Abstract
The problem of detecting DNA motifs with functional relevance in real biological sequences is difficult due to a number of biological, statistical and computational issues and also because of the lack of knowledge about the structure of searched patterns. Many algorithms are implemented in fully automated processes, which are often based upon a guess of input parameters from the user at the very first step. In this paper, we present a novel method for the detection of seeded DNA motifs, composed by regions with a different extent of variability. The method is based on a multi-step approach, which was implemented in a motif searching web tool (MOST). Overrepresented exact patterns are extracted from input sequences and clustered to produce motifs core regions, which are then extended and scored to generate seeded motifs. The combination of automated pattern discovery algorithms and different display tools for the evaluation and selection of results at several analysis steps can potentially lead to much more meaningful results than complete automation can produce. Experimental results on different yeast and human real datasets proved the methodology to be a promising solution for finding seeded motifs. MOST web tool is freely available at http://telethon.bio.unipd.it/bioinfo/MOST.
INTRODUCTION
Transcriptional control mechanisms have been investigated in different organisms for at least three decades. Nevertheless, our understanding of how regulatory information is encoded in DNA sequence is still fragmentary. Even knowing the sequence of region(s) controlling the expression of a gene, it is very difficult to formulate reliable predictions about its tissue-specific or developmental stage-specific expression (1).
Since comparative genome analysis revealed a surprising constancy in genetic content among Eukaryotes, it has been recently suggested that biological complexity of organisms could arise more from increased elaboration of gene expression regulation than from an increased number of genes in genomes (2). Given the combinatorial nature of transcription regulation, with an estimation of as many as 3000 transcription factors in humans, the regulatory complexity of the human genome is considerable. On the other hand, experimental studies of transcriptional regulation are time consuming and, in general, focused on single genes. In the post-genomic era, a major challenge is represented by deciphering expression regulation of thousands of annotated genes in genomes, which could be achieved by combining computational analysis of large-scale expression data and functional information on genes with knowledge of complete genomic sequences.
Gene expression is controlled by specific interactions between regulatory proteins, transcription factors and short sequences in the regulatory regions of genes to which they bind. Control regions are modular and the regulatory output of a sequence depends on the specific combination of its elements as well as, partially, on the order and on the orientation in which they occur.
The search for common elements in upstream regions of genes known to have common biological function and/or expression could be a valuable tool for the discovery of novel transcription factor binding sites. It could be reasonably assumed that genes with similar expression are frequently co-regulated and that genes with related function are often similarly expressed and, possibly, regulated in a coordinate way. Moreover, it is known that, in general, tissue-specific or developmental stage-specific gene expression is regulated by a relatively small number of transcription factors. Available data on gene expression and function could be used to select sets of genes putatively co-regulated, which could be searched for common sequence elements in their regulatory regions, by pattern discovery techniques.
The problem of identifying novel regulatory short DNA sequences (‘patterns’) within DNA sequences is not trivial. Given a set of k unaligned sequences, a distance measure d and a threshold value t for d, a typical problem in pattern discovery is to find all patterns that occur in at least q sequences out of k within distance t from the sequence (3). These patterns are conserved but not exact, since they may include nucleotide mutations, insertions or deletions.
Statistical measures of overrepresentation, such as Z-scores, have also been proposed to find candidate motif patterns within and among sequences. For exact patterns, an optimal linear time and space solution for the extraction of surprising solid words (i.e. no mismatches or insertions or deletions are allowed) has been achieved by the annotation of a suffix tree with expected and counted number of occurrences (4,5).
Recently, algorithms based on dynamic programming have been proposed, to compute expectation of patterns with mismatches, with a fixed or variable length, to relax the solid words constraint (6). Another Z-score-based algorithm is implemented in YMF (7), where flexibility of the pattern is achieved by considering a broader alphabet including also degenerate symbols.
Comparison of some different pattern discovery approaches (8) showed that the structure of the planted motif plays an important role in the performance of the algorithms and that there is room for consistent improvement for pattern discovery software. In fact, most programs perform better on synthetic than on real sequences and are more efficient when analysing sequences of yeast rather than of higher Eukaryotes (9).
Regulatory sequence elements are in general from 5 to 25 nt long (3,10), often separated by unconserved sequences. A recent systematic analysis of consensus sequences describing sequence elements binding transcription factors limited to the mammalians and vertebrates subsets of TRANSFAC data (11) showed that the length of the most frequent motifs was 12 bp. As already known (10), it also showed that different regulatory elements binding the same transcription factor are very similar but not identical and that variable positions are more rare in the central part of regulatory elements. More in detail, 80% of considered motifs showed only invariant positions in their central regions and no central asymmetry was observed in the distribution of variable positions (12).
