) Immunopolarization of CD4+ and CD8+ T cells to type-1-like is associated with melanocyte loss in human vitiligo. Lab Invest 83:683–695 [DOI] [PubMed] [Google Scholar]
- Xu YY, Ye DQ, Tong ZC, Hao JH, Jin J, Shen SF, Li CR, Zhang XJ (2002) An epidemiological survey for four skin diseases in Anhui [In Chinese]. Chin J Dermatol 35:406–407 [Google Scholar]
- Zamani M, Spaepen M, Sghar SS, Huang C, Westerhof W, Nieuweboer-Krobotova L, Cassiman JJ (2001) Linkage and association of HLA class II genes with vitiligo in a Dutch population. Br J Dermatol 145:90–94 10.1046/j.1365-2133.2001.04288.x [DOI] [PubMed] [Google Scholar]
- Zhang XJ, He PP, Wang ZX, Zhang J, Li YB, Wang HY, Wei SC, Chen SY, Xu SJ, Jin L, Yang S, Huang W (2002) Evidence for a major psoriasis susceptibility locus at 6p21 (PSORS1) and a novel candidate region at 4q31 by genome-wide scan in Chinese Hans. J Invest Dermatol 119:1361–1366 10.1046/j.1523-1747.2002.19612.x [DOI] [PubMed] [Google Scholar]
- Zhang XJ, Liu HS, Liang YH, Sun LD, Wang JY, Yang S, Liu JB, Gao M, He PP, Cui Y, Yang Q (2004a) Association of HLA class I alleles with vitiligo in Chinese Hans. J Dermatol Sci 35:165–168 10.1016/j.jdermsci.2004.05.003 [DOI] [PubMed] [Google Scholar]
- Zhang XJ, Liu JB, Gui JP, Li M, Xiong QG, Wu HB, Li JX, Yang S, Wang HY, Gao M, Yang J, Yang Q (2004b) Characteristics of genetic epidemiology and genetic models for vitiligo. J Am Acad Dermatol 51:383–390 10.1016/j.jaad.2003.12.044 [DOI] [PubMed] [Google Scholar]