Abstract
Assembly of new chromatin during S phase requires the histone chaperone complexes CAF-1 (Cac2p, Msi1p and Rlf2p) and RCAF (Asf1p plus acetylated histones H3 and H4). Cells lacking CAF-1 and RCAF are hypersensitive to DNA-damaging agents, such as methyl methanesulfonate and camptothecin, suggesting a possible defect in double-strand break (DSB) repair. Assays developed to quantitate repair of defined, cohesive-ended break structures revealed that DSB-induced plasmid:chromosome recombination was reduced ∼10-fold in RCAF/CAF-1 double mutants. Recombination defects were similar with both chromosomal and plasmid targets in vivo, suggesting that inhibitory chromatin structures were not involved. Consistent with these observations, ionizing radiation-induced loss of heterozygosity was abolished in the mutants. Nonhomologous end-joining (NHEJ) repair proficiency and accuracy were intermediate between wild-type levels and those of NHEJ-deficient yku70 and rad50 mutants. The defects in NHEJ, but not homologous recombination, could be rescued by deletion of HMR-a1, a component of the a1/alpha2 transcriptional repressor complex. The findings are consistent with the observation that silent mating loci are partially derepressed. These results demonstrate that defective assembly of nucleosomes during new DNA synthesis compromises each of the known pathways of DSB repair and that the effects can be indirect consequences of changes in silenced chromatin structure.
INTRODUCTION
DNA double-strand breaks (DSBs) can arise within cells by several different mechanisms. Exogenous sources include physical and chemical DNA-damaging agents, such as ionizing radiation, bleomycin and methyl methanesulfonate (MMS). Most such agents produce multiple types of DNA damage, but cause cell killing primarily because of unrepaired DSBs (1,2). Endogenous sources of DSBs include intracellular nucleases, chemicals such as highly reactive free radicals derived from oxygen metabolism, and processes leading to arrest and collapse of DNA replication forks (3–6). DNA strand breaks are also associated with at-risk sequence motifs, such as inverted repeats and triplet repeats (7,8). Consequences of DSBs include arrest at damage-associated cell cycle checkpoints, loss of cell viability and elevated DNA instability, including increases in mutation, recombination and chromosome rearrangements.
The two major DSB repair pathways, nonhomologous end-joining (NHEJ) and homologous recombination, are highly conserved in eukaryotes. In the budding yeast Saccharomyces cerevisiae Rad50, Mre11 and Xrs2 form a nuclease complex, RMX, that is active in both the recombination and NHEJ pathways (9). The recombination pathway also requires Rad51, Rad52, Rad54, Rad55, Rad57, Rad59, the Rfa complex (single-stranded binding protein) and other accessory proteins (9,10). These proteins form specific associations with each other in vitro and in vivo and mediate DNA strand annealing and exchange.
Proteins involved in homology-independent repair by the NHEJ pathway include the RMX, Yku70/Yku80, Dnl4/Lif1/Nej1 and Sir2/Sir3/Sir4 complexes (5,11–14). Past studies have suggested that these complexes are able to bind to DNA ends either directly or indirectly, though their exact roles in NHEJ remain unclear (5,15).
An essential step in the completion of S phase of the cell cycle is the reassembly of histone octamers onto newly replicated DNA and most new nucleosome assembly occurs at this time. Two histone chaperone complexes, CAF-1 and RCAF, are known to be involved in replication-coupled nucleosome assembly in yeast and in other eukaryotes. CAF-1 consists of at least three components, referred to as Rlf2, Cac2 and Msi1 in S.cerevisiae (16–18). Inactivation of any of these genes leads to partial alleviation of silencing at telomeric and mating type loci, indicating a role for CAF-1 in maintenance of repressive chromatin structures (17,19–24). Further evidence suggesting a role for the complex during S-phase replication comes from the observations that CAF-1 binds to and co-localizes with the DNA polymerase clamp protein PCNA (proliferating cell nuclear antigen) and replication factor C in vivo (11,25,26).
The RCAF complex was initially identified as a factor in Drosophila embryo extracts displaying activity in an SV40-based DNA replication/chromatin assembly assay (17). This complex, consisting of the highly conserved protein Asf1 combined with partially acetylated forms of histones H3 and H4, was found to act with CAF-1 in the assembly of chromatin onto newly synthesized DNA (17,27). Inactivation of the yeast ASF1 gene results in hypersensitivity to DNA-damaging agents and increased chromosome loss (17,24,28–30). Recent experiments have demonstrated that Asf1 can form associations with Rad53, a mediator of cell cycle checkpoint responses to DNA damage, and with Hir1, a trans-acting regulator of histone gene expression (24,29,31). Although Asf1 binds to Rad53 in undamaged cells, the available data are consistent with a role for Asf1 in nucleosome assembly and DNA repair, but not in cell cycle checkpoint signaling (29).
RCAF works synergistically with CAF-1 in assays of nucleosome assembly, possibly acting as a histone donor in the process (16,17). Yeast cells lacking both complexes (e.g. rlf2 asf1 double mutants) are viable, but grow slowly and have greater defects in the assembly of silenced chromatin at the mating loci and at telomeres than single mutants. Past studies have also suggested a role in repair of damaged chromosomal DNA. For example, rlf2 mutants (CAF-1−) exhibit slight sensitivity to ultraviolet (UV), possibly due to impacts on the RAD6/RAD18 pathway of post-replication repair (32), and asf1 (RCAF−) cells were previously found to be moderately sensitive to UV and the chemical DNA-damaging agents MMS and HU (17,25,29,30,32). Furthermore, double mutant strains that have both RLF2 and ASF1 inactivated are more sensitive to UV and MMS than either single mutant (17).
Although not generally incorporated into DSB repair models, chromatin assembly and remodeling are likely to have an impact on the rejoining of broken chromosomal DNA ends. In the current study we have assessed the role of replication-coupled nucleosome assembly in the repair of chemical and radiation-induced DSBs inside cells and in the repair of a defined, site-specific DSB by the NHEJ and homologous recombination pathways.
MATERIALS AND METHODS
Yeast cells and growth media
Yeast strains used for this work are shown in Table 1. Most gene disruptions were accomplished using PCR fragment-mediated deletion disruption as described previously (33) in conjunction with plasmid-encoded genes providing resistance to Geneticin/G418 (Life Technologies, Inc.), hygromycin B (Boehringer Mannheim) or nourseothricin (Hans-Knoell Institute for Natural Products Research). For the selection of resistant yeast cells, antibiotics were added to rich (YPDA) plates as described previously (34). Gene disruptions involving HIS3, LEU2 or hisG-URA3-hisG markers were performed using deletion plasmids as described previously (33).
Table 1.