A shortcoming of pattern discovery approaches could be found in the a priori establishment of the ‘quorum’ and of the search parameters, such as pattern length and number of allowed wildcards or distance of occurrences from the model. In addition, it is well known that patterns with biological significance could be subtle, with a ‘borderline’ statistical significance (13). Increasing the distance and/or decreasing the quorum produces the output of very large number of false positives and, at the end, too many results. A promising solution to the problem of output explosion could be the use of biological knowledge and human judgement before, during and after applying pattern discovery algorithms. This could be achieved with a flexible method integrating different pattern discovery procedures to be completed step by step in an interactive way. The method presented in this paper, and implemented in the web tool MOST (motif searching web tool), is based on several analysis steps leading to extraction and visualization of putative regulatory motifs.
MATERIALS AND METHODS
Seeded motifs
Regulatory sequence motifs are often composed by regions with different levels of variability. We define a seeded motif as a motif in which it is possible to identify at least two regions with a different level of variability. In particular, in this paper we focus on symmetrical distributions, hence to regulatory motifs that are characterized by a core region (central, less variable) and two side regions (more variable). The motif M can then be seen as a composition, as follows:
where cl is the core region of length l, while si, i = 1, 2 are the side regions of length l′i. In principle, l′1 can differ from l′2, but for the sake of simplicity we will consider them to be equal in the following sections. To identify seeded motifs, we developed a multi-step approach.
Identification of surprising exact patterns
The first step consists of the identification of exact patterns (solid words). A word is considered surprising if its score, computed according to some selected scoring function, is greater than a given threshold. The scoring function used in the evaluation of surprise measures the difference between counted and expected frequencies of words among sequences. In particular, we used the following scores:
where Seqobs and Seqexp are the counted and expected number of sequences, respectively, in which the word w occurs, and Occobs and Occexp are the counted and expected number of occurrences in the considered group of sequences.
Building the motif core
Motifs core regions are weakly variable but not necessarily completely invariant. Thus, surprising solid words extracted from the input sequences are grouped in clusters of similar words. This is achieved by a ‘k-means’ like clustering algorithm. The measure of similarity is given by the likelihood of belonging to the given cluster. Each cluster is represented by the profile matrix of the words that belong to the cluster. We indicate with wi[pos] the symbol in position pos of word wi and with Mj [symbol][position] the value of the profile matrix of cluster Cj in correspondence of a given symbol and position.
The probability for a word wi to belong to a given cluster Cj is calculated according to the following metric:
that is a measure of how likely is to see a symbol in any position of the word wi in the corresponding position of the cluster Cj profile. If the value of Tij is greater than a given threshold then the word can be assigned to all the clusters for which the relation holds, or, alternatively, to the cluster with maximum probability, depending on the user preference.
To evaluate the quality of the clustering the square-error criterion is used:
where E is the sum of the square error for all the words considered in each cluster. The term |w − mi| needs some clarification. Dealing with words, we identify the mean of the cluster with the profile. So the distance between a word and the centre of the cluster it belongs to is given by the complement to 1 of the probability that the word belongs to the cluster: 1 − Tij. The process iterates until the error converges or a maximum number of iterations is reached.
Side regions selection
Once the core motifs have been identified, the contiguous regions lying at the left and right side of them are taken into consideration for further processing. As expected, by simply considering all the possible extensions, consensus of extended motif would show a structure like NNNN-CoreConsensus-NNNN. To remove the noise, we select a subgroup of extended words producing an ‘optimal’ motif consensus sequence, according to some heuristics. Two scoring functions, called Fixed Value function (FV) and MultiValue (MV) function, were developed, which are both based on the observation that the core regions of functional motifs are often much more conserved than boundary regions.
The weight assigned to each position i is a function of both the nucleotide that occurs at that position i (e.g. A) and its location in the extended word (e.g. 2 nt from the core region).
Let w = w1w2…wm, with wi ∈ Σ, be an extended word of the cluster Ck. The score of the cluster Ck is produced as follows. To the most frequent nucleotide symbol in each position (e.g. A) is assigned a maximum weight maxi, which depends on the localization of the position in the extended word. The second most frequent nucleotide is given a smaller value, and so on.
Let fi(s) be the function assigning a fraction of the maximum weight to each nucleotide (symbol s of the alphabet) depending on its frequency with respect to frequency of the other symbols at the same position i. For example, if A is the most frequent symbol at position 5, and C the second most frequent symbol at position 5, we could have f5(A) = 1.0 and f5(C) = 0.85. In general, the weight corresponding to a position i is given by:
For positions in the core region the value of maxi is equal to 1. The two scoring functions differ in the way in which the maximum weight is assigned to lateral positions.
The FV function assigns a unique maximum value for all side region positions, which is given by a fixed positive value lower than 1.
The MV function assigns to each position in lateral regions a maximum value lower than 1, which decreases with distance from the core region:
where h(i) < h(i + 1) for left side regions, and h(i) > h(i + 1) for right side regions.
