Abstract
Waterborne enteric viruses threaten both human and animal health. These pathogens are host specific and cause a wide range of diseases and symptoms in humans or other animals. While considerable research has documented the risk of enteric viruses to human health from contact with contaminated water, the current bacterial indicator-based methods for evaluation of water quality are often ineffectual proxies for pathogenic viruses. Additionally, relatively little work has specifically investigated the risk of waterborne viruses to animal health, and this risk currently is not addressed by routine water quality assessments. Nonetheless, because of their host specificity, enteric viruses can fulfill a unique role both for assessing health risks and as measures of contamination source in a watershed, yet the use of animal, as well as human, host-specific viruses in determining sources of fecal pollution has received little attention. With improved molecular detection assays, viruses from key host groups can be targeted directly using PCR amplification or hybridization with a high level of sensitivity and specificity. A multispecies viral analysis would provide needed information for controlling pollution by source, determining human health risks based on assessments of human virus loading and exposure, and determining potential risks to production animal health and could indicate the potential for the presence of other zoonotic pathogens. While there is a need to better understand the prevalence and environmental distribution of nonhuman enteric viruses, the development of improved methods for specific and sensitive detection will facilitate the use of these microbes for library-independent source tracking and water quality assessment tools.
INTRODUCTION
Enteric viruses may be present naturally in aquatic environments or, more commonly, are introduced through human activities such as leaking sewage and septic systems, urban runoff, agricultural runoff, and, in the case of estuarine and marine waters, sewage outfall and vessel wastewater discharge. Over 100 types of pathogenic viruses are excreted in human and animal wastes (108). These viruses can be transported in the environment through groundwater, estuarine water, seawater, rivers, aerosols emitted from sewage treatment plants, insufficiently treated water, drinking water, and private wells that receive treated or untreated wastewater either directly or indirectly (9, 93, 99, 131, 146, 169). These viruses, collectively known as enteric viruses, usually are transmitted via the fecal-oral route and primarily infect and replicate in the gastrointestinal tract of the host. Enteric viruses are shed in extremely high numbers in the feces of infected individuals, typically between 105 and 1011 virus particles per gram of stool (45).
Commonly studied groups of enteric viruses belong to the families Picornaviridae (polioviruses, enteroviruses, coxsakieviruses, hepatitis A virus, and echoviruses), Adenoviridae (adenoviruses), Caliciviridae (noroviruses, caliciviruses, astroviruses, and small round-structured viruses), and Reoviridae (reoviruses and rotaviruses). Enteric virus groups that are considered to be emerging waterborne pathogens, based on their cellular and molecular structures that make them resistant to current water treatment processes, include circoviruses (consisting of torque tenovirus and torque tenovirus-like virus; these are nonenveloped viruses with single-stranded circular DNA and are resistant to heat inactivation), picobirnaviridae (small nonenveloped viruses with bisegmented double-stranded RNA that are extremely resistant to UV light inactivation), parvoviruses (the smallest known enteric viruses, with single-stranded RNA and high heat resistance), and polyomaviruses (including JC virus, BK virus, and simian virus 40; these are nonenveloped double-stranded DNA viruses that have been found to be very heat stable but are less resistant to chlorination than enteroviruses) (12, 20, 43, 155, 166).
Although enteric virus infections are associated primarily with diarrhea and self-limiting gastroenteritis in humans, they may also cause respiratory infections, conjunctivitis, hepatitis, and diseases that have high mortality rates, such as aseptic meningitis, encephalitis, and paralysis in immunocompromised individuals (90). In addition, some enteric viruses have been linked to chronic diseases such as myocarditis and insulin-dependent diabetes (60, 90). Enteric virus infections in animals such as cattle and swine are normally asymptomatic but can lead to abortion, neurological disorders, and mortality (80, 83, 97, 105).
Enteric viruses can be transmitted by food, water, fomites, and human contact. In addition to causing acute diseases, they are of public health concern due to their low infectious dose (65). For example, the probability of infection from exposure to one rotavirus is 31%, and no more than 1 PFU is required to cause infection in 1% of healthy adults with no antibody to the virus (135). Haas et al. (65) concluded that the risk of infection when consuming viruses in drinking water is 10- to 10,000-fold greater than that for pathogenic bacteria at similar exposures (16, 65). Because of the potential for contamination from a variety of sources, enteric viruses in water are of particular concern. Since the 1980s, with significant advancements in the area of environmental virology, enteric viruses have been recognized as the causative agents in many nonbacterial gastroenteritis cases and outbreaks (16). Enteric viruses have been isolated from and linked to outbreaks originating from contaminated drinking water sources, recreational waters (e.g., waters for swimming, canoeing, surfing, etc.), urban rivers, and shellfish harvested from contaminated waters (25, 37, 79, 93, 102, 115, 125). Between 1975 and 1979, water, followed by shellfish, was reported to be the main vehicle in outbreaks of vehicle-associated viral disease in the United States (28). Several reports indicate that only a fraction of waterborne disease incidences are ever reported; Craun suggested in 1991 that fewer than half of waterborne outbreaks occurring in the United States are investigated and reported (32). Nonpotable water, such as seawater, is also important; enteric viruses are able to persist for extended periods in the marine environment, which increases the probability of human exposure by recreational contact and accumulation in shellfish (102). Because shellfish are filter feeders, the concentration of viruses accumulated in their edible tissues may be much higher than that in the surrounding water (1). Consumption of shellfish harvested from enteric virus-contaminated waters often has led to human outbreaks (17, 18, 102).
In many countries, including the United States, regulators are still relying solely on bacterial indicators such as enterococci and fecal coliform and total coliform bacteria to assess the microbiological quality of water; however, bacterial indicators do not always reflect the risk from many important pathogens, such as viruses, stressed pathogenic bacteria (viable but nonculturable), and protozoa (54, 79, 118). Infectious enteric viruses have been isolated from aquatic environments that are in compliance with bacterial indicator standards, and there have been several virus-related outbreaks linked to ingestion of waters that met fecal coliform standards (32, 106). One of the major drawbacks in using fecal coliform bacteria and other traditional indicators (e.g., enterococci) is that these indicators may be found in both human and animal feces and naturally in soils. Furthermore, they may regrow in the environment after being excreted from their host (148). The ability to identify the dominant sources of fecal pollutants in aquatic environments has become increasingly important in water quality management and remediation; however, tracking the host source of bacterial indicators in environmental waters is impossible without laborious and extensive assays such as multiple antibiotic resistance profiling and ribotyping (39, 120). Complicating matters, studies have shown that in coastal and marine waters traditional bacterial indicators generally die off quickly compared to viruses and protozoa (15, 147).
Viral pathogens, because of their host specificity, have been suggested as one of the most promising tools to determine the sources of fecal contaminants in aquatic environments and may be used in conjunction with bacterial indicators to assess water quality and improve public health surveillance (48, 107). Pathogenic viruses are generally more resistant than bacterial indicators during conventional wastewater treatment such as chlorination and filtration and are able to withstand lipid solvents (51, 79, 152). In the environment, enteric viruses can survive under a wide pH range (pH 3 to 10) and for extended periods at low temperatures (90). Viruses have been reported to survive and remain infective for up to 130 days in seawater, for up to 120 days in freshwater and sewage, and for up to 100 days in soil at 20 to 30°C (79, 159). These survival periods surpass those reported for fecal coliform and other indicator bacteria in similar environments (109). Finally, because viruses have an obligate host requirement, there is no potential for regrowth in the environment. In general, enteric viruses show great potential to be used as water quality indicators to assess the risks associated with infectious virus transmission as well as to identify the dominant source of fecal contamination in waters.
Here we present information on the role of enteric viruses as pathogens of both humans and other animals and the risks presented from waterborne exposure. Specific discussions are limited to the enterovirus and adenovirus groups, which are among the most well studied in terms of waterborne contamination. The potential for these host-specific pathogens to be used as an additional tool in water quality studies and fecal source tracking is also highlighted.
ENTERIC VIRUSES AS PATHOGENS
Enteric viruses represent a diverse group. Most of the mammalian viruses, such as picornaviruses, rotaviruses, and noroviruses, are nonenveloped RNA viruses, while adenoviruses and polyomaviruses (for which transmission via the fecal-oral route has yet to be proven) are the only groups with double-stranded DNA (11). The impracticability of monitoring for the presence of all viral pathogens has led to the concept of an indicator organism. Two of the most studied groups of enteric viruses as potential water quality indicators are the enteroviruses and adenoviruses. While the occurrence of human enteric viruses in the environment and their role in waterborne transmission have been studied extensively, little information is available on environmental transmission of enteric viruses in animals, and additional research is needed to elucidate the environmental pathways of these viruses and their roles in the transmission of diseases in animals.
Enteroviruses
Enteroviruses consist of poliovirus, coxsackieviruses, echoviruses, and the numbered enteroviruses. As of 2003, 89 serotypes of enteroviruses have been identified and ratified by the Executive Committee of the International Committee on Taxonomy of Viruses (22). Enteroviruses are single-stranded RNA viruses with an icosahedral capsid ranging from 20 to 30 nm in diameter. About 70% (62 serotypes) of nonpoliovirus enteroviruses have been associated with human infections, and 30% have been associated with animal infections (80). Enteroviral infections in humans are reported to peak in summer and early fall, which also coincides with increased water recreational activities and water contact (90).
Enteroviruses as human pathogens.
Enteroviruses can cause a wide spectrum of diseases in humans. All enteroviruses are transmitted by the fecal-oral route, but clinical outcomes may go beyond gastroenteritis, as some viruses travel from the intestinal tract to other organs. Polioviruses usually infect their host by attacking the central nervous system and cause paralysis in victims (poliomyelitis). Coxsackieviruses have been associated with not only respiratory system infections and gastroenteritis but also insulin-dependent diabetes and heart diseases, such as myocarditis and pericarditis (60, 90, 162). Echoviruses are generally less infectious than other enteroviruses and are usually associated with the common cold and respiratory diseases. The numbered enteroviruses (enterovirus types 68 to 71) have not been studied extensively but have been isolated from patients with bronchiolitis, conjunctivitis, meningitis, and paralysis resembling poliomyelitis (90).
Enteroviruses as animal pathogens.
Animal-specific enterovirus infections in hosts such as cattle and pigs are often asymptomatic but may cause diseases ranging from diarrhea to reproductive failure and neurological disorders (Table 1) (92, 97). Two bovine enteroviruses (BEV), three porcine enteroviruses (PEV), 11 porcine teschoviruses (PTV) (10 were formerly classified as porcine enteroviruses), and 1 ovine enterovirus have been identified (87).
TABLE 1.
Host species, related diseases, and prevalence of animal-specific enteroviruses and adenoviruses in hosts and aquatic environments
| Virus type | Specific virus | Host species | Related diseases | Notes | Detection in water | Reference(s) |
|---|---|---|---|---|---|---|
| Enterovirus | Bovine enterovirus | Cattle | Diarrhea, abortion | Yes; detected in watering tanks, pastures, runoff streams, and river water | 97 | |
| Porcine enterovirus/teschovirus | Pig | Teschen-Talfan disease, pneumonia, polioencephalomyelitis, vesicular diseases, myocarditis, diarrhea, fertility disorders, and dermal lesions | Prevalence of 65% in pigs and wild hogs | Yes; detected in ponds, sewage, and river water | 23, 36, 42, 46, 70, 80, 88 | |
| Swine vesicular disease virus | Pig | High contagious lesions in pigs that are indistinguishable from those caused by foot-and-mouth disease | A porcine variant of human coxsackievirus B5 | No; however, studies have shown environmental transmission | 23, 34, 50 | |
| Adenovirus | Bovine adenovirus | Cattle | Keratoconjunctivitis, acute febrile disease, pneumonenteritis, and acute and fatal enteric diseases | No; however, has been detected in bovine fecal samples | 78, 96, 105 | |
| Porcine adenovirus | Pig | Mild diarrhea and respiratory signs; encephalitis and pneumoenteritis suspected | No; however, has been detected in swine fecal samples | 68, 78, 85, 105 | ||
| Chicken embryo lethal orphan adenovirus | Chicken | Not linked with any clinical sign | Widespread among chicken populations | Information not available | 30 | |
| Egg drop syndrome virus | Chicken | Reduction in egg production; loss of egg color and thin-shelled eggs | Hens in laying period | Information not available | 74, 156 | |
| Hemorrhagic enteritis virus | Turkey | Intestinal hemorrhages accompanied by immunosuppression | Information not available | 73, 160 |
Based on a study in Maryland, BEV have a prevalence of 76% in farmed cattle. While they are usually nonpathogenic, BEV have been linked to diarrhea and abortions in some infected cattle (97). PEV have a prevalence of 65% in pigs and wild hogs (23). PEV and PTV have been identified as the etiologic agents of the neurological disorder known as Teschen-Talfan disease, polioencephalomyelitis, vesicular diseases, myocarditis, pneumonia, diarrhea, fertility disorders, and dermal lesions in swine (42, 46, 70, 88).