Yeast strains used in this study
| Strain | Genotype | Reference |
|---|---|---|
| BY4742 | MATα ura3Δ0 leu2Δ0 his3Δ1 lys2Δ0 | (38) |
| YB146 | MATa ura3-52 his3-Δ200 lys2-801 ade2-101 trp1-Δ1 | |
| gal3 trp1::[his3-Δ3′::HOcs his3-Δ5′] | (67) | |
| YLKL644 | BY4742, Δcac2::HygBr | This work |
| YLKL645 | BY4742, Δrlf2::HygBr | This work |
| YLKL646 | BY4742, Δmsi1::HygBr | This work |
| YLKL652 | BY4742, Δyku70::HIS3 | This work |
| YLKL687 | BY4742, Δrad51::LEU2 | This work |
| YLKL689 | BY4742, Δrad52::LEU2 | This work |
| YLKL728 | BY4742, Δasf1::G418r | This work |
| YLKL729 | BY4742, Δasf1::G418r, Δrlf2::HygBr | This work |
| YLKL730 | BY4742, Δasf1::G418r, Δcac2::HygBr | This work |
| YLKL753 | YB146, Δasf1::G418r, Δrlf2::HygBr | This work |
| YLKL755 | YB146, Δasf1::G418r, Δcac2::HygBr | This work |
| YLKL762 | BY4742, his3Δ0::Natr | This work |
| YLKL763 | BY4742, Δhmr-a1::Natr | This work |
| YLKL764 | BY4742, Δasf1::G418r, Δrlf2::HygBr Δhmr-a1::Natr | This work |
| YLKL765 | BY4742, Δasf1::G418r, Δrlf2::HygBr his3Δ0::Natr | This work |
| YLKL774 | YLKL762, Δrad51::LEU2 | This work |
| YLKL778 | YLKL762, Δrad50::hisG | This work |
Cell survival and loss of heterozygosity assays
For standard survival assays using replica-pronging techniques, wild-type and mutant strains were grown in YPDA broth in 96 well microtiter plates. Cells were subsequently pronged onto YPDA plates with or without DNA-damaging agents following 10-fold serial dilution. MMS (Fluka), hydroxyurea (HU) (Life Technologies, Inc.) or bleomycin (Sigma–Aldrich) were added to media after autoclaving. For YPDA plates containing camptothecin (Sigma–Aldrich), the drug was added at 10 µg/ml after buffering the growth medium with 25 mM HEPES, pH 7.2. To assess gamma radiation survival requiring sister chromatid recombination, 107 cells from logarithmically growing haploid cell cultures were exposed to 0, 40 or 80 krads, diluted serially and spread onto YPDA plates. Colonies were counted after 3–4 days of incubation at 30°C. To monitor gamma radiation resistance due primarily to recombination between homologous chromosomes, diploid strains were created by crossing BY4742 × YB146 (ASF1 RLF2) or YLKL729 × YLKL753 (asf1 rlf2) (Table 1). The diploid cells were grown for 2 days to stationary phase in YPDA broth, analyzed microscopically to confirm unbudded cells (i.e. G1 cells), irradiated and spread onto YPDA plates. Four independent cultures of each strain were assessed for the low dose experiments depicted in Figure 2B. For the high dose experiments (inset graph in Figure 2B), two cultures were used for each strain and the results averaged. For the measurement of loss of heterozygosity (LOH) frequencies, involving recombination within diploid ADE2/ade2 cells to form colonies containing both Ade+ (white) and Ade− (red) sectors, cells from two stationary phase cultures of each strain were spread to 40–50 YPDA plates and results combined. Plates were incubated for up to 5 days at 30°C to maximize red color development. Two independent isolates of each gene disrupted strain were tested using the haploid chemical and radiation sensitivity assays and gave comparable results. Subsequent tests of DSB repair used multiple cultures of one independent isolate characterized in the mutagen survival experiments.
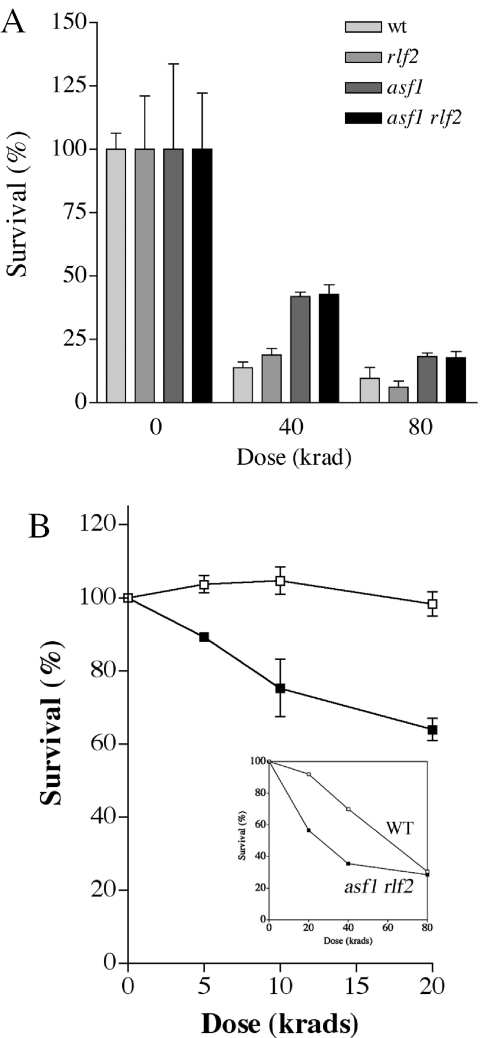
Figure 2.
Impact of inactivation of replication-coupled chromatin assembly genes on sensitivity to damage induced by ionizing radiation. (A) Logarithmically growing haploid cell cultures were exposed to gamma radiation and subsequently spread onto rich (YPDA) plates. (B) Stationary phase diploid cells were irradiated as for (A). The inset graph in (B) indicates a separate experiment performed using high radiation doses. Open squares, wild-type diploid cells; filled squares, RCAF/CAF-1 double mutant cells.
Nonhomologous end-joining and ends-in homologous recombination assays
Plasmid-based NHEJ repair assays were performed by LiAc transformation as described previously (33) using uncut or Nco1-cut plasmid p315URA3 (CEN/ARS URA3 LEU2) and selection for Leu+ transformant colonies. p315URA3 was generated by inserting the 1.2 kb HindIII fragment containing URA3 from YEp24 into the HindIII site of plasmid pRS315 (35). NcoI cleaves uniquely within the URA3 gene and mutational events associated with NHEJ repair were assessed by replica-plating Leu+ colonies to synthetic plates lacking uracil. In all experiments transformation efficiencies for broken plasmid DNAs were normalized to efficiencies for uncut centromeric control plasmids performed concurrently with the same competent yeast cell preparations.
‘Ends-in’ recombination proficiencies of cells were assessed by transformation with the integrating vector pLKL37Y (HIS3 URA3) that had been cut inside HIS3 with BclI. To allow digestion by the methylation-sensitive enzyme BclI, plasmid DNA was prepared from dam− Escherichia coli SCS110 cells (Stratagene). After BclI digestion and transformation, His+ colonies formed by recombinational integration of the plasmid into the his3-Δ1 locus were scored. Repair efficiencies (measured as transformants per microgram of DNA) were normalized to those for uncut CEN/ARS plasmid DNA (pRS313). Results presented are the mean ± SD of 3–5 experiments for each strain.
For recombination assays involving exchange between linearized plasmid DNA and either a chromosomal target or a plasmid target, pRS303 (HIS3) was used after cleavage with BclI. To generate the target plasmid used in the assays, the his3-Δ1 locus on chromosome XV of BY4742 was PCR amplified using primers 5′BamHIS3 (ATGCGTACGGATCCGCCTCCTCTAGTACACTCTATATT) and 3′BamHIS3 (ATGCGTACGGATCCGCAGCTTTAAATAATCGGTGTCAC). The resulting PCR fragment was digested with BamHI and cloned into BamHI-cut YCp50 (36) to create plasmid pLKL71Y (CEN/ARS URA3 his3-Δ1). Host cells for the assays were BY4742 cells in which his3-Δ1 had been converted to a complete deletion of the HIS3 gene by insertion of Natr (encoding resistance to nourseothricin). Strain YLKL762 and repair-deficient derivatives were used for the experiments (Table 1). To confirm that BclI-cut pRS303 DNA efficiently recombined in vivo with pLKL71Y after transformation, 20 His+ transformant colonies were replica-plated to synthetic plates containing 5-fluoroorotic acid (5-FOA) (ZymoResearch). The resulting Ura− cells were all His−, indicating that loss of the URA3 plasmid was always associated with loss of HIS3 as expected if both markers were on the plasmid.
RESULTS
Role of replication-coupled nucleosome assembly in the repair of chemical and radiation-induced DSBs
The sensitivity of RCAF/CAF-1 double mutants (asf1 rlf2 cells) to MMS, which is thought to produce strand breaks by indirect mechanisms after DNA methylation (37), suggested a possible role for the complexes in repair of DSBs (17). This led us to investigate the role of CAF-1 and RCAF in each of the major DSB repair pathways. A series of four single mutants and all three CAF-1 RCAF double mutants containing deletions of RLF2, CAC2, MSI1 and/or ASF1 were created in the haploid strain BY4742 (Table 1). This strain background was used previously to create an ordered library of yeast gene deletion mutants (28,38,39). The resistance of cells lacking RCAF (asf1Δ) or CAF-1 (rlf2Δ, cac2Δ or msi1Δ strains) to MMS, hydroxyurea (HU) and camptothecin was assessed by performing serial dilutions (10-fold) and pronging cells as shown in Figure 1A. HU is an inhibitor of ribonucleotide reductase and camptothecin is a widely used inhibitor of topoisomerase I that creates DSBs by a distinctly different mechanism than MMS or HU (3). DSB repair-deficient rad52 mutants were included as controls.