For a given word w, belonging to a given cluster, we calculate the global score:
For each cluster Cj we compute the maximum score maxCj that can be achieved by a word that perfectly matches the consensus string of the cluster. The threshold used to select the words is a fraction of this maximum score:
The parameter p is a percentage value that the user will input at the appropriate step. If the score value is greater than a given threshold, then the extended word is kept, otherwise it is discarded.
RESULTS
MOST web tool
We developed a multi-step semi-automatic tool for the identification of seeded motif (MOST; http://telethon.bio.unipd.it/bioinfo/MOST/). MOST was implemented mainly using Java programming language and Java Servlet technology, but it also includes C++ and Perl modules.
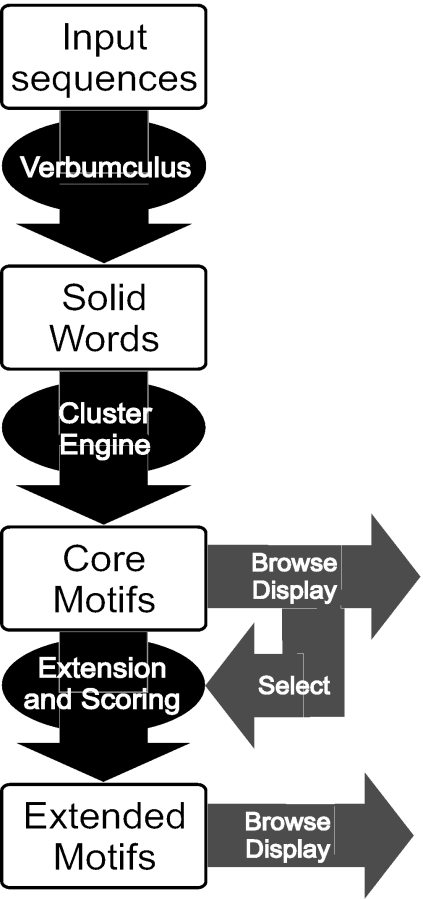
For each of the three main phases of analysis of MOST (exact patterns extraction, clustering, extension and scoring), different parameters could be set. In addition, at different points the user is allowed to browse results and to select those that seem interesting and promising for subsequent analysis. This is achieved by different tabular representations of results as well as by a graphical interface for visualization of motifs positions in the input sequences. MOST architecture is shown in Figure 1.
Figure 1.
Multi-Step architecture of MOST.
Solid word extraction is the first step of the analysis, and it is critical for the success of motif detection. In our implementation, we used the Verbumculus Software Tool (5) developed in C++. Verbumculus is based on a suffix tree data structure that makes possible to extract overrepresented exact patterns from a set of input sequences. The annotation of the tree with the calculated values of expectation and variance is performed in overall linear time. For a detailed description of the algorithm, we refer to the cited bibliography (4,5). For our purposes we used a simplified version of Verbumculus, by limiting the choice of scores to those related to multiple sequences analysis, that are the most significant in this context, and extracting only fixed length words. The threshold value is a critical parameter, as it affects the following processing phases, and must be chosen carefully. The results of the first step are summarized in a table of solid words. It is possible to sort the results according to score, expected and observed number of occurrences, and expected and observed number of sequences in which words occur. The order of the results will be crucial in the initialization of the clusters in the next step.
The third step requires the user to input clustering parameters, such as number of surprising words to cluster, maximum number of clusters and threshold for assigning a word to a cluster. Moreover, it must be selected whether a specific word can be assigned to several clusters or exclusively to a single cluster. If the number of words to consider (Nmax) is smaller than the output size of the previous step, then only the first Nmax words will be clustered. The maximum number of clusters sets the size of the output.
The results of the clustering phase are shown in a summary table reporting, for each cluster: profile, total number of occurrences, total number of sequences in which at least one cluster word appear and core motif consensus quality. By clicking on a motif consensus sequence, details related to the corresponding cluster are displayed in another window (cluster profile matrix, list of words included in the cluster and, for each solid word, number of occurrences and number of sequences in which the word is found). In order to help the user to appropriately select thresholds and focus on the most interesting clusters, we developed a tool to visualize the position in the sequences of up to 10 different clusters. We limited the number of concurrent visualizations to avoid the confusion that could arise from the usage of several similar colours. The positions at which the words of the considered cluster occur in the input sequences are represented by coloured triangles placed at the beginning of each occurrence. Visualization in both direct and reverse strand is allowed. Results can be sorted by consensus quality, number of occurrences, number of sequences in which the core motif occurs and number of strings pertaining to the core motif cluster. Some results, which are considered noise or not interesting, can be deleted. They will not be considered in the further analysis step of motif cores extension and selection.