Swine vesicular disease virus, a porcine variant of human coxsackievirus B5, causes lesions in pigs that are indistinguishable from those caused by foot-and-mouth disease virus, an aphthovirus (50). Because swine vesicular disease is highly contagious and difficult to eradicate and there is no effective vaccine, control measures often necessitate the slaughter of infected and contacted animals, which leads to severe economic losses (50). Transmission of this disease includes direct contact among infected animals and environmental contamination (34).
Adenoviruses
Adenoviruses were first isolated from humans and identified as the causative agent of epidemic febrile respiratory disease among military recruits in the 1950s (75, 132). Adenoviruses are nonenveloped, range from 90 to 100 nm in diameter, and consist of double-stranded DNA (84). All adenoviruses with human or mammalian hosts are classified under genus Mastadenovirus (78).
In 1998, adenoviruses were included in the “Candidate Contaminant List” as part of the Safe Drinking Water Act by the U.S. Environmental Protection Agency, and they are one of only four virus groups on the list (the three others are caliciviruses, coxsackieviruses, and echoviruses) (157). Adenoviruses are included because of their public health implications and their frequent occurrence in many aquatic environments. In addition, adenoviruses have been shown to be up to 60 times more resistant to UV irradiation than RNA viruses, such as enteroviruses and hepatitis A virus. Because they are double stranded, an undamaged DNA strand in adenoviruses may serve as a template for repair by host enzymes; furthermore, these viruses have a high molecular weight that may also impart increased UV resistance (56, 110, 130).
Adenoviruses as human pathogens.
Fifty-one serotypes of human adenoviruses (HAdV) have been identified (64). Human adenoviruses are the second most important viral pathogen of childhood gastroenteritis after rotavirus (31). They have been cited to cause symptomatic infections in several organ systems, including the respiratory system (pharyngitis, acute respiratory disease, and pneumonia), eye (conjunctivitis), gastrointestinal tract (gastroenteritis), central nervous system (meningoencephalitis), and genitalia (urethritis and cervicitis) (31, 84). Human adenovirus types 40 and 41 have been associated with gastroenteritis in children, while human adenovirus type 4 is linked to persistent epidemics of acute respiratory disease in the United States (33, 107). Transmission includes the fecal-oral route and inhalation of aerosols (79). The viruses are shed for extended periods in feces, urine, and respiratory secretions of infected persons (31).
In contrast to the notion that only those adenoviruses that infect the intestinal tract of the host will be excreted in feces, adenoviruses type 5, the nonenteric adenovirus strain that accounts for 11% of clinical adenovirus cases reported to World Health Organization, is also frequently detected in aquatic environments (93, 151).
Adenoviruses as animal pathogens.
Humans are not the only host for adenoviruses; animal-specific adenoviruses infect a wide range of species, including other mammals, birds, reptiles, amphibians, and fish (Table 1) (133). Five porcine adenoviruses (PAdV), five bovine adenoviruses (BAdV), and six ovine adenoviruses have been classified under the genus Mastadenovirus (5). Most adenoviruses infecting fowl have been classified as Aviadenovirus (5).
Infection with PAdV is usually asymptomatic, although cases of mild diarrhea or mild respiratory signs in swine have been noted (68). PAdV also has been isolated from pigs with encephalitis and pneumoenteritis (78, 85). Some BAdV, such as BAdV-3, have been shown to replicate in cattle and produce mild or no clinical signs, but several other serotypes have been linked to keratoconjunctivitis, acute febrile disease, pneumonenteritis, and acute and fatal enteric diseases in calves (78, 96, 105).
Among avian adenoviruses, chicken embryo lethal orphan adenovirus, classified as the type 1 fowl adenovirus, is widespread among chicken populations but has never been associated with serious disease and does not induce clinical signs when experimentally inoculated in chickens (30). Avian adenoviruses that often induce clinical signs or cause fatalities in avian species include hemorrhagic enteritis virus (infecting turkeys and causing intestinal hemorrhages accompanied with immunosuppression) and egg drop syndrome virus (infecting only hens in the laying period, causing thin-shelled eggs with loss of color and a reduction in egg production) (73, 74, 160).
Animals infected by adenoviruses have been shown to excrete the infectious viruses through their feces and can potentially become infected through the ingestion of fecally contaminated water or food (78, 156).
DETECTION OF ENTEROVIRUSES AND ADENOVIRUSES IN AQUATIC ENVIRONMENTS
Monitoring for the presence of human enteric viruses in environmental waters began in the 1940s, and this work has been applied to the detection of both human and animal viruses for monitoring microbial water quality and possibly tracking major sources of water pollution (62). In early studies on the occurrence of human enteric viruses in aquatic environments, cell culture was the most widely used technique for detection and isolation of infectious enteric viruses. Other viral detection methods typically used for clinical samples, such as radioimmunoassay, immunofluorescence, complement fixation, and enzyme-linked immunosorbent assay, were either too costly or lacked the sensitivity to detect viruses in environmental samples (60). The basic steps of virological analysis of environmental waters are sampling, virus concentration (and purification), and detection with cell culture assays or, more recently, molecular methods such as PCR and hybridization.
Virus Concentration Methods
Because the levels of enteric viruses in natural environments often are low, large volumes of water (up to thousands of liters) are frequently concentrated before analysis by inoculation on cultured host cells or by molecular methods (61, 100, 161). Different types of filters and filtration methods, such as cartridge filters (electropositive or electronegative), glass fiber filters, glass wool filters, vortex flow filtration, tangential flow filtration, and acid flocculation, traditionally have been used to collect and concentrate viral particles from water samples (53, 60, 79, 101, 121). Because of the small size of viral particles, mechanical filtration is often not possible; therefore, adsorption-elution methods are employed. These involve manipulation of charges on the virus surface, using pH changes to maximize their adsorption to charged filters (101, 121). Adsorption-elution of viruses with an electropositive filter (i.e., 1MDS Zetapor Virosorb [CUNO, Meriden, CN]) is one of the most commonly used techniques and is the method for recovery of enteric viruses from drinking water that is designated by the Information Collection Rule of the U.S. Environmental Protection Agency (158). These filters require no manipulation of pH because most enteric viruses are negatively charged at ambient pH (101). However, electropositive filters are easily clogged and have low recovery rates for viruses in marine water; the presence of salt and alkalinity of seawater cause low absorption of viruses to the filters (104).
Electronegative filters show higher virus recoveries from marine water and waters of high turbidity than do electropositive filters (44, 86, 101, 104). Under ambient conditions, enteric viruses are negatively charged and will adsorb to a negatively charged membrane only in the presence of Mg2+ (i.e., salt), other multivalent cations, or, more commonly, under acidic conditions (when their net charge becomes positive) (145, 164). Katayama et al. (86) developed a modified virus concentration method with a rate of high virus recovery from seawater (up to 73% of poliovirus was recovered from 1 liter) and minimal inhibitory effects. This method uses adsorption with a type HA, negatively charged membrane (Millipore, Billerica, MA) rather than a cartridge as used in traditional methods (101). After the sample is filtered and viruses adsorbed to the membrane, an acid rinse step is used to remove cations (i.e., salt) and other inhibitors while keeping viruses attached to the membrane. In addition, an inorganic eluting medium (NaOH) that has fewer inhibitory effects in PCR assays than the commonly used organic eluting medium (beef extract) is used (69, 86); however, a high-pH beef extract solution is the most widely used medium to elute absorbed viruses from cartridge filters and has worked well with cell culture assays (4, 136, 140). In PCR, the use of NaOH as an eluent provides a good alternative to other methods that attempt to remove PCR inhibitors from beef extract solution, such as resin treatments, polyethylene glycol (PEG) precipitation-resuspension techniques, immunomagnetic capture, and glass purification, which can be expensive and complicated (86).
For improved recovery of viruses from freshwater, such as groundwater, river water, and tap water, Haramoto et al. (69) modified the virus concentration method of Katayama et al. (86) by precoating a type HA, negatively charged membrane (Millipore, Billerica, MA) with AlCl3 prior to filtering samples, yielding a mean poliovirus recovery of 109% from 10 liters of seeded MilliQ water.
Ultrafiltration methods such as vortex flow filtration (VFF) and tangential flow filtration (TFF) are alternatives to adsorption-elution techniques and have been shown to be efficient in recovering viruses from marine water (a recovery rate of 72% for T2 bacteriophage in seeded samples with VFF) (60, 126). Both filtration devices utilize a flow pattern that forces water through a cylindrical filter with pressure while keeping and retaining particles from filters to avoid clogging (126). These methods require minimal manipulation of water; samples can be processed under natural pH, and an elution step is not needed (79). The typical volume of water processed is 20 liters, which is concentrated to ∼50 ml (60). TFF requires prefiltration of water samples to remove plankton and suspended solids. VFF has been shown to be more time-efficient because prefiltration of samples is not required, and it has a higher viral recovery rate than TFF, but it tends to concentrate more PCR inhibitors with the viruses (79). However, both VFF and TFF are less cost- and time-effective than adsorption-elution because of the high cost of equipment and limitations on the volume of sample that can be concentrated at one time.
Concentrated or eluted water samples usually are further concentrated and purified to reduce the final volume of samples to 1 or 2 ml for processing (69, 79, 86, 101, 118). Commonly used secondary concentration methods include organic flocculation (recommended by the Information Collection Rule of the U.S. Environmental Protection Agency), PEG precipitation, and centrifugal ultrafiltration (ultraconcentration based on a molecular weight cutoff, such as with Centriprep YM-30 or YM-50 concentrator columns [Millipore, Billerica, MA]) (79, 86, 101, 158).
In organic flocculation, buffered beef extract is used to precipitate viruses from concentrated samples by reducing the pH to 3.5. The precipitate is then centrifuged to form a pellet before being dissolved in sodium phosphate (158). The PEG precipitation procedure consists of precipitating viral particles by addition of 0.5 M NaCl and 7% PEG to beef extract with constant stirring for 2 h at 4°C followed by centrifugation. The virus pellet is then resuspended in Tris-buffered saline (44). Again, the use of beef extract in these procedures has been reported to cause inhibitory effects in PCR assays (4, 136). Ultrafiltration concentration methods do not require manipulation of samples and have shown a high virus recovery; seeded MilliQ water samples concentrated by Centriprep YM-50 filter units give a mean polioviruses recovery of 74% (69).
Virus Detection Methods
Cell culture assays.
Concentrated samples can be either extracted for viral nucleic acid analysis (PCR amplification) or inoculated onto common cell lines, such as buffalo green monkey kidney (BGM) cells, MA104 cells, RD cells, A549 cells, FRhK-4 cells, CaCo-2 cells, Madin-Darby bovine kidney (MDBK) cells, and pig kidney (PK-15 A) cells, specific to each virus type for quantification and isolation of infectious viruses (38, 97, 101, 127). The cell culture technique was the most widely used technique to determine the occurrence of infectious enteroviruses in environmental samples before the development of molecular methods such as PCR in the late 1980s and is still the best method to isolate and determine the infectivity of viruses from environmental samples (Table 2). After inoculation of a chosen cell line, flasks are evaluated for the presence of damaged cells or rounding of cells and sloughing of the monolayer (cytopathogenic effects [CPE]) as evidence for viral infection.
TABLE 2.
