Abstract
The ataxia telangiectasia mutant (ATM) protein kinase regulates the cell's response to DNA damage through the phosphorylation of proteins involved in cell-cycle checkpoints and DNA repair. However, the signal-transduction pathway linking DNA strand breaks to activation of ATM's kinase activity is not clearly defined. Here, we demonstrate that DNA damage induces the rapid acetylation of ATM. This acetylation depends on the Tip60 histone acetyltransferase (HAT). Suppression of Tip60 blocks the activation of ATM's kinase activity and prevents the ATM-dependent phosphorylation of p53 and chk2. Further, inactivation of Tip60 sensitizes cells to ionizing radiation. ATM forms a stable complex with Tip60 through the conserved FATC domain of ATM. The interaction between ATM and Tip60 is not regulated in response to DNA damage. Instead, the HAT activity of the ATM–Tip60 complex is specifically activated by DNA damage. Furthermore, this activation of Tip60 by DNA damage and the recruitment of the ATM–Tip60 complex to sites of DNA damage is independent of ATM's kinase activity. The results demonstrate that the Tip60 HAT plays a key role in the activation of ATM's kinase activity in response to DNA damage.
Keywords: FATC domain, TRRAP, NuA4, chromatin remodeling
The product of the ataxia telangiectasia (AT) gene, the AT mutant (ATM) protein kinase, is a key component of the signal-transduction pathway activated by DNA damage (1). In response to DNA strand breaks, ATM's kinase activity is upregulated, and ATM becomes autophosphorylated on Ser 1981 (2). ATM autophosphorylation initiates the conversion of the inactive ATM dimer to an active, monomeric ATM. ATM then phosphorylates multiple proteins involved in the DNA damage response, including nbs1, p53, chk2, and SMC1 (1, 3). These phosphorylated proteins, in turn, regulate the two key responses to DNA damage: the activation of cell-cycle checkpoints and the initiation of DNA repair.
Although the downstream signaling pathways activated by ATM are well characterized, the mechanism by which DNA strand breaks are detected and how this leads to activation of ATM's kinase activity are less clear. The Mre11/Rad50/Nbs1 (MRN) complex, a DNA-binding complex involved in the detection and repair of DNA damage, may play a role in regulating ATM activity. MRN functions as an adaptor protein that links activated ATM with its target proteins [including chk2 (4) and SMC1 (3)] to allow productive phosphorylation (5, 6). Biochemical studies have shown that the DNA-dependent activation of ATM's kinase activity requires the functional MRN complex (7, 8). However, although several studies have demonstrated that activation of ATM's kinase activity in cells lacking functional MRN complex is reduced (6, 9), other studies have shown that ATM activation is relatively normal in MRN-defective cells (5, 10). Furthermore, other proteins, including the p18 tumorsuppressor protein (11), have been shown to participate in ATM activation. Thus, although the MRN complex may regulate ATM activation under certain conditions, additional protein factors may also be required for ATM activation in vivo.
Recent studies have shown that the repair of DNA double-strand breaks requires the remodeling of chromatin structure (12–14). Furthermore, ATM can be activated by changes in chromatin structure that occur independently of DNA damage (2), leading to the proposal that ATM activation is mediated by alterations in chromatin structure at sites of DNA damage rather than through direct activation of ATM by strand breaks (1, 2). Chromatin remodeling is linked to the posttranslational modification of histones through phosphorylation, methylation, and acetylation (15). Histone acetyltransferases (HATs) can acetylate both histones and several nonhistone proteins (13, 15). Here, we examined whether Tip60, a HAT previously implicated in the DNA-damage response, provides the link between DNA strand breaks in chromatin and the activation of ATM. The results indicate that DNA damage induces rapid acetylation of ATM by a mechanism that depends on the Tip60 HAT. Suppression of Tip60 blocks activation of ATM's kinase activity and sensitizes cells to ionizing radiation (IR). Furthermore, the FATC domain of ATM mediates the interaction between ATM and Tip60, and the HAT activity of this ATM–Tip60 complex is specifically activated by DNA damage. These results indicate that the Tip60 HAT is a key component of the signal-transduction pathway that links the detection of DNA damage to the activation of the ATM protein kinase.
Materials and Methods
Cells and Antibodies. GM5849 AT cells, 293T, HeLaTip60wt or HAT-deficient HeLaTip60mt cells (containing mutations Q377E/G380E), and HeLaATM601 cells are described in refs. 16–18. Small interfering RNAs (siRNAs) T3 (GGAAGCUGCUGAUCGAGUUUU), T4 (GACGUAAGAACAAGAGUUAUU) and GFP (Dharmacon Research, Golden, CO) were transfected into cells with Lipofectamine 2000 (Invitrogen). Antibodies used were ATM antibodies 5C2 and 2C1 (Genetex, San Antonio, TX), phospho-Ser 1981 (Rockland, Gilbertsville, PA), anti-acetyllysine or Tip60 antibody (Upstate Biotechnology, Lake Placid, NY), p53 (Calbiochem), and γH2AX and ATM antibody PC116 (Oncogene Science) or chk2 (Cell Signaling Technology, Beverly, MA). Cell survival and immunofluorescence were as described in ref. 19.