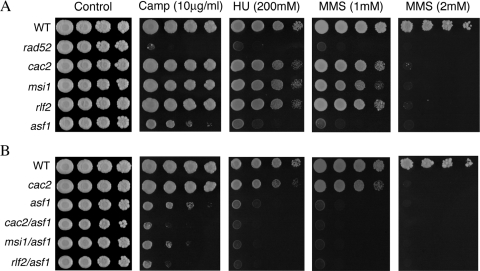
Figure 1.
(A and B) Sensitivity of chromatin assembly mutants to S-phase clastogens camptothecin (camp), hydroxyurea (HU) and methyl methanesulfonate (MMS). Overnight cultures of each strain were serially diluted 10-fold and pronged to YPDA plates containing the indicated concentrations of each drug.
asf1 mutants exhibited sensitivity to each of the drugs, but were not as sensitive as recombination-deficient rad52 cells. In contrast, the three CAF-1 single mutants exhibited only a slight inhibition at doses that reduced the growth of asf1 mutants by more than two orders of magnitude. At a higher dose of MMS (2 mM), inhibition of all three CAF-1 single mutants relative to wild-type cells was apparent (Figure 1A). A previous study using 0.01% MMS (∼1 mM) concluded that rlf2 mutants are essentially resistant to this agent (17), but the results in Figure 1 suggest that higher doses may have been required to see an effect. The three CAF-1 RCAF double mutants exhibited greater inhibition in the presence of each chemical than any of the single mutants. This was most evident for the plates containing 10 µg/ml camptothecin and was also readily apparent when lower doses of each drug were used (Figure 1B and data not shown).
The resistance of cells to gamma radiation, which produces DSBs by both free radical attack (via ionization of water) and direct deposition of energy onto DNA, was also assessed. Interestingly, logarithmically growing haploid asf1 cells and asf1 rlf2 double mutants were slightly more resistant to radiation doses of 40 and 80 krads than wild-type cells (Figure 2A). asf1 and asf1 rlf2 cell cultures have previously been shown to contain cells that primarily have an elevated, G2 phase content of DNA (17,24,28). Since the resistance of haploid yeast cells to ionizing radiation is primarily due to sister chromatid recombination occurring in late S and G2 phase (28), the resistance of asf1 and asf1 rlf2 mutants is likely due to the population being enriched for such cells.
Unlike haploids, diploid cells have the potential for recombinational repair of DSBs using homologous chromosomes as well as sister chromatids. To address the impact of the combined CAF-1 and RCAF complexes on repair between homologous chromosomes, we examined the survival of wild-type and mutant cells grown to stationary phase. In stationary phase diploid cells (mostly G1) survival after exposure to low doses of gamma radiation is primarily dependent upon recombinational repair between homologous chromosomes. The ability of cells lacking RCAF and CAF-1 to effect this type of repair was assessed in Figure 2B. Results for asf1 rlf2/asf1 rlf2 diploid mutants are presented in the figure. Results with another RCAF/CAF-1 mutant, asf1 cac2/asf1 cac2, gave similar results (data not shown) and other mutant combinations were not tested. At low doses of radiation (0–20 krads), the mutants consistently exhibited greater killing than wild-type cells. This increased sensitivity was only detected at low doses, as the wild-type and mutant curves converged at ∼20% survival at 80 krads (Figure 2B, inset graph). This percentage of resistant cells corresponded to the approximately 20–25% of cells that were budded (mostly G2 phase), determined by phase contrast microscopy. These survival data suggest that inefficient nucleosome assembly associated with new DNA synthesis reduces recombinational repair between homologous chromosomes, but not between sister chromatids.
Repair of a single, cohesive-ended DSB by homologous recombination and NHEJ is reduced in the chromatin assembly mutants
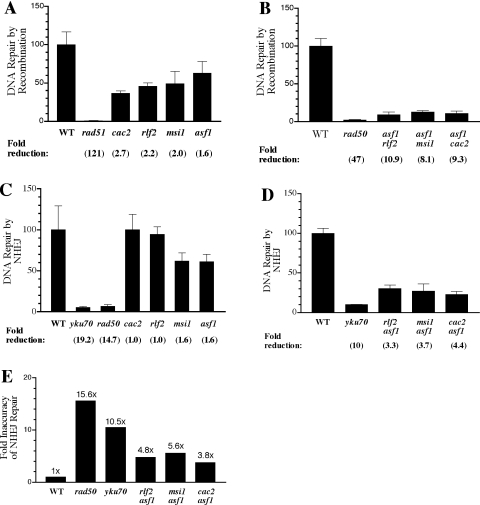
To assess directly the role of CAF-1/RCAF-mediated nucleosome assembly in the two major pathways of DSB repair, recombination between homologous DNAs and repair by NHEJ were assessed separately using DNA substrates that contain a single, site-specific DSB with complementary ends. As shown in Figure 3A, the recombination assays employed an integrating plasmid containing both HIS3 and URA3. After the creation of a DSB in the HIS3 gene by digestion with BclI and subsequent transformation into yeast cells, His+ Ura+ recombinants were identified that had arisen by recombination with the his3-Δ1 allele on chromosome XV. An advantage of this ‘ends-in’ type of assay is that a large window of recombination proficiencies can be assessed. For example, rad50, mre11 and xrs2 mutants typically exhibit only modest deficiencies of 3- to 5-fold in spontaneous or DSB-induced mitotic recombination assays (5,9,40), but have up to 50-fold reductions in this assay (depending on the specific plasmid constructs used). For each assay, transformation efficiencies of linearized plasmid DNA were normalized to those for uncut CEN/ARS plasmids transformed into the same competent cell preparations (40). As shown in Figure 4A and B, site-specific recombination was reduced 47- and 121-fold, respectively, in control recombination pathway-deficient rad50 and rad51 strains. Frequencies were only slightly reduced in CAF-1 or RCAF single mutants (ranging from 1.6- to 2.7-fold relative to wild type), but the three double mutant strains (rlf2 asf1, cac2 asf1 and msi1 asf1) exhibited strong reductions in recombination (8- to 11-fold) (Figure 4B).
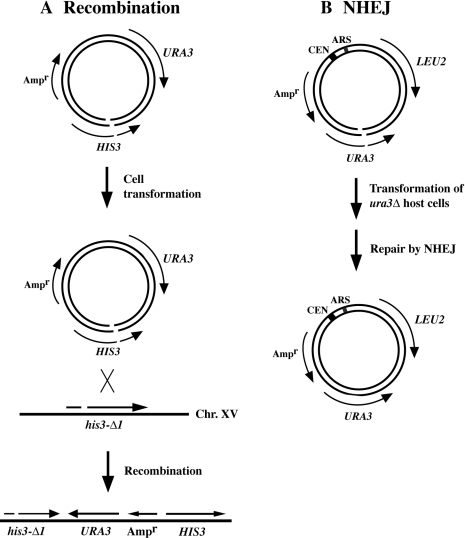
Figure 3.
Assay systems developed to monitor homologous recombination (A) and NHEJ repair efficiency and accuracy (B). For each assay, repair of plasmid DNA containing a cohesive-ended, site-specific DSB within the coding region of a gene (HIS3 or URA3) was monitored after transformation of haploid yeast cells.
Figure 4.
