Abstract
The importance of regulatory control in metabolic processes is widely acknowledged, and several enquiries (both local and global) are being made in understanding regulation at various levels of the metabolic hierarchy. The wealth of biological information has enabled identifying the individual components (genes, proteins, and metabolites) of a biological system, and we are now in a position to understand the interactions between these components. Since phenotype is the net result of these interactions, it is immensely important to elucidate them not only for an integrated understanding of physiology, but also for practical applications of using biological systems as cell factories. We present some of the recent “-omics” approaches that have expanded our understanding of regulation at the gene, protein, and metabolite level, followed by analysis of the impact of this progress on the advancement of metabolic engineering. Although this review is by no means exhaustive, we attempt to convey our ideology that combining global information from various levels of metabolic hierarchy is absolutely essential in understanding and subsequently predicting the relationship between changes in gene expression and the resulting phenotype. The ultimate aim of this review is to provide metabolic engineers with an overview of recent advances in complementary aspects of regulation at the gene, protein, and metabolite level and those involved in fundamental research with potential hurdles in the path to implementing their discoveries in practical applications.
INTRODUCTION
Metabolic engineering is the rational alteration of the genetic architecture of an organism to achieve a specific phenotype (8, 197). Conventionally, the first step in the rational alteration process is identifying the “rate-limiting step” in a given metabolic process based on carbon mass-flux distribution. The solution to overcoming this bottleneck has been either overexpressing the heterologous gene(s) responsible for affecting the rate-limiting step(s) or inactivating the inefficient pathway(s) that contributes to by-product formation (143, 152, 184). While this strategy has enjoyed moderate success, more complex aspects of metabolism cannot be altered as desired by manipulating the metabolic gene(s), such as, for example, increasing the pH tolerance or expanding the range of consumable substrates in microorganisms. This is because cells have evolved to comprise a complex network of regulatory mechanisms that counteract the genetic mutation by employing alternative pathways for continued robust performance. Careful analysis of the regulatory control mechanisms governing the shift in metabolic processes will facilitate the application of metabolic engineering with greater efficiency. We define efficiency as the ability to impart the desired phenotype to the cell with minimal impact on the rest of the metabolism.
While the focus of metabolic engineering is shifting towards engineering the regulatory control mechanisms (53), the exact nature and operation of these control systems are not yet clearly understood. It is generally accepted that the control of metabolic processes is hierarchical and originates at the level of transcription (induction-repression mechanism and mRNA degradation), moving on to translation (protein activation and proteolysis) and enzyme activity (allostery) or usually a combination of them (such as signaling cascades). The presence of several feedback loops among these regulatory processes makes their organization and functioning very complicated. Consequently, accurately predicting the cellular response of any genetic (or environmental) perturbation is an extremely convoluted procedure and should take into consideration as many regulatory constraints as possible.
The significant advances in genome sequencing, transcription, and protein and metabolite profiling have not translated into successful metabolic engineering applications, mainly due to the limitations in our understanding of how these components work in unison to produce the desired trait in the cell. We are still far from understanding regulatory phenomena from a global perspective. These high-throughput techniques have the potential to disclose extremely useful information, but they provide a snapshot from only one stage of transfer of information from gene to function (Fig. 1) while possibly missing the cause and effect relationships from other stages.
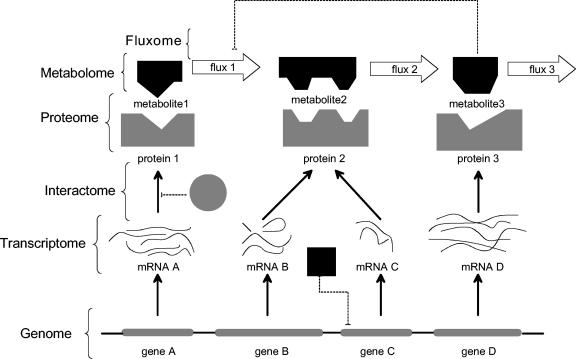
FIG. 1.
Organization of the various -omes in a hierarchical fashion. The comprehensive DNA sequence in a cell is the genome, which consists of coding regions, shown as gray bars. The coding process begins with the expression of DNA to the respective RNA species, which consists of the transcriptome. The transcription process is dictated by several factors, including the interaction of proteins and metabolites with DNA and the presence/absence of the required cellular machinery. The mRNA species are translated to form proteins (proteome). As shown in the figure, there may not exist a one-to-one correspondence between proteins and genes. The various interactions between proteins, DNA, and metabolites, called the interactome, is the key determinant of any cellular process. The solid arrows represent the flow of biological information, while the dashed lines show possible interactions between various cellular components. Proteins are the functional entities that carry out the actual metabolic process by interconverting metabolites (metabolome). Any observed phenotype such as growth and product formation is the net result of all these cellular events. Therefore, capturing information at just one stage of the process (transcription, translation, etc.) will not reveal the cause and effect relationships between cellular components. It is necessary for the metabolic engineer to understand these relationships in order to accurately design and control biological systems.
A holistic approach towards understanding these complex regulatory phenomena would reveal several previously unknown interactions between genes, proteins, and metabolites, facilitating truly rational cellular perturbations in the practice of metabolic engineering. In accordance with this ideology, we first introduce recent advances in the study of regulation using genome, proteome, and metabolome analysis and provide a very brief overview of some of the methods used in these analyses, followed by innovative approaches to integrate global information from these “-omes.” We also present a perspective on the influence of these new discoveries in regulation on metabolic engineering with the hope that researchers investigating one stage of global information may be acquainted with information from complementary stages (Fig. 1).
REGULATION AT THE TRANSCRIPTIONAL LEVEL
Contribution of Microarrays
The genome defines the phenotype, and all phenotypic changes due to a perturbation ultimately originate at the transcription level. The ability to measure mRNA abundance (118) merely reveals the end effects of the global metabolic control machinery on transcription by identifying differentially expressed genes. The mechanism of transcriptional regulation that leads to the differential expression of genes could partially be explained based on a priori information available regarding the genes as well as results from metabolic studies using strains with appropriate genetic manipulation. Among the early efforts towards this goal was identifying genes that were differentially expressed in Saccharomyces cerevisiae in response to a metabolic shift from growth on glucose to diauxic growth on glucose and ethanol (44). Identifying similarities in the transcriptional profile, the role of many previously uncharacterized genes was predicted, based on the assumption that coexpressed genes are coregulated.
Another classic example of experimental design to understand transcriptional regulation using gene expression measurements was to elucidate the mechanism of tryptophan metabolism in Escherichia coli, which involved a combination of genetic mutation and growth conditions (108). While demonstrating a greater role for the trp, mtr, and aro operons in the regulation of tryptophan metabolism, the results also revealed that the genes involved in the biosynthesis of arginine are sensitive to tryptophan starvation. The results presented in these papers demonstrated the tremendous potential of microarrays and prompted the widespread use of microarrays (Fig. 2). The exponential increase in publications that utilized microarrays demonstrates the growing popularity of this technology.
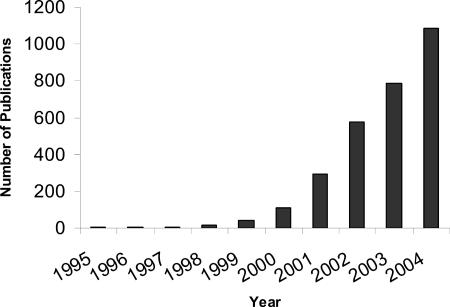
FIG. 2.
Exponential increase in the application of microarray technology in research. The statistics are taken from the papers in the Pubmed database. Only papers including the words microarray, oligonucleotide array, or global gene expression in the title or as key words were taken into account.
Among the multitude of applications of microarrays, the most relevant ones are in the study of global regulators. Prior to global transcription analysis, the effect of global regulators on the transcriptional changes of large gene sets has been reported using classical genetic and biochemical methods. In E. coli these regulators, such as catabolite repressor protein and leucine-responsive regulator protein (LRP) influence the expression of stimulons, each of which comprises several operons with common functionality (153). LRP is a DNA-binding dual regulator of the Lrp regulon and branched-chain amino acid transport in E. coli. Genomewide differential expression between lrp and lrp+ strains revealed a correlation of metabolism with the nutritional state along with discovering several new genes that are regulated by Lrp (85). Similarly, global transcriptional changes as a consequence of deleting lhrA, another global regulator, revealed changes in genes involved in flagellation and chemotaxis that were not known to be regulated by lhrA (123).
Another class of global regulators is DNA architectural proteins involved in the condensation of the bacterial chromosome, which include members such as heat-stable nucleoid-structuring (H-NS) proteins and integration host factors. These kinds of regulators bind to sequence-specific degenerate DNA sites and control DNA replication, recombination, and even transcription (36, 45, 51, 64, 65, 157, 179). Therefore, they affect the expression of several operons and genes and, unlike most other global regulators, are not affected by metabolic regulators.
The H-NS protein assumes a universal role in suppressing genes responsible for a large number of functions, since it binds to bends in DNA structure, commonly found at promoter sites. Based on genomic analysis, broad functionalities such as adaptation of E. coli to high-pressure stress, entry into transition phase, and drug resistance have recently been attributed to the H-NS protein (91, 156). In addition to playing a key role in DNA architecture, the integration host factor and histone-like protein from E. coli strain U93, both members of the DNABII family of DNA-binding proteins, were found to regulate more than 120 genes (5). Other examples where global gene expression analysis contributed to existing knowledge on regulatory information is in the adaptation of E. coli to stationary-phase conditions (205) and the transition between aerobic and anaerobic conditions (182).
Two-component regulatory systems are an integral part of adaptive responses in bacteria, yeasts, and plants (78, 235). These highly evolved regulatory systems contain a histidine kinase and a response regulator. Upon receiving the signal (change in environmental condition, such as nitrogen, oxygen, or phosphorus levels, for example), the histidine kinase is autophosphorylated and transfers the phophoryl group to its cognate response protein. The response protein usually binds to the DNA controlling the expression of the target gene.
It has been reported that in E. coli there are 29 histidine kinases and 32 response regulators (148). Until the evolution of transcriptomics as a routine method to study gene expression, these two-component systems were largely studied independently of the others. However, comprehensive transcriptome analyses of the two-component systems in E. coli and Bacillus subtilis revealed significant cross talk between the regulatory systems, such as, for example, the YxjML and YvqEC systems in B. subtilis (111) and ArcAB and EnvZ/OmpR systems in E. coli (160), indicating that these systems might share the same signaling mechanism. Recently, the Snf3/Rgt2-Rgt1 glucose-sensing pathway in Saccharomyces cerevisiae was discovered to function in conjunction with the Snf1-Mig1 glucose-sensing pathway (99).
Even in S. cerevisiae, extensive applications of microarrays have been reported, and for the first time genomewide responses to several environmental and genetic perturbations of great research interest were studied. These initial transcriptomic applications relied on existing knowledge to confirm some of the results as a means of validating new discoveries. For example, the application of microarrays to the classical study of aging and cell cycle identified several previously known genes in addition to discovering several new ones. Although the cell division cycle in yeast is known to regulate the expression of several histone genes (74), the transcriptional changes in the genome were followed in synchronized yeast cells during various stages of the cell cycle (31, 145, 195). About 7% of the genome oscillated with the cell cycle, and every chromosome contained at least one cell cycle-dependant gene. By correlating the expression of the oscillating gene with the stage of the cell cycle, Cho et al. discovered hundreds of transcripts exhibiting rhythmic expression trends exhibiting close periodicity to the cell cycle (31). Based on the cell cycle stage, these genes were grouped into different clusters, and analyzing the upstream sequences of genes from the same cluster revealed binding sites for several known as well as unknown transcription factors, indicating the involvement of additional transcription factors in regulating cell cycle. Considering that a large number of human proteins have high homology to yeast proteins, this research could have important applications in understanding human aging. Further analysis using Fourier time series refined the list of cell cycle-regulated genes by removing some identified by Cho et al. and adding almost 500 new ones (195).
Another area of immense interest that global gene expression profiling has made considerable contribution to is in the development of effective drugs, particularly antifungals. Azole-based drugs are the most popular antifungals in the market, which act by inhibiting ergosterol biosynthesis (245). Using S. cerevisiae as the model organism in which the functioning of ergosterol pathway was evaluated, the quality of antifungals is being improved. The genes in this pathway are transcriptionally regulated by mutations in other genes, causing sterol limitation (6, 103, 125). On the other hand, when these genes are overexpressed, the products of genes such as CYB5, COX3, and RPL27 exhibit allosteric sensitivity to azoles (119, 216), suggesting novel mechanisms for resistance and regulation. Analysis of gene expression changes in response to mutations in the ergosterol pathway in S. cerevisiae exposed to antifungals revealed several unexpected changes in genes related to mitochondria and oxidative stress (10). Several applications of global expression systems to analyze antifungal properties have been reported recently to predict and characterize physiological changes in the organism (2, 13, 32, 40, 41, 164, 193, 242, 246).
Analysis of Gene Expression Data
The increasing application of global gene expression methods (Fig. 2) to address a variety of microbial studies results in accumulating huge amounts of data within a short time span, which has inherent noise in it. Therefore, robust statistical methods need to be implemented not only to design gene expression experiments but also to establish confidence criteria for the inferences drawn from the data. Consequently, any introduction to gene expression technology will remain incomplete without addressing the statistical difficulties involved in obtaining useful information from them.
Before presenting some of the recent computational models to interpret transcriptional data, it is appropriate to note that there are two kinds of data that are commonly generated depending on the kind of method used, although both are based on the fundamental base-pairing ability of the nucleotides. The conventional terminology is to refer to robotically printed sets of PCR products or conventionally synthesized oligonucleotides on glass slides as microarrays (49), while high-density arrays of oligonucleotides that are synthesized in situ using photolithography are referred to as GeneChips (131, 133), although here we refer to both as microarrays. These two methods have become popular after the genomes of many microorganisms (and higher eukaryotes) were completely sequenced.
Prior to the availability of complete sequences, cDNA clones from cDNA banks were PCR amplified and robotically printed onto glass slides, which were used to study gene expression (136, 186). A schematic representation of these three techniques is illustrated in Fig. 3. On the other hand, in the photolithography technique, which is popularized by Affymetrix, synthetic linkers are adhered to a glass surface using photosensitive groups, and a light mask is used to direct light to specific areas on the glass to remove the exposed groups. A new mask is used to direct coupling at other sites, and the process is repeated until the desired sequence and length of the oligonucleotide is synthesized. This method, very similar to the production of computer chips, is very efficient in high-throughput generation of identical arrays. However, this method is very expensive when the new genes need to be added. A slightly modified version of this method that resolves this issue is the ink-jet printing of 60-mer oligonucleotides (18, 83). This method can generate new arrays or modify the gene content by reprogramming the synthesis of the new set of oligonucleotide sequences.
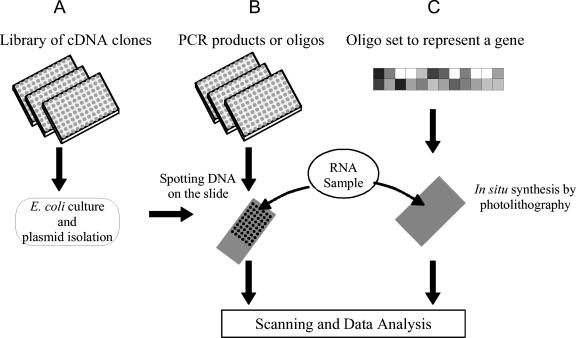
FIG. 3.
Three techniques of preparing microarrays. A) Early microarray platform involved obtaining a library of cDNA clones, which are inserted in a plasmid, expressed in E. coli. These inserts are then purified and spotted on a glass slide. B) In the postsequencing era, long oligonucleotides (about 60 bp) are designed for binding in the nonconserved region of the gene sequences, or commonly, the entire gene is PCR amplified and the purified product is spotted on the glass slide. In both A and B, the DNA bound to the glass slide will act as a template to which the target gene will uniquely hybridize. C) Several short oligonucleotides (about 20 to 25 bp long) are designed per gene along with one mismatch to serve as a negative control. These oligonucleotides are synthesized in situ using the photolithographic method. This platform offers very high quality data.
The availability of the genome sequences for several model organisms has facilitated several researches to PCR-amplify genes (either a selected few or the entire list of open reading frames) from chromosomal DNA or design oligonucleotides to develop arrays that are very specifically suited for their purpose. Transcriptional profiling by global gene expression technology is a paradigm of the convergence of several technologies, such as DNA sequencing and amplification, synthesis of oligonucleotides, fluorescence biochemistry, and computational statistics. Numerous reviews have been published that describe the methodologies and analytics behind these two methods. The primary focus of review in this section will be on the algorithms and statistical methods used to make biologically relevant inferences from data generated using either of these two methods published in the past 3 years and to develop the issues introduced in these reviews as well as highlighting new ones.