Comparison of common methods for the detection of enteric viruses from environmental sources
| Method | Advantages | Disadvantages | References |
|---|---|---|---|
| Cell culture | Infectivity can be determined; provides quantitative data | Long processing time (takes days to weeks); relatively more expensive than conventional PCR; not all viruses can grow on cultured cells | 101, 149 |
| PCR (RT-PCR) | Rapid; increased sensitivity and specificity compared to cell culture | Presence or absence only (nonquantitative); inhibitors present in environmental samples may interfere with PCR amplification; infectivity cannot be determined | 61, 99 |
| Nested PCR (semi/heminested) | Increased sensitivity compared to conventional PCR; can replace PCR confirmation steps, such as hybridization | Potential risk of carryover contamination when transferring PCR products | 79, 127, 161 |
| Multiplex PCR | Several types, groups or species of viruses can be detected in a single reaction; saves time and cost | Difficult to achieve equal sensitivity for all targeted virus species, groups, or types; may produce nonspecific amplification in environmental samples | 49, 57 |
| Real-time PCR | Provides quantitative data; confirmation of PCR products is not required (saves time); can be done in a closed system, which reduces risk of contamination compared to nested PCR | Expensive equipment; occasionally less sensitive than conventional PCR and nested PCR | 7, 40, 120 |
| ICC-PCR | Improves detection of infectious viral pathogens compared to conventional cell culture; detects viruses that do not produce CPE in cell culture; provides results in half the time required for conventional cell culture | Less time-efficient and more costly than direct PCR detection; carryover detection of DNA of inactivated viruses inoculated onto cultured cells is possible | 26, 59, 89 |
The major drawback to the cell culture assay is that it is laborious and time-consuming; it requires days to weeks of incubation and several passages to confirm both positive and negative results. In addition, some samples may be cytotoxic but appear as CPE on cells. A universal cell line that can be used for culturing all enteric viruses has not been established, and there are many viruses that cannot be detected through cell culture assay because they either do not produce CPE, are extremely slow growing, or do not grow on established cell lines (26, 101, 128). For example, adenoviruses, which are one of the most important human pathogens and are often detected in greater numbers than enteroviruses in wastewater, are slow growing, often do not produce CPE, and are consistently underestimated when fast-growing enteroviruses are present (77, 151). Likewise, noroviruses, one of the major causative agents for viral gastroenteritis and food-borne outbreaks, cannot be propagated in cell culture (69).
Molecular assays.
With both concentrated samples and infected cultured cells, viral nucleic acids can be extracted and purified to remove cell debris and inhibitors before being amplified and detected by PCR (61, 101). One of the most widely used methods for viral nucleic acid extraction and purification was developed by Boom et al. (13) and is based on guanidium thiocyanate extraction and use of silica columns to bind and wash nucleic acids. This method is rapid, easy to use, and efficient in removing inhibitors (79, 128). Casas et al. (24) developed an extraction method with the use of guanidium thiocyanate and an inorganic solvent to purify both viral RNA and DNA in a single extraction step. Extraction kits based on modifications of these methods are available commercially (61, 79, 101). Other methods for viral nucleic acid extraction and purification include proteinase K treatment followed by phenol-chloroform extraction and ethanol precipitation, sonication, and heat treatment (2, 19, 26, 58, 94, 113).
(i) PCR.
Molecular techniques have been used extensively to detect enteric viruses from environmental samples since the early 1990s. Molecular viral detection assays, such as PCR and hybridization, usually are based on the detection of a part of the viral genome that is highly conserved with broad homology within a specific group of viruses (3, 35). PCR-based assays offer several advantages over cell culture assays in detecting viral pathogens from environmental samples. PCR is rapid, highly sensitive, and specific if a well-designed assay is developed. PCR viral detection is less laborious and time-consuming and also more specific and sensitive than cell culture (Table 2) (27, 66, 79). Results from PCR assays can be obtained within 24 h of sampling, compared to days or weeks of incubation for cell culture assay (62, 120). PCR is also capable of differentiating specific viruses (79, 86, 127). For example, PCR primers can be designed to target whole virus orders (e.g., enteroviruses or adenoviruses) or may be specific to a single type of virus (e.g., poliovirus) or tailored for virus serotypes within a host group (e.g., humans, cattle, and pigs) (64, 168).
PCR is also highly sensitive and is capable of detecting viruses that are present in low numbers in environmental samples and either are difficult to grow in cultured cells or replicate without producing CPE (26, 101, 128). The high level of sensitivity in PCR assays has indicated that cell culture detection alone may underestimate the true level of contamination in environmental sources. Pina et al. (127) suggested that PCR has led to higher rates of detection of adenoviruses in environmental samples. Borchardt et al. (14) detected enteric viruses (enteroviruses, rotavirus, Norwalk-like virus [norovirus], and hepatitis A virus) from 4 (8%) of 50 household wells by PCR, while no virus was detected by cell culture. Unlike with cell culture, however, the infectivity of viruses detected by molecular methods is often unknown.
While PCR detection methods offer a high level of sensitivity, this property may also increase the risk for false-positive results due to low levels of contamination. In order to reduce false positive rates, stringent quality control measures, such as using aerosol-resistant pipette tips or positive-displacement pipettors, decontamination of instruments between experiments, and physical separation of pre- and post-PCR products, are required in processing the samples to prevent cross-contamination and ensure the quality of PCR products. Likewise, false-negative results may also be a problem when inhibitors in environmental samples are present. Humic and fulvic acids, heavy metals, and phenolic compounds may inhibit the activity of polymerase enzyme (149, 167, 170). Additional manipulations, including resin treatments, polyethylene glycol precipitation-resuspension, immunomagnetic capture, and glass purification are sometimes required to remove inhibitors (16). Additives may also be used in PCR, directly, to reduce the effects of inhibitory compounds.
Among the problems with traditional PCR has been the inability to enumerate viruses. Recently, conventional PCR has been modified to improve specificity, sensitivity and efficiency but also to quantify the number of viruses detected (Table 2). Some variations of conventional PCR include nested PCR, multiplex PCR, and real-time PCR (for quantification). Seminested PCR and nested PCR assays increase the sensitivity and specificity of PCR with the use of an internal primer or primer set and are sometimes used as a confirmation step. Nested PCR assays for adenoviruses as reported by Allard et al. (3) and Van Heerden et al. (161) were shown to have increased sensitivities compared to conventional PCR, with detection limits of one adenovirus particle and 10−2 PFU, respectively. However, nested PCR has been shown to have a high probability of carryover contamination when PCR products from the first round of PCR are transferred to the reaction mixture for the nested PCR (79, 86).
(ii) Multiplex PCR.
The application of multiplex PCR (where several sets of primers against several targets are included in a single PCR) may save time and costs because several types of viruses can be detected in a single PCR assay (49). The development of a multiplex PCR assay, however, is not easy and requires careful optimization of reaction mixtures and PCR conditions (49, 57, 153). The original effort of Fout et al. (49) to develop a multiplex PCR assay that would detect five enteric viruses (i.e., enteroviruses, reovirus, rotavirus, hepatitis A virus, and Norwalk virus) was not successful, and instead two multiplex PCR assays had to be developed to detect the five targeted virus groups. Fout et al. (49) also noted that even with optimal conditions, enteroviruses and Norwalk virus were not amplified as efficiently as other virus groups. A multiplex PCR developed by Green and Lewis (57) to detect enterovirus, rotavirus, and hepatitis A worked well in analyzing seeded samples, but a large number of nonspecific PCR products were formed when environmental samples were analyzed; secondary PCR was required to confirm positive samples.
(iii) Real-time PCR.
Real-time PCR provides quantitative data for the presence of enteric viral genomes in environmental samples with the use of a fluorescent dye, such as SYBR Green (Molecular Probes, Eugene, OR), that will bind to amplified cDNA or with fluorochrome-tagged probes that fluoresce when bound to complementary sequences in the amplified region. The procedure is less time-consuming because a confirmation step such as agarose gel electrophoresis and additional hybridization are generally not required. The entire analysis can be done in a closed system, which may reduce the potential for contamination. Real-time PCR assays have shown detection sensitivities comparable to or greater than those of conventional PCR in several studies (7, 40). Beuret (7) reported that real-time reverse transcription-PCR (RT-PCR) detection of norovirus and enteroviruses in seeded samples showed an increased sensitivity of 1 and 2 orders of magnitude, respectively, compared to the conventional RT-PCR protocol. The real-time RT-PCR assay developed by Donaldson et al. (40) for detection of enteroviruses showed a detection limit of 9.3 viral particles ml−1 for seawater and 155 viral particles g−1 for sponge. However, the cost of a real-time PCR instrument is still substantially more than that of a conventional PCR instrument, and in some cases, real-time PCR has been shown to be less sensitive than conventional RT-PCR and nested PCR (120). Noble et al. (120) reported that human adenovirus 40 was detected by real-time PCR in only two of the four samples positive for adenoviruses by conventional nested PCR; none of the samples that were positive for enteroviruses by conventional RT-PCR was detected by real-time RT-PCR.
(iv) Integrated cell culture PCR.
While PCR-based methods offer many advantages in sensitivity, specificity, and efficiency over cell culture, they still cannot provide information on the infectivity of viruses detected with the reliability of cell culture. To address this, several studies have combined cell culture and PCR and have reported that this method improves the specific detection of infectious enteric virus from environmental samples. The hypothesis behind this method is that after inoculation of a cell line, only infectious viruses, if present, will propagate; the cells can then be extracted and tested for viruses by PCR before CPE is noted. This is also appropriate for viruses that do not produce CPE but still infect and grow in a cell line. Chapron et al. (26) noted that an integrated cell culture-RT-nested PCR (ICC-RT-PCR) procedure provided increased sensitivity compared to the conventional cell culture method (CPE only). By ICC-RT-PCR, 68.9% of samples were positive for an infectious virus, compared to 17.2% determined by traditional cell culture (26). Detection of infectious adenoviruses also showed significant improvement with this method; the percentage of positive environmental samples (including sewage, sludge, river, and shellfish samples) increased from 28.6% by conventional cell culture to 50% by ICC-PCR (59). ICC-(RT)-PCR also increases the frequency at which viruses are detected from environmental samples that normally have very low levels of infectious enteric viruses, including potable water (93). In Korea, 65.2% (15 of 23) of tap water samples were positive for infectious enteric viruses by integrated cell culture and (RT)-multiplex nested PCR, compared to a detection rate of below 10% from similar studies in the 1980s and early 1990s (55, 93). With this method, infectious adenoviruses which do not usually produce CPE were detected in 39.1% (9 of 23) of tap water samples; enteroviruses and adenoviruses were detected simultaneously in 21.7% (5) samples (93). ICC-PCR can also produce results in a shorter period than traditional cell culture (i.e., ≤3 days) (59). However, recent work by Ko et al. (89) suggests that carryover of nucleic acids of inactivated viruses inoculated onto cultured cells might result in a false-positive result from samples containing no infectious viruses. To address this, Ko et al. (89) developed an ICC-RT-PCR-based assay to detect viral mRNA rather than DNA in the case of adenoviruses; mRNA is only transcribed by infectious adenoviruses during replication. After exposure to different doses of UV radiation, adenovirus DNA was detected consistently in inoculated cell culture lysate by PCR even when adenovirus mRNA could no longer be detected (89).
Assessing viral infectivity.
While information derived from direct molecular detection assays for viruses can indicate contamination in an area, data on viral infectivity is needed to make determinations about health risks. Therefore, understanding the relationship between viral persistence (as detected by molecular methods) and infectivity (as detected as CPE in cell culture) is critical to ultimately using molecularly derived data for health protection. Several recent studies have provided evidence that degradation of viral nucleic acids (particularly RNA) is well correlated with loss of infectivity (based on loss of CPE in cell culture), even when the viral genome is more persistent than infectious viruses (41, 144, 154, 165). However, because RNA degrades relatively rapidly in the environment (in a few minutes) compared to DNA, viruses that are no longer infectious because of damage to the capsid also experience damage to the RNA on the same time scale, thus becoming undetectable by both cell culture and RT-PCR (98). Wetz et al. (165) showed that the detection rate for polioviruses varied little between cell culture and RT-PCR in unfiltered (natural) seawater. Tsai et al. (154) showed that naked enteroviral RNA could not be detected by RT-PCR and dot blot hybridization after 2 days of incubation at both 4°C and 23°C in unfiltered seawater. Skraber et al. (144) observed that although the poliovirus genome has a higher persistence than an infectious poliovirus, the loss in detection of the viral genome is directly correlated to the disappearance of infectious virus, suggesting that viral nucleic acids may indeed serve as an efficient indicator for infectious viruses in aquatic environments.