Western Blot and Kinase Assays. Cell lysis and immunoprecipitation were as described in ref. 19. For kinase assays, immunoprecipitates were washed once in kinase buffer (10 mM Hepes, pH 7.4/10 mM MgCl2/50 mM NaCl/10 mM MnCl2) and incubated in 50 μl of kinase buffer containing 50 μM ATP, p53 peptide (2 μg of EPPLSQEAFADLWKK), and 10 μCi of [γ-32P]ATP (1 Ci = 37 GBq) for 30 min at 30°C. Reactions were terminated with 30% acetic acid (20 μl), spotted onto P81 paper, washed in 15% acetic acid, air-dried, and counted.
HAT Assays. Extracts were immunoprecipitated as above, except that the high salt wash was omitted. Immunoprecipitates were washed twice in HAT assay buffer (50 mM Tris, pH 8/10% glycerol/0.1 mM EDTA/1 mM DTT), and incubated in 60 μl of HAT assay buffer containing acetyl-CoA (100 μM) and biotinylated histone H4 peptide (0.5 μg) for 30 min at 30°C. An aliquot of the reaction was immobilized onto streptavidin plates and acetylation detected by using a HAT ELISA according to the manufacturer's instructions (Upstate Biotechnology).
ATM Constructs. Point mutations were inserted by site-directed mutagenesis to create restriction sites for SpeI (nucleotide 9279: A9281T/G9282A) and EcoR1 (nucleotide 9373: A8378C) in the ATM cDNA. The C terminus of ATM was removed by SpeI/EcoR1 digestion, and oligonucleotides with overhanging SpeI/EcoR1 sites encoding the indicated mutations were inserted.
Results
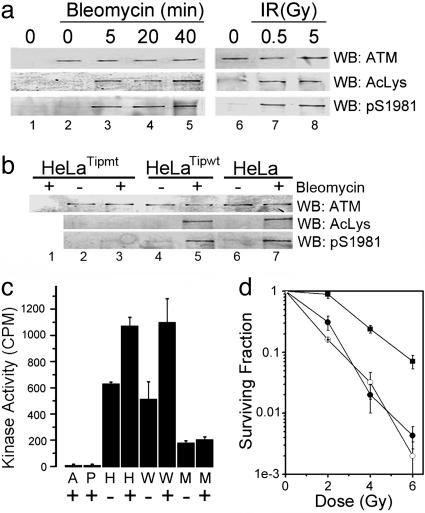
Initially, we determined whether ATM was acetylated in response to DNA damage. Acetylation of ATM was examined by using a previously characterized acetyllysine-specific antibody (20). Both bleomycin and low doses of IR (0.5 Gy) induced rapid autophosphorylation and acetylation of ATM (Fig. 1a). The time course and dose-dependence of ATM acetylation and Ser 1981 autophosphorylation were indistinguishable. Activation of ATM by DNA damage, therefore, correlates with the rapid acetylation and autophosphorylation of the ATM protein. The Tip60 HAT participates in the repair of DNA strand breaks (14, 16) and the exchange of phospho-H2Av at sites of DNA damage (12). To determine whether Tip60 was involved in the acetylation of ATM, HeLa cells overexpressing wild-type Tip60 (Tip60wt) or a dominant-negative HAT-deficient Tip60 (Tip60mt) were used. The overexpressed Tip60mt represents ≈80% of the cellular Tip60 (see Fig. 6f, which is published as supporting information on the PNAS web site) (16). ATM was efficiently acetylated and autophosphorylated in HeLa or HeLaTip60wt cells. In contrast, the autophosphorylation and acetylation of ATM in HeLaTip60mt cells was greatly reduced (Fig. 1b), implying that ATM activation requires functional Tip60 protein. To confirm this interpretation, the intrinsic kinase activity of ATM was measured. HeLa cells and HeLaTip60wt cells retained normal activation of ATM's kinase activity in response to bleomycin (Fig. 1c). HeLaTip60mt cells had decreased basal kinase activity that was not activated by exposure to bleomycin. ATM's kinase activity is required for ATM to regulate cell survival after exposure to IR (17). The failure to fully activate ATM in HeLaTip60mt cells implies that these cells should be more sensitive to IR. Fig. 1d demonstrates that HeLaTip60mt cells have increased sensitivity to IR, compared with HeLaTip60wt cells. Furthermore, the radiosensitivity of HeLaTip60mt cells is comparable with that seen in HeLaATM601 cells, in which ATM expression is stably silenced by short hairpin RNAi (18) (Fig. 1d).
Fig. 1.
Activation of ATM requires Tip60's HAT activity. (a) HeLa cells were exposed to bleomycin (5 μM) or IR (30 min). Cell extracts were immunoprecipitated with IgG (lane 1) or ATM antibody (lanes 2–8). Western blotting (WB) was used to detect ATM, acetylated ATM (AcLys), or autophosphorylated ATM (phospho-Ser 1981). (b) HeLa cells, HeLa cells expressing Tip60wt, or a HAT-inactive Tip60mt were examined for acetylation and phosphorylation of ATM after exposure to bleomycin (5 μM for 30 min). (c) ATM kinase activity was measured in HeLa (H), HeLaTip60wt (W) or HeLaTip60mt (M) cells. Controls using IgG (A) or in which the peptide was omitted (P) were included. Shown are results (±SEM) (n = 6). (d) HeLa cells were irradiated and clonogenic cell-survival assays carried out. In HeLaATM601 cells, ATM expression is silenced by stable expression of ATM-specific short hairpin RNAi (18). ▪, HeLaTip60wt; •, HeLaTip60mt; ○, HeLaATM601. Shown are results (±SEM) (n = 6).