Analysis of microarray data has developed to be a very attractive field of research for statisticians. Due to the high noise-to-signal ratio inherent in gene expression data, at least three biologically independent replicates are usually recommended when publishing the data (120). Several normalization methods have been proposed to eliminate noise from the data, particularly for cDNA microarray data analysis. The noise sources in cDNA microarray experiments include the efficiency of dye incorporation, regional hybridization, differential spot quality, variation in experimental conditions during hybridization, scanner settings, etc. Housekeeping genes are frequently used as internal standards for normalization, but since their expression cannot be assumed a priori, external RNA standards were developed for normalization (48).
One of the commonly used statistical methods to account for local variations arising due to improper labeling is the locally weighted scatter plot smoothing (LOWESS) algorithm (14). A smoothing parameter between 0 and 1 is chosen, and a line is fitted to the data based on weighted least squares so that the effect of outliers is minimized. While this has been the most popular method of smoothing data from cDNA microarrays, there are other modifications that have been developed, such as fitting a regression model using background intensities, followed by LOWESS smoothing (243), or using cubic splines to fit the data (237). As an alternative to these computationally intensive methods, identifying a regression function from the data using a wavelet regression was reported to produce reliable results faster (230).
The two fundamental goals of all data analysis methods are to identify genes that are differentially expressed in the test sample relative to the reference sample and recognize patterns in gene expression that correlate with the phenotype. A third and emerging goal of several recent gene expression enquiries is to elucidate the cause and effect relationship between gene expression and regulatory networks in metabolic pathway analysis.
The first goal of identifying differentially expressed genes from transcription data was originally based on using a cutoff value for the change of a gene in the test sample compared to the reference sample. This method is not only statistically insufficient but also does not consider the variations arising as a part of a multiprocess experimentation. Consequently, analysis merely on the basis of a fixed threshold ratio will increase the proportion of false positives. A better approach would be to rank genes according to the expression data obtained from replicates and the selection of a cutoff value for rejecting the null hypothesis that the expression of a particular gene has not changed.
The availability of replicated data allows reliable ranking of the genes using common statistical methods such as Student's t test (or its variations), analysis of variance (104-106), Bayesian method (9, 134), or Mann-Whitney test (238). An important issue of debate that arises using these approaches is the value of the cutoff. There needs to be a balance between type I error (appearance of false positives) and type II error (appearance of false negatives). Even at a relatively stringent cutoff value of 0.05, a data set consisting of 5,000 genes allows the selection of 5,000 × 0.05 = 250 genes, irrespective of their true expression value. This problem could partly solved by using Bonferroni correction for the cutoff values.
The second goal of identifying gene expression patterns enables discovering new regulatory mechanisms. The common methods used for this purpose are unsupervised methods such as K-means clustering (207), hierarchical clustering (50), and self-organizing maps (213). The clusters obtained by these methods are highly dependant on the clustering technique and the distance metric used to calculate the clusters. Moreover, the number of reliable clusters has always been a matter of debate. The biggest drawback of these methods is that they classify genes and experimental conditions as disjoint variables. Since genes and the conditions in which they are expressed are interdependent on each other, the conventional clustering methods do not enable making inferences between genes and conditions. Some of these issues are resolved in other unsupervised dimension reduction methods, which primarily aid in reducing the data to a more manageable size while preserving its nature.
Among the dimension reduction methods, principal component analysis (PCA) is the most applied to gene expression data analysis (199). Other less-used dimension reduction approaches, such as independent component analysis (ICA) (130) and correspondence factor analysis (54), have also been applied to analyze gene expression data. As the name indicates, these unsupervised learning methods work with a defined metric between expression patterns without prior knowledge on functional gene classification.
Although the clustering algorithms and self-organizing maps have been implemented with considerable success (33, 163, 213), their biggest drawback is the lack of gene functionality. This demerit is addressed using supervised learning techniques such as support vector machines. This is a supervised learning technique which begins with a set of genes that share a common function. Additionally, another set of unclassified genes is also defined. These two sets are projected into higher dimensional space, where they are linearly separable. The algorithm finds hyperplane in this space where the separation between the data points is maximal. Now the “intelligent” support vector machine has the ability to distinguish between new genes belonging to either of the two sets based on their features. Therefore, this method uses prior knowledge about gene functionality to determine which functional category a new gene is likely to belong in.
As evident from the description, this supervised method is invaluable in classifying open reading frames whose function is not known (24, 122, 221). For further information about supervised learning algorithms for analyzing gene expression data, the reader is referred to a recent review by Kuo et al. (115). Other mathematically more complex supervised methods such as weighted voting (116) and k-nearest neighbors (151, 218) are more suited for gene classification and prediction. More recently, gene expression analyses are aimed at revealing regulatory causes for the changes in gene expression and associating these changes with physiology.
The idea behind formulating gene networks and subnetworks is essentially to identify those genes that are commonly bound by the same transcription factor. Since the output of a microarray experiment is the result of the interplay between transcription factors and genes, this aspect has been the focus of recent data analysis methods. Since one gene is under the control of multiple transcription factors, the amount of control from each transcription factor is not easy to quantify.
Associating transcription with binding information for 106 transcription factors, Bar-Joseph et al. clustered coexpressed genes to reconstruct regulatory networks in Saccharomyces cerevisiae (11). They identified established interactions as well as discovering new interactions that they used to construct regulatory models. Liao et al. developed a similar approach called network component analysis to quantify the strength of interactions between genes and transcription factors (128). The interactions were modeled as a two-layered network, with transcription factors consisting of the first layer and the genes in the next layer and the interactions between the two layers as edges. Implementing this technique to a glucose-to-acetate diauxic shift in E. coli, 16 transcription factors were found to be significantly involved in the transition.
The biggest advantage of this method is that it does not assume independence or orthogonality of genes, unlike ICA or PCA, respectively. While these reports demonstrated the use of gene expression microarrays to study the regulation of specific pathways at the transcriptional level, they still do not account for regulatory effects brought about by proteins and metabolites interacting with DNA, and therefore, such an approach would not be feasible in higher organisms with a greater level of complexity. As pointed out by Nielsen, the percentage of genes that encode nonmetabolic functions (particularly regulatory functions) increases with increasing cellular complexity (155). In order to reveal regulatory phenomena based only on changes in gene expression, detailed information about interactions between genes and their transcription factor proteins must be elucidated.
DNA-Protein Interactions: Impact on Transcription
The availability of genome sequences as well as methods to identify the transcriptional regulator binding sites in the cell make it possible to discover sets of target genes that are bound in vivo by each transcriptional regulator in a more physiological sense (159). This principle was used to develop technology that directly maps the global interactions between transcription factors and their DNA binding sites using chromatin immunoprecipitation (94, 121, 129, 178). DNA fragments from cells grown under controlled experimental conditions that are bound to the transcriptional regulators are recovered by a chromatin immunoprecipitation assay using an antibody specific to the protein of interest and are hybridized to DNA microarrays that contain the complete set of intergeneic regions. The strength of hybridization intensity signal of a particular gene reflects binding of the transcriptional regulator to the promoter site of that gene.
This second-generation application of microarrays reveals the network of genes that are bound by one or more transcriptional regulators and presents a very powerful experimental methodology into revealing the first step in transcriptional regulation by identifying gene sets that are bound by the same transcription regulators. While this method can only map the probable protein-DNA interaction loci within 1 to 2 kilobases, it also fails to distinguish between positive and negative regulation. Clearly, the key step in this method is the accurate detection of transcription regulator binding sites, which is predominantly achieved by computational predictions (132, 140, 144, 181, 204).
Based on known regulatory information gleaned from biochemistry, gene expression, and chromatin immunoprecipitation results, Luscombe et al. demonstrated that the strength of interactions between transcription factors and genes is context dependent in S. cerevisiae (137). Studying the changes in gene expression patterns in response to changes in cell cycle, sporulation, diauxic shift, DNA damage, and stress, they concluded that a few transcription factors are always involved in regulation, while others depend on the stimulus, thus constantly reprogramming the regulatory network. Only a few target genes are expressed under a specific condition. One of the ramifications of this conclusion, based on over 7,000 interactions between genes and transcription factors in S. cerevisiae, is that one must use caution when extrapolating the interactions and regulatory mechanisms identified under condition to another.
Posttranscriptional Regulation: Role of mRNA
mRNA synthesis by gene transcription, its translation, and subsequent degradation, all processes involving several proteins, has a global regulatory effect on cellular processes. Many of these regulatory proteins were first identified in E. coli and subsequently in other organisms. More recently, E. coli has become the focus of research upon the discovery of new posttranscriptional regulatory features of small noncoding RNAs. Subsequently, these small noncoding RNA regulators were found to be ubiquitous and to regulate multiple functions by base pairing with the target gene. For example, they were shown to regulate metabolic and chaperone functions in E. coli (59, 141, 142), quorum sensing in Vibrio (124), and developmental functions in Drosophila (4).
For any RNA to function as a regulator, it must be transcribed only under specific conditions and have specific base-pairing capability that is limited only during the presence of the activating signal (66, 67). In eukaryotic cells, the transcribed RNA is transported from the nucleus into the cytoplasm by proteins that bind and export the message in the form of a messenger ribonucleoprotein (mRNP) complex (200). There is one gene in yeast (S. cerevisiae) that encodes the mRNP export receptor (192), two in Caenorhabditis elegans, and four in humans (239). After the transport, mRNPs regulate several complex cellular processes. Immunoprecipitation of the mRNA transport components followed by genomewide transcription analyses and reverse transcription-PCR have been used to systematically identify the localization of mRNAs in S. cerevisiae and identify genes that are associated with these components (194, 203). The regulatory role of mRNPs in the translation is further discussed in the section on translational regulation.
Another aspect of posttranslational regulation that is emerging into prominence is the degradation of the transcribed gene (mRNA). Cells have the capability to regulate the level of various mRNA species by differential rates of degradation of each mRNA species. mRNA degradation was originally thought to be a salvage mechanism until the discovery of transcriptional regulation mechanism for RNase III in synthesizing proteins that are recruited for the degradation process in E. coli (12). While most of the bacterial proteins responsible for mRNA degradation are autoregulated by cleaving a stem-loop structure upstream of the ribosome-binding site (12, 95, 162), mammalian RNA decay mechanisms remain poorly characterized. With a wide variation in the stabilities of different mRNA species, ranging from a few minutes to many hours (107) in addition to their dynamically changing half-lives, mRNA degradation-mediated gene regulation clearly occupies a very important role in the hierarchy of control mechanisms. Various regulatory aspects of mRNA degradation and their mechanisms are reviewed in detail elsewhere (117, 236). In addition to regulating gene expression, the mRNA degradation process also has evolved to check the fidelity of the information processing by degrading incorrectly processed mRNA transcripts that lack a stop codon (57, 223).
Microarrays have found a new niche in analyzing mRNA degradation patterns from a global perspective in an effort to understand the associated regulatory mechanisms (15, 68, 73). The dynamics of mRNA degradation have been studied using time course experiments following transcription inhibition (15, 69, 172, 231). In all these reports, degradation dynamics seem to be closely related to the physiological function of the end product because there is a positive correlation between the stability of mRNA and the function of its corresponding protein. Recently, Mohanty and Kushner discovered a protective role for RNase II in safeguarding specific mRNAs from the activity of other nucleases using genomewide transcript analysis of rnb (encoding RNase II) mutant of E. coli (149). In the same study, they also concluded that in spite of accounting for only 10% of exonuclease activity in E. coli, polynucleotide phosphorylase is more important in the degradation of mRNAs than RNase II (149). In fact, a greater role for polynucleotide phosphorylase in the degradation process was later revealed by its participation in forming an assembly with other proteins, known as degradosome (16). Since enolase (which converts 2-phosphoglycerate to phosphoenol pyruvate) has been shown to be a part of the degradosome assembly in E. coli (170), and the genes of the glycolysis and cysteine biosynthesis pathways (which originates from 3-phosphoglycerate) respond similarly to mutations in the degradosome complex, it is now believed that the expression of these genes is modulated by the degradosome activity (16, 170).
Yet another example of posttranscriptional regulation that directly affects central carbon metabolism and is therefore of immense relevance to metabolic engineers is the control of glucose uptake. In addition to the degradosome assembly, small noncoding RNA has recently been found to be involved in the posttranscriptional control of glucose uptake in E. coli (100, 222). As the details of mRNA regulation become available, we can anticipate more of this kind of unexpected regulatory connection in the future.
CONTRIBUTION OF PROTEOMICS TO UNDERSTANDING REGULATION
Even though global changes in gene expression provide deep insights into understanding transcriptional control, proteins have to be recruited to perform the process since they are the actual functional units. Therefore, knowledge of protein abundance reveals the extent to which regulatory proteins and transcription binding factors participate in the resulting change in gene expression profile. Since gene function is heavily associated with proteins, analysis of proteins will divulge more information on protein function and the pathways they act on. Moreover, although proteins are the end products of genes, there is no one-to-one correspondence between the number of proteins and the number of genes.
The initiation of translation and its subsequent regulation largely depend on the ribosome-binding site. Upon receiving a signal, the regulatory proteins bind to the promoters and recruit RNA polymerase enzymes to the transcription start site. For a detailed description of the various regulatory mechanisms during translation in prokaryotes, see the recent review by Schlax and Worhunsky (189). While the conventional trend in analyzing proteomes using two-dimensional gel electrophoresis has had a good turnover of information, the greatest setback in this method is that it is heavily biased towards proteins expressed at high concentrations (70). Different staining methods have been developed to improve the accuracy and the sensitivity of protein detection and quantification (167, 220), yet proteins expressed at low concentrations may not be detected accurately. Since several regulatory proteins are present at extremely low concentrations, the need to develop other sensitive high-throughput methods for accurate protein detection and quantification is widely acknowledged. Moreover, the dynamic nature of protein synthesis and consequent modifications, identification, and quantification of proteins alone may not be sufficient.
Alongside the continuing efforts to develop reliable methods to quantify the proteome, an important advancement in our understanding of function is the global identification of protein localization in the cell (61, 84). Information about the localization of a protein reveals its function, activation state, and potential interactions with other proteins, particularly in eukaryotic cells, which are compartmentalized. For example, in S. cerevisiae, 82 new proteins were discovered in the nucleolus and were predicted to be involved in ribosomal function, and in general, the localization results had 80% agreement with the data in the Saccharomyces Genome Database (84). This study confirmed previously known protein-protein interactions in addition to identifying new ones such as those between cell structure and morphology.
Localization of proteins depends on cell signaling events and their state of activation, which depend on the environmental conditions. Such intercompartmental translocation of proteins triggers new signals. Among the various methods used to study protein localization, variants of green fluorescent protein are commonly used to tag the protein for visualization using a light microscope (38, 84).
Translational Regulation and Functional Proteomics
A major portion of adaptation responses to environmental changes in bacteria are carried out by the proteins, and therefore, adaptive regulation is very closely linked to ribosome synthesis. Although ribosomal concentration has been shown to be a function of growth rate in bacteria (110, 185), the control of bacterial rRNA synthesis has been a topic of much debate (43, 247, 248). However, it is generally agreed that bacteria optimize the synthesis of ribosomes and biosynthetic precursors to grow at an optimum rate using the available nutrients and under the environmental conditions provided (43). This balance is achieved by the translational control of ppGpp synthase (encoded by the spoT gene) in E. coli. However, the metabolic signal that triggers this regulation remains to be discovered.
Translation regulation is more complex in eukaryotes and involves several mRNA-binding proteins, which, together with the specific mRNA species itself, constitute the mRNP complex (101). In addition to the mRNA degradation mechanisms, these regulatory RNPs often play an important role in the efficiency of translation as well as subsequent localization of the translated product. As with all other aspects of regulation, the study of translation regulation by mRNP complexes has started with a small component set, and global analyses came into focus with microarray and immunoprecipiration technology (101, 168, 211).
This kind of analysis, we believe, is the birth of an entirely new approach to study global regulation and is known as ribonomics (21, 212). For the first time, it was shown that the mRNA-binding proteins are very selective in the transcripts to which they bind and multiple proteins may bind to a single transcript (75, 90, 101, 211, 212). For example, the proteins HuR (135), HuB (211), αCP2 (226), and Lhp1 (90) were found to bind specifically to their target mRNA species to form an mRNP module. The coexpression of genes that belong to the same functional category observed in microarray experiments over several conditions is largely attributed to the specificity with which the mRNA-binding proteins bind to these genes.