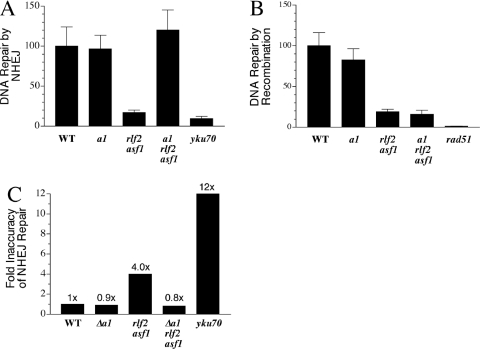
Effects of inactivation of RCAF and CAF-1 genes on repair by recombination and NHEJ. Repair efficiencies of wild-type cells were normalized to 100% in graphs A–D. In part E, the relative inaccuracy of NHEJ repair in wild-type cells (∼1 per 200 transformants) was set at 1.0. Numbers underneath graphs refer to fold reductions relative to wild-type cells.
The impact of inactivation of replication-coupled chromatin assembly on the other DSB repair pathway, NHEJ, was assessed using a plasmid-based assay designed to assess both efficiency and accuracy of repair by end-joining (Figure 3B). Briefly, a centromeric plasmid containing LEU2 and URA3 was linearized by cleavage at the NcoI site in URA3. This region of the plasmid lacks homology with cellular DNA since the host strains have a deletion of the URA3 gene (ura3-Δ0). After transfer into cells, transformant colonies arise by recombination-independent repair of the broken molecules to form stable, single-copy plasmids in the nucleus. This system also permits monitoring of the accuracy of NHEJ repair by determining the number of stable Leu+ colonies that are Ura+ versus Ura−. Repaired plasmids in some NHEJ mutants (e.g. ku and sir strains) have high frequencies of associated small deletions near the joined ends (5). NHEJ efficiencies were near-wild type in each of the four nucleosome assembly single mutants (<2-fold effect) under conditions where NHEJ-deficient rad50 and yku70 cells exhibited reductions of 15- and 19-fold, respectively (Figure 4C). In contrast, each of the three CAF-1 RCAF double mutants showed a consistent, modest reduction in NHEJ repair of 3- to 4-fold (Figure 4D). In addition, the accuracy of NHEJ repair was reduced in each of the double mutants (Figure 4E). Approximately 1 in 200 Leu+ transformants of wild-type cells contained an associated mutation in URA3 (resulting in Leu+ Ura− colonies). This level of mutagenic repair (normalized to 1.0 in Figure 4E) was increased 10- to 15-fold in yku70 and rad50 mutants (Figure 4E). ura3 gene mutations were increased 4- to 5-fold in each of the chromatin assembly double mutants, an intermediate level consistent with the modest reductions in NHEJ repair efficiency in the same strains.
Transcriptional silencing at telomeres, rRNA and the mating loci HML and HMR is partially derepressed in rlf2 (cac1) mutants and more strongly reduced in cells lacking both CAF-1 and RCAF (e.g. rlf2 asf1 mutants) (17,20,22,23,41). Reduced silencing at the mating loci also occurs in sir2, sir3 and sir4 mutants, leading to production of the a1/α2 transcriptional repressor, an inhibitor of NEJ1 gene expression, and reduced efficiency of NHEJ repair (11–14). To test if the partial reduction in end-joining in the three CAF-1 RCAF double mutants was due to the derepression of HMR-a1 and HML-α2, the HMR-a1 gene was deleted in the MATα strain background used above and levels of DSB repair by NHEJ and homologous recombination were assessed separately as before. As shown in Figure 5A, the defect in end-joining observed in rlf2 asf1 double mutants was completely abolished when the expression of the a1/α2 repressor complex was prevented (in a1 rlf2 asf1 cells). In contrast, the large decrease in ends-in recombination seen in these mutants was not affected by deletion of HMR-a1 (Figure 5B). The intermediate (4-fold) reduction in accuracy of NHEJ repair seen in the double mutants was also restored to wild-type levels in the a1-deleted mutants (Figure 5C). These results demonstrate that the reduction of NHEJ, but not homologous recombination, in the chromatin assembly mutants is an indirect consequence of derepression of silencing at the mating loci.
Figure 5.
(A–C) Inactivation of mating factor gene a1 suppresses defects in NHEJ repair, but not recombination, in RCAF CAF-1 double mutants. Assays were performed as for Figure 4.
Recombination defects are not altered when the target lacks higher order chromatin structure
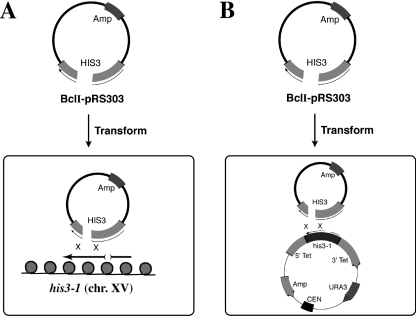
The recombination assays employed here involve recombination between a broken DNA molecule and an intact chromosome in vivo. Chromosomes contain highly ordered local nucleosome arrays that are packaged into more complex structures, e.g. 30 nm fibers and higher order fibers and loops (18,42). In contrast, small circular plasmid DNAs propagated in the nucleus contain nucleosomes, but lack higher order protein–DNA folding (43). It is possible that the strong recombination defects of the CAF-1/RCAF mutants are caused by specific impairment of one or more steps in the homologous recombination pathway, potentially through reduced assembly of nucleosomes onto newly repaired and replicated DNA. However, there might also be indirect effects on repair due to alteration of higher order chromatin structures, possibly reducing access of enzymes of the recombination machinery to the DNA. To address potential effects resulting from changes in higher order chromatin structure, another recombination assay system was devised that involved targeting the homologous DNA fragment containing a DSB to either a chromosomal locus or to the same target gene present only on a single-copy plasmid in the cell. As depicted in Figure 6A and B, a DNA fragment generated by BclI-cleavage of the integrating plasmid pRS303 (within HIS3) was transformed into either his3-Δ1 host cells to monitor chromosomal targeting or into HIS3-deleted cells containing his3-Δ1 on the centromeric plasmid pLKL71Y to assess plasmid targeting.
Figure 6.
Recombination assay systems used to assess the role of higher order chromatin structure in the recombination defects associated with RCAF CAF-1 double mutants. (A) System used to assess repair of plasmid DNA (pRS303) containing a single cohesive-ended DSB by homologous recombination with chromosome XV to form His+ colonies. (B) Method used for assessment of recombination with an intracellular plasmid target (pLKL71Y). The host strain used in (B) had a deletion of the entire HIS3 coding sequence on chromosome XV.
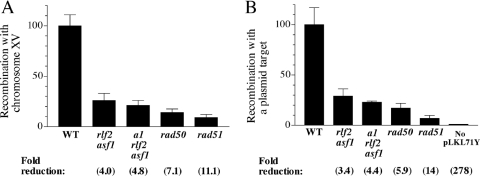
Recombination with the chromosomal target was reduced 7- and 11-fold in rad50 and rad51 mutants, respectively (Figure 7A). Recombination was reduced 4- to 5-fold in rlf2 asf1 double mutants in the same assay (and also in the rlf2 asf1 hmr-a1 mutants included as an additional control). In assays using a plasmid target (Figure 7B), rad50 and rad51 controls were down 6- and 14-fold, respectively, and once again recombination in rlf2 asf1 mutants was consistently reduced by ∼4-fold. We confirmed that the linearized pRS303 DNA recombined in vivo with pLKL71Y by replica-plating His+ tranformants to plates containing 5-FOA acid and demonstrating that loss of the URA3 plasmid was always associated with loss of HIS3, as expected if both markers were on the plasmid (see Materials and Methods). As an additional control for these experiments, HIS3-deleted host cells without the his3-Δ1 target plasmid were also assayed. Mean His+ frequencies were 280-fold lower than in cells with a plasmid target, indicating a baseline level for the assays (Figure 7B). These rare transformants likely arise by recircularization and recombination-independent integration of the HIS3 fragment into chromosomal DNA. The nearly identical decreases in recombination at plasmid and chromosomal DNA targets in the nucleosome assembly mutants suggest that the defects are not caused by changes in higher order chromosome packing.
Figure 7.