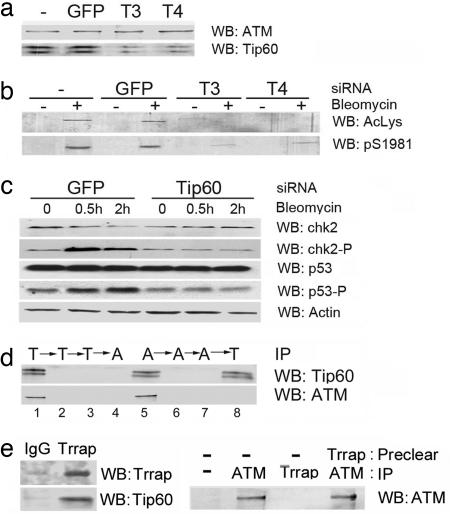
To control for the potential nonspecific effects of the dominant-negative Tip60mt mutant, siRNAs were used to target the 2 alternatively spliced forms of Tip60 present in HeLa cells. Two different siRNAs decreased expression of Tip60 without affecting the level of ATM (Fig. 2a) or other proteins (Fig. 2c). This reduction in Tip60 protein levels greatly reduced the autophosphorylation and acetylation of ATM in response to bleomycin (Fig. 2b). A nonspecific siRNA targeting GFP was without effect. Next, we examined whether Tip60 was required for ATM to phosphorylate the p53 and chk2 proteins. Both chk2 and p53 were phosphorylated in response to bleomycin in control cells (Fig. 2c). However, Tip60 depletion by siRNA inhibited the ATM-dependent phosphorylation of both p53 and chk2.
Fig. 2.
Depletion of Tip60 by siRNA attenuates ATM activation. (a) The 293T cells were untreated (-), transfected with siRNA targeting GFP, or Tip60 (T3 and T4). ATM and Tip60 were detected by Western analysis. (b) The 293T cells were transfected with GFP siRNA or Tip60 siRNAs T3 or T4. At 48 h, cells were exposed to bleomycin (5 μM for 30 min), immunoprecipitated with ATM antibody and ATM acetylation (AcLys), and autophosphorylation (phospho-Ser 1981) was measured. (c) The 293T cells were transiently transfected with GFP or Tip60 siRNA. At 48 h, cells were exposed to solvent (-) or bleomycin (5 μM). p53, chk2, or their phosphorylated isoforms were detected by Western analysis. (d) HeLa cell extracts were immunodepleted of Tip60 by three consecutive immunoprecipitations with Tip60 antibody (T, lanes 1–3), followed by immunoprecipitation with ATM antibody (A, lane 4) to detect unbound ATM. The opposite experiment, in which extracts were immunodepleted of ATM (A, lanes 5–7), followed by immunoprecipitation with Tip60 antibody (T, lane 8) to detect unbound Tip60, was also carried out. (e Left) Cell extracts were immunoprecipitated with IgG or TRRAP antibody, and levels of TRRAP and Tip60 were detected by Western blot analysis. (Right) Cell extracts were immunodepleted with IgG (-) or TRRAP antibody and immunoprecipitated with IgG (-), ATM antibody, or TRRAP antibody. ATM was measured by Western blot analysis.
The results indicate that functional Tip60 is required for the activation of ATM's kinase activity and for the ability of cells to survive DNA damage. The MRN complex may function as the upstream activator of ATM under some conditions (6, 8). However, we did not see any significant change in the acetylation or autophosphorylation of ATM in cells with mutations in the nbs1 or mre11 protein subunits of the MRN complex (Fig. 6a). The MRN complex in our experiments is, therefore, not essential for the acetylation of ATM. Next, we considered how Tip60 activates the ATM protein. The levels of Tip60 protein are unaltered by exposure to bleomycin, and immunoprecipitation experiments demonstrate that ATM and Tip60 can be specifically coprecipitated (Fig. 6 b and c). Interestingly, the association between ATM and Tip60 was not altered by DNA damage, and the interaction was resistant to ethidium bromide, indicating that DNA was not required for this association (Fig. 6 c and d). ATM and Tip60, therefore, exist as a preformed complex in cells, in the absence of a DNA-damage signal. To determine the stoichiometric relationship between ATM and Tip60, immunodepletion experiments were carried out. Whole-cell extracts were subjected to three consecutive immunoprecipitations with Tip60 antibody, removing all of the Tip60 (Fig. 2d Upper, lanes 1–3) and associated ATM (Lower). Subsequent immunoprecipitation with ATM antibody failed to detect any further ATM protein (Fig. 2d, lane 4). In the reverse experiment, extracts were subjected to three consecutive immunoprecipitations with ATM antibody, removing all of the ATM (Fig. 2d Upper, lanes 5–7) and associated Tip60 (Lower). Subsequent immunoprecipitation with Tip60 antibody yielded significant amounts of Tip60 but no further ATM protein (Fig. 2d, lane 8). Examination of the whole-cell extracts after immunodepletion demonstrates that the ATM and Tip60 antibodies removed essentially all of the soluble ATM from the whole-cell extracts (Fig. 6e). Fig. 2d demonstrates that all of the immunodetectable, soluble ATM was complexed with Tip60. The majority of ATM is soluble (>90% (21), implying that the majority of ATM exists in association with Tip60. Tip60 is the HAT component of the NuA4 complex (13, 22), which contains the ATM homologue TRRAP. Although several protein components of NuA4 have been implicated in DNA repair (14, 23), ATM has not been identified as a component of the NuA4 complex (22). To determine whether NuA4 and ATM constitute distinct Tip60 complexes, cell extracts were immunoprecipitated with antibodies to either ATM or TRRAP. TRRAP antibodies immunoprecipitated TRRAP and associated Tip60 (Fig. 2e). However, immunoprecipitated TRRAP was not associated with ATM (Fig. 2e), and immunodepletion of TRRAP complexes from whole-cell extracts did not remove significant amounts of ATM protein. We interpret this to indicate that cells contain at least two distinct Tip60 complexes: the NuA4–Tip60 complex and the ATM–Tip60 complex.