Ribonomics is still an emerging discipline and holds the promise for providing invaluable information on the posttranscriptional fate of eukaryotic RNA, as already demonstrated (75, 211, 212), and has established a foundation for accurate identification of targets for metabolic manipulation in eukaryotes. The lack of such information thus far has been an important reason why the metabolic engineering of higher eukaryotes is still uncharted territory. Currently, robust methods exist for the isolation of mRNA complexes by immunoprecipitation or chromatography followed by the identification of target genes and proteins by genomic approaches, but the critical step of identifying functional relationships among the target proteins is still in a bottleneck. The readers are directed to an excellent review by Hieronymus and Silver for more detailed descriptions of advances in the area of mRNP systems biology (76).
Signal transduction pathways.
The significance of the interaction of proteins with DNA is best reflected in the signal transduction pathways. Signal transduction is a very important mechanism by which the cell exercises its regulatory impact depending on the perturbation. The signal transduction pathways communicate extracellular conditions to the cell interior using a signal (usually a metabolite). Alternative phosphorylation and dephosphorylation of the intermediate proteins (usually kinases) transfers the signal to the transcription factor, which ultimately binds to the DNA to bring about transcriptional changes (Fig. 4).
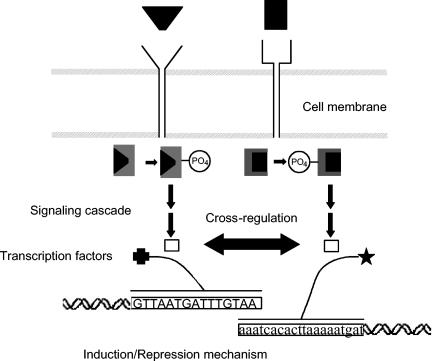
FIG. 4.
Representation of interaction between two signal transduction pathways. Upon external stimulation, the signaling molecule (usually the stimulus itself) binds to the outer membrane receptor proteins. The structural changes that these proteins undergo trigger the phosphorylation of kinases which are activated to trigger the transfer of signal to the transcription factors that bind to specific binding targets upstream of genes, thereby regulating their expression. Such two-component signal transduction systems have so far been studied in isolation, but recent -omic approaches have provided ample evidence for the existence of interactions between these systems.
One such very important signal transduction pathway in facultative anaerobes such as E. coli is the aerobic respiratory control system, a two-component system comprising a membrane-bound sensor kinase (arcB) and its cognate cytosolic response regulator (arcA) (93). The sensor kinase undergoes ATP-dependent autophosphorylation at a conserved histidine position upon stimulation by an external signal, which is believed to be the redox state of the cell (60). The activated kinase transfers the signal to the ArcA protein, which physically binds upstream of more than 30 operons, regulating their expression based on the redox.
Therefore, in order to establish a desired phenotype, rather than manipulating the genes that are directly involved, it may be more efficient to manipulate the action of the signal transduction pathways that govern the expression of these genes. For example, inactivating the action of the Arc regulatory system seems an efficient alternative to relieve the repression of several genes of aerobic respiration to overexpressing these genes. The elegant mechanism of these pathways has been studied in isolation from one another, and with the advent of global techniques, an overlap and interaction between signal transduction pathways has become evident, as revealed, for example, by the cross talk in the two glucose-signaling pathways in S. cerevisiae (99). In fact, the Arc system of E. coli has also been reported to interact with the EnvZ/OmpR osmoresponsive system (146). Under anaerobic conditions, the Arc regulatory system participates in the control of porin synthesis (by the ompC and ompF genes), which was believed to be solely controlled by OmpR.
Protein-Protein Interactions
An important goal of functional genomics is the identification of functional modules from genomewide information. Since microarrays cannot contribute to the knowledge of protein action, protein-protein interactions play a crucial role in elucidating the nature of these mechanisms. Recently, innovative methods for a comprehensive analysis of protein interaction events and signaling pathways have been implemented to provide additional information, such as the high-throughput yeast two-hybrid systems (92, 219, 241), mass spectrometry-based protein analysis (1, 17, 174), protein arrays (154, 229, 234, 250), and fluorescence-based interaction assays (81, 82). For a detailed review on the methodologies and applications of various screening tools in proteomic research, the reader is directed to Templin et al. (208).
Among these methods, the yeast two-hybrid system is the most established genetic method to study proteomes and is based on the modular organization of the eukaryotic transcription regulators, consisting of a DNA-interacting domain and a transcription-activating domain (58). Several thousand interactions with the protein of interest were identified by the transcription of reporter genes (92, 219). Parallel to unraveling the interactome using the yeast two-hybrid system, mass spectrometry-based protein interactions were evaluated at a global scale in S. cerevisiae (58, 77), which enabled predicting new cellular functions for about 350 proteins whose orthologues have relevance in human disease (58).
Using multidimensional liquid chromatography combined with mass spectrometry, 131 membrane-bound proteins with at least three transmembrane domains were identified (232). The principle underlying this method, called MudPIT (MultiDimensional Protein Identification Technology), is to digest the immunoprecipitates with several proteases and analyze the resulting peptide fragments using liquid chromatography-mass spectrometry (232).
In spite of the significant progress made by using the yeast two-hybrid system and mass spectrometry, there exists a fundamental difference in the nature of protein interactions identified by the two methods: the yeast two-hybrid system identifies binary interactions, while mass spectrometry-based techniques address the formation of protein complexes. The proteome research community is beginning to acknowledge that comprehensive interactions in the yeast proteome could therefore be revealed by combining the information from these two approaches (29). The accuracy of predicting protein-protein interactions could then be assisted by the localization profiles to increase confidence in the predictions (114).
In a more physiological sense, protein-protein interactions in S. cerevisiae were used to assign a phenotypic definition to the interactions based on the recovery after exposure to DNA-damaging agents (180). Since it requires a wide range of cellular activities to prevent cell death upon exposure to mutagens, this experimental setup provided an extremely conducive environment to study protein interactions. Several new phenotypic features of protein-protein interactions were discovered along with the observation that these networks are much more complex than the metabolic networks.
In contrast to clustering genes, clustering protein interactions would reveal modules which have similar functionalities and would therefore be more closely associated in bringing out a response. The protein interactions were transformed into a weighted network, with the weights representing the experimentally determined confidence levels for a particular interaction (169). Using this approach, Pereira-Leal et al. clustered the protein interactions into functional modules in S. cerevisiae, many of which agreed with previously determined results and additionally found several new interactions between proteins (169). The fact that most of the new interactions predicted by this model were between proteins that were localized in the same compartment gives more credibility to this method. They went on to successfully reconstruct the signal transduction in the cell wall biogenesis pathway. Computational approaches taking into account the protein-protein interactions to identifying the formation of protein complexes during the S. cerevisiae cell cycle revealed protein subunits that are expressed only at a certain stage of the cell cycle and are under the control of the regulatory protein Cdc28p (39).
Protein arrays.
Considering the pivotal functional role proteins play in defining the phenotype, it is important to quantify protein abundance as well as activity. In the lines of DNA microarrays, protein arrays are rapidly becoming powerful high-throughput tools to identify proteins, monitor their expression, and elucidate their function and interactions within them and, more importantly, the posttranslational changes that they undergo. However unlike the microarrays used for transcription, which are based on simple nucleotide hybridization, proteins have much more complicated binding schemes. Moreover, there are only four nucleotides that comprise a DNA molecule, while proteins are made up of several more building blocks. These drawbacks in addition to issues related to protein stability make protein arrays much more challenging than DNA microarrays.
In the most popular kind of protein microarray, antibodies prepared for specific proteins (or epitopes) are spotted on a slide and incubated with cell extracts that are quantitatively labeled with fluorescent markers. The bound proteins can subsequently be detected using the same instrumentation that is used for conventional DNA microarrays (25, 72, 229). Another kind of protein array using recombinant protein probes spotted onto a slide has also been used to study protein interactions in yeast (138, 139, 173, 250). Zhu et al. detected global protein interactions by probing the yeast proteome with biotinylated calmodulin on a proteome chip containing 5,800 open reading frames in nanowells (249). Using this technology, several known calcineurins and kinases were identified in addition to 33 new proteins that can potentially bind to calmodulin.
This elegant tool facilitated the global analysis of protein interactions with phospholipids for the first time and paves the way for the next generation of protein arrays which can present global functional data for thousands of genes in higher eukaryotes and even humans. There are several excellent reviews published recently that describe the principles and applications of protein arrays in greater detail, which also serve as a resource for several cross-references, and the reader is encouraged to refer to them (25, 190, 208-210).
In spite of these advances, the fundamental aspect that currently limits the advancement of proteomics (in contrast to genomics) is the lack of protein amplification mechanisms analogous to PCR. Therefore, only those proteins that are produced naturally in large quantities or by recombinant techniques can be analyzed. Although there is no established proteomics technology to detect all the desired aspects of proteins, aggressive research in the area of proteomics reflects the pivotal role that proteins play in executing metabolic control. It is expected that proteomics will continue to be in the forefront of functional genomics research and contribute to several key discoveries. However, one caveat of proteomic research is the current thrust on data generation, while the next, more important, step of data interpretation, validation, and integration with metabolism is still in a bottleneck (20, 166).
INTEGRATIVE APPROACHES: SYSTEMS BIOLOGY
Systems biology is the study of how various cellular components function and result in a biological property or the phenotype. The current trend in systems biology originated from the integrative physiology that has been practiced for almost three decades (30, 86, 240). However, the transformation of biology from a descriptive science to a quantitative science profoundly expanded our framework to understand cells and has taken systems biology far ahead of classical integrative physiology. With the availability of the components list for several model organisms, reductionist approaches such as gene expression analysis and protein analysis are already giving way to systems biology (79, 80).
The digital organization of the genetic information (22, 102) defines the analog metabolic processes. The fundamental tenet of systems biology is to decipher the nature and mechanism of this relationship. With the direction biology has been progressing, systems biology is regarded as studying gene expression, protein abundance, and intracellular metabolite profiles at a global scale and developing mathematical models to integrate these components to predict the phenotype. The obvious complexity involved in this process makes it a daunting task.
Quantifying gene expression is the most important aspect of systems biology. Preliminary efforts to integrate global information involved comparing and directly correlating gene expression and the abundance of the corresponding gene product (protein). The absence of correlation between mRNA transcript level and the corresponding protein level in Haemophilus influenzae exposed to antibiotics (62), high cell density in cultures of E. coli (244), Bacillus subtilis subjected to peroxide stress (150), exponentially growing S. cerevisiae (71), cells of S. cerevisiae exposed to lithium (23), and hybridoma cells subjected to glucose-induced metabolic shift (112) or in the human liver tissue (3) reflect the manifestation of significant posttranscriptional regulatory control. These reports do provide a wealth of information to further our understanding of biological systems.
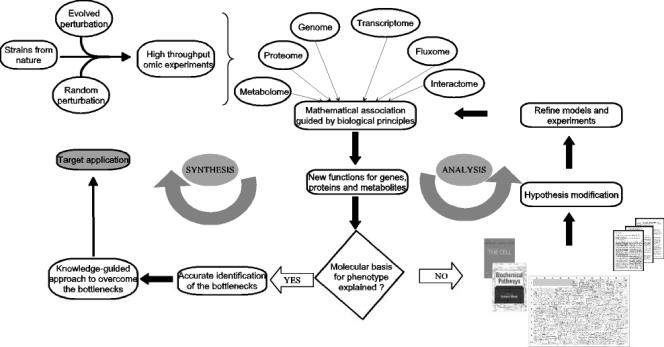
Global information from different stages of metabolic hierarchy needs to be integrated using mathematical and statistical methods to make new discoveries as well as to refine the existing knowledge. Figure 5 illustrates a schematic flowchart of our ideology to achieve a truly rational strain design with minimal perturbations. The preliminary models of individual components such as gene expression, protein networks, and signal transduction pathways are descriptive. Based on these descriptive or rather simple quantitative models, experiments are performed to assess the systemic response due to a perturbation in one of the components. The likely disparity between experimental observation and initial model prediction leads to modification of the model and design of the next round of experiments to validate the model predictions in an iterative manner. Experimental agreement of these new discoveries and predictions indicates fundamental understanding of the phenotype of interest, which enables efficient strain design. Referring to the available literature, metabolic pathways and fundamental biochemistry can correct any disparities in the predictions. Modifying the initial hypothesis and performing the association iteratively will ultimately reveal the control mechanisms (Fig. 5).
FIG. 5.
Analysis and synthesis in metabolic engineering as advocated by systems biology. Associating global information from strains with little-known physiology or from model organisms leads to new discoveries while refining existing knowledge. Experimental validation of these inferences will guide the fundamental understanding of microbial physiology as well as strain design for a purposeful end. However, often the model predictions and preliminary hypothesis do not agree with experimental observations. Referring to the available literature, prior knowledge about basic biochemistry and metabolic pathways leads to modifying the hypothesis, possibly requiring further experimentation. Performing the association again under the modified framework of knowledge should correct any disparities between predictions and experiments. Such iterative procedures are rapidly gaining prominence in systems biology.
Models and Predictions in Systems Biology
The primary step in understanding any biological entity from a systems perspective is to identify its structural organization, such as the gene interactions and biochemical networks, followed by the dynamic interactions between them. Characterization of biological networks requires detailed maps elucidating proteins, RNAs, promoters, and other macromolecules. Towards this broad goal, metabolic networks (96), regulatory networks (121), and protein interaction networks (228) have already begun to be established. These maps are commonly represented as a static set of nodes to represent the components (RNA, proteins, macromolecules, transcription factors, etc) of the network and edges to represent the interactions (activation/inhibition or induction/repression, etc.) between them.
For example, using a bipartite graphic visualization, Patil and Nielsen showed similarities in metabolic network patterns and transcriptional responses that led to the identification of “reporter metabolites” in S. cerevisiae which represent the hub of regulatory action (165). Similarly, topological analysis of metabolism in 43 organisms revealed hierarchical modularity in the network organization (175). Using the path of shortest length in a graph theory approach, Said et al. identified that the toxicity-modulating proteins in S. cerevisiae have more interactions with other proteins, leading to a greater degree of metabolic adaptation upon modulating the functioning of these proteins (180). This result has direct implications on many human degenerative disorders such as cancer and even aging. The authors demonstrate that the protein interaction network is much more complex than the metabolic network, consistent with the knowledge that signaling pathways and regulatory networks have more complex organizational structure than the metabolic network. Although only protein interactions were studied, deeper regulatory aspects could have been revealed by also including protein interactions with DNA, particularly since the study focused on the recovery of S. cerevisiae from DNA-damaging agents.
As opposed to the representation of biological networks as graphs that reflect only the static properties of a system, de Lichtenberg et al. recently reported the dynamics of protein interactions during the yeast cell cycle (39). They used previously published gene expression data from different stages of the cell cycle (31, 195) and integrated it with a network of physically interacting proteins from public databases such as MIPS (147) and discovered that most of the protein complexes are comprised of both constitutively and just-in-time expressed proteins.
Currently, the mathematical models that represent cellular components and their interactions either compromise the specificity or lack the sensitivity. This is due to several reasons, such as a limitation in biological information available and lack of mathematical rules to integrate the available information. Learning how the structure changes in response to various conditions and more importantly what makes the system respond in this fashion will enable identifying precise targets for metabolic engineering (109). Established protocols are not immediately available to guide the merger of global information from the various -omes indicated in Fig. 5.
Ideker et al. compared the global changes in the expression of mRNA and proteins in S. cerevisiae in response to a series of perturbations in the GAL regulatory system (87). They used the yeast galactose metabolic model as a prototype and studied the global responses in response to genetic and environmental perturbations. The key feature of this study that is missing from the previous comparisons was that the authors also considered protein interactions with other proteins and with DNA in their model. Not surprisingly, the expression of those genes that are linked by physical interactions exhibited a higher degree of correlation with corresponding protein levels. Information about protein-protein interactions in S. cerevisiae (191, 219) facilitates the integration of the resulting mRNA and protein responses with known physical interactions to discover and/or refine gene functions.
Since it is the proteins that actually execute the genetic program, mapping global interactions between proteins or the “interactome” in single-celled (219) and multicellular (126) organisms is particularly valuable in revealing the signal transduction pathways which play an integral part in overall regulation. Such a comprehensive mapping of the prokaryotic interactome has not yet been reported to our knowledge. These reports on transcriptome-proteome-interactome analysis communicate a unified theme, suggesting strong posttranscriptional as well as posttranslational control of metabolism.
Ihmels et al. developed an integrated analysis methodology, called the signature algorithm, for S. cerevisiae, which analyzes patterns in gene expression changes over a large number of data sets with various conditions to establish proximity between genes in terms of their expression under various conditions (88). Although this work did not incorporate changes in the metabolic profile as that of Ideker et al. did, physiological changes were used to provide functionalities to genes based on similarity profiles. The premise of organizing genes into transcription modules is that genes that are expressed similarly under a large variety of conditions are more likely to be coregulated than those clustered based on fewer conditions. This method was then used to study various cellular functions as well as the global transcription program. For example, applying this method to an S. cerevisiae data set, genes with previously unknown (or speculated) function such as YGR067C, YGL186C, and YJL1200C were identified with the regulation of the glyoxylate shunt, purine transport, and lysine biosynthesis, respectively (89).
