Abstract
Single-gene disorders with “simple” Mendelian inheritance do not always imply that there will be an easy prediction of the phenotype from the genotype, which has been shown for a number of metabolic disorders. We propose that moonlighting enzymes (i.e., metabolic enzymes with additional functional activities) could contribute to the complexity of such disorders. The lack of knowledge about the additional functional activities of proteins could result in a lack of correlation between genotype and phenotype. In this review, we highlight some notable and recent examples of moonlighting enzymes and their possible contributions to human disease. Because knowledge and cataloging of the moonlighting activities of proteins are essential for the study of cellular function and human physiology, we also review recently reported and recommended methods for the discovery of moonlighting activities.
Introduction
We now recognize that single-gene disorders, with deceptively simple Mendelian inheritance patterns, do not always show evidence of predictable relationships between genotypes and phenotypes (Scriver and Waters 1999; Dipple and McCabe 2000a, 2000b). This has been observed in the case of metabolic disorders such as glycerol kinase deficiency (GKD [MIM 307030]), phenylketonuria (MIM 261600), and Gaucher disease (MIM 230800), in which the mutations identified in the genomes of affected patients do not correlate clearly with their symptoms. This is a source of disappointment to clinical geneticists, since it presents a serious challenge to the concept that genotype can predict the clinical phenotype (Dipple and McCabe 2000b).
Previous reviews from our group have proposed three reasons for the absence of a clear genotype-phenotype relationship in such cases: the presence of indeterminate ranges in protein activities (Dipple and McCabe 2000b), the existence of modifier genes or loci that modulate the phenotype (Dipple and McCabe 2000a), and the role of systems dynamics (including flux through related metabolic pathways) in imparting a phenotype that is not easily deduced from the genotype (Dipple et al. 2001a, 2001b; Clipsham et al. 2002; Clipsham and McCabe 2003).
In this review, we point out an additional factor that could contribute to the complexity of metabolic disorders—the existence of moonlighting enzymes. “Moonlighting,” a term coined by Jeffery (1999), refers to the phenomenon that many metabolic enzymes can exhibit unrelated functional activities within or outside the cell. The definition of moonlighting proteins excludes proteins that result from alternative mRNA splicing, posttranslational modifications, and gene fusions; proteins capable of utilizing multiple substrates (e.g., aldolase); and proteins catalyzing multiple steps in the same metabolic pathway (e.g., phosphofructokinase/fructose-2,6-bisphosphatase); nevertheless, there are a few dozen moonlighting proteins (Jeffery 2003b).
Examples of moonlighting activities include, but are not limited to, transcriptional regulation and apoptosis. If the gene of interest in a metabolic single-gene disorder encodes a metabolic enzyme whose moonlighting activities are as yet unknown, and if one or more of these moonlighting activities contribute to the phenotype of the disorder, then this could result in a phenotype that cannot be explained by the loss of the metabolic activity of the enzyme, thus causing an apparent uncorrelation between genotype and phenotype.
Here, we illustrate moonlighting proteins with specific examples and discuss their possible roles in disease. We also review methods of discovery of moonlighting activities and emphasize the need for reliable discovery methods.
Examples of Moonlighting Proteins
In this section, we highlight selected examples of metabolic enzymes that have demonstrated moonlighting activities. It should be noted that we present only a representative sample of the numerous metabolic enzymes that display additional functional activities. Previous reviews provide exhaustive lists (Ramasarma 1994; Jeffery 1999, 2003a, 2003b, 2004b), and we have chosen to draw attention only to moonlighting proteins that are not covered in those reviews (aldolase, enolase, glycerol kinase, and glycogen synthase kinase 3β), have additional moonlighting activities elucidated since those reviews were published (aconitase and glyceraldehyde-3-phosphate dehydrogenase), and/or are notable for the variety of moonlighting activities they exhibit (glyceraldehyde-3-phosphate dehydrogenase and phosphoglucose isomerase).
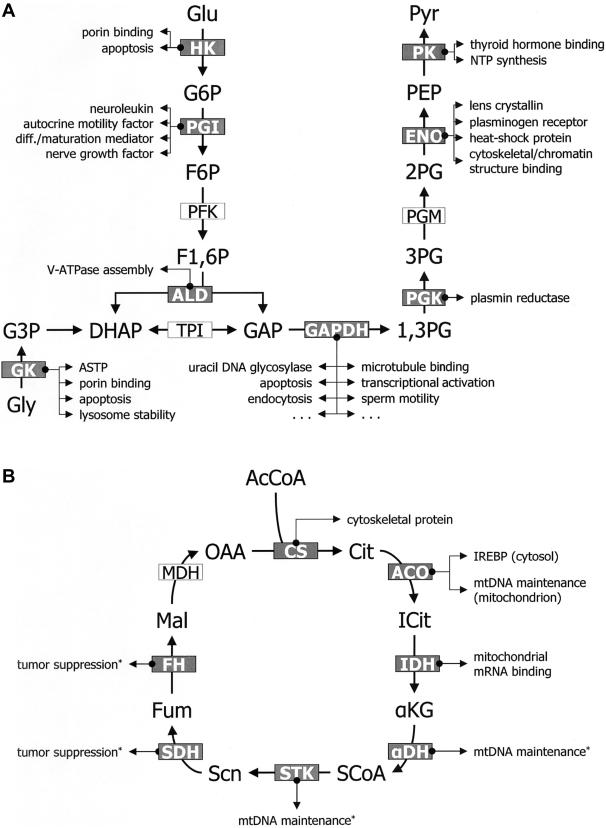
Figure 1 illustrates the high prevalence of moonlighting among familiar enzymes. A metabolic map of glycolysis (fig. 1A) reveals that 7 of the 10 glycolytic enzymes exhibit various moonlighting activities. In addition, other metabolic enzymes closely linked to glycolysis (e.g., glycerol kinase and fructose-1,6-bisphosphatase) also moonlight. Similarly, figure 1B shows that at least seven of the eight enzymes of the tricarboxylic acid (TCA) cycle have moonlighting or suspected-moonlighting activities.
Figure 1.
High prevalence of moonlighting enzymes. Most metabolic enzymes in familiar pathways—for example, glycolysis (and glycerol metabolism) (A) and the TCA cycle (B)—exhibit moonlighting activities. Moonlighting enzymes are shown in white font in gray boxes, and their known moonlighting activities are listed on the side. Suspected moonlighting activities are marked with an asterisk (*). Metabolite abbreviations are as follows: 1,3PG = 1,3-bisphosphoglycerate; 2PG = 2-phosphoglycerate; 3PG = 3-phosphoglycerate; AcCoA = acetyl coenzyme A; Cit = citrate; DHAP = dihydroxyacetone phosphate; F1,6P = fructose-1,6-phosphate; F6P = fructose-6-phosphate; Fum = fumarate; G3P = glycerol-3-phosphate; G6P = glucose-6-phosphate; GAP = glyceraldehyde-3-phosphate; Glu = glucose; Gly = glycerol; ICit = isocitrate; Mal = malate; OAA = oxaloacetate; PEP = phosphoenolpyruvate; Pyr = pyruvate; SCoA = succinyl coenzyme A; Scn = succinate; aKG = α-ketoglutarate. Enzyme abbreviations are as follows: ACO = aconitase; ALD = aldolase; CS = citrate synthase; ENO = enolase; FH = fumarate hydratase; HK = hexokinase; IDH = isocitrate dehydrogenase; MDH = malate dehydrogenase; PFK = phosphofructokinase; PGK = phosphoglycerate kinase; PGM = phosphoglycerate mutase; PK = pyruvate kinase; STK = succinate thiokinase; TPI = triose phosphate isomerase; aDH = α-ketoglutarate dehydrogenase. diff. = differentiation. Enzymes shown here but not discussed in the main text (and their corresponding references) are HK (Fiek et al. 1982; Linden et al. 1982; Ostlund et al. 1983), PGK (Lay et al. 2000, 2002), CS (Numata et al. 1991; Numata 1996), IDH (Elzinga et al. 1993; Chen et al. 2005), aDH (Chen et al. 2005), and STK (Chen et al. 2005).
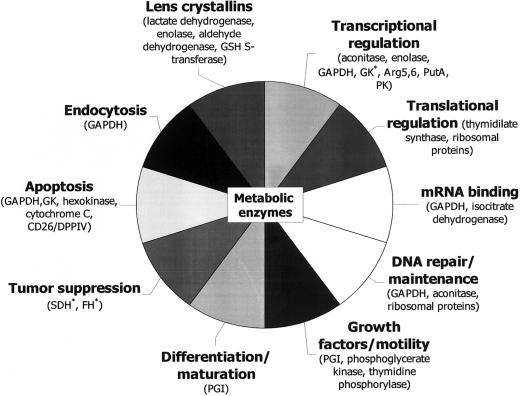
The diversity of nonenzymatic additional functions exhibited by moonlighting enzymes is shown in figure 2. The moonlighting activities range from signal transduction events, such as transcriptional regulation and apoptosis; to growth and motility; to structural functions, such as those of lens crystallins.
Figure 2.