Similar reductions in recombination proficiency using chromosomal (A) or intracellular plasmid (B) targets in RCAF CAF-1 double mutants. For experiments performed in (B), recombination events were targeted to the his3-Δ1 plasmid pLKL71Y. ‘No pLKL71Y’ indicates the background of His+ cells produced when the host strain YLKL762 (his3-Δ0) did not contain a target plasmid.
Altered spontaneous and radiation-induced recombination between homologous chromosomes in RCAF CAF-1 mutants
Diploid G1 phase cells lacking RCAF and CAF-1 are more sensitive to ionizing radiation than wild-type cells (Figure 2B), suggesting a defect in DSB repair involving recombination between homologous chromosomes. To address this possibility more directly, spontaneous and gamma radiation-induced LOH at the ADE2 locus was monitored in normal and chromatin assembly-deficient diploid cells. Several previous studies have demonstrated that spontaneous and damage-induced LOH events (such as ADE2/ade2→ade2/ade2) occur primarily by homologous recombination that is initiated during G1 phase in yeast cells (44–47). ADE2/ade2 heterozygotes grow as white colonies, but homozygous ade2/ade2 cells are red (or pink) due to the accumulation of a pigmented adenine biosynthetic intermediate. Thus, early LOH recombination events are detectable as sectored colonies that contain both white and red colored regions. In wild-type cells, the frequency of spontaneous LOH events was low (≤0.01%; only one event detected out of ∼10 000 colonies examined), but was several hundred fold higher after exposure to 40 or 80 krads (becoming 4–6% of all colonies) (Table 2). RCAF/CAF-1 mutants exhibited a much higher spontaneous level than wild-type cells, 0.6%, that did not increase after exposure to radiation (remaining at ∼0.5%). This striking lack of induced LOH in the mutant cells is consistent with the site-specific DSB recombination assays and points to a strong requirement for replication-mediated chromatin assembly for each of these types of DSB repair events.
Table 2.
Spontaneous and ionizing radiation-induced LOH frequenciesa
| Red–white sectored colonies | |||
|---|---|---|---|
| Strain | 0 krad | 40 krads | 80 krads |
| ≤0.01% (1/9541) | 6.4% (36/562) | 3.7% (59/1591) | |
| 0.58% (48/8239) | 0.39% (3/764) | 0.46% (12/2637) | |
aSpontaneous and gamma radiation-induced loss of heterozygosity (LOH) at ADE2 was assessed in stationary phase diploid cells. Numerals in parentheses indicate number of colonies exhibiting red (ade2/ade2) sectors and the total number of colonies examined.
DISCUSSION
We have demonstrated an important role for the replication-coupled nucleosome assembly complexes CAF-1 and RCAF in repair of DNA DSBs that arise by different mechanisms. The reduced chromatin assembly in RCAF/CAF-1 double mutants was found to be associated with markedly decreased repair by both the recombination and nonhomologous end-joining pathways.
Although earlier studies did not detect MMS sensitivity in CAF-1 deletion strains, we observed enhanced killing in all three CAF-1 single mutants (rlf2, cac2 and msi1) when higher doses of MMS were employed (up to 2 mM). This result suggests a modest requirement for the replication-coupled nucleosome assembly provided by CAF-1 alone for the repair of DNA alkylation-induced damage. This conclusion is supported by the previously reported MMS-sensitivity of an unusual msi1 (cac3) truncation mutant (48). The results are also in accord with work demonstrating direct association of CAF-1 and PCNA with each other and with damaged DNA (25,26,49). Recent demonstration that gross chromosome rearrangements are increased in rlf2 mutants and reach high levels after exposure to MMS provides another indication that chromosome stabilization mechanisms are compromised in CAF-1 mutants (50).
Haploid strains lacking the RCAF complex (asf1 cells) were more sensitive to each of the three S-phase clastogens than CAF-1 mutants, consistent with previous results (17,29,30). Recent work (29,31) has suggested that molecules of Asf1 are normally sequestered in the cell by association with Rad53 protein, but are released after exposure to DNA-damaging agents. In this scheme, the released Asf1 then works jointly with acetylated forms of histones H3 and H4 (forming the RCAF complex) to mediate assembly of new nucleosomes onto DNA. It is likely that this latter process involves the coordinated actions of RCAF with CAF-1, based on observations that Drosophila RCAF can stimulate the nucleosome assembly activity of CAF-1 in vitro (possibly acting as a histone donor) and that a subunit of CAF-1 binds to and co-localizes in vivo with Asf1 (16,17,27). Although Asf1 binds to the checkpoint kinase Rad53, the protein does not appear to play an important role in cell cycle checkpoint pathways (29).
Repair of radiation-induced DSBs in haploids is primarily restricted to homologous recombination between sister chromatids in late S/G2 phase (1,2). Since logarithmically growing haploid cells deficient in chromatin assembly were resistant to gamma radiation, we conclude that recombinational repair involving sister chromatids is not greatly affected. Additional support for this conclusion comes from a recent study that detected elevated sister chromatid exchange in asf1 single mutants (51), which is consistent with the observation of increased radiation resistance. In contrast to results with gamma radiation, the RCAF/CAF-1 mutants exhibited strong sensitivity to MMS, HU and camptothecin. The gamma survival experiments involved a single brief exposure to radiation while the chemical clastogen-induced DSBs were continuously generated in cells that were cycling (in rich nutrient media supplemented with the mutagen). The latter process leads to DSB formation directly in S phase of each cell cycle, the time when most new chromatin is assembled (16), and this timing may the basis for the stronger sensitivity. The possible importance of structural differences between DSB ends produced indirectly by the chemicals and the ‘dirty’ end structures generated more directly by radiation (primarily resulting from hydroxyl radical-mediated reactions) is unclear (1,5). In contrast to log phase haploid cells, the G1 population present in stationary phase diploid cells lacking RCAF and CAF-1 was radiation sensitive. This result suggests a defect in DSB-induced recombination between homologous chromosomes and is also consistent with the results of more direct recombination assays involving plasmid DNA targeting and loss of heterozygosity (see below). Interestingly, inactivation of the RDH54, SGS1 or SRS2 genes produces a similar phenotype, i.e. reduction of recombination between homologous chromosomes, but not between sister chromatids (9,52). In the latter case, the reduction was ascribed to an inability to complete interhomolog strand exchange events once they had been initiated (52,53).
Consistent with the mutagen survival results, RCAF/CAF-1 double mutants were found to have reduced ability to repair DNA with a defined DSB structure. Specifically, all three combinations of RCAF and CAF-1 mutations (involving asf1 combined with either rlf2, cac2 or msi1) were found to be defective in repair of a site-specific DSB by both the NHEJ and recombination pathways. The modest defect in NHEJ efficiency and accuracy appears to be indirect, caused by partial derepression of silencing at HMRa to permit synthesis of the a1/α2 complex, a repressor of the end-joining gene NEJ1 (11–14). This conclusion is supported by the demonstration that deletion of the a1 gene restored NHEJ proficiency in asf1 rlf2 mutants and by previous reports of a partial loss of silencing at the mating loci in these mutants (16,19,20). The proposed mechanism of reduced NHEJ repair is analogous to that suggested for sir2, sir3 and sir4 mutants, which exhibit a strong reduction in silencing at HMRa and HMLα and in NHEJ repair. This indirect impairment suggests that DNA replication with associated nucleosome reassembly is not an intermediate step in repair by the NHEJ pathway.
Each of the RCAF/CAF-1 mutants exhibited an ∼10-fold reduction of ends-in recombinational repair of a defined DSB with complementary overhangs. Deletion of the HMR-a1 gene did not restore recombination proficiency, an indication that the reduction in recombination was not due to altered transcription at the mating type loci. This reduced capacity for recombinational repair of a defined DSB is consistent with the subsequent observation that radiation-induced LOH was abolished in the mutants. Past studies have indicated that both spontaneous and damage-induced LOH events (involving conversion from diploid ADE2/ade2 cells to ade2/ade2 cells in the current study) occur primarily by homologous recombination initiated during G1 phase, with a small fraction of phenotypically Ade− cells arising by alternative mechanisms such as chromosome loss or mutation at much lower frequency (44–47). Interestingly, although the >100-fold increase in recombination between homologous chromosomes induced by radiation was abolished in the mutants, spontaneous LOH events were elevated. Reduction of replication-coupled chromatin assembly has recently been shown to lead to increases in spontaneous chromosome loss and mutation (30,50,54). Since LOH phenotypes may still be generated by chromosome loss or arm loss when recombination is defective, the increased frequency observed in the mutants is likely due to such events.