Parameters for the extension of core motifs and selection of extended motifs must be set, namely the single side extension length, the function to be used to score lateral nucleotide positions and the threshold percentage of the maximum score that has to be reached by a word in order to be kept in the cluster. A summary table gives then information about extended motifs: profile matrix, number of strings included in the group determining the motif, number of occurrences of extended motif, number of sequences in which motif occurs and consensus quality. Motif consensus sequence is linked to detailed information about motif composition. Sorting and visualization of motifs location is also possible at this final stage.
Benchmark tests
Experimental evaluation of the power of the method was conducted with different datasets and under various testing conditions, in order to study the influence of specific search parameters on results. Different groups of promoter sequences, for which it is known which regulatory signals should be detected, were used as positive control datasets. In particular, we analysed with MOST:
a public yeast benchmark dataset, developed by Tompa et al. (9) for a systematic assessment of pattern discovery tools; each of the eight groups of sequences contained some instances of a given signal.
a custom produced dataset, consisting in a group of 37 human promoter sequences, subgroups of which contained some instances of one of nine different signals (Mixed signals dataset).
Yeast datasets
We considered eight yeast datasets, constructed by Tompa et al. (9), such as groups of sequences containing known instances of signals at known positions. Datasets included three different types of sequences: three groups of real yeast genomic promoter sequences containing known transcription factors binding sites (yst04r, yst05r and yst08r), four groups of randomly chosen yeast genomic promoter sequences in which the binding sites were planted (yst01g, yst02g, yst06g and yst09g) and one group of sequences randomly generated according to a Markov chain of order 3, constructed from yeast promoter sequences, in which the binding sites were planted (yst03m). We analysed such datasets by using the following conditions: solid words of 6 bp represented in a number of sequences at least twice than expected were searched in both strands of DNA sequences [z2(w) score set to 2.0]. Obtained solid words were clustered by using the algorithm assigning a word to a unique cluster, and the similarity threshold was set to 80%. Resulting clusters were used as input to the ‘extension and selection’ phase, for which the MV function was adopted with a 95% scoring threshold. The analysis was repeated by using the same settings but with the 80% scoring threshold for the MV function of the ‘extension and selection’ phase.
For each dataset (e.g. yst01g: nine sequences of 1000 bp containing seven instances of the yst01 signal), the sensitivity of each experiment (proportion of signal instances detected) and of different phases has been evaluated. First of all we searched, in the list of overrepresented extracted solid words, all substrings of length six of the known instances of the signal included in the sample of promoters. A signal instance has been considered detected in the list of overrepresented solid words if at least one of its substrings was contained in the list.
Then, in order to evaluate how well extracted motifs represent known signal, we recorded the proportion of known instances of the signal represented in ‘core motifs’ clusters and in ‘extended motifs’. Moreover, for both the clustering and the ‘extension and selection’ phases, the maximum number of signal instances represented in a unique cluster (maximal cluster) was recorded, together with the number of maximal clusters. Results of MOST evaluation on yeast datasets are reported in Table 1.
Table 1.
Results of MOST evaluation on yeast datasets
| Dataset | Yst01g | yst02g | yst03m | yst04r | yst05r | yst06g | yst08r | yst09g | Total | Average |
|---|---|---|---|---|---|---|---|---|---|---|
| Number of sequences | 8 | 4 | 8 | 6 | 3 | 7 | 11 | 16 | ||
| Sequence length | 1000 | 500 | 500 | 1000 | 500 | 500 | 1000 | 1000 | ||
| Number of known signals | 6 | 5 | 18 | 7 | 4 | 7 | 14 | 13 | 47 | |
| Phase 1: solid words extracted | 65 | 255 | 286 | 162 | 337 | 214 | 88 | 40 | ||
| Phase 2: clusters (80% similarity threshold) | 50 | 126 | 141 | 84 | 157 | 111 | 46 | 31 | ||
| Phase 3A: ext. clusters (95% threshold for the MV function) | 50 | 123 | 137 | 81 | 154 | 106 | 44 | 29 | ||
| Phase 3B: ext. clusters (80% threshold for the MV function) | 50 | 126 | 141 | 84 | 157 | 110 | 46 | 31 | ||
| Phase 1: signals found in solid words | 2 | 5 | 14 | 6 | 4 | 7 | 10 | 12 | 38 | |