A representative sample of the wide variety of moonlighting activities of common metabolic enzymes. Suspected moonlighting activities are marked with an asterisk (*). FH = fumarate hydratase; PK = pyruvate kinase; GSH = glutathione. Moonlighting proteins shown here but not discussed in the main text (and their corresponding references) are CD26/dipeptidyl peptidase IV (CD26/DPPIV) (Boonacker and Van Noorden 2003) and cytochrome C, ribosomal proteins, thymidilate synthase, and thymidine phosphorylase (Jeffery 2003b).
Aconitase
Aconitase is an enzyme of the TCA cycle, and it catalyzes the reaction that converts isocitrate to α-ketoglutarate (fig. 1B). Cytosolic aconitase also plays a role in iron metabolism by functioning as an iron-responsive element-binding protein (IREBP), which regulates the mRNAs corresponding to ferritin, which is an iron-sequestration protein, and the transferrin receptor, a protein involved in iron uptake (Kennedy et al. 1992; Rouault et al. 1992). The aconitase and IREBP activities of this protein are mutually exclusive and are dictated by cytosolic iron levels. When iron levels in the cytosol are low, aconitase loses its enzymatic activity and functions as an IREBP (Kennedy et al. 1992). A recent study found an inverse correlation between the iron-responsive element-binding activity and the nonheme iron content in the livers of various mammals (Starzynski et al. 2004). The IREBP functions by binding to the iron-responsive element, which is located in the 5′ UTR of ferritin mRNA and in the 3′ UTR of transferrin receptor mRNA. This binding regulates the translation initiation of ferritin mRNA or the half-life of the transferrin receptor mRNA, thus exercising control of iron metabolism (Basilion et al. 1994; Jeffery 1999). Furthermore, it has been shown recently that, in yeast, mitochondrial aconitase is essential for mtDNA maintenance and that this activity is independent of catalytic activity or flux through aconitase (Chen et al. 2005).
Aldolase
Aldolase is a glycolytic enzyme that catalyzes the cleavage of fructose-1,6-bisphosphate into dihydroxyacetone phosphate and glyceraldehyde phosphate (fig. 1A). Lu et al. (2001, 2004) reported that aldolase binds to vacuolar H+-ATPase (V-ATPase) in yeast, and adolase was found to be associated with intact V-ATPase in bovine kidney microsomes and in osteoclast-containing mouse marrow cultures. V-ATPases are essential for the acidification of intracellular compartments and for proton secretion from the plasma membrane in kidney epithelial cells and osteoclasts. Aldolase has been reported elsewhere to bind to actin (Arnold and Pette 1970).
Enolase
Enolase catalyzes the conversion of 2-phosphoglycerate to phosphoenolpyruvate in glycolysis (fig. 1A). It has been reported to function as a plasminogen receptor (Miles et al. 1991; Dudani et al. 1993; Nakajima et al. 1994) and a heat-shock protein and to bind to cytoskeletal and chromatin structures (Pancholi 2001). Enolase serves as a crystallin in the eye lens (Wistow et al. 1988). Recently, Wang et al. (2005) identified α-enolase as a nuclear DNA-binding protein in the zona fasciculata of the human adrenal cortex. The transfection of an α-enolase expression vector into human adenocortical cells enhanced the promoter activity of the type II 3β-hydroxysteroid dehydrogenase gene, which indicates that enolase may have a functional role in regulating the expression of this gene (Wang et al. 2005).
Glyceraldehyde-3-Phosphate Dehydrogenase
Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) has been discovered to have several moonlighting activities (Sirover 1999)—it has at least 10 distinct, confirmed nonenzymatic activities apart from its enzymatic function of converting glyceraldehyde-3-phosphate to 1,3-diphosphoglycerate (fig. 1A). GAPDH is the same protein as uracil DNA glycosylase, which repairs DNA by excising uracil (Meyer-Siegler et al. 1991). It is also involved in endocytosis (Robbins et al. 1995), microtubule bundling (Kumagai and Sakai 1983), phosphate group transfer (Kawamoto and Caswell 1986; Duclos-Vallee et al. 1998; Engel et al. 1998), nuclear tRNA export (Singh and Green 1993), and vesicular transport (Tisdale 2001). GAPDH plays a structural role in the eye, where it acts as a lens crystallin (Jeffery 1999). It also binds to biomolecules, including RNA (Ryazanov 1985), RNA polymerase (Mitsuzawa et al. 2005), and diadenosine tetraphosphate (Ap4A) (Baxi and Vishwanatha 1995), one of whose functions is to decrease blood insulin levels (Rusing and Verspohl 2004). Recently, GAPDH has also been demonstrated to be important in sperm motility and, therefore, in male fertility (Miki et al. 2004).
Moreover, GAPDH plays a significant role in apoptosis. This is apparent from a mounting body of indirect and direct evidence (Berry and Boulton 2000; Tatton et al. 2000). For example, GAPDH binds to Ap4A, which is involved in apoptosis (Vartanian 2003; Wang et al. 2003). Low extracellular K+ levels in cerebellar granule cell cultures that induced apoptosis also resulted in an increase in GAPDH mRNA and protein (Sunaga et al. 1995). Furthermore, treatment with GAPDH antisense oligonucleotides affected both GAPDH levels and apoptosis, establishing a direct link between the two (Sunaga et al. 1995; Ishitani et al. 1997). Although the lowering of extracellular K+ levels can result in both apoptosis and necrosis (Berry and Boulton 2000), GAPDH antisense treatment affected apoptosis only and did not prevent necrosis (Ishitani et al. 1997).
The apoptotic activity of GAPDH involves the translocation of the protein into the nucleus (Sawa et al. 1997; Shashidharan et al. 1999) and is independent of both its glycolytic and uracil DNA glycosylase activities (Berry and Boulton 2000). Although the exact mechanism of nuclear translocation and subsequent initiation of apoptosis are not yet well understood (Chuang et al. 2005), some recent evidence has suggested a potential mode of action. This includes the binding of GAPDH to a nuclear localization signal–containing protein, Siah, to initiate nuclear translocation and an apparent change in the structure of GAPDH after its nuclear translocation (Chuang et al. 2005).
Also, GAPDH was recently shown to be involved in the formation of Lewy body–like inclusions in cultured cells and the concomitant apoptosis (Tsuchiya et al. 2005). Lewy bodies are intraneuronal proteinaceous cytoplasmic inclusions and are a pathological characteristic of Parkinson disease, which is linked to apoptosis of dopaminergic cells (Tsuchiya et al. 2005).
Glycerol Kinase
Glycerol kinase (GK) catalyzes the conversion of glycerol to glycerol-3-phosphate (fig. 1A) and is known to be an important enzyme in glycerol metabolism. Independently, Okamoto et al. (1993) cloned the gene responsible for the ATP-stimulated translocation protein (ASTP) from rat liver, and Huq et al. (1996) found that ASTP had 99% homology to the amino acid sequence of mouse GK. ASTP is an ATP-stimulated factor that enhances the nuclear binding of the activated glucocorticoid-receptor complex (Okamoto et al. 1984). Additionally, GK/ASTP has been shown to bind to histones (Okamoto et al. 1989), to interact with porin or the voltage-dependent anion channel on the outer surface of the outer mitochondrial membrane (McCabe 1983; Ostlund et al. 1983), and to be a lysosomal stabilization factor (Arai et al. 2002). Recently, our group has shown that GK may have a role in apoptosis (Martinez and McCabe 2004).
Glycogen Synthase Kinase 3β
Glycogen synthase kinase 3 (GSK3) was initially discovered as a kinase involved in the regulation of glucose metabolism, in which it phosphorylates glycogen synthase A to glycogen synthase B. In mammals, it occurs as two closely related isoforms, GSK3α and GSK3β. The GSK3β isoform is highly expressed in neural tissue. It has functions in various neuronal signal-transduction pathways and is emerging as a promising drug target for CNS therapies (Bhat et al. 2004). Most recently, GSK3β has been reported to play a critical role in the establishment and maintenance of neuronal polarity (Jiang et al. 2005; Yoshimura et al. 2005). In particular, it regulates collapsin-response mediator protein 2, which is crucial for axon outgrowth and for determining of the fate of the axon and dendrites and, therefore, in establishing and maintaining neuronal polarity (Yoshimura et al. 2005).
Lens Crystallins
Many crystallins present in the lens of the eye as refractive proteins have been identified as well-known metabolic enzymes, such as lactate dehydrogenase, arginosuccinate lyase, glutathione-S-transferase, enolase, and aldehyde dehydrogenase (Ramasarma 1994; Piatigorsky 1998b, 2003; Jeffery 1999). A recent study that analyzed the proteomes of human lens epithelial cells reported high abundances of the aforementioned enzymes among the proteins present, along with the glycolytic enzymes aldolase, phosphoglycerate kinase, and triose phosphate isomerase (Wang-Su et al. 2003). Lens crystallins are believed to have been recruited as structural proteins during the evolution of the eye (Piatigorsky 1998a, 1998b, 2003). Some crystallins may also serve additional functions. For instance, aldehyde dehydrogenase is postulated to function as a UV filter in the human lens, because of its binding to nicotinamide adenine dinucleotide (Bateman et al. 2003).