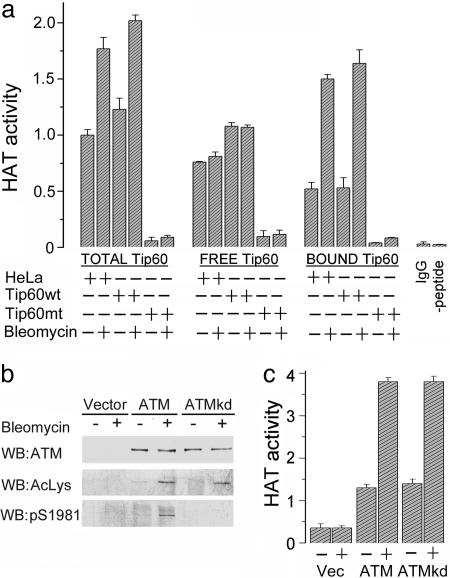
Because neither Tip60 protein levels nor the association of Tip60 with ATM is altered by DNA damage, we inferred that Tip60's HAT activity, itself, must be activated by DNA damage. The HAT activity of the ATM–Tip60 complex and of the remaining, non-ATM-associated Tip60, was measured. In Fig. 3a, the total cellular Tip60 HAT activity in HeLa and HeLaTip60wt cells was increased after exposure to bleomycin. As expected, Tip60 HAT activity was minimal in HeLaTip60mt cells. The free (non-ATM-associated) Tip60 activity was not activated by bleomycin. In contrast, there was specific activation of the HAT activity of the ATM–Tip60 complex after exposure to bleomycin (Fig. 3a, bound), demonstrating that only the Tip60 associated with ATM is responsive to DNA damage.
Fig. 3.
DNA damage activates Tip60's HAT activity. (a) To obtain total Tip60 HAT activity, HeLa cells were immunoprecipitated with Tip60 antibody. To obtain bound Tip60 HAT activity, cells were immunoprecipitated with ATM antibody. Free Tip60 was then obtained by immunoprecipitating ATM-depleted extracts with Tip60 antibody. Washed immunoprecipitates were incubated with a peptide derived from the N terminus of histone H4 and acetylation monitored by using a modified ELISA. Controls in which IgG was substituted for Tip60 antibody or the peptide was omitted from the assay are included. (b) ATM-negative GM5849 cells expressing vector, ATM, or ATMkd were exposed to solvent (-) or bleomycin (5 μM for 30 min) and ATM protein levels, ATM acetylation, and ATM autophosphorylation were measured. (c) GM5849 cells expressing vector, ATM, or ATMkd were exposed to bleomycin and ATM immunoprecipitated with ATM antibody. ATM-associated HAT activity was measured as above. Shown are results (±SEM) (n = 6).
ATM phosphorylates multiple proteins (1, 5), suggesting that ATM may phosphorylate and activate Tip60's HAT activity. To exclude this possibility, we determined whether Tip60 activation required ATM kinase activity. DNA-damage-inducible autophosphorylation and acetylation of ATM was restored in AT cells expressing exogenous ATM (Fig. 3b). In contrast, kinase-inactive ATM (ATMkd) cells failed to undergo Ser 1981 autophosphorylation, but retained inducible acetylation. Furthermore, the Tip60 HAT activity associated with both ATM and ATMkd was activated by DNA damage (Fig. 3c). Therefore, both the acetylation of ATM and the activation of Tip60's HAT activity are independent of ATM's kinase activity. Tip60 activation is, therefore, not regulated by ATM's kinase activity.
ATM's kinase domain is wedged between two conserved domains termed the FAT and FATC domains (24, 25). The FAT/kinase-domain/FATC structure is present in several ATM-like proteins, including ATM, DNA-PKcs, Atr, and TRRAP (25), with the C-terminal FATC region exhibiting the highest level of sequence homology. TRRAP is a key component of the NuA4 transcriptional complex (26), and the C terminus of TRRAP may mediate the association of Tip60 with the NuA4 complex (27). Because both TRRAP and ATM associate with Tip60 and share a common C-terminal-domain structure, we examined whether the highly conserved FATC domain of ATM was required for the formation of the ATM–Tip60 complex.