During mating type switching (a DSB-induced gene conversion process), some normally essential recombination proteins become dispensable when plasmid substrates are substituted for whole chromosomes in vivo (55,56). This suggests that an important function of some repair proteins is to promote access to chromosomal DNA for critical recombination enzymes, such as Rad52. The target molecule in the ends-in recombination assays employed here was a chromosome. Changing the intracellular target to a small plasmid did not abolish the recombination defect, indicating that the decreased DNA exchange in the mutants was not due to a change in higher order chromatin structure. The result also implies that access to the target DNA is not impaired in the mutants. We note the interesting possibility that nucleosomes may form on the linear DNA transformed into cells prior to or in conjunction with 5′→3′ strand rescission and initiation of recombination. Though unlikely to involve RCAF/CAF-1 because the transformed DNA fragment lacks an origin and should not replicate, it is possible that reduced availability of nucleosomes for assembly onto the donor DNA (the broken plasmid) influences the efficiency of subsequent recombination reactions.
In support of the results presented in this work, some correlations between the level and/or modified state of histones and homologous recombination have been reported previously. A variant of the core histone H2A called H2AX is phosphorylated at sites of induced DSBs and its absence leads to reduced levels of immunoglobulin class switching, a form of homologous recombination (57,58). In addition, the wrapping of DNA around histones helps to maintain the duplex in a negatively supercoiled state that favors strand unpairing and promotes formation of D-loops, thought to be intermediates in strand exchange (10,59,60). A general reduction in the number of nucleosomes on chromosomal DNA, a likely consequence of loss of RCAF and CAF-1, might lead to a decrease in negative supercoils and make D-loop formation more energetically unfavorable. In support of the idea that reduced nucleosome density is detrimental to DNA metabolism, a modest reduction of histone levels (presumably leading to an increase in nucleosome-free segments and possibly local changes in superhelical density) causes inhibition of meiosis in diploid cells and larger histone reductions result in mitotic cell cycle arrest (61,62). Furthermore, we note that the presence of the linker histone H1 in yeast cells is inhibitory to some types of recombinational repair (63) and that repositioning of whole nucleosomes to cover recombination signal sequences at mouse cell V(D)J junctions can lead to reduced levels of recombinational switching (64).
Models for recombinational repair of DSBs typically propose a sequence of events that includes resection of the DSB ends to produce 3′ overhangs, a homology search, strand exchange with new DNA synthesis, branch migration and then resolution followed by ligation (10,59,60). It is possible that loss of RCAF and CAF-1 affects one or more specific steps in the pathway. For example, RCAF and CAF-1 work synergistically in the assembly of chromatin onto newly replicated DNA (and bind to the polymerase-associated PCNA complex and replication factor C) and have previously been implicated in post-replication repair of UV-induced lesions via the RAD6 pathway (25,26,32,65). Since an intermediate step in recombinational repair involves DNA synthesis and branch migration, reduced chromatin reassembly in RCAF/CAF-1 mutants might lead to formation of DNA regions that are repaired, but nucleosome-depleted. Formation of such regions is detrimental to the cell, though the precise mechanisms involved remain unclear (61,62). In addition, Rad54 is a conserved ATPase enzyme with homology to Swi/Snf-like chromatin remodeling proteins that appears to function directly in DNA recombination along with Rad51, Rad52 and several other proteins (10,59). The enzyme stimulates homologous strand pairing by Rad51 and does so more efficiently with chromatin-associated DNA than with naked DNA (59,66). The possibility that altered DNA-nucleosome topology in the mutants might produce changes in affinity or processivity of specific recombination enzymes such as Rad54 awaits further investigation.
In summary, we have demonstrated that both major pathways of DSB repair are compromised when assembly of nucleosomes associated with new DNA synthesis is reduced. Defects in NHEJ repair were shown to be indirect effects of alteration of the repressive chromatin structures that mediate transcriptional silencing. The data are consistent with a post-replicative repair function for CAF-1 and RCAF in homologous recombination, possibly involving deposition of new histone octamers after DNA synthesis associated with strand exchange. In combination with other data, especially the recent demonstration of a role for phosphorylated histone H2AX at DSBs (57,58), these findings solidify the concept that new DNA repair models must incorporate descriptions of both repair enzymes and nucleosomal structures at sites of DNA lesions.
Acknowledgments
L.K.L. was supported in part by Research Corporation grant #CC5767. Support was also provided in part by a Department of Energy interagency award (DE-AI02-99ER62749) to M.A.R. Funding to pay the Open Access publication charges for this article was provided by NIEHS.
Conflict of interest statement. None declared.
REFERENCES
- 1.Obe G., Johannes C., Schulte-Frohlinde D. DNA double-strand breaks induced by sparsely ionizing radiation and endonucleases as critical lesions for cell death, chromosomal aberrations, mutations and oncogenic transformation. Mutagenesis. 1992;7:3–12. doi: 10.1093/mutage/7.1.3. [DOI] [PubMed] [Google Scholar]
- 2.Resnick M.A., Martin P. The repair of double-strand breaks in the nuclear DNA of Saccharomyces cerevisiae and its genetic control. Mol. Gen. Genet. 1976;143:119–129. doi: 10.1007/BF00266917. [DOI] [PubMed] [Google Scholar]
- 3.Arnaudeau C., Lundin C., Helleday T. DNA double-strand breaks associated with replication forks are predominantly repaired by homologous recombination involving an exchange mechanism in mammalian cells. J. Mol. Biol. 2001;307:1235–1245. doi: 10.1006/jmbi.2001.4564. [DOI] [PubMed] [Google Scholar]
- 4.Karthikeyan G., Santos J.H., Graziewicz M.A., Copeland W.C., Isaya G., Van Houten B., Resnick M.A. Reduction in frataxin causes progressive accumulation of mitochondrial damage. Human Mol. Genet. 2003;12:3331–3342. doi: 10.1093/hmg/ddg349. [DOI] [PubMed] [Google Scholar]
- 5.Lewis L.K., Resnick M.A. Tying up loose ends: nonhomologous end-joining in Saccharomyces cerevisiae. Mutat. Res. 2000;451:71–89. doi: 10.1016/s0027-5107(00)00041-5. [DOI] [PubMed] [Google Scholar]
- 6.Pastink A., Eeken J.C., Lohman P.H. Genomic integrity and the repair of double-strand DNA breaks. Mutat. Res. 2001;480–481:37–50. doi: 10.1016/s0027-5107(01)00167-1. [DOI] [PubMed] [Google Scholar]
- 7.Freudenreich C.H., Kantrow S.M., Zakian V.A. Expansion and length-dependent fragility of CTG repeats in yeast. Science. 1998;279:853–856. doi: 10.1126/science.279.5352.853. [DOI] [PubMed] [Google Scholar]
- 8.Lobachev K.S., Gordenin D.A., Resnick M.A. The Mre11 complex is required for repair of hairpin-capped double-strand breaks and prevention of chromosome rearrangements. Cell. 2002;108:183–193. doi: 10.1016/s0092-8674(02)00614-1. [DOI] [PubMed] [Google Scholar]