Phosphoglucose Isomerase
Phosphoglucose isomerase (PGI) catalyzes the second step in glycolysis, the conversion of glucose-6-phosphate to fructose-6-phosphate (fig. 1A). Mammalian PGI also functions as a neurotrophic factor, a neuroleukin (Chaput et al. 1988; Faik et al. 1988), an autocrine motility factor (Watanabe et al. 1996), and a nerve growth factor (Gurney et al. 1986), as well as a differentiation and maturation mediator (Xu et al. 1996). Interestingly, not all these activities are exhibited by PGI in other organisms. For example, bacterial or yeast PGI does not exhibit autocrine motility factor activity (Amraei and Nabi 2002).
Pyruvate Kinase
Pyruvate kinase mediates the final step of glycolysis—the conversion of phosphoenolpyruvate to pyruvate (fig. 1A). As a metabolic enzyme, it is a homotetramer in almost all organisms (Munoz and Ponce 2003). However, it can exist as a monomer, and its monomeric form in mammalian muscle is a thyroid hormone–binding protein that regulates the transcriptional responses of the thyroid-hormone receptor (Ashizawa et al. 1991, 1992). Additionally, it can synthesize nucleotide triphosphate (NTP) under anaerobic conditions in bacteria (Saeki et al. 1974; Sundin et al. 1996).
Switching between Functions
One or more factors may be responsible for the function exhibited by a moonlighting protein. A partial list of such factors includes cellular sublocalization, expression in different cell types, presence inside or outside the cell, oligomeric state, substrate or ligand concentration, binding sites, and phosphorylation. Although a similar list was presented in an earlier publication by Jeffery (1999), it is revisited here with recently elucidated examples.
Cellular Sublocalization
GAPDH functions as an enzyme outside the nucleus but has an apoptotic function in the nucleus. Similarly, GK plays an enzymatic role outside the nucleus and an ASTP role within the nucleus. Aconitase functions as an enzyme in the mitochondrion and the cytosol; however, its compartmentation determines the nature of its moonlighting activity—it functions as an IREBP in the cytosol and has an mtDNA maintenance function in the mitochondrion.
Expression in Different Cell Types
Enolase functions as a glycolytic enzyme in most cell types but also as a structural crystallin in the eye lens. Furthermore, the DNA-binding function of enolase in the human adrenal cortex was observed only in the zona fasciculata and not in the zona reticularis (Wang et al. 2005).
Presence Inside or Outside the Cell
Ku70/Ku80 is a protein complex involved in DNA strand–break recognition and repair within the cell, but it participates in proteolytic processes outside the cell (Muller et al. 2005).
Oligomeric State
Pyruvate kinase exhibits metabolic activity as a tetramer and thyroid hormone–binding activity as a monomer.
Concentration of Ligand or Substrate
As mentioned above, iron levels determine whether aconitase functions as an enzyme or as an IREBP.
Binding Sites
Moonlighting proteins may employ different binding or catalytic sites for different functions. For example, ceruloplasmin, an oxidase that participates in copper metabolism, moonlights as a copper-independent glutathione peroxidase (Bielli and Calabrese 2002), and it employs distinct catalytic sites to bind to the substrates of either function. Similarly, mammalian InsP6 kinase (an inositol phosphate kinase) binds to the guanine nucleotide exchange factor, and this binding does not require the catalytic site of the enzyme (Shears 2004).
Phosphorylation
It is likely that phosphorylation is sometimes responsible for switching between functions. There is recent evidence that phosphorylation of PGI at the residue Ser185 by protein kinase CK2 causes it to lose its enzymatic activity but does not affect its function as an autocrine motility factor (Yanagawa et al. 2005). However, there are instances in which phosphorylation is critical for both activities of a protein, as in the case of aconitase, in which loss of phosphorylation at Ser711 results in loss of both the aconitase and the IREBP activities (Fillebeen et al., in press).
Why Do Moonlighting Activities Exist?
There are two nonexclusive hypotheses about the existence of moonlighting activities: that these activities simply evolved and were not lost and that they offer definite advantages to the cell or organism (Jeffery 1999).
Evolution
Since many moonlighting proteins are familiar metabolic enzymes (fig. 1) and are present in a wide variety of organisms that have evolved over billions of years, it is likely that the additional activities of the proteins evolved, possibly to make use of unused sites on the protein surface. It is notable that active sites usually do not occupy a large fraction of the protein surface area, leaving considerable solvent-exposed surface that can be recruited for other functions (Jeffery 1999).
A recent study of lens crystallins (Piatigorsky 2003) provides interesting insights into the evolution of moonlighting activities. Ancestral enzymes may have been recruited as crystallins, following a gene duplication (with one gene encoding a protein with enzymatic activity and the other gene encoding a lens-specific crystallin) or a regulatory change (high expression of the ancestral protein in the lens), or both. Gene duplication is evidenced by the presence of two genes encoding α-crystallin/heat-shock protein (αA and αB), which are situated on separate chromosomes in mice and humans. The protein encoded by the αB-crystallin gene has remained a heat-shock protein, is expressed constitutively, and is inducible by stress. In contrast, αA-crystallin is expressed predominantly in the lens and is not stress inducible. However, certain other crystallins, such as ɛ-crystallin/lactate dehydrogenase and γ-crystallin/α-enolase, are encoded by single-copy genes, indicating that a transcriptional regulatory change conducive to high expression in the lens must have preceded the evolution of the refractory role for the protein. The fact that many crystallin genes are regulated by similar transcription factors (e.g., Pax-6, retinoic acid receptors, maf, Sox, AP-1, and cAMP response element B) reinforces the idea that the common feature of lens-specific expression may have played an important role in the recruitment of diverse enzymes as lens crystallins (Piatigorsky 2003).
Advantages to the Cell or Organism
Moonlighting activities can also be advantageous to the cell. One simple reason is that they reduce the number of proteins that the organism has to synthesize—thus, less energy is expended for DNA replication, and the genome is more compact (Jeffery 1999). The cell can benefit from the increased complexity with relatively fewer genes. Additionally, moonlighting can serve to coordinate related cellular activities or to switch between pathways (Jeffery 1999). For example, the interaction of aldolase with V-ATPase links two related pathways: the ATP-producing glycolytic pathway and the ATP-hydrolyzing proton pump (Lu et al. 2004; Sautin et al. 2005). The use of moonlighting activities to switch between proximate pathways is exemplified by mitochondrial aconitase (Chen et al. 2005) and rat liver mitochondrial Lon protease, an ATP-dependent protease that degrades certain mitochondrial proteins but that is also a mitochondrial chaperone (Jeffery 1999).
Moonlighting Proteins and Human Disease
As mentioned above, moonlighting proteins can add to the complexity of single-gene disorders. In a majority of cases, there is as yet no confirmed evidence that the nonmetabolic activity of a moonlighting enzyme encoded by a disease gene is responsible for the phenotype of a disorder. Nevertheless, there are strong indicators that this may be occurring, as discussed below.
GAPDH
We noted above that GAPDH plays a role in apoptosis. Excessive apoptosis is involved in many neurodegenerative diseases, such as Huntington disease (MIM 143100), Alzheimer disease (MIM 104300 and 104310), Parkinson disease (MIM 168601), and brain ischemia (Chuang et al. 2005), and recent findings support the role of GAPDH in these diseases. For example, nuclear-aggregated GAPDH was found in neurons of affected areas in postmortem brains of individuals who had Alzheimer disease (Tsuchiya et al. 2004). A proteomic analysis of neurofibrillary tangles in Alzheimer disease by mass spectroscopy identified GAPDH to be associated with neurofibrillary tangles (Wang et al., in press). A linkage analysis of patients with late-onset Alzheimer disease (MIM 104310) revealed an association with SNPs in the GAPDH gene, indicating that this gene is a risk factor for the disease (Li et al. 2004). Furthermore, potential drugs for treatment of Alzheimer disease, such as tetrahydroaminoacridine and ONO-1603, suppress GAPDH expression and its nuclear translocation in rat brain neurons undergoing apoptosis in cultures (Katsube et al. 1996, 1999).
These results suggest a role for GAPDH-induced apoptosis in Alzheimer disease. The role of GAPDH in Parkinson disease is indicated by its participation in the formation of Lewy body–like inclusions in cultured cells and subsequent apoptosis (Tsuchiya et al. 2005). For a full review that identifies the role of GAPDH in the aforementioned neurodegenerative diseases, see the article by Chuang et al. (2005).
Xanthine Oxidoreductase
Xanthine oxidoreductase (XOR) is the rate-limiting enzyme in purine catabolism. Vorbach et al. (2002) noted that XOR is expressed at very high levels in the lactating mammary epithelium and suggested that it may serve a structural role. They confirmed this hypothesis by studying XOR-deficient mice. XOR+/− females, although they appeared healthy, were unable to maintain lactation, and their pups died of starvation. Further investigations showed that the mammary epithelium collapsed in the knockout mice, and this resulted in premature involution of the mammary gland. XOR−/− mice, on the other hand, were runted and did not survive beyond 6 wk. Although this shows that XOR is necessary for survival, it does not indicate whether the protein's metabolic function (purine catabolism) or moonlighting function (structural role in mammary epithelium) is responsible for survival.