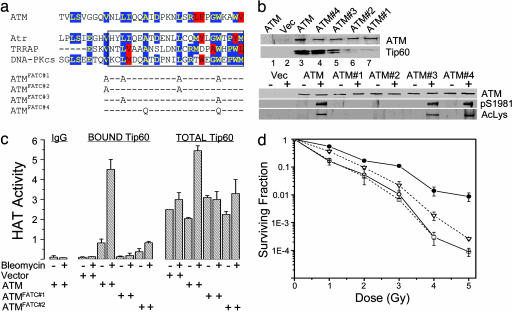
Several conserved amino acids in the FATC domain were mutated (Fig. 4a) and the effect on ATM–Tip60 complex formation assessed (Fig. 4b). Mutations ATMFATC#1 and ATMFATC#2 greatly reduced the interaction between ATM and Tip60 and the autophosphorylation and acetylation of ATM in vivo. ATMFATC#3 also decreased association between ATM and Tip60 and reduced both the acetylation and autophosphorylation of ATM. Mutation ATMFATC#4 had minimal impact.
Fig. 4.
Formation of the ATM–Tip60 complex requires the FATC domain of ATM. (a) FATC domains of ATM, Atr, TRRAP, and DNA-PKcs, comprising the last 33 amino acids at the C terminus of each of the proteins. Mutations inserted into the FATC domain are shown. blue box, identical; red box, similar. (b Upper) GM5849 cells expressing mutations in the FATC domain were immunoprecipitated with ATM antibody and the levels of ATM and associated Tip60 protein measured. (Lower) GM5849 cells expressing the indicated construct were exposed to solvent (-) or bleomycin (5 μM for 30 min). ATM protein, ATM acetylation, and ATM autophosphorylation were detected by Western blot analysis. (c) To obtain total Tip60 HAT activity, cells were immunoprecipitated with Tip60 antibody. To obtain bound Tip60 HAT activity, cells were immunoprecipitated with ATM antibody. Controls using IgG or omitting the peptide are shown. (d) GM5849 cells expressing the indicated construct were irradiated and colony formation assay carried out. ○, vector; •, ATM; □,= ATMFATC#1; ▿, ATMFATC#2. Shown are results (±SEM) (n = 3). Error bars are shown when larger than symbol size.
The mutations in the FATC domain that abolish the interaction between ATM and Tip60 will release Tip60 into the general pool of cellular Tip60. In Fig. 3a, only the Tip60 associated with ATM was activated by DNA damage, raising the question of whether Tip60 is still activated, even though it was no longer associated with ATM. The activation of Tip60 in AT cells expressing ATM, ATMFATC#1, or ATMFATC#2 was measured. ATMFATC#1 had no detectable HAT activity, whereas ATMFATC#2 retained low levels of bleomycin-induced HAT activity (Fig. 4c). In AT cells expressing ATM, the total cellular Tip60 HAT activity was increased several-fold by DNA damage (Fig. 4c). In contrast, the total cellular Tip60 was not significantly increased in either AT cells or AT cells expressing ATMFATC#1. There was a small but significant increase in Tip60 HAT activity in ATMFATC#2 cells, consistent with the low levels of Tip60 associated with this mutant (Fig. 4b). These results also were reflected in the ability of each construct to complement the increased radiosensitivity of AT cells (Fig. 4d). ATMFATC#1, which has no detectable Tip60 HAT activity, did not alter the radiosensitivity of AT cells, whereas ATMFATC#2, which retains residual Tip60 HAT activity, retained some functional activity. Although in the specific experiment shown in Fig. 4b, ATMFATC#2 did not display detectable Ser 1981 phosphorylation by Western blot analysis, the more sensitive immunohistochemistry data revealed low levels of phospho-Ser 1981 autophosphorylation of ATMFATC#2 after exposure to IR (Fig. 5b). Therefore, for each of the mutations, there was a direct correlation between the disruption of the ATM–Tip60 complex, and the loss of ATM activation, as monitored by ATM acetylation, autophosphorylation associated Tip60 HAT activity, and radiosensitivity. Furthermore, the results from Fig. 4 demonstrate that only the Tip60 associated with ATM is activated in response to DNA damage.
Fig. 5.
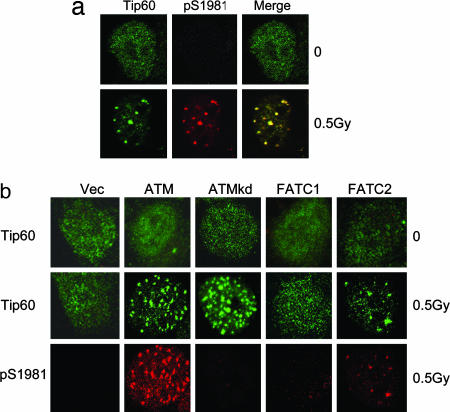
ATM and Tip60 colocalize to IRIF. (a) HeLa cells expressing HA-Tip60 were irradiated and allowed to recover for 20 min. Cells were fixed and immunostained with HA antibody (to detect Tip60) or phospho-Ser 1981 antibody to detect activated ATM. (b) GM5849 AT cells expressing ATM, ATMkd, ATMFATC#1 (FATC1), or ATMFATC#2 (FATC2) were irradiated (0.5 Gy) and allowed to recover for 20 min. Cells were fixed and immunostained with HA antibody (to detect Tip60) and phospho-Ser 1981 antibody (to detect activated ATM).