OCCURRENCE OF ENTEROVIRUSES AND ADENOVIRUSES IN AQUATIC ENVIRONMENTS
Infectious enteric viruses, especially enteroviruses and adenoviruses, have been isolated from various types of water, including groundwater, treated sewage, marine water, rivers, streams, and drinking water, under various environmental conditions (79, 91, 93, 95, 99, 139). Factors controlling the occurrence, survival, and distribution of enteric viruses in the environment include host excretion, water temperature, susceptibility to sunlight inactivation, virus attachment to suspended solids, and other environmental variables such as nutrient concentrations, presence of predators or grazers, rainfall, and stream flow (100, 151).
Host Excretion
Many researchers have observed peaks in both human enteric virus infections and excretion in summer and early fall in temperate climates, which also coincide with increased water recreational activities and human-water contact (90, 116, 138, 161). In tropical climates, human enteric viruses, especially enteroviruses, are isolated throughout the year and in some cases are more prevalent during rainy seasons (153). Not surprisingly, there appears to be a connection between environmental and clinical isolates in a given year within specific geographic areas. A study that compared clinical and sewage isolates of enteroviruses from Milwaukee, Wis., collected between 1994 and 2002 found that the predominant clinical serotype was most often also the predominant sewage serotype for that year (138). For example, in 1998, echovirus 30 accounted for 50.0% of sewage isolates and 46.1% of clinical cases, and in 1990, 79.7% of sewage isolates and 60.3% of clinical cases were echovirus 11 (138). However, clinical cases did not precede environmental detection in all cases; often viruses found in sewage in the spring were highly predictive of clinical strains that predominated during the following summer (138). One of the explanations for this phenomenon is that a “new” serotype or strain may cause asymptomatic or mild infections that do not require clinical attention in the earlier season (i.e., spring), but as that serotype becomes predominant during peak infection season (i.e., summer), clinical cases of that serotype are identified and diagnosed (138).
In a study examining the occurrence of animals viruses from farm livestock waste and surrounding soil and water, both bovine and porcine enteroviruses were found to be excreted at higher rates by young animals, e.g., piglets and calves (36).
Water Temperature
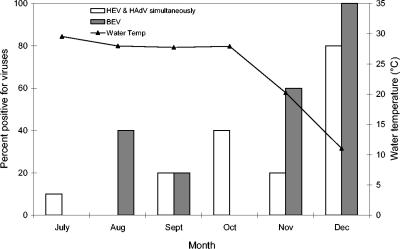
While peaks in clinical cases and sewage isolates for enteric viruses are often observed in late summer and early fall, Green and Lewis (57) isolated a higher number of infectious enteroviruses from raw sewage and final effluent during the winter months. In several studies, enteric viruses have been reported to survive longer and occur more frequently at lower temperatures in natural environments (i.e., seawater, river, and groundwater) (57, 99, 100, 165). High temperatures can damage the virus capsid or nucleic acids, which might prevent adsorption of the virus to its host and may inactivate enzymes required for replication (8). In a year-long survey of the occurrence of adenoviruses in drinking water in South Africa, adenovirus detection peaked in July (winter in South Africa), when up to 30% and 60% of treated and raw water samples were positive for adenoviruses, respectively (161). Lipp et al. (100) detected enteroviruses from an estuary in southwest Florida only when the water temperature was below 23°C. In an in vitro study, enhanced poliovirus survival and detection were observed at 22°C compared to 30°C in seawater (165). In artificial seawater, viruses were detected by RT-PCR for at least 60 days at 22°C but for only 30 days at 30°C (165). Similarly, Gantzer et al. (52) showed that in seawater, it took 671 days to inactivate 90% of poliovirus and hepatitis A virus at 4°C and only 25 days at 25°C. In a study evaluating both human and bovine enteric viruses by PCR in a mixed-use estuary, Fong et al. (48) found that all virus types were correlated with cool water temperatures (Fig. 1).
FIG. 1.
Percentage of samples positive for human enteroviruses and human adenoviruses (detected simultaneously), as well as bovine enteroviruses, by month versus the mean monthly water temperature (°C) along the lower Altamaha River, Georgia, between July and December 2002 (n = 5). (Adapted from reference 48 with permission.)
Sunlight Inactivation
After temperature, UV radiation is the most common factor leading to virus inactivation. Sinton et al. (142) found that bacteriophage inactivation rates in sunlight are 10 times higher than their inactivation rates in the dark. This is consistent with the findings by Johnson et al. (81), who observed 90% and 99.9% inactivation of polioviruses in marine water after 24 h of incubation in the dark and exposure to sunlight, respectively. Despite their susceptibility to UV radiation, viruses are more resilient than many other pathogens and indicator bacteria to that effect because of their low molecular weight (i.e., lower target density) (51, 81). Furthermore, viruses with double-stranded DNA or RNA (e.g., double-stranded DNA adenoviruses) are extremely stable when exposed to UV because their undamaged DNA or RNA strand may serve as a template for repair by host enzymes (56, 152).
Adsorption to Suspended Solids and Sediment
Association of viruses with solids is believed to increase their persistence in natural environments by offering protection from enzymes, other degrading factors, and UV inactivation (52, 57, 94, 111).
In a comparative virus-soil sorption study, poliovirus 1 (i.e., enterovirus) was shown to be the most sorptive to all soil types tested, followed by Norwalk virus (norovirus) and the F+ RNA coliphage MS2 (111). Green and Lewis (57) reported that enteroviruses and hepatitis A virus could be detected throughout the year in sediment in the immediate vicinity of a sewage outfall even though enterovirus concentrations peaked in the wastewater during winter months. Likewise, during wet weather when contaminants were loaded into an estuary, Ferguson et al. (47) could isolate enteric viruses in both water and sediment samples, but during the dry season, viruses persisted only in the sediment. Furthermore, during the same dry season, Ferguson et al. (47) isolated enteroviruses from sediment samples collected as far as 23 km from the sewage overflow point. Because of frequent detections of pathogenic microorganisms in solids, Brookes et al. (21) suggested taking into account the survival and accumulation of microbes in sediments as well as the likelihood of their resuspension and redistribution by natural and anthropogenic disturbance when assessing water quality issues related to public health risk.
Aquatic Environmental Factors
In certain cases, aquatic environmental factors (i.e., nutrient concentrations, predators, and dissolved oxygen) might have a significant effect, in addition to water temperature and other factors, on virus survival. In a study comparing in vitro and in situ survival in seawater, viruses survived significantly longer at lower temperatures in laboratory conditions, but despite a similar temperature range in field studies, there was no significant difference in poliovirus survival between seasons in natural water (163). Likewise, Wetz et al. (165) showed that virus survival at both 22°C and 30°C in unfiltered natural seawater was much shorter than survival in filtered seawater or artificial seawater at either temperature. Finally, sampling season (summer versus winter) was shown to have a greater effect than incubation temperature on survival of both infectious poliovirus and poliovirus RNA (144). Survival of infectious poliovirus 1 seeded in river water collected during winter was greater than that of fecal coliform bacteria regardless of incubation temperatures, whereas the opposite was observed in water collected during the summer (144). Therefore, in addition to temperature, UV effects, and adsorption to solids, viral persistence in natural waters may be strongly related to predation by flagellates, extracellular proteases, nucleases, and other enzymes (117, 119, 150). Salinity has not been shown to have a direct effect on virus survival, although accelerated inactivation of fecal indicators in higher salinities has been reported (15, 52, 94, 147).
Overall, factors that influence the occurrence and survival of enteric viruses in waters, such as water temperature, suspended solids, turbulence, sunlight intensity, host excretion, nutrient content of water, and predation, have been extensively studied, and these parameters should be included when attempting to predict the presence of viral pathogens in the environment.
ADENOVIRUSES AND ENTEROVIRUSES AS WATER QUALITY INDICATORS AND MICROBIAL SOURCE TRACKING TOOLS
The enteroviruses were the first enteric virus group studied in fecally contaminated waters, in the 1940s, and they continued to be among the most well studied (62). Enteroviruses are included in the European Union regulations governing water quality as a parameter for evaluating viral pollution of a water body because they can easily be isolated and quantified as PFU in cell culture, and vaccine-related poliovirus is prevalent in contaminated waters (103, 129). Studies conducted in Europe and other parts of the world recently have suggested including adenoviruses as an index of pollution of human origin in waters because they have been shown to be more persistent and present in greater numbers than enteroviruses in sewage and contaminated aquatic environments (77, 79, 91, 127, 152). Muniain-Mujika et al. (114) studied the prevalence of viral pathogens in shellfish from three sites in Spain with different levels of fecal contamination; 47%, 19%, and 24% of their samples were positive for HAdV, human enteroviruses (HEV), and hepatitis A virus, respectively. They proposed using HAdV as a molecular index for viral contamination in shellfish because HAdV was detected in all samples that were positive for HEV and hepatitis A virus (114). In addition, Pina et al. (127) reported that the presence of human adenoviruses in sewage samples is highly correlated to the presence of hepatitis A viruses and human-specific bacteriophages, such as those infecting Bacteroides fragilis HSP40.
In many watersheds experiencing water quality deterioration, pollutants originating from nonpoint sources such as urban runoff, forests, wildlife, and agricultural runoff (including contamination by manure application and unrestricted access of livestock and wildlife to rivers and streams) are difficult to identify for proper management and remediation planning. Enteroviruses and adenoviruses, just as other types of viruses, have a narrow host range. For example, human enteroviruses infect only humans and cannot cause infection in cattle or fowl and vice versa. Because human and animal enteric viruses are excreted in large number in infected hosts, a fecal pollution tracking method based on the host specificity of viruses has been hypothesized as a useful indicator system for the presence of contaminants originating from specific sources (e.g., human sewage, cattle, or swine farms) (80, 97, 105, 120). In 1995, Metcalf et al. (112) were among the first researchers to hypothesize that molecular detection techniques based on host specificity of viral pathogens in environmental samples would allow the determination of the sources of contaminants and improve surveillance for public health; however, this system has not been widely used, and most methods for tracking pollution sources rely on microbial indicators (i.e., Escherichia coli or enterococci).
Microbial Source Tracking Methods
Microbial source tracking methods currently being used can be categorized into four basic groups (Table 3): (i) genotypic library-based methods, i.e., ribotyping, repetitive extragenic palindromic-PCR, and pulsed-field gel electrophoresis, which differentiate sources of pollutants by matching the genetic patterns of isolated bacteria to a library with bacterial isolates from known sources; (ii) phenotypic library-based methods, i.e., antibiotic resistance analysis and carbon source utilization, which differentiate sources of pollutants by matching the growth pattern of a bacterium, such as E. coli or enterococci, on a suite of antibiotics or carbon sources with those of isolates from a library of sources; (iii) the use of library-independent bacterial host-specific markers, i.e., identifying host-specific genetic markers of bacteria, such as host-specific Bacteroides-Prevotella 16S rRNA gene markers, with terminal restriction fragment length polymorphism or length heterogeneity PCR; and (iv) direct measurement of viral pathogens and bacteriophages with different host groups, e.g., HEV, HAdV, BEV, and the F+ RNA coliphages (63, 82, 122, 123, 137, 141).
TABLE 3.