- 9.Symington L.S. Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol. Mol. Biol. Rev. 2002;66:630–670. doi: 10.1128/MMBR.66.4.630-670.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sung P., Trujillo K.M., Van Komen S. Recombination factors of Saccharomyces cerevisiae. Mutat. Res. 2000;451B:257–275. doi: 10.1016/s0027-5107(00)00054-3. [DOI] [PubMed] [Google Scholar]
- 11.Frank-Vaillant M., Marcand S. NHEJ regulation by mating type is exercised through a novel protein, Lif2p, essential to the ligase IV pathway. Genes Dev. 2001;15:3005–3012. doi: 10.1101/gad.206801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kegel A., Sjostrand J.O., Astrom S.U. Nej1p, a cell type-specific regulator of nonhomologous end-joining in yeast. Curr. Biol. 2001;11:1611–1617. doi: 10.1016/s0960-9822(01)00488-2. [DOI] [PubMed] [Google Scholar]
- 13.Ooi S.L., Shoemaker D.D., Boeke J.D. A DNA microarray-based genetic screen for nonhomologous end-joining mutants in Saccharomyces cerevisiae. Science. 2001;294:2552–2556. doi: 10.1126/science.1065672. [DOI] [PubMed] [Google Scholar]
- 14.Valencia M., Bentele M., Vaze M.B., Herrmann G., Kraus E., Lee S.E., Schar P., Haber J.E. NEJ1 controls non-homologous end joining in Saccharomyces cerevisiae. Nature. 2001;414:666–669. doi: 10.1038/414666a. [DOI] [PubMed] [Google Scholar]
- 15.Imai S., Johnson F.B., Marciniak R.A., McVey M., Park P.U., Guarente L. Sir2: an NAD-dependent histone deacetylase that connects chromatin silencing, metabolism, and aging. Cold Spring Harb. Symp. Quant. Biol. 2000;65:297–302. doi: 10.1101/sqb.2000.65.297. [DOI] [PubMed] [Google Scholar]
- 16.Ridgway P., Almouzni G. CAF-1 and the inheritance of chromatin states: at the crossroads of DNA replication and repair. J. Cell Sci. 2000;113:2647–2658. doi: 10.1242/jcs.113.15.2647. [DOI] [PubMed] [Google Scholar]
- 17.Tyler J.K., Adams C.R., Chen S.R., Kobayashi R., Kamakaka R.T., Kadonaga J.T. The RCAF complex mediates chromatin assembly during DNA replication and repair. Nature. 1999;402:555–560. doi: 10.1038/990147. [DOI] [PubMed] [Google Scholar]
- 18.Vaquero A., Loyola A., Reinberg D. The constantly changing face of chromatin. Sci. Aging Knowledge Environ. 2003;14:RE4. doi: 10.1126/sageke.2003.14.re4. [DOI] [PubMed] [Google Scholar]
- 19.Enomoto S., McCune-Zierath P.D., Gerami-Nejad M., Sanders M.A., Berman J. RLF2, a subunit of yeast chromatin assembly factor-I, is required for telomeric chromatin function in vivo. Genes Dev. 1997;11:358–370. doi: 10.1101/gad.11.3.358. [DOI] [PubMed] [Google Scholar]
- 20.Enomoto S., Berman J. Chromatin assembly factor I contributes to the maintenance, but not the re-establishment, of silencing at the yeast silent mating loci. Genes Dev. 1998;12:219–232. doi: 10.1101/gad.12.2.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaufman P.D., Kobayashi R., Stillman B. Ultraviolet radiation sensitivity and reduction of telomeric silencing in Saccharomyces cerevisiae cells lacking chromatin assembly factor-I. Genes Dev. 1997;11:345–357. doi: 10.1101/gad.11.3.345. [DOI] [PubMed] [Google Scholar]
- 22.Monson E.K., de Bruin D., Zakian V.A. The yeast Cac1 protein is required for the stable inheritance of transcriptionally repressed chromatin at telomeres. Proc. Natl Acad. Sci. USA. 1997;94:13081–13086. doi: 10.1073/pnas.94.24.13081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smith J.S., Caputo E., Boeke J.D. A genetic screen for ribosomal DNA silencing defects identifies multiple DNA replication and chromatin-modulating factors. Mol. Cell. Biol. 1999;19:3184–3197. doi: 10.1128/mcb.19.4.3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sutton A., Bucaria J., Osley M.A., Sternglanz R. Yeast ASF1 protein is required for cell cycle regulation of histone gene transcription. Genetics. 2001;158:587–596. doi: 10.1093/genetics/158.2.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moggs J.G., Grandi P., Quivy J.P., Jonsson Z.O., Hubscher U., Becker P.B., Almouzni G. A CAF-1-PCNA-mediated chromatin assembly pathway triggered by sensing DNA damage. Mol. Cell. Biol. 2000;20:1206–1218. doi: 10.1128/mcb.20.4.1206-1218.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shibahara K., Stillman B. Replication-dependent marking of DNA by PCNA facilitates CAF-1-coupled inheritance of chromatin. Cell. 1999;96:575–585. doi: 10.1016/s0092-8674(00)80661-3. [DOI] [PubMed] [Google Scholar]
- 27.Tyler J.K., Collins K.A., Prasad-Sinha J., Amiott E., Bulger M., Harte P.J., Kobayashi R., Kadonaga J.T. Interaction between the Drosophila CAF-1 and ASF1 chromatin assembly factors. Mol. Cell. Biol. 2001;21:6574–6584. doi: 10.1128/MCB.21.19.6574-6584.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bennett C.B., Lewis L.K., Karthikeyan G., Lobachev K.S., Jin Y.H., Sterling J.F., Snipe J.R., Resnick M.A. Genes required for ionizing radiation resistance in yeast. Nature Genet. 2001;29:426–434. doi: 10.1038/ng778. [DOI] [PubMed] [Google Scholar]
- 29.Emili A., Schieltz D.M., Yates J.R., Hartwell L.H. Dynamic interaction of DNA damage checkpoint protein Rad53 with chromatin assembly factor Asf1. Mol. Cell. 2001;7:13–20. doi: 10.1016/s1097-2765(01)00150-2. [DOI] [PubMed] [Google Scholar]
- 30.Le S., Davis C., Konopka J.B., Sternglanz R. Two new S-phase-specific genes from Saccharomyces cerevisiae. Yeast. 1997;13:1029–1042. doi: 10.1002/(SICI)1097-0061(19970915)13:11<1029::AID-YEA160>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 31.Hu F., Alcasabas A.A., Elledge S.J. Asf1 links Rad53 to control of chromatin assembly. Genes Dev. 2001;15:1061–1066. doi: 10.1101/gad.873201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Game J.C., Kaufman P.D. Role of Saccharomyces cerevisiae chromatin assembly factor-I in repair of ultraviolet radiation damage in vivo. Genetics. 1999;151:485–497. doi: 10.1093/genetics/151.2.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lewis L.K., Westmoreland J.W., Resnick M.A. Repair of endonuclease-induced double-strand breaks in Saccharomyces cerevisiae: essential role for genes associated with nonhomologous end-joining. Genetics. 1999;152:1513–1529. doi: 10.1093/genetics/152.4.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goldstein A.L., McCusker J.H. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast. 1999;15:1541–1553. doi: 10.1002/(SICI)1097-0061(199910)15:14<1541::AID-YEA476>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 35.Sikorski R.S., Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rose M.D., Broach J.R. Cloning genes by complementation in yeast. In: Guthrie C., Fink G.R., editors. Methods in Enzymology. Vol. 194. San Diego, CA: Academic Press, Inc.; 1991. pp. 195–229. [DOI] [PubMed] [Google Scholar]
- 37.Xiao W., Chow B.L., Rathgeber L. The repair of DNA methylation damage in Saccharomyces cerevisiae. Curr. Genet. 1996;30:461–468. doi: 10.1007/s002940050157. [DOI] [PubMed] [Google Scholar]