| Phase 2: signals found in clusters | 2 | 5 | 14 | 6 | 4 | 7 | 10 | 12 | 38 | |
| Phase 3A: signals found in ext. clusters | 0 | 5 | 10 | 7 | 3 | 5 | 7 | 6 | 30 | |
| Phase 3B: signals found in ext. clusters | 1 | 5 | 16 | 7 | 4 | 7 | 9 | 11 | 40 | |
| Phase 1 | 0.33 | 1.00 | 0.78 | 0.86 | 1.00 | 1.00 | 0.71 | 0.92 | 0.83 | |
| Phase 2 Sensitivity | 0.33 | 1.00 | 0.78 | 0.86 | 1.00 | 1.00 | 0.71 | 0.92 | 0.83 | |
| Phase 3A | 0.00 | 1.00 | 0.56 | 1.00 | 0.75 | 0.71 | 0.50 | 0.46 | 0.67 | |
| Phase 3B | 0.17 | 1.00 | 0.89 | 1.00 | 1.00 | 1.00 | 0.64 | 0.85 | 0.84 | |
| Phase 2 | ||||||||||
| Maximum number of signals per cluster | 1 | 4 | 9 | 5 | 3 | 6 | 8 | 8 | 28 | |
| Number of maximal clusters | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | ||
| Sensitivity | 0.17 | 0.80 | 0.50 | 0.71 | 0.75 | 0.86 | 0.57 | 0.62 | 0.63 | |
| Phase 3A | ||||||||||
| Maximum number of signals per cluster | 0 | 1 | 5 | 4 | 3 | 5 | 4 | 3 | 18 | |
| Number of maximal clusters | - | 11 | 3 | 2 | 1 | 1 | 1 | 4 | ||
| Sensitivity | 0.00 | 0.20 | 0.28 | 0.57 | 0.75 | 0.71 | 0.29 | 0.23 | 0.42 | |
| Phase 3B | ||||||||||
| Maximum number of signals per cluster | 1 | 3 | 8 | 3 | 3 | 6 | 4 | 7 | 24 | |
| Number of maximal clusters | 1 | 1 | 1 | 3 | 1 | 1 | 2 | 1 | ||
| Sensitivity | 0.17 | 0.60 | 0.44 | 0.43 | 0.75 | 0.86 | 0.29 | 0.54 | 0.54 |
Datasets are identified by the names originally used by Tompa et al. For each dataset, the total number of sequences included the length of promoter sequences, and the number of signals included is reported. Rows from four to seven describe results obtained by MOST different analysis steps and, for the third step, with different conditions. The following eight rows show the number and the proportion (sensitivity) of known signals per dataset represented in results of the previously described analysis steps. In the last part of the table, the number and the proportion (sensitivity) of known signals represented in the maximal cluster are shown, for each dataset.
In average, 83% of signal instances were represented in extracted solid words and then in clusters corresponding to core motifs, also when adopting the quite stringent threshold for the overrepresentation score (2.0). It should be noticed that, for each dataset, among different clusters obtained, it exists an unique cluster, corresponding to a motif model, containing words representing in average the 63% of signal instances (for four out of eight datasets, in the maximal cluster more than 71% of signal instances are represented). After the ‘extension and selection’ phase for generation of extended seeded motifs, the number and the dimension of clusters slightly decreases and the average sensitivity associated to the maximal cluster decreases to 0.42 and to 0.54 for the 95% and the 80% thresholds, respectively. This behaviour was expected for signals with short instances, such those in Yeast benchmark dataset. Nevertheless, these experiments showed that short signals can be quite efficiently found by MOST already after the completion of the clustering phase which leads to the extraction of core motifs.
Mixed signals dataset
A group of human gene promoter sequences containing each one at least one transcription factor binding site, which specific activity has been experimentally proven (as recorded in TRANSFAC database), has been analysed by MOST as positive control dataset. Promoter sequences were localized after the most probable transcription start site prediction, according to spliced expressed sequence tag data and Acembly gene boundaries.
In total, 37 genomic sequences of 550 bp have been considered, containing 45 known transcription factors binding elements, pertaining to 8 different transcription factors (AP-1, c-Myc, CREB, CRE-BP1, CTF, E2F, E2F-1 and HIF-1) (Table 2). Two couples of transcription factors recognized similar sequences (CREB and CRE-BP1, E2F and E2F-1, respectively). In the 37 sequences, the number of known instances per signal ranged from four to nine; the minimum length of signal instances was 6, the average was 8.5 and the mode 8.
Table 2.
Description of transcription factors binding sites, whose activity was experimentally proven, represented in the group of human promoter sequences composing the Mixed signals benchmark dataset
| Transcription factor | Sequence elements | |||
|---|---|---|---|---|
| Number | Average length | Minimum length | Maximum length | |
| AP-1 | 9 | 9.6 | 7 | 17 |
| c-Myc | 4 | 8.0 | 6 | 14 |
| CREB | 4 | 8.5 | 8 | 10 |
| CRE-BP1 | 7 | 8.3 | 7 | 10 |
| CTF | 4 | 6.8 | 6 | 7 |
| E2F | 5 | 9.0 | 8 | 12 |
| E2F-1 | 5 | 9.8 | 8 | 12 |
| HIF-1 | 7 | 7.4 | 6 | 8 |
For each transcription factor, the number of binding sites in the considered group of sequences, their average, minimum and maximum length are reported.