Comparison of methods for microbial source tracking in aquatic environments
| Method | Advantages | Disadvantages | References |
|---|---|---|---|
| Genotypic, library based | |||
| Ribotyping | Quantitative; highly sensitive and reproducible; classifies isolates from multiple sources | Large isolate database required, geographically specific; labor-intensive and time-consuming; high percentage of inconclusive results | 71, 124 |
| Pulsed-field gel electrophoresis | Sensitive, discriminative, and reproducible; quantitative | Labor-intensive and time-consuming; may be too sensitive for discriminating multiple sources | 82, 122 |
| Phenotypic, library based | |||
| Antibiotic resistance analysis | Rapid; classifies isolates from multiple animal sources | Large isolate database required, geographically specific; isolates have to show antibiotic resistance to be typed; antibiotic resistance traits are not stable; no consensus on combination and dose of antibiotics used | 67, 123 |
| Library and culture independent (bacterial host-specific markers) | |||
| Terminal restriction fragment length polymorphism, length heterogeneity PCR | Rapid and easy to perform; no database or cultivation required; high accuracy in differentiating human and nonhuman sources | Survival and distribution of molecular markers in aquatic environments are not well studied; expensive equipment; currently applicable to only a limited number of host groups | 6, 10 |
| Direct measurement of host-specific viruses | |||
| PCR for viral pathogens | Library independent and directly relates to health risk; rapid and straightforward; detects conserved regions of a viral genome, may not have geographical limits | Nonquantitative in conventional PCR; requires more sensitive detection methods; limited knowledge of prevalence of animal-specific viruses in aquatic environments; serotyping is expensive and time-consuming | 40, 100, 120 |
| PCR and phage typing (e.g., F+ RNA coliphage) | Subgroups are well-correlated to sources; straightforward | Serotyping is expensive and time-consuming; low survival in marine and tropical waters; may proliferate in sewage; exceptions in association of coliphage subgroup and host group have been noted | 76, 134 |
Library-based methods have the advantages of being quantitative, highly sensitive, and reproducible and may be used to classify isolates from multiple sources (67, 124). The major drawback of library-based methods is the requirement of a large isolate database, which can be extremely labor-intensive and time-consuming and may be geographically specific (63, 71, 137, 141). In addition, these methods have shown high false-positive rates when tested with spiked samples, and in the case of antibiotic resistance analysis, bacterial isolates have to show antibiotic resistance to be typed (Table 3) (63, 137, 141).
In contrast to library-based methods, the use of bacterial host-specific markers does not require a reference library or a cultivation step, and testing is rapid and easy to perform with PCR (although nonquantitative) and has been reported to have very low false-positive and false-negative rates when differentiating human and nonhuman sources of contaminants from spiked samples (63, 137, 141). However, the survival and distribution of bacterial host-specific markers in aquatic systems have not been extensively studied, and this method is currently applicable to only a limited number of host groups (137).
The F+ RNA coliphages are the most extensively studied and well-characterized phages for use in source tracking (137, 143). Detection, enumeration, and subtyping of the F+ RNA coliphages are easy to perform and straightforward; however, their low occurrence in human feces and other aquatic environments despite their frequent detection in wastewater indicates that the phages might be able to proliferate in sewage (72). In addition, their survival rate in marine and tropical waters is varied, and exceptions to the associations between coliphage subgroup and particular host group have been reported (e.g., subgroup II and III coliphages [human specific] have been isolated from pigs) (29, 134).
Detection of host-specific viral pathogens with molecular assays, such as PCR and its variants, is less laborious and time-consuming than library-based microbial source tracking techniques because a reference database is not required and results generally can be obtained in a relatively short period (hours to a day) (120). Through different primer sets that target enteric viruses within a specific host group, PCR detection can be used to directly identify the major source of contamination in environmental samples. In addition, primers are usually developed from conserved regions of the viral genome that show high stability and do not change appreciably with time or environmental conditions (120). Although enteric viruses occur in low numbers in some aquatic environments, recent modifications and improvements in viral concentration, extraction, and detection techniques have allowed detection from waters that generally have very low number of viruses, such as potable water (93). Furthermore, the occurrence, survival, and transport of theses viruses under different environmental conditions have been well characterized (16, 60, 127).
Recent studies reveal that tracking of HEV and HAdV shows promising and reliable results in indicating the presence of human sewage and discriminating between human and nonhuman pollution sources in environmental waters (120, 127). In one study, PCR detection of human enteric viruses from mixed fecal samples of human and nonhuman origins showed that HAdV is more specific than HEV in that it picked up most samples with human feces but none of the solely non-human-origin samples (127).
Animal-specific enteroviruses and adenoviruses have also shown great potential as indicators for fecal contamination of animal origin (36, 48, 80, 97, 105). Derbyshire and Brown isolated porcine enteroviruses from 2 of 26 surface runoff samples from a pig slurry application site and from 1 of 33 surface water samples collected on pig farms (36). They also found that a porcine enterovirus could remain viable in surface soil for at least 8 days during summer. In addition, bovine enteroviruses were isolated from two out of seven cattle feedlot runoff samples in the same study (36). In a study that examined the prevalence of BEV in a closed herd of cattle, other animals on the premises, and environmental samples in the area, Ley et al. (97) found BEV in feces of 76% of cattle, 38% of white-tailed deer, and one of three geese. BEV were also isolated from streams and rivers that received runoff from the farm and in oysters collected in the rivers. Ley et al. (97) concluded that with additional analyses of BEV in animals from other areas, BEV might serve as marker for bovine fecal contamination. In addition, BEV were detected by PCR in 11 out of 30 water samples from an estuary influenced by agricultural activities upstream (48). Jiménez-Clavero et al. (80) detected PTV RNA in water and fecal samples from five pig farms located in different parts of Spain but not in fecal samples from other animals. In addition, Jiménez-Clavero et al. (80) also suggested that PCR-based identification of virus species was a more reliable and sensitive marker than conventional chemical water quality indicators such as nitrate and nitrite readings (80). PTV RNA was detected as far as 3 km downstream from the discharge, where the impacts on nitrates and nitrites were no longer observed (80). Maluquer de Motes et al. (105) reported a high prevalence of bovine BAdV and PAdV in animal feces, but all human sewage-contaminated samples tested negative. These studies suggest that while animal-specific viruses have been shown to have a high prevalence in aquatic environments directly influenced by the particular animal source, the analysis of a large number of samples from different geographical areas is necessary to validate the application of animal-specific viruses for identifying the source of fecal contamination (80, 97, 105).
CONCLUSIONS
Enteric viruses are important pathogens that are frequently isolated from waters directly or indirectly influenced by fecal contamination and have been associated with many waterborne outbreaks (28, 60, 79, 100, 127). Given that traditional bacterial indicators have been shown to be inappropriate for viruses and other pathogens and that direct detection methods now exist for easy analysis of viral pathogens, direct surveillance for pathogens may be warranted to better protect public health (100, 109). Factors that have been shown to affect the occurrence and survival of viruses can be incorporated into models that predict the levels of viral contamination in specific types of water and can contribute to efforts to control contamination. In addition, the stringent host specificity of enteric viruses suggests that they can be good library-independent indicators for identifying sources of water pollution. Molecular detection (e.g., by PCR and hybridization) of viral pathogens is rapid and highly specific and sensitive, and with the use of quantitative (real-time) PCR, concentrations of viral pathogens in environmental samples can be determined. PCR assays can be developed based on genotypic differences between viruses with different host groups and can be used to better characterize sources of contamination in aquatic environment so that an appropriate and cost-effective water quality remediation plan can be developed. However, additional research to study the prevalence and distribution of animal-specific viruses in environmental waters is required to validate the use of these viruses for source tracking purposes.
REFERENCES
- 1.Abad, F. X., R. M. Pinto, R. Gajardo, and A. Bosch. 1997. Viruses in mussels: public health implications and depuration. J. Food Prot. 60:677-681. [DOI] [PubMed] [Google Scholar]
- 2.Albert, M., and L. Schwartzbrod. 1991. Recovery of enterovirus from primary sludge using three elution concentration procedures. Water Sci. Technol. 24:225-228. [Google Scholar]
- 3.Allard, A., B. Albinsson, and G. Wadell. 1992. Detection of adenoviruses in stools from healthy persons and patients with diarrhea by two-step polymerase chain reaction. J. Med. Virol. 37:149-157. [DOI] [PubMed] [Google Scholar]
- 4.Arnal, C., V. Ferre-Aubineau, B. Besse, B. Mignotte, L. Schwartzbrod, and S. Billaudel. 1999. Comparison of seven RNA extraction methods on stool and shellfish samples prior to hepatitis A virus amplification. J. Virol. Methods 77:17-26. [DOI] [PubMed] [Google Scholar]
- 5.Benko, M., P. Elo, K. Ursu, W. Ahne, S. E. LaPatra, D. Thomson, and B. Harrach. 2002. First molecular evidence for the existence of distinct fish and snake adenoviruses. J. Virol. 76:10056-10059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bernhard, A. E., T. Goyard, M. T. Simonich, and K. G. Field. 2003. Application of a rapid method for identifying fecal pollution sources in a multi-use estuary. Water Res. 37:909-913. [DOI] [PubMed] [Google Scholar]
- 7.Beuret, C. 2004. Simultaneous detection of enteric viruses by multiplex real-time RT-PCR. J. Virol. Methods 115:1-8. [DOI] [PubMed] [Google Scholar]
- 8.Bitton, G. 1980. Introduction to environmental virology. Wiley-Interscience, New York, N.Y.
- 9.Bitton, G., and C. P. Gerba. 1984. Groundwater pollution microbiology. Wiley & Sons, New York, N.Y.
- 10.Boehm, A. B., J. A. Fuhrman, R. D. Mrse, S. B. Grant. 2003. Tiered approach for identification of a human fecal pollution source at a recreational beach: case study at Avalon Bay, Catalina Island, California. Environ. Sci. Technol. 37:673-680. [DOI] [PubMed] [Google Scholar]
- 11.Bofill-Mas, S., M. Formiga-Cruz, P. Clemente-Casares, F. Calafell, and R. Girones. 2001. Potential transmission of human polyomaviruses through the gastrointestinal tract after exposure to virions or viral DNA. J. Virol. 75:10290-10299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bofill-Mas, S., S. Pina, and R. Girones. 2000. Documenting the epidemiologic patterns of polyomaviruses in human populations by studying their presence in urban sewage. Appl. Environ. Microbiol. 66:238-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boom, R., C. J. A. Sol, M. M. M. Salimans, C. L. Jansen, P. M. E. Wertheim-Dillen, and J. Van Der Noordaa. 1990. Rapid and simple method for purification of nucleic acids. J. Clin. Microbiol. 28:495-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Borchardt, M. A., P. D. Bertz, S. K. Spencer, and D. A. Battigelli. 2003. Incidence of enteric viruses in groundwater from household wells in Wisconsin. Appl. Environ. Microbiol. 69:1172-1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bordalo, A. A., R. Onrassami, C. Dechsakulwatana. 2002. Survival of faecal indicator bacteria in tropical estuarine waters (Bangpakong River, Thailand). J. Appl. Microbiol. 93:864-871. [DOI] [PubMed] [Google Scholar]
- 16.Bosch, A. 1998. Human enteric viruses in the water environment. Int. Microbiol. 1:191-196. [PubMed] [Google Scholar]
- 17.Bosch, A., F. Lucena, J. M. Diez, R. Gajardo, M. Blasi, J. Jofre. 1991. Waterborne viruses associated with hepatities outbreak. J. Am. Water Works Assoc. 83:80-83. [Google Scholar]
- 18.Bosch, A., G. Sanchez, F. Le Guyader, H. Vanaclocha, L. Haugarreau, and R. M. Pinto. 2001. Human enteric viruses in Coquina clams associated with a large hepatitis A outbreak. Water Sci. Technol. 43:61-65. [PubMed] [Google Scholar]
- 19.Bosch, A., R. M. Pinto, C. Villena, and F. X. Abad. 1997. Persistence of human astrovirus in fresh and marine water. Water Sci. Technol. 35:243-247. [Google Scholar]
- 20.Brauniger, S., J. Peters, U. Borchers, M. Kao, and U. Borchers. 2000. Further studies on thermal resistance of bovine parvovirus against moist and dry heat. Int. J. Hyg. Environ. Health 203:71-75. [DOI] [PubMed] [Google Scholar]
- 21.Brookes, J. D., J. Antenucci, M. Hipsey, M. D. Burch, N. J. Ashbolt, and C. Ferguson. 2004. Fate and transport of pathogens in lakes and reservoirs. Environ. Int. 30:741-759. [DOI] [PubMed] [Google Scholar]
- 22.Büchen-Osmond, C. 28 June 2002, posting date. 52.0.1. Enterovirus. [Online.] http://www.ncbi.nlm.nih.gov/ICTVdb/ICTVdB/52010000.htm.