Activated ATM forms IR-induced foci (IRIF) and colocalizes with other DNA-damage-response proteins, such as H2AX (28). ATM and Tip60 colocalized to IRIF after exposure to low (0.5 Gy) doses of radiation (Fig. 5a). Tip60 also colocalized with γH2AX after exposure to IR (see Fig. 7, which is published as supporting information on the PNAS web site). Tip60 is, therefore, rapidly recruited to sites of DNA damage. We next examined whether the formation of Tip60 IRIF depended on ATM. In AT cells, no ATM or Tip60 foci were seen, whereas AT cells complemented with wild-type ATM displayed both ATM and Tip60 IRIF (Fig. 5b). No Tip60 or ATM foci were detected with the ATMFATC#1 mutant, consistent with the inability of this construct to associate with Tip60. However, a small number of Tip60 and ATM IRIF were detected in the ATMFATC#2 mutant cells, consistent with the low levels of Tip60 HAT activity and the small decrease in radiosensitivity seen with ATMFATC#2 (Fig. 4). These results clearly demonstrate that ATM and Tip60 must be associated for both the activation of Tip60 (Fig. 4) and the formation of IRIF (Fig. 5b). Next, we determined whether ATM's kinase activity was required for the formation of IRIF. Surprisingly, the kinase-inactive ATM, which retains DNA-damage-induced Tip60 activation (Fig. 3), is also permissive for the formation of Tip60 IRIF. Thus, ATM's kinase activity is not required for the recruitment of the ATM–Tip60 complex to sites of DNA damage.
Discussion
The results demonstrate a key role for Tip60 in the cell's response to DNA damage. The rapid activation of Tip60's HAT activity and the recruitment of Tip60 to sites of DNA damage defines Tip60 as a DNA-damage-response protein, consistent with reports demonstrating that Tip60 is essential for the acetylation of histone H4 (14), the repair of DNA strand breaks (14, 16), and the removal of phospho-H2Av after DNA damage (12). The rapid acetylation of ATM occurs within the same time frame as the phosphorylation of H2AX (29) and the autophosphorylation of ATM (2), placing ATM acetylation among the earliest events to occur in response to DNA strand breaks. The observation that ATM activation requires Tip60's HAT activity implies that acetylation of ATM is essential for activation of the ATM protein kinase. However, identification of the acetylated lysine residues will be required to determine how acetylation regulates ATM.
Cells contain multiple Tip60 complexes, including the NuA4–Tip60 chromatin remodeling complex (13, 22) and a smaller complex termed picNuA4 (16, 30). No interaction between ATM and the NuA4 subunit TRRAP was detected (Fig. 2e), and ATM has not been previously identified as a component of NuA4 (16, 22). In addition, only the Tip60 associated with ATM was activated by DNA damage (Fig. 3). Although several protein components of NuA4 have been implicated in DNA repair (14, 23), the results indicate that NuA4–Tip60 and ATM–Tip60 represent distinct functional Tip60 complexes. Therefore, despite both complexes using Tip60 to provide HAT activity, NuA4 and ATM will likely have distinct functions in the DNA-damage response. For example, the Arp4 subunit of yeast NuA4 targets NuA4 to γH2AX after DNA damage (23). The recruitment of NuA4 to γH2AX may be responsible for chromatin remodeling at sites of DNA damage, including the acetylation and removal of phospho-H2Av by Tip60 (12). The ATM–Tip60 complex may activate cellular signaling pathways directed toward regulating cell-cycle progression and related ATM-dependent processes. Furthermore, because ATM phosphorylates H2AX, the ATM–Tip60 complex must act upstream of the NuA4–Tip60 complex in the DNA-damage response.
The FATC domain at the C terminus of ATM functions as the binding domain for the Tip60 HAT. However, no direct interaction between the FATC domain of ATM and Tip60 has been observed (B.D.P., unpublished observation). Previous reports indicate that the association of Tip60 with NuA4 requires TRRAP (27) and is mediated by the epc1 protein (30). Tip60 may, therefore, be linked to the FATC domain of ATM by one or more intermediary proteins. The C-terminal FATC domains of the TRRAP, ATM, DNA-PKcs, and Atr proteins are highly conserved (Fig. 4), predicting that Atr and DNA-PKcs are also associated with Tip60 or a related HAT and that Atr and DNA-PKcs may be acetylated in response to DNA damage. Interestingly, truncating mutations that delete part or all of the FATC domain of ATM have been reported from patients with AT and B cell chronic lymphocytic leukemia (31, 32). Our results indicate that these deletions would block ATM–Tip60 association and compromise ATM activation, accounting for the classical AT phenotypes described in those patients (32).