- 38.Brachmann C.B., Davies A., Cost G.J., Caputo E., Li J., Hieter P., Boeke J.D. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 39.Giaever G., Chu A.M., Ni L., Connelly C., Riles L., Veronneau S., Dow S., Lucau-Danila A., Anderson K., Andre B., et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418:387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- 40.Lewis L.K., Storici F., Van Komen S., Calero S., Sung P., Resnick M.A. Role of the nuclease activity of Saccharomyces cerevisiae Mre11 in repair of DNA double-strand breaks in mitotic cells. Genetics. 2004;166:1701–1713. doi: 10.1534/genetics.166.4.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Meijsing S.H., Ehrenhofer-Murray A.E. The silencing complex SAS-I links histone acetylation to the assembly of repressed chromatin by CAF-I and Asf1 in Saccharomyces cerevisiae. Genes Dev. 2001;15:3169–3182. doi: 10.1101/gad.929001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Horn P.J., Peterson C.L. Chromatin higher order folding: wrapping up transcription. Science. 2002;297:1824–1827. doi: 10.1126/science.1074200. [DOI] [PubMed] [Google Scholar]
- 43.Shen C.H., Clark D.J. DNA sequence plays a major role in determining nucleosome positions in yeast CUP1 chromatin. J. Biol. Chem. 2001;276:35209–35216. doi: 10.1074/jbc.M104733200. [DOI] [PubMed] [Google Scholar]
- 44.Acuna G., Wurgler F.E., Sengstag C. Reciprocal mitotic recombination is the predominant mechanism for the loss of a heterozygous gene in Saccharomyces cerevisiae. Environ. Mol. Mutagen. 1994;24:307–316. doi: 10.1002/em.2850240408. [DOI] [PubMed] [Google Scholar]
- 45.Nakai S., Mortimer R.K. Studies on the genetic mechanism of radiation-induced mitotic segregation in yeast. Mol. Gen. Genet. 1969;103:329–338. doi: 10.1007/BF00383483. [DOI] [PubMed] [Google Scholar]
- 46.Petes T.D., Malone R.E., Symington L.S. Recombination in yeast. In: Broach J.R., Pringle J.R., Jones E.W., editors. The Molecular Biology of the Yeast Saccharomyces: Genome Dynamics, Protein Synthesis and Energetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1991. pp. 407–521. [Google Scholar]
- 47.Roman H., Ruzinski M.M. Mechanisms of gene conversion in Saccharomyces cerevisiae. Genetics. 1990;124:7–25. doi: 10.1093/genetics/124.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Qian Z., Huang H., Hong J.Y., Burck C.L., Johnston S.D., Berman J., Carol A., Liebman S.W. Yeast Ty1 retrotransposition is stimulated by a synergistic interaction between mutations in chromatin assembly factor I and histone regulatory proteins. Mol. Cell. Biol. 1998;18:4783–4792. doi: 10.1128/mcb.18.8.4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Martini E., Roche D.M., Marheineke K., Verreault A., Almouzni G. Recruitment of phosphorylated chromatin assembly factor 1 to chromatin after UV irradiation of human cells. J. Cell Biol. 1998;143:563–575. doi: 10.1083/jcb.143.3.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Myung K., Pennaneach V., Kats E.S., Kolodner R.D. Saccharomyces cerevisiae chromatin-assembly factors that act during DNA replication function in the maintenance of genome stability. Proc. Natl Acad. Sci. USA. 2003;100:6640–6645. doi: 10.1073/pnas.1232239100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Prado F., Cortes-Ledesma F., Aguilera A. The absence of the yeast chromatin assembly factor Asf1 increases genomic instability and sister chromatid exchange. EMBO Rep. 2004;5:497–502. doi: 10.1038/sj.embor.7400128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gangloff S., Soustelle C., Fabre F. Homologous recombination is responsible for cell death in the absence of the Sgs1 and Srs2 helicases. Nature Genet. 2000;25:192–194. doi: 10.1038/76055. [DOI] [PubMed] [Google Scholar]
- 53.Liberi G., Maffioletti G., Lucca C., Chiolo I., Baryshnikova A., Cotta-Ramusino C., Lopes M., Pellicioli A., Haber J.E., Foiani M. Rad51-dependent DNA structures accumulate at damaged replication forks in sgs1 mutants defective in the yeast ortholog of BLM RecQ helicase. Genes Dev. 2005;19:339–350. doi: 10.1101/gad.322605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ramey C.J., Howar S., Adkins M., Linger J., Spicer J., Tyler J.K. Activation of the DNA damage checkpoint in yeast lacking the histone chaperone anti-silencing function. Mol Cell. Biol. 2004;24:10313–10327. doi: 10.1128/MCB.24.23.10313-10327.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bartsch S., Kang L.E., Symington L.S. RAD51 is required for the repair of plasmid double- stranded DNA gaps from either plasmid or chromosomal templates. Mol. Cell. Biol. 2000;20:1194–1205. doi: 10.1128/mcb.20.4.1194-1205.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sugawara N., Ivanov E.L., Fishman-Lobell J., Ray B.L., Wu X., Haber J.E. DNA structure-dependent requirements for yeast RAD genes in gene conversion. Nature. 1995;373:84–86. doi: 10.1038/373084a0. [DOI] [PubMed] [Google Scholar]
- 57.Bassing C.H., Alt F.W. H2AX may function as an anchor to hold broken chromosomal DNA ends in close proximity. Cell Cycle. 2004;3:149–153. doi: 10.4161/cc.3.2.689. [DOI] [PubMed] [Google Scholar]
- 58.Reina-San-Martin B., Difilippantonio S., Hanitsch L., Masilamani R.F., Nussenzweig A., Nussenzweig M.C. H2AX is required for recombination between immunoglobulin switch regions but not for intra-switch region recombination or somatic hypermutation. J. Exp. Med. 2003;197:1767–1778. doi: 10.1084/jem.20030569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jaskelioff M., Van Komen S., Krebs J.E., Sung P., Peterson C.L. Rad54p is a chromatin remodeling enzyme required for heteroduplex DNA joint formation with chromatin. J. Biol. Chem. 2003;278:9212–9218. doi: 10.1074/jbc.M211545200. [DOI] [PubMed] [Google Scholar]
- 60.Osman F., Subramani S. Double-strand break-induced recombination in eukaryotes. Prog. Nucleic Acid Res. Mol. Biol. 1998;58:263–299. doi: 10.1016/s0079-6603(08)60039-2. [DOI] [PubMed] [Google Scholar]
- 61.Hanlon S.E., Norris D.N., Vershon A.K. Depletion of H2A-H2B dimers in Saccharomyces cerevisiae triggers meiotic arrest by reducing IME1 expression and activating the BUB2-dependent branch of the spindle checkpoint. Genetics. 2003;164:1333–1344. doi: 10.1093/genetics/164.4.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kim U.J., Han M., Kayne P., Grunstein M. Effects of histone H4 depletion on the cell cycle and transcription of Saccharomyces cerevisiae. EMBO J. 1988;7:2211–2219. doi: 10.1002/j.1460-2075.1988.tb03060.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Downs J.A., Kosmidou E., Morgan A., Jackson S.P. Suppression of homologous recombination by the Saccharomyces cerevisiae linker histone. Mol. Cell. 2003;11:1685–1692. doi: 10.1016/s1097-2765(03)00197-7. [DOI] [PubMed] [Google Scholar]
- 64.Baumann M., Mamais A., McBlane F., Xiao H., Boyes J. Regulation of V(D)J recombination by nucleosome positioning at recombination signal sequences. EMBO J. 2003;22:5197–5207. doi: 10.1093/emboj/cdg487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Franco A.A., Lam W.M., Burgers P.M., Kaufman P.D. Histone deposition protein Asf1 maintains DNA replisome integrity and interacts with replication factor C. Genes Dev. 2005;19:1365–1375. doi: 10.1101/gad.1305005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Alexiadis V., Kadonaga J.T. Strand pairing by Rad54 and Rad51 is enhanced by chromatin. Genes Dev. 2002;16:2767–2771. doi: 10.1101/gad.1032102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fasullo M., Bennett T., AhChing P., Koudelik J. The Saccharomyces cerevisiae RAD9 checkpoint reduces the DNA damage-associated stimulation of directed translocations. Mol. Cell. Biol. 1998;18:1190–1200. doi: 10.1128/mcb.18.3.1190. [DOI] [PMC free article] [PubMed] [Google Scholar]