- 23.Callens, M., and K. De Clercq. 1999. Highly sensitive detection of swine vesicular disease virus based on a single tube RT-PCR system and DIG-ELISA detection. J. Virol. Methods 77:87-99. [DOI] [PubMed] [Google Scholar]
- 24.Casas, I., L. Powell, P. E. Klapper, and G. M. Cleator. 1995. New method for the extraction of viral RNA and DNA from cerebrospinal fluid for use in the polymerase chain reaction assay. J. Virol. Methods 53:25-36. [DOI] [PubMed] [Google Scholar]
- 25.Cecuk, D., V. Kruzic, B. Turkovic, and M. Gree. 1993. Human viruses in the coastal environment of a Croatian harbor. Rev. Epidemiol. Santé 41:487-493. [PubMed] [Google Scholar]
- 26.Chapron, C. D., N. A. Ballester, J. H. Fontaine, C. N. Frades, and A. B. Margolin. 2000. Detection of astroviruses, enteroviruses, and adenovirus types 40 and 41 in surface waters collected and evaluated by the information collection rule and an integrated cell culture-nested PCR procedure. Appl. Environ. Microbiol. 66:2520-2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chung, H., L. Jaykus, and M. Sobsey. 1996. Detection of human enteric viruses in oysters by in vivo and in vitro amplification of nucleic acids. Appl. Environ. Microbiol. 62:3772-3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cliver, D. O. 1984. Significance of water and the environment in the transmission of viral disease, p. 30-42. In J. L. Melnick (ed.), Enteric viruses in water, vol. 15. Karger, Basel, Switzerland. [Google Scholar]
- 29.Cole, D., S. C. Long, and M. D. Sobsey. 2003. Evaluation of F+ RNA and DNA coliphages as source-specific indicators of fecal contamination in surface waters. Appl. Environ. Microbiol. 69:6507-6514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cook, J. K. A. 1974. Pathogenicity of avian adenoviruses for day-old chicks. J. Comp. Pathol. 84:505. [DOI] [PubMed] [Google Scholar]
- 31.Crabtree, K. D., C. P. Gerba, J. B. Rose, and C. N. Haas. 1997. Waterborne adenovirus: a risk assessment. Water Sci. Technol. 35:1-6. [Google Scholar]
- 32.Craun, G. F. 1991. Causes of waterborne outbreaks in the United States. Water Sci. Technol. 24:17-20. [Google Scholar]
- 33.Cruz, J. R., P. Cáceres, F. Cano, J. Flores, A. Bartlett, and B. Torún. 1990. Adenovirus types 40 and 41 and rotaviruses associated with diarrhea in children from Guatemala. J. Clin. Microbiol. 28:1780-1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dekker, A., P. Moonen, E. A. de Boer-Luijtze, and C. Terpstra. 1995. Pathogenesis of swine vesicular disease after exposure of pigs to an infected environment. Vet. Microbiol. 45:243-250. [DOI] [PubMed] [Google Scholar]
- 35.De Leon, R., C. Shieh, R. S. Baric, and M. D. Sobsey. 1990. Presented at the Proceedings of the 1990 Water Quality Technology Conference, Denver, Colo.
- 36.Derbyshire, J. B., and E. G. Brown. 1978. Isolation of animal viruses from farm livestock waste, soil and water. J. Hyg. 81:295-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dewailly, E., C. Poirier, F. M. Meyer. 1986. Health hazards associated with windsurfing on polluted waters. Am. J. Public Health 76:690-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Doherty, M., D. Todd, N. McFerran, and E. M. Hoey. 1999. Sequence analysis of a porcine enterovirus serotype 1 isolate: relationships with other picornaviruses. J. Gen. Virol. 80:1929-1941. [DOI] [PubMed] [Google Scholar]
- 39.Dombek, P. E., L. K. Johnson, S. T. Zimmerley, and M. J. Sadowsky. 2000. Use of repetitive DNA sequences and the PCR to differentiate Escherichia coli isolates from human and animal sources. Appl. Environ. Microbiol. 66:2572-2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Donaldson, K. A., D. W. Griffin, and J. H. Paul. 2002. Detection, quantitation and identification of enteroviruses from surface waters and sponge tissue from the Florida Keys using real-time RT-PCR. Water Res. 36:2505-2514. [DOI] [PubMed] [Google Scholar]
- 41.Dubois, E., F. LeGuyader, L. Haugarreau, H. Kopecka, M. Cormier, and M. Pommepuy. 1997. Molecular epidemiological survey of rotaviruses in sewage by reverse transcriptase seminested PCR and restriction fragment length polymorphism assay. Appl. Environ. Microbiol. 63:1794-1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dunne, H. W., J. L. Gobble, J. F. Hokanson, D. C. Kradel, and G. R. Bubash. 1965. Porcine reproductive failure associated with a newly identified “SMEDI” group of picorna viruses. Am. J. Vet. Res. 26:1284-1297. [PubMed] [Google Scholar]
- 43.Engelbrecht, R. S., M. J. Weber, B. L. Salter, and C. A. Schmidt. 1980. Comparative inactivation of viruses by chlorine. Appl. Environ. Microbiol. 40:249-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Enriquez, C. E., and C. P. Gerba. 1995. Concentration of enteric adenovirus 40 from tap, sea and waste water. Water Res. 29:2554-2560. [Google Scholar]
- 45.Farthing, M. J. G. 1989. Viruses and the gut. Smith Kline & French, Walwyn Garden City, Hertfordshire, United Kingdom.
- 46.Fenner, F. J., E. P. J. Gibbs, F. A. Murphy, R. Rott, M. J. Studdert, and D. O. White. 1993. Veterinary virology, 2nd ed., p. 416-419. Academic Press, San Diego, Calif.
- 47.Ferguson, C. M., B. G. Coote, N. J. Ashbolt, and I. M. Stevenson. 1996. Relationships between indicators, pathogens and water quality in an estuarine system. Water Res. 30:2045-2054. [Google Scholar]
- 48.Fong, T.-T., D. W. Griffin, and E. K. Lipp. 2005. Molecular assays for targeting human and bovine enteric viruses in coastal waters and application for library-independent source tracking. Appl. Environ. Microbiol. 71:2070-2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fout, G. S., B. C. Martinson, M. W. N. Moyer, and D. R. Dahling. 2003. A multiplex reverse transcription-PCR method for detection of human enteric viruses in groundwater. Appl. Environ. Microbiol. 69:3158-3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fry, E. E., N. J. Knowles, J. W. I. Newman, G. Wilsden, Z. Rao, A. M. Q. King, and D. I. Stuart. 2003. Crystal structure of swine vesicular disease virus and implications for host adaptation. J. Virol. 77:5475-5486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fujioka, R. S., and B. S. Yoneyama. 2002. Sunlight inactivation of human enteric viruses and fecal bacteria. Water Sci. Technol. 46:291-295. [PubMed] [Google Scholar]
- 52.Gantzer, C., E. Dubois, J.-M. Crance, S. Billaudel, H. Kopecka, L. Schwartzbrod, M. Pommepuy, and F. Le Guyader. 1998. Influence of environmental factors on the survival of enteric viruses in seawater. Oceanol. Acta 21:983-992. [Google Scholar]
- 53.Gantzer, C., S. Senouci, A. Maul, Y. Levi, and L. Schwartzbrod. 1999. Enterovirus detection from wastewater by RT-PCR and cell culture. Water Sci. Technol. 40:105-109. [Google Scholar]
- 54.Geldenhuys, J. C., and P. D. Pretorius. 1989. The occurrence of enteric viruses in polluted water, correlation to indicator organisms and factors influencing their numbers. Water Sci. Technol. 21:105-109. [Google Scholar]
- 55.Gerba, C. P., and J. B. Rose. 1990. Viruses in source and drinking water, p. 380-396. In G. A. McFeters (ed.), Drinking water microbiology. Springer, New York, N.Y.
- 56.Gerba, C. P., D. M. Gramos, and N. Nwachuku. 2002. Comparative inactivation of enteroviruses and adenovirus 2 by UV light. Appl. Environ. Microbiol. 68:5167-5169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Green, D. H., and G. D. Lewis. 1999. Comparative detection of enteric viruses in wastewaters, sediments and oysters by reverse transcription PCR and cell culture. Water Res. 33:1195-1200. [Google Scholar]
- 58.Green, D. H., and G. D. Lewis. 1995. Enzymatic amplification of enteric viruses from wastewaters. Water Sci. Technol. 31:329-336. [Google Scholar]
- 59.Greening, G. E., J. Hewitt, and G. D. Lewis. 2002. Evaluation of integrated cell culture-PCR (C-PCR) for virological analysis of environmental samples. J. Appl. Microbiol. 93:745-750. [DOI] [PubMed] [Google Scholar]
- 60.Griffin, D. W., K. A. Donaldson, J. H. Paul, and J. B. Rose. 2003. Pathogenic human viruses in coastal waters. Clin. Microbiol. Rev. 16:129-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Griffin, D. W., C. J. Gibson, E. K. Lipp, K. Riley, J. H. Paul, and J. B. Rose. 1999. Detection of viral pathogens by reverse transcriptase PCR and of microbial indicators by standard methods in the canals of the Florida Keys. Appl. Environ. Microbiol. 65:4118-4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Griffin, D. W., E. K. Lipp, M. R. McLaughlin, and J. B. Rose. 2001. Marine recreation and public health microbiology: quest for the ideal indicator. BioScience 51:817-825. [Google Scholar]
- 63.Griffith, J. H., S. B. Weisberg, and C. D. McGee. 2003. Evaluation of microbial source tracking methods using mixed fecal sources in aqueous test samples. J. Water Health 1:141-151. [PubMed] [Google Scholar]
- 64.Gu, Z., S. W. Belzer, C. S. Gibson, M. J. Bankowski, and R. T. Hayden. 2003. Multiplexed, real-time PCR for quantitative detection of human adenovirus. J. Clin. Microbiol. 41:4636-4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Haas, C. N., J. B. Rose, C. P. Gerba, and R. Regli. 1993. Risk assessment of viruses in drinking water. Risk Anal. 13:545-552. [DOI] [PubMed] [Google Scholar]
- 66.Hafliger, D., M. Gligen, Luthy, J., and P. Hubner. 1997. Seminested RT-PCR systems for small round structured viruses and enteric viruses detection in seafood. Int. J. Food Microbiol. 37:27-36. [DOI] [PubMed] [Google Scholar]
- 67.Hagedorn, C., S. L. Robinson, J. R. Filtz, S. M. Grubbs, T. A. Angier, and R. B. Reneau, Jr. 1999. Determining sources of fecal pollution in a rural Virginia watershed with antibiotic resistance patterns in fecal streptococci. Appl. Environ. Microbiol. 65:5522-5531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hammond, J. M., E. S. Jansen, C. J. Morrissy, A. L. M. Hodgson, and M. A. Johnson. 2003. Protection of pigs against ‘in contact’ challenge with classical swine fever following oral or subcutaneous vaccination with a recombinant porcine adenovirus. Virus Res. 97:151-157. [DOI] [PubMed] [Google Scholar]
- 69.Haramoto, E., H. Katayama, and S. Ohgaki. 2004. Detection of noroviruses in tap water in Japan by means of a new method for concentrating enteric viruses in large volumes of freshwater. Appl. Environ. Microbiol. 70:2154-2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Harding, J. D. J., J. T. Done, and G. F. Kershaw. 1965. A transmissible polio-encephalomyelitis of pigs (Talfan disease). Vet. Rec. 69:824-832. [Google Scholar]
- 71.Hartel, P. G., J. D. Summer, J. L. Hill, J. V. Collins, J. A. Entry, and W. I. Segars. 2002. Geographic variability of Escherichia coli ribotypes from animals in Idaho and Georgia. J. Environ. Qual. 31:1273-1278. [DOI] [PubMed] [Google Scholar]
- 72.Havelaar, A. H., W. M. Pot-Hogeboom, K. Furuse, R. Pot, and M. P. Hormann. 1990. F-specific RNA bacteriophages and sensitive host strains in faeces and wastewater of human and animal origin. J. Appl. Bacteriol. 69:30-37. [DOI] [PubMed] [Google Scholar]
- 73.Hess, M., H. Blocker, and P. Brandt. 1997. The complete nucleotide sequence of the egg drop syndrome virus: an intermediate between Mastadenoviruses and Aviadenoviruses. Virology 238:145-156. [DOI] [PubMed] [Google Scholar]
- 74.Hess, M., R. Raue, and H. M. Hafez. 1999. PCR for specific detection of haemorrhagic enteritis virus of turkeys, an avian adenovirus. J. Virol. Methods 81:199-203. [DOI] [PubMed] [Google Scholar]
- 75.Hilleman, M. R., and J. H. Werner. 1954. Recovery of new agents from patients with acute respiratory illness. Proc. Soc. Exp. Biol. Med. 85:183-188. [DOI] [PubMed] [Google Scholar]
- 76.Hsu, F., Y. Shieh, J. van Duin, M. Beekwilder, and M. Sobsey. 1995. Genotyping male-specific RNA coliphages by hybridization with oligonucleotide probes. Appl. Environ. Microbiol. 61:3960-3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Irving, L. G., F. A. Smith. 1981. One-year survey of enteroviruses, adenoviruses, and reoviruses isolated from effluent at an activated-sludge purification plant. Appl. Environ. Microbiol. 41:51-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ishibashi, M., and H. Yasue. 1984. Adenoviruses of animals, p. 497-562. In H. S. Ginsberg (ed.), The adenoviruses. Plenum Press, New York, N.Y.