One of the very surprising discoveries made by Ihmels et al. was that about 63% of the isozyme pairs were not coregulated (89). Since isozymes serve in redundancy or amplification of the same metabolic function, they are expected to be regulated similarly. An experimental validation of one such prediction of isozymes not being coregulated was that of the two glutamate dehydrogenases, encoded by GDH1 and GDH3. In a completely independent work, these isozymes were demonstrated to be nonredundant and their expression is carbon source dependent (42). This result agrees very nicely with the work of Kafri et al. on identifying the nature of backup functions that genes perform (98). They argue that genes that are similarly expressed do not back each other up in the event of a mutation but rather through a transcriptional reprogramming mechanism that S. cerevisiae has evolved. Paralogs for the mutated genes are activated only when the gene in question is inactivated. Although the authors did not discuss this aspect, this result might provide some clues to the nature of silent mutations.
The hundreds of components in the cell are organized into modules and interact dynamically with one another. The consequent phenotype is a reflection of these dynamic interactions. Although there is no clear boundary between these modules, the probability of interaction of a component with k other components, p(k), has been shown to decrease according to the power law k−2.2 (96). However, few widely connected components, such as ATP, for example, connect a large portion of metabolism and result in an integrated module-free metabolic network. This dilemma has been resolved by demonstrating that metabolic networks are organized in highly connected modules that operate in conjunction with each other in a hierarchical manner (175). Elucidating the principles that govern the nature and function of these individual modules may be possible with help from engineering, life sciences and computer applications and is indeed the essence of functional genomics.
FUNCTIONAL GENOMICS PERSPECTIVE OF METABOLIC FLUX
Having introduced the technology that is available for enhancing our knowledge of regulation and the recent discoveries in regulation, the remainder of the article will focus on the issue of how this additional knowledge can be implemented in metabolic engineering applications. For a metabolic engineer, pathway flux remains the yardstick by which the physiology of an organism is gauged. The ability to accurately measure pathway flux at a fine resolution using radiolabeled substrates (201, 202) facilitated the use of metabolic fluxes in characterizing phenotypes by quantitative physiological analysis, based on which conclusions are drawn regarding in vivo carbon flow. Since a change in metabolic flux due to any perturbation is a direct ramification of genomic and proteomic changes, effective gene or protein manipulation to mediate a metabolic step requires deeper insights into the cause and effect relationships between a perturbation and the level of expression of genes and proteins (155). Metabolic flux analysis, which is a quantitative description of the phenotype, is conspicuously missing from the prior attempts to combine -omics information from different stages of genetic information pipeline.
Constraint-Based Network Models
One of the most popular tools in recent metabolic engineering applications is the development of a stoichiometric model to represent the biological network (176). Such a model is built by connecting the metabolites and the reactions they participate in through a matrix, which is then used as a set of biochemical constraints in the linear programming to optimize the reaction rates (fluxes) given an objective function (usually the biomass formation). Genome-scale models have been constructed for many model organisms (47, 55, 187) with more being developed for several other microbes (personal communication, J. Nielsen). These models have emerged to serve the fundamental needs to assess the metabolic capabilities of microorganisms and predict carbon flux distribution, which guides further metabolic engineering strategies. These models have also been invaluable in predicting the impact of gene deletions (46, 52, 56) with about 70% to 80% accuracy.
Based on the stoichiometric representation of the biological network, elementary modes were calculated using convex analysis (196). An elementary mode is a unique vector that reflects the minimum number of independent reactions needed for the system to exist as a functional unit. Using elementary mode analysis, it was found that four unique pathways could efficiently produce biomass and energy under different levels of oxygen limitation in E. coli, and depending on the amount of glucose consumed, it is possible to quantify the contribution of each pathway to the overall flux (26, 27).
A subset of the elementary modes is the set of extreme pathways which represent the edges of the solution space and reflect the biochemical capabilities of the system (188). The algorithms used to calculate the elementary modes and extreme pathways are essentially the same, differing only in their incorporation of reversible reactions. The former method accounts for the reaction directionality by a set of rules, while the latter separates them into forward and reverse reactions. Nevertheless, these two approaches provide key information about the degree of pathway utilization that could be used for targeted gene manipulations.
One of the fundamental reasons for the deviation of predictions of stoichiometric models from experimental observations is that these models do not incorporate any kinetic and regulatory information. Kinetic models require more detailed dynamic interactions that bring about the reactions and other parameters that make them more complicated but also more reliable. This inherent drawback in the stoichiometric models has been addressed by incorporating regulatory features as constraints in addition to stoichiometric constraints (35). The fluxes for which the corresponding genes and enzymes are repressed/induced (or inhibited/activated) are constrained by assigning 0 or 1 using the Boolean approach to reflect their participation in the overall physiology.
Subsequently, Covert et al. expanded the regulation-constrained stoichiometric model to the genome scale (34). To this end, the authors developed a robust in silico E. coli strain that is well characterized and used it as a model organism to incorporate regulatory aspects in an iterative fashion. Simulated results from the final model were used to compare the response of E. coli to various levels of oxygenation, with 98% agreement with the experimental results. Although the iterative Boolean approach of imposing regulatory constraints in a flux balance model increased the prediction capability, the regulation in situ is not either 0 or 1, but rather there is a gradual change in the dynamics of the mechanism. In these lines, we can expect in the future such in silico models with regulatory constraints for several other strains with increasing level of complexity.
The evolution of the concept of the “fluxome” as a high-throughput tool to capture the degree of global metabolic pathway utilization suggests that metabolic fluxes will be used extensively in the context of global data integration and analysis (183). Using these methods, it is easier now than ever to harness strains from nature to perform novel biological processes. The fundamental analysis and synthesis aspects in designing and engineering metabolic networks after integrating global information are illustrated in Fig. 5. Novel strains that exhibit the potential for bioprocess applications are obtained from nature. Since the physiology of these strains is not likely to be clear, subjecting the strains to the iterative cycle of global analysis and data integration and drawing inferences will reveal information that can be used in synthesizing a metabolic network with a purposeful end.
Disparity in Gene Expression and Metabolic Flux
At this juncture, we draw attention to an important consideration in associating metabolic fluxes with the transcriptome. Metabolic fluxes, as calculated using the physicochemical stoichiometric models, are predictions based on a few inputs that are obtained experimentally and are largely dependant on problem formulation and the nature of the constraints used even though the biological accuracy of these predictions using stoichiometric flux balance models has been experimentally validated (46). The values of gene expression changes are experimentally determined values, albeit with some inherent level of noise (217). Since the values of metabolic fluxes are estimated using a small number of measured fluxes as inputs and are subsequently used to draw inferences on cellular physiology, they should always be backed by complementing evidence, such as metabolic network analysis using labeled substrates or transcription profile, for example, particularly for organisms with lesser known physiology. Such organisms could be studied based on the cellular architecture of other model organisms.
Recently, a transcription profile was evaluated in S. cerevisiae growing at a steady state in chemostats on various carbon sources and compared with metabolic fluxes (37) to highlight the role of transcriptional regulation in controlling metabolism. The values of metabolic fluxes were determined by flux balance analysis using a genome-scale stoichiometric matrix (55). This constraint-based linear optimization method of estimating metabolic fluxes had the inherent drawback of not being able to account for the repression effects that some sugars, such as glucose, may have on metabolism. However, chemostats offered the unique advantage of maintaining subrepressing residual concentrations of the carbon sources while attaining a steady state.
Surprisingly, the expression of very few genes varied in response to growth on various carbon sources in spite of a widespread change in the estimated values of fluxes, as expected (a total of 180 genes exhibited varied expression in response to growth on four carbon sources). Enzymes, which are the ultimate gene products, affect metabolic flux and, depending on the physiological conditions and intracellular concentration of metabolites, control its magnitude. They can even bring about the reversal of flux under extreme conditions. While synthesis of a particular enzyme may be essential for metabolism, the direction of the reaction it mediates is dictated purely by physiological and thermodynamic conditions. The transcription profile merely reported the absence of significant change in the expression of most genes except those that are directly involved in the uptake of the carbon source. Hence, it can be concluded that except for the enzymes that are specifically required to metabolize a particular sugar, all other proteins are comparable in abundance. The metabolic flux profile, on the other hand, specified the direction of carbon flow. As a result, the changes in gene expression are not as prevalent as those in metabolic fluxes during growth on different carbon sources.
Krömer et al. performed a comprehensive study comparing the intracellular metabolite concentrations, metabolic fluxes, and gene expression to study lysine production and metabolism in Corynebacterium glutamicum (113). The study indicated that the concentrations of intracellular amino acids have complex profiles and reflect a change in physiology much earlier than what can be detected by measuring extracellular products. They also suggested that the excretion of alanine and valine from pyruvate is a phenomenon of overflow metabolism, arising due to the down-regulation of the central metabolism. The combined analysis revealed no change in the expression of lysine biosynthetic genes despite a sevenfold increase in the pathway flux to lysine, signifying not only the action of posttranscriptional control but also that the metabolic capabilities of pathways are usually not limited by the expression of their mRNAs. An important inference from this observation is that overexpressing a gene(s) is not always the approach to enhance metabolic flux.
As was observed from the work of Daran-Lapujade et al. (37), the possible metabolic flux reversal patterns prohibited quantitatively correlating gene expression ratios and corresponding metabolic flux ratios, although the data sets could be transformed into qualitative ordinal representations. One solution to this shortcoming is to profile the transcription in a set of diverse strains to identify those genes that correlate with experimentally determined robust parameters such as product formation (7).
Gene fragment microarrays were used to correlate gene expression with lovastatin production in engineered strains of Aspergillus terreus carrying mutations in genes that directly affect the production of this metabolite. The results from the association were used to identify the triggered promoters that correlated with lovastatin production and assisting the construction of strains with enhanced lovastatin production (7). This metabolic engineering approach demonstrated the implementation of -omics approaches to engineer the phenotype, even in strains whose genome has not been completely sequenced.
While gene fragment microarray has inherent limitations, associating transcription with the synthesis of just two metabolites narrows the realm of fundamental understanding of metabolic processes that lead to just these metabolites. Metabolic fluxes provide useful information regarding the degree of pathway utilization, which is the result of all the regulation machinery. Therefore, analyzing the flux profile from a global control perspective provides the underlying principles behind the changes brought about by a perturbation and will lead to more accurate identification of the “rate-limiting step” in the synthesis of a desired metabolite or the regulatory events that lead to by-product formation.
The presence of multiple events that control the rate of metabolic pathways (particularly of conserved pathways) often contributes to the inherent complexity (28). In light of this feature of the metabolic network structure, one possible approach to overcome regulatory effects is to uncouple metabolism from regulation by deleting global regulators. A recent example that implemented this approach was deleting the csrA gene in E. coli (206). The product of this gene, carbon storage regulator protein, binds several species of mRNA molecules responsible for carbon metabolism, specifically repressing pckA and pps (encoding phosphoenolpyruvate carboxykinase and phosphoenolpyruvate synthase), thus inhibiting the formation of phosphoenolpyruvate, an essential precursor of aromatic amino acid biosynthesis. Not surprisingly, an independent study reported increased titers for phenylalanine in E. coli strains in which the pps gene was overexpressed due to increased phosphoenolpyruvate availability (227). Similarly, disrupting three repressors of galactose uptake (Gal6p, Gal80p, and Mig1p) in S. cerevisiae increased galactose consumption (161).
Yet another example of manipulating global regulators to achieve a metabolic goal is the construction of an artificial feedback loop using acetylphosphate as the signal molecule (53). Using NtrC as the sensor and the regulator in the absence of its natural sensing protein, NtrB, to sense inefficient glucose metabolism, the expression of the rate-controlling idi and pps genes was controlled under the supervision of NtrC. This closed-loop dynamic metabolic controller was used to enhance lycopene synthesis in E. coli.
Protein-RNA Fusions for In Vitro Metabolic Engineering
More recently, functional protein chips that have the ability to carry out enzymatic reactions have been used to optimize metabolic pathway utilization (97). The functional protein chips take advantage of the catalytic ability of mRNA-protein chimeric molecules. The mRNA end of the chimeric molecule hybridizes to the homologous DNA that is immobilized on a matrix, while its corresponding functional protein end catalyzes the reaction. The chimeric molecule was prepared by ligating DNA to the mRNA and translated in vitro and gel purified. The two key issues are the specificity of hybridization of the chimeric mRNA with immobilized DNA and the retention of the catalytic ability of the protein after in vitro translation. Although these potential drawbacks were tested using luciferase as a model enzyme with acceptable results, the flexibility of this tool remains to be tested for other combinations of mRNA-protein fusions. The authors used this “metabolism-on-a-chip” system to conclude that the optimal ratio of the two branch point enzymes phosphoglucomutase and trehalose-6-phosphate synthase, be 1:5 for maximal trehalose synthesis.
In a related development, another RNA-based application to activate or repress genes, a riboregulatory system, was developed in which a series of RNA switches very precisely control gene expression by binding to the ribosome-binding site (90). These riboswitches, which take advantage of the regulatory role of RNA, aid in designing alternative gene circuits. The readers are referred to Liao for a deeper introduction to these two new developments in pathway engineering (127).
RANK OF METABOLOME IN REGULATORY HIERARCHY
Cells control the concentrations of their intracellular metabolites very rigidly. There is normally a very low tolerance for variations in metabolite concentrations for a given physiological state. Stated conversely, a change in the concentration of a metabolite beyond the tolerance level induces a change in cell physiology. Therefore, similar to the transcriptome and proteome, the metabolome also presents a snapshot of the physiological state of the cell, and measuring changes in the concentrations of intracellular metabolites would reveal an aspect of regulation (such as allosteric inhibition/activation, metabolite-DNA binding, etc.) which cannot be studied by any other approaches described. Metabolome profiling also presents a more complete representation of metabolism by defining the thermodynamic equilibrium of a reaction. Therefore, metabolite profiling is now considered an important part of systems biology, offering a complementary role to genomics and proteomics (214, 215, 233). However, this field is still in its infancy, mostly due to the lack of analytical techniques.
In comparison to more than 6,000 protein-coding genes in S. cerevisiae (63) and more than 4,200 in E. coli (19), there are only about 600 metabolites in S. cerevisiae (158) and E. coli (177), although plants have a very large number of metabolites. Metabolic control analysis established that changes in the enzyme concentrations in vivo have a greater impact on the concentration of metabolites than on metabolic fluxes (53), a concept that has recently been verified experimentally (171).
The goal of any metabolome experiment is to quantify the level of all intracellular metabolites in a cell, tissue, or organism. This is currently not possible due to the lack of a robust, automated, reproducible analytical technique. The popular technology to identify and quantify metabolites is to use a combination of gas chromatography and mass spectrometry or liquid chromatography and mass spectrometry. Nonvolatile, polar metabolites are derivatized to enable separation in the gas chromatography column. In the same study, changes in the metabolic profile in response to a silent mutation were compared to those in response to a mutation of a characterized gene, and probable functions were assigned to the silent genes (171).
A comprehensive metabolome profile is required to detect patterns of change, but it is not known a priori the mutations of which characterized gene(s) to be used for comparison. There are two major hurdles in developing high-throughput metabolite measurement techniques: rapid turnover (on the order of 2 to 3 s) of metabolites, requiring robust sample extracting techniques that give reproducible results, and the development of analytical techniques. Currently, it is possible to accurately quantify and identify about 50 amino and organic acids simultaneously (224, 225) and further research is required to reliably quantify the comprehensive metabolome, although several hundred can be monitored qualitatively. A comprehensive review on metabolome analysis has been published recently which describes the current state of the field in addition to providing a summary on the various analytical methods currently used and forecasting the future direction of metabolomics (225).
CONCLUSIONS
We are currently witnessing a transition in the approach to microbial physiology from traditional macroscopic procedures to a molecular approach. Research in the field of metabolic engineering is primarily driven by end use and the quest for fundamental understanding. Such research, characterized by Pasteur as “use-inspired basic research,” drives technology and vice versa (198). Truly comprehensive approaches to metabolic engineering lie at the union of pure basic research and use-inspired basic research. Since such comprehensive approaches seem to be the future trend in studying physiology, it is necessary to establish a common platform to enable effective information exchange between different research groups.