Current data indicate that the activation of ATM's kinase leads to the autophosphorylation of Ser 1981 and subsequent dissociation of the inactive ATM dimer into active ATM monomers (2). We propose that the initial step in the ATM DNA repair pathway is up-regulation of Tip60's HAT activity. This up-regulation of Tip60 HAT activity does not require a functional ATM kinase domain, indicating that Tip60 is upstream of ATM's kinase activity. However, up-regulation of Tip60's HAT activity requires that it be associated with ATM, indicating a kinase-independent function for ATM in Tip60 activation. For example, ATM may bring Tip60 to sites of DNA damage, or ATM may associate with regulatory proteins at or near sites of DNA damage. One potential mechanism for this activation step would be through the interaction of Tip60's chromodomain with methylated lysine residues on histones (33). Previous work has shown that methylated histones at sites of DNA damage can recruit the 53BP1 protein (34). Tip60's chromodomain could interact with methylated histones that are exposed at the site of DNA damage. This interaction would activate Tip60's HAT activity, and ATM would become acetylated at one or more lysine residues. The acetylation of ATM by Tip60 may facilitate dimer–monomer transitions or may activate ATM's kinase activity, allowing for autophosphorylation of ATM. The fully activated ATM–Tip60 complex is then free to phosphorylate key target proteins. These results are consistent with previous observations that ATM activation requires changes in chromatin structure (2). Tip60, therefore, provides the essential link to explain how changes in chromatin structure can contribute to the activation of the ATM protein kinase. Although some studies have indicated that the MRN complex participates in the activation of ATM (6–9), we found that, consistent with other reports (5, 10), the acetylation and autophosphorylation of ATM did not require a functional MRN complex.
The recruitment of ATM to IRIF is thought to occur through a direct interaction between activated ATM and MRN (6, 8). However, the kinase-inactive ATM–Tip60 complex was efficiently recruited to IRIF. The autophosphorylation and activation of ATM's kinase activity are, therefore, not required for the recruitment of ATM to IRIF. Recruitment of ATM to IRIF could, therefore, either depend on ATM acetylation or be mediated by conformational changes in MRN after DNA damage.
In conclusion, the results demonstrate that the Tip60 HAT is a key component of the signal-transduction pathway that links the detection of DNA strand breaks to the activation of the ATM protein. Furthermore, this activated ATM kinase remains associated with the activated Tip60. Cells exposed to DNA damage will, therefore, contain an activated ATM–Tip60 complex with dual protein kinase and protein acetyltransferase activity. The ATM–Tip60 complex may, therefore, regulate the cell's response to DNA damage through both phosphorylation and acetylation of key target proteins.
Supplementary Material
Acknowledgments
We thank Alan D'Andrea (Dana–Farber Cancer Institute) for helpful comments and critical reading of the manuscript and for NBS cells, Yoshihiro Nakatani (Dana–Farber Cancer Institute) for the HeLaTip60 cells, Matthew Weitzman (Salk Institute, La Jolla, CA) for the A-TLD cells, and Craig Robson (University of Newcastle-upon-Tyne, U.K.) for the HA-Tip60 vector. This work was supported by National Cancer Institute grants (to B.D.P.), the Ataxia-Telangiectasia Childrens Project, and the Associates of the Joint Center for Radiation Therapy Foundation.
Author contributions: B.D.P. designed research; S.Y., X.J., S.C., and N.F. performed research; S.Y., X.J., and B.D.P. analyzed data; and B.D.P. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: AT, ataxia telangiectasia; ATM, AT mutant; ATMkd, kinase-inactive ATM; HAT, histone acetyltransferase; IR, ionizing radiation; IRIF, IR-induced foci; MRN, mre11/rad50/nbs1; siRNA, small interfering RNA.
References
- 1.Bakkenist, C. J. & Kastan, M. B. (2004) Cell 118, 9-17. [DOI] [PubMed] [Google Scholar]
- 2.Bakkenist, C. J. & Kastan, M. B. (2003) Nature 421, 499-506. [DOI] [PubMed] [Google Scholar]