- 79.Jiang, S., R. Noble, and W. P. Chui. 2001. Human adenoviruses and coliphages in urban runoff-impacted coastal waters of Southern California. Appl. Environ. Microbiol. 67:179-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jimenez-Clavero, M. A., C. Fernandez, J. A. Ortiz, J. Pro, G. Carbonell, J. V. Tarazona, N. Roblas, and V. Ley. 2003. Teschoviruses as indicators of porcine fecal contamination of surface water. Appl. Environ. Microbiol. 69:6311-6315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Johnson, D. C., C. E. Enriquez, I. L. Pepper, T. L. Davis, C. P. Gerba, and J. B. Rose. 1997. Survival of giardia, cryptosporidium, poliovirus and salmonella in marine waters. Water Sci. Technol. 35:261-268. [Google Scholar]
- 82.Johnson, J. M., S. D. Weagant, K. C. Jinneman, and J. L. Bryant. 1995. Use of pulsed-field gel electrophoresis for epidemiological study of Escherichia coli O157:H7 during a food-borne outbreak. Appl. Environ. Microbiol. 61:2806-2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kaku, Y., A. Sarai, and Y. Murakami. 2001. Genetic reclassification of porcine enteroviruses. J. Gen. Virol. 82:417-424. [DOI] [PubMed] [Google Scholar]
- 84.Kapikian, A. Z., and R. G. Wyatt. 1992. Textbook of pediatric infectious diseases, 3rd ed., vol. 1, p. 667-676. W. B. Saunders Co., Philadelphia, Pa.
- 85.Kasza, L. 1966. Isolation of an adenovirus from the brain of a pig. Am. J. Vet. Res. 27:751-758. [PubMed] [Google Scholar]
- 86.Katayama, H., A. Shimasaki, and S. Ohgaki. 2002. Development of a virus concentration method and its application to detection of enterovirus and Norwalk virus from coastal seawater. Appl. Environ. Microbiol. 68:1033-1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.King, A. M. Q., F. Brown, P. Christian, T. Hovi, T. Hyypiä, N. J. Knowles, S. M. Lemon, P. D. Minor, A. C. Palmenberg, T. Skern, and, G., Stanway. 2000. Picornaviridae, p. 657-678. In M. H. V. Van Regenmortel, C. M. Fauquet, D. H. L. Bishop, C. H. Calisher, E. B. Carsten, M. K. Estes, S. M. Lemon, J. Maniloff, M. A. Mayo, D. J. McGeoch, C. R. Pringle, and R. B. Wickner. (ed.), Virus taxonomy. Seventh report of the International Committee for the Taxonomy of Viruses. Academic Press, San Diego, Calif.
- 88.Knowles, N. J. 1988. The association of group III porcine enteroviruses with epithelial tissue. Vet. Rec. 122:441-442. [DOI] [PubMed] [Google Scholar]
- 89.Ko, G., T. L. Cromeans, and M. D. Sobsey. 2003. Detection of infectious adenovirus in cell culture by mRNA reverse transcription-PCR. Appl. Environ. Microbiol. 69:7377-7384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kocwa-Haluch, R. 2001. Waterborne enteroviruses as a hazard for human health. Polish J. Environ. Studies 10:485-487. [Google Scholar]
- 91.Krikelis, V., P. Markoulatos, N. Spyrou, and C. Serie. 1985. Detection of indigenous enteric viruses in raw sewage effluents of the city of Athens, Greece, during a two year survey. Water Sci. Technol. 17:159-164. [Google Scholar]
- 92.Krumbholz, A., R. Wurm, O. Scheck, E. Birch-Hirschfeld, R. Egerer, A. Henke, P. Wutzler, and R. Zell. 2003. Detection of porcine teschoviruses and enteroviruses by LightCycler real-time PCR. J. Virol. Methods 113:51-63. [DOI] [PubMed] [Google Scholar]
- 93.Lee, S. H., and S. J. Kim. 2002. Detection of infectious enteroviruses and adenoviruses in tap water in urban areas in Korea. Water Res. 36:248-256. [DOI] [PubMed] [Google Scholar]
- 94.Le Guyader, F., M. L. Dincher, D. Menard, L. Schwartzbrod, and M. Pommepuy. 1994. Comparative study of the behaviour of poliovirus in sterile seawater using RT-PCR and cell culture. Mar. Poll. Bull. 28:723-726. [Google Scholar]
- 95.Le Guyader, F., D. Menard, M. Pommepuy, and H. Kopecka. 1995. Use of RT seminested PCR to assess viral contamination in Caribbean Rivers (Martinique). Water Sci. Technol. 31:391-394. [Google Scholar]
- 96.Lehmkuhl, H. D., M. H. Smith, and R. E. Dierks. 1975. A bovine adenovirus type 3: isolation, characterization, and experimental infection in calves. Arch. Virol. 48:39-46. [DOI] [PubMed] [Google Scholar]
- 97.Ley, V., J. Higgins, and R. Fayer. 2002. Bovine enteroviruses as indicators of fecal contamination. Appl. Environ. Microbiol. 68:3455-3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Limsawat, S., and S. Ohgaki. 1997. Fate of liberated viral RNA in wastewater determined by PCR. Appl. Environ. Microbiol. 63:2932-2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lipp, E. K., J. L. Jarrell, D. W. Griffin, J. Lukasik, J. Jacukiewicz, and J. B. Rose. 2002. Preliminary evidence for human fecal contamination in corals of the Florida Keys, USA. Mar. Poll. Bull. 44:666-670. [DOI] [PubMed] [Google Scholar]
- 100.Lipp, E. K., R. Kurz, R. Vincent, C. Rodriguez-Palacios, S. R. Farrah, and J. B. Rose. 2001. The effects of seasonal variability and weather on microbial fecal pollution and enteric pathogens in a subtropical estuary. Estuaries 24:266-276. [Google Scholar]
- 101.Lipp, E. K., J. Lukasik, and J. B. Rose. 2001. Human enteric viruses and parasites in the marine environment. Methods Microbiol. 30:559-588. [Google Scholar]
- 102.Lipp, E. K., and J. B. Rose. 1997. The role of seafood in foodborne diseases in the United States of America. Rev. Sci. Tech. Office Int. Epizooties 16:620-640. [DOI] [PubMed] [Google Scholar]
- 103.Lucena, F., L. Schwartzbrod, and A. Bosch. 1986. The effect of a mass poliomyelitis vaccination program on the occurrence of enterovirus in seawater. Zentbl. Bakteriol. Hyg. B 183:67-69. [PubMed] [Google Scholar]
- 104.Lukasik, J., T. M. Scott, D. Andryshak, and S. R. Farrah. 2000. Influence of salts on virus adsorption to microporous filters. Appl. Environ. Microbiol. 66:2914-2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Maluquer de Motes, C., P. Clemente-Casares, A. Hundesa, M. Martin, and R. Girones. 2004. Detection of bovine and porcine adenoviruses for tracing the source of fecal contamination. Appl. Environ. Microbiol. 70:1448-1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.McAnulty, J. M., G. L. Rubin, C. T. Carvan, W. J. Huntley, G. Grohmann, and R. Hunter. 1993. An outbreak of Norwalk-like gastroenteritis associated with contaminated drinking water at a caravan park. Aust. J. Public Health 17:36-41. [DOI] [PubMed] [Google Scholar]
- 107.McNeil, K. M., R. M. Hendrix, J. L. Lindner, R. R. Benton, S. C. Monteith, M. A. Tuchscherer, G. C. Gray, and J. C. Gaydos. 1999. Large, persistent epidemic of adenovirus type 4-associated acute respiratory disease in U.S. army trainess. Emerg. Infect. Dis. 5:798-801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Melnick, J. L. 1984. Etiologic agents and their potential for causing waterborne virus diseases, p. 1-16. In J. L. Melnick (ed.), Enteric viruses in water, vol. 15. Karger, Basel, Switzerland. [Google Scholar]
- 109.Melnick, J. L., and C. P. Gerba. 1989. The ecology of enteroviruses in natural waters. Crit. Rev. Environ. Control 10:65. [Google Scholar]
- 110.Meng, Q. S., and C. P. Gerba. 1996. Comparative inactivation of enteric adenovirus, poliovirus and coliphages by ultraviolet irradiation. Water Res. 30:2665-2668. [Google Scholar]
- 111.Meschke, J. S., and M. D. Sobsey. 1998. Comparative adsorption of Norwalk virus, poliovirus 1 and F+ RNA coliphage MS2 to soils suspended in treated wastewater. Water Sci. Technol. 38:187-189. [PubMed] [Google Scholar]
- 112.Metcalf, T. G., J. L. Melnick, and M. K. Estes. 1995. Environmental virology: from detection of virus in sewage and water by isolation to identification by molecular biology—a trip of over 50 years. Annu. Rev. Microbiol. 49:461-487. [DOI] [PubMed] [Google Scholar]
- 113.Monpoeho, S., A. Maul, B. Mignotte-Cadiergues, L. Schwartzbrod, S. Billaudel, and V. Ferre. 2001. Best viral elution method available for quantification of enteroviruses in sludge by both cell culture and reverse transcription-PCR. Appl. Environ. Microbiol. 67:2484-2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Muniain-Mujika, I., M. Calvo, F. Lucena, and R. Girones. 2003. Comparative analysis of viral pathogens and potential indicators in shellfish. Int. J. Food Microbiol. 83:75-85. [DOI] [PubMed] [Google Scholar]
- 115.Muscillo, M., F. A. Aulicino, A. M. Patti, P. Orsini, L. Volterra, and G. M. Farra. 1994. Molecular techniques for the identification of enteric viruses in marine waters. Water Res. 28:1-7. [Google Scholar]
- 116.Nairn, C., and G. B. Clements. 1999. A study of enterovirus isolations in Glasgow from 1977 to 1997. J. Med. Virol. 58:304-312. [PubMed] [Google Scholar]
- 117.Noble, R. T., and J. A. Fuhrman. 1999. Breakdown and microbial uptake of marine viruses and other lysis products. Aquat. Microb. Ecol. 20:1-11. [Google Scholar]
- 118.Noble, R. T., and J. A. Fuhrman. 2001. Enteroviruses detected by reverse transcriptase polymerase chain reaction from the coastal waters of Santa Monica Bay, California: low correlation to bacterial indicator levels. Hydrobiologia 460:175-184. [Google Scholar]
- 119.Noble, R. T., and J. A. Fuhrman. 1997. Virus decay and its causes in coastal waters. Appl. Environ. Microbiol. 63:77-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Noble, R. T., S. M. Allen, A. D. Blackwood, W. Chu, S. C. Jiang, G. L. Lovelace, M. D. Sobsey, J. R. Stewart, and D. A. Wait. 2003. Use of viral pathogens and indicators to differentiate between human and non-human fecal contamination in a microbial course tracking comparison study. J. Water Health 1:195-207. [PubMed] [Google Scholar]
- 121.Pallin, R., A. P. Wyn-Jones, B. M. Place, and N. F. Lightfoot. 1997. The detection of enteroviruses in large volume concentrates of recreational waters by the polymerase chain reaction. J. Virol. Methods 67:57-67. [DOI] [PubMed] [Google Scholar]
- 122.Parveen, S., N. C. Hodge, R. E. Stall, S. R. Farrah, and M. L. Tamplin. 2001. Phenotypic and genotypic characterization of human and nonhuman Escherichia coli. Water Res. 35:379-386. [DOI] [PubMed] [Google Scholar]
- 123.Parveen, S., R. Murphree, L. Edmiston, C. Kaspar, K. Portier, and M. Tamplin. 1997. Association of multiple-antibiotic-resistance profiles with point and nonpoint sources of Escherichia coli in Apalachicola Bay. Appl. Environ. Microbiol. 63:2607-2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Parveen, S., K. M. Portier, K. Robinson, L. Edmiston, and M. L. Tamplin. 1999. Discriminant analysis of ribotype profiles of Escherichia coli for differentiating human and nonhuman sources of fecal pollution. Appl. Environ. Microbiol. 65:3142-3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Patti, A. M., F. A. Aulicino, A. L. Santi, M. Muscillo, P. Orsini, C. Bellucci, G. La Rosa, I. Mastroeni, and L. Volterra. 1996. Enteric virus pollution of Tyrrhenian areas. Water Air Soil Pollut. 88:261-267. [Google Scholar]
- 126.Paul, J. H., S. C. Jiang, and J. B. Rose. 1991. Concentration of viruses and dissolved DNA from aquatic environments by vortex flow filtration. Appl. Environ. Microbiol. 57:2197-2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Pina, S., M. Puig, F. Lucena, J. Jofre, and R. Girones. 1998. Viral pollution in the environment and in shellfish: human adenovirus detection by PCR as an index of human viruses. Appl. Environ. Microbiol. 64:3376-3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Pommepuy, M., and F. Le Guyader. 1998. Molecular approaches to measuring microbial marine pollution. Curr. Opin. Biotechnol. 9:292-299. [DOI] [PubMed] [Google Scholar]
- 129.Puig, M., J. Jofre, F. Lucena, A. Allard, G. Wadell, and R. Girones. 1994. Detection of adenoviruses and enteroviruses in polluted waters by nested-PCR amplification. Appl. Environ. Microbiol. 60:2963-2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Roessler, P. F., and B. F. Severin. 1996. Ultraviolet light disinfection of water and wastewater, p. 313-368. In C. J. Hurst (ed.), Modeling disease transmission and its prevention by disinfection. Cambridge University Press, Cambridge, United Kingdom.