The generation of high-throughput global data that will be used in the integrated methodologies will prove to be an expensive venture and will undeniably require extensive knowledge about computer modeling, physiology, and metabolism as well as excellent technical skills in measuring gene and protein expression and metabolic flux analysis. While the current trend of generating of high-throughput data is increasingly popular, we believe that extremely useful information could still be extracted from the data that are already generated, as illustrated recently (41). Such a multidisciplinary approach paves the way to establishing strong symbiotic research collaborations. On the whole, we can expect great advances in the field of metabolic engineering as a consequence of elucidating the several previously unknown regulatory properties using such holistic approaches.
Acknowledgments
We are grateful to the anonymous reviewers for their critical appraisal of the manuscript and excellent suggestions that gave the manuscript its final shape.
REFERENCES
- 1.Aebersold, R., and M. Mann. 2003. Mass spectrometry-based proteomics. Nature 422:198-207. [DOI] [PubMed] [Google Scholar]
- 2.Agarwal, A. K., P. D. Rogers, S. R. Baerson, M. R. Jacob, K. S. Barker, J. D. Cleary, L. A. Walker, D. G. Nagle, and A. M. Clark. 2003. Genome-wide expression profiling of the response to polyene, pyrimidine, azole, and echinocandin antifungal agents in Saccharomyces cerevisiae. J. Biol. Chem. 278:34998-35015. [DOI] [PubMed] [Google Scholar]
- 3.Anderson, L., and J. Seilhamer. 1997. A comparison of selected mRNA and protein abundances in human liver. Electrophoresis 18:533-537. [DOI] [PubMed] [Google Scholar]
- 4.Aravin, A. A., M. Lagos-Quintana, A. Yalcin, M. Zavolan, D. Marks, B. Snyder, T. Gaasterland, J. Meyer, and T. Tuschl. 2003. The small RNA profile during Drosophila melanogaster development. Dev. Cell 5:337-350. [DOI] [PubMed] [Google Scholar]
- 5.Arfin, S. M., A. D. Long, E. T. Ito, L. Tolleri, M. M. Riehle, E. S. Paegle, and G. W. Hatfield. 2000. Global gene expression profiling in Escherichia coli K12. The effects of integration host factor. J. Biol. Chem. 275:29672-29684. [DOI] [PubMed] [Google Scholar]
- 6.Arthington-Skaggs, B. A., D. N. Crowell, H. Yang, S. L. Sturley, and M. Bard. 1996. Positive and negative regulation of a sterol biosynthetic gene (ERG3) in the postsqualene portion of the yeast ergosterol pathway. FEBS Lett. 392:161-165. [DOI] [PubMed] [Google Scholar]
- 7.Askenazi, M., E. M. Driggers, D. A. Holtzman, T. C. Norman, S. Iverson, D. P. Zimmer, M. E. Boers, P. R. Blomquist, E. J. Martinez, A. W. Monreal, T. P. Feibelman, M. E. Mayorga, M. E. Maxon, K. Sykes, J. V. Tobin, E. Cordero, S. R. Salama, J. Trueheart, J. C. Royer, and K. T. Madden. 2003. Integrating transcriptional and metabolite profiles to direct the engineering of lovastatin-producing fungal strains. Nat. Biotechnol. 21:150-156. [DOI] [PubMed] [Google Scholar]
- 8.Bailey, J. E. 1991. Toward a science of metabolic engineering. Science 252:1668-1675. [DOI] [PubMed] [Google Scholar]
- 9.Baldi, P., and A. D. Long. 2001. A Bayesian framework for the analysis of microarray expression data: regularized t-test and statistical inferences of gene changes. Bioinformatics 17:509-519. [DOI] [PubMed] [Google Scholar]
- 10.Bammert, G. F., and J. M. Fostel. 2000. Genome-wide expression patterns in Saccharomyces cerevisiae: comparison of drug treatments and genetic alterations affecting biosynthesis of ergosterol. Antimicrob. Agents Chemother. 44:1255-1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bar-Joseph, Z., G. K. Gerber, T. I. Lee, N. J. Rinaldi, J. Y. Yoo, F. Robert, D. B. Gordon, E. Fraenkel, T. S. Jaakkola, R. A. Young, and D. K. Gifford. 2003. Computational discovery of gene modules and regulatory networks. Nat. Biotechnol. 21:1337-1342. [DOI] [PubMed] [Google Scholar]
- 12.Bardwell, J. C., P. Regnier, S. M. Chen, Y. Nakamura, M. Grunberg-Manago, and D. L. Court. 1989. Autoregulation of RNase III operon by mRNA processing. EMBO J. 8:3401-3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barker, K. S., M. M. Pearson, and P. D. Rogers. 2003. Identification of genes differentially expressed in association with reduced azole susceptibility in Saccharomyces cerevisiae. J. Antimicrob. Chemother. 51:1131-1140. [DOI] [PubMed] [Google Scholar]
- 14.Berger, J. A., S. Hautaniemi, A. K. Jarvinen, H. Edgren, S. K. Mitra, and J. Astola. 2004. Optimized LOWESS normalization parameter selection for DNA microarray data. BMC Bioinformatics 5:194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bernstein, J. A., A. B. Khodursky, P. H. Lin, S. Lin-Chao, and S. N. Cohen. 2002. Global analysis of mRNA decay and abundance in Escherichia coli at single-gene resolution using two-color fluorescent DNA microarrays. Proc. Natl. Acad. Sci. USA 99:9697-9702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bernstein, J. A., P. H. Lin, S. N. Cohen, and S. Lin-Chao. 2004. Global analysis of Escherichia coli RNA degradosome function using DNA microarrays. Proc. Natl. Acad. Sci. USA 101:2758-2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blagoev, B., I. Kratchmarova, S. E. Ong, M. Nielsen, L. J. Foster, and M. Mann. 2003. A proteomics strategy to elucidate functional protein-protein interactions applied to EGF signaling. Nat. Biotechnol. 21:315-318. [DOI] [PubMed] [Google Scholar]
- 18.Blanchard, A. P., and L. Hood. 1996. Sequence to array: probing the genome's secrets. Nat. Biotechnol. 14:1649. [DOI] [PubMed] [Google Scholar]
- 19.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 20.Bodovitz, S., and T. Joos. 2004. The proteomics bottleneck: strategies for preliminary validation of potential biomarkers and drug targets. Trends Biotechnol. 22:4-7. [DOI] [PubMed] [Google Scholar]
- 21.Bourdeau, V., G. Ferbeyre, M. Pageau, B. Paquin, and R. Cedergren. 1999. The distribution of RNA motifs in natural sequences. Nucleic Acids Res. 27:4457-4467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brazhnik, P., A. de la Fuente, and P. Mendes. 2002. Gene networks: how to put the function in genomics. Trends Biotechnol. 20:467-472. [DOI] [PubMed] [Google Scholar]
- 23.Bro, C., B. Regenberg, G. Lagniel, J. Labarre, M. Montero-Lomeli, and J. Nielsen. 2003. Transcriptional, proteomic, and metabolic responses to lithium in galactose-grown yeast cells. J. Biol. Chem. 278:32141-32149. [DOI] [PubMed] [Google Scholar]
- 24.Brown, M. P., W. N. Grundy, D. Lin, N. Cristianini, C. W. Sugnet, T. S. Furey, M. Ares, Jr., and D. Haussler. 2000. Knowledge-based analysis of microarray gene expression data by using support vector machines. Proc. Natl. Acad. Sci. USA 97:262-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cahill, D. J., and E. Nordhoff. 2003. Protein arrays and their role in proteomics. Adv. Biochem. Eng Biotechnol. 83:177-187. [DOI] [PubMed] [Google Scholar]
- 26.Carlson, R., and F. Srienc. 2004. Fundamental Escherichia coli biochemical pathways for biomass and energy production: creation of overall flux states. Biotechnol. Bioeng. 86:149-162. [DOI] [PubMed] [Google Scholar]
- 27.Carlson, R., and F. Srienc. 2004. Fundamental Escherichia coli biochemical pathways for biomass and energy production: identification of reactions. Biotechnol. Bioeng. 85:1-19. [DOI] [PubMed] [Google Scholar]
- 28.Cascante, M., L. G. Boros, B. Comin-Anduix, P. de Atauri, J. J. Centelles, and P. W. Lee. 2002. Metabolic control analysis in drug discovery and disease. Nat. Biotechnol. 20:243-249. [DOI] [PubMed] [Google Scholar]
- 29.Causier, B. 2004. Studying the interactome with the yeast two-hybrid system and mass spectrometry. Mass Spectrom. Rev. 23:350-367. [DOI] [PubMed] [Google Scholar]
- 30.Chauvet, G. A. 1993. Hierarchical functional organization of formal biological systems: a dynamical approach. I. The increase of complexity by self-association increases the domain of stability of a biological system. Phil. Trans. R. Soc. Lond. B Biol. Sci. 339:425-444. [DOI] [PubMed] [Google Scholar]
- 31.Cho, R. J., M. J. Campbell, E. A. Winzeler, L. Steinmetz, A. Conway, L. Wodicka, T. G. Wolfsberg, A. E. Gabrielian, D. Landsman, D. J. Lockhart, and R. W. Davis. 1998. A genome-wide transcriptional analysis of the mitotic cell cycle. Mol. Cell. 2:65-73. [DOI] [PubMed] [Google Scholar]
- 32.Clarke, P. A., R. T. Poele, and P. Workman. 2004. Gene expression microarray technologies in the development of new therapeutic agents. Eur. J. Cancer. 40:2560-2591. [DOI] [PubMed] [Google Scholar]
- 33.Covell, D. G., A. Wallqvist, A. A. Rabow, and N. Thanki. 2003. Molecular classification of cancer: unsupervised self-organizing map analysis of gene expression microarray data. Mol. Cancer Ther. 2:317-332. [PubMed] [Google Scholar]
- 34.Covert, M. W., E. M. Knight, J. L. Reed, M. J. Herrgard, and B. O. Palsson. 2004. Integrating high-throughput and computational data elucidates bacterial networks. Nature 429:92-96. [DOI] [PubMed] [Google Scholar]
- 35.Covert, M. W., C. H. Schilling, and B. Palsson. 2001. Regulation of gene expression in flux balance models of metabolism. J. Theor. Biol. 213:73-88. [DOI] [PubMed] [Google Scholar]
- 36.Craig, N. L., and H. A. Nash. 1984. E. coli integration host factor binds to specific sites in DNA. Cell 39:707-716. [DOI] [PubMed] [Google Scholar]
- 37.Daran-Lapujade, P., M. L. Jansen, J. M. Daran, G. W. van, J. H. de Winde, and J. T. Pronk. 2004. Role of transcriptional regulation in controlling fluxes in central carbon metabolism of Saccharomyces cerevisiae. A chemostat culture study. J. Biol. Chem. 279:9125-9138. [DOI] [PubMed] [Google Scholar]
- 38.Davis, T. N. 2004. Protein localization in proteomics. Curr. Opin. Chem. Biol. 8:49-53. [DOI] [PubMed] [Google Scholar]
- 39.de Lichtenberg, U., L. J. Jensen, S. Brunak, and P. Bork. 2005. Dynamic complex formation during the yeast cell cycle. Science 307:724-727. [DOI] [PubMed] [Google Scholar]
- 40.De Backer, Marianne D., T. Ilyina, X. J. Ma, S. Vandoninck, W. H. Luyten, and B. H. Vanden. 2001. Genomic profiling of the response of Candida albicans to itraconazole treatment using a DNA microarray. Antimicrob. Agents Chemother. 45:1660-1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.De Backer, Marianne D., R. L. Thurmond, A. A. Carmen, and W. H. Luyten. 2002. Gene-expression-based responses to drug treatment. Drug News Perspect. 15:155-165. [DOI] [PubMed] [Google Scholar]
- 42.DeLuna, A., A. Avendano, L. Riego, and A. Gonzalez. 2001. NADP-glutamate dehydrogenase isoenzymes of Saccharomyces cerevisiae. Purification, kinetic properties, and physiological roles. J. Biol. Chem. 276:43775-43783. [DOI] [PubMed] [Google Scholar]
- 43.Dennis, P. P., M. Ehrenberg, and H. Bremer. 2004. Control of rRNA synthesis in Escherichia coli: a systems biology approach. Microbiol. Mol. Biol. Rev. 68:639-668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.DeRisi, J. L., V. R. Iyer, and P. O. Brown. 1997. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science 278:680-686. [DOI] [PubMed] [Google Scholar]
- 45.Ditto, M. D., D. Roberts, and R. A. Weisberg. 1994. Growth phase variation of integration host factor level in Escherichia coli. J. Bacteriol. 176:3738-3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Edwards, J. S., R. U. Ibarra, and B. O. Palsson. 2001. In silico predictions of Escherichia coli metabolic capabilities are consistent with experimental data. Nat. Biotechnol. 19:125-130. [DOI] [PubMed] [Google Scholar]
- 47.Edwards, J. S., and B. O. Palsson. 1999. Systems properties of the Haemophilus influenzae Rd metabolic genotype. J. Biol. Chem. 274:17410-17416. [DOI] [PubMed] [Google Scholar]
- 48.Eickhoff, B., B. Korn, M. Schick, A. Poustka, and J. van der Bosch. 1999. Normalization of array hybridization experiments in differential gene expression analysis. Nucleic Acids Res. 27:e33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Eisen, M. B., and P. O. Brown. 1999. DNA arrays for analysis of gene expression. Methods Enzymol. 303:179-205. [DOI] [PubMed] [Google Scholar]
- 50.Eisen, M. B., P. T. Spellman, P. O. Brown, and D. Botstein. 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 95:14863-14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ellenberger, T., and A. Landy. 1997. A good turn for DNA: the structure of integration host factor bound to DNA. Structure 5:153-157. [DOI] [PubMed] [Google Scholar]
- 52.Famili, I., J. Forster, J. Nielsen, and B. O. Palsson. 2003. Saccharomyces cerevisiae phenotypes can be predicted by using constraint-based analysis of a genome-scale reconstructed metabolic network. Proc. Natl. Acad. Sci. USA 100:13134-13139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Farmer, W. R., and J. C. Liao. 2000. Improving lycopene production in Escherichia coli by engineering metabolic control. Nat. Biotechnol. 18:533-537. [DOI] [PubMed] [Google Scholar]
- 54.Fellenberg, K., N. C. Hauser, B. Brors, A. Neutzner, J. D. Hoheisel, and M. Vingron. 2001. Correspondence analysis applied to microarray data. Proc. Natl. Acad. Sci. USA 98:10781-10786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Forster, J., I. Famili, P. Fu, B. O. Palsson, and J. Nielsen. 2003. Genome-scale reconstruction of the Saccharomyces cerevisiae metabolic network. Genome Res. 13:244-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Forster, J., I. Famili, B. O. Palsson, and J. Nielsen. 2003. Large-scale evaluation of in silico gene deletions in Saccharomyces cerevisiae. OMICS 7:193-202. [DOI] [PubMed] [Google Scholar]
- 57.Frischmeyer, P. A., A. van Hoof, K. O’Donnell, A. L. Guerrerio, R. Parker, and H. C. Dietz. 2002. An mRNA surveillance mechanism that eliminates transcripts lacking termination codons. Science 295:2258-2261. [DOI] [PubMed] [Google Scholar]
- 58.Gavin, A. C., M. Bosche, R. Krause, P. Grandi, M. Marzioch, A. Bauer, J. Schultz, J. M. Rick, A. M. Michon, C. M. Cruciat, M. Remor, C. Hofert, M. Schelder, M. Brajenovic, H. Ruffner, A. Merino, K. Klein, M. Hudak, D. Dickson, T. Rudi, V. Gnau, A. Bauch, S. Bastuck, B. Huhse, C. Leutwein, M. A. Heurtier, R. R. Copley, A. Edelmann, E. Querfurth, V. Rybin, G. Drewes, M. Raida, T. Bouwmeester, P. Bork, B. Seraphin, B. Kuster, G. Neubauer, and G. Superti-Furga. 2002. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415:141-147. [DOI] [PubMed] [Google Scholar]
- 59.Geissmann, T. A., and D. Touati. 2004. Hfq, a new chaperoning role: binding to messenger RNA determines access for small RNA regulator 6. EMBO J. 23:396-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Georgellis, D., O. Kwon, and E. C. Lin. 2001. Quinones as the redox signal for the arc two-component system of bacteria. Science 292:2314-2316. [DOI] [PubMed] [Google Scholar]
- 61.Ghaemmaghami, S., W. K. Huh, K. Bower, R. W. Howson, A. Belle, N. Dephoure, E. K. O'shea, and J. S. Weissman. 2003. Global analysis of protein expression in yeast. Nature 425:737-741. [DOI] [PubMed] [Google Scholar]
- 62.Gmuender, H., K. Kuratli, P. K. Di, C. P. Gray, W. Keck, and S. Evers. 2001. Gene expression changes triggered by exposure of Haemophilus influenzae to novobiocin or ciprofloxacin: combined transcription and translation analysis. Genome Res. 11:28-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Goffeau, A., B. G. Barrell, H. Bussey, R. W. Davis, B. Dujon, H. Feldmann, F. Galibert, J. D. Hoheisel, C. Jacq, M. Johnston, E. J. Louis, H. W. Mewes, Y. Murakami, P. Philippsen, H. Tettelin, and S. G. Oliver. 1996. Life with 6000 genes. Science 274:546-567. [DOI] [PubMed] [Google Scholar]
- 64.Goodman, S. D., N. J. Velten, Q. Gao, S. Robinson, and A. M. Segall. 1999. In vitro selection of integration host factor binding sites. J. Bacteriol. 181:3246-3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Goodrich, J. A., M. L. Schwartz, and W. R. McClure. 1990. Searching for and predicting the activity of sites for DNA binding proteins: compilation and analysis of the binding sites for Escherichia coli integration host factor (IHF). Nucleic Acids Res. 18:4993-5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gottesman, S. 2004. The small RNA regulators of Escherichia coli: roles and mechanisms. Annu. Rev. Microbiol. 58:303-328. [DOI] [PubMed] [Google Scholar]
- 67.Gottesman, S., G. Storz, C. Rosenow, N. Majdalani, F. Repoila, and K. M. Wassarman. 2001. Small RNA regulators of translation: mechanisms of action and approaches for identifying new small RNAs. Cold Spring Harb. Symp. Quant. Biol. 66:353-362. [DOI] [PubMed] [Google Scholar]
- 68.Grigull, J., S. Mnaimneh, J. Pootoolal, M. D. Robinson, and T. R. Hughes. 2004. Genome-wide analysis of mRNA stability using transcription inhibitors and microarrays reveals posttranscriptional control of ribosome biogenesis factors. Mol. Cell. Biol. 24:5534-5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gutierrez, R. A., R. M. Ewing, J. M. Cherry, and P. J. Green. 2002. Identification of unstable transcripts in Arabidopsis by cDNA microarray analysis: rapid decay is associated with a group of touch- and specific clock-controlled genes. Proc. Natl. Acad. Sci. USA 99:11513-11518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gygi, S. P., G. L. Corthals, Y. Zhang, Y. Rochon, and R. Aebersold. 2000. Evaluation of two-dimensional gel electrophoresis-based proteome analysis technology. Proc. Natl. Acad. Sci. USA 97:9390-9395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gygi, S. P., Y. Rochon, B. R. Franza, and R. Aebersold. 1999. Correlation between protein and mRNA abundance in yeast. Mol. Cell. Biol. 19:1720-1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Haab, B. B., M. J. Dunham, and P. O. Brown. 2001. Protein microarrays for highly parallel detection and quantitation of specific proteins and antibodies in complex solutions. Genome Biol. 2:RESEARCH0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.He, F., X. Li, P. Spatrick, R. Casillo, S. Dong, and A. Jacobson. 2003. Genome-wide analysis of mRNAs regulated by the nonsense-mediated and 5′ to 3′ mRNA decay pathways in yeast. Mol. Cell 12:1439-1452. [DOI] [PubMed] [Google Scholar]
- 74.Hereford, L. M., M. A. Osley, T. R. Ludwig, and C. S. McLaughlin. 1981. Cell-cycle regulation of yeast histone mRNA. Cell 24:367-375. [DOI] [PubMed] [Google Scholar]
- 75.Hieronymus, H., and P. A. Silver. 2003. Genome-wide analysis of RNA-protein interactions illustrates specificity of the mRNA export machinery. Nat. Genet. 33:155-161. [DOI] [PubMed] [Google Scholar]
- 76.Hieronymus, H., and P. A. Silver. 2004. A systems view of mRNP biology. Genes Dev. 18:2845-2860. [DOI] [PubMed] [Google Scholar]
- 77.Ho, Y., A. Gruhler, A. Heilbut, G. D. Bader, L. Moore, S. L. Adams, A. Millar, P. Taylor, K. Bennett, K. Boutilier, L. Yang, C. Wolting, I. Donaldson, S. Schandorff, J. Shewnarane, M. Vo, J. Taggart, M. Goudreault, B. Muskat, C. Alfarano, D. Dewar, Z. Lin, K. Michalickova, A. R. Willems, H. Sassi, P. A. Nielsen, K. J. Rasmussen, J. R. Andersen, L. E. Johansen, L. H. Hansen, H. Jespersen, A. Podtelejnikov, E. Nielsen, J. Crawford, V. Poulsen, B. D. Sorensen, J. Matthiesen, R. C. Hendrickson, F. Gleeson, T. Pawson, M. F. Moran, D. Durocher, M. Mann, C. W. Hogue, D. Figeys, and M. Tyers. 2002. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415:180-183. [DOI] [PubMed] [Google Scholar]
- 78.Hoch, J. A., and T. J. Silhavy. 1995. Two-component signal transduction. ASM Press, Washington, D.C.