However, previous and recent reports provide strong indications that XOR plays a moonlighting structural role in the mammary gland. For example, XOR protein levels selectively increased in mammary tissue during pregnancy and lactation (McManaman et al. 1999, 2002), and XOR in human mammary epithelial cells had low specific activities despite high mRNA levels (Page et al. 1998; Harrison 2004), suggesting that XOR may be playing a nonenzymatic role.
Recent findings have also shown that XOR structurally interacts with milk-fat globule proteins, including butyrophilin and adipophilin (McManaman et al. 2004; Ogg et al. 2004). Ogg et al. (2004) have suggested that the butyrophilin-XOR interaction may contribute to the formation of a protein complex that stabilizes the milk droplet.
PGI
Deficiency of PGI is an autosomal recessive genetic disorder (MIM 172400), which manifests as hemolytic anemia but can also include neurological defects (Kugler et al. 1998). This is consistent with the enzyme’s moonlighting role as a neuroleukin. A severe form of this disorder has been reported in which the mutations A59C (H20P) and T1016C (L339P) cause substitutions by prolines, with the consequent misfolding of the protein and the loss of both the enzymatic and neuroleukin activities. However, a less severe form of this disorder involved the mutations A1166G (H389R) and C1549G (L517V) located at the subunit surface, which affected the enzyme activity of PGI but not the neuroleukin activity. Not surprisingly, the latter mutations were not associated with neurological defects (Kugler et al. 1998). This example clearly illustrates that moonlighting activities of an enzyme can help to explain the apparent lack of genotype-phenotype correlation in a disease involving deficiency of the enzyme.
ERCC2
Mutations in the excision-repair, complementing defective, in Chinese hamster, 2 gene (ERCC2 [MIM 126340], formerly known as the “xeroderma pigmentosum group D” gene), result in three disorders with markedly different clinical features (Lehmann 2001). Xeroderma pigmentosum (XP [MIM 278730]) is characterized by several skin abnormalities that range from excessive freckling to multiple skin cancers, with a high risk of skin cancer in affected individuals. Cockayne syndrome (MIM 216400) is characterized by severe cachectic dwarfism, mental retardation, microcephaly, and retinal and skeletal abnormalities. Trichothiodystrophy (TTD [MIM 601675]) is characterized by sulfur-deficient brittle hair, mental retardation, unusual facies, ichthyotic skin, and reduced stature, as well as sun sensitivity, but there are no reports of skin cancer. In addition, the XP-causing alleles of the gene were not observed in individuals with TTD, and vice versa (Petrini 2000).
The distinct phenotypes are now explained by the finding that ERCC2 encodes a protein that has two distinct functions. It functions as a DNA helicase involved in the repair of DNA damaged by exposure to ultraviolet light, and it is also a subunit of TFIIH, a basal transcription factor (Lehmann 2001). XP is caused principally by defects in nucleotide-excision repair—that is, by the enzymatic (helicase) function of the gene product. In contrast, TTD and Cockayne syndrome are caused by defects in transcription (Lehmann 2003).
GK
GKD is a disorder that occurs because of mutations, deletions, or insertions in the GK gene (MIM 300474) on Xp21 (Dipple et al. 2001b). It can occur either as complex GKD, a contiguous-gene-deletion syndrome, or as isolated GKD. Isolated GKD can be symptomatic or asymptomatic (Sjarif et al. 2000; Dipple et al. 2001b). Thus far, there has been no satisfactory correlation between genotype and phenotype in patients with isolated GKD (Sargent et al. 2000; Dipple et al. 2001b). In fact, the exact same mutation has been observed in a symptomatic and an asymptomatic individual (Sargent et al. 2000), and this discrepancy has also been observed in two brothers with the identical mutation (Blomquist et al. 1996). The GK enzymatic activities in lymphoblastoid cell lines or fibroblasts were similar for the symptomatic and asymptomatic individuals. Also, mapping of the individuals' GK mutations to the Escherichia coli three-dimensional protein structure did not distinguish the symptomatic from the asymptomatic individuals (Dipple et al. 2001b).
As noted above, GK is a moonlighting enzyme, and it is likely that one of its additional functional activities, rather than its enzymatic activity, may help explain the GKD symptomatic phenotype. In particular, the role of GK in transcriptional regulation through nuclear translocation of the glucocorticoid receptor complex, or the role of GK in apoptosis, could provide clues to explain the complexity of GKD.
However, moonlighting might not by itself account for the perplexing fact that individuals or siblings with the same mutation exhibit different phenotypes. This could be a result of the action of modifier genes, as postulated elsewhere (Dipple and McCabe 2000a). Also, it is likely that the modifier genes interact with the moonlighting function of the protein, rather than the enzymatic function.
Succinate Dehydrogenase and Fumarate Hydratase
Succinate dehydrogenase (SDH [MIM 602690]) and fumarate hydratase (or fumarase [MIM 136850]) catalyze consecutive steps in the TCA cycle. Recently, it was discovered that a gene that predisposes to inherited multiple uterine leiomyomata (fibroids) (MIM 150800) and papillary renal cell cancer (MIM 650839) is the same as the gene encoding fumarate hydratase (Tomlinson et al. 2002). Along the same lines, germline SDH mutations were shown to be associated with paraganglioma (MIM 168000) and pheochromocytoma (MIM 171300) (Astuti et al. 2001). These findings suggest that these enzymes, directly or indirectly, play specific tumor-suppressing roles, which supports the potential existence of moonlighting activities for these metabolic enzymes, since the tumor suppression cannot be explained by the metabolic activity of the enzymes (Jeffery 2003a).
However, firm evidence is still lacking, and the investigation of the nature of the tumor-suppressing mechanism should shed more light on this interesting finding. A recent study showed that, at least in the case of SDH, metabolic activity may be indirectly responsible for tumor suppression; succinate, which accumulates as a result of SDH deficiency, inhibits hypoxia-inducible factor 1α (HIF-1α) prolyl hydroxylases, thereby stabilizing HIF-1α, which causes angiogenesis and tumor growth (Selak et al. 2005).
Aconitase
There is a myopathy (MIM 255125) that is associated with aconitase and SDH deficiency and in which the affected individual has an abnormal deposition of iron in the mitochondria (Haller et al. 1991; Hall et al. 1993). This affected individual experienced exertional muscle fatigue, dyspnea, and cardiac palpitations. Hall et al. (1993) have speculated that the dynamic Fe-S center of mitochondrial aconitase may respond to mitochondrial iron levels, as is seen with IREBP/aconitase. Although there is no evidence that the moonlighting function of aconitase is responsible for the myopathy, a recent article by Lipinski et al. (2005) shows that the IREBP function of this protein can have far-reaching effects, including DNA damage. Specifically, Lipinski et al. (2005) showed that increases in cellular iron (due to an increase in IREBP activity and a decrease in aconitase activity) may result in DNA damage induced by hydrogen peroxide through the Fenton reaction.
Elucidation of Moonlighting Activities
Serendipity
Serendipity has been a major contributor to the identification of moonlighting activities thus far. For example, the 10 moonlighting activities of GAPDH listed above were reported independently by 13 different research groups in five countries (see citations in the “Glyceraldehyde-3-Phosphate Dehydrogenase” section). Furthermore, 11 of those 13 research groups identified an unknown protein as GAPDH, so this enzyme was not even the initially intended subject of the study. A glimpse at articles that first reported moonlighting activities of many other proteins also reveals that many of the findings were unforeseen.
Nevertheless, the possible role of moonlighting proteins in metabolic disorders and the consequent urgency to identify them necessitates systematic methods of discovery. Below are some methods that have been reported and recommended.
Mass Spectrometry
Mass spectrometry (MS) is a powerful technique in proteomics. Jeffery (in press) has suggested the use of MS to identify potential moonlighting proteins. MS can provide two principal indicators of moonlighting activities. First, the presence of a protein in an unexpected location in the cell, in an unexpected cell type, or in an unexpected multiprotein complex could point to possible moonlighting activities. For example, large amounts of the glycolytic enzymes aldolase, phosphoglycerate kinase, and triose phosphate isomerase were detected in the proteomes of human lens epithelial cells by use of MS (Wang-Su et al. 2003), and this result indicates a possible role for these proteins in the lens. Second, if there are high protein expression levels that do not correlate with an enzyme’s measured metabolic activity, that may indicate that the protein is performing a function distinct from its known metabolic activity (Jeffery, in press).
Since MS can be used in a high-throughput fashion, it holds promise for detection of possible moonlighting activities. However, it can provide only an indication, and further experimentation is essential to confirm whether the protein actually moonlights and to determine the nature of the moonlighting activity.
Proteome Arrays
A recent study reported that Arg5,6—a mitochondrial enzyme involved in arginine biosynthesis—binds to DNA and regulates transcription in yeast (Hall et al. 2004). That study employed a proteome array; a microarray containing 6,500 protein preparations of 5,800 yeast proteins was probed with genomic yeast double-stranded, fluorescently labeled DNA. Arg5,6 was identified among the positives found. Since transcriptional regulation by a moonlighting protein can play a potentially important role in disease, this technique of discovery is valuable, subject to the easy availability of proteome arrays for higher organisms.