- 131.Rose, J. B., R. L. Mullinax, S. N. Singh, M. V. Yates, and C. P. Gerba. 1987. Occurrence of rotaviruses and enteroviruses in recreational waters of Oak Creek, Arizona. Water Res. 21:1375-1381. [Google Scholar]
- 132.Rowe, W. P., R. J. Huebner, L. K. Gilmore, R. H. Parrot, and T. G. Ward. 1953. Isolation of a cytopathic agent from human adenoids undergoing spontaneous degradation in tissue culture. Proc. Soc. Exp. Biol. Med. 84:570-573. [DOI] [PubMed] [Google Scholar]
- 133.Russell, W. C., and M. Benk. 1999. Animal viruses, p. 14-21. In A. Granoff and R. G. Webster (ed.), Encyclopedia of virology, 2nd ed. Academic Press, London, United Kingdom.
- 134.Schaper, M., J. Jofre, M. Uys, and W. O. K. Grabow. 2002. Distribution of genotypes of F-specific RNA bacteriophages in human and non-human sources of faecal pollution in South Africa and Spain. J. Appl. Microbiol. 92:657-668. [DOI] [PubMed] [Google Scholar]
- 135.Schiff, G. M., G. M. Stefanovic, B. Young, and J. K. Pennekamp. 1984. Minimum human infectious dose of enteric viruses (echovirus-12) in drinking water, p. 222-228. In J. L. Melnick (ed.), Enteric viruses in water, vol. 15. Karger, Basel, Switzerland. [Google Scholar]
- 136.Schwab, K. J., R. De Leon, and M. D. Sobsey. 1995. Concentration and purification of beef extract mock eluates from water samples for the detection of enteroviruses, hepatitis A virus, and Norwalk virus by reverse transcription-PCR. Appl. Environ. Microbiol. 61:531-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Scott, T. M., J. B. Rose, T. M. Jenkins, S. R. Farrah, and J. Lukasik. 2002. Microbial source tracking: current methodology and future directions. Appl. Environ. Microbiol. 68:5796-5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Sedmak, G., D. Bina, and J. MacDonald. 2003. Assessment of an enterovirus sewage surveillance system by comparison of clinical isolates with sewage isolates from Milwaukee, Wisconsin, collected August 1994 to December 2002. Appl. Environ. Microbiol. 69:7181-7187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Shieh, Y.-S. C., R. S. Baric, and M. D. Sobsey. 1997. Detection of low levels of enteric viruses in metropolitan and airplane sewage. Appl. Environ. Microbiol. 63:4401-4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Shieh, Y.-S. C., D. Wait, L. Tai, and M. D. Sobsey. 1995. Methods to remove inhibitors in sewage and other fecal wastes for enterovirus detection by the polymerase chain reaction. J. Virol. Methods 54:51-66. [DOI] [PubMed] [Google Scholar]
- 141.Simpson, J. M., J. W. Santo Domingo, and D. J. Reasoner. 2002. Microbial source tracking: state of the science. Environ. Sci. Technol. 36:5279-5288. [DOI] [PubMed] [Google Scholar]
- 142.Sinton, L. W., C. H. Hall, P. A. Lynch, and R. J. Davies-Colley. 2002. Sunlight inactivation of fecal indicator bacteria and bacteriophages from waste stabilization pond effluent in fresh and saline waters. Appl. Environ. Microbiol. 68:1122-1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Sinton, L. W., R. K. Finlay, and A. J. Reid. 1996. A simple membrane filtration-elution method for the enumeration of F-RNA, F-DNA and somatic coliphages in 100 ml water samples. J. Microbiol. Methods 25:257-269. [Google Scholar]
- 144.Skraber, S., B. Gassilloud, L. Schwartzbrod, and C. Gantzer. 2004. Survival of infectious poliovirus-1 in river water compared to the persistence of somatic coliphages, thermotolerant coliforms and poliovirus-1 genome. Water Res. 38:2927. [DOI] [PubMed] [Google Scholar]
- 145.Sobsey, M. D., C. Wallis, M. Henderson, and J. L. Melnick. 1973. Concentration of enteroviruses from large volumes of water. Appl. Microbiol. 26:529-534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Sobsey, M. D., P. A. Shields, F. S. Hauchman, R. L. Hazard, and L. W. Caton. 1986. Survival and transport of hepatitis A virus in soils, groundwater and wastewater. Water Sci. Technol. 18:97-106. [Google Scholar]
- 147.Solic, M., and N. Krstulovic. 1992. Separate and combined effects of solar radiation, temperature, salinity, and pH on the survival of faecal coliforms in seawater. Mar. Poll. Bull. 24:411-416. [Google Scholar]
- 148.Springthorpe, V. S., C. L. Loh, W. J. Robertson, and S. A. Sattar. 1993. In situ survival of indicator bacteria, MS-2 phages and human pathogenic viruses in river water. Water Sci. Techonol. 27:413-420. [Google Scholar]
- 149.Straub, T. M., I. L. Pepper, and C. P. Gerba. 1995. Removal of PCR inhibiting substances in sewage sludge amended soil. Water Sci. Technol. 31:311-315. [Google Scholar]
- 150.Suttle, C. A., and F. Chen. 1992. Mechanisms and rates of decay of marine viruses in seawater. Appl. Environ. Microbiol. 58:3721-3729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Tani, N., Y. Dohi, N. Jurumatani, and K. Yonemasu. 1995. Seasonal distribution of adenoviruses, enteroviruses and reoviruses in urban river water. Microbiol. Immunol. 39:577-580. [DOI] [PubMed] [Google Scholar]
- 152.Thurston-Enriquez, J. A., C. N. Haas, J. Jacangelo, K. Riley, and C. P. Gerba. 2003. Inactivation of feline calicivirus and adenovirus type 40 by UV radiation. Appl. Environ. Microbiol. 69:577-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Tsai, Y. L., M. D. Sobsey, L. R. Sangermano, and C. J. Palmer. 1993. Simple method of concentrating enteroviruses and hepatitis A virus from sewage and ocean water for rapid detection by reverse transcriptase-polymerase chain reaction. Appl. Environ. Microbiol. 59:3488-3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Tsai, Y. L., B. Tran, and C. J. Palmer. 1995. Analysis of viral RNA persistence in seawater by reverse transcriptase-PCR. Appl. Environ. Microbiol. 61:363-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Urlings, H. A., G. F. de Boer, D. J. van Roozelaar, and G. Koch. 1993. Inactivation of chicken anaemia virus in chickens by heating and fermentation. Vet. Q. 15:85-88. [DOI] [PubMed] [Google Scholar]
- 156.Ursula, H., S. E. D. Khalaf, and E. F. Kaleta. 1982. Studies on the persistence and excretion of egg drop syndrome 1976 virus in chickens. Avian Pathol. 11:441-452. [DOI] [PubMed] [Google Scholar]
- 157.U.S. Environmental Protection Agency. 1998. Drinking water contamination candidate list. Fed. Regulat. 63:10274-10287. [Google Scholar]
- 158.U.S. Environmental Protection Agency. 1996, posting date. ICR microbial laboratory manual. EPA 600R95178. U.S. Environmental Protection Agency. [Online.]. http://www.epa.gov/nerlcwww/icrmicro.pdf.
- 159.U.S. Environmental Protection Agency. 1992. Manual on guidelines for water reuse. EPA/625/R-92/004. Center for Environmental Reservation Information, Cincinnati, Ohio.
- 160.van den Hurk, J. V. 1992. Characterization of the structural proteins of hemorrhagic enteritis virus. Arch. Virol. 126:195-213. [DOI] [PubMed] [Google Scholar]
- 161.Van Heerden, J., M. M. Ehlers, W. B. Van Zyl, and W. O. K. Grabow. 2003. Incidence of adenoviruses in raw and treated water. Water Res. 37:3704-3708. [DOI] [PubMed] [Google Scholar]
- 162.Wagenknecht, L. E., J. M. Roseman, and W. H. Herman. 1991. Increased incidence of insulin-dependent diabetes mellitus following an epidemic of coxsackievirus B5. Am. J. Epidemiol. 133:1024-1031. [DOI] [PubMed] [Google Scholar]
- 163.Wait, D. A., and M. D. Sobsey. 2001. Comparative survival of enteric viruses and bacteria in Atlantic Ocean seawater. Water Sci. Technol. 43:139-142. [PubMed] [Google Scholar]
- 164.Wallis, C., and J. L. Melnick. 1967. Concentration of enteroviruses on membrane filters. J. Virol. 1:472-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Wetz, J. J., E. K. Lipp, D. W. Griffin, J. Lukasik, D. Wait, M. D. Sobsey, T. M. Scott, and J. B. Rose. 2004. Presence, infectivity, and stability of enteric viruses in seawater: relationship to marine water quality in the Florida Keys. Mar. Poll. Bull. 48:698-704. [DOI] [PubMed] [Google Scholar]
- 166.Wilhelmi, I., E. Roman, and A. Sanchez-Fauquier. 2003. Viruses causing gastroenteritis. Clin. Microbiol. Infect. 9:247-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Wilson, I. 1997. Inhibition and facilitation of nucleic acid amplification. Appl. Environ. Microbiol. 63:3741-3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Xu, W., M. C. McDonough, and D. D. Erdman. 2000. Species-specific identification of human adenoviruses by a multiplex PCR assay. J. Clin. Microbiol. 38:4114-4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Yates, M. V., C. P. Gerba, and L. M. Kelley. 1985. Virus persistence in groundwater. Appl. Environ. Microbiol. 49:778-781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Young, C., R. Burghoff, L. Keim, V. Minak-Bernero, J. Lute, and S. Hinton. 1993. Polyvinylpyrrolidone-agarose gel electrophoresis purification of polymerase chain reaction-amplifiable DNA from soils. Appl. Environ. Microbiol. 59:1972-1974. [DOI] [PMC free article] [PubMed] [Google Scholar]