- 79.Hood, L. 2002. A personal view of molecular technology and how it has changed biology. J. Proteome Res. 1:399-409. [DOI] [PubMed] [Google Scholar]
- 80.Hood, L. 2003. Systems biology: integrating technology, biology, and computation. Mech. Ageing Dev. 124:9-16. [DOI] [PubMed] [Google Scholar]
- 81.Hu, C. D., Y. Chinenov, and T. K. Kerppola. 2002. Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol. Cell 9:789-798. [DOI] [PubMed] [Google Scholar]
- 82.Hu, C. D., and T. K. Kerppola. 2003. Simultaneous visualization of multiple protein interactions in living cells using multicolor fluorescence complementation analysis. Nat. Biotechnol. 21:539-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hughes, T. R., M. Mao, A. R. Jones, J. Burchard, M. J. Marton, K. W. Shannon, S. M. Lefkowitz, M. Ziman, J. M. Schelter, M. R. Meyer, S. Kobayashi, C. Davis, H. Dai, Y. D. He, S. B. Stephaniants, G. Cavet, W. L. Walker, A. West, E. Coffey, D. D. Shoemaker, R. Stoughton, A. P. Blanchard, S. H. Friend, and P. S. Linsley. 2001. Expression profiling using microarrays fabricated by an ink-jet oligonucleotide synthesizer. Nat. Biotechnol. 19:342-347. [DOI] [PubMed] [Google Scholar]
- 84.Huh, W. K., J. V. Falvo, L. C. Gerke, A. S. Carroll, R. W. Howson, J. S. Weissman, and E. K. O'shea. 2003. Global analysis of protein localization in budding yeast. Nature 425:686-691. [DOI] [PubMed] [Google Scholar]
- 85.Hung, S. P., P. Baldi, and G. W. Hatfield. 2002. Global gene expression profiling in Escherichia coli K12. The effects of leucine-responsive regulatory protein. J. Biol. Chem. 277:40309-40323. [DOI] [PubMed] [Google Scholar]
- 86.Iberall, A. S. 1977. A field and circuit thermodynamics for integrative physiology. I. Introduction to the general notions. Am. J. Physiol. 233:R171-R180. [DOI] [PubMed] [Google Scholar]
- 87.Ideker, T., V. Thorsson, J. A. Ranish, R. Christmas, J. Buhler, J. K. Eng, R. Bumgarner, D. R. Goodlett, R. Aebersold, and L. Hood. 2001. Integrated genomic and proteomic analyses of a systematically perturbed metabolic network. Science 292:929-934. [DOI] [PubMed] [Google Scholar]
- 88.Ihmels, J., G. Friedlander, S. Bergmann, O. Sarig, Y. Ziv, and N. Barkai. 2002. Revealing modular organization in the yeast transcriptional network. Nat. Genet. 31:370-377. [DOI] [PubMed] [Google Scholar]
- 89.Ihmels, J., R. Levy, and N. Barkai. 2004. Principles of transcriptional control in the metabolic network of Saccharomyces cerevisiae. Nat. Biotechnol. 22:86-92. [DOI] [PubMed] [Google Scholar]
- 90.Inada, M., and C. Guthrie. 2004. Identification of Lhp1p-associated RNAs by microarray analysis in Saccharomyces cerevisiae reveals association with coding and noncoding RNAs. Proc. Natl. Acad. Sci. USA 101:434-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ishii, A., T. Oshima, T. Sato, K. Nakasone, H. Mori, and C. Kato. 2004. Analysis of hydrostatic pressure effects on transcription in Escherichia coli by DNA microarray procedure. Extremophiles 9:65-73. [DOI] [PubMed]
- 92.Ito, T., T. Chiba, and M. Yoshida. 2001. Exploring the protein interactome using comprehensive two-hybrid projects. Trends Biotechnol. 19:S23-S27. [DOI] [PubMed] [Google Scholar]
- 93.Iuchi, S., and E. C. Lin. 1988. arcA (dye), a global regulatory gene in Escherichia coli mediating repression of enzymes in aerobic pathways. Proc. Natl. Acad. Sci. USA 85:1888-1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Iyer, V. R., C. E. Horak, C. S. Scafe, D. Botstein, M. Snyder, and P. O. Brown. 2001. Genomic binding sites of the yeast cell-cycle transcription factors SBF and MBF. Nature 409:533-538. [DOI] [PubMed] [Google Scholar]
- 95.Jain, C., and J. G. Belasco. 1995. RNase E autoregulates its synthesis by controlling the degradation rate of its own mRNA in Escherichia coli: unusual sensitivity of the rne transcript to RNase E activity. Genes Dev. 9:84-96. [DOI] [PubMed] [Google Scholar]
- 96.Jeong, H., B. Tombor, R. Albert, Z. N. Oltvai, and A. L. Barabasi. 2000. The large-scale organization of metabolic networks. Nature 407:651-654. [DOI] [PubMed] [Google Scholar]
- 97.Jung, G. Y., and G. Stephanopoulos. 2004. A functional protein chip for pathway optimization and in vitro metabolic engineering. Science 304:428-431. [DOI] [PubMed] [Google Scholar]
- 98.Kafri, R., A. Bar-Even, and Y. Pilpel. 2005. Transcription control reprogramming in genetic backup circuits. Nat. Genet. 37:295-299. [DOI] [PubMed] [Google Scholar]
- 99.Kaniak, A., Z. Xue, D. Macool, J. H. Kim, and M. Johnston. 2004. Regulatory network connecting two glucose signal transduction pathways in Saccharomyces cerevisiae. Eukaryot. Cell 3:221-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kawamoto, H., T. Morita, A. Shimizu, T. Inada, and H. Aiba. 2005. Implication of membrane localization of target mRNA in the action of a small RNA: mechanism of post-transcriptional regulation of glucose transporter in Escherichia coli. Genes Dev. 19:328-338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Keene, J. D., and S. A. Tenenbaum. 2002. Eukaryotic mRNPs may represent posttranscriptional operons. Mol. Cell. 9:1161-1167. [DOI] [PubMed] [Google Scholar]
- 102.Kell, D. B. 2002. Genotype-phenotype mapping: genes as computer programs. Trends Genet. 18:555-559. [DOI] [PubMed] [Google Scholar]
- 103.Kennedy, M. A., R. Barbuch, and M. Bard. 1999. Transcriptional regulation of the squalene synthase gene (ERG9) in the yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta. 1445:110-122. [DOI] [PubMed] [Google Scholar]
- 104.Kerr, M. K. 2003. Linear models for microarray data analysis: hidden similarities and differences. J. Comput. Biol. 10:891-901. [DOI] [PubMed] [Google Scholar]
- 105.Kerr, M. K., and G. A. Churchill. 2001. Experimental design for gene expression microarrays. Biostatistics 2:183-201. [DOI] [PubMed] [Google Scholar]
- 106.Kerr, M. K., M. Martin, and G. A. Churchill. 2000. Analysis of variance for gene expression microarray data. J. Comput. Biol. 7:819-837. [DOI] [PubMed] [Google Scholar]
- 107.Khodursky, A. B., and J. A. Bernstein. 2003. Life after transcription-revisiting the fate of messenger RNA. Trends Genet. 19:113-115. [DOI] [PubMed] [Google Scholar]
- 108.Khodursky, A. B., B. J. Peter, N. R. Cozzarelli, D. Botstein, P. O. Brown, and C. Yanofsky. 2000. DNA microarray analysis of gene expression in response to physiological and genetic changes that affect tryptophan metabolism in Escherichia coli. Proc. Natl. Acad. Sci. USA 97:12170-12175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kitano, H. 2002. Computational systems biology. Nature 420:206-210. [DOI] [PubMed] [Google Scholar]
- 110.Kjeldgaard, N. O., O. Maaloe, and M. Schaechter. 1958. The transition between different physiological states during balanced growth of Salmonella typhimurium. J. Gen. Microbiol. 19:607-616. [DOI] [PubMed] [Google Scholar]
- 111.Kobayashi, K., M. Ogura, H. Yamaguchi, K. Yoshida, N. Ogasawara, T. Tanaka, and Y. Fujita. 2001. Comprehensive DNA microarray analysis of Bacillus subtilis two-component regulatory systems. J. Bacteriol. 183:7365-7370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Korke, R., A. Rink, T. K. Seow, M. C. Chung, C. W. Beattie, and W. S. Hu. 2002. Genomic and proteomic perspectives in cell culture engineering. J. Biotechnol. 94:73-92. [DOI] [PubMed] [Google Scholar]
- 113.Kromer, J. O., O. Sorgenfrei, K. Klopprogge, E. Heinzle, and C. Wittmann. 2004. In-depth profiling of lysine-producing Corynebacterium glutamicum by combined analysis of the transcriptome, metabolome, and fluxome. J. Bacteriol. 186:1769-1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kumar, A., S. Agarwal, J. A. Heyman, S. Matson, M. Heidtman, S. Piccirillo, L. Umansky, A. Drawid, R. Jansen, Y. Liu, K. H. Cheung, P. Miller, M. Gerstein, G. S. Roeder, and M. Snyder. 2002. Subcellular localization of the yeast proteome. Genes Dev. 16:707-719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kuo, W. P., E. Y. Kim, J. Trimarchi, T. K. Jenssen, S. A. Vinterbo, and L. Ohno-Machado. 2004. A primer on gene expression and microarrays for machine learning researchers. J. Biomed. Inform. 37:293-303. [DOI] [PubMed] [Google Scholar]
- 116.Kurokawa, Y., R. Matoba, H. Nagano, M. Sakon, I. Takemasa, S. Nakamori, K. Dono, K. Umeshita, N. Ueno, S. Ishii, K. Kato, and M. Monden. 2004. Molecular prediction of response to 5-fluorouracil and interferon-alpha combination chemotherapy in advanced hepatocellular carcinoma. Clin. Cancer Res. 10:6029-6038. [DOI] [PubMed] [Google Scholar]
- 117.Kushner, S. R. 2002. mRNA decay in Escherichia coli comes of age. J. Bacteriol. 184:4658-4665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lander, E. S. 1999. Array of hope. Nat. Genet. 21:3-4. [DOI] [PubMed] [Google Scholar]
- 119.Launhardt, H., A. Hinnen, and T. Munder. 1998. Drug-induced phenotypes provide a tool for the functional analysis of yeast genes. Yeast 14:935-942. [DOI] [PubMed] [Google Scholar]
- 120.Lee, M. L., F. C. Kuo, G. A. Whitmore, and J. Sklar. 2000. Importance of replication in microarray gene expression studies: statistical methods and evidence from repetitive cDNA hybridizations. Proc. Natl. Acad. Sci. USA 97:9834-9839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Lee, T. I., N. J. Rinaldi, F. Robert, D. T. Odom, Z. Bar-Joseph, G. K. Gerber, N. M. Hannett, C. T. Harbison, C. M. Thompson, I. Simon, J. Zeitlinger, E. G. Jennings, H. L. Murray, D. B. Gordon, B. Ren, J. J. Wyrick, J. B. Tagne, T. L. Volkert, E. Fraenkel, D. K. Gifford, and R. A. Young. 2002. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science 298:799-804. [DOI] [PubMed] [Google Scholar]
- 122.Lee, Y., and C. K. Lee. 2003. Classification of multiple cancer types by multicategory support vector machines using gene expression data. Bioinformatics 19:1132-1139. [DOI] [PubMed] [Google Scholar]
- 123.Lehnen, D., C. Blumer, T. Polen, B. Wackwitz, V. F. Wendisch, and G. Unden. 2002. LrhA as a new transcriptional key regulator of flagella, motility and chemotaxis genes in Escherichia coli. Mol. Microbiol. 45:521-532. [DOI] [PubMed] [Google Scholar]
- 124.Lenz, D. H., K. C. Mok, B. N. Lilley, R. V. Kulkarni, N. S. Wingreen, and B. L. Bassler. 2004. The small RNA chaperone Hfq and multiple small RNAs control quorum sensing in Vibrio harveyi and Vibrio cholerae. Cell 118:69-82. [DOI] [PubMed] [Google Scholar]
- 125.Li, L., and J. Kaplan. 1996. Characterization of yeast methyl sterol oxidase (ERG25) and identification of a human homologue. J. Biol. Chem. 271:16927-16933. [DOI] [PubMed] [Google Scholar]
- 126.Li, S., C. M. Armstrong, N. Bertin, H. Ge, S. Milstein, M. Boxem, P. O. Vidalain, J. D. Han, A. Chesneau, T. Hao, D. S. Goldberg, N. Li, M. Martinez, J. F. Rual, P. Lamesch, L. Xu, M. Tewari, S. L. Wong, L. V. Zhang, G. F. Berriz, L. Jacotot, P. Vaglio, J. Reboul, T. Hirozane-Kishikawa, Q. Li, H. W. Gabel, A. Elewa, B. Baumgartner, D. J. Rose, H. Yu, S. Bosak, R. Sequerra, A. Fraser, S. E. Mango, W. M. Saxton, S. Strome, H. S. Van Den, F. Piano, J. Vandenhaute, C. Sardet, M. Gerstein, L. Doucette-Stamm, K. C. Gunsalus, J. W. Harper, M. E. Cusick, F. P. Roth, D. E. Hill, and M. Vidal. 2004. A map of the interactome network of the metazoan C. elegans. Science 303:540-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Liao, J. C. 2004. Custom design of metabolism. Nat. Biotechnol. 22:823-824. [DOI] [PubMed] [Google Scholar]
- 128.Liao, J. C., R. Boscolo, Y. L. Yang, L. M. Tran, C. Sabatti, and V. P. Roychowdhury. 2003. Network component analysis: reconstruction of regulatory signals in biological systems. Proc. Natl. Acad. Sci. USA 100:15522-15527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Lieb, J. D., X. Liu, D. Botstein, and P. O. Brown. 2001. Promoter-specific binding of Rap1 revealed by genome-wide maps of protein-DNA association. Nat. Genet. 28:327-334. [DOI] [PubMed] [Google Scholar]
- 130.Liebermeister, W. 2002. Linear modes of gene expression determined by independent component analysis. Bioinformatics 18:51-60. [DOI] [PubMed] [Google Scholar]
- 131.Lipshutz, R. J., D. Morris, M. Chee, E. Hubbell, M. J. Kozal, N. Shah, N. Shen, R. Yang, and S. P. Fodor. 1995. Using oligonucleotide probe arrays to access genetic diversity. BioTechniques. 19:442-447. [PubMed] [Google Scholar]
- 132.Liu, X. S., D. L. Brutlag, and J. S. Liu. 2002. An algorithm for finding protein-DNA binding sites with applications to chromatin-immunoprecipitation microarray experiments. Nat. Biotechnol. 20:835-839. [DOI] [PubMed] [Google Scholar]
- 133.Lockhart, D. J., H. Dong, M. C. Byrne, M. T. Follettie, M. V. Gallo, M. S. Chee, M. Mittmann, C. Wang, M. Kobayashi, H. Horton, and E. L. Brown. 1996. Expression monitoring by hybridization to high-density oligonucleotide arrays. Nat. Biotechnol. 14:1675-1680. [DOI] [PubMed] [Google Scholar]
- 134.Long, A. D., H. J. Mangalam, B. Y. Chan, L. Tolleri, G. W. Hatfield, and P. Baldi. 2001. Improved statistical inference from DNA microarray data using analysis of variance and a Bayesian statistical framework. Analysis of global gene expression in Escherichia coli K12. J. Biol. Chem. 276:19937-19944. [DOI] [PubMed] [Google Scholar]
- 135.Lopez, D. S., I, M. Zhan, A. Lal, X. Yang, and M. Gorospe. 2004. Identification of a target RNA motif for RNA-binding protein HuR. Proc. Natl. Acad. Sci. USA 101:2987-2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Lou, X. J., M. Schena, F. T. Horrigan, R. M. Lawn, and R. W. Davis. 2001. Expression monitoring using cDNA microarrays. A general protocol. Methods Mol. Biol. 175:323-340. [DOI] [PubMed] [Google Scholar]
- 137.Luscombe, N. M., M. M. Babu, H. Yu, M. Snyder, S. A. Teichmann, and M. Gerstein. 2004. Genomic analysis of regulatory network dynamics reveals large topological changes. Nature 431:308-312. [DOI] [PubMed] [Google Scholar]