Bioinformatic Approaches
Sequence-analysis algorithms, such as those used in BLAST, PROSITE, EMOTIF, PROTLOC, and many other programs, rely on sequence motifs to identify protein function (Jeffery 2004b). A recent study attempted to identify moonlighting activities of common moonlighting proteins by employing 11 different sequence-analysis algorithms (Gomez et al. 2003). It was observed that success rates were only moderate at best. For GAPDH, 7 of 11 algorithms identified the enzymatic function, and only 3 of 11 algorithms identified the uracil DNA glycosylase function. Among the algorithm-based programs, PSI-BLAST performed reasonably well in identifying moonlighting activities.
Network component analysis (NCA) is another bioinformatic approach that identifies possible new functions by recognizing novel connectivities between genes and transcription factors (Liao et al. 2003; Kao et al. 2004). NCA has been used with gene-expression microarray data from E. coli and yeast to uncover hidden regulatory signals and possible new network relationships when only partial knowledge of the underlying network topology exists. In E. coli, ∼70% of randomly selected transcription-factor networks could be identified by NCA (Liao et al. 2003). NCA provides a powerful approach to mining microarray data and to gaining insight into new network connectivities and, therefore, novel putative functions.
Insights from the Structure-Function Relationships of Moonlighting Proteins
Many recent studies have focused on the elucidation of the structure-function relationships of moonlighting proteins. For example, a recent study of aconitase/IREBP (Fillebeen et al., in press) reported that, in HEK293 cells, phosphorylation of this protein at the residue Ser711 had devastating effects on both the enzymatic and IREBP activities, which indicates that Ser711 is a critical residue for the control of the activities of this protein. However, the exact mechanism by which phosphorylation controls the activities of this protein is yet to be elucidated.
The insights into multifunctionality obtained from three-dimensional crystal structures of moonlighting proteins (reviewed by Jeffery [2004a]) are of particular interest. For example, Lee et al. (2003) reported the crystal structure of PutA proline dehydrogenase, which moonlights as a transcriptional regulator of its own gene. Lee et al. (2003) noted that the structure contains a helical domain with three helices arranged in a helix-turn-helix pattern found in other DNA-binding domains, and they suggested that this could represent the DNA-binding domain responsible for transcriptional repression, although further experiments are needed to confirm this supposition (Jeffery 2004a).
Lee et al. (2004) investigated functional domains of the Brevibacillus thermophilus Lon protease (Bt-Lon), which is involved in the degradation of damaged and short-lived proteins, in ATPase and chaperone-like activities, and in DNA binding. This protein includes an N-terminal domain, a central sensor- and substrate-discrimination domain, and a C-terminal protease domain. Lee et al. (2004) prepared seven mutants of Bt-Lon, each of which lacked one or more domains. The truncation of the N-terminal domain led to the failure of oligomerization and the inactivation of proteolytic, ATPase, and chaperone-like activities, suggesting that oligomerization is essential for the catalytic and chaperone-like activities. Furthermore, gel-mobility shift assays indicated that the substrate-discrimination domain is involved in DNA binding.
Summary and Outlook
In this review, we highlighted some key and recently reported moonlighting enzymes that exhibit an astounding variety of additional functional activities. We also summarized mechanisms employed by moonlighting proteins to switch between functions, as well as hypotheses about the existence of moonlighting proteins.
The moonlighting activities of familiar metabolic enzymes include signal-transduction events such as transcriptional regulation and apoptosis, and it is likely that the moonlighting activities of these enzymes may contribute to the complex phenotype of a given disorder. It is very likely that the complex phenotypes observed in many single-gene disorders that cannot be explained solely by the loss of the enzymatic activity of the protein might be explained by a moonlighting activity of the protein.
The identification and cataloging of all activities of proteins in the proteome is therefore essential for understanding human genetic diseases. Thus, high-throughput methods of identification of moonlighting functions are required. Although most moonlighting activities reported to date were recognized by serendipity, the use of systematic methods is slowly gaining prominence, and we reviewed above some methodologies that hold promise in this area.
Acknowledgments
The research that led to the formulation of these models was supported in part by National Institute on Diabetes and Digestive and Kidney Diseases, National Institutes of Health (NIH), grant K08 DK60055 (to K.M.D.); National Institute of General Medical Science, NIH, grant R01 GM067929 (to K.M.D.); and National Institute of Child Health and Human Development, NIH, grant R01 HD22563 (to E.R.B.M.).
Electronic-Database Information
The URL for data presented herein is as follows:
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/
References
- Amraei M, Nabi IR (2002) Species specificity of the cytokine function of phosphoglucose isomerase. FEBS Lett 525:151–155 [DOI] [PubMed] [Google Scholar]
- Arai K, Yasuda N, Isohashi F, Okamoto K, Ohkuma S (2002) Inhibition of weak-base amine-induced lysis of lysosomes by cytosol. J Biochem (Tokyo) 132:529–534 [DOI] [PubMed] [Google Scholar]
- Arnold H, Pette D (1970) Binding of aldolase and triosephosphate dehydrogenase to F-actin and modification of catalytic properties of aldolase. Eur J Biochem 15:360–366 [DOI] [PubMed] [Google Scholar]
- Ashizawa K, Fukuda T, Cheng SY (1992) Transcriptional stimulation by thyroid hormone of a cytosolic thyroid hormone binding protein which is homologous to a subunit of pyruvate kinase M1. Biochemistry 31:2774–2778 [DOI] [PubMed] [Google Scholar]
- Ashizawa K, McPhie P, Lin KH, Cheng SY (1991) An in vitro novel mechanism of regulating the activity of pyruvate kinase M2 by thyroid hormone and fructose 1,6-bisphosphate. Biochemistry 30:7105–7111 [DOI] [PubMed] [Google Scholar]
- Astuti D, Latif F, Dallol A, Dahia PL, Douglas F, George E, Skoldberg F, Husebye ES, Eng C, Maher ER (2001) Gene mutations in the succinate dehydrogenase subunit SDHB cause susceptibility to familial pheochromocytoma and to familial paraganglioma. Am J Hum Genet 69:49–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basilion JP, Rouault TA, Massinople CM, Klausner RD, Burgess WH (1994) The iron-responsive element-binding protein: localization of the RNA-binding site to the aconitase active-site cleft. Proc Natl Acad Sci USA 91:574–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman OA, Purkiss AG, van Montfort R, Slingsby C, Graham C, Wistow G (2003) Crystal structure of η-crystallin: adaptation of a class 1 aldehyde dehydrogenase for a new role in the eye lens. Biochemistry 42:4349–4356 [DOI] [PubMed] [Google Scholar]
- Baxi MD, Vishwanatha JK (1995) Uracil DNA-glycosylase/glyceraldehyde-3-phosphate dehydrogenase is an Ap4A binding protein. Biochemistry 34:9700–9707 [DOI] [PubMed] [Google Scholar]
- Berry MD, Boulton AA (2000) Glyceraldehyde-3-phosphate dehydrogenase and apoptosis. J Neurosci Res 60:150–154 [DOI] [PubMed] [Google Scholar]
- Bhat RV, Budd Haeberlein SL, Avila J (2004) Glycogen synthase kinase 3: a drug target for CNS therapies. J Neurochem 89:1313–1317 [DOI] [PubMed] [Google Scholar]
- Bielli P, Calabrese L (2002) Structure to function relationships in ceruloplasmin: a ‘moonlighting’ protein. Cell Mol Life Sci 59:1413–1427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blomquist HK, Dahl N, Gustafsson L, Hellerud C, Holme E, Holmgren G, Matsson L, von Zweigbergk M (1996) Glycerol kinase deficiency in two brothers with and without clinical manifestations. Clin Genet 50:375–379 [DOI] [PubMed] [Google Scholar]