We evaluated the sensitivity of the first step of MOST, with different measures and conditions adopted for the extraction of overrepresented solid words from input sequences. The z1(w) score (observed occurrences/expected occurrences) and the z2(w) score (observed sequences/expected sequences) were tested. Experiments were conducted with different stringency of the corresponding threshold, which ranged from 1.0 to 3.0 (Table 3).
Table 3.
Results of MOST evaluation on Mixed signals dataset
| No. of 6 bp words | TP | FN | Sensitivity | |
| Occobs/Occexp | ||||
| 1.0 | 1164 | 41 | 4 | 0.91 |
| 1.1 | 1152 | 41 | 4 | 0.91 |
| 1.3 | 929 | 37 | 8 | 0.82 |
| 1.5 | 710 | 36 | 9 | 0.80 |
| 1.7 | 560 | 35 | 10 | 0.78 |
| 2.0 | 382 | 29 | 16 | 0.64 |
| 2.2 | 334 | 27 | 18 | 0.60 |
| 2.5 | 221 | 27 | 18 | 0.60 |
| 3.0 | 109 | 4 | 41 | 0.09 |
| Seqobs/Seqexp | ||||
| 1.0 | 1173 | 34 | 11 | 0.76 |
| 1.1 | 999 | 34 | 11 | 0.76 |
| 1.3 | 773 | 34 | 11 | 0.76 |
| 1.5 | 579 | 34 | 11 | 0.76 |
| 1.7 | 469 | 28 | 17 | 0.62 |
| 2.0 | 246 | 21 | 24 | 0.47 |
| 2.2 | 293 | 18 | 27 | 0.40 |
| 2.5 | 102 | 4 | 41 | 0.09 |
| 3.0 | 48 | 1 | 44 | 0.02 |
Experiments on MOST first step: identification of surprising words. The sensitivity of MOST first phase, carried out with different overrepresentation measures, was evaluated. All the 6 bp sequences representing known binding sites, or all of the substrings of binding sites whose length exceeded six, were searched in the list of 6 nt strings extracted as overrepresented, according to different measures and different thresholds (first column). The sensitivity has been calculated as the number of known sites represented in the list over the total number of known sites [sensitivity = TP/(TP+FN); TP, true positives; FN, false negatives].
The sensitivity has been evaluated again as the proportion of signal instances detected and was calculated for each given signal or for the whole group of signals. We calculated the sensitivity of MOST first step under different conditions over the whole group of mixed signals. Results of these experiments are reported in Table 3. The value of 0.91 was reached, when the z1(w) score was adopted and the threshold was set to 1, producing the extraction of 1164 six nucleotides long solid words represented at least as expected in the considered promoter sequences.
The results of the second analysis phase of MOST were then considered in order to evaluate how well clusters of solid words (extracted motifs) represent specific groups of instances of given signals. The sensitivity was calculated analytically for each of the nine signals.
The clustering has been performed under different conditions starting from the previously cited group of 1164 solid words. Cluster analysis was conducted by adopting the algorithm assigning words to one or more individual clusters. In different experiments, the similarity threshold was set to 60 and 80%, without using an upper bound for the number of clusters, thus obtaining 55 and 272 clusters, respectively. Results of clustering experiments consist of sets of words, each corresponding to an extracted motif, associated to a matrix of nucleotides frequencies in motif positions, and to a consensus sequence. For each experiment, the maximum number of known instances of each specific signal (e.g. AP-1 group of nine signal instances) represented in a unique cluster has been recorded (Table 4). The maximum number of sequences represented in a single cluster, over the total number of sequence elements, pertaining to the same factor and found in the 1164 overrepresented words dataset (resulting from the phase 1) was obtained. The average sensitivity resulted 0.85 when an 80% clustering threshold was adopted, whereas it was 0.56 with a 60% similarity threshold. Results obtained with the 80% threshold (272 clusters) are quite good: more than 85% of sequence elements pertaining to a specific transcription factor are represented in at least one of the obtained clusters.
Table 4.