- 138.MacBeath, G. 2002. Protein microarrays and proteomics. Nat. Genet. 32(Suppl.):526-532. [DOI] [PubMed] [Google Scholar]
- 139.MacBeath, G., and S. L. Schreiber. 2000. Printing proteins as microarrays for high-throughput function determination. Science 289:1760-1763. [DOI] [PubMed] [Google Scholar]
- 140.Madan, B. M., and S. A. Teichmann. 2003. Evolution of transcription factors and the gene regulatory network in Escherichia coli. Nucleic Acids Res. 31:1234-1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Majdalani, N., S. Chen, J. Murrow, J. K. St, and S. Gottesman. 2001. Regulation of RpoS by a novel small RNA: the characterization of RprA. Mol. Microbiol. 39:1382-1394. [DOI] [PubMed] [Google Scholar]
- 142.Majdalani, N., D. Hernandez, and S. Gottesman. 2002. Regulation and mode of action of the second small RNA activator of RpoS translation, RprA. Mol. Microbiol. 46:813-826. [DOI] [PubMed] [Google Scholar]
- 143.Martin, V. J., D. J. Pitera, S. T. Withers, J. D. Newman, and J. D. Keasling. 2003. Engineering a mevalonate pathway in Escherichia coli for production of terpenoids. Nat. Biotechnol. 21:796-802. [DOI] [PubMed] [Google Scholar]
- 144.Martinez-Antonio, A., and J. Collado-Vides. 2003. Identifying global regulators in transcriptional regulatory networks in bacteria. Curr. Opin. Microbiol. 6:482-489. [DOI] [PubMed] [Google Scholar]
- 145.Mata, J., R. Lyne, G. Burns, and J. Bahler. 2002. The transcriptional program of meiosis and sporulation in fission yeast. Nat. Genet. 32:143-147. [DOI] [PubMed] [Google Scholar]
- 146.Matsubara, M., S. I. Kitaoka, S. I. Takeda, and T. Mizuno. 2000. Tuning of the porin expression under anaerobic growth conditions by his-to-Asp cross-phosphorelay through both the EnvZ-osmosensor and ArcB-anaerosensor in Escherichia coli. Genes Cells. 5:555-569. [DOI] [PubMed] [Google Scholar]
- 147.Mewes, H. W., C. Amid, R. Arnold, D. Frishman, U. Guldener, G. Mannhaupt, M. Munsterkotter, P. Pagel, N. Strack, V. Stumpflen, J. Warfsmann, and A. Ruepp. 2004. MIPS: analysis and annotation of proteins from whole genomes. Nucleic Acids Res. 32:D41-D44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Mizuno, T. 1997. Compilation of all genes encoding two-component phosphotransfer signal transducers in the genome of Escherichia coli. DNA Res. 4:161-168. [DOI] [PubMed] [Google Scholar]
- 149.Mohanty, B. K., and S. R. Kushner. 2003. Genomic analysis in Escherichia coli demonstrates differential roles for polynucleotide phosphorylase and RNase II in mRNA abundance and decay. Mol. Microbiol. 50:645-658. [DOI] [PubMed] [Google Scholar]
- 150.Mostertz, J., C. Scharf, M. Hecker, and G. Homuth. 2004. Transcriptome and proteome analysis of Bacillus subtilis gene expression in response to superoxide and peroxide stress. Microbiology 150:497-512. [DOI] [PubMed] [Google Scholar]
- 151.Murakawa, K., M. Tada, M. Takada, E. Tamoto, G. Shindoh, K. Teramoto, A. Matsunaga, K. Komuro, M. Kanai, A. Kawakami, Y. Fujiwara, N. Kobayashi, K. Shirata, N. Nishimura, S. Okushiba, S. Kondo, J. Hamada, H. Katoh, T. Yoshiki, and T. Moriuchi. 2004. Prediction of lymph node metastasis and perineural invasion of biliary tract cancer by selected features from cDNA array data. J. Surg. Res. 122:184-194. [DOI] [PubMed] [Google Scholar]
- 152.Nawrath, C., Y. Poirier, and C. Somerville. 1994. Targeting of the polyhydroxybutyrate biosynthetic pathway to the plastids of Arabidopsis thaliana results in high levels of polymer accumulation. Proc. Natl. Acad. Sci. USA 91:12760-12764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Neidhardt, F. C., J. L. Ingraham, and M. Schaechter. 1990. Physiology of the bacterial cell: a molecular approach. Sinauer Associates, Sunderland, Mass.
- 154.Newman, J. R., and A. E. Keating. 2003. Comprehensive identification of human bZIP interactions with coiled-coil arrays. Science 300:2097-2101. [DOI] [PubMed] [Google Scholar]
- 155.Nielsen, J. 2003. It is all about metabolic fluxes. J. Bacteriol. 185:7031-7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Nishino, K., and A. Yamaguchi. 2004. Role of histone-like protein H-NS in multidrug resistance of Escherichia coli. J. Bacteriol. 186:1423-1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Oberto, J., K. Drlica, and J. Rouviere-Yaniv. 1994. Histones, HMG, HU, IHF: Meme combat. Biochimie 76:901-908. [DOI] [PubMed] [Google Scholar]
- 158.Oliver, S. G., M. K. Winson, D. B. Kell, and F. Baganz. 1998. Systematic functional analysis of the yeast genome. Trends Biotechnol. 16:373-378. [DOI] [PubMed] [Google Scholar]
- 159.Orlando, V. 2000. Mapping chromosomal proteins in vivo by formaldehyde-crosslinked-chromatin immunoprecipitation. Trends Biochem. Sci. 25:99-104. [DOI] [PubMed] [Google Scholar]
- 160.Oshima, T., H. Aiba, Y. Masuda, S. Kanaya, M. Sugiura, B. L. Wanner, H. Mori, and T. Mizuno. 2002. Transcriptome analysis of all two-component regulatory system mutants of Escherichia coli K-12. Mol. Microbiol. 46:281-291. [DOI] [PubMed] [Google Scholar]
- 161.Ostergaard, S., L. Olsson, M. Johnston, and J. Nielsen. 2000. Increasing galactose consumption by Saccharomyces cerevisiae through metabolic engineering of the GAL gene regulatory network. Nat. Biotechnol. 18:1283-1286. [DOI] [PubMed] [Google Scholar]
- 162.Ow, M. C., Q. Liu, B. K. Mohanty, M. E. Andrew, V. F. Maples, and S. R. Kushner. 2002. RNase E levels in Escherichia coli are controlled by a complex regulatory system that involves transcription of the rne gene from three promoters. Mol. Microbiol. 43:159-171. [DOI] [PubMed] [Google Scholar]
- 163.Papadimitriou, S., and S. D. Likothanassis. 2004. Kernel-based self-organized maps trained with supervised bias for gene expression data analysis. J. Bioinform. Comput. Biol. 1:647-680. [DOI] [PubMed] [Google Scholar]
- 164.Parveen, M., M. K. Hasan, J. Takahashi, Y. Murata, E. Kitagawa, O. Kodama, and H. Iwahashi. 2004. Response of Saccharomyces cerevisiae to a monoterpene: evaluation of antifungal potential by DNA microarray analysis. J. Antimicrob. Chemother. 54:46-55. [DOI] [PubMed] [Google Scholar]
- 165.Patil, K. R., and J. Nielsen. 2005. Uncovering transcriptional regulation of metabolism by using metabolic network topology. Proc. Natl. Acad. Sci. USA 102:2685-2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Patterson, S. D., and R. H. Aebersold. 2003. Proteomics: the first decade and beyond. Nat. Genet. 33(Suppl.):311-323. [DOI] [PubMed] [Google Scholar]
- 167.Patton, W. F. 2002. Detection technologies in proteome analysis. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 771:3-31. [DOI] [PubMed] [Google Scholar]
- 168.Penalva, L. O., S. A. Tenenbaum, and J. D. Keene. 2004. Gene expression analysis of messenger RNP complexes. Methods Mol. Biol. 257:125-134. [DOI] [PubMed] [Google Scholar]
- 169.Pereira-Leal, J. B., A. J. Enright, and C. A. Ouzounis. 2004. Detection of functional modules from protein interaction networks. Proteins 54:49-57. [DOI] [PubMed] [Google Scholar]
- 170.Py, B., C. F. Higgins, H. M. Krisch, and A. J. Carpousis. 1996. A DEAD-box RNA helicase in the Escherichia coli RNA degradosome. Nature 381:169-172. [DOI] [PubMed] [Google Scholar]
- 171.Raamsdonk, L. M., B. Teusink, D. Broadhurst, N. Zhang, A. Hayes, M. C. Walsh, J. A. Berden, K. M. Brindle, D. B. Kell, J. J. Rowland, H. V. Westerhoff, K. van Dam, and S. G. Oliver. 2001. A functional genomics strategy that uses metabolome data to reveal the phenotype of silent mutations. Nat. Biotechnol. 19:45-50. [DOI] [PubMed] [Google Scholar]
- 172.Raghavan, A., R. L. Ogilvie, C. Reilly, M. L. Abelson, S. Raghavan, J. Vasdewani, M. Krathwohl, and P. R. Bohjanen. 2002. Genome-wide analysis of mRNA decay in resting and activated primary human T lymphocytes. Nucleic Acids Res. 30:5529-5538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Ramachandran, N., E. Hainsworth, B. Bhullar, S. Eisenstein, B. Rosen, A. Y. Lau, J. C. Walter, and J. LaBaer. 2004. Self-assembling protein microarrays. Science 305:86-90. [DOI] [PubMed] [Google Scholar]
- 174.Ranish, J. A., E. C. Yi, D. M. Leslie, S. O. Purvine, D. R. Goodlett, J. Eng, and R. Aebersold. 2003. The study of macromolecular complexes by quantitative proteomics. Nat. Genet. 33:349-355. [DOI] [PubMed] [Google Scholar]
- 175.Ravasz, E., A. L. Somera, D. A. Mongru, Z. N. Oltvai, and A. L. Barabasi. 2002. Hierarchical organization of modularity in metabolic networks. Science 297:1551-1555. [DOI] [PubMed] [Google Scholar]
- 176.Reed, J. L., and B. O. Palsson. 2003. Thirteen years of building constraint-based in silico models of Escherichia coli. J. Bacteriol. 185:2692-2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Reed, J. L., T. D. Vo, C. H. Schilling, and B. O. Palsson. 2003. An expanded genome-scale model of Escherichia coli K-12 (iJR904 GSM/GPR). Genome Biol. 4:R54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Ren, B., F. Robert, J. J. Wyrick, O. Aparicio, E. G. Jennings, I. Simon, J. Zeitlinger, J. Schreiber, N. Hannett, E. Kanin, T. L. Volkert, C. J. Wilson, S. P. Bell, and R. A. Young. 2000. Genome-wide location and function of DNA binding proteins. Science 290:2306-2309. [DOI] [PubMed] [Google Scholar]
- 179.Rouquette, C., M. C. Serre, and D. Lane. 2004. Protective role for H-NS protein in IS1 transposition. J. Bacteriol. 186:2091-2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Said, M. R., T. J. Begley, A. V. Oppenheim, D. A. Lauffenburger, and L. D. Samson. 2004. Global network analysis of phenotypic effects: protein networks and toxicity modulation in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 101:18006-18011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Salgado, H., S. Gama-Castro, A. Martinez-Antonio, E. az-Peredo, F. Sanchez-Solano, M. Peralta-Gil, D. Garcia-Alonso, V. Jimenez-Jacinto, A. Santos-Zavaleta, C. Bonavides-Martinez, and J. Collado-Vides. 2004. RegulonDB (version 4.0): transcriptional regulation, operon organization and growth conditions in Escherichia coli K-12. Nucleic Acids Res. 32(database issue):D303-D306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Salmon, K. A., S. P. Hung, N. R. Steffan, R. Krupp, P. Baldi, G. W. Hatfield, and R. P. Gunsalus. 2005. Global gene expression profiling in Escherichia coli K12: the effects of oxygen availability and ArcA. J. Biol. Chem. 280:15084-15096. [DOI] [PubMed] [Google Scholar]
- 183.Sauer, U. 2004. High-throughput phenomics: experimental methods for mapping fluxomes. Curr. Opin. Biotechnol. 15:58-63. [DOI] [PubMed] [Google Scholar]
- 184.Sauer, U., V. Hatzimanikatis, J. E. Bailey, M. Hochuli, T. Szyperski, and K. Wuthrich. 1997. Metabolic fluxes in riboflavin-producing Bacillus subtilis. Nat. Biotechnol. 15:448-452. [DOI] [PubMed] [Google Scholar]
- 185.Schaechter, M., O. Maaloe, and N. O. Kjeldgaard. 1958. Dependency on medium and temperature of cell size and chemical composition during balanced grown of Salmonella typhimurium. J. Gen. Microbiol. 19:592-606. [DOI] [PubMed] [Google Scholar]
- 186.Schena, M., D. Shalon, R. W. Davis, and P. O. Brown. 1995. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science 270:467-470. [DOI] [PubMed] [Google Scholar]
- 187.Schilling, C. H., M. W. Covert, I. Famili, G. M. Church, J. S. Edwards, and B. O. Palsson. 2002. Genome-scale metabolic model of Helicobacter pylori 26695. J. Bacteriol. 184:4582-4593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Schilling, C. H., J. S. Edwards, D. Letscher, and B. O. Palsson. 2000. Combining pathway analysis with flux balance analysis for the comprehensive study of metabolic systems. Biotechnol. Bioeng. 71:286-306. [PubMed] [Google Scholar]
- 189.Schlax, P. J., and D. J. Worhunsky. 2003. Translational repression mechanisms in prokaryotes. Mol. Microbiol. 48:1157-1169. [DOI] [PubMed] [Google Scholar]
- 190.Schweitzer, B., P. Predki, and M. Snyder. 2003. Microarrays to characterize protein interactions on a whole-proteome scale. Proteomics 3:2190-2199. [DOI] [PubMed] [Google Scholar]
- 191.Schwikowski, B., P. Uetz, and S. Fields. 2000. A network of protein-protein interactions in yeast. Nat. Biotechnol. 18:1257-1261. [DOI] [PubMed] [Google Scholar]
- 192.Segref, A., K. Sharma, V. Doye, A. Hellwig, J. Huber, R. Luhrmann, and E. Hurt. 1997. Mex67p, a novel factor for nuclear mRNA export, binds to both poly(A)+ RNA and nuclear pores. EMBO J. 16:3256-3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Shaw, K. J., N. Miller, X. Liu, D. Lerner, J. Wan, A. Bittner, and B. J. Morrow. 2003. Comparison of the changes in global gene expression of Escherichia coli induced by four bactericidal agents. J. Mol. Microbiol. Biotechnol. 5:105-122. [DOI] [PubMed] [Google Scholar]
- 194.Shepard, K. A., A. P. Gerber, A. Jambhekar, P. A. Takizawa, P. O. Brown, D. Herschlag, J. L. DeRisi, and R. D. Vale. 2003. Widespread cytoplasmic mRNA transport in yeast: identification of 22 bud-localized transcripts using DNA microarray analysis. Proc. Natl. Acad. Sci. USA 100:11429-11434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Spellman, P. T., G. Sherlock, M. Q. Zhang, V. R. Iyer, K. Anders, M. B. Eisen, P. O. Brown, D. Botstein, and B. Futcher. 1998. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol. Biol. Cell. 9:3273-3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Stelling, J., S. Klamt, K. Bettenbrock, S. Schuster, and E. D. Gilles. 2002. Metabolic network structure determines key aspects of functionality and regulation. Nature 420:190-193. [DOI] [PubMed] [Google Scholar]
- 197.Stephanopoulos, G., and J. J. Vallino. 1991. Network rigidity and metabolic engineering in metabolite overproduction. Science 252:1675-1681. [DOI] [PubMed] [Google Scholar]
- 198.Stokes, D. E. 1997. Pasteur's quadrant: basic science and technological innovation. Brookings Institution Press, Washington, D.C.