- 3.Kim, S. T., Xu, B. & Kastan, M. B. (2002) Genes Dev. 16, 560-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buscemi, G., Savio, C., Zannini, L., Micciche, F., Masnada, D., Nakanishi, M., Tauchi, H., Komatsu, K., Mizutani, S., Khanna, K., et al. (2001) Mol. Cell. Biol. 21, 5214-5222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kitagawa, R., Bakkenist, C. J., McKinnon, P. J. & Kastan, M. B. (2004) Genes Dev. 18, 1423-1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Uziel, T., Lerenthal, Y., Moyal, L., Andegeko, Y., Mittelman, L. & Shiloh, Y. (2003) EMBO J. 22, 5612-5621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee, J. H. & Paull, T. T. (2004) Science 304, 93-96. [DOI] [PubMed] [Google Scholar]
- 8.Lee, J. H. & Paull, T. T. (2005) Science 308, 551-554. [DOI] [PubMed] [Google Scholar]
- 9.Carson, C. T., Schwartz, R. A., Stracker, T. H., Lilley, C. E., Lee, D. V. & Weitzman, M. D. (2003) EMBO J. 22, 6610-6620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Falck, J., Coates, J. & Jackson, S. P. (2005) Nature 434, 605-611. [DOI] [PubMed] [Google Scholar]
- 11.Park, B. J., Kang, J. W., Lee, S. W., Choi, S. J., Shin, Y. K., Ahn, Y. H., Choi, Y. H., Choi, D., Lee, K. S. & Kim, S. (2005) Cell 120, 209-221. [DOI] [PubMed] [Google Scholar]
- 12.Kusch, T., Florens, L., Macdonald, W. H., Swanson, S. K., Glaser, R. L., Yates, J. R., Abmayr, S. M., Washburn, M. P. & Workman, J. L. (2004) Science 306, 2084-2087. [DOI] [PubMed] [Google Scholar]
- 13.Carrozza, M. J., Utley, R. T., Workman, J. L. & Cote, J. (2003) Trends Genet. 19, 321-329. [DOI] [PubMed] [Google Scholar]
- 14.Bird, A. W., Yu, D. Y., Pray-Grant, M. G., Qiu, Q., Harmon, K. E., Megee, P. C., Grant, P. A., Smith, M. M. & Christman, M. F. (2002) Nature 419, 411-415. [DOI] [PubMed] [Google Scholar]
- 15.Peterson, C. L. & Laniel, M. A. (2004) Curr. Biol. 14, R546-51. [DOI] [PubMed] [Google Scholar]
- 16.Ikura, T., Ogryzko, V. V., Grigoriev, M., Groisman, R., Wang, J., Horikoshi, M., Scully, R., Qin, J. & Nakatani, Y. (2000) Cell 102, 463-473. [DOI] [PubMed] [Google Scholar]
- 17.Turenne, G. A., Paul, P., Laflair, L. & Price, B. D. (2001) Oncogene. 20, 5100-5110. [DOI] [PubMed] [Google Scholar]
- 18.Chen, S., Wang, G., Makrigiorgos, G. M. & Price, B. D. (2004) Biochem. Biophys. Res. Commun. 317, 1037-1044. [DOI] [PubMed] [Google Scholar]
- 19.Fernandes, N., Sun, Y., Chen, S., Paul, P., Shaw, R. J., Cantley, L. C. & Price, B. D. (2005) J. Biol. Chem. 280, 15158-15164. [DOI] [PubMed] [Google Scholar]
- 20.Yao, Y. L., Yang, W. M. & Seto, E. (2001) Mol. Cell. Biol. 21, 5979-5991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Andegeko, Y., Moyal, L., Mittelman, L., Tsarfaty, I., Shiloh, Y. & Rotman, G. (2001) J. Biol. Chem. 276, 38224-38230. [DOI] [PubMed] [Google Scholar]
- 22.Doyon, Y., Selleck, W., Lane, W. S., Tan, S. & Cote, J. (2004) Mol. Cell. Biol. 24, 1884-1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Downs, J. A., Allard, S., Jobin-Robitaille, O., Javaheri, A., Auger, A., Bouchard, N., Kron, S. J., Jackson, S. P. & Cote, J. (2004) Mol. Cell 16, 979-990. [DOI] [PubMed] [Google Scholar]
- 24.Keith, C. T. & Schreiber, S. L. (1995) Science 270, 50-51. [DOI] [PubMed] [Google Scholar]
- 25.Bosotti, R., Isacchi, A. & Sonnhammer, E. L. (2000) Trends Biochem. Sci. 25, 225-227. [DOI] [PubMed] [Google Scholar]
- 26.Doyon, Y. & Cote, J. (2004) Curr. Opin. Genet. Dev. 14, 147-154. [DOI] [PubMed] [Google Scholar]
- 27.Park, J., Kunjibettu, S., McMahon, S. B. & Cole, M. D. (2001) Genes Dev. 15, 1619-1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stiff, T., O'Driscoll, M., Rief, N., Iwabuchi, K., Lobrich, M. & Jeggo, P. A. (2004) Cancer Res. 64, 2390-2396. [DOI] [PubMed] [Google Scholar]
- 29.Rogakou, E. P., Pilch, D. R., Orr, A. H., Ivanova, V. S. & Bonner, W. M. (1998) J. Biol. Chem. 273, 5858-5868. [DOI] [PubMed] [Google Scholar]
- 30.Boudreault, A. A., Cronier, D., Selleck, W., Lacoste, N., Utley, R. T., Allard, S., Savard, J., Lane, W. S., Tan, S. & Cote, J. (2003) Genes Dev. 17, 1415-1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schaffner, C., Stilgenbauer, S., Rappold, G. A., Dohner, H. & Lichter, P. (1999) Blood 94, 748-753. [PubMed] [Google Scholar]
- 32.Laake, K., Jansen, L., Hahnemann, J. M., Brondum-Nielsen, K., Lonnqvist, T., Kaariainen, H., Sankila, R., Lahdesmaki, A., Hammarstrom, L., Yuen, J., et al. (2000) Hum. Mutat. 16, 232-246. [DOI] [PubMed] [Google Scholar]
- 33.Brehm, A., Tufteland, K. R., Aasland, R. & Becker, P. B. (2004) BioEssays. 26, 133-140. [DOI] [PubMed] [Google Scholar]
- 34.Huyen, Y., Zgheib, O., Ditullio, R. A., Jr., Gorgoulis, V. G., Zacharatos, P., Petty, T. J., Sheston, E. A., Mellert, H. S., Stavridi, E. S. & Halazonetis, T. D. (2004) Nature 432, 406-411. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.