Results of MOST evaluation on Mixed signals dataset
| Clustering similarity threshold | Transcription factor | Known instances | ||||
|---|---|---|---|---|---|---|
| Maximum per cluster | Phase 1 found | Total | Maximum per cluster/phase 1 found | Maximum per cluster/total | ||
| 80% | AP-1 | 7 | 8 | 9 | 0.88 | 0.78 |
| C-MYC | 4 | 4 | 4 | 1.00 | 1.00 | |
| CREB | 3 | 4 | 4 | 0.75 | 0.75 | |
| CREB-BP1 | 6 | 6 | 7 | 1.00 | 0.86 | |
| CTF | 3 | 4 | 4 | 0.75 | 0.75 | |
| E2F | 4 | 4 | 5 | 1.00 | 0.80 | |
| E2F-1 | 3 | 5 | 5 | 0.60 | 0.60 | |
| HIF-1 | 5 | 6 | 7 | 0.83 | 0.71 | |
| Total | 35 | 41 | 45 | Average 0.85 | Average 0.78 | |
| 60% | AP-1 | 1 | 8 | 9 | 0.13 | 0.11 |
| C-MYC | 1 | 4 | 4 | 0.25 | 0.25 | |
| CREB | 3 | 4 | 4 | 0.75 | 0.75 | |
| CREB-BP1 | 7 | 6 | 6 | 1.00 | 0.86 | |
| CTF | 2 | 4 | 4 | 0.50 | 0.50 | |
| E2F | 3 | 4 | 5 | 0.75 | 0.60 | |
| E2F-1 | 4 | 5 | 5 | 0.80 | 0.80 | |
| HIF-1 | 2 | 6 | 7 | 0.33 | 0.29 | |
| Total | 23 | 41 | 45 | Average 0.56 | Average 0.52 | |
Experiments on MOST second step: clustering of exact patterns for building the motif core. In two clustering experiments, core motifs were built by grouping 1164 surprising words with similarity threshold set to 60 and 80%, respectively. The number of known sequence elements pertaining to each specific group (e.g. AP-1 group of nine elements) represented in obtained clusters is reported.
Solid words pertained to these 272 core motifs clusters were further considered and used as input for the third analysis phase. The ‘extension and selection’ analysis was performed by extending selected motif cores of 1 nt on each side both with a 95% and with a 80% score threshold, adopting the MV extension function. The maximum number of sequences represented in a single ‘extended motif’ cluster, over the total number of sequence elements, pertaining to the same factor and found in the 1164 words overrepresented solid words dataset, was calculated. The average of obtained values was 0.58 for experiments with the 95% threshold and 0.68 when an 80% threshold was applied.
DISCUSSION
For the development of our methodology and software, we introduced a model of seeded sequence motif. We focused on motifs with a central region showing low variability and two more variable lateral regions. This choice, motivated by biological knowledge, should facilitate the discovery of sequence motifs with true biological function in the regulation of gene expression. The surprising solid words dataset that is produced by the first sieving step consists of a subset of words that are overrepresented in the DNA sequences under consideration, according to the selected measure and thresholds. The size of this dataset deeply influences the results of clustering and obtained core motifs. It is expected that, when analyzing appropriate groups of promoter sequences, the sharing of solid words among different promoters is significantly higher than in random sequences of the same composition and length. Actually the user could do different attempts of building a set of surprising solid words by different measures. It is also possible to extract all solid words appearing in the considered sequences at least exactly as expected and to postpone to the following analysis steps the identification of most interesting words. In fact, the obtained surprising solid words can be ordered according to different parameters and a selected subset of surprising words can be used as input for the following steps. In the MOST web tool implementation, the maximum number of extracted solid words is bounded by an internal limit on the maximum size of the output file. Properties of core motifs obtained by clustering solid words may be deeply modulated according to input size (number of words clustered) and to its characteristics, to the setting of clustering parameters (maximum number of clusters, word assignment to single or multiple clusters) and to the selected similarity threshold. The group of such obtained core motifs could be further analysed in order to select a ‘particularly interesting’ subset of core motifs to be used for the following analysis phases. The selection could be performed on the basis of different properties of motifs (e.g. motif occurrences, consensus quality, sequence composition, absolute and relative position in the promoters) by exploiting the provided facilities for the manipulation of clusters, allowing reordering and deletion of core motifs. Moreover, graphical display of motif occurrences in input sequences can be used to identify in which sequences they are found and may facilitate the inspection of relative positions of different elements. Analysis can be possibly repeated with different settings or on a more homogeneous dataset, such as a group of promoters sharing one or more specific motifs. The extension and scoring phase allows to identify, from each group of solid words representing a core motifs, a restricted number of extended words which optimize the consensus sequence of the extended motif. Identified motifs generally are less variable in the core region but, depending on the settings of the extension and scoring phase, homogeneously variable motifs or motifs with poorly variable side regions could also be identified.
Our testing of performances of the method on different benchmark datasets gave quite positive results. The first dataset consisted in eight different groups of yeast sequences of length ranging from 500 to 1000 bp, containing known instances of signals at known positions. In this way, we tested the efficiency of MOST in finding signals of different strength in more or less numerous groups of promoter sequences. The second benchmark dataset, based on real human sequences, was designed to be fairly representative of those possibly analysed by future MOST users. In particular, genomic sequences of gene promoters were selected such that each of them contained at least one element known to bind one of the nine different transcription factors, by experimental evidence. Promoter sequences were localized by TSS annotation via integration between information on alignment between genomic DNA and cDNA, which is the most widely used system and which was proven to lead to the identification of reasonably well functional promoters (14). The Mixed signals benchmark dataset comprised sequences containing binding sites for different transcription factors. Moreover, binding sites were of various lengths, and more than one known binding site is included in some sequences. Different groups of sequences containing at least one instance of a specific motif were merged with other groups of sequences containing instances of different motifs, thus obtaining that positive control signals were dispersed and appeared only in a fraction of sequences under consideration.
A limitation of some motif extraction tools is that they request too many parameters as input of a single step, and their output is determined mostly on the basis of this large number of parameters. Changing one parameter value means that complete processing has to be performed again. Hence, the possibility to browse partial results and refine the analysis should save considerable amount of time and may allow the production of more meaningful results. Based upon the above considerations, a modular architecture was the natural choice for the design of MOST. Moreover, the modular design of MOST makes easy the possible extension of the tool in the future, or the modification of the implementation or the settings of a specific module. In particular, more scoring functions are actually under study and will be possibly added in the future. The possibility to merge clusters or delete words within each cluster could also be considered.
Acknowledgments
The authors thank Professor Alberto Apostolico for helpful discussion on method and results. Financial support of the University of Padova to G.A.D. and of AIRC (Associazione Italiana Ricerca sul Cancro) to S.B. is gratefully acknowledged. S.B. is recipient of a postdoctoral fellowship of the University of Padova, A.C. of an AIRC fellowship. Funding to pay the Open Access publication charges for this article was provided by Department of Biology, University of Padua, Italy.
Conflict of interest statement. None declared.
REFERENCES
- 1.Bucher P. Regulatory elements and expression profiles. Curr. Opin. Struct. Biol. 1999;9:400–407. doi: 10.1016/S0959-440X(99)80054-2. [DOI] [PubMed] [Google Scholar]
- 2.Levine M., Tjian R. Transcription regulation and animal diversity. Nature. 2003;424:147–151. doi: 10.1038/nature01763. [DOI] [PubMed] [Google Scholar]
- 3.Brazma A., Jonassen I., Vilo J., Ukkonen E. Predicting gene regulatory elements in silico on a genomic scale. Genome Res. 1998;8:1202–1215. doi: 10.1101/gr.8.11.1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Apostolico A., Bock M.E., Lonardi S., Xu X. Efficient detection of unusual words. J. Comput. Biol. 2000;7:71–94. doi: 10.1089/10665270050081397. [DOI] [PubMed] [Google Scholar]
- 5.Apostolico A., Bock M.E., Lonardi S. Monotony of surprise and large-scale quest for unusual words. J. Comput. Biol. 2003;10:283–311. doi: 10.1089/10665270360688020. [DOI] [PubMed] [Google Scholar]
- 6.Apostolico A., Pizzi C. Monotone scoring of pattern with mismatches. Proceedings of the Seventh Workshop on Algorithms in Bioinformatics (WABI 2004); Bergen, Norway. Springer; 2004. pp. 87–98. LNCS3240, Springer. [Google Scholar]
- 7.Sinha S., Tompa M. A statistical method for finding transcription factor binding sites. Proc. Int. Conf. Intell. Syst. Mol. Biol. 2000;8:344–354. [PubMed] [Google Scholar]
- 8.Sinha S., Tompa M. YMF: a program for discovery of novel transcription factor binding sites by statistical overrepresentation. Nucleic Acids Res. 2003;31:3586–3588. doi: 10.1093/nar/gkg618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tompa M., Li N., Bailey T.L., Church G.M., De Moor B., Eskin E., Favorov A.V., Frith M.C., Fu Y., Kent W.J., et al. Assessing computational tools for the discovery of transcription factor binding sites. Nat. Biotechnol. 2005;23:137–144. doi: 10.1038/nbt1053. [DOI] [PubMed] [Google Scholar]
- 10.Werner T., Fessele S., Maier H., Nelson P.J. Computer modeling of promoter organization as a tool to study transcriptional coregulation. FASEB J. 2003;17:1228–1237. doi: 10.1096/fj.02-0955rev. [DOI] [PubMed] [Google Scholar]
- 11.Wingender E., Dietze P., Karas H., Knuppel R. TRANSFAC: a database on transcription factors and their DNA binding sites. Nucleic Acids Res. 1996;24:238–241. doi: 10.1093/nar/24.1.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bortoluzzi S., Coppe A., Bisognin A., Pizzi C., Danieli G.A. A multistep bioinformatic approach detects putative regulatory elements in gene promoters. BMC Bioinformatics. 2005;6:121. doi: 10.1186/1471-2105-6-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Price A., Ramabhadran S., Pevzner P.A. Finding subtle motifs by branching from sample strings. Bioinformatics. 2003;19(Suppl. 2):ii149–ii155. doi: 10.1093/bioinformatics/btg1072. [DOI] [PubMed] [Google Scholar]
- 14.Trinklein N.D., Aldred S.J., Saldanha A.J., Myers R.M. Identification and functional analysis of human transcriptional promoters. Genome Res. 2003;13:308–312. doi: 10.1101/gr.794803. [DOI] [PMC free article] [PubMed] [Google Scholar]