- 199.Stoyanova, R., T. D. Querec, T. R. Brown, and C. Patriotis. 2004. Normalization of single-channel DNA array data by principal component analysis. Bioinformatics 20:1772-1784. [DOI] [PubMed] [Google Scholar]
- 200.Stutz, F., and E. Izaurralde. 2003. The interplay of nuclear mRNP assembly, mRNA surveillance and export. Trends Cell Biol. 13:319-327. [DOI] [PubMed] [Google Scholar]
- 201.Szyperski, T. 1998. 13C-NMR, MS and metabolic flux balancing in biotechnology research. Q. Rev. Biophys. 31:41-106. [DOI] [PubMed] [Google Scholar]
- 202.Szyperski, T. 1995. Biosynthetically directed fractional 13C-labeling of proteinogenic amino acids. An efficient analytical tool to investigate intermediary metabolism. Eur. J. Biochem. 232:433-448. [DOI] [PubMed] [Google Scholar]
- 203.Takizawa, P. A., J. L. DeRisi, J. E. Wilhelm, and R. D. Vale. 2000. Plasma membrane compartmentalization in yeast by messenger RNA transport and a septin diffusion barrier. Science 290:341-344. [DOI] [PubMed] [Google Scholar]
- 204.Tan, K., L. A. McCue, and G. D. Stormo. 2005. Making connections between novel transcription factors and their DNA motifs. Genome Res. 15:312-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Tani, T. H., A. Khodursky, R. M. Blumenthal, P. O. Brown, and R. G. Matthews. 2002. Adaptation to famine: a family of stationary-phase genes revealed by microarray analysis. Proc. Natl. Acad. Sci. USA 99:13471-13476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Tatarko, M., and T. Romeo. 2001. Disruption of a global regulatory gene to enhance central carbon flux into phenylalanine biosynthesis in Escherichia coli. Curr. Microbiol. 43:26-32. [DOI] [PubMed] [Google Scholar]
- 207.Tavazoie, S., J. D. Hughes, M. J. Campbell, R. J. Cho, and G. M. Church. 1999. Systematic determination of genetic network architecture. Nat. Genet. 22:281-285. [DOI] [PubMed] [Google Scholar]
- 208.Templin, M. F., D. Stoll, J. Bachmann, and T. O. Joos. 2004. Protein microarrays and multiplexed sandwich immunoassays: what beats the beads? Comb. Chem. High Throughput Screen. 7:223-229. [DOI] [PubMed] [Google Scholar]
- 209.Templin, M. F., D. Stoll, M. Schrenk, P. C. Traub, C. F. Vohringer, and T. O. Joos. 2002. Protein microarray technology. Trends Biotechnol. 20:160-166. [DOI] [PubMed] [Google Scholar]
- 210.Templin, M. F., D. Stoll, J. M. Schwenk, O. Potz, S. Kramer, and T. O. Joos. 2003. Protein microarrays: promising tools for proteomic research. Proteomics 3:2155-2166. [DOI] [PubMed] [Google Scholar]
- 211.Tenenbaum, S. A., C. C. Carson, P. J. Lager, and J. D. Keene. 2000. Identifying mRNA subsets in messenger ribonucleoprotein complexes by using cDNA arrays. Proc. Natl. Acad. Sci. USA 97:14085-14090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Tenenbaum, S. A., P. J. Lager, C. C. Carson, and J. D. Keene. 2002. Ribonomics: identifying mRNA subsets in mRNP complexes using antibodies to RNA-binding proteins and genomic arrays. Methods 26:191-198. [DOI] [PubMed] [Google Scholar]
- 213.Toronen, P., M. Kolehmainen, G. Wong, and E. Castren. 1999. Analysis of gene expression data using self-organizing maps. FEBS Lett. 451:142-146. [DOI] [PubMed] [Google Scholar]
- 214.Trethewey, R. N. 2001. Gene discovery via metabolic profiling. Curr. Opin. Biotechnol. 12:135-138. [DOI] [PubMed] [Google Scholar]
- 215.Trethewey, R. N., A. J. Krotzky, and L. Willmitzer. 1999. Metabolic profiling: a Rosetta Stone for genomics? Curr. Opin. Plant Biol. 2:83-85. [DOI] [PubMed] [Google Scholar]
- 216.Truan, G., J. C. Epinat, C. Rougeulle, C. Cullin, and D. Pompon. 1994. Cloning and characterization of a yeast cytochrome b5-encoding gene which suppresses ketoconazole hypersensitivity in a NADPH-P-450 reductase-deficient strain. Gene 142:123-127. [DOI] [PubMed] [Google Scholar]
- 217.Tu, Y., G. Stolovitzky, and U. Klein. 2002. Quantitative noise analysis for gene expression microarray experiments. Proc. Natl. Acad. Sci. USA 99:14031-14036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Tung, W. L., and C. Quek. 2005. GenSo-FDSS: a neural-fuzzy decision support system for pediatric ALL cancer subtype identification using gene expression data. Artif. Intell. Med. 33:61-88. [DOI] [PubMed] [Google Scholar]
- 219.Uetz, P., L. Giot, G. Cagney, T. A. Mansfield, R. S. Judson, J. R. Knight, D. Lockshon, V. Narayan, M. Srinivasan, P. Pochart, A. Qureshi-Emili, Y. Li, B. Godwin, D. Conover, T. Kalbfleisch, G. Vijayadamodar, M. Yang, M. Johnston, S. Fields, and J. M. Rothberg. 2000. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature 403:623-627. [DOI] [PubMed] [Google Scholar]
- 220.Unlu, M. 1999. Difference gel electrophoresis. Biochem. Soc. Trans. 27:547-549. [DOI] [PubMed] [Google Scholar]
- 221.Valentini, G. 2002. Gene expression data analysis of human lymphoma using support vector machines and output coding ensembles. Artif. Intell. Med. 26:281-304. [DOI] [PubMed] [Google Scholar]
- 222.Vanderpool, C. K., and S. Gottesman. 2004. Involvement of a novel transcriptional activator and small RNA in post-transcriptional regulation of the glucose phosphoenolpyruvate phosphotransferase system. Mol. Microbiol. 54:1076-1089. [DOI] [PubMed] [Google Scholar]
- 223.van Hoof, A., P. A. Frischmeyer, H. C. Dietz, and R. Parker. 2002. Exosome-mediated recognition and degradation of mRNAs lacking a termination codon. Science 295:2262-2264. [DOI] [PubMed] [Google Scholar]
- 224.Villas-Boas, S. G., D. G. Delicado, M. Akesson, and J. Nielsen. 2003. Simultaneous analysis of amino and nonamino organic acids as methyl chloroformate derivatives using gas chromatography-mass spectrometry. Anal. Biochem. 322:134-138. [DOI] [PubMed] [Google Scholar]
- 225.Villas-Boas, S. G., S. Mas, M. Akesson, J. Smedsgaard, and J. Nielsen. 19August2004, posting date. Mass spectrometry in metabolome analysis. Mass Spectrom. Rev. [Online.] [DOI] [PubMed]
- 226.Waggoner, S. A., and S. A. Liebhaber. 2003. Identification of mRNAs associated with alphaCP2-containing RNP complexes. Mol. Cell. Biol. 23:7055-7067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Wahl, A., M. El Massaoudi, D. Schipper, W. Wiechert, and R. Takors. 2004. Serial 13C-based flux analysis of an L-phenylalanine-producing E. coli strain using the sensor reactor. Biotechnol. Prog. 20:706-714. [DOI] [PubMed] [Google Scholar]
- 228.Walhout, A. J., R. Sordella, X. Lu, J. L. Hartley, G. F. Temple, M. A. Brasch, N. Thierry-Mieg, and M. Vidal. 2000. Protein interaction mapping in C. elegans using proteins involved in vulval development. Science 287:116-122. [DOI] [PubMed] [Google Scholar]
- 229.Walter, G., K. Bussow, D. Cahill, A. Lueking, and H. Lehrach. 2000. Protein arrays for gene expression and molecular interaction screening. Curr. Opin. Microbiol. 3:298-302. [DOI] [PubMed] [Google Scholar]
- 230.Wang, J., J. Z. Ma, and M. D. Li. 2004. Normalization of cDNA microarray data using wavelet regressions. Comb. Chem. High Throughput Screen. 7:783-791. [DOI] [PubMed] [Google Scholar]
- 231.Wang, Y., C. L. Liu, J. D. Storey, R. J. Tibshirani, D. Herschlag, and P. O. Brown. 2002. Precision and functional specificity in mRNA decay. Proc. Natl. Acad. Sci. USA 99:5860-5865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Washburn, M. P., D. Wolters, and J. R. Yates III. 2001. Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat. Biotechnol. 19:242-247. [DOI] [PubMed] [Google Scholar]
- 233.Weckwerth, W. 2003. Metabolomics in systems biology. Annu. Rev. Plant Biol. 54:669-689. [DOI] [PubMed] [Google Scholar]
- 234.Weiner, H., T. Faupel, and K. Bussow. 2004. Protein arrays from cDNA expression libraries. Methods Mol. Biol. 264:1-13. [DOI] [PubMed] [Google Scholar]
- 235.West, A. H., and A. M. Stock. 2001. Histidine kinases and response regulator proteins in two-component signaling systems. Trends Biochem. Sci. 26:369-376. [DOI] [PubMed] [Google Scholar]
- 236.Wilusz, C. J., M. Wormington, and S. W. Peltz. 2001. The cap-to-tail guide to mRNA turnover. Nat. Rev. Mol. Cell. Biol. 2:237-246. [DOI] [PubMed] [Google Scholar]
- 237.Workman, C., L. J. Jensen, H. Jarmer, R. Berka, L. Gautier, H. B. Nielser, H. H. Saxild, C. Nielsen, S. Brunak, and S. Knudsen. 2002. A new non-linear normalization method for reducing variability in DNA microarray experiments. Genome Biol. 3:research0048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Wu, T. D. 2001. Analysing gene expression data from DNA microarrays to identify candidate genes. J. Pathol. 195:53-65. [DOI] [PubMed] [Google Scholar]
- 239.Yang, J., H. P. Bogerd, P. J. Wang, D. C. Page, and B. R. Cullen. 2001. Two closely related human nuclear export factors utilize entirely distinct export pathways. Mol. Cell. 8:397-406. [DOI] [PubMed] [Google Scholar]
- 240.Yates, F. E. 1979. Integrative physiology as physical biology. Am. J. Physiol. 236:R239-R240. [DOI] [PubMed] [Google Scholar]
- 241.Yatherajam, G., L. Zhang, S. M. Kraemer, and L. A. Stargell. 2003. Protein-protein interaction map for yeast TFIID. Nucleic Acids Res. 31:1252-1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Yin, D., B. Fox, M. L. Lonetto, M. R. Etherton, D. J. Payne, D. J. Holmes, M. Rosenberg, and Y. Ji. 2004. Identification of antimicrobial targets using a comprehensive genomic approach. Pharmacogenomics 5:101-113. [DOI] [PubMed] [Google Scholar]
- 243.Yoon, D., S. G. Yi, J. H. Kim, and T. Park. 2004. Two-stage normalization using background intensities in cDNA microarray data. BMC Bioinformatics 5:97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Yoon, S. H., M. J. Han, S. Y. Lee, K. J. Jeong, and J. S. Yoo. 2003. Combined transcriptome and proteome analysis of Escherichia coli during high cell density culture. Biotechnol. Bioeng. 81:753-767. [DOI] [PubMed] [Google Scholar]
- 245.Yoshida, Y., and Y. Aoyama. 1987. Interaction of azole antifungal agents with cytochrome P-45014DM purified from Saccharomyces cerevisiae microsomes. Biochem. Pharmacol. 36:229-235. [DOI] [PubMed] [Google Scholar]
- 246.Zhang, L., Y. Zhang, Y. Zhou, S. An, Y. Zhou, and J. Cheng. 2002. Response of gene expression in Saccharomyces cerevisiae to amphotericin B and nystatin measured by microarrays. J. Antimicrob. Chemother. 49:905-915. [DOI] [PubMed] [Google Scholar]
- 247.Zhang, X., and H. Bremer. 1995. Control of the Escherichia coli rrnB P1 promoter strength by ppGpp. J. Biol. Chem. 270:11181-11189. [DOI] [PubMed] [Google Scholar]
- 248.Zhang, X., P. Dennis, M. Ehrenberg, and H. Bremer. 2002. Kinetic properties of rrn promoters in Escherichia coli. Biochimie 84:981-996. [DOI] [PubMed] [Google Scholar]
- 249.Zhu, H., J. F. Klemic, S. Chang, P. Bertone, A. Casamayor, K. G. Klemic, D. Smith, M. Gerstein, M. A. Reed, and M. Snyder. 2000. Analysis of yeast protein kinases using protein chips. Nat. Genet. 26:283-289. [DOI] [PubMed] [Google Scholar]
- 250.Zhu, H., and M. Snyder. 2003. Protein chip technology. Curr. Opin. Chem. Biol. 7:55-63. [DOI] [PubMed] [Google Scholar]