- Boonacker E, Van Noorden CJ (2003) The multifunctional or moonlighting protein CD26/DPPIV. Eur J Cell Biol 82:53–73 [DOI] [PubMed] [Google Scholar]
- Chaput M, Claes V, Portetelle D, Cludts I, Cravador A, Burny A, Gras H, Tartar A (1988) The neurotrophic factor neuroleukin is 90% homologous with phosphohexose isomerase. Nature 332:454–455 [DOI] [PubMed] [Google Scholar]
- Chen XJ, Wang X, Kaufman BA, Butow RA (2005) Aconitase couples metabolic regulation to mitochondrial DNA maintenance. Science 307:714–717 [DOI] [PubMed] [Google Scholar]
- Chuang DM, Hough C, Senatorov VV (2005) Glyceraldehyde-3-phosphate dehydrogenase, apoptosis, and neurodegenerative diseases. Annu Rev Pharmacol Toxicol 45:269–290 [DOI] [PubMed] [Google Scholar]
- Clipsham R, McCabe ER (2003) DAX1 and its network partners: exploring complexity in development. Mol Genet Metab 80:81–120 [DOI] [PubMed] [Google Scholar]
- Clipsham R, Zhang YH, Huang BL, McCabe ER (2002) Genetic network identification by high density, multiplexed reversed transcriptional (HD-MRT) analysis in steroidogenic axis model cell lines. Mol Genet Metab 77:159–178 [DOI] [PubMed] [Google Scholar]
- Dipple KM, McCabe ER (2000a) Modifier genes convert “simple” Mendelian disorders to complex traits. Mol Genet Metab 71:43–50 [DOI] [PubMed] [Google Scholar]
- ——— (2000b) Phenotypes of patients with “simple” Mendelian disorders are complex traits: thresholds, modifiers, and systems dynamics. Am J Hum Genet 66:1729–1735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dipple KM, Phelan JK, McCabe ER (2001a) Consequences of complexity within biological networks: robustness and health, or vulnerability and disease. Mol Genet Metab 74:45–50 [DOI] [PubMed] [Google Scholar]
- Dipple KM, Zhang YH, Huang BL, McCabe LL, Dallongeville J, Inokuchi T, Kimura M, Marx HJ, Roederer GO, Shih V, Yamaguchi S, Yoshida I, McCabe ER (2001b) Glycerol kinase deficiency: evidence for complexity in a single gene disorder. Hum Genet 109:55–62 [DOI] [PubMed] [Google Scholar]
- Duclos-Vallee JC, Capel F, Mabit H, Petit MA (1998) Phosphorylation of the hepatitis B virus core protein by glyceraldehyde-3-phosphate dehydrogenase protein kinase activity. J Gen Virol 79:1665–1670 [DOI] [PubMed] [Google Scholar]
- Dudani AK, Cummings C, Hashemi S, Ganz PR (1993) Isolation of a novel 45 kDa plasminogen receptor from human endothelial cells. Thromb Res 69:185–196 [DOI] [PubMed] [Google Scholar]
- Elzinga SD, Bednarz AL, van Oosterum K, Dekker PJ, Grivell LA (1993) Yeast mitochondrial NAD+-dependent isocitrate dehydrogenase is an RNA-binding protein. Nucleic Acids Res 21:5328–5331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel M, Seifert M, Theisinger B, Seyfert U, Welter C (1998) Glyceraldehyde-3-phosphate dehydrogenase and Nm23-H1/nucleoside diphosphate kinase A: two old enzymes combine for the novel Nm23 protein phosphotransferase function. J Biol Chem 273:20058–20065 [DOI] [PubMed] [Google Scholar]
- Faik P, Walker JI, Redmill AA, Morgan MJ (1988) Mouse glucose-6-phosphate isomerase and neuroleukin have identical 3′ sequences. Nature 332:455–457 [DOI] [PubMed] [Google Scholar]
- Fiek C, Benz R, Roos N, Brdiczka D (1982) Evidence for identity between the hexokinase-binding protein and the mitochondrial porin in the outer membrane of rat liver mitochondria. Biochim Biophys Acta 688:429–440 [DOI] [PubMed] [Google Scholar]
- Fillebeen C, Caltagirone A, Martelli A, Moulis JM, Pantopoulos K. IRP1 Ser-711 is a phosphorylation site, critical for regulation of RNA-binding and aconitase activities. Biochem J (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez A, Domedel N, Cedano J, Pinol J, Querol E (2003) Do current sequence analysis algorithms disclose multifunctional (moonlighting) proteins? Bioinformatics 19:895–896 [DOI] [PubMed] [Google Scholar]
- Gurney ME, Heinrich SP, Lee MR, Yin HS (1986) Molecular cloning and expression of neuroleukin, a neurotrophic factor for spinal and sensory neurons. Science 234:566–574 [DOI] [PubMed] [Google Scholar]
- Hall DA, Zhu H, Zhu X, Royce T, Gerstein M, Snyder M (2004) Regulation of gene expression by a metabolic enzyme. Science 306:482–484 [DOI] [PubMed] [Google Scholar]
- Hall RE, Henriksson KG, Lewis SF, Haller RG, Kennaway NG (1993) Mitochondrial myopathy with succinate dehydrogenase and aconitase deficiency: abnormalities of several iron-sulfur proteins. J Clin Invest 92:2660–2666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haller RG, Henriksson KG, Jorfeldt L, Hultman E, Wibom R, Sahlin K, Areskog NH, et al (1991) Deficiency of skeletal muscle succinate dehydrogenase and aconitase: pathophysiology of exercise in a novel human muscle oxidative defect. J Clin Invest 88:1197–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison R (2004) Physiological roles of xanthine oxidoreductase. Drug Metab Rev 36:363–375 [DOI] [PubMed] [Google Scholar]
- Huq AH, Lovell RS, Sampson MJ, Decker WK, Dinulos MB, Disteche CM, Craigen WJ (1996) Isolation, mapping, and functional expression of the mouse X chromosome glycerol kinase gene. Genomics 36:530–534 [DOI] [PubMed] [Google Scholar]
- Ishitani R, Sunaga K, Tanaka M, Aishita H, Chuang DM (1997) Overexpression of glyceraldehyde-3-phosphate dehydrogenase is involved in low K+-induced apoptosis but not necrosis of cultured cerebellar granule cells. Mol Pharmacol 51:542–550 [DOI] [PubMed] [Google Scholar]
- Jeffery CJ (1999) Moonlighting proteins. Trends Biochem Sci 24:8–11 [DOI] [PubMed] [Google Scholar]
- ——— (2003a) Moonlighting proteins: old proteins learning new tricks. Trends Genet 19:415–417 [DOI] [PubMed] [Google Scholar]
- ——— (2003b) Multifunctional proteins: examples of gene sharing. Ann Med 35:28–35 [DOI] [PubMed] [Google Scholar]
- ——— (2004a) Molecular mechanisms for multitasking: recent crystal structures of moonlighting proteins. Curr Opin Struct Biol 14:663–668 [DOI] [PubMed] [Google Scholar]
- ——— (2004b) Moonlighting proteins: complications and implications for proteomics research. Drug Discov Today Targets 3:71–78 [Google Scholar]
- ——— Mass spectrometry and the search for moonlighting proteins. Mass Spectrom Rev (in press) [DOI] [PubMed] [Google Scholar]
- Jiang H, Guo W, Liang X, Rao Y (2005) Both the establishment and the maintenance of neuronal polarity require active mechanisms: critical roles of GSK-3β and its upstream regulators. Cell 120:123–135 [DOI] [PubMed] [Google Scholar]
- Kao KC, Yang YL, Boscolo R, Sabatti C, Roychowdhury V, Liao JC (2004) Transcriptome-based determination of multiple transcription regulator activities in Escherichia coli by using network component analysis. Proc Natl Acad Sci USA 101:641–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsube N, Sunaga K, Aishita H, Chuang DM, Ishitani R (1999) ONO-1603, a potential antidementia drug, delays age-induced apoptosis and suppresses overexpression of glyceraldehyde-3-phosphate dehydrogenase in cultured central nervous system neurons. J Pharmacol Exp Ther 288:6–13 [PubMed] [Google Scholar]
- Katsube N, Sunaga K, Chuang D-M, Ishitani R (1996) ONO-1603, a potential antidementia drug, shows neuroprotective effects and increases m3-muscarinic receptor mRNA levels in differentiating rat cerebellar granule neurons. Neurosci Lett 214:151–154 [DOI] [PubMed] [Google Scholar]
- Kawamoto RM, Caswell AH (1986) Autophosphorylation of glyceraldehydephosphate dehydrogenase and phosphorylation of protein from skeletal muscle microsomes. Biochemistry 25:657–661 [DOI] [PubMed] [Google Scholar]
- Kennedy MC, Mende-Mueller L, Blondin GA, Beinert H (1992) Purification and characterization of cytosolic aconitase from beef liver and its relationship to the iron-responsive element binding protein. Proc Natl Acad Sci USA 89:11730–11734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kugler W, Breme K, Laspe P, Muirhead H, Davies C, Winkler H, Schroter W, Lakomek M (1998) Molecular basis of neurological dysfunction coupled with haemolytic anaemia in human glucose-6-phosphate isomerase (GPI) deficiency. Hum Genet 103:450–454 [DOI] [PubMed] [Google Scholar]
- Kumagai H, Sakai H (1983) A porcine brain protein (35 K protein) which bundles microtubules and its identification as glyceraldehyde 3-phosphate dehydrogenase. J Biochem 93:1259–1269 [DOI] [PubMed] [Google Scholar]
- Lay AJ, Jiang XM, Daly E, Sun L, Hogg PJ (2002) Plasmin reduction by phosphoglycerate kinase is a thiol-independent process. J Biol Chem 277:9062–9068 [DOI] [PubMed] [Google Scholar]
- Lay AJ, Jiang XM, Kisker O, Flynn E, Underwood A, Condron R, Hogg PJ (2000) Phosphoglycerate kinase acts in tumour angiogenesis as a disulphide reductase. Nature 408:869–873 [DOI] [PubMed] [Google Scholar]
- Lee AY, Hsu CH, Wu SH (2004) Functional domains of Brevibacillus thermoruber Lon protease for oligomerization and DNA binding: role of N-terminal and sensor and substrate discrimination domains. J Biol Chem 279:34903–34912 [DOI] [PubMed] [Google Scholar]
- Lee Y-H, Nadaraia S, Gu D, Becker DF, Tanner JJ (2003) Structure of the proline dehydrogenase domain of the multifunctional PutA flavoprotein. Nat Struct Biol 10:109–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann AR (2001) The xeroderma pigmentosum group D (XPD) gene: one gene, two functions, three diseases. Genes Dev 15:15–23 [DOI] [PubMed] [Google Scholar]
- ——— (2003) DNA repair-deficient diseases, xeroderma pigmentosum, Cockayne syndrome and trichothiodystrophy. Biochimie 85:1101–1111 [DOI] [PubMed] [Google Scholar]
- Li Y, Nowotny P, Holmans P, Smemo S, Kauwe JSK, Hinrichs AL, Tacey K, et al (2004) Association of late-onset Alzheimer’s disease with genetic variation in multiple members of the GAPD gene family. Proc Natl Acad Sci USA 101:15688–15693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao JC, Boscolo R, Yang YL, Tran LM, Sabatti C, Roychowdhury VP (2003) Network component analysis: reconstruction of regulatory signals in biological systems. Proc Natl Acad Sci USA 100:15522–15527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linden M, Gellerfors P, Nelson BD (1982) Pore protein and the hexokinase-binding protein from the outer membrane of rat liver mitochondria are identical. FEBS Lett 141:189–192 [DOI] [PubMed] [Google Scholar]
- Lipinski P, Starzynski RR, Drapier J-C, Bouton C, Bartlomiejczyk T, Sochanowicz B, Smuda E, Gajkowska A, Kruszewski M (2005) Induction of iron regulatory protein 1 RNA-binding activity by nitric oxide is associated with a concomitant increase in the labile iron pool: implications for DNA damage. Biochem Biophys Res Commun 327:349–355 [DOI] [PubMed] [Google Scholar]
- Lu M, Holliday LS, Zhang L, Dunn WA Jr, Gluck SL (2001) Interaction between aldolase and vacuolar H+-ATPase: evidence for direct coupling of glycolysis to the ATP-hydrolyzing proton pump. J Biol Chem 276:30407–30413 [DOI] [PubMed] [Google Scholar]
- Lu M, Sautin YY, Holliday LS, Gluck SL (2004) The glycolytic enzyme aldolase mediates assembly, expression, and activity of vacuolar H+-ATPase. J Biol Chem 279:8732–8739 [DOI] [PubMed] [Google Scholar]
- Martinez JA, McCabe ERB (2004) Apoptosis in glycerol kinase deficiency: investigations in Drosophila melangaster. Paper presented at the American Society of Human Genetics Annual Meeting, Toronto, October 26–30 [Google Scholar]
- McCabe ER (1983) Human glycerol kinase deficiency: an inborn error of compartmental metabolism. Biochem Med 30:215–230 [DOI] [PubMed] [Google Scholar]
- McManaman JL, Neville MC, Wright RM (1999) Mouse mammary gland xanthine oxidoreductase: purification, characterization, and regulation. Arch Biochem Biophys 371:308–316 [DOI] [PubMed] [Google Scholar]
- McManaman JL, Palmer CA, Anderson S, Schwertfeger K, Neville MC (2004) Regulation of milk lipid formation and secretion in the mouse mammary gland. Adv Exp Med Biol 554:263–279 [DOI] [PubMed] [Google Scholar]
- McManaman JL, Palmer CA, Wright RM, Neville MC (2002) Functional regulation of xanthine oxidoreductase expression and localization in the mouse mammary gland: evidence of a role in lipid secretion. J Physiol 545:567–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer-Siegler K, Mauro DJ, Seal G, Wurzer J, deRiel JK, Sirover MA (1991) A human nuclear uracil DNA glycosylase is the 37-kDa subunit of glyceraldehyde-3-phosphate dehydrogenase. Proc Natl Acad Sci USA 88:8460–8464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki K, Qu W, Goulding EH, Willis WD, Bunch DO, Strader LF, Perreault SD, Eddy EM, O’Brien DA (2004) Glyceraldehyde 3-phosphate dehydrogenase-S, a sperm-specific glycolytic enzyme, is required for sperm motility and male fertility. Proc Natl Acad Sci USA 101:16501–16506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miles LA, Dahlberg CM, Plescia J, Felez J, Kato K, Plow EF (1991) Role of cell-surface lysines in plasminogen binding to cells: identification of α-enolase as a candidate plasminogen receptor. Biochemistry 30:1682–1691 [DOI] [PubMed] [Google Scholar]
- Mitsuzawa H, Kimura M, Kanda E, Ishihama A (2005) Glyceraldehyde-3-phosphate dehydrogenase and actin associate with RNA polymerase II and interact with its Rpb7 subunit. FEBS Lett 579:48–52 [DOI] [PubMed] [Google Scholar]
- Muller C, Paupert J, Monferran S, Salles B (2005) The double life of the Ku protein: facing the DNA breaks and the extracellular environment. Cell Cycle 4:438–441 [DOI] [PubMed] [Google Scholar]
- Munoz ME, Ponce E (2003) Pyruvate kinase: current status of regulatory and functional properties. Comp Biochem Physiol B Biochem Mol Biol 135:197–218 [DOI] [PubMed] [Google Scholar]
- Nakajima K, Hamanoue M, Takemoto N, Hattori T, Kato K, Kohsaka S (1994) Plasminogen binds specifically to α-enolase on rat neuronal plasma membrane. J Neurochem 63:2048–2057 [DOI] [PubMed] [Google Scholar]
- Numata O (1996) Multifunctional proteins in Tetrahymena: 14-nm filament protein/citrate synthase and translation elongation factor-1α. Int Rev Cytol 164:1–35 [DOI] [PubMed] [Google Scholar]
- Numata O, Takemasa T, Takagi I, Hirono M, Hirano H, Chiba J, Watanabe Y (1991) Tetrahymena 14-nm filament-forming protein has citrate synthase activity. Biochem Biophys Res Commun 174:1028–1034 [DOI] [PubMed] [Google Scholar]
- Ogg SL, Weldon AK, Dobbie L, Smith AJH, Mather IH (2004) Expression of butyrophilin (Btn1a1) in lactating mammary gland is essential for the regulated secretion of milk-lipid droplets. Proc Natl Acad Sci USA 101:10084–10089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto K, Hirano H, Isohashi F (1993) Molecular cloning of rat liver glucocorticoid-receptor translocation promoter. Biochem Biophys Res Commun 193:848–854 [DOI] [PubMed] [Google Scholar]
- Okamoto K, Isohashi F, Horiuchi M, Sakamoto Y (1984) An ATP-stimulated factor that enhances the nuclear binding of “activated” receptor-glucocorticoid complex. Biochem Biophys Res Commun 121:940–945 [DOI] [PubMed] [Google Scholar]
- Okamoto K, Isohashi F, Ueda K, Sakamoto Y (1989) Properties of an adenosine triphosphate-stimulated factor that enhances the nuclear binding of activated glucocorticoid-receptor complex: binding to histone-agarose. Endocrinology 124:675–680 [DOI] [PubMed] [Google Scholar]
- Ostlund AK, Gohring U, Krause J, Brdiczka D (1983) The binding of glycerol kinase to the outer membrane of rat liver mitochondria: its importance in metabolic regulation. Biochem Med 30:231–245 [DOI] [PubMed] [Google Scholar]
- Page S, Powell D, Benboubetra M, Stevens CR, Blake DR, Selase F, Wolstenholme AJ, Harrison R (1998) Xanthine oxidoreductase in human mammary epithelial cells: activation in response to inflammatory cytokines. Biochim Biophys Acta 1381:191–202 [DOI] [PubMed] [Google Scholar]
- Pancholi V (2001) Multifunctional α-enolase: its role in diseases. Cell Mol Life Sci 58:902–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrini JH (2000) When more is better. Nat Genet 26:257–258 [DOI] [PubMed] [Google Scholar]
- Piatigorsky J (1998a) Gene sharing in lens and cornea: facts and implications. Prog Retin Eye Res 17:145–174 [DOI] [PubMed] [Google Scholar]
- ——— (1998b) Multifunctional lens crystallins and corneal enzymes: more than meets the eye. Ann N Y Acad Sci 842:7–15 [DOI] [PubMed] [Google Scholar]
- ——— (2003) Crystallin genes: specialization by changes in gene regulation may precede gene duplication. J Struct Funct Genomics 3:131–137 [PubMed] [Google Scholar]
- Ramasarma T (1994) One protein—many functions. Curr Sci 67:24–29 [Google Scholar]
- Robbins AR, Ward RD, Oliver C (1995) A mutation in glyceraldehyde 3-phosphate dehydrogenase alters endocytosis in CHO cells. J Cell Biol 130:1093–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouault TA, Haile DJ, Downey WE, Philpott CC, Tang C, Samaniego F, Chin J, Paul I, Orloff D, Harford JB, Klausner RD (1992) An iron-sulfur cluster plays a novel regulatory role in the iron-responsive element binding protein. Biometals 5:131–140 [DOI] [PubMed] [Google Scholar]
- Rusing D, Verspohl EJ (2004) Influence of diadenosine tetraphosphate (Ap4A) on lipid metabolism. Cell Biochem Funct 22:333–338 [DOI] [PubMed] [Google Scholar]
- Ryazanov AG (1985) Glyceraldehyde-3-phosphate dehydrogenase is one of the three major RNA-binding proteins of rabbit reticulocytes. FEBS Lett 192:131–134 [DOI] [PubMed] [Google Scholar]
- Saeki T, Hori M, Umezawa H (1974) Pyruvate kinase of Escherichia coli: its role in supplying nucleoside triphosphates in cells under anaerobic conditions. J Biochem 76:631–637 [DOI] [PubMed] [Google Scholar]
- Sargent CA, Kidd A, Moore S, Dean J, Besley GT, Affara NA (2000) Five cases of isolated glycerol kinase deficiency, including two families: failure to find genotype:phenotype correlation. J Med Genet 37:434–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sautin YY, Lu M, Gaugler A, Zhang L, Gluck SL (2005) Phosphatidylinositol 3-kinase-mediated effects of glucose on vacuolar H+-ATPase assembly, translocation, and acidification of intracellular compartments in renal epithelial cells. Mol Cell Biol 25:575–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawa A, Khan AA, Hester LD, Snyder SH (1997) Glyceraldehyde-3-phosphate dehydrogenase: nuclear translocation participates in neuronal and nonneuronal cell death. Proc Natl Acad Sci USA 94:11669–11674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scriver CR, Waters PJ (1999) Monogenic traits are not simple: lessons from phenylketonuria. Trends Genet 15:267–272 [DOI] [PubMed] [Google Scholar]
- Selak MA, Armour SM, Mackenzie ED, Boulahbel H, Watson DG, Mansfield KD, Pan Y, Simon MC, Thompson CB, Gottlieb E (2005) Succinate links TCA cycle dysfunction to oncogenesis by inhibiting HIF-α prolyl hydroxylase. Cancer Cell 7:77–85 [DOI] [PubMed] [Google Scholar]
- Shashidharan P, Chalmers-Redman RM, Carlile GW, Rodic V, Gurvich N, Yuen T, Tatton WG, Sealfon SC (1999) Nuclear translocation of GAPDH-GFP fusion protein during apoptosis. Neuroreport 10:1149–1153 [DOI] [PubMed] [Google Scholar]
- Shears SB (2004) How versatile are inositol phosphate kinases? Biochem J 377:265–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R, Green MR (1993) Sequence-specific binding of transfer RNA by glyceraldehyde-3-phosphate dehydrogenase. Science 259:365–368 [DOI] [PubMed] [Google Scholar]
- Sirover MA (1999) New insights into an old protein: the functional diversity of mammalian glyceraldehyde-3-phosphate dehydrogenase. Biochim Biophys Acta 1432:159–184 [DOI] [PubMed] [Google Scholar]
- Sjarif DR, Ploos van Amstel JK, Duran M, Beemer FA, Poll-The BT (2000) Isolated and contiguous glycerol kinase gene disorders: a review. J Inherit Metab Dis 23:529–547 [DOI] [PubMed] [Google Scholar]
- Starzynski RR, Gralak MA, Smuda E, Lipinski P (2004) A characterization of the activities of iron regulatory protein 1 in various farm animal species. Cell Mol Biol Lett 9:651–664 [PubMed] [Google Scholar]
- Sunaga K, Takahashi H, Chuang DM, Ishitani R (1995) Glyceraldehyde-3-phosphate dehydrogenase is over-expressed during apoptotic death of neuronal cultures and is recognized by a monoclonal antibody against amyloid plaques from Alzheimer’s brain. Neurosci Lett 200:133–136 [DOI] [PubMed] [Google Scholar]
- Sundin GW, Shankar S, Chugani SA, Chopade BA, Kavanaugh-Black A, Chakrabarty AM (1996) Nucleoside diphosphate kinase from Pseudomonas aeruginosa: characterization of the gene and its role in cellular growth and exopolysaccharide alginate synthesis. Mol Microbiol 20:965–979 [DOI] [PubMed] [Google Scholar]
- Tatton WG, Chalmers-Redman RM, Elstner M, Leesch W, Jagodzinski FB, Stupak DP, Sugrue MM, Tatton NA (2000) Glyceraldehyde-3-phosphate dehydrogenase in neurodegeneration and apoptosis signaling. J Neural Transm Suppl:77–100 [DOI] [PubMed] [Google Scholar]
- Tisdale EJ (2001) Glyceraldehyde-3-phosphate dehydrogenase is required for vesicular transport in the early secretory pathway. J Biol Chem 276:2480–2486 [DOI] [PubMed] [Google Scholar]
- Tomlinson IP, Alam NA, Rowan AJ, Barclay E, Jaeger EE, Kelsell D, Leigh I, et al (2002) Germline mutations in FH predispose to dominantly inherited uterine fibroids, skin leiomyomata and papillary renal cell cancer. Nat Genet 30:406–410 [DOI] [PubMed] [Google Scholar]
- Tsuchiya K, Tajima H, Kuwae T, Takeshima T, Nakano T, Tanaka M, Sunaga K, Fukuhara Y, Nakashima K, Ohama E, Mochizuki H, Mizuno Y, Katsube N, Ishitani R (2005) Pro-apoptotic protein glyceraldehyde-3-phosphate dehydrogenase promotes the formation of Lewy body-like inclusions. Eur J Neurosci 21:317–326 [DOI] [PubMed] [Google Scholar]
- Tsuchiya K, Tajima H, Yamada M, Takahashi H, Kuwae T, Sunaga K, Katsube N, Ishitani R (2004) Disclosure of a pro-apoptotic glyceraldehyde-3-phosphate dehydrogenase promoter: anti-dementia drugs depress its activation in apoptosis. Life Sciences 74:3245–3258 [DOI] [PubMed] [Google Scholar]
- Vartanian AA (2003) Gelsolin and plasminogen activator inhibitor-1 are Ap3A-binding proteins. Ital J Biochem 52:9–16 [PubMed] [Google Scholar]
- Vorbach C, Scriven A, Capecchi MR (2002) The housekeeping gene xanthine oxidoreductase is necessary for milk fat droplet enveloping and secretion: gene sharing in the lactating mammary gland. Genes Dev 16:3223–3235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Woltjer RL, Cimino PJ, Pan C, Montine KS, Zhang J, Montine TJ. Proteomic analysis of neurofibrillary tangles in Alzheimer disease identifies GAPDH as a detergent-insoluble paired helical filament tau binding protein. FASEB J (in press) [DOI] [PubMed] [Google Scholar]
- Wang W, Wang L, Endoh A, Hummelke G, Hawks CL, Hornsby PJ (2005) Identification of α-enolase as a nuclear DNA-binding protein in the zona fasciculata but not the zona reticularis of the human adrenal cortex. J Endocrinol 184:85–94 [DOI] [PubMed] [Google Scholar]
- Wang Y, Chang CF, Morales M, Chiang YH, Harvey BK, Su TP, Tsao LI, Chen S, Thiemermann C (2003) Diadenosine tetraphosphate protects against injuries induced by ischemia and 6-hydroxydopamine in rat brain. J Neurosci 23:7958–7965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang-Su ST, McCormack AL, Yang S, Hosler MR, Mixon A, Riviere MA, Wilmarth PA, Andley UP, Garland D, Li H, David LL, Wagner BJ (2003) Proteome analysis of lens epithelia, fibers, and the HLE B-3 cell line. Invest Ophthalmol Vis Sci 44:4829–4836 [DOI] [PubMed] [Google Scholar]
- Watanabe H, Takehana K, Date M, Shinozaki T, Raz A (1996) Tumor cell autocrine motility factor is the neuroleukin/phosphohexose isomerase polypeptide. Cancer Res 56:2960–2963 [PubMed] [Google Scholar]
- Wistow GJ, Lietman T, Williams LA, Stapel SO, de Jong WW, Horwitz J, Piatigorsky J (1988) τ-Crystallin/α-enolase: one gene encodes both an enzyme and a lens structural protein. J Cell Biol 107:2729–2736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Seiter K, Feldman E, Ahmed T, Chiao JW (1996) The differentiation and maturation mediator for human myeloid leukemia cells shares homology with neuroleukin or phosphoglucose isomerase. Blood 87:4502–4506 [PubMed] [Google Scholar]
- Yanagawa T, Funasaka T, Tsutsumi S, Raz T, Tanaka N, Raz A (2005) Differential regulation of phosphoglucose isomerase/autocrine motility factor activities by protein kinase CK2 phosphorylation. J Biol Chem 280:10419–10426 [DOI] [PubMed] [Google Scholar]
- Yoshimura T, Kawano Y, Arimura N, Kawabata S, Kikuchi A, Kaibuchi K (2005) GSK-3β regulates phosphorylation of CRMP-2 and neuronal polarity. Cell 120:137–149 [DOI] [PubMed] [Google